Abstract
ATM and p53 are critical regulators of the cellular DNA damage response, and function as potent tumor suppressors. In cells undergoing ionizing radiation, ATM is activated by double-strand DNA breaks and phosphorylates the amino terminus of p53 at serine residue 18. We have previously generated mice bearing an amino acid substitution at this position (p53S18A) and documented a role for p53 phosphorylation in DNA damage-induced apoptosis. In this present study, we have crossed Eμmyc transgenic mice with our p53S18A mice to explore a role for ATM-p53 signaling in response to oncogene-induced tumorigenesis. Similar to DNA damage induced by ionizing radiation, expression of c-Myc in pre-B cells induces p53 serine18 phosphorylation and Puma expression to promote apoptosis. Eμmyc transgenic mice develop B cell lymphoma more rapidly when heterozygous or homozygous for p53S18A alleles. However, Eμmyc-induced tumorigenesis in p53S18A mice is slower than that observed in Eμmyc mice deficient for either p53 or ATM, indicating that both p53-induced apoptosis and p53-induced growth arrest contribute to suppression of B cell lymphoma formation in Eμmyc mice. These findings further reveal that oncogene expression and DNA damage activate the same ATM-p53 signaling cascade in vivo to regulate apoptosis and tumorigenesis.
Keywords: Myc, p53, ATM, B-cell, tumorigenesis
Introduction
The p53 transcription factor activates the expression of genes encoding negative regulators of growth in cells exposed to various types of stress, including DNA damage, hypoxia, or inappropriate growth stimulation due to oncogene activation (1). These p53 target genes include Cdkn1a (p21), 14-3-3σ, cyclin G, and other genes whose products inhibit cell proliferation, as well as Bax, Puma, Noxa, and other p53 target genes that encode pro-apoptotic proteins (2). Activation of these genes by p53 blocks proliferation or induces apoptosis in damaged cells, thereby preventing the accumulation of oncogenic mutations and subsequent tumorigenesis. The importance of p53 in controlling cell growth and in tumor suppression is underscored by the fact that mutations in p53 or in other genes encoding proteins that govern p53 function is the most common mechanistic step in the development of human cancer (3).
The activity of p53 is tightly regulated during normal cell growth by Mdm2, an E3 ligase that binds and ubiquitinates the p53 protein to regulate p53 cellular localization and stability (4). Regulation of p53 activity is also mediated by other, numerous post-translational modifications of the p53 protein, including acetylation, neddylation, sumoylation, and phosphorylation; perhaps the best understood of these p53 modifications (5,6). In response to DNA damage, the ATM (mutated in ataxia telangiectasia) kinase becomes activated and phosphorylates p53 on serine residue 15 (p53 Ser18 in mice). This phosphorylation is reduced in ATM-mutant mice and is proposed to upregulate the p53 DNA damage response, as p53 activity is reduced in ATM-deficient mouse and human cells treated with ionizing radiation (IR) (7-12). To explore the significance of this p53 phosphorylation in vivo, we generated a p53 knock-in mouse model in which the serine residue at p53 position 18 is replaced by alanine (13). Previous analysis of primary fibroblasts isolated from this p53S18A model (p53S18A/S18A) revealed that phosphorylation of serine18 did not alter p53 protein stabilization or inhibit the ability of p53 to induce cell cycle arrest in fibroblasts following DNA damage. However, apoptosis was compromised in p53S18A thymocytes treated with IR, indicating that this phosphorylation regulates p53-induced apoptosis in response to DNA damage. Further analysis of this model revealed that p53S18A homozygous mice developed spontaneous tumors, albeit with a very delayed onset of cancer relative to p53-null mice (14). In addition, B cell lymphomas were the predominant tumor type observed in the p53S18A mice, whereas T cell lymphomas are most frequently seen in mice deleted for either p53 (15-17) or ATM (18-20). Collectively, these studies reveal that phosphorylation of p53 serine 18 upregulates DNA damage-induced apoptosis in thymocytes and can contribute to p53-mediated tumor suppression. However, some p53 functions may not be dependent upon ATM phosphorylation of p53 serine 18, and it is unclear from these studies if phosphorylation of p53 serine 18 is critical in regulating the p53 response in cells exposed to stress other than DNA damage.
The Eμmyc transgenic mouse is a well-established model of oncogene-induced tumorigenesis. Eμmyc transgenic mice overexpress c-Myc in progenitor B cells and rapidly develop non-Hodgkin's B-cell lymphoma. (21). Inappropriate levels of Myc in the pre-malignant B cells of Eμmyc mice results in the activation of the ARF–Mdm2–p53 signaling pathway and induces p53-dependent apoptosis, and B cell tumors that arise in this mouse model display either alterations in p19Arf expression or Mdm2 expression, mutation of the p53 gene, or loss of Puma, the pro-apoptotic p53-upregulated modulator of apoptosis (22, 23, 24).
To determine if phosphorylation of p53 serine residue 18 also plays a role in regulating p53 functions in the cellular response to activated oncogenes, we placed the Eμmyc transgene on a p53S18A background and examined B cell proliferation, apoptosis, and tumorigenesis. The results indicate that ATM phosphorylation of p53 serine 18 is a critical regulatory step in promoting B cell apoptosis and in suppressing B cell lymphomagenesis induced by Myc expression.
Results and Discussion
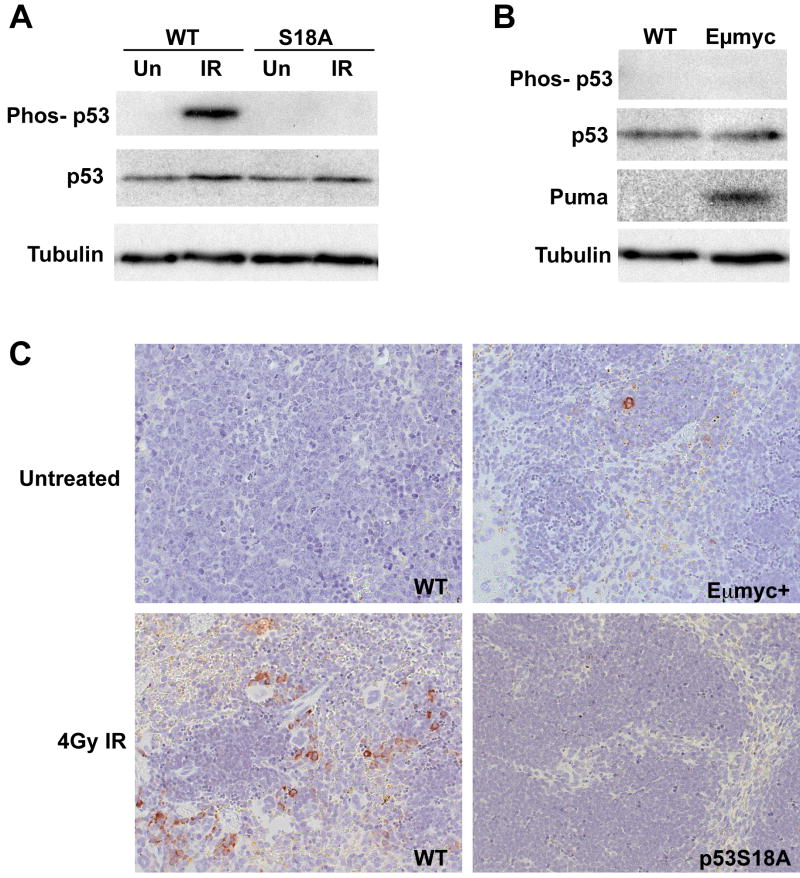
In order to examine ATM phosphorylation of p53 serine residue 18 in mouse spleen, we whole-body irradiated p53-wildtype (WT) or p53S18A mice (C57Bl/6 inbred background) with 4Gy ionizing radiation (IR) and recovered the spleens at 4 hours post-IR treatment (Figure 1A). A slight increase in p53 levels was observed at this early time point in IR-treated mice relative to non-treated mice, but no difference was detected in total p53 levels between WT and p53S18A mice regardless of IR treatment. This result indicates that phosphorylation of p53 serine 18 by ATM does not alter p53 levels in spleen, in agreement with our earlier finding in fibroblasts that modification of this p53 residue does not impact p53 stability (13). Western analysis of splenic extracts was also performed using an antibody that specifically recognizes p53 when phosphorylated at serine residue 18. As expected, DNA damage induced by IR treatment results in phosphorylation of p53 serine 18 in WT spleen, whereas this p53 phosphorylation event cannot occur in the spleen of IR-treated p53S18A mice. Phosphorylation of p53 serine 18 cannot be detected in the spleen of Eμmyc transgenic mice (C57Bl/6 strain) by western analysis (Figure 1B). However, these mice do display an increase in Puma levels in the spleen, consistent with a model for Myc-induction of p53 transcriptional activity and p53-mediated apoptosis. Spleen samples were isolated from 5-6 week old WT mice or pre-malignant, Eμmyc transgenic mice, and histologic staining of these tissue sections was performed using the p53 phopho-serine 18 antibody (Figure 1C-top panels). No p53 phosphorylation was seen in the WT tissue, whereas sporadic staining was observed in the Eμmyc transgenic spleen. This modest level of p53 phospho-serine 18 staining in the Eμmyc+ spleen likely accounts for the undetected (by western analysis) level of p53 phosphorylation in the Eμmyc+ sample (Figure 1B). In contrast, IR-treatment of WT mice resulted in robust p53 phospho-serine 18 staining, whereas no staining was observed in the spleen of irradiated p53S18A mice (Figure 1C-bottom panels). These results reveal that DNA damage results in p53 serine 18 phosphorylation in spleen, and that p53 serine 18 phosphorylation and Puma activation can be detected in the spleen of Eμmyc transgenic mice, albeit at a very low level. These in vivo results are in keeping with published results documenting increased p53 serine18 phosphorylation levels in B cells transduced with exogenous Myc (25).
Figure 1.
Myc overexpression induces p53 serine 18 phosphorylation and upregulates Puma expression and p53-dependent apoptosis in B cells. (A) Western analysis of spleen samples from wildtype (WT) or p53S18A mice either untreated (Un) or 4 hours after whole body irradiation with 4Gy (IR). Phosphorylation of p53 at serine 18 occurs in response to DNA damage in spleen. (B) Western analysis of untreated spleens reveals upregulation of Puma expression in Eμmyc transgenic mice. (C) Histochemical staining of representative sections of pre-malignant spleens using a p53-serine18 phospho-specific antibody. Samples were isolated from untreated wildtype mice and Eμmyc transgenic mice, and from IR-treated wildtype mice or p53S18A homozygous mice. Sporadic p53 phosphorylation is detected in Eμmyc transgenic mouse spleen.
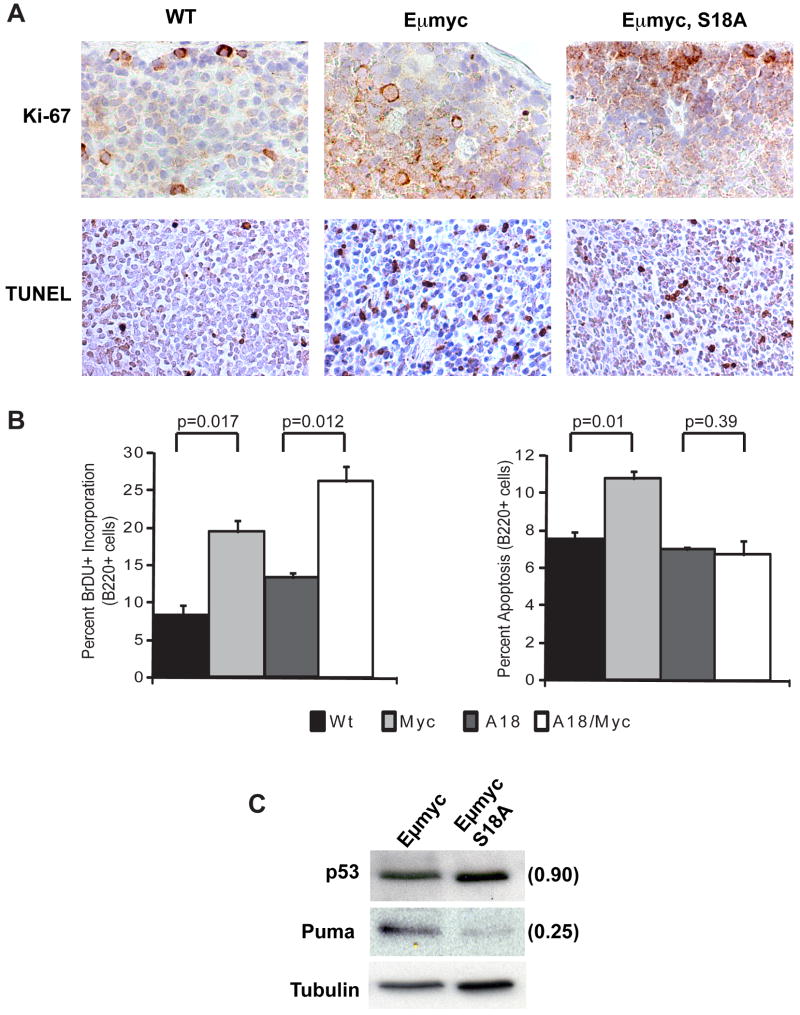
To determine if phosphorylation of p53 serine 18 regulates the response of p53 to inappropriate Myc expression in vivo, we placed the Eμmyc transgene on a p53S18A background. Eμmyc transgenic mice (C57Bl/6 strain) were mated with congenic p53S18A/S18A mice to obtain Eμmyc transgenic, p53S18A/+ male mice, which were crossed with female p53S18A/S18A mice to obtain littermates heterozygous or homozygous for the p53S18A allele, and that contained or lacked the Eμmyc transgene. All genotypes were recovered from these matings in the expected Mendelian ratios (data not shown). Spleen samples were isolated from 5-6 week old, pre-malignant, Eμmyc transgenic mice that were either p53-wt or homozygous for p53S18A, and tissue sections were analyzed using an antibody against the proliferation marker Ki-67 or by performing TUNEL staining for apoptosis (Figure 2A). Cell proliferation appeared to be increased in the spleens of Eμmyc transgenic mice and Eμmyc transgenic, p53S18A mice relative to the levels observed in WT mice. Apoptosis was also upregulated in the spleen of Eμmyc transgenic mice relative to WT samples. However, less TUNEL staining was seen in the Eμmyc transgenic spleen when p53 was not capable of being phosphorylated at serine reside 18 (compare EμMyc staining to EμMyc, S18A staining). These data suggest that phosphorylation of p53 serine residue 18 in spleen regulates p53 apoptosis induced by Myc expression.
Figure 2.
Myc expression induces B cell proliferation as well as B cell apoptosis that is regulated by p53 serine 18 phosphorylation. (A) Histochemical staining of representative sections of pre-malignant spleens isolated from WT mice, from Eμmyc transgenic mice, and from Eμmyc transgenic, p53S18A mice. Ki-67 staining indicates that Myc expression upregulates cell proliferation in vivo regardless of p53 status, whereas TUNEL staining reveals that Myc expression upregulates splenic apoptosis in a p53 serine 18 phosphorylation-dependent manner. (B) B cell growth in Eμmyc transgenic mice. Left panel: Myc-induced proliferation is not regulated by p53 serine18 phosphorylation in B cells. Average values for proliferation are derived from four mice per genotype, with bars representing SD. P values are given above brackets. Right panel: Myc-induced apoptosis is regulated by p53 Serine 18 phosphorylation. Average values for apoptosis are derived from four mice per genotype, with bars representing SD. P values are given above brackets. (C) Western analysis of p53 and Puma levels in premalignant spleens harvested from Eμmyc transgenic mice, or Eμmyc transgenic, p53S18A mice. Tubulin was used as a loading control. Values for protein amounts in Eμmyc transgenic, p53S18A mice are given in parentheses, as determined by densitometry, with p53 or Puma levels in Eμmyc transgenic spleens adjusted for Tubulin value and set at 1.
To confirm these findings in the more relevant cell population, we examined the effects of Myc expression on B220+ cells in the p53S18A model. BrDU uptake in sorted B220+ cells harvested from the spleens of pre-malignant mice. Eμmyc transgenic mice displayed a 2-fold increase in the percentage of BrDU positive, B220+ spleen cells relative to wildtype controls (Figure 2B- left). As expected, Eμmyc expression increased the proliferation of the p53-wildtype B cells (26). However, the relative increase in the proliferation rate of Eμmyc transgenic, p53-wildtype cells (2.2-fold) was similar to that observed in the Eμmyc transgenic, p53S18A cells (2.1-fold). Therefore, while Myc increases B220+ cell proliferation in mice, this increase in B-cell proliferation is not regulated by p53 serine18 phosphorylation.
To explore the effects of Myc on B-cell apoptosis in p53S18A mice, we isolated B220+ cells from the spleens of pre-malignant Eμmyc mice and performed annexin V staining (Figure2B– right). Expression of Myc increased the rate of spontaneous apoptosis in B220+ cells, in keeping with previous reports indicating that Myc induces ATM/p53-dependent apoptosis in Eμmyc transgenic mice (22, 23, 27). Although no difference was observed in the frequency of cells undergoing apoptosis in non-transgenic p53S18A mice and wt mice, the increase in B cell apoptosis documented in the Eμmyc transgenic, p53-wt model was not observed in Eμmyc transgenic, p53S18A mice. Thus, in contrast to our proliferation results in B cells, p53-mediated apoptosis induced by Myc expression in B cells is dependent upon phosphorylation of p53 serine18.
To confirm that Myc-induced apoptosis is compromised in p53S18A mice, we examined Puma expression levels in pre-malignant, age-matched Eμmyc, p53-WT mouse spleen and Eμmyc, p53S18A mouse spleen (Figure 2C). Puma, a BH3-only protein that activates Bax and Bak, is a major mediator of p53-induced apoptosis (28), and we have reported previously that p53S18A thymocytes and splenocytes display reduced Puma levels after exposure to IR (13,14), indicating that p53-induction of Puma following DNA damage is dependent upon p53 serine18 phosphorylation. Western blot analysis of spleen extracts harvested from 5-week old mice revealed little difference in p53 levels between Eμmyc transgenic mice, or Eμmyc transgenic mice homozygous for the p53S18A allele (as adjusted for control anti-tubulin staining). In contrast, Puma levels are reduced by 75% in Eμmyc transgenic, p53S18A pre-malignant cells. These results indicate that Myc overexpression (in pre-B cells) induces apoptosis that requires p53 serine18 phosphorylation and p53 activation of Puma expression. As loss of Puma is frequently observed in B cell lymphomas arising in EμMyc mice (24) our data suggest that p53S18A mice would be more susceptible to Myc-driven lymphomagenesis.
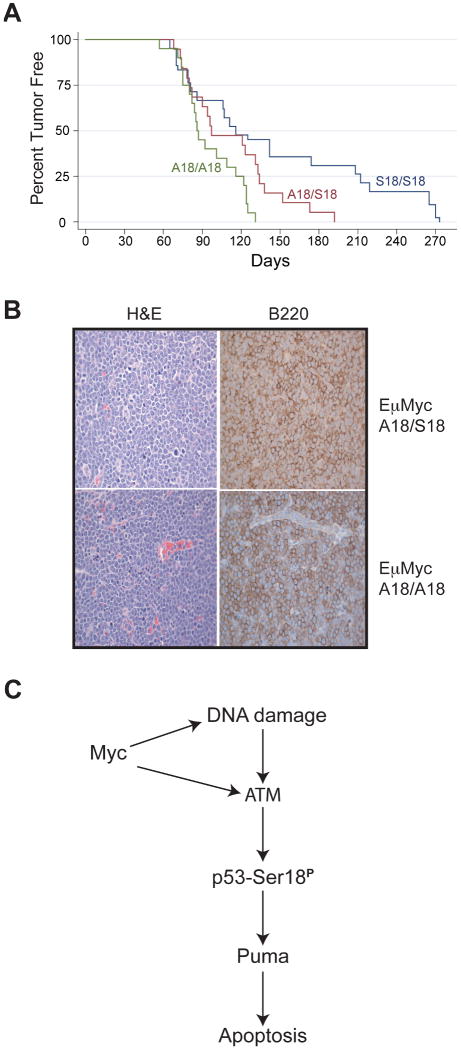
To determine if serine18 phosphorylation regulates p53 suppression of Myc-induced tumorigenesis, cohorts of Eμmyc transgenic mice that were either p53 wildtype (S18/S18), heterozygous for the p53S18A allele (A18/S18), or homozygous for the modified serine to alanine p53 allele (A18/A18) were established and surveyed for tumor formation. Tumorigenesis in the Eμmyc transgenic, p53-wildtype mice (n=42) displayed kinetics similar to those described previously for Eμmyc transgenic mice (Figure 3A), with a mean time to tumorigenesis of 116 days (29, 30). Eμmyc mice heterozygous for the p53S18A mutation (n=19) had an increased rate of tumor onset and a mean time to tumorigenesis of 96 days, whereas Eμmyc mice homozygous for the p53S18A mutation (n=20) developed tumors even more rapidly, with a mean time to tumorigenesis of 87 days. Log-rank analysis indicates that Eμmyc mice heterozygous for the serine18 mutation (p53S18/A18) or homozygous for the p53Ser18 mutation (p53A18/A18) have reduced survival compared to Eμmyc, p53-wildtype mice (p= 0.023, and 0.0013, respectively), and Eμmyc transgenic, p53A18/A18 are reduced in their survival relative to Eμmyc transgenic, p53S18/A18 mice (p= 0.033). All mice in the three cohorts developed cancer, and presented with enlarged spleens and lymph nodes. Since mice presenting with hallmarks of disease typically die within a few days due to B cell lymphomagenesis, and since we did not find any tumors in asymptomatic mouse, the time to tumorigenesis reflects the time of tumor onset and not the rate of disease progression. Tumors were isolated by necropsy, fixed, stained, and were classified by morphology as B cell lymphomas. To confirm the tumor type, select tumors were fixed and immunostained for B220 antigens (Figure 3B). All mice developed B220+ lymphomas, as expected, since Eμmyc mice rapidly develop B cell lymphomas (21). Notably, Eμmyc transgenic, p53S18A mice develop B cell lymphomas more rapidly than Eμmyc transgenic, p53-wildtype mice, indicating that phosphorylation of p53 at serine18 regulates p53 suppression of oncogene-induced tumorigenesis.
Figure 3.
Myc-induced, B cell tumorigenesis is suppressed by p53 serine18 phosphorylation in mice. (A) Kaplan-Meier survival curve of Eμmyc transgenic mice that are either wildtype for p53 (S18/S18), heterozygous for the p53S18A allele (S18/A18), or homozygous for the p53S18A allele (A18/A18). (B) Representative tumor sections from Eμmyc transgenic mice heterozygous for the p53S18A allele (S18/A18) or homozygous for the p53S18A allele (A18/A18) stained with hematoxylin and eosin, or with B220 antibody. (C) Model for Myc activation of p53 tumor suppression. Inappropriate levels of Myc may activate ATM, p53 Serine18 phosphorylation, and p53-mediated apoptosis either by inducing DNA damage within the cell or by directly activating ATM-p53 signaling.
It is interesting to note that the mean time to tumorigenesis for Eμmyc transgenic mice on the p53S18A background (87 days) is delayed relative to the mean time to tumorigenesis for Eμmyc transgenic mice on an ATM-null background (69 days) (23) or on a p53-heterozygous background (35 days) (25). Furthermore, Eμmyc transgenic, p53-null mice cannot be recovered through normal breeding strategies due to the very rapid onset of neoplasia in Eμmyc transgenic, p53-heterozygous mice. In contrast, Eμmyc, p53S18A mice are easily recovered by breeding. As these mice are viable and also delayed in tumorigenesis relative to p53-haploinsufficient, Eμmyc transgenic mice, some p53 tumor-suppressing functions must still exist in p53S18A mice to slow the onset of the disease. That p53S18A retains some ability to suppress Myc induced tumorigenesis is in agreement with our previous findings that p53S18A mice form spontaneous tumors more slowly than p53–null mice (13, 14). As p53S18A mice are compromised in Puma induction and apoptosis (Figure 2A, C), the ability of the p53S18A allele to inhibit Myc upregulation of B cell proliferation (Figure 2B) must also be an important facet of p53 suppression of Myc-induced tumorigenesis.
ATM and p53 are critical regulators of the cellular DNA damage response, and both proteins function as potent tumor suppressors. In cells undergoing DNA damage, ATM activation induces phosphorylation of p53 at serine residue 18, and we have documented previously that p53 serine18 phosphorylation promotes DNA damage-induced apoptosis in murine thymocytes and splenocytes (13). However, the role of p53 phosphorylation in modulating p53 activity in response to oncogene expression is not well understood. Interestingly, deregulation of oncogene expression has been proposed to induce DNA damage in human cells (31, 32). In keeping with this paradigm, ATM deficiency has been found to impair Myc-induced apoptosis and augment tumorigenesis in mouse epithelial cells (33) and mouse B cells (25, 27). However, oncogene-induced DNA damage was not detected by phopho-H2AX staining in pre-malignant, Eμmyc-expressing B cells (25). Therefore, it is unclear if Myc overexpression activates p53 tumor suppressor functions by inducing DNA damage and ATM-p53 signaling in the Eμmyc mouse model, or if Myc induces p53 activity in B cells by some other mechanism. It has been established that Myc can activate p53 levels and activity by upregulating the levels of p19Arf in mice (22, 34), and other oncogenes such as Ras have been recently found to induce p53 in an ATM-independent manner (35). Thus, it is possible that inappropriate oncogene expression may induce p53 tumor suppression in mice without invoking a DNA damage-response signaling pathway (Figure 2C). However, if Myc expression is not causing DNA damage, then the p53 signaling pathway induced by Myc expression shares several mechanistic steps with DNA damage-induced p53 activation, as the results of our study strongly supports a role for ATM-p53 signaling in Myc activation of p53 tumor suppression.
Methods
Mice and tumor assays
The generation of p53S18A mice and of Eμmyc transgenic mice has been described previously (13, 21). All mice were backcrossed to C57Bl/6 strain for a minimum of 10 generations. The p53S18A mice and Eμmyc transgenic mice were intercrossed to generate Eμmyc transgenic mice that were p53-wildtype, p53S18A/+, or p53S18A/S18A. Cohorts of each line of mice were collected and aged in order to perform a tumor assay. Mice displaying obvious tumors or signs of reduced vitality were euthanized, necropsied, and tissues were harvested and fixed in 10% formalin. All mouse tumors were classified by morphology to be B cell lymphomas, regardless of p53 status. Mice were maintained and used in accordance with both federal guidelines and those established by the Institutional Animal Care and Use Committee at the University of Massachusetts Medical School.
Western analysis, immunohistochemistry, and antibodies
Protein extracts were generated as described previously (13), using antibodies against p53 (1:1 mix of AB-1 and AB-3, Calbiochem), Puma (4976, Cell Signaling) or α–Tubulin (T5168, Sigma). Spleens (pre-neoplastic) were isolated from 6-8 week old mice, and a portion of each tissue was fixed in 10% phosphate-buffered formalin, embedded in paraffin, sectioned, and stained with an antibody that recognizes mouse p53 phospho-serine 18 (Cell Signaling). TUNEL staining and Ki-67 staining of spleen sections was performed as described previously (36). Tumors were isolated from the cohorts of mice as they developed, fixed in 10% phosphate-buffered formalin, and paraffin-embedded sections were stained with hemotoxylin and eosin or with an antibody to CD45R/B220 (BD Pharmingen, diluted 1/50) before being analyzed by microscopy. Images of stained spleens or tumor sections were obtained using an Axioskop (Zeiss) with 40× optic lens and a 3008 Prog/Res digital camera coupled to a MacIntosh G4 computer, using Photoshop 4 software (Adobe).
Proliferation and apoptosis assays
Proliferation of B220+ cells in wildtype (Wt), Eμmyc transgenic (Myc+), p53S18A/S18A (p53S18A), and Eμmyc transgenic, p53S18A/S18A mice (p53S18A, Myc+) mice was determined by injecting 6-8 week old mice with 50μg BrDU/gram body weight. After 4 hours, the spleens were harvested, made into single cell suspensions, stained with anti-BrDU-PE, and B220-APC, and analyzed by FACS. Proliferating cells were scored as those double positive for B220 and BrDU. Apoptosis of B220+ cells in wildtype (Wt), Eμmyc transgenic (Myc+), p53S18A/S18A (p53S18A), or Eμmyc transgenic, p53S18A/S18A mice (p53S18A, Myc+) mice was determined by harvesting spleens from 6-8 week old mice which were made into single cell suspensions, stained with annexin V-FITC and B220-APC, and analyzed by FACS. Cells undergoing apoptosis were scored as those staining double positive for annexin V and B220+. Flow cytometry was performed on a 3-laser 12-color LSR II (BD Biosciences, San Jose CA) and analyzed with FlowJo software (Tree Star, Ashland, OR).
Acknowledgments
We thank Heather L. Armata and Kathleen Hoover for assistance with the mouse colony, David S. Garlick for histologic analysis of tumors, and C. C. Hsieh for statistical analysis. We also thank C Baron for help with figure preparations. This work was supported by a grant from the National Institutes of Health to SNJ (CA077735). HKS was supported in part by a grant from the National Ataxia Foundation. CME is a Leukemia and Lymphoma Society Scholar.
References
- 1.Harris SL, Levine AJ. The p53 pathway: positive and negative feedback loops. Oncogene. 2005;24:2899–2908. doi: 10.1038/sj.onc.1208615. [DOI] [PubMed] [Google Scholar]
- 2.Vousden KH, Lu X. Live or let die: the cell's response to p53. Nat Rev Cancer. 2002;2:594–604. doi: 10.1038/nrc864. [DOI] [PubMed] [Google Scholar]
- 3.Soussi T, Beroud C. Assessing TP53 status in human tumours to evaluate clinical outcome. Nat Rev Cancer. 2001;1:233–240. doi: 10.1038/35106009. [DOI] [PubMed] [Google Scholar]
- 4.Iwakuma T, Lozano G. MDM2, an introduction. Mol Cancer Res. 2003;1:993–1000. [PubMed] [Google Scholar]
- 5.Giaccia AJ, Kastan MB. The complexity of p53 modulation: emerging patterns from divergent signals. Genes Dev. 1998;12:2973–2983. doi: 10.1101/gad.12.19.2973. [DOI] [PubMed] [Google Scholar]
- 6.Appella E, Anderson CW. Post-translational modifications and activation of p53 by genotoxic stresses. Eur J Biochem. 2001;268:2764–2772. doi: 10.1046/j.1432-1327.2001.02225.x. [DOI] [PubMed] [Google Scholar]
- 7.Banin S, Moyal L, Shieh S, et al. Enhanced phosphorylation of p53 by ATM in response to DNA damage. Science. 1998;281:1674–1677. doi: 10.1126/science.281.5383.1674. [DOI] [PubMed] [Google Scholar]
- 8.Canman CE, Lim DS, Cimprich KA, et al. Activation of the ATM kinase by ionizing radiation and phosphorylation of p53. Science. 1998;281:1677–1679. doi: 10.1126/science.281.5383.1677. [DOI] [PubMed] [Google Scholar]
- 9.Chao C, Saito S, Anderson CW, Appella E, Xu Y. Phosphorylation of murine p53 at ser-18 regulates the p53 responses to DNA damage. Proc Natl Acad Sci U S A. 2000;97:11936–11941. doi: 10.1073/pnas.220252297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Khanna KK, Keating KE, Kozlov S, et al. ATM associates with and phosphorylates p53: mapping the region of interaction. Nat Genet. 1998;20:398–400. doi: 10.1038/3882. [DOI] [PubMed] [Google Scholar]
- 11.Siliciano JD, Canman CE, Taya Y, Sakaguchi K, Appella E, Kastan MB. DNA damage induces phosphorylation of the amino terminus of p53. Genes Dev. 1997;11:3471–3481. doi: 10.1101/gad.11.24.3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nakagawa K, Taya Y, Tamai K, Yamaizumi M. Requirement of ATM in phosphorylation of the human p53 protein at serine 15 following DNA double-strand breaks. Mol Cell Bio. 1999;19:2828–2834. doi: 10.1128/mcb.19.4.2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sluss HK, Armata H, Gallant J, Jones SN. Phosphorylation of serine 18 regulates distinct p53 functions in mice. Mol Cell Biol. 2004;24:976–984. doi: 10.1128/MCB.24.3.976-984.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Armata HL, Garlick DS, Sluss HK, Jones SN. The ataxia telangiectasia-mutated target site Ser18 is required for p53-mediated tumor suppression. Cancer Res. 2007;67:11696–11703. doi: 10.1158/0008-5472.CAN-07-1610. [DOI] [PubMed] [Google Scholar]
- 15.Donehower LA, Harvey M, Slagle BL, et al. Mice deficient for p53 are developmentally normal but susceptible to spontaneous tumours. Nature. 1992;356:215–221. doi: 10.1038/356215a0. [DOI] [PubMed] [Google Scholar]
- 16.Harvey M, McArthur MJ, Montgomery CA, Jr, Butel JS, Bradley A, Donehower LA. Spontaneous and carcinogen-induced tumorigenesis in p53-deficient mice. Nat Genet. 1993;5:225–229. doi: 10.1038/ng1193-225. [DOI] [PubMed] [Google Scholar]
- 17.Jones SN, Sands AT, Hancock AR, et al. The tumorigenic potential and cell growth characteristics of p53-deficient cells are equivalent in the presence or absence of Mdm2. Proc Natl Acad Sci U S A. 1996;93:14106–14111. doi: 10.1073/pnas.93.24.14106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu Y, Ashley T, Brainerd EE, Bronson RT, Meyn MS, Baltimore D. Targeted disruption of ATM leads to growth retardation, chromosomal fragmentation during meiosis, immune defects, and thymic lymphoma. Genes Dev. 1996;10:2411–2422. doi: 10.1101/gad.10.19.2411. [DOI] [PubMed] [Google Scholar]
- 19.Elson A, Wang Y, Daugherty CJ, Morton CC, et al. Pleiotropic defects in ataxia-telangiectasia protein-deficient mice. Proc Natl Acad Sci U S A. 1996;93:13084–13089. doi: 10.1073/pnas.93.23.13084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Barlow C, Hirotsune S, Paylor R, et al. Atm-deficient mice: a paradigm of ataxia telangiectasia. Cell. 1996;86:159–171. doi: 10.1016/s0092-8674(00)80086-0. [DOI] [PubMed] [Google Scholar]
- 21.Adams JM, Harris AW, Pinkert CA, et al. The c-myc oncogene driven by immunoglobulin enhancers induces lymphoid malignancy in transgenic mice. Nature. 1985;318:533–538. doi: 10.1038/318533a0. [DOI] [PubMed] [Google Scholar]
- 22.Eischen CM, Weber JD, Roussel MF, Sherr CJ, Cleveland JL. Disruption of the ARF-Mdm2-p53 tumor suppressor pathway in Myc-induced lymphomagenesis. Genes Dev. 1999;13:2658–2669. doi: 10.1101/gad.13.20.2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schmitt CA, McCurrach ME, de Stanchina E, Wallace-Brodeur RR, Lowe SW. INK4a/ARF mutations accelerate lymphomagenesis and promote chemoresistance by disabling p53. Genes Dev. 1999;13:2670–2677. doi: 10.1101/gad.13.20.2670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Garrison SP, Jeffers JR, Yang C, et al. Selection against PUMA gene expression in Myc-driven B-cell lymphomagenesis. Mol Cell Biol. 2008;28:5391–5402. doi: 10.1128/MCB.00907-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maclean KH, Kastan MB, Cleveland JL. Atm deficiency affects both apoptosis and proliferation to augment Myc-induced lymphomagenesis. Mol Cancer Res. 2007;5:705–711. doi: 10.1158/1541-7786.MCR-07-0058. [DOI] [PubMed] [Google Scholar]
- 26.Langdon WY, Harris AW, Cory S, Adams JM. The c-myc oncogene perturbs B lymphocyte development in E-mu-myc transgenic mice. Cell. 1986;47:11–18. doi: 10.1016/0092-8674(86)90361-2. [DOI] [PubMed] [Google Scholar]
- 27.Shreeram S, Hee WK, Demidov ON, et al. Regulation of ATM/p53-dependent suppression of myc-induced lymphomas by Wip1 phosphatase. J Exp Med. 2006;203:2793–2799. doi: 10.1084/jem.20061563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jeffers JR, Parganas E, Lee Y, et al. Puma is an essential mediator of p53-dependent and -independent apoptotic pathways. Cancer Cell. 2003;4:321–328. doi: 10.1016/s1535-6108(03)00244-7. [DOI] [PubMed] [Google Scholar]
- 29.Alt JR, Greiner TC, Cleveland JL, Eischen CM. Mdm2 haplo-insufficiency profoundly inhibits Myc-induced lymphomagenesis. EMBO J. 2003;22:1442–1450. doi: 10.1093/emboj/cdg133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Eischen CM, Alt JR, Wang P. Loss of one allele of ARF rescues Mdm2 haploinsufficiency effects on apoptosis and lymphoma development. Oncogene. 2004;23:8931–8940. doi: 10.1038/sj.onc.1208052. [DOI] [PubMed] [Google Scholar]
- 31.Bartek J, Bartkova J, Lukas J. DNA damage signalling guards against activated oncogenes and tumour progression. Oncogene. 2007;26:7773–7779. doi: 10.1038/sj.onc.1210881. [DOI] [PubMed] [Google Scholar]
- 32.Gorgoulis VG, Vassiliou LV, Karakaidos P, et al. Activation of the DNA damage checkpoint and genomic instability in human precancerous lesions. Nature. 2005;434:907–913. doi: 10.1038/nature03485. [DOI] [PubMed] [Google Scholar]
- 33.Pusapati RV, Rounbehler RJ, Hong S, et al. ATM promotes apoptosis and suppresses tumorigenesis in response to Myc. Proc Natl Acad Sci U S A. 2006;103:1446–1451. doi: 10.1073/pnas.0507367103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zindy F, Eischen CM, Randle DH, et al. Myc signaling via the ARF tumor suppressor regulates p53-dependent apoptosis and immortalization. Genes Dev. 1998;12:2424–2433. doi: 10.1101/gad.12.15.2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Efeyan A, Murga M, Martinez-Pastor B, et al. Limited role of murine ATM in oncogene-induced senescence and p53-dependent tumor suppression. PLoS ONE. 2009;4:e5475. doi: 10.1371/journal.pone.0005475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Steinman H, Sluss H, Sands A, et al. Absence of p21 partially rescues Mdm4 loss and uncovers an antiproliferative effect of Mdm4 on cell growth. Oncogene. 2004;23:303–306. doi: 10.1038/sj.onc.1206925. [DOI] [PubMed] [Google Scholar]