Abstract
To understand possible causative roles of apoptosis gene regulation in age-related hearing loss (presbycusis), apoptotic gene expression patterns in the CBA mouse cochlea of four different age and hearing loss groups were compared, using GeneChip and real-time (qPCR) microarrays. GeneChip transcriptional expression patterns of 318 apoptosis-related genes were analyzed. Thirty eight probes (35 genes) showed significant differences in expression. The significant gene families include Caspases, B-cell leukemia/lymphoma2 family, P53, Cal-pains, Mitogen activated protein kinase family, Jun oncogene, Nuclear factor of kappa light chain gene enhancer in B-cells inhibitor-related and tumor necrosis factor-related genes. The GeneChip results of 31 genes were validated using the new TaqMan® Low Density Array (TLDA). Eight genes showed highly correlated results with the GeneChip data. These genes are: activating transcription factor3, B-cell leukemia/lymphoma2, Bcl2-like1, caspase4 apoptosis-related cysteine protease 4, Calpain2, dual specificity phosphatase9, tumor necrosis factor receptor superfamily member12a, and Tumor necrosis factor superfamily member13b, suggesting they may play critical roles in inner ear aging.
Keywords: Apoptosis, Cochlea, Hearing loss, Gene expression, PCR, Presbycusis
Introduction
Cell death can be broadly classified into three types: Apoptosis, oncosis (necrosis), and cell death with autophagy. Apoptosis (derived from Greek; apo means off and ptosis means falling) is an endogenous programmed cell death (PCD). It requires active participation of the cell itself, leaving a dead cell with intact plasma membrane and cellular organelles in addition to a reduction in cellular volume (pyknosis) and chromatin condensation that proceeds later to fragmented nuclei (karyorhexis). In a later phase, the plasma membrane shows budding, but it prevents the release of any factor that affects neighboring cells [1–3]. Apoptosis plays an important function in regulating organogenesis and maintaining normal cellular homeostasis during embryonic development and in adult organisms, respectively. Reports estimate that either too little or too much apoptosis can contribute to a significant number of medical illnesses, including oncogenesis [4, 5].
On the contrary, necrosis is unintentional (accidental) cell death, usually in response to an outside offense to the cell, such as toxins or inflammation. This can cause swelling of the dying cell, rupture of the plasma membrane and release of cytoplasmic content that may affect surrounding cells. Cell death with autophagy is typically characterized by engulfment of cellular contents in autophagosomes inside the cytoplasm [3, 6–8]. Mixed apoptotic and necrotic cell death can be found as the pathogenesis of the same disease. Some evidence supports that different types of cell death can share common mechanisms [3, 9–16].
Apoptosis has multiple pathways that differ according to tissue type and pathological condition. These pathways have been identified and broadly classified into two main types: extrinsic and intrinsic pathways. The extrinsic pathways include death-receptor and survival-factor-withdrawal pathways. The first is activated by stimulation of certain membranous receptors like TNF-alpha and Fas receptors while the latter involves activation of c-Jun and JNK; by reactive oxygen species (ROS), inflammatory cytokines, mixed lineage kinases, radiation or excitotoxicity. Both pathways subsequently activate certain cascades of factors that ultimately lead to cell death through their effect on mitochondrial membrane stability (increase in Bid, Bax, Bak, Noxa, Puma or Hrk, and decrease in Bcl and Bcl-xL families) and activation of caspases [8, 17–19]. Intrinsic apoptotic pathways are caused either by DNA damage or stress to the endoplasmic reticulum. DNA damage causes release of P53 that leads to mitochondrial membrane dysfunction, while endoplasmic reticulum stress causes calcium (Ca++) accumulation and calpain activation that can lead either to activation of caspases or lysosomes rupture, cathepsins release [2, 20, 21], or PARP-1 cleavage and ultimately DNA damage. In addition, Ca++ may activate c-Jun and JNK pathways and start the extrinsic survival factor withdrawal apoptotic pathway. In both pathways, cytochrome C is released with activation of down-stream caspases and cell death. Some exceptions for which apoptosis do not require caspase activation include the release of factors like Endo G and AIF from the mitochondria. These factors can bypass caspase activation and cause cellular damage and apoptosis [3, 22, 23].
The cochlea is a complex hydro-electro-chemical-mechanical system consisting of different structures that work together for effective sound processing and auditory perception. These structures include the inner and the outer hair cells, their supporting cells and the stria vascularis, which produces the potassium-rich endolymph underlying the endocochlear potential required for sensory transduction. All these types of cells are vital for sound transduction and initial processing of sound signals. Damage or death of one or more types of these cells with age is the main cause of age-related hearing loss (presbycusis).
Generally in the nervous system, aging can effect expression for apoptotic pathway genes so it is important to investigate aging changes in cochlear apoptotic gene expression and the possible roles in inner ear cell death. It is hypothesized that certain apoptotic pathways will show significant transcriptional gene expression changes with age and hearing loss, and that these will be correlated with functional hearing loss phenotype measures.
Material and methods
The four CBA mouse subject groups, the auditory brainstem response recordings (ABR thresholds), the otoacoustic emissions (DPOAE amplitudes), and gene microarray methodologies were the same as those of Tadros et al. [24].
Animal model
CBA/CaJ mice were bred in-house and housed according to institutional protocol, with original breeding pairs obtained from Jackson Laboratories. All animals had similar environmental and non-ototoxic history. The CBA mouse is a model organism for presbycusis. The young adult group was used as the baseline group for gene expression data analyses (e.g., calculation of fold changes). Functional hearing measurements were obtained prior to sacrifice similar to our previous investigations on all animals [25–27]. The sample set is segregated into four groups based upon age, DPOAE and ABR measurements, as given in Table 1: young adult control with good hearing (N = 8, 4 males and 4 females, age = 3.5 ± 0.4 months), middle-aged with good hearing (N = 17, 8 males and 9 females, age = 12.3 ± 1.3 months), old with mild presbycusis (N = 9, 4 males and 5 females, age = 27.7 ± 3.4 months) and old with severe presbycusis (N = 6, 2 males and 4 females, age = 30.6 ± 1.9 months).
Table 1.
(A) Mean auditory brainstem response (ABR) thresholds (dB SPL) for the four CBA mouse groups, and (B) mean distortion product otoacoustic emission (DPOAE) amplitudes (dB SPL) for the same four subject groups
| Young | Middle-age | Old mild HL | Old severe HL | |
|---|---|---|---|---|
| (A) | ||||
| ABR/3 kHz | 53.3 | 64.4 | 78.9 | 85.0 |
| ABR/6 kHz | 26.1 | 33.2 | 58.3 | 73.3 |
| ABR/12 kHz | 8.3 | 15.3 | 37.8 | 48.3 |
| ABR/24 kHz | 14.4 | 21.5 | 45.0 | 66.7 |
| ABR/32 kHz | 21.1 | 30.6 | 59.4 | 71.7 |
| ABR/48 kHz | 25.0 | 35.3 | 67.8 | 73.3 |
| (B) | ||||
| DPOAE/5,600 Hz | 0.7 | 4.7 | 1.4 | −8.5 |
| DPOAE/6,107 Hz | 1.7 | 5.5 | −9.8 | −21.7 |
| DPOAE/6,660 Hz | 12.5 | 13.7 | 4.9 | −18.5 |
| DPOAE/7,262 Hz | 12.2 | 13.1 | 2.7 | −21.0 |
| DPOAE/7,920 Hz | 21.9 | 23.1 | 8.1 | −14.9 |
| DPOAE/8,636 Hz | 17.4 | 17.5 | 2.7 | −19.0 |
| DPOAE/9,418 Hz | 20.8 | 20.8 | 10.0 | −15.5 |
| DPOAE/10,270 Hz | 24.9 | 25.3 | 10.5 | −13.7 |
| DPOAE/11,200 Hz | 22.0 | 21.4 | 8.0 | −15.1 |
| DPOAE/12,214 Hz | 19.2 | 22.2 | 14.8 | −3.9 |
| DPOAE/13,319 Hz | 32.0 | 31.1 | 22.3 | −0.4 |
| DPOAE/14,525 Hz | 27.6 | 26.7 | 16.8 | −11.2 |
| DPOAE/15,839 Hz | 21.8 | 25.5 | 9.9 | −18.3 |
| DPOAE/17,273 Hz | 24.1 | 25.6 | 10.3 | −21.0 |
| DPOAE/18,836 Hz | 30.3 | 27.9 | 11.2 | −18.2 |
| DPOAE/20,541 Hz | 21.0 | 20.6 | 7.1 | −15.3 |
| DPOAE/22,400 Hz | 15.2 | 14.5 | 3.5 | −19.4 |
| DPOAE/24,427 Hz | 6.7 | 6.0 | −5.4 | −21.5 |
| DPOAE/26,638 Hz | 8.0 | 5.9 | −13.8 | −22.6 |
| DPOAE/29,049 Hz | 20.1 | 18.8 | 10.3 | −19.1 |
| DPOAE/31,678 Hz | 16.0 | 14.1 | 1.0 | −21.2 |
| DPOAE/34,546 Hz | 10.1 | 7.9 | −2.5 | −23.1 |
| DPOAE/37,672 Hz | 11.2 | 7.7 | 2.2 | −22.1 |
| DPOAE/41,082 Hz | 10.1 | 4.9 | −7.2 | −23.6 |
| DPOAE/44,800 Hz | 12.3 | 8.0 | −0.8 | −19.8 |
All animal procedures were approved by the University of Rochester (Rochester, NY) Medical Center Animal Resource Review Committee.
Functional hearing assessment
Distortion product otoacoustic emissions (DPOAEs)
Ipsilateral acoustic stimulation and simultaneous measurement of distortion-product otoacoustic emissions were accomplished with the TDT BioSig III system. Stimuli were digitally synthesized at 200 kHz using SigGen software applications with the ratio of frequency 2 (F2) to frequency 1 (F1) constant at 1.25; L1 was equal to 65 dB sound pressure level (SPL) and L2 was equal to 55 dB SPL as calibrated in a coupler simulating the mouse ear canal. After synthesis, F1, F2, and the wideband noise were each passed through an RP2.1 D/A converter to PA5 programmable attenuators. Following attenuation, the signals went to ED1 speaker drivers which fed into the EC1 electrostatic loudspeakers coupled to the ear canal through short, flexible tubes with rigid plastic tapering tips. For DPOAE measurements, resultant ear canal sound pressure was recorded with an ER10B+ low-noise microphone and probe (Etymotic) housed in the same coupler as the F1 and F2 speakers. The output of the ER10B+ amplifier was put into an MA3 microphone amplifier, whose output went to an RP2.1 A/D converter for sampling at 200 kHz. A fast Fourier transform (FFT) was performed with TDT BioSig software on the resultant waveform. The magnitude of F1, F2, 2f1-f2 distortion product, and the noise floor of the frequency bins surrounding the 2f1-f2 components were measured from the FFT. The procedure was repeated for geometric mean frequencies ranging from 5.6 to 44.8 kHz (eight frequencies per octave) to adequately assess the neuroethologically functional range of mouse hearing.
Before data acquisition, individual mice were microscopically examined for evidence of external ear canal and middle ear obstruction. Mice with clearly visualized, healthy tympanic membranes were included. Mice were anesthetized with a mixture of ketamine and xylazine (120 and 10 mg/kg body weight, respectively) by intraperitoneal injection before all experimental sessions. All recording sessions were completed in a soundproof acoustic chamber (lined with Sonex) with body temperature maintained with a heating pad. Before recording, the operating microscope (Zeiss) was used to place the stimulus probe and microphone in the test ear, with the recording session duration limited by depth of anesthesia. Duration of testing was approximately 1 h per animal.
Auditory brainstem responses (ABRs)
Auditory brainstem responses were measured in response to tone pips of 3, 6, 12, 24, 32, and 48 kHz presented at a rate of 11 bursts/s. Auditory brainstem responses were recorded with subcutaneous platinum needle electrodes placed at the vertex (noninverting input), right-side mastoid prominence (inverted input), and tail (indifferent site). Electroencephalographic (EEG) activity was differentially amplified (50 or 100×) (Grass Model P511 EEG amplifier), then put into an analogue-to-digital converter (AD1, Tucker-Davis [TDT]) and digitized at 50 kHz. Each averaged response was based on 300–500 stimulus repetitions recorded over 10-millisec epochs. Contamination by muscle and cardiac activities was prevented by rejecting data epochs in which the single-trace electroencephalogram contained peak-to-peak amplitudes exceeding 50 μV. During this procedure, 5.0 mg/10.0 g body weight general anesthetic, Avertin (Tribromoethanol, delivered IP), was used to immobilize the mice. Normal body temperature was maintained at 38°C with a Servo heating pad. The ABR was recorded in a small sound-attenuating chamber.
Microarray gene expression processing
GeneChip
One Affymetrix M430A high-density oligonucleotide array set (A) was used for each cochlear sample. Each array contains 22,600 probe sets analyzing the expression of over 14,000 mouse genes. Eleven pairs of 25-mer oligonucleotides that span the coding region of the genes represent each gene. Each probe pair consists of a perfect match sequence that is complementary to the mRNA target and a mismatch sequence that has a single base pair mutation in a region critical for target hybridization; this sequence serves as a control for non-specific hybridization. Sequences used in the design of the array were selected from GenBank, dbEST, and RefSeq.
Samples preparation
RNA extraction
After completing the functional hearing assessments of each animal, it was sacrificed and the cochlear soft tissue, including the organ of Corti, supporting cells and lateral wall of two cochleae from each subject were dissected from the temporal bone, pooled together, and were homogenized in 1 ml of Trizol reagent per 50–100 mg of tissue using a polytron power homogenizer. The total RNA was isolated from the tissue homogenates of each sample using a modified Trizol protocol (Gibco BRL). Each sample was centrifuged at 12,000g for 10 min at 4°C and the clear supernatant was transferred to a new tube and incubated for 5 min at 15–30°C to permit the complete dissociation of nucleoprotein complexes. 0.2 ml of chloroform per each ml of Trizol reagent was added and the tube was shaken vigorously by hand for 15 s, then incubated at 15–30°C for 2 min and centrifuged at 12,000g for 15 min at 4°C. The aqueous phase was transferred to a new tube, 0.5 ml of isopropyl alcohol per 1 ml Trizol reagent, then incubated at 15–30°C for 10 min and centrifuged at 12,000g for 10 min at 4°C. The supernatant was separated and the RNA pellet was washed once with 1 ml 75% ethanol (EtOH) for each 1 ml Trizol reagent. The sample was mixed by vortex and then centrifuged at 7500g for 5 min at 4°C. The new RNA pellet was air-dried, dissolved in 10–20 μl of RNase-free water and incubated at 42°C for 5 min. The RNA quality was assessed by Agilent Bioanalyzer 2100, and absorbance measurements at A260/A280 using the nanodrop.
cDNA synthesis
For gene array analysis, cDNA synthesis was performed with 20 μg of total RNA using the Superscript Choice cDNA Synthesis Kit (Invitrogen). For qPCR, nuGen cDNA reagents kit was used to generate a high fidelity cDNA, which was modified at the 3′ end to contain an initiation site for T7 RNA polymerase. Detailed protocol is found in www.nugeninc.com.
In vitro transcription (IVT) and fragmentation
Clean up of double-stranded cDNA was done according to the Affymetrix GeneChip Expression analysis protocol. Synthesis of Biotin-labeled cRNA was performed by adding 1 μg of cDNA to 10× IVT labeling buffer, IVT labeling NTP mix, IVT labeling enzyme mix and RNase-free water, then, incubated at 37°C for 16 h. The Biotin-labeled cRNA was cleaned up according to the Affymetrix GeneChip expression analysis protocol and a 20 μg of full-length cRNA from each sample was fragmented by adding 5× fragmentation buffer and RNase-free water, followed by incubation at 94°C for 35 min. The standard fragmentation procedure produces a distribution of RNA fragment sizes from approximately 35–200 bases. After the fragmentation, cDNA, full-length cRNA and fragmented cRNA were analyzed by electrophoresis using the Agilent Bioanalyzer 2100 to assess the appropriate size distribution prior to microarray hybridization.
Target hybridization, washing, staining and scanning
GeneChip M430A probe arrays (Affymetrix) were hybridized, washed and stained according to the manufacturer’s instructions in a fluidics station. The arrays were scanned using a Hewlett Packard confocal laser scanner and visualized using GeneChip 5.1 software. Three data files were created, namely image data (.dat), cell intensity data (.cel) and expression probe analysis data (.chp) files. Detailed protocols for sample preparation and target labeling assays for expression analysis can be found at http://www.Affymetrix.com.
Real-time PCR (qPCR)
TaqMan® Low Density Array (TLDA) custom-designed format (48 plates) from Applied Biosystems, Inc. (CA) were used as duplicates for each gene expression per sample. Total cDNA concentration of 100 ng of each sample, RNase-free water and 50 μl of TaqMan® Universal PCR Master Mix were loaded into each reservoir, a total volume of 100 μl/reservoir. Each plate was centrifuged and sealed. Each plate had 18S-specific primer/probe as an endogenous control. The Applied Biosystems 7900HT Real-Time PCR system was used to perform the real-time reaction.
The reactions’ thermal cycle conditions were adjusted as 10 min initial setup at 95°C, followed by 40 cycles, each of which consisted of 15 s denaturing at 95°C and 1 min annealing/extend at 60°C. The results of each plate were analyzed using ABI PRISM software to calculate the CT value of each well and compare these values in studied gene wells with endogenous control wells. In this investigation, real-time PCR was done to confirm and quantify gene expression of 31 out of 35 significant apoptosis-related genes in the cochlea of different age groups of CBA mice as revealed by the GeneChip microarrays. The primer/probes of the other four genes were not available for the custom-designed TLDA. Detailed information about protocols is found in www.appliedbiosystems.com.
Statistical analyses
GeneChip expression analysis
After assessing chip quality, the Affymetrix GeneChip Operating Software (GCOS) automatically generates the (.cel) image file from the (.dat) data file. The signal log ratio of each sample determined the difference in expression of the studied gene in that sample from the mean expression of that gene in all samples from the young adult mice. A signal log ratio of 1.0 indicates an increase of the transcript level by 2-fold and −1.0 indicates a decrease by 2-fold. A signal log ratio of zero indicates no change.
For the 318 apoptosis-related probe genes on GeneChip, one-way Analysis of Variance (ANOVA) was used to compare between the signal log ratio values of the different subject groups. In addition, fold changes of all samples were calculated from signal log ratios using the following equations:
Real time PCR analysis
The threshold cycle (CT) values were measured to detect the threshold of each of the 31 significant and 18S genes in all cochleae samples [28, 29]. Each sample was measured in duplicate and normalized to the reference 18S gene expression. The CT value of each well was determined and the average of the two wells of each sample was calculated. For samples that showed no expression of the test gene, the results of the sample are not considered in the calculation.
Delta CT (ΔCT) for test gene of each sample was calculated using the equation:
Delta delta CT (ΔΔCT) was calculated using the following equation:
The fold change in the test gene expression was finally calculated from the formula:
A statistical evaluation of qPCR results was performed using one-way ANOVA to compare between ΔCT for gene expression in young adult age, middle age, old age mild presbycusis, and old age severe presbycusis groups.
For the significantly different genes on the GeneChip and qPCR, linear regression analyses were employed to find correlations between the signal log ratio values or fold change and the functional hearing measurements. These measurements were the distortion product otoacoustic emission (DPOAE) amplitudes at low frequencies (5.6–14.5 kHz), mid frequencies (15.8–29.0 kHz) and high frequencies (31.6–44.8 kHz), in addition to auditory brainstem response thresholds (ABR) at 3, 6, 12, 24, 32 and 48 kHz. The GraphPad Prism 4 software was used to perform the one-way ANOVA and the linear regression statistics.
Results
Microarray gene expression
The GeneChip results contain the gene expression values of more than 14,000 mouse genes. We selected 318 apoptosis-related genes to compare the levels of cochlear expression of these genes in different age/hearing loss mouse subject groups. Thirty eight GeneChip probes representing 35 genes showed statistical significance between different age groups (in three cases, there were two probe sets for the same genes). These 35 genes may play a role in cochlear presbycusis through coding proteins that are constituents of different apoptotic pathways, whose activity levels change with age or hearing loss. For example, extrinsic death receptor pathways may be affected by changes of the expression of the tumor necrosis factor receptor superfamily, member 12a (Tnfrsf12a), tumor necrosis factor superfamily, member 13b (Tnfsf13b), Tnf receptor-associated factor 3 (Traf3), rel oncogene (Rela), nuclear factor of kappa light chain gene enhancer in B-cells inhibitor, alpha (Nfkbia), nuclear factor of kappa light chain gene enhancer in B-cells inhibitor, beta (Nfkbib), CASP8 and FADD-like apoptosis regulator (Cflar, also known as Cflip), CD40 antigen (Cd40), C1qTNF-related protein-3 (C1qtnf3) and C1qTNF-related protein-5 (C1qtnf5). Extrinsic survival factor withdrawal pathways may be affected by Jun oncogene (Jun), Mitogen activated protein kinase kinase kinase 1 (Map3k1), Mitogen activated protein kinase kinase kinase 11 (Map3k11), Mitogen activated protein kinase 10 (Mapk10), Dual specificity phosphatase 9 (Dusp9), Activating transcription factor 3 (Atf3) and BH3 interacting domain, Harakiri, BCL2 interacting protein, apoptosis agonist (Hrk). Intrinsic pathway-related genes including Calpain 2 (Capn2), Calpain 7 (Capn7) and P53 may also play a role in apoptotic changes with presbycusis. Data of the present study show that in addition to these pathways, mitochondrial factors including the B-cell leukemia/lymphoma (Bcl) family may have a significant relation to presbycusis. These genes include B-cell leukemia/lymphoma 2 transcript variant 1 (Bcl2), B-cell leukemia/lymphoma 2 related protein A1d (Bcl2a1a), Bcl2-like 1 (bcl2l1), Bcl2-like 2 (Bcl2l2), BCL2-like 11 (also known as Bim or Bod) (apoptosis facilitator), transcript variant 1 (Bcl2l11), B-cell leukemia/lymphoma 3 (Bcl3), B-cell CLL/lymphoma 7B (Bcl7b), Myeloid cell leukemia sequence 1 (Mcl1) and Solute carrier family 25 (mitochondrial carrier, brain), member 14 (Slc25a14). Lastly, some downstream pathway genes significantly change their expression with age and hearing loss. These include cytochrome C, somatic (Cycs), Apoptotic peptidase activating factor 1 (Apaf1), Caspase-1 or IL-1B converting enzyme (Casp1), Caspase 4 apoptosis-related cysteine protease (Casp4), Caspase 7 (Casp7) and Caspase 9 (Casp9). In Table 2, the 38 significant apoptosis-related probes are classified according to their type of expression (up- or down-regulation) and their pathway type.
Table 2.
Statistical data for the 38 probes (35 genes) that showed significant gene expression differences (main effects) for the cochlear samples of all the subject groups from the gene microarrays
| Probeset name | Symbol | P-value; F |
|---|---|---|
| Probeset up-regulated with age/hearing loss | ||
| Tumor necrosis factor receptor superfamily, member 12a | Tnfrsf12a | 0.001**; 8.33 |
| Tumor necrosis factor receptor superfamily, member 12a | Tnfrsf12a | 0.001**; 8.84 |
| Tumor necrosis factor superfamily, member 13b | Tnfsf13b | 0.036*; 3.73 |
| Rel oncogene | Rela | 0.035*; 3.78 |
| Nuclear factor of kappa light chain gene enhancer in B-cells inhibitor, alpha | Nfkbia | 0.002**; 7.92 |
| Nuclear factor of kappa light chain gene enhancer in B-cells inhibitor, beta | Nfkbib | 0.034*; 3.80 |
| CASP8 and FADD-like apoptosis regulator | Cflar/Cflip | 0.04*; 3.6 |
| CD40 antigen | Cd40 | 0.042*; 3.55 |
| Jun oncogene | Jun | 0.0125*; 5.12 |
| Mitogen activated protein kinase kinase kinase 1 | Map3k1 | 0.047*; 4.4 |
| Mitogen activated protein kinase kinase kinase 11 | Map3k11 | 0.028*; 4.05 |
| Dual specificity phosphatase 9 | Dusp9/Mkp4 | 0.042*; 3.56 |
| Activating transcription factor 3 | Atf3 | 0.0001***; 12.85 |
| C1q and tumor necrosis factor related protein 3 | C1qtnf3 | 0.024*; 4.25 |
| C1q and tumor necrosis factor related protein 5 | C1qtnf5 | 0.016*; 4.81 |
| Transformation related protein 53 | P53 | 0.014*; 4.97 |
| BH3 interacting domain, Harakiri, BCL2 interacting protein, apoptosis agonist | Hrk | 0.005**; 6.39 |
| B-cell leukemia/lymphoma 2 (Bcl2), transcript variant 1 | Bcl2 | 0.042*; 3.53 |
| B-cell leukemia/lymphoma 2 related protein A1d | Bcl2a1a | 0.001**; 8.83 |
| B-cell leukemia/lymphoma 2 related protein A1d | Bcl2a1a | < 0.0001***; 31.11 |
| Bcl2-like 1 | Bcl2l1 | 0.0135*; 5.01 |
| Bim/Bod/BCL2-like 11 (apoptosis facilitator), transcript variant 1 | Bcl2l11 | 0.016*; 4.80 |
| Bim/Bod/BCL2-like 11 (apoptosis facilitator), transcript variant 1 | Bcl2l11 | 0.043*; 3.52 |
| B-cell leukemia/lymphoma 3 | Bcl3 | 0.039*; 3.62 |
| Myeloid cell leukemia sequence 1 | Mcl1 | 0.033*; 3.84 |
| Caspase-1/IL-1B converting enzyme | Casp1 | 0.029*; 4.03 |
| Caspase-9 | Casp9 | 0.036*; 3.73 |
| Caspase 4/11, apoptosis-related cysteine protease | Casp4/11 | 0.008**; 5.70 |
| Apoptotic peptidase activating factor 1 | Apaf1 | 0.034*; 3.82 |
| Cytochrome c, somatic | Cycs | 0.018*; 4.61 |
| Probeset down-regulated with age/hearing loss | ||
| Tnf receptor-associated factor 3 | Traf3 | 0.03*; 3.96 |
| Mitogen activated protein kinase 10 | Mapk10 | 0.024*; 4.23 |
| Calpain 2 | Capn2 | 0.002**; 7.725 |
| Calpain 7 | Capn7 | 0.049*; 3.36 |
| Bcl2-like 2 | Bcl2l2 | 0.002**; 8.02 |
| B-cell CLL/lymphoma 7B | Bcl7b | 0.034*; 3.82 |
| Caspase 7 | Casp7 | 0.035*; 3.78 |
| Solute carrier family 25 (mitochondrial carrier, brain), member 14 | Slc25a14 | 0.009**; 5.6 |
F The F ratio for the main effects of the ANOVAs
0.01 < P < 0.05
0.001 < P < 0.01
0.0001 <P <0.001
qPCR
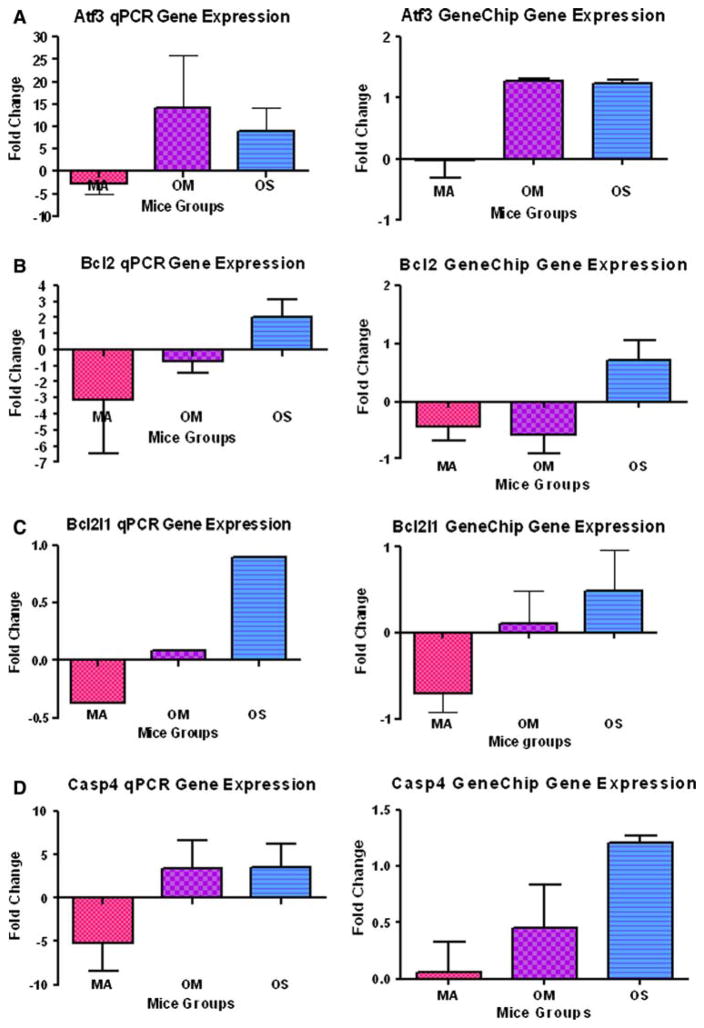
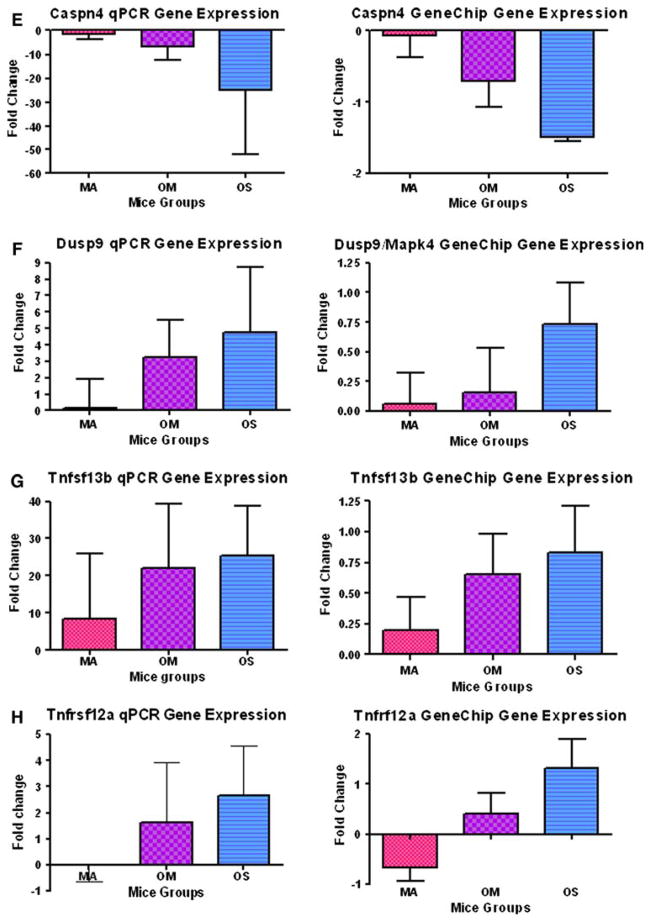
The qPCR is used as a quantitative verification test for gene expression values obtained from high throughput techniques. In this investigation, 8 genes out of the 35 genes showing statistical changes in the microarray, showed comparable results for both the GeneChip and the qPCR. Comparability of results was defined as cases where fold changes of gene expression have similar quantitative patterns for the three older subject groups in both GeneChip and qPCR results. We considered the results not comparable if the direction of regulation of gene expression of any subject group was different, compared to the young normal hearing group, between the two different techniques. The eight genes showing comparable changes are: activating transcription factor 3 (Atf3), B-cell leukemia/lymphoma 2 (Bcl2), Bcl2-like 1 (Bcl2l1), caspase 4 apoptosis-related cysteine protease (Casp4), Calpain 2 (Capn2), dual specificity phosphatase 9 (Dusp9), Tumor necrosis factor receptor superfamily member 12a (Tnfrsf12a), and Tumor necrosis factor superfamily member 13b (Tnfsf13b) (Fig. 1a–h). Comparing the gene expressions of the middle age, old age with mild hearing loss, and old age with severe hearing loss subject groups, all these genes except Capn2 showed upregulation with age and hearing loss. Six genes (Atf3, Bcl2, Bcl2l1, Casp4, Capn2 and Tnfrsf12a) showed down-regulation of the middle age group relative to the young adult baseline group, followed by upregulation of the old age groups with hearing loss.
Fig. 1.
The averages of fold changes of gene expression of the eight genes where GeneChip and real time PCR results in middle age; old mild hearing loss; and old severe hearing loss subject groups were the same. a Atf3. b Bcl2. c Bcl2l1. d Casp4/11. e Capn2. f Dusp9/Mkp4. g Tnfsf13b. h Tnfrsf12a. Comparing the gene expression between the three older mouse groups; note that all of these cell death-related genes were upregulated with age and hearing loss; except Capn2. Error bars are SEM. MA = middle age group; OM = old mild hearing loss group; and OS = old severe hearing loss group. Statistical significance of these gene expression changes can be found in Table 2
Correlation between gene expression and hearing measurements
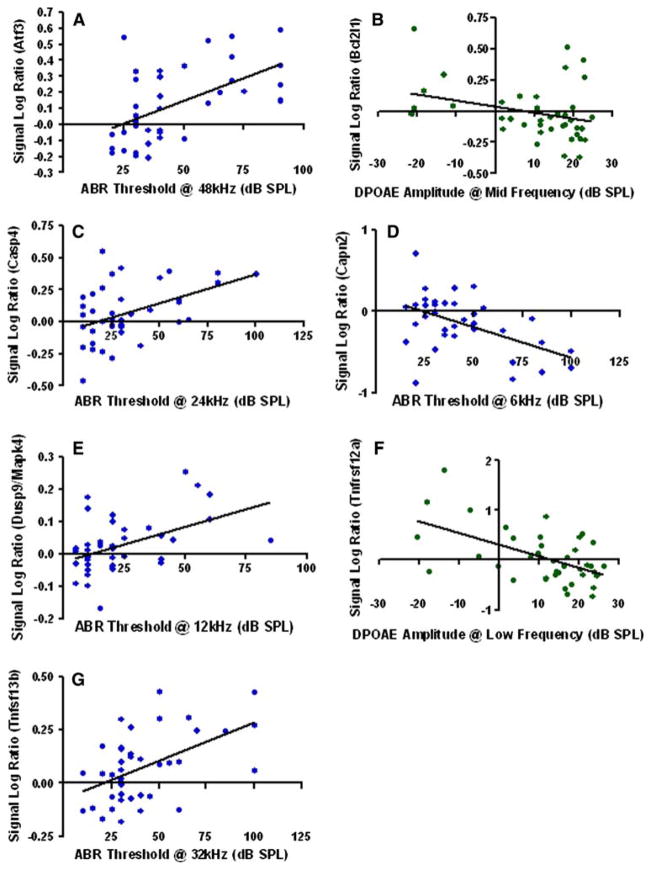
Linear regression tests were used to analyze the correlations between hearing measurements (ABR and DPOAE) and gene expression for both GeneChip and qPCR. Table 3 shows the correlation findings between the GeneChip expression data (signal log ratios of the 35 significant genes) with the functional hearing measurements. Regression plots utilized data of all the mice in the study. Notice in Table 3 that most of the qPCR-significant genes (as Atf3, Casp4, Capn2, Dusp9, Tnfrsf12a and Tnfsf13b) showed strong correlations between the genomic data and the hearing measurements. For instance, note that for the gene Bcl2ala, very strong correlations were discovered, with r values ranging from 0.61 to 0.77. Similarly, for gene Slc25a14, r values ranged from 0.45 to 0.65. Specific examples of some statistically significant correlations are given graphically in Fig. 2a–g.
Table 3.
The correlations, represented by r (correlation coefficient) and P values, between the GeneChip® expressions (as signal log ratios) of the 38 significant probes (35 genes) and the functional hearing measurements of all mice of the present study
| Symbol | ABR/3 kHz (r; P value) |
ABR/6 kHz (r; P value) |
ABR/12 kHz (r; P value) |
ABR/24 kHz (r; P value) |
ABR/32 kHz (r; P value) |
ABR/48 kHz (r; P value) |
DPOAE low freq. (r; P value) |
DPOAE mid freq. (r; P value) |
DPOAE high freq. (r; P value) |
|---|---|---|---|---|---|---|---|---|---|
| Probeset up-regulated with age/hearing loss | |||||||||
| Bcl2 | 0.06; 0.71 | 0.04; 0.81 | 0.16; 0.48 | 0.08; 0.64 | 0.12; 0.45 | 0.07; 0.68 | 0.30; 0.06 | 0.21; 0.19 | 0.19; 0.25 |
| Bcl2a1a | 0.20; 0.21 | 0.36; 0.021* | 0.40; 0.011* | 0.32; 0.048* | 0.36; 0.021* | 0.355; 0.025* | 0.53; 0.0004*** | 0.44; 0.0042** | 0.38; 0.016* |
| Bcl2a1a | 0.61; < 0.0001*** | 0.75; < 0.0001*** | 0.69; < 0.0001*** | 0.73; < 0.0001*** | 0.67; < 0.0001*** | 0.76; < 0.0001*** | 0.71; < 0.0001*** | 0.77; < 0.0001*** | 0.75; < 0.0001*** |
| Bcl2l1 | 0.17; 0.28 | 0.23; 0.16 | 0.12; 0.47 | 0.21; 0.18 | 0.12; 0.45 | 0.15; 0.34 | 0.31; 0.051 | 0.32; 0.042* | 0.31; 0.049* |
| Bcl2l11 | 0.18; 0.27 | 0.33; 0.036* | 0.34; 0.029* | 0.32; 0.044* | 0.249; 0.12 | 0.24; 0.134 | 0.335; 0.035* | 0.29; 0.069 | 0.185; 0.25 |
| Bcl2l11 | 0.046; 0.78 | 0.148; 0.36 | 0.13; 0.419 | 0.123; 0.45 | 0.104; 0.53 | 0.086; 0.597 | 0.217; 0.179 | 0.16; 0.322 | 0.086; 0.598 |
| Bcl3 | 0.155; 0.34 | 0.23; 0.148 | 0.211; 0.19 | 0.337; 0.03* | 0.318; 0.045* | 0.292; 0.068 | 0.357; 0.024* | 0.364; 0.021* | 0.326; 0.04* |
| Mcl1 | 0.254; 0.11 | 0.26; 0.1 | 0.147; 0.365 | 0.183; 0.259 | 0.131; 0.419 | 0.167; 0.303 | 0.27; 0.092 | 0.26; 0.105 | 0.22; 0.171 |
| Casp1 | 0.014; 0.93 | 0.133; 0.41 | 0.102; 0.53 | 0.182; 0.26 | 0.156; 0.34 | 0.1899; 0.24 | 0.123; 0.45 | 0.143; 0.38 | 0.129; 0.428 |
| Casp9 | 0.283; 0.08 | 0.3; 0.057 | 0.32; 0.044* | 0.22; 0.168 | 0.322; 0.043* | 0.284; 0.076 | 0.321; 0.044* | 0.308; 0.053 | 0.272; 0.089 |
| Casp4/11 | 0.179; 0.27 | 0.35; 0.028* | 0.495; 0.001** | 0.451; 0.004** | 0.6; < 0.0001*** | 0.6; < 0.0001*** | 0.51; 0.0009*** | 0.472; 0.002** | 0.497; 0.001** |
| Apaf1 | 0.158; 0.33 | 0.37; 0.018* | 0.456; 0.003** | 0.385; 0.014* | 0.392; 0.012* | 0.349; 0.027* | 0.44; 0.0044** | 0.44; 0.0044** | 0.42; 0.007** |
| P53 | 0.055; 0.74 | 0.229; 0.16 | 0.345; 0.029* | 0.321; 0.043* | 0.289; 0.07 | 0.211; 0.19 | 0.365; 0.02* | 0.366; 0.02* | 0.362; 0.022* |
| Jun | 0.176; 0.28 | 0.049; 0.77 | 0.013; 0.94 | 0.01; 0.95 | 0.142; 0.38 | 0.22; 0.17 | 0.042; 0.799 | 0.068; 0.68 | 0.105; 0.52 |
| Map3k1 | 0.24; 0.138 | 0.35; 0.027* | 0.397; 0.011* | 0.323; 0.042* | 0.419; 0.007** | 0.51; 0.0009*** | 0.246; 0.125 | 0.271; 0.091 | 0.252; 0.116 |
| Map3k11 | 0.115; 0.48 | 0.183; 0.26 | 0.113; 0.488 | 0.192; 0.235 | 0.216; 0.18 | 0.203; 0.21 | 0.333; 0.036* | 0.238; 0.14 | 0.206; 0.202 |
| Dusp9/Mkp4 | 0.27; 0.089 | 0.43; 0.006** | 0.47; 0.0023** | 0.42; 0.0069** | 0.489; 0.001** | 0.497; 0.001** | 0.48; 0.0016** | 0.40; 0.0097** | 0.367; 0.02* |
| Atf3 | 0.423; 0.0065** | 0.379; 0.016* | 0.426; 0.0062** | 0.426; 0.0062** | 0.518; 0.0006*** | 0.547; 0.0003*** | 0.447; 0.0039** | 0.473; 0.0021** | 0.493; 0.0012** |
| Hrk | 0.219; 0.17 | 0.395; 0.01* | 0.389; 0.013* | 0.325; 0.041* | 0.369; 0.019* | 0.379; 0.016* | 0.495; 0.001** | 0.458; 0.003** | 0.464; 0.0026** |
| Tnfrsf12a | 0.44; 0.004** | 0.5; 0.001** | 0.46; 0.0026** | 0.49; 0.0012** | 0.42; 0.0076** | 0.44; 0.0043** | 0.52; 0.0007*** | 0.46; 0.0028** | 0.41; 0.0087** |
| Tnfrsf12a | 0.5; 0.001** | 0.5; 0.0003*** | 0.47; 0.002** | 0.498; 0.001** | 0.43; 0.0056** | 0.47; 0.0022** | 0.55; 0.0002*** | 0.49; 0.0011** | 0.44; 0.0041** |
| Tnfsf13b | 0.2; 0.199 | 0.34; 0.011* | 0.43; 0.0057** | 0.46; 0.0029** | 0.52; 0.0007*** | 0.47; 0.0023** | 0.45; 0.0036** | 0.446; 0.004** | 0.487; 0.0014** |
| Rela | 0.056; 0.73 | 0.077; 0.64 | 0.055; 0.74 | 0.113; 0.49 | 0.002; 0.99 | 0.082; 0.62 | 0.126; 0.44 | 0.156; 0.39 | 0.103; 0.53 |
| Nfkbia | 0.31; 0.054 | 0.237; 0.14 | 0.25; 0.12 | 0.27; 0.092 | 0.316; 0.047* | 0.37; 0.019* | 0.35; 0.026* | 0.359; 0.023* | 0.301; 0.059 |
| Nfkbib | 0.174; 0.28 | 0.27; 0.097 | 0.235; 0.145 | 0.24; 0.136 | 0.166; 0.31 | 0.136; 0.403 | 0.302; 0.058 | 0.26; 0.104 | 0.218; 0.177 |
| Cflar/Cflip | 0.09; 0.583 | 0.124; 0.45 | 0.249; 0.122 | 0.225; 0.163 | 0.355; 0.025* | 0.355; 0.025* | 0.28; 0.077 | 0.237; 0.142 | 0.23; 0.153 |
| Cd40 | 0.33; 0.036* | 0.5; 0.001** | 0.47; 0.0022** | 0.47; 0.0024** | 0.223; 0.166 | 0.317; 0.046* | 0.34; 0.032* | 0.41; 0.0088** | 0.344; 0.0297* |
| C1qtnf3 | 0.224; 0.16 | 0.159; 0.33 | 0.097; 0.55 | 0.1; 0.95 | 0.003; 0.99 | 0.053; 0.75 | 0.22; 0.17 | 0.224; 0.16 | 0.232; 0.15 |
| C1qtnf5 | 0.098; 0.55 | 0.205; 0.21 | 0.305; 0.056 | 0.387; 0.014* | 0.417; 0.008** | 0.315; 0.048* | 0.426; 0.006** | 0.392; 0.012* | 0.339; 0.032* |
| Cycs | 0.054; 0.11 | 0.029; 0.86 | 0.065; 0.69 | 0.0005; 0.99 | 0.102; 0.53 | 0.129; 0.429 | 0.09; 0.582 | 0.044; 0.786 | 0.012; 0.943 |
| Probeset down-regulated with age/hearing loss | |||||||||
| Bcl2l2 | 0.245; 0.13 | 0.41; 0.009** | 0.48; 0.0019** | 0.445; 0.004** | 0.48; 0.0019** | 0.53; 0.0004*** | 0.49; 0.0015** | 0.498; 0.0011** | 0.455; 0.0032** |
| Bcl7b | 0.159; 0.33 | 0.247; 0.12 | 0.393; 0.012* | 0.44; 0.0049** | 0.467; 0.002** | 0.449; 0.0037** | 0.378; 0.016* | 0.398; 0.011* | 0.429; 0.0058** |
| Casp7 | 0.065; 0.69 | 0.238; 0.14 | 0.282; 0.078 | 0.299; 0.061 | 0.287; 0.072 | 0.204; 0.207 | 0.352; 0.026* | 0.264; 0.100 | 0.218; 0.177 |
| Slc25a14 | 0.46; 0.003** | 0.5; 0.0008*** | 0.5; 0.0009*** | 0.52; 0.0006*** | 0.57; 0.0001*** | 0.65; < 0.0001*** | 0.45; 0.0034** | 0.49; 0.0014** | 0.48; 0.0016** |
| Capn2 | 0.41; 0.009** | 0.5; 0.0009*** | 0.56; 0.0002*** | 0.5; 0.001** | 0.43; 0.005** | 0.44; 0.004** | 0.51; 0.0007*** | 0.49; 0.0014** | 0.38; 0.0153* |
| Capn7 | 0.167; 0.3 | 0.17; 0.29 | 0.083; 0.61 | 0.113; 0.49 | 0.02; 0.9 | 0.048; 0.77 | 0.199; 0.22 | 0.187; 0.248 | 0.167; 0.31 |
| Mapk10 | 0.256; 0.11 | 0.44; 0.004** | 0.46; 0.0076** | 0.49; 0.0015** | 0.456; 0.003** | 0.397; 0.011* | 0.436; 0.0049** | 0.408; 0.009** | 0.363; 0.021* |
| Traf3 | 0.24; 0.13 | 0.33; 0.038* | 0.271; 0.09 | 0.278; 0.083 | 0.266; 0.097 | 0.284; 0.076 | 0.268; 0.094 | 0.283; 0.077 | 0.254; 0.114 |
0.01 < P < 0.05
0.001 <P <0.01
0.0001 < P < 0.001
Fig. 2.
a–g Representative examples of the correlation between signal log ratios of different gene expressions in cochlear GeneChip samples and hearing measurements (ABR and DPOAE) for different frequencies. Low-frequency and mid-frequency DPOAEs in f and b represent the average DPOAE amplitudes in the frequency ranges of 5.6–14.5 kHz; and 15.8–29.0 kHz; respectively. Statistical significance of these; including P values and r (correlation coefficient) values can be found in Table 3
Discussion
Gene expression changes for apoptotic pathways in the cochlea have been investigated and these new findings point to the importance of different pathways in playing a role in cell death in the aging cochlea, as shown in Fig. 3. Relative to noise-, aminoglycoside- and cisplatin-induced hair cell damage, the age-related gene expression changes discovered in the present investigation appear to share a downstream final pathway via the release of cytochrome C into the cytoplasm and casp3 activation [30, 31]. Major upstream caspase activation, such as casp8 and casp9, has also been reported, but the protective effect of their inhibition, especially casp8, seems to be minor. Intracellular upstream events involving caspase activation, including JNK and c-jun activation have been explored and a protective effect of their inhibition was found for aminoglycoside- and noise-induced, but not cisplatin-induced, hair cell death [31–37]. P53 suppression was found to have a protective effect for cisplatin-induced apoptosis in both cochlear and vestibular hair cells [31, 38]. Data supported a protective effect of Bcl2 in the inner ear and demonstrated the translocation of Bax from cytoplasm to the mitochondria in hair cells of rats treated with cisplatin. Similar findings were reported in aged gerbils where immunohistochemical studies showed decreased expression of Bcl2 and increased expression of Bax in hair cells [39]. Other factors, such as mitochondrial uncoupling proteins (UCPs) [40] and nuclear factor kappaB (NF-κB) [41] play roles in protecting cochlear cells from amino-glycoside- and noise-induced damage, respectively.
Fig. 3.
A simplified schematic diagram for different apoptotic pathways showing the position of each gene or group of genes in these pathways; highlighting the importance that these results may have for future therapeutic manipulations of the expression of some of the significant genes to reduce apoptosis in aging sensory systems. Genes whose expression was significantly altered with age and hearing loss in the present investigation are indicated with red font (Please see on-line version for color coding)
GeneChip results are considered semi-quantitative and must be confirmed by qPCR for firm conclusions. The percentage of GeneChip results confirmed by qPCR varies across studies. In the present investigation, after validation of the GeneChip results with a novel qPCR array, the gene expression changes of eight genes were confirmed. These genes were Tnfrsf12a, Tnfsf13b, Dusp9, Atf3, Bcl2, Bcl2l1, Casp4 and Calp2 (Fig. 3). The discovery that most of the qPCR validated genes showed strong correlations between their transcriptional expression changes and the functional hearing tests (ABR and DPOAE), adds more implications for focusing on possible therapeutic applications of the observed expression changes of these genes on neurodegenerative pathways involved in cochlear hearing loss.
Tumor necrosis factor (TNF) and TNF receptor family
Research efforts for the past two decades have revealed the existence of a superfamily of 18 genes encoding 19 TNF ligands that signal through 30 receptors. One ligand has been found to bind to as many as five different receptors. This superfamily is associated with several disease conditions including acute and chronic inflammatory and genetic diseases [42–44]. TNF family and their corresponding receptors play important roles in cell death or survival, proliferation and maturation. A specific characteristic of all members of the TNF superfamily is that, through recruitment of TNF receptor-associated factors (TRAFs), they all activate NF-κB, a transcription factor that causes suppression of apoptosis, cell survival and proliferation. Some TNF ligands can activate both apoptosis and anti-apoptosis cell survival pathways according to certain conditions. The balance between these two types of pathways gives this superfamily the ability to act as switches for cell death or survival [43, 45].
The cytokine tumor necrosis factor-like weak inducer of apoptosis (TWEAK) is a member of the TNF superfamily. In addition to its known characteristic of inducing cell proliferation, survival and migration, TWEAK can also induce apoptosis [46–52]. Tumor necrosis factor receptor superfamily, member 12a (Tnfrsf12a), also known as Tweak receptor (TweakR) or fibroblast growth factor-inducible 14 receptor (Fn14 receptor), a type I transmembrane protein, is currently the only known receptor to bind TWEAK.
Tnfrsf12a is expressed by endothelial cells and fibroblasts in a variety of adult tissues as heart, kidney, lung and placenta [43, 53–55]. Recent studies showed that this receptor is involved in the pathogenesis of several human diseases as atherosclerosis, stroke, rheumatoid arthritis and cancer [52, 55]. Up-regulation of both TWEAK and its receptor may induce neurodegeneration through mediation of NF-κB which can play a pro-apoptotic role in addition to its well-known role as an anti-apoptotic factor [50, 56–60].
In the present investigation, we report for the first time that the Tnfrsf12a is expressed in CBA mouse cochlea and this expression is up-regulated with and correlated to age and hearing loss. The up-regulation of this receptor may play a significant role in induction of cochlear cell death and age-related hearing loss.
Tumor necrosis factor superfamily, member 13b (Tnfsf13b) (also known as B lymphocyte stimulator (BLyS), B-cell activation factor (BAFF), TNF homologue that activates apoptosis (THANK), TNF and Apol-related leukocyte-expressed ligand 1 (TALL1 or zTNF4) has typical structural characteristics of a TNF superfamily member. Tnfsf13b binds to three types of receptors, all of which are type III transmembrane proteins: Transmembrane activator and calcium modulator ligand interactor (TACI; Tnfrsf13b), B-cell maturation antigen (BCMA) and BAFF receptor (BAFF-R; Tnfrsf13c). Tnfsf13b is expressed in T cells, monocytes, macrophages and dendritic cells [61–64] while its receptors are expressed in B and T cells. Strictly regulated expression of Tnfsf13b is essential to maintain B cell survival without triggering autoimmunity through stimulation of the anti-apoptotic effect of NF-κB [65, 66]. Overexpression of Tnfsf13b has been reported in autoimmune diseases as in systemic lupus erythematosus [67, 68]. Our results indicate that Tnfsf13b has significantly higher expression in old aged hearing loss groups. This may indicate the autoimmune factor in age-related hearing loss.
MAPK and dual-specificity MAPK phosphatase (MKP) families
Mitogen-activated protein kinases (MAPKs) are a family of kinases that transduce signals from cell membrane to nucleus in response to a wide range of stimuli, including stress. MAPK signaling pathway can modulate gene expression, mitosis, proliferation, motility, metabolism and programmed cell death. MAPKs are classified into four subfamilies: Extracellular signal-regulated kinases (ERKs), c-Jun N-terminal kinases (JNKs), p38-MAPKs and ERK5, also known as Big MAP kinase 1 (BMK1). Different subfamily members control different apoptotic signaling pathways, relaying different types of stimulus information, which can lead either to cell death or survival [69–76]. Because the magnitude and duration of MAPK activation has a crucial effect on physiological outcomes [77, 78], negative regulatory mechanisms by dephosphorylation of either threonine or tyrosine residues is extremely important. In mammalian cells, dual-specificity MAPK phosphatases (MKPs) are a subfamily of ten catalytically active proteins that form part of a complex regulatory network controlling the MAPK pathway [76, 79].
DUSP9 is a cytoplasmic extracellular signal regulated kinase (ERK)-specific MKP that may also be able to inactivate p38 MAPK [76, 79–84]. The DUSP9 gene is on the X chromosome. Deletion of this gene causes a lethal embryonic abnormality, indicating that DUSP9 performs an essential role during placental development. Though DUSP9 does not lead to a significant changes in the basal phosphorylation state of MAPKs in adult kidney and testis [84] or play any role to control JNK activation in response to oxidative stress [85], it is possible that it is required to regulate MAPK signaling in adult tissue during undefined conditions of stress [76]. Our investigation shows that there is a significant upregulation of DUSP9 expression in CBA mouse cochlea with age and hearing loss. This may be explained as a defensive mechanism to block MAPK signaling and extrinsic apoptotic pathways due to stress in presbycusis.
Activating transcription factor 3 (Atf3)
Activating transcription factor 3 (Atf3) is a member of the CREB (cyclic adenosine monophosphate-responsive element binding protein)/ATF family. It is induced in vivo in response to seizure, toxic chemicals, mechanical injury and ischemia or in vitro as exposure to growth-stimulating factors, cytokines, homocysteine, oxidized low density lipoprotein, tumor necrosis factor α (TNFα) [86–92], UV, anisomycine and cyclohexamide [88, 93–95], suggesting that Atf3 plays a role in the stress-responsive pathway. Previous reports indicated that JNK signaling pathways are involved in the induction of Atf3, however, further investigations showed that interleukin 6 (IL-6) and p53 also play a role in Atf3 induction [88, 96, 97]. Atf3 has different activities as a homodimer or heterodimers. As a homodimer, Atf3 acts as a transcriptional repressor. Atf3 forms heterodimers with Atf2 [98], c-Jun [99], JunB [100], and JunD [101, 102]. In its heterodimeric form, Atf3 can act as activator or repressor according to the promoter context [88].
The target promoters of Atf3 are CHOP (also known as GADD153) [103, 104] and Atf3 itself which may explain the transient nature of its induction. Atf3 and c-Jun have been implicated in the survival, repair and neurite outgrowth, but they may also trigger apoptosis under certain conditions by repressing the CRE (Cyclic-AMP response element)-dependent gene expression of cell survival factors [105–109]. As Atf3 is specifically induced in sensory and motor neurons after nerve injury, it may be considered as a neuronal marker of nerve injury in the nervous system [110].
Our results show that Atf3 expression is upregulated with age and hearing loss in the CBA mouse cochlea. This may be a sign of cellular stress, induction of stress-response apoptotic pathways, or a tool for cell survival.
B-cell leukemia/lymphoma family (Bcl2)
The Bcl2 family consists of approximately 15 members. The family can be divided into anti-apoptotic and pro-apoptotic subgroups that share one or more Bcl2 homology domains (BH1–BH4). The anti-apoptotic members [Bcl2, Bcl-xL (the long isoform encoded by Bcl2l1), Bcl-W (Bcl2l2), Mcl1 and A1/Bfl1 (Bcl2a1)] all have BH1 and BH2 domains. They are thought to exert their effects by stabilizing the mitochondrial membrane potential and preventing the release of cytochrome C and apoptosis inducing factor (AIF). The release of cytochrome C into the cytoplasm would result in activation of an apoptosome complex composed of APAF1, cytochrome C and desoxy-ATP. This complex can activate casp9 which then cleaves and activates downstream effector caspases as casp3. Activated casp3 initiates cell death by degrading different cell proteins and DNA fragments through the cleavage of CAD (Caspase activated DNase). Bcl2 and Bcl-xL can block cell death caused by a wide variety of apoptotic stimuli as chemotherapeutic drugs, ultraviolet radiation, heat shock, free radicals, calcium ions, TNF and interleu-kin-3 [111–113]. The pro-apoptotic members [Bax, Bak, Bok, Bid, Bad, Bcl-xS (the short isoform encoded by Bcl2l1), Bim (Bcl2l11), Bik, Blk and Hrk] all have the BH3 domain which is necessary for their pro-apoptotic effect. Interaction between anti-apoptotic and pro-apoptotic members participates in the determination of the sensitivity of a cell to apoptosis [114–117].
While Bcl-xL (Bcl2l1) appears to be the most dominant anti-apoptotic factor in developing central nervous system and adult neurons, both Bcl2 and Bcl-xL are expressed in embryonic neurons [118]. Bcl-xL expression levels have been found to be high prior to birth, undetectable in neonatal tissue, low in young adult brain, and elevated again in the human aged brain [119], while over-expression of Bcl2 prevents neomycin-induced hearing loss in adult mouse [120]. Our findings showing the higher expressions of Bcl-xL and Bcl2 in aged mice are consistent with these previous findings and suggest protective effects of these genes during cochlear aging. The significantly higher expression of these genes with age and hearing loss in CBA mouse cochlea may be a defensive response to increased exposure to age-related oxidative stress.
Caspase family
Caspases are a family of cysteine proteases that play a critical role in mammalian apoptosis, activation of cytokines and induction of inflammation. To date, 11 human and 10 murine caspases have been identified. They can be subdivided into two main groups: Upstream or initiator caspases (Casp1, 2, 4, 5, 8, 9, 10, 12, 13) and downstream or effector caspases (Casp3, 6, 7). Upstream caspases are responsible for caspase activation or regulation of inflammatory processes and they have long N-terminal prodomains [Caspase recruitment domains (CARDs) or death effector domains (DEDs)]. Downstream caspases cause the actual demolition of the cell and they tend to have short or absent domains [121]. Another classification of caspases is Interleukin-1β converting enzyme (ICE) caspases (Human Casp1, 4, 5, 13, 14 and mouse casp11 and 12) and CED-3 caspases (Casp3, 6, 7, 8, 9 and 10) [122].
ICE (Casp1) family plays an important role in induction of cellular inflammation through interleukin-1β (IL-1β) and interleukin-18 (IL-18) production in addition to their role at the onset of apoptosis [123]. Casp11 is the murine homolog of human casp4 with 59% identical amino acid sequence [124]. Casp11 acts as an upstream activator of Casp1 as it does not process pro-IL-1β directly, but only through activation of Casp1. Mice deficient in Casp11 have a similar phenotype as Casp1−/− mice while overexpression of either Casp1 or 11 induces apoptosis which can be prevented by Bcl2. Unlike Casp1, Casp11 mediates the lipopolysaccharide (LPS) neurotoxicity [125] and regulates cell migration [126]. In Fas- and TNF-mediated apoptosis, the Casp1 proteases family seem to act downstream of Casp8 and Casp10 and upstream to the Casp3 proteases family [127]. Human Casp4 may also be involved in a different apoptotic pathway as it has been found that to be activated by agents that cause stress of the endoplasmic reticulum (ER), inducing activation of Casp3 [124].
Our results show that Casp4 (also known as Casp11) is upregulated in CBA mouse cochlea with age and hearing loss. This may play an important causative role for presbycusis through activation of Casp3. Recent studies showed that hearing loss due to cisplatin ototoxicity can be prevented by Casp3 and Casp9 inhibitors [128]. Therefore, inhibition of human Casp4 or mouse Casp11 may also play a role in slowing down or prevention of age-related hearing loss.
Calpain family
Calpains are cytoplasmic cysteine proteases similar to caspases. Fourteen human genes have been identified as members of the calpain family. These genes express several tissue-specific isoforms (n-calpains and two ubiquitous isoforms [μ-calpain (Calp1) and m-calpain (Calp2)]. Calp1 binds to calcium with relatively high affinity while Calp2 binds to calcium in relatively low affinity [129–131]. Unlike caspases, they require calcium ions for activity and have no sequence preferences at cleavage sites [130]. In addition, calpains have different substrates than caspases, including IκBα [145], calpastatin, p53, Casp9, and Casp12. Calpains play an important role in apoptosis and pathological conditions. Calp1 and Calp2 are well-documented factors in pathogenesis of acute neurological injuries and Alzheimer’s disease.
Calp2 is involved in different apoptotic pathways. In the endoplasmic reticulum (ER) stress pathway, ER stress causes release of calcium that leads to activation of Calp2 which in turn causes cleavage of Procasp12 to generate active Casp12. Casp12 activates the downstream Casp3, 6 and 7 leading to cell death. Indeed, this apoptotic pathway is one of the pathways responsible for kanamycin-induced cochlear hair cell death [132]. In a different apoptotic pathway, Calp2 causes fragmentation of Bcl-xL (Bcl2l1) and Bcl2 in addition to cleavage of Bax to increase the Bax/bcl2 ratio, a step that mediates cytochrome C release, activates Casp3 and initiates the apoptotic execution.
In this investigation, we have found that Calp2 is down-regulated with age and hearing loss. This down-regulation may play a role in the previously reported up-regulation of Bcl-xL and Bcl2 as a protective mechanism against apoptosis. The Calp2 down-regulation, in addition to the non-significant change of the other calpains, may exclude the endoplasmic reticulum stress apoptotic pathway as a cause of presbycusis.
Age changes in the brain
Gene expression profiling of the aging brain has revealed some provocative similarities with the results of the present investigation of the sensory cells of the inner ear. Specifically, expression changes of mouse genes involved in inflammatory responses, oxidative stress and neurotrophic support have been observed, which can activate a variety of apoptotic pathways [133, 134]. It is interesting to note that some evidence suggests that an enriched environment or caloric restriction in mice can decrease the age-related upregulation of apoptosis-related genes. A study of gene expression changes in the aging human retina also had some parallels with the current investigation, as changes in genes involved in energy metabolism, stress responses, cell growth, and neuronal transmission/signaling were observed [135].
Conclusion
The present investigation suggests that apoptosis in cochlear age-related hearing loss is induced by more than one pathway. The significant changes in gene expression related to extrinsic apoptotic pathways exceeded those of the intrinsic pathways. Therapeutic strategies to decrease the rate of progression or to prevent age-related hearing loss could focus on extrinsic pathways; particularly the death receptor pathway and survival factor withdrawal pathway. For example, antagonists of TNF-related factors such as TWEAK and BAFF or a TweakR blocker could be administered to inhibit apoptosis in the aging inner ear. Therapeutic maneuvering of MKP4, MAPK and c-Jun might also help achieve this goal. In addition, overexpression of Bcl2 or Bcl-xL and/or induction of caspase-4 antagonists may have important biotherapeutic impacts.
Acknowledgments
The research was funded by NIH Grant P01 AG09524 from the Nat. Inst. on Aging, and P30 DC05409 from the Nat. Inst. on Deafness & Comm. Disorders.
References
- 1.Melino G, Knight RA, Nicotera P. How many ways to die?. How many different models of cell death? Cell Death Differ . 2005;12:1457–1462. doi: 10.1038/sj.cdd.4401781. [DOI] [PubMed] [Google Scholar]
- 2.Stoka V, Turk V, Turk B. Lysosomal cysteine cathepsins: signaling pathways in apoptosis. Biol Chem . 2007;388:555–560. doi: 10.1515/BC.2007.064. [DOI] [PubMed] [Google Scholar]
- 3.Yasuhara S, Asai A, Sahani N, Martyn JAJ. Mitochondria, endoplasmic reticulum, and alternative pathways of cell death in critical illness. Crit Care Med. 2007;35(9):488–495. doi: 10.1097/01.CCM.0000278045.91575.30. [DOI] [PubMed] [Google Scholar]
- 4.Fischer U, Schulze-Osthoff K. Apoptosis-based therapies and drug targets. Cell Death Differ . 2005;12:942–961. doi: 10.1038/sj.cdd.4401556. [DOI] [PubMed] [Google Scholar]
- 5.Elmore S. Apoptosis: a review of programmed cell death. Toxicol Pathol. 2007;35(4):495–516. doi: 10.1080/01926230701320337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clarke PG. Developmental cell death: morphological diversity and multiple mechanisms. Anat Embryol (Berl) . 1990;181:195–213. doi: 10.1007/BF00174615. [DOI] [PubMed] [Google Scholar]
- 7.Ihara T, Yamamoto T, Sugamata M, Okumura H, Ueno Y. The process of ultrastructural changes from nuclei to apoptotic body. Virchows Arch . 1998;433:443–447. doi: 10.1007/s004280050272. [DOI] [PubMed] [Google Scholar]
- 8.LeDoux SP, Druzhyna NM, Hollensworth SB, Harrison JF, Wilson GL. Mitochondrial and repair: a critical player in the response of cells of the CNS to genotoxic insults. Neuroscience . 2007;145:1249–1259. doi: 10.1016/j.neuroscience.2006.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vaux DL, Korsmeyer SJ. Cell death in development. Cell . 1999;96:245–254. doi: 10.1016/S0092-8674(00)80564-4. [DOI] [PubMed] [Google Scholar]
- 10.Mathiasen IS, Sergeev IN, Bastholm L, Elling F, Norman AW, Jaattela M. Calcium and calpain as key mediators of apoptosis-like death induced by vitamin D compounds in breast cancer cells. J Biol Chem . 2002;277:30738–30745. doi: 10.1074/jbc.M201558200. [DOI] [PubMed] [Google Scholar]
- 11.Haeberlein SLB. Mitochondrial function in apoptotic neuronal cell death. Neurochem Res. 2004;29(3):521–530. doi: 10.1023/B:NERE.0000014823.74782.b7. [DOI] [PubMed] [Google Scholar]
- 12.Leyton L, Quest AF. Supramolecular complex formation in cell signaling and disease; an update on a recurrent theme in cell life and death. Biol Res. 2004;37:29–43. doi: 10.4067/s0716-97602004000100004. [DOI] [PubMed] [Google Scholar]
- 13.Liu X, Van Vleet T, Schnellmann RG. The role of calpain in oncotic cell death. Annu Rev Pharmacol Toxicol . 2004;44:349–370. doi: 10.1146/annurev.pharmtox.44.101802.121804. [DOI] [PubMed] [Google Scholar]
- 14.Hetz CA, Hunn M, Rojas P, Torres V, Leyton L, Quest AF. Caspase-dependent initiation of apoptosis and necrosis by the Fas receptor in lymphoid cells: onset of necrosis is associated with delayed ceramide increase. J Cell Sci . 2002;115:4671–4683. doi: 10.1242/jcs.00153. [DOI] [PubMed] [Google Scholar]
- 15.Hetz CA, Torres V, Quest AF. Beyond apoptosis: nonapoptotic cell death in physiology and disease. Biochem Cell Biol. 2005;83(5):579–588. doi: 10.1139/o05-065. [DOI] [PubMed] [Google Scholar]
- 16.Demarchi F, Bertoli C, Copetti T, Tanida I, Brancolini C, Eskelinen EL, et al. Calpain is required for macroautophagy in mammalian cells. J Cell Biol . 2006;175:595–605. doi: 10.1083/jcb.200601024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Skulachev VP. Why are mitochondria involved in apoptosis? Permeability transition pores and apoptosis as selective mechanisms to eliminate superoxide-producing mitochondria and cells. FEBS Lett . 1996;397:7–10. doi: 10.1016/0014-5793(96)00989-1. [DOI] [PubMed] [Google Scholar]
- 18.Ferri KF, Kroemer G. Mitochondria—the suicide organelles. Bioessays . 2001;23:111–115. doi: 10.1002/1521-1878(200102) 23:2<111::AID-BIES1016>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 19.Josa A, Susin SA, Daugas E, Stanford WL, Cho SK, Li CYJ, et al. Essential role of the mitochondrial apoptosis-inducing factor in programmed cell death. Nature . 2001;410:549–554. doi: 10.1038/35069004. [DOI] [PubMed] [Google Scholar]
- 20.Guicciardi ME, Deussing J, Miyoshi H, Bronk SF, Svingen PA, Peters C, et al. Cathepsin B contributes to TNF-α mediated hepatocyte apoptosis by promoting mitochondrial release of cytochrome c. J Clin Invest . 2000;106:1127–1137. doi: 10.1172/JCI9914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stoka V, Turk B, Schendel SL, Kim TH, Cirman T, Snipas SJ, et al. Lysosomal protease pathways to apoptosis. Cleavage of bid, not procaspases, is the most likely route. J Biol Chem . 2001;276:3149–3157. doi: 10.1074/jbc.M008944200. [DOI] [PubMed] [Google Scholar]
- 22.Hail N, Jr, Carter BZ, Konopleva M, Andreeff M. Apoptosis effector mechanisms: a requiem performed in different keys. Apoptosis . 2006;11:889–904. doi: 10.1007/s10495-006-6712-8. [DOI] [PubMed] [Google Scholar]
- 23.Falschlehner C, Emmerich CH, Gerlach B, Walczak H. TRAIL signaling: decisions between life and death. Int J Biochem Cell Biol . 2007;39:1462–1475. doi: 10.1016/j.biocel.2007.02.007. [DOI] [PubMed] [Google Scholar]
- 24.Tadros SF, D’Souza M, Zettel ML, Zhu X, Frisina RD. Glutamate-related gene expression changes with age in the mouse auditory midbrain. Brain Res . 2007;1127:1–9. doi: 10.1016/j.brainres.2006.09.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jacobson M, Kim S, Romney J, Zhu X, Frisina RD. Contralateral suppression of distortion-product otoacoustic emissions declines with age: a comparison of findings in CBA mice with human listeners. Laryngoscope. 2003;113(10):1707–1713. doi: 10.1097/00005537-200310000-00009. [DOI] [PubMed] [Google Scholar]
- 26.Guimaraes P, Zhu X, Cannon T, Kim S, Frisina RD. Sex differences in distortion product otoacoustic emissions as a function of age in CBA mice. Hear Res. 2004;192(1–2):83–89. doi: 10.1016/j.heares.2004.01.013. [DOI] [PubMed] [Google Scholar]
- 27.Varghese GI, Zhu X, Frisina RD. Age-related declines in distortion product otoacoustic emissions utilizing pure tone contralateral stimulation in CBA/CaJ mice. Hear Res . 2005;209:60–67. doi: 10.1016/j.heares.2005.06.006. [DOI] [PubMed] [Google Scholar]
- 28.Giulietti A, Overbergh L, Valckx D, Decallonne B, Bouillon R, Mathieu C. An overview of real-time quantitative PCR: applications to quantify cytokine gene expression. Methods . 2001;4:386–401. doi: 10.1006/meth.2001.1261. [DOI] [PubMed] [Google Scholar]
- 29.Baik SY, Jung KH, Choi MR, Yang BH, Kim SH, Lee JS, et al. Fluoxetine-induced up-regulation of 14-3-3zeta and tryptophan hydroxylase levels in RBL-2H3 cells. Neurosci Lett . 2005;374:53–57. doi: 10.1016/j.neulet.2004.10.047. [DOI] [PubMed] [Google Scholar]
- 30.Van De Water TR, Lallemend F, Eshraghi AA, Ahsan S, He J, Guzman J, et al. Caspases, the enemy within, and their role in oxidative stress-induced apoptosis of inner ear sensory cells. Otol Neurotol . 2004;25:627–632. doi: 10.1097/00129492-200407000-00035. [DOI] [PubMed] [Google Scholar]
- 31.Cheng AG, Cunningham LL, Ruble EW. Mechanisms of hair cell death and protection. Curr Opin Otolaryngol Head Neck Surg . 2005;13:343–348. doi: 10.1097/01.moo.0000186799.45377.63. [DOI] [PubMed] [Google Scholar]
- 32.Ylikoski J, Xing-Qun L, Virkkala J, Pirvola U. Blockade of c-Jun N-terminal kinase pathway attenuates gentamicin-induced cochlear and vestibular hair cell death. Hear Res . 2002;163:71–81. doi: 10.1016/S0378-5955(01)00380-X. [DOI] [PubMed] [Google Scholar]
- 33.Wang J, Van De Water TR, Bonny C, De Ribaupierre F, Puel JL, Zine A. A peptide inhibitor of c-Jun N-terminal kinase protects against both aminoglycoside and acoustic trauma-induced auditory hair cell death and hearing loss. J Neurosci. 2003;23:8596–8607. doi: 10.1523/JNEUROSCI.23-24-08596.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Scarpidis U, Madnani D, Shoemaker C, Fletcher CH, Kojima K, Eshraghi AA, et al. Arrest of apoptosis in auditory neurons: implications for sensorineural preservation in cochlear implantation. Otol Neurotol . 2003;24:409–417. doi: 10.1097/00129492-200305000-00011. [DOI] [PubMed] [Google Scholar]
- 35.Matsui JI, Gale JE, Warchol ME. Critical signaling events during the aminoglycoside-induced death of sensory hair cells in vitro. J Neurobiol. 2004;61(2):250–266. doi: 10.1002/neu.20054. [DOI] [PubMed] [Google Scholar]
- 36.Zine A, Van De Water TR. The MAPK/JNK signalling pathway offers potential therapeutic targets for the prevention of acquired deafness. Curr Drug Target CNS Neurol Disord. 2004;3(4):325–332. doi: 10.2174/1568007043337166. [DOI] [PubMed] [Google Scholar]
- 37.Sugahara K, Rubel EW, Cunningham LL. JNK signaling in neomycin-induced vestibular hair cell death. Hear Res . 2006;221:128–135. doi: 10.1016/j.heares.2006.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang M, Liu W, Ding D, Salvi R. Pifithrin-α suppresses p53 and protects cochlear and vestibular hair cells from cisplatin-induced apoptosis. Neuroscience . 2003;120:191–205. doi: 10.1016/S0306-4522(03)00286-0. [DOI] [PubMed] [Google Scholar]
- 39.Alam SA, Oshima T, Suzuki M, Kawase T, Takasaka T, Ikeda K. The expression of apoptosis-related proteins in the aged cochlea of Mongolian gerbils. Laryngoscope . 2001;111:528–534. doi: 10.1097/00005537-200103000-00026. [DOI] [PubMed] [Google Scholar]
- 40.Kitahara T, Li-Korotky HS, Balaban CD. Regulation of mitochondrial uncoupling proteins in mouse inner ear ganglion cells in response to systemic kanamycin challenge. Neuroscience . 2005;135:639–653. doi: 10.1016/j.neuroscience.2005.06.056. [DOI] [PubMed] [Google Scholar]
- 41.Lang H, Schulte BA, Zhou D, Smythe N, Spicer SS, Schmiedt RA. Nuclear factor kappaB deficiency is associated with auditory nerve degeneration and increased noise-induced hearing loss. J Neurosci. 2006;26(13):3541–3550. doi: 10.1523/JNEUROSCI.2488-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bodmer JL, Schneider P, Tschopp J. The molecular architecture of the TNF superfamily. Trends Biochem Sci. 2002;27(1):19–26. doi: 10.1016/S0968-0004(01)01995-8. [DOI] [PubMed] [Google Scholar]
- 43.Gaur U, Aggarwal BB. Regulation of proliferation, survival and apoptosis by members of the TNF superfamily. Biochem Pharmacol . 2003;66:1403–1408. doi: 10.1016/S0006-2952 (03)00490-8. [DOI] [PubMed] [Google Scholar]
- 44.Aggarwal BB. Signaling pathways of the TNF superfamily: a double-edged sword. Nat Rev Immunol. 2003;3(9):745–756. doi: 10.1038/nri1184. [DOI] [PubMed] [Google Scholar]
- 45.Ch’en PFT, Xu XG, Liu XS, Liu Y, Song CJ, Screaton GR, et al. Characterization of monoclonal antibodies to the TNF and TNF receptor families. Cell Immunol . 2005;236:78–85. doi: 10.1016/j.cellimm.2005.08.010. [DOI] [PubMed] [Google Scholar]
- 46.Schneider P, MacKay F, Steiner V, Hofmann K, Bodmer J, Holler N, et al. BAFF, a novel ligand of the tumor necrosis factor family, stimulates B cell growth. J Exp Med . 1999;189:1747–1756. doi: 10.1084/jem.189.11.1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wilson CA, Browning JL. Death of HT29 adenocarcinoma cells induced by TNF family receptor activation is caspase-independent and displays features of both apoptosis and necrosis. Cell Death Differ . 2002;9:1321–1333. doi: 10.1038/sj.cdd.4401107. [DOI] [PubMed] [Google Scholar]
- 48.Nakayama M, Ishidoh K, Kayagaki N, Kojima Y, Yamaguchi N, Nakano H, et al. Multiple pathways of TWEAK-induced cell death. J Immunol. 2002;168:734–743. doi: 10.4049/jimmunol.168.2.734. [DOI] [PubMed] [Google Scholar]
- 49.Nakayama M, Ishidoh K, Kojima Y, Harada N, Kominami E, Okumura K, et al. Fibroblast growth factor-inducible 14 mediates multiple pathways of TWEAK-induced cell death. J Immunol. 2003;170:341–348. doi: 10.4049/jimmunol.170.1.341. [DOI] [PubMed] [Google Scholar]
- 50.Potrovita I, Zhang W, Burkly L, Hahm K, Lincecum J, Wang MZ, et al. Tumor necrosis factor-like weak inducer of apoptosis-induced neurodegeneration. J Neurosci. 2004;24(38):8237–8244. doi: 10.1523/JNEUROSCI.1089-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Baxter FO, Came PJ, Abell K, Kedjouar B, Huth M, Rajewsky K, et al. IKKβ/2 induces TWEAK and apoptosis in mammary epithelial cells. Development . 2006;133:3485–3494. doi: 10.1242/dev.02502. [DOI] [PubMed] [Google Scholar]
- 52.Winkles JA, Tran NL, Brown SAN, Stains N, Cunliffe HE, Berens ME. Role of TWEAK and Fn14 in tumor biology. Front Biosci . 2007;12:2761–2771. doi: 10.2741/2270. [DOI] [PubMed] [Google Scholar]
- 53.Feng SL, Guo Y, Factor VM, Thorgeirsson SS, Bell DW, Testa JR, et al. The Fn14 immediate-early response gene is induced during liver regeneration and highly expressed in both human and murine hepatocellular carcinomas. Am J Pathol. 2000;156:1253–1261. doi: 10.1016/S0002-9440(10)64996-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Meighan-Mantha RL, Hsu DK, Guo Y, Brown SAN, Feng SLY, Peifley KA, et al. The mitogen-inducible Fn14 gene encodes a type I transmembrane protein that modulates fibroblast adhesion and migration. J Biol Chem . 1999;274:33166–33176. doi: 10.1074/jbc.274.46.33166. [DOI] [PubMed] [Google Scholar]
- 55.Wiley SR, Winkles JA. TWEAK, a member of the TNF superfamily, is a multifunctional cytokine that binds the TweakR/Fn14 receptor. Cytokine Growth Factor Rev. 2003;14(3–4):241–249. doi: 10.1016/S1359-6101(03)00019-4. [DOI] [PubMed] [Google Scholar]
- 56.Kim SH, Lee WH, Kwon BS, Oh GT, Choi YH, Park JE. Tumor necrosis factor receptor superfamily 12 may destabilize atherosclerotic plaques by inducing matrix metalloproteinases. Jpn Circ J . 2001;65:136–138. doi: 10.1253/jcj.65.136. [DOI] [PubMed] [Google Scholar]
- 57.Han S, Yoon K, Lee K, Kim K, Jang H, Lee NK, et al. TNF-related weak inducer of apoptosis receptor, a TNF receptor superfamily member, activates NF-κB through TNF receptor-associated factors. Biochem Biophys Res Commun . 2003;305:789–796. doi: 10.1016/S0006-291X(03)00852-0. [DOI] [PubMed] [Google Scholar]
- 58.Saitoh T, Nakayama M, Nakano H, Yagita H, Yamamoto N, Yamaoka S. TWEAK induces NF-κB2 p100 processing and long lasting NF-κB activation. J Biol Chem. 2003;278(38):36005–36012. doi: 10.1074/jbc.M304266200. [DOI] [PubMed] [Google Scholar]
- 59.He L, Grammer AC, Wu X, Lipsky PE. TRAF3 forms heterotrimers with TRAF2 and modulates its ability to mediate NF-κB activation. J Biol Chem. 2004;279(53):55855–55865. doi: 10.1074/jbc.M407284200. [DOI] [PubMed] [Google Scholar]
- 60.Pachiappan A, Thwin MM, Manikandan J, Gopalakrishnakone P. Glial inflammation and neurodegeneration induced by candoxin, a novel neurotoxin from Bungarus candidus venom: global gene expression analysis using microarray. Toxicon . 2005;46:883–899. doi: 10.1016/j.toxicon.2005.08.017. [DOI] [PubMed] [Google Scholar]
- 61.Moore PA, Belvedere O, Orr A, Pieri K, LaFleur DW, Feng P, et al. BLyS: member of the tumor necrosis factor family and B lymphocyte stimulator. Science . 1999;285:260–263. doi: 10.1126/science.285.5425.260. [DOI] [PubMed] [Google Scholar]
- 62.Schneider P, Schwenzer R, Haas E, Muhlenbeck F, Schubert G, Scheurich P, et al. TWEAK can induce cell death via endogenous TNF and TNF receptor 1. Eur J Immunol . 1999;29:1785–1792. doi: 10.1002/(SICI)1521-4141(199906)29:06<1785::AID-IMMU1785>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 63.Nardelli B, Belvedere O, Roschke V, Moore PA, Olsen HS, Migone TS, et al. Synthesis and release of B-lymphocyte stimulator from myeloid cells. Blood . 2001;97:198–204. doi: 10.1182/blood.V97.1.198. [DOI] [PubMed] [Google Scholar]
- 64.Kawasaki A, Tsuchiya N, Fukazawa T, Hashimoto H, Tokunaga K. Analysis on the association of human BLYS (BAFF, TNFSF13B) polymorphisms with systemic lupus erythematosus and rheumatoid arthritis. Genes Immun . 2002;3:424–429. doi: 10.1038/sj.gene.6363923. [DOI] [PubMed] [Google Scholar]
- 65.Kern C, Cornuel JF, Billard C, Tang R, Rouillard D, Stenou V, et al. Involvement of BAFF and APRIL in the resistance to apoptosis of B-CLL through an autocrine pathway. Blood . 2004;103:679–688. doi: 10.1182/blood-2003-02-0540. [DOI] [PubMed] [Google Scholar]
- 66.Ramanujam M, Davidson A. The current status of targeting BAFF/BLyS for autoimmune diseases. Arthritis Res Ther . 2004;6:197–202. doi: 10.1186/ar1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gross JA, Johnston J, Murdi S, Enselman R, Dillon SR, Madden K, et al. TACI and BCMA are receptors for a TNF homologue implicated in B-cell autoimmune disease. Nature . 2000;404:995–999. doi: 10.1038/35010115. [DOI] [PubMed] [Google Scholar]
- 68.Croker JA, Kimberly RP. SLE: challenges and candidates in human disease. Trends Immunol. 2005;26(11):580–586. doi: 10.1016/j.it.2005.09.001. [DOI] [PubMed] [Google Scholar]
- 69.Davis RJ. Signal transduction by the JNK group of MAP kinases. Cell . 2000;103:239–252. doi: 10.1016/S0092-8674(00)00116-1. [DOI] [PubMed] [Google Scholar]
- 70.Chang L, Karin M. Mammalian MAP kinase signaling cascades. Nature. 2001;410:37–40. doi: 10.1038/35065000. [DOI] [PubMed] [Google Scholar]
- 71.Pearson G, Robinson F, Beers Gibson T, Xu BE, Karandikar M, Berman K, et al. Mitogen-activated protein (MAP) kinase pathways: regulation and physiological functions. Endocr Rev. 2001;22:153–183. doi: 10.1210/edrv.22.2.0428. [DOI] [PubMed] [Google Scholar]
- 72.Johnson GL, Lapadat R. Mitogen-activated protein kinase pathways mediated by ERK, JNK, and p38 protein kinases. Science. 2002;298:1911–1912. doi: 10.1126/science.1072682. [DOI] [PubMed] [Google Scholar]
- 73.Theodosiou A, Ashworth A. MAP kinase phospatases. Genome Biol. 2002;3(7):1–10. doi: 10.1186/gb-2002-3-7-reviews3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wada T, Penninger JM. Mitogen-activated protein kinases in apoptosis regulation. Oncogene. 2004;23:2838–2849. doi: 10.1038/sj.onc.1207556. [DOI] [PubMed] [Google Scholar]
- 75.Dickinson RJ, Keyse SM. Diverse physiological functions for dual-specificity MAP kinase phosphatases. J Cell Sci. 2006;119(22):4607–4615. doi: 10.1242/jcs.03266. [DOI] [PubMed] [Google Scholar]
- 76.Marshall CJ. Specificity of receptor tyrosine kinase signaling: transient versus sustained extracellular signal-regulated kinase activation. Cell. 1995;80:179–185. doi: 10.1016/0092-8674(95)90401-8. [DOI] [PubMed] [Google Scholar]
- 77.Sabbagh W, Jr, Flatauer LJ, Bardwell AJ, Bardwell L. Specificity of MAP kinase signaling in yeast differentiation involves transient versus sustained MAPK activation. Mol Cell. 2001;8:683–691. doi: 10.1016/s1097-2765(01)00322-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ebisuya M, Kondoh K, Nishida E. The duration, magnitude and compartmentalization of ERK MAP kinase activity: mechanisms for providing signaling specificity. J Cell Sci. 2005;118:2997–3002. doi: 10.1242/jcs.02505. [DOI] [PubMed] [Google Scholar]
- 79.Owens DM, Keyse SM. Differential regulation of MAP kinase signaling by dual-specificity protein phosphatases. Oncogene. 2007;26:3203–3213. doi: 10.1038/sj.onc.1210412. [DOI] [PubMed] [Google Scholar]
- 80.Groom L, Sneddon AA, Alessi DR, Dowd S, Keyse SM. Differential regulation of the MAP, SAP and RK/p38 kinases by Pyst1, a novel cytosolic dual-specificity phosphatase. EMBO J. 1996;15:3621–3632. [PMC free article] [PubMed] [Google Scholar]
- 81.Camps M, Nichols A, Gillieron C, Antonsson B, Muda M, Chabert C, et al. Catalytic activation of the phosphatase MKP-3 by ERK2 mitogen-activated protein kinase. Science. 1998;280:1262–1265. doi: 10.1126/science.280.5367.1262. [DOI] [PubMed] [Google Scholar]
- 82.Dowd S, Sneddon AA, Keyse SM. Isolation of the human genes encoding the Pyst1 and Pyst2 phosphatases: characterisation of Pyst2 as a cytosolic dual-specificity MAP kinase phosphatase and its catalytic activation by both MAP and SAP kinases. J Cell Sci. 1998;111:3389–3399. doi: 10.1242/jcs.111.22.3389. [DOI] [PubMed] [Google Scholar]
- 83.Dickinson RJ, Williams DJ, Slack DN, Williamson J, Seternes OM, Keyse SM. Characterization of a murine gene encoding a developmentally regulated cytoplasmic dual-specificity mitogen-activated protein kinase phosphatase. Biochem J. 2002;364:145–155. doi: 10.1042/bj3640145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Christie GR, Williams DJ, MacIsaac F, Dickinson RJ, Rosewell I, Keyse SM. The dual-specificity protein phosphatase DUSP9/MKP-4 is essential for placental function but it is not required for normal embryonic development. Mol Cell Biol. 2005;25(18):8323–8333. doi: 10.1128/MCB.25.18.8323-8333.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Teng CH, Huang WN, Meng TC. Several dual specificity phosphatases coordinate to control the magnitude and duration of JNK activation in signaling response to oxidative stress. J Biol Chem. 2007;282(39):28395–28407. doi: 10.1074/jbc.M705142200. [DOI] [PubMed] [Google Scholar]
- 86.Liang G, Wolfgang CD, Chen BPC, Chen TH, Hai T. ATF3 gene, genomic organization, promoter, and regulation. J Biol Chem. 1996;271(3):1695–1701. doi: 10.1074/jbc.271.3.1695. [DOI] [PubMed] [Google Scholar]
- 87.McCully KS. Homocysteine and vascular disease. Nat Med. 1996;2:386–389. doi: 10.1038/nm0496-386. [DOI] [PubMed] [Google Scholar]
- 88.Hai T, Wolfgang CD, Marsee DK, Allen AE, Sivaprasad U. ATF3 and stress responses. Gene Expr. 1999;7(4–6):321–335. [PMC free article] [PubMed] [Google Scholar]
- 89.Cai Y, Zhang C, Nawa T, Aso T, Tanaka M, Oshiro S, et al. Homocysteine-responsive ATF3 gene expression in human vascular endothelial cells: activation of c-Jun NH2-terminal kinase and promoter response element. Blood. 2000;96:2140–2148. [PubMed] [Google Scholar]
- 90.Kawauchi J, Zhang C, Nobori K, Hashimoto Y, Adachi MT, Noda A, et al. Transcriptional repressor activating transcription factor 3 protects human umbilical vein endothelial cells from tumor necrosis factor-α-induced apoptosis through down-regulation of p53 transcription. J Biol Chem. 2002;277:39025–39034. doi: 10.1074/jbc.M202974200. [DOI] [PubMed] [Google Scholar]
- 91.Nawa T, Nawa MT, Adachi MT, Uchimura I, Schimokawa R, Fujisawa K, et al. Expression of transcriptional repressor ATF3/LRF1 in human atherosclerosis: colocalization and possible involvement in the cell death of vascular endothelial cells. Atherosclerosis. 2002;161:281–291. doi: 10.1016/s0021-9150(01)00639-6. [DOI] [PubMed] [Google Scholar]
- 92.Inoue K, Zama T, Kamimoto T, Aoki R, Ikeda Y, Kimura H, et al. TNFα-induced ATF3 expression is bidirectionally regulated by the JNK and ERK pathways in vascular endothelial cells. Genes Cells. 2004;9:59–70. doi: 10.1111/j.1356-9597.2004.00707.x. [DOI] [PubMed] [Google Scholar]
- 93.Mahadevan LC, Edwards DR. Signaling and superinduction. Nature. 1991;349:747–748. doi: 10.1038/349747c0. [DOI] [PubMed] [Google Scholar]
- 94.Kyriakis JM, Banerjee P, Nikolakaki E, Dai T, Rubie EA, Ahmed MF, et al. The stress-activated protein kinase subfamily of c-Jun kinases. Nature. 1994;369:156–160. doi: 10.1038/369156a0. [DOI] [PubMed] [Google Scholar]
- 95.Wolfgang CD, Liang G, Okamoto Y, Allen AE, Hai T. Transcriptional autorepression of the stress-inducible gene ATF3. J Biol Chem. 2000;275(22):16865–16870. doi: 10.1074/jbc.M909637199. [DOI] [PubMed] [Google Scholar]
- 96.Cressman DE, Greenbaum LE, deAngelis RA, Cilibarto G, Furth EE, Poli V, et al. Liver failure and defective hepatocyte regeneration in interleukin-6-deficient mice. Science. 1996;274:1379–1383. doi: 10.1126/science.274.5291.1379. [DOI] [PubMed] [Google Scholar]
- 97.Zhang C, Gao C, Kawauchi J, Hashimoto Y, Tsuchida N, Kitajima S. Transcriptional activation of the human stress-inducible transcriptional repressor ATF3 gene promoter by p53. Biochem Biophys Res Comm. 2002;297:1302–1310. doi: 10.1016/s0006-291x(02)02382-3. [DOI] [PubMed] [Google Scholar]
- 98.Hai T, Liu F, Coukos WJ, Green MR. Transcription factor ATF cDNA clones: an extensive family of leucine zipper proteins able to selectively form DNA-binding heterodimers. Genes Dev. 1989;3:2083–2090. doi: 10.1101/gad.3.12b.2083. [DOI] [PubMed] [Google Scholar]
- 99.Hai T, Curran T. Cross-family dimerization of transcription factors Fos/Jun and ATF/CREB alters DNA binding specificity. Proc Natl Acad Sci USA. 1991;88:3720–3724. doi: 10.1073/pnas.88.9.3720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Hsu JC, Bravo R, Taub R. Interactions among LRF-1, JunB, c-Jun and c-Fos define a regulatory program in the G1 phase of liver regeneration. Mol Cell Biol. 1992;12:4654–4665. doi: 10.1128/mcb.12.10.4654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Chu HM, Tan Y, Kobierski LA, Balsam LB, Comb MJ. Activating transcription factor-3 stimulates 3′, 5′-cyclic adenosine monophosphate-dependent gene expression. Mol Endocrinol. 1994;8:59–68. doi: 10.1210/mend.8.1.8152431. [DOI] [PubMed] [Google Scholar]
- 102.Fawcett TW, Martindale JL, Guyton KZ, Hai T, Holbrook NJ. Complexes containing activating transcription factor (ATF)/cAMP-responsive-element-binding protein (CREB) interact with the CCAAT-enhancer-binding protein (C/EBP)-ATF composite site to regulate Gadd153 expression during the stress response. Biochem J. 1999;339:135–141. [PMC free article] [PubMed] [Google Scholar]
- 103.Chen BPC, Wolfgang CD, Hai T. Analysis of ATF3, a transcription factor induced by physiological stresses and modulated by gadd153/Chop10. Mol Cell Biol. 1996;16(3):1157–1168. doi: 10.1128/mcb.16.3.1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Wolfgang CD, Chen BPC, Martindale JL, Holbrook NJ, Hai T. Gadd153/Chop10, a potential target gene of the transcriptional repressor ATF3. Mol Cell Biol. 1997;17:6700–6707. doi: 10.1128/mcb.17.11.6700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Mashima T, Udagawa S, Tsuruo T. Involvement of transcriptional repressor ATF3 in acceleration of caspase protease activation during DNA damaging agent-induced apoptosis. J Cell Physiol. 2001;188:352–358. doi: 10.1002/jcp.1130. [DOI] [PubMed] [Google Scholar]
- 106.Nakagomi S, Suzuki Y, Namikawa K, Kiryu-Seo S, Kiyama H. Expression of the activating transcription factor 3 prevents c-Jun N-terminal kinase-induced neuronal death by promoting heat shock protein 27 expression and Akt activation. J Neurosci. 2003;23(12):5187–5196. doi: 10.1523/JNEUROSCI.23-12-05187.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Pearson AG, Gary CW, Pearson JF, Greenwood JM, During MJ, Dragunow M. ATF3 enhances c-Jun-mediated neurite sprouting. Mol Brain Res. 2003;120:38–45. doi: 10.1016/j.molbrainres.2003.09.014. [DOI] [PubMed] [Google Scholar]
- 108.Vulg AS, Teuling E, Haasdijk ED, French P, Hoogenraad CC, Jaarsma D. ATF3 expression precedes death of spinal motoneurons in amyotrophic lateral sclerosis-SOD1 transgenic mice and correlates with c-Jun phosphorylation, CHOP expression, somato-dendritic ubiquitination and Golgi fragmentation. Eur J Neurosci. 2005;22:1881–1894. doi: 10.1111/j.1460-9568.2005.04389.x. [DOI] [PubMed] [Google Scholar]
- 109.Seijffers R, Allchorne AJ, Woolf CJ. The transcription factor ATF-3 promotes neurite outgrowth. Mol Cell Neurosci. 2006;32:143–154. doi: 10.1016/j.mcn.2006.03.005. [DOI] [PubMed] [Google Scholar]
- 110.Tsujino H, Kondo E, Fukuoka T, Dai Y, Tokunaga A, Miki K, et al. Activating transcription factor 3 (ATF3) induction by axotomy in sensory and motoneurons: a novel neuronal marker of nerve injury. Mol Cell Neurosci. 2000;15:170–182. doi: 10.1006/mcne.1999.0814. [DOI] [PubMed] [Google Scholar]
- 111.McDonnell TJ, Beham A, Sarkiss M, Andersen MM, Lo P. Importance of the Bcl-2 family in cell death regulation. Cell Mol Life Sci. 1996;52(10–11):1008–1017. doi: 10.1007/BF01920110. [DOI] [PubMed] [Google Scholar]
- 112.Tang L, Tron VA, Reed JC, Mah KJ, Krajewska M, Li G, et al. Expression of apoptosis regulators in cutaneous malignant melanoma. Clin Cancer Res. 1998;4:1865–1871. [PubMed] [Google Scholar]
- 113.Pena JC, Thompson CB, Recant W, Vokes EE, Rudin CM. Bcl-xL and Bcl-2 expression in squamous cell carcinoma of the head and neck. Cancer. 1999;85(1):164–170. doi: 10.1002/(sici)1097-0142(19990101)85:1<164::aid-cncr23>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- 114.Nagy ZS, Esiri MM. Apoptosis-related protein expression in the hippocampus in Alzheimer’s disease. Neurobiol Aging. 1997;18(6):565–571. doi: 10.1016/s0197-4580(97)00157-7. [DOI] [PubMed] [Google Scholar]
- 115.Kuan CY, Roth KA, Flavell RA, Rakic P. Mechanisms of programmed cell death in the developing brain. Trends Neurosci. 2000;23:291–297. doi: 10.1016/s0166-2236(00)01581-2. [DOI] [PubMed] [Google Scholar]
- 116.Hopkins-Donaldson S, Cathomas R, Simoes-Wust AP, Kurtz S, Belyanskya L, Stahel RA, et al. Induction of apoptosis and chemosensitization of mesothelioma cells by Bcl-2 and Bcl-xL antisense treatment. Int J Cancer. 2003;106:160–166. doi: 10.1002/ijc.11209. [DOI] [PubMed] [Google Scholar]
- 117.Rodemann HP, Blaese MA. Responses of normal cells to ionizing radiation. Semin Radiat Oncol. 2007;17:81–88. doi: 10.1016/j.semradonc.2006.11.005. [DOI] [PubMed] [Google Scholar]
- 118.Levin LA, Schlamp CL, Spieldoch RL, Geszvain KM, Nickells RW. Identification of the bcl-2 family of genes in the rat retina. Invest Ophthalmol Vis Sci. 1997;38(12):2545–2553. [PubMed] [Google Scholar]
- 119.Drache B, Diehl GE, Beyreuther K, Perlmutter LS, Konig G. Bcl-xl-specific antibody labels activated microglia associated with Alzheimer’s disease and other pathological states. J Neurosci Res. 1997;47:98–108. [PubMed] [Google Scholar]
- 120.Cunningham LL, Matsui JI, Warchol ME, Rubel EW. Overexpression of Bcl-2 prevents neomycin-induced hair cell death and caspase-9 activation in the adult mouse utricle in vitro. J Neurobiol. 2004;60:89–100. doi: 10.1002/neu.20006. [DOI] [PubMed] [Google Scholar]
- 121.Martinon F, Tschopp J. Inflammatory caspases: linking an intracellular innate immune system to autoinflammatory diseases. Cell. 2004;117:561–574. doi: 10.1016/j.cell.2004.05.004. [DOI] [PubMed] [Google Scholar]
- 122.Monack DM, Navarre WW, Falkow S. Salmonella-induced macrophage death: the role of caspase-1 in death and inflammation. Microbes Infect. 2001;3:1201–1212. doi: 10.1016/s1286-4579(01)01480-0. [DOI] [PubMed] [Google Scholar]
- 123.Siegmund B. Interleukin-1β converting enzyme (caspase-1) in intestinal inflammation. Biochem Pharm. 2002;64:1–8. doi: 10.1016/s0006-2952(02)01064-x. [DOI] [PubMed] [Google Scholar]
- 124.Lamkanfi M, Declercq W, Saelens X, Vandenabeele P. Alice in caspase land. A phylogenetic analysis of caspases from worm to man. Cell Death Differ. 2002;9:358–361. doi: 10.1038/sj.cdd.4400989. [DOI] [PubMed] [Google Scholar]
- 125.Arai H, Furuya T, Yasuda T, Miura M, Mizuno Y, Mochizuki H. Neurotoxic effects of lipopolysaccharide on nigral dopaminergic neurons are mediated by microglial activation, interleukin-1β, and expression of caspase-11 in mice. J Biol Chem. 2004;279:51637–51653. doi: 10.1074/jbc.M407328200. [DOI] [PubMed] [Google Scholar]
- 126.Li J, Brieher WM, Scimone ML, Kang SJ, Zhu H, Yin H, et al. Caspase-11 regulates cell migration by promoting Aip1-Cofilin-mediated actin depolymerization. Nat Cell Biol. 2007;9(3):276–286. doi: 10.1038/ncb1541. [DOI] [PubMed] [Google Scholar]
- 127.Enari M, Talanian RV, Wong WW, Nagata S. Sequential activation of ICE-like and CPP32-like proteases during Fas-mediated apoptosis. Nature. 1996;380:723–726. doi: 10.1038/380723a0. [DOI] [PubMed] [Google Scholar]
- 128.Wang J, Ladrech S, Pujol R, Brabet P, Van De Water TR, Puel JL. Caspase inhibitors, but not c-Jun NH2-terminal kinase inhibitor treatment, prevent cisplatin-induced hearing loss. Cancer Res. 2004;64:9217–9224. doi: 10.1158/0008-5472.CAN-04-1581. [DOI] [PubMed] [Google Scholar]
- 129.Chan SL, Mattson MP. Caspase and calpain substrates: roles in synaptic plasticity and cell death. J Neurosci Res. 1999;58:167–190. [PubMed] [Google Scholar]
- 130.Han Y, Weinman S, Boldogh I, Walker RK, Brasier AR. Tumor necrosis factor-α-inducible IκBα proteolysis mediated by cytosolic m-calpain. J Biol Chem. 1999;274:787–794. doi: 10.1074/jbc.274.2.787. [DOI] [PubMed] [Google Scholar]
- 131.Porn-Ares MI, Samali A, Orrenius S. Cleavage of the calpain inhibitor, calpastatin, during apoptosis. Cell Death Differ. 1998;5:1028–1033. doi: 10.1038/sj.cdd.4400424. [DOI] [PubMed] [Google Scholar]
- 132.Jiang H, Sha SH, Forge A, Schacht J. Caspase-independent pathways of hair cell death induced by kanamycin in vivo. Cell Death Differ. 2006;13:20–30. doi: 10.1038/sj.cdd.4401706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Lee CK, Weindruch R, Prolla TA. Gene-expression profile of the ageing brain in mice. Nat Genet. 2000;25:294–297. doi: 10.1038/77046. [DOI] [PubMed] [Google Scholar]
- 134.Jiang CH, Tsien JZ, Schultz PG, Yinghe H. The effects of aging on gene expression in the hypothalamus and cortex of mice. Proc Natl Acad Sci USA. 2001;98:1930–1934. doi: 10.1073/pnas.98.4.1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Yoshida S, Yashar BM, Hiriyanna S, Swaroop A. Microarray analysis of gene expression in the aging human retina. Invest Ophthalmol Vis Sci. 2002;43:2554–2560. [PubMed] [Google Scholar]