Abstract
Reemerged Plasmodium vivax malaria in South Korea has not yet been eradicated despite continuous governmental efforts. It has rather become an endemic disease. Our study aimed to determine the genetic diversity in P. vivax merozoite surface protein-1 (PvMSP-1) and circumsporozoite protein (PvCSP) genes over an extended period after its reemergence to its current status. Sequence analysis of PvMSP-1 gene sequences from the 632 P. vivax isolates during 1996–2007 indicates that most isolates recently obtained were different from isolates obtained in the initial reemergence period. There was initially only one subtype (recombinant) present but its subtypes have varied since 2000; six MSP-1 subtypes were recently found. A similar variation was observed by CSP gene analysis; a new CSP subtype was found. Understanding genetic variation patterns of the parasite may help to analyze trends and assess extent of endemic malaria in South Korea.
Introduction
With an estimated burden of 70–80 million cases annually, Plasmodium vivax is the most widely distributed human malaria parasite. It is endemic in tropical and subtropical countries of Asia, including the Korean peninsula, the South Pacific, Central and South America, the Middle East, and North Africa.1 The Korean peninsula has been politically divided into two countries (South Korea; the Republic of Korea and North Korea; the Democratic People's Republic of Korea) since 1945. After the Korean War, a cease-fire line was formed in 1953, and it became the international border between these two countries. The demilitarized zone (DMZ) on the border makes control of malaria difficult.
Until the late 1970s, P. vivax malaria was highly endemic in the Korean peninsula.2 Subsequently, malaria was eradicated in South Korea with efforts led by the World Health Organization and the National Malaria Eradication Program.3 However, in 1993, indigenous malaria reemerged in a soldier who had never been abroad and who had served only in the DMZ between South and North Korea.4 Plasmodium vivax malaria outbreaks in South Korea have been correlated with those in North Korea. Since the reemergence of malaria, numbers of indigenous malaria cases have been increasing and decreasing. The patient population shifted to include most non-military personnel. Thus, malaria is endemic and once again has become a serious public health threat in South Korea.
Plasmodium vivax merozoite surface protein-1 (PvMSP-1), an immune dominant antigen expressed on the surface of the malaria merozoite,5 is a valuable polymorphic marker for detection of malaria parasites. The PvMSP-1 gene is organized into several variable blocks flanked by 10 conserved sections. The variable blocks have a dimorphic pattern.6 Block 5, the region encompassed by the interspecies conserved block (ICB)5 and ICB6, shows a dimorphic pattern of sequences that have little homology. In particular, a variable poly-glutamine (poly-Q) region is characteristic of the Belem type. Interallelic recombinations between the two types are frequently seen.7 This polymorphic marker is useful for genetic epidemiological surveys where P. vivax is endemic. The analysis of PvMSP-1 polymorphisms enhances the data collected in P. vivax-related epidemiological and molecular biological surveys.8
We investigated the extent of block 5 PvMSP-1 polymorphism among isolates obtained in South Korea to characterize the genetic structure and composition of the P. vivax parasite population in South Korea. Since 2000,9 no data have been published on the genetic composition of the PvMSP-1 gene in South Korea. Analysis of the PvMSP-1 sequence in Korean clinical isolates is warranted.
The central repetitive domain from the P. vivax circumsporozoite surface protein (PvCSP), the most abundant polypeptide on the sporozoite surface, varies in sequence and length among Plasmodium spp. The classic P. vivax VK210 strain has a PvCSP amino sequence that includes a GDRAA/DGQPA repeat.10 A variant form, VK247, later identified in Thailand, possesses an ANGAGNQPG amino acid repeat within the amino acid tandem repeat region.11 All PvCSP variant genotypes have a worldwide distribution.12,13
We conducted the present study to analyze sequences of these two genes from isolates obtained during 1999–2007 and to identify changes in the genetic diversity of the reemerged P. vivax population in South Korea. This study surveyed previous prevalence and evaluated current prevalence and is useful because it might predict future prevalence.
Materials and Methods
Malaria incidence.
Counts of reported malaria cases for South Korea for 1999–2008 were compiled from Korea National Notifiable Infectious Disease Surveillance System. Counts for North Korea were cited from the World Health Organization World Malaria Report 2008 for 1999–2006. Data for 2007–2008 were cited from the Donor Report, Malaria Control Program, North Korea, which was submitted by World Health Organization. The spot map was generated in March 2009 by using data of malaria cases reported to the Korea National Notifiable Infectious Disease Surveillance System.
Blood samples.
Blood was collected from 632 patients and packed cells and plasma were separated and frozen at −75°C at the Korea National Institutes of Health until used. Except for 1997, all cases during 1996–2007 were confirmed in randomly selected blood samples obtained from indigenous patients. The presence of P. vivax was confirmed by light microscopic examination of thin and thick blood smears stained with Field's stain. All patients provided informed consent before blood samples were obtained. The study was reviewed and approved by the National Institutes of Health, Korea Centers for Disease Control and Prevention.
Preparation of DNA.
Plasmodium vivax DNA was extracted from a 200-μL sample of each blood sample using the proteinase K digestion technique. The extracted DNA was purified with phenol-chloroform or using a QIAamp® DNA Mini Kit (Qiagen, Valencia, CA) following the manufacturer's instructions.
Genotyping of P. vivax isolates at the PvMSP-1 locus.
The amplified region of the PvMSP-1 gene included the sequence between ICBs 5 and 6. The amplification used primers MSP-1A (5′-GAGCCCTACTACTTGATGGTCC-3′) and MSP-1B (5′-CCTTCTGGTACAGCTCAATG-3′).9 Amplification was performed in a 20-μL reaction mixture containing the parasite DNA, 1× reaction buffer, 2.5 mM MgCl2, 0.2 mM of each dNTP, 6 pmol of each primer, and 1 unit of HF DNA polymerase (Bioneer, Seoul, South Korea and Takara, Shiga, Japan). Polymerase chain reaction (PCR) cycling conditions were an initial denaturation at 95°C for 7 minutes; followed by 35 cycles of denaturation at 94°C for 1 minute, annealing at 58°C for 1 minute, and extension at 72°C for 1 minute; and a final extension at 72°C for 7 minutes. Amplification was performed in a GeneAmp PCR System 9600 DNA thermal cycler (Applied Biosystems, Foster City, CA). The PCR product was subjected to electrophoresis on a 1% agarose gel.
Genotyping P. vivax isolates for the PvCSP locus.
A PCR was used to amplify the PvCSP gene with the oligonucleotide primers CSP3 (5′-CCAGATGACGAGGAAGGAGATGC-3′) and CSP4 (5′-TCTTTCACAGACTTTTCATTTGGG-3′). Amplification was performed in a 20-μL reaction volume containing 6 pmol of each primer, 0.2 mM dNTPs, 1× PCR buffer, 2.5 mM MgCl2, parasite DNA, 1× reaction buffer, and 1 unit of Taq DNA polymerase (Bioneer and Takara). The PCR cycling conditions were an initial denaturation at 95°C for 7 minutes; 35 cycles of denaturation at 94°C for 1 minute, annealing at 58°C for 1 minute, and extension at 72°C for 1 minute; and a final extension at 72°C for 7 minutes. Amplification was preformed in the aforementioned DNA thermal cycler (Applied Biosystems). The PCR products were fractionated by electrophoresis on a 1% agarose gel containing ethidium bromide.
Purification of PCR products and DNA cloning and sequencing of PvMSP-1 and PvCSP gene segments.
The PCR products were purified using a QIAquick Gel Extraction Kit (Qiagen) according to the manufacturer's instructions. After amplified fragments were gel purified, and PCR products were sequenced using an ABI 3730XL DNA Analyzer automated sequencer (Applied Biosystems). Sequencing reactions were performed using the primers from ICB5-ICB6 or using CSP3-CSP4 primers. In some cases, we could not obtain the sequence information directly from the PCR products. For these instances, the sequence was obtained with universal primers after cloning the segment using a TOPO-TA cloning kit (pCR®2.1-TOPO; Invitrogen, Carlsbad, CA).
Sequence analysis and phylogeny.
Parasite gene sequences obtained from the blood samples of the malaria patients in this study were compared with previously published PvMSP-1 gene sequences. ExPASy Molecular Biology (http://us.expasy.org/) was used to convert nucleotide sequences into amino acid sequences. WBioEdit version 7.0 software (www.mbio.ncsu.edu/BioEdit/BioEdit.html), the CLUSTAL X program,14 and EditSeq 3.88 (DNASTAR, Madison, WI) software were used to align and edit the data. MEGA 2.115 was used for phylogenetic analysis.
Results
Prevalence of the reemerged Plasmodium vivax malaria in Korea.
The number of indigenous malaria cases was increased three to five times annually and peaked in 2000 at 4,183 cases. The number of patients decreased slightly until 2004 but increased again in 2005 (1,369 cases), 2006 (2,051 cases), and 2007 (2,227 cases) (Table 1). The risk areas have varied and gradually expanded to the near urban areas during past 15 years (Figure 1). As shown in Figure 2 and Table 1, the incidence rate of malaria in North Korea is considerably higher than that in South Korea (Figure 2 and Table 1).
Table 1.
Comparison of malaria incidence between Republic of Korea and Democratic People's Republic of Korea, 1999–2008
| Year | Republic of Korea | Democratic People's Republic of Korea | Incidence (95% confidence interval) | |||||
|---|---|---|---|---|---|---|---|---|
| Population (×1,000) | No. of cases | Incidence (/100,000) | Population (×1,000) | No. of cases | Incidence (/100,000) | |||
| 1999 | 46,617 | 3,674 | 7.88 | 22,082 | 7,980 | 36.14 | 4.6 | (4.4–4.8) |
| 2000 | 47,008 | 4,183 | 8.90 | 22,175 | 73,742 | 332.55 | 37.4 | (36.2–38.6) |
| 2001 | 47,357 | 2,556 | 5.40 | 22,253 | 296,540 | 1,332.58 | 246.9 | (237.5–256.7) |
| 2002 | 47,622 | 1,799 | 3.78 | 22,369 | 241,190 | 1,078.23 | 285.4 | (272.5–299.0) |
| 2003 | 47,859 | 1,171 | 2.45 | 22,522 | 60,559 | 268.89 | 109.9 | (103.7–116.4) |
| 2004 | 48,039 | 864 | 1.80 | 22,709 | 33,803 | 148.85 | 82.8 | (77.4–88.5) |
| 2005 | 48,138 | 1,369 | 2.84 | 22,928 | 11,507 | 50.19 | 17.6 | (16.7–18.7) |
| 2006 | 48,297 | 2,051 | 4.25 | 23,079 | 9,353 | 40.53 | 9.5 | (9.1–10.0) |
| 2007 | 48,456 | 2,227 | 4.60 | 23,200 | 7,436 | 32.05 | 7.0 | (6.7–7.3) |
| 2008 | 48,607 | 1,052 | 2.16 | 23,298 | 23,409 | 100.48 | 46.4 | (43.6–49.4) |
Figure 1.
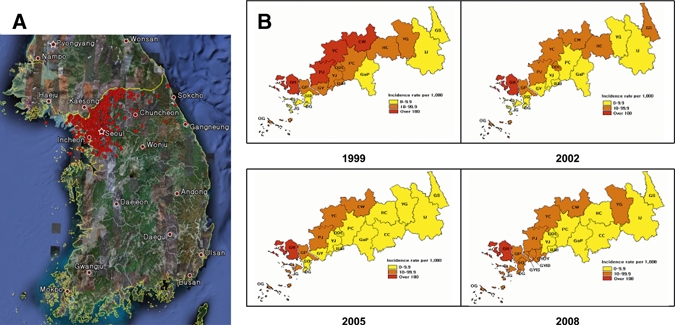
High-risk areas for malaria in Republic of Korea in 1999, 2002, 2005, and 2008. A, Spot map of the occurrence of Plasmodium vivax malaria. Active local transmission may occur near the Demilitarized Zone (DNZ). South Korea and North Korea are separated by the yellow national boundary line with the DMZ adjacent to the line. B, Major risk area of P. vivax malaria in South Korea. Results are plotted in yellow, orange, and red, which represent < 10.0, 10.1–99.9, and > 100.0 patients/1,000 population, respectively. The subsets by year (1999, 2000, 2005, and 2008) represent 23 high-risk areas in South Korea that showed epidemiological changes. GH = Ganghwa-gun; DG = Dong-gu; SG = Seo-gu; OG = Ongjin-gun; JG = Jung-gu, Gyeonggi-do Province; GaP = Gapyeong-gun; GY = Goyang-si; GYDY = Goyang-si Deogyang-gu; GYID = Goyang-si Ilsan-Donggu; GYIS = Goyang-si Ilsan-Seogu; GIP = Gimpo-si; DDC = Dongducheon-si; YJ = Yangju-gun; YC = Yeoncheon-gun; UJB = Uijeongbu-si; PC = Pocheon-gun; PJ = Paju-si, Gangwon-do Province; GS = Goseong-gun; YG = Yanggu-gun; IJ = Inje-gun; CW = Cheorwon-gun; CC = Chuncheon-si; HC = Hwacheon-gun.
Figure 2.
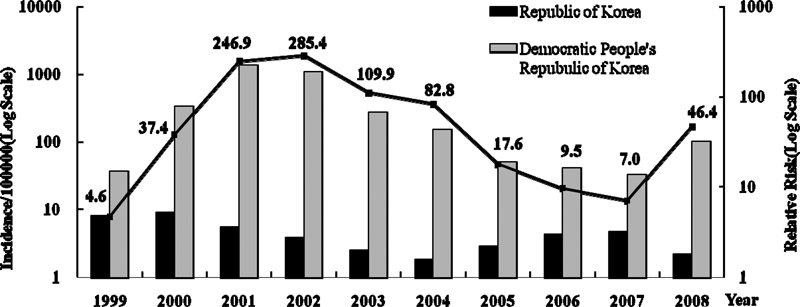
Malaria incidence rate and relative risk for the Republic of Korea and the Democratic People's Republic of Korea, 1999–2008.
PvMSP-1 gene analysis.
Overall, six PvMSP-1 subtypes were identified in genomic parasite DNA obtained from the 632 patients examined in our study (Figure 3 and Table 2). Salvador-1 (Sal-1) was dominant strain in South Korea and included the most variant polymorphic region of PvMSP-1. The four Sal-1 subtypes were designated S-R (VIAAT; reference strain; AAN86207), S-a (VTTVQ), S-b (QVIAAQ), and S-c (QAIAAQ). In these Sal-1 subtypes, we identified five amino acid substitutions (V/A, I/T, A/T, A/V, E/Q) at different positions and only one glutamine insertion (Figure 3).
Figure 3.

Comparison of amino acid sequences of Plasmodium vivax merozoite surface protein-1 (PvMSP-1) from South Korea with those from worldwide strains. Amino acid positions indicated by shaded regions depict non-synonymous substitutions and poly-Q residues in isolates from the Republic of Korea.
Table 2.
Plasmodium vivax circumsporozoite protein polymorphisms based on repeat regions, Republic of Korea
Subtypes derived from the Belem strain had variable numbers of glutamine resides in the polyglutamine (poly-Q) region. All three Belem subtypes showed a different number of poly-Q repeats and continuous three amino acid substitutions (ESMIA/QAMIT). These Belem subtypes were designated B-1 (QAMIT-14PolyQ), B-2 (ESMIA-19PolyQ), and B-R (ESMIT-23PolyQ; reference strain; AAN86208) (Figure 3).
Comparison of the recombinant Sal-1 and Belem strains detected only one subtype of the recombinant strain, which displayed a forward and reverse sequence pattern similar to the Sal I and Belem strains, respectively. In addition to these sources of diversity, 10 glutamine residue repeats were present in the recombinant strain of Korean isolates (Figure 3).
DNA sequence polymorphism analyses were performed using samples obtained over a 10-year period. Except for 1997, genomic P. vivax DNA samples were examined from 106 patients during 1996–1999. Most of these isolates from Korea evident during this period belonged to one recombinant strain. However, in 2000, one case of Sal-1 subtype S-b was detected among 44 samples obtained from different regions. In 2003, Belem strain subtype B-1 was detected in one of 47 genomic DNA samples obtained from patients. The samples analyzed until 1999 were confirmed as single strain infection. From 2000 through 2002, all Sal-1 strains were confirmed as S-b. Since 2003, the sequence patterns obtained from the total genomic parasite DNA in our study became more diverse. The Sal-1 strain was distributed almost equally among the recombinant strains identified (Figure 4A). We observed two PvMSP-1 strains, Sal-1 and Belem. These strains showed non-synonymous amino acid substitutions and sequence patterns over the study period. The Sal-1 strain diverged by adding the S-a subtype in 2003, and a new subtype of the Belem strain was apparent only in two cases in 2006.
Figure 4.
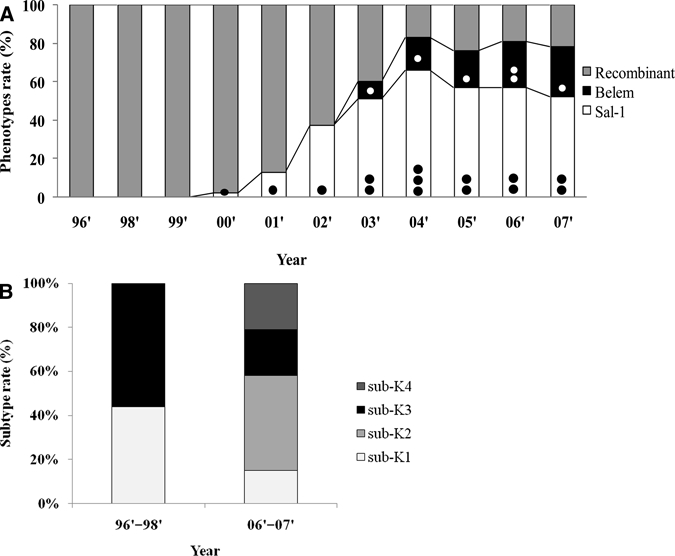
Frequency of Plasmodium vivax merozoite surface protein-1 (PvMSP-1) and P. vivax circumsporozoite protein (PvCSP) strains in the Republic of Korea during the study period. A, Between 1996 and 2000, most isolates differed from recombinant to Sal-1 strains. Black circles indicate subtypes of the Sal-1 strain and white circles indicate subtypes of the Belem strain. • = only S-b subtype; •• = S-a and S-b subtypes; ••• = S-a, S-b, and S-c subtypes; ○ = only B-1 subtype; ○○ = B-1 and B-2 subtypes. B, PvCSP subtypes of isolates from the Republic of Korea that diversified during 1996–2007.
The prevalence of the S-a subtype steadily increased, with the S-a and S-b subtypes contributing similarly to the total Sal-1 strain population. By 2007, cases with the S-a subtype exceeded cases with the S-b subtype. In contrast, the Belem B-1 subtype remained dominant. During 2004–2007, we estimated that the average proportions of the recombinant, Belem, and Sal-1 strains were 20.5%, 21.5%, and 58%, respectively (Figure 4A).
PvCSP gene analysis.
The classic P. vivax VK210 strain has a PvCSP sequence that includes a GDRAA/DGQPA amino acid repeat. A variant form, VK247, later identified in Thailand, has an ANGAGNQPG amino acid repeat in the tandem repeat amino acid region.16 We analyzed our samples for the presence of the VK210 repeat. Of our patient blood samples, 79 samples were positive for malaria from 1996–1998 and 2006–2007. All samples were positive for P. vivax. Each amino acid sequence group was representative of one or more strains (Table 2). All sequences were incorporated the GDRA(A/D)(P/A)A repeating motif 5, 10, 12, or 13 times. Furthermore, the most common amino acid sequence variation was the A/D polymorphism. We detected four VK210 subtypes (sub-K1–subK4). Another Korean subtype previously reported17 was included in our study as subK5 (Figure 4B and Table 2). In previous cases of malaria, the major subtypes were sub-K1 (44%) and sub-K3 (56%). However, with reemergence of malaria, the subtype distribution expanded to include sub-K1 (15%), sub-K2 (43%), sub-K3 (21%), and sub-K4 (21%) (Figure 4B).
Phylogenic tree of PvMSP-1.
Phylogenies were constructed for the PvMSP-1 gene (Figure 5). Subtypes of Belem strains (B-1 and B-2) identified in this study, were similar to Belem reference strain (B-R) (AAN86208), with 89% and 94% sequence homology, respectively. Variation of similarity with the Sal-1 reference subtype S-R (AAN86207) was also 97% with S-a and as much as 98% with S-b and S-c (Figure 5). The Sal-1 subtypes (S-a, S-b, and S-c) identified in this study were similar to those from other countries. S-a and S-c were similar (98%) to the Iran isolate AAM53559 and the Thailand isolate BAA18985, respectively, and S-b had extensive similarity (99%) with the Brazil isolate AAN86237. The Turkey isolate (AD38672 and the Iran isolate AAO27788 were identical to the Korean B-2 isolate. Similarity with the Korean B-1 isolate was 95% for the Turkey isolate CAD38667 and 96% for the Brazil isolate ABJ53045. The Korean recombinant strain was identical to the North Korean isolate AAG53740.
Figure 5.

Plasmodium vivax merozoite surface protein-1 (PvMSP-1) gene distance tree constructed by using the phylogenic tree neighbor-method. PvMSP-1 gene sequences of strains obtained from the Republic of Korea and from GenBank were analyzed. Numbers on branches indicate bootstrap proportions from 1,000 replicates. Scale bar indicates 0.1 amino acids substitutions per site and is proportional to the genetic difference (%).
Discussion
Since the reemergence of malaria in South Korea, the annual incidence of malaria has repeatedly increased and decreased. However, for as yet unknown reasons, in 2008, the number of cases decreased. Possible explanations include climate factors, such as an increase in rainfall, and strengthening of control programs. However, there was a large increase in malaria incidence in North Korea (Figure 2 and Table 1). Thus, reemerged malaria is still a threat in the Korean peninsula.
This two-peak incident pattern might be characteristic of endemic malaria. Since 1993, when malaria reemerged, the distribution of risk areas has varied (Figure 1) and the portion of patients has changed. The area of increased malaria risk initially changes from the eastern region of Korea to the western region along the DMZ and then expanded south. These changes indicate one of the characteristics of endemic malaria.
In the present study, we investigated P. vivax genetic diversity based on the PvMSP-1 and PvCSP genes to assess reemergence of P. vivax in Korea during 1996–2007. Our aim was to determine genetic diversity, which can show whether P. vivax malaria in Korea has become heterogeneous. Genetic studies of P. vivax malaria in Korea have been performed by using several parasitic proteins, including CSP,17 Duffy-binding protein,18 apical membrane antigen-1,19 18S ribosomal RNA,20 and MSP-1.9 However, these studies focused on simple polymorphism analysis for each gene over a short period.
PvMSP-1 has been used as a polymorphic marker to study genetic structure of P. vivax populations and is also a valuable epidemiologic marker in malaria-endemic areas.6,21–23 We analyzed the extent of genetic polymorphism and diversity of the P. vivax population in Korea by using PvMSP-1 and observed a changing diversity of the population annually during 1996–2007.
Interestingly, only one P. vivax malaria strain (the recombinant strain) was observed during 1996–1999 (Figure 4A). The sequence pattern of 10 poly-Q repeats is consistent with that of a previous study9 and there have been no variations. Another polymorphism, the Sal-1 strain, was observed in 2000. The Belem strain of P. vivax was not found at that time.
We identified the Belem strain with two alleles in 2003. All three strains of P. vivax (Sal-1, Belem, and recombinant) have been observed in Korea since 2003 (Figure 4A). Since 2004, the proportion of Sal-1 and Belem strain has greatly increased, and the recombinant strain has steadily decreased. This strain diversity pattern (Sal-1 > Belem > recombinant) continues at the present time (Figure 4A). Moreover, more variant subtypes within the Sal-1, Belem, and recombinant strains emerged during 2003–2007 (Figure 4A). We identified six allelic subtypes: S-a, S-b, and S-c (Sal-1); B-1 and B-2 (Belem); and one recombinant subtype (Figure 4A).
Since 2003, the number of Sal-1 variants has expanded to include S-a, S-b, and S-c. Cases with S-a now outnumber cases with S-b. With the continued increase in P. vivax malaria in Korea, the diversity of the parasite has steadily increased. Since peaking in 2000, diversity based on the PvMSP-1 gene has become complex. This finding is supported by our similar variation patterns of PvCSP and PvMSP-1 genes (Figure 4). In the PvCSP gene analysis, only two subtypes (sub-K1 and sub-K3) were observed in samples obtained before 2000. In the recent samples, subtypes sub-K1, sub-K2, sub-K3, and sub-K4 were evident. Sub-K2 and sub-K4 were newly found in the present study (Table 2 and Figure 4B).
Despite the current trend, Korea is not a country with a high risk for contracting malaria. On the basis of PvMSP-1 gene diversity, we observed six subtypes. Previous studies that examined PvMSP-1 gene diversity in Brazil, Columbia, and India, all of which have a high risk for malaria, documented 9–13 subtypes.24–26 Currently, allelic subtype variation seems to be restricted to six subtypes. However, continuous expansion of malaria in Korea may generate more complex variants with more subtypes. In addition, the increased influx of international travelers and foreign workers from regions where malaria is endemic (such as Southeast Asia, the Middle East, and Latin America) might increase diversity of allelic variation.27
We constructed phylogenetic trees for PvMSP-1 gene sequences obtained in our study (Figure 5). Among published data, the recombinant strain from South Korea was identical to the strain from North Korea published in GenBank. The Sal-1 strain from Korea, including the three subtypes, was similar to those from Iran, India, and Azerbaijan. Belem subtypes isolated in Korea were clustered with isolates from Iran, Brazil, and Turkey. These results are consistent with similar genetic polymorphisms in isolates from geographically diverse regions with similar latitudes or countries in Asia in which malaria is endemic.
Conversely, since the reemergence of malaria in Korea, only the recombinant PvMSP-1 strain was found until 2000. These results are consistent with those of a previous study9 of a malaria outbreak in the DMZ during 1996–1997. In this study, all samples sequenced were the recombinant strain that contained 10 poly-Q residues. In addition, we recently examined MSP-1 diversity in malaria samples from North Korea and found that most of these samples were also the recombinant strain, and that the sequences including the Q repeat region are identical. There are reports indicating that initial transmission of malaria in South Korea may have originated from mosquitoes infected in North Korea, with subsequent spread through the DMZ and secondary and tertiary transmission in South Korea.2,28 These data suggest that the malaria outbreak in South Korea is linked to malaria in North Korea, at least at the initial time of reemergence. Information concerning parasite polymorphisms in malaria patients in North Korea would be useful in assessing this possibility.
Phylogenic and geographical results suggest that malaria prevalence in South Korea may have been influenced by other countries. Moreover, the numbers of foreigners immigrating to Korea and imported cases of malaria have increased over the past 15 years. We are now monitoring imported P. vivax malaria cases in South Korea.
In conclusion, analysis of PvMSP-1 and PvCSP genes in isolates from Korea suggests that malaria parasites are becoming progressively more genetically diverse and supports the idea that P. vivax malaria is endemic in Korea. Understanding the genetic variation patterns of the parasite may help to analyze trends and assess the extent of malaria endemic in Korea. Moreover, genetic characterization of natural populations of P. vivax in Korea is crucial for development of antimalarial vaccines or new drugs.
Footnotes
Financial support: This study was supported by a grant of The Korea Foundation for International Cooperation of Science and Technology provided by the Korean Ministry of Science and Technology (No. K2071900000608-D010000600) and an intramural fund provided by Korea National Institute of Health (No. 4837-302-210-13).
Authors' addresses: Yien-Kyoung Choi, Kyung-Mi Choi, Yeon-Joo Kim, Byeong-Chul Lee, Shin-Hyung Cho, and Jung-Yeon Kim, Divisions of Malaria and Parasitic Diseases, National Institute of Health, Korea CDC, Seoul, Republic of Korea, E-mails: jykim-malaria@nih.go.kr. Mi-Hyun Park, Department of Biological Science, Inha University College of Medicine, Incheon, Republic of Korea. Eun-Gyu Lee, Divisions of Infectious Disease Surveillance, Korea CDC, Seoul, Republic of Korea. Ho-Gun Rhie and Ho-Sa Lee, Institute of Global Environment and Department of Biology, Kyung Hee University, Seoul, Republic of Korea. Joo-Shil Lee, Center for Immunology and Pathology, National Institute of Health, Korea CDC, Seoul, Republic of Korea. Tong-Soo Kim, Department of Parasitology, Inha University College of Medicine, Incheon, Republic of Korea.
References
- 1.Mendis K, Sina BJ, Marchesini P, Carter R. The neglected burden of Plasmodium vivax malaria. Am J Trop Med Hyg. 2001;64:97–106. doi: 10.4269/ajtmh.2001.64.97. [DOI] [PubMed] [Google Scholar]
- 2.Lee HW, Shin EH, Cho SH, Lee HI, Kim CL, Lee WG, Moon SU, Lee JS, Lee WJ, Kim TS. Detection of vivax sporozoites naturally infected in Anopheline mosquitoes from endemic areas of northern parts of Gyeonggi-do (province) in Korea. Korean J Parasitol. 2002;40:75–81. doi: 10.3347/kjp.2002.40.2.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chai JY. Re-emerging Plasmodium vivax malaria in the Republic of Korea. Korean J Parasitol. 1999;37:129–143. doi: 10.3347/kjp.1999.37.3.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chai IH. Occurrence of tertian malaria in a male patient who has never been abroad. Korean J Parasitol. 1994;32:195–200. doi: 10.3347/kjp.1994.32.3.195. [DOI] [PubMed] [Google Scholar]
- 5.Leclerc MC, Gauthier C, Villegas L, Urdaneta L. Genetic diversity of merozoite surface protein-1 gene of Plasmodium vivax isolates in mining villages of Venezuela (Bolivar State) Acta Trop. 2005;95:26–32. doi: 10.1016/j.actatropica.2005.03.007. [DOI] [PubMed] [Google Scholar]
- 6.Del Portillo HA, Longacre S, Khouri E, David PH. Primary structure of the merozoite surface antigen 1 of Plasmodium vivax reveals sequences conserved between different Plasmodium species. Proc Natl Acad Sci USA. 1991;88:4030–4034. doi: 10.1073/pnas.88.9.4030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Putaporntip C, Jongwutiwes S, Tanabe K, Thaithong S. Interallelic recombination in the merozoite surface protein-1 (MSP-1) gene of Plasmodium vivax from Thai isolates. Mol Biochem Parasitol. 1997;84:49–56. doi: 10.1016/s0166-6851(96)02786-7. [DOI] [PubMed] [Google Scholar]
- 8.Zakeri S, Djadid ND, Zeinali S. Sequence heterogeneity of the merozoite surface protein-1 gene (MSP-1) of Plasmodium vivax wild isolates in southeastern Iran. Acta Trop. 2003;88:91–97. doi: 10.1016/s0001-706x(03)00192-x. [DOI] [PubMed] [Google Scholar]
- 9.Lim CS, Kim SH, Kown SI, Song JW, Song KJ, Lee KN. Analysis of Plasmodium vivax merosoite surface protein-1 gene sequences from resurgent Korean isolates. Am J Trop Med Hyg. 2000;62:261–265. doi: 10.4269/ajtmh.2000.62.261. [DOI] [PubMed] [Google Scholar]
- 10.Arnot DE, Barnwell JW, Tam JP, Nussenzweig V, Nussenzweig RS, Enea V. Circumsporozoite protein of Plasmodium vivax: gene cloning and characterization of the immunodominant epitope. Science. 1985;230:815–818. doi: 10.1126/science.2414847. [DOI] [PubMed] [Google Scholar]
- 11.Rosenberg R, Wirtz RA, Lanar DE, Sattabongkot J, Hall T, Waters AP, Prasittisuk C. Circumsporozoite protein heterogeneity in the human malaria parasite Plasmodium vivax. Science. 1989;245:973–976. doi: 10.1126/science.2672336. [DOI] [PubMed] [Google Scholar]
- 12.Gonzalez JM, Hurtado S, Arevalo-Herrera M, Herrera S. Variants of Plasmodium vivax circumsporozoite protein (VK210 and VK247) in Colombian isolates. Mem Inst Oswaldo Cruz. 2001;96:709–712. doi: 10.1590/s0074-02762001000500023. [DOI] [PubMed] [Google Scholar]
- 13.Zakeri S, Abouie-Mehrizi A, Djadid ND, Snounou G. Circumsporozoite protein gene diversity among temperate and tropical Plasmodium vivax isolates from Iran. Trop Med Int Health. 2006;11:729–737. doi: 10.1111/j.1365-3156.2006.01613.x. [DOI] [PubMed] [Google Scholar]
- 14.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. The Cluster X Windows interface. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kumar S, Tamura K, Jakobsen IB, Nei M. Molecular Evolutionary Genetics Analysis Software. Tempe, AZ: Arizona State University; 2001. (MEGA2). [DOI] [PubMed] [Google Scholar]
- 16.Bonilla JA, Validum L, Cummings R, Palmer CJ. Genetic diversity of Plasmodium vivax PvCSP and PvMSP1 in Guyana, South America. Am J Trop Med Hyg. 2006;75:830–835. [PubMed] [Google Scholar]
- 17.Lim CS, Kim YK, Lee KN, Kim SH, Hoffman KJ, Song KJ, Song JW. The analysis of circumsporozoite protein gene sequences from South Korean isolates of Plasmodium vivax. Ann Trop Med Parasitol. 2001;95:229–235. doi: 10.1080/00034980120053997. [DOI] [PubMed] [Google Scholar]
- 18.Kho WG, Chung JY, Sim EJ, Kim DW, Chung WC. Analysis of polymorphic regions of Plasmodium vivax Duffy binding protein of Korean isolates. Korean J Parasitol. 2001;39:143–150. doi: 10.3347/kjp.2001.39.2.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chung JY, Chun EH, Chun JH, Kho WG. Analysis of the Plasmodium vivax apical membrane antigen-1 gene from reemerging Korean isolates. Parasitol Res. 2003;90:325–329. doi: 10.1007/s00436-002-0777-2. [DOI] [PubMed] [Google Scholar]
- 20.Chai JY, Park YK, Guk SM, Oh KH, Oh MD, Lee SH, Kim HS, Wataya Y. A trial for a DNA diagnosis of Plasmodium vivax malaria recently reemerging in the Republic of Korea using microtiter plate hybridization assay. Am J Trop Med Hyg. 2000;63:80–84. doi: 10.4269/ajtmh.2000.63.80. [DOI] [PubMed] [Google Scholar]
- 21.Kolakovich KA, Ssengoba A, Wojcik K, Tsuboi T, Al-Yaman F, Alpers M, Adams JH. Plasmodium vivax: favored gene frequencies of the merozoite surface protein-1 and the multiplicity of infection in a malaria endemic region. Exp Parasitol. 1996;83:11–18. doi: 10.1006/expr.1996.0044. [DOI] [PubMed] [Google Scholar]
- 22.Figtree M, Pasay CJ, Slade R, Cheng Q, Cloonan N, Walker J, Saul A. Plasmodium vivax synonymous frequencies, evolution and population structure deduced from diversity in AMA-1 and MSP-1 genes. Mol Biochem Parasitol. 2000;108:53–66. doi: 10.1016/s0166-6851(00)00204-8. [DOI] [PubMed] [Google Scholar]
- 23.Cui L, Escalante AA, Imwong M, Snounou G. The genetic diversity of Plasmodium vivax populations. Trends Parasitol. 2003;19:220–226. doi: 10.1016/s1471-4922(03)00085-0. [DOI] [PubMed] [Google Scholar]
- 24.Santos-Ciminera PD, Alecrim MGC, Roberts DR, Quinnan GV., Jr Molecular epidemiology of Plasmodium vivax in the State of Amazonas, Brazil. Acta Trop. 2007;102:38–46. doi: 10.1016/j.actatropica.2007.02.013. [DOI] [PubMed] [Google Scholar]
- 25.Amanda M, Sujatha S, Gul A, Asif M, Marcela E, Mauricio C, Silvia B, Virander SC, Pawan M. Inter-allelic recombination in the Plasmodium vivax merozoite surface protein 1 gene among Indian and Colombian isolates. Malar J. 2004;3:4–10. doi: 10.1186/1475-2875-3-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moon SU, Lee HW, Kim JY, Na BK, Cho SH, Lin K, Sohn WM, Kim TS. High frequency of genetic diversity of Plasmodium vivax field isolates in Myanmar. Acta Trop. 2009;109:30–36. doi: 10.1016/j.actatropica.2008.09.006. [DOI] [PubMed] [Google Scholar]
- 27.Romi R, Sabatineli G, Majori G. Could malaria reappear in Italy? Emerg Infect Dis. 2001;7:915–919. doi: 10.3201/eid0706.010601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shin EH, Guk SM, Kim HJ, Lee SH, Chai JY. Trends in parasitic diseases in Republic of Korea. Trends Parasitol. 2008;24:143–150. doi: 10.1016/j.pt.2007.12.003. [DOI] [PubMed] [Google Scholar]


