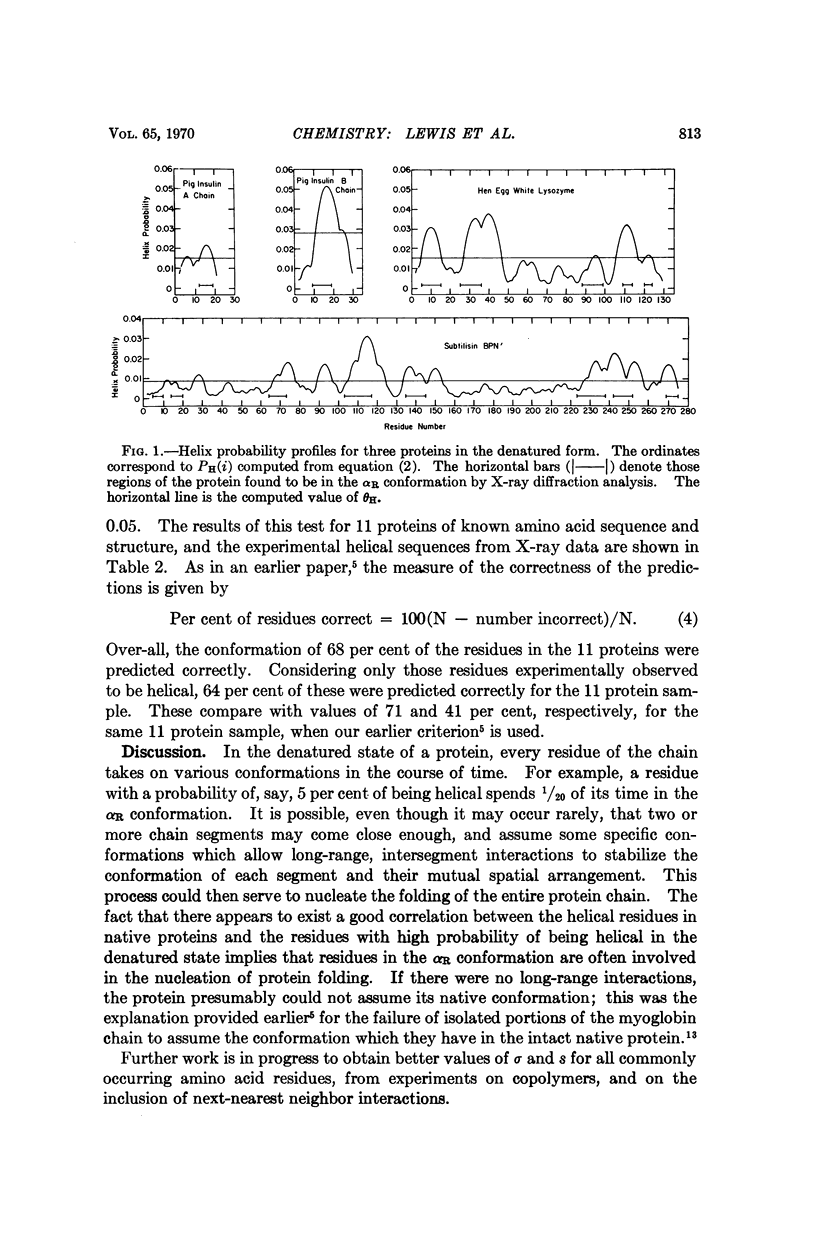
Abstract
The Zimm-Bragg formulation for the one-dimensional Ising model is applied to denatured proteins in order to compute helix probability profiles with different σ and s parameters for the various amino acids; the latter are in principle determinable from melting curves for helix-coil transitions in random copolymers of amino acids. Using a tentative assignment of σ and s values, we found a correlation for the propensity of a residue to be helical in the denatured protein and its occurrence in a helical region in the globular structure of the corresponding native protein. Thus, these incipient helical regions in the denatured chain may serve to nucleate the folding to form the native protein. Short-range interactions appear to determine the tendency for a residue to be helical or not, whereas long-range interactions may serve to carry out the nucleation and refolding processes.
Full text
PDF




Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Blake C. C., Koenig D. F., Mair G. A., North A. C., Phillips D. C., Sarma V. R. Structure of hen egg-white lysozyme. A three-dimensional Fourier synthesis at 2 Angstrom resolution. Nature. 1965 May 22;206(4986):757–761. doi: 10.1038/206757a0. [DOI] [PubMed] [Google Scholar]
- Drenth J., Jansonius J. N., Koekoek R., Swen H. M., Wolthers B. G. Structure of papain. Nature. 1968 Jun 8;218(5145):929–932. doi: 10.1038/218929a0. [DOI] [PubMed] [Google Scholar]
- Epand R. M., Scheraga H. A. The influence of long-range interactions on the structure of myoglobin. Biochemistry. 1968 Aug;7(8):2864–2872. doi: 10.1021/bi00848a024. [DOI] [PubMed] [Google Scholar]
- Guzzo A. V. The influence of amino-acid sequence on protein structure. Biophys J. 1965 Nov;5(6):809–822. doi: 10.1016/S0006-3495(65)86753-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kartha G., Bello J., Harker D. Tertiary structure of ribonuclease. Nature. 1967 Mar 4;213(5079):862–865. doi: 10.1038/213862a0. [DOI] [PubMed] [Google Scholar]
- Kotelchuck D., Dygert M., Scheraga H. A. The influence of short-range interactions on protein conformation. 3. Dipeptide distributions in proteins of known sequence and structure. Proc Natl Acad Sci U S A. 1969 Jul;63(3):615–622. doi: 10.1073/pnas.63.3.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotelchuck D., Scheraga H. A. The influence of short-range interactions on protein conformation. I. Side chain-backbone interactions within a single peptide unit. Proc Natl Acad Sci U S A. 1968 Dec;61(4):1163–1170. doi: 10.1073/pnas.61.4.1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipscomb W. N., Hartsuck J. A., Reeke G. N., Jr, Quiocho F. A., Bethge P. H., Ludwig M. L., Steitz T. A., Muirhead H., Coppola J. C. The structure of carboxypeptidase A. VII. The 2.0-angstrom resolution studies of the enzyme and of its complex with glycyltyrosine, and mechanistic deductions. Brookhaven Symp Biol. 1968 Jun;21(1):24–90. [PubMed] [Google Scholar]
- Matthews B. W., Sigler P. B., Henderson R., Blow D. M. Three-dimensional structure of tosyl-alpha-chymotrypsin. Nature. 1967 May 13;214(5089):652–656. doi: 10.1038/214652a0. [DOI] [PubMed] [Google Scholar]
- Perutz M. F., Muirhead H., Cox J. M., Goaman L. C. Three-dimensional Fourier synthesis of horse oxyhaemoglobin at 2.8 A resolution: the atomic model. Nature. 1968 Jul 13;219(5150):131–139. doi: 10.1038/219131a0. [DOI] [PubMed] [Google Scholar]
- Prothero J. W. Correlation between the distribution of amino acids and alpha helices. Biophys J. 1966 May;6(3):367–370. doi: 10.1016/S0006-3495(66)86662-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiffer M., Edmundson A. B. Use of helical wheels to represent the structures of proteins and to identify segments with helical potential. Biophys J. 1967 Mar;7(2):121–135. doi: 10.1016/S0006-3495(67)86579-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright C. S., Alden R. A., Kraut J. Structure of subtilisin BPN' at 2.5 angström resolution. Nature. 1969 Jan 18;221(5177):235–242. doi: 10.1038/221235a0. [DOI] [PubMed] [Google Scholar]



