Abstract
A natural focus of plague exists in the Western Usambara Mountains of Tanzania. Despite intense research, questions remain as to why and how plague emerges repeatedly in the same suite of villages. We used human plague incidence data for 1986–2003 in an ecological-niche modeling framework to explore the geographic distribution and ecology of human plague. Our analyses indicate that plague occurrence is related directly to landscape-scale environmental features, yielding a predictive understanding of one set of environmental factors affecting plague transmission in East Africa. Although many environmental variables contribute significantly to these models, the most important are elevation and Enhanced Vegetation Index derivatives. Projections of these models across broader regions predict only 15.5% (under a majority-rule threshold) or 31,997 km2 of East Africa as suitable for plague transmission, but they successfully anticipate most known foci in the region, making possible the development of a risk map of plague.
Introduction
Plague is a zoonotic disease caused by the bacillus Yersinia pestis. Plague bacteria circulate in small mammal hosts and are transmitted through adult fleas, cannibalism, or (potentially) contaminated soil.1 The disease is enzootic in wild rodent species and in diverse biotopes under wide ranges of environmental circumstances.2
Plague is a rapidly progressing serious illness in humans that is likely to be fatal if untreated or inappropriately treated.1,2 It remains a public-health concern in many countries but most particularly in east-central Africa and Madagascar with > 10,000 cases reported in the last decade.3,4 Human plague occurs in seasonal pulses and shows a geographic distribution in circumscribed foci assumed to be correlated with distributions of dominant reservoirs and vectors and their ecology.2 Although several such foci are known, a recent World Health Organization report concluded that additional foci remain to be discovered, so it is unknown how many people live in plague-risk areas.5
Studies of plague ecology and epidemiology in Africa have generally been focused at microscales, examining host–vector–parasite systems and human social-activity patterns within single foci.6–8 These studies have nonetheless been unable to elucidate environmental factors shaping plague distributions at regional scales; a few studies have attempted to link plague distribution and the distributions of plague hosts and vectors with environmental factors,9–13 but only two of these studies have focused on Africa. In a previous study, we developed ecological-niche models for Africa, exploring environmental conditions appropriate for plague occurrence and finding a broad potential distributional area of plague across Africa.12 However, because the resolution used in that initial study was coarse (~10 km), its local public-health utility was limited. A fine-scale model for human plague was recently developed for the West Nile region of Uganda.13 A logistic regression model explained 74% of plague-incidence variation, and plague incidence was predicted at parish-level scale based on environmental variables, such as remotely sensed variables associated with differences in soil and vegetation; this study revealed that plague cases in the region were more common above than below 1300 m.
In the Western Usambara Mountains, Lushoto District, northeastern Tanzania, plague has been a public-health problem since the area's first documented outbreak in 1980.14 The rodent and flea species involved in plague transmission there have not been identified, and questions focus on why plague reemerges in the same suite of villages.6,15,16 Earlier, long-term microscale research comparing distributions of rodent and flea populations and examining socio-cultural factors favorable to plague transmission in plague-positive and plague-negative villages could not explain the details of the spatio-temporal distribution of the disease in the region.6,8,15–17
In this study, we aim to (1) explore the geographic distribution and coarse-resolution ecology of human plague in Lushoto District of Tanzania, (2) identify environmental conditions correlated with human-plague occurrences in Lushoto, (3) reflect on predictability of plague transmission in the region, and finally, (4) extend our local Lushoto model across East Africa. To this end, we developed ecological-niche models (ENM) based on human-plague case-occurrence data of Lushoto and project them across the broader region. This study takes advantage of relatively fine-resolution satellite imagery (250 m spatial resolution) that combines the broad generality of global-extent, satellite-based data with the specificity of detecting relatively fine-resolution features of landscapes.
Materials and Methods
Human-plague occurrence data.
Data on human-plague cases in the Western Usambara Mountains (Figure 1) between October 1986 and December 2003 were obtained from records at the Lushoto District Hospital. Although most patients were diagnosed and treated in local health centers or dispensaries, the District Medial Officer was always contacted when plague was suspected; a health officer was then sent to investigate the case and add the record to the database. Plague diagnosis was most frequently based on clinical manifestations, including typical buboes, chest pains accompanied by coughing with blood in the sputum, headache, chills, and malaise; a few human cases were diagnosed presumptively by microscopic observations of bipolar staining of a blood smear. Bacteriological culturing and isolation of Y. pestis was performed for several suspected cases during plague outbreaks in 1980 and 1991, confirming plague as the causative agent.14,18 Since 1998, polymerase chain reaction (PCR) and enzyme-linked immunosorbent assay (ELISA) techniques were also used to confirm the presence of plague bacteria in human-serum samples.19 Davis and others15 calculated mean plague-incidence rates and case frequency per village for the 49 villages recording at least one plague case during 1986–2003 and found a positive correlation between the two variables. Geographic coordinates were recorded with a global positioning system (GPS) receiver (spatial precision finer than 30 m) at village offices for 48 of 49 plague villages, as well as for 57 other villages located in the same region but without recorded plague cases during the same time period. These villages without reported plague cases were used as plague-absence points in model validation exercises; setting and size were similar for plague-positive and plague-negative villages (inhabited area typically < 1 km2). For the purpose of exploration of plague incidences and their relationship to environmental dimensions, we developed models based on different subsets of occurrence points, from high-incidence models to low-to-high incidence models. More specifically, we used three sets of presence points seeking to predict where plague will occur: (1) the high-incidence subset with seven villages with high-plague incidence rates (4.17–10.46 cases/1,000 inhabitants); (2) the moderate-to-high incidence subset with 20 villages with moderate and high plague-incidence rates (1.91–10.46 cases/1,000 inhabitants); and (3) the low-to-high incidence subset with all 48 villages experiencing ≥ 1 case (0.02–10.46 cases/1,000 inhabitants).
Figure 1.
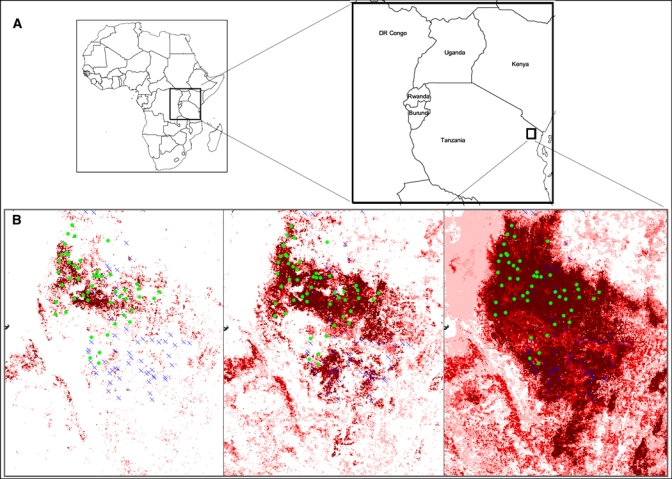
(A) Predictions based on three different sets of occurrence points. The study (training) region, the Lushoto District located in northeast Tanzania (small rectangle) within the projection region (large rectangle), is located in East Africa. (B) Predictions of the Lushoto ecological-niche model based on 7 high-incidence villages, 20 moderate-to-high incidence villages, and 48 low-to-high incidence villages are shown respectively from left to right. This figure appears in color at www.ajtmh.org.
To test the performance of our best-performing Lushoto model in predicting potential for plague transmission across broader areas (Figure 1), locations of known human-plague occurrences between 1970 and 2008 in Tanzania, Kenya, Uganda, and the Democratic Republic of the Congo (DRC) were compiled through an extensive literature search.4 For six reported plague outbreaks, sufficient spatial information was available to assign geographic coordinates (spatial precision finer than 1 km) using gazetteer data and hardcopy maps. This level of precision matches reasonably closely to that of the satellite imagery used, such that the georeferencing will place plague occurrence points reliably within (or at least close to) the corresponding map grid cell from which they came. Fine-resolution georeferencing is not desirable because of uncertainty regarding the actual site of infection, which is not necessarily in the residence of the afflicted person.
Environmental datasets.
We assembled raster-format environmental datasets for ENM development summarizing (1) vegetation and its phenology and (2) topography. All environmental datasets were projected in geographic coordinates and generalized to a resolution of 0.0021° or about 250 m. We used Enhanced Vegetation Index (EVI) layers obtained from the Moderate Resolution Imaging Spectroradiometer (MODIS) sensor from February 18, 2000 to February 17, 2001 (native resolution of 0.0021° or about 250 m), and downloaded from the Land Processes Distributed Archive Center (available from https://lpdaac.usgs.gov/). We calculated nine derivatives from the 23 raw 16-day EVI layers: mean and standard deviation across the year, mean across the first rainy period (R1), mean across the second rainy period (R2), mean across the first dry period (D1), mean across the second dry period (D2), seasonality defined as [(R1 + R2)/2 − (D1 + D2)/2)], and local landscape heterogeneity based on 3 × 3 pixel windows and 51 × 51 pixel windows (i.e., standard deviation of pixels within windows surrounding the central pixel). To establish rainy and dry periods, we analyzed general rainfall patterns across the study area based on the seasonal pattern over the 23 raw EVI layers, which may be slightly offset from the seasonality of the rainfall per se. Data layers summarizing elevation, slope, aspect, and compound topographic index were derived from the digital elevation model of the Shuttle Radar Topography Mission (SRTM; native resolution = 90 m).20
To avoid complications from multicollinearity, we reduced the number of variables for inclusion in the models. We calculated Spearman rank correlations among the 13 coverages and removed heterogeneity based on 51 × 51 pixel windows, mean EVI across the R2, and mean EVI across the D2, because they showed high correlations (ρ > 0.8) with other variables.
Ecological-niche modeling.
Ecological niches are defined as the set of coarse-resolution environmental conditions under which a species can maintain populations without immigration.21,22 Known plague occurrences were related to the environmental data layers to develop a quantitative picture of their ecological distribution.22 Because our only indication of plague occurrence in Lushoto is human-case occurrences, we subsume all of the factors that accompany its transmission to and detection and reporting in humans in the ecological niche of plague.22 We used the Genetic Algorithm for Rule-Set Prediction (GARP), which uses an evolutionary-computing genetic algorithm to develop conditional rules describing the ecological niche.23 Although early evaluations cast doubt on GARP's predictive ability,24 refined recent analyses indicate better performance,25,26 and GARP has served well in previous ENM analyses of disease systems.27–29 All modeling in this study was carried out in DesktopGarp (freely available from http://www.nhm.ku.edu/desktopgarp/).
Within GARP processing, occurrence data points are subdivided as follows: 25% are set aside for developing rules (training data), 50% for filtering models based on error statistics (extrinsic testing data), and 25% for model refinement within GARP (intrinsic testing data), all contrasted with 1,250 pseudo-absence points created by random sampling from areas lacking known presences. GARP works in an iterative process of rule selection, evaluation, testing, and incorporation or rejection. Initially, a method is chosen from four basic rule types (atomic rules, bioclimatic rules, range rules, or logistic regression).23 Specific operators designed to mimic chromosomal evolution (e.g., crossing-over among rules, point mutations, deletions, etc.) then modify the initial rules. After each modification, rule quality is evaluated (to maximize both significance and predictive accuracy), based on the intrinsic testing data, and a size-limited set of rules is retained. Because rules are tested based on independent data (intrinsic testing data), performance values reflect expected performance of the rule to estimate true rule performance more reliably. The final result are rules that have evolved (hence, the genetic algorithm) for maximum significance and predictive ability that can be projected onto the broader landscape to identify a potential geographic distribution.23
To optimize model performance, we developed 100 replicate models based on independent random subsamples from available occurrences. A best subset of 10 models was chosen on the basis of omission (leaving out true potential distributional areas) and commission (including areas not potentially suitable) statistics calculated from the extrinsic testing data.30 Specifically, we used a relative omission threshold, in which the 20% of the models with lowest omission rates were retained. We then chose the 10 models having intermediate levels of commission (i.e., the central 50% of the commission-index distribution among the 20 low-omission models). The 10 models selected by this procedure were summed to produce a final map.
ENM validation.
To test model predictability, we implemented a jackknife procedure developed by Pearson and others31 to test model significance. Each occurrence locality was removed one time from the set of occurrence points, and a model was built using the remaining n − 1 localities. Thus, n separate predictions were built for testing with one of the observed localities excluded in each case. For each prediction, different thresholds for distinguishing between presence and absence predictions were applied (based on the training localities), and predictive performance was assessed based on the ability of each model to predict the single locality excluded from the training dataset. Taking into account the proportion of overall area predicted as suitable (equivalent to the predictive performance of a random model), a P value was calculated across the set of jackknife predictions and tests. For more information on how this probability was calculated, we refer to the paper of Pearson and others31; an easy-to-use software package to calculate this probability is freely available at http://www3.interscience.wiley.com/journal/117963712/suppinfo
To test the ability of the different ENMs (i.e., high-incidence, moderate-to-high incidence, and low-to-high incidence models) to predict plague presence versus plague absence accurately across the Western Usambara Mountains, we evaluated commission (false positive) and omission (false negative) rates using the 48 plague presence locations and the 57 absence points. Models must reach an acceptable compromise between false-negative and false-positive predictions; models predicting absence everywhere would have zero false positives but a high level of false negatives, and models predicting presence everywhere would show the reverse. In our risk-mapping exercises, for reasons discussed below, we accord more weight to false-negative predictions. No real rule exists for choosing a particular threshold for predicting an area as suitable for plague, so we defined three different thresholds: predicted plague-presence areas were areas predicted by ≥ 1 of 10 best replicate models (T1), ≥ 6 of 10 best replicate models (T6), or 10 of 10 best replicate models (T10). We chose the best threshold based on associated error statistics.
To test model transferability to broader regions, we evaluated the predicted distributional area for plague based on the independent set of observed plague occurrences across Tanzania, Kenya, Uganda, and DRC described above. To this end, we projected our best niche model (based on moderate-to-high incidence villages) across relevant parts of East Africa (Tanzania, southern Kenya, eastern DRC, and Uganda). Cumulative binomial probabilities were used to assess the degree to which observed levels of agreement exceeded expectations under a null hypothesis of no association between prediction and spatially segregated testing points.30 This spatial stratification allows some degree of confidence that at least some part of the pattern documented does not result from the effects of spatial autocorrelation. To produce a plague-risk map for East Africa, we constructed a new ENM using the set of uncorrelated environmental coverages with the best subset of Lushoto occurrence points (20 villages with moderate-to-high incidence) and the six locations from surrounding areas to which geographic coordinates were assigned.
Characterization and comparison of ecological niches.
To assess contributions of particular environmental dimensions to the niche models, we used a jackknife procedure.32 We developed models using combinations of 9 of 10 environmental coverages (N − 1 layer models); similarly, each coverage was included systematically in analyses to evaluate the explanatory ability of each on its own (single-layer models).32 Inspecting model performance based on these two sets of coverages in relation to omission error rates then provides a sensitivity analysis, summarizing contributions of each to model predictivity.32,33
To compare ecological conditions under which plague is transmitted to humans in Lushoto, we combined GARP predictions based on different subsets with the 10 environmental data layers into composite grids. The associated attribute tables effectively list all unique environmental combinations represented across the landscape with associated model predictions, which were exported and used to construct scatter plots.
Results
Three sets of models were built using different subsets of occurrence points based on plague-case incidence rates (7-, 20-, and 48-point locations, respectively); differences between them can be seen clearly in the results (Figure 1). The model based on the seven high-incidence villages predicted a small proportion (2.7% under T6 or the region predicted as plague-suitable area by ≥ 6 of 10 replicate models) of the training region to have potential plague presence. In contrast, the model based on the 48 presence points predicted almost the whole mountainous area to have plague present (36.6% under T6). In between these two extremes, the model based on 20 moderate-to-high villages predicted 11.2% under T6 of the training region as potential plague distributional area.
The jackknife procedure showed that predictions of plague-case distributions were always better than random expectations (P < 0.01). To compare the ability of the different models, we evaluated commission and omission error rates based on 48 plague-presence and 57 plague-absence points (Table 1). In high-incidence models, omission rates were very high in many plague-positive villages in the predicted absence region, whereas commission errors were low (i.e., most plague-negative villages were correctly predicted as absent). However, models based on all 48 plague-presence locations showed low omission rates and high commission rates. Because a good model balances omission and commission rates, the models based on moderate-to-high incidence villages were the best at meeting this condition (Figure 2).
Table 1.
Omission and commission errors for three different thresholds
| High-incidence model | Moderate-to-high incidence model | Low-to-high incidence model | |
|---|---|---|---|
| Omission | |||
| T1 | 16/48 (33.3%) | 4/48 (8.3%) | 0/48 (0.0%) |
| T6 | 33/48 (68.8%) | 8/48 (16.7%) | 0/48 (0.0%) |
| T10 | 39/48 (81.3%) | 21/48 (43.8%) | 1/48 (2.1%) |
| Commission | |||
| T1 | 10/57 (17.5%) | 39/57 (68.4%) | 56/57 (98.2%) |
| T6 | 3/57 (5.3%) | 23/57 (40.3%) | 49/57 (86.0%) |
| T10 | 2/57 (3.5%) | 11/57 (19.3%) | 35/57 (61.4%) |
The results of omission (true presence and predicted absence) and commission (true absence and predicted presence) errors for three models based on 7 high-incidence villages, 20 moderate-to-high incidence villages, and 48 low-to-high incidence villages (≥ 1 case reported), respectively. The numbers of sites that were incorrectly classified (false positives and false negatives) and the percentages for each of these categories are presented. Models are evaluated using 48 presence and 57 absence points at three different thresholds: predicted plague-presence areas were taken as areas predicted by ≥ 1 of 10 best replicate models (T1), ≥ 6 of 10 best replicate models (T6), and 10 of 10 best replicate models (T10). For example, a plague-positive village that is predicted by 6 of 10 replicate models is considered as predicted plague-presence area at T1 and T6, but not at T10.
Figure 2.
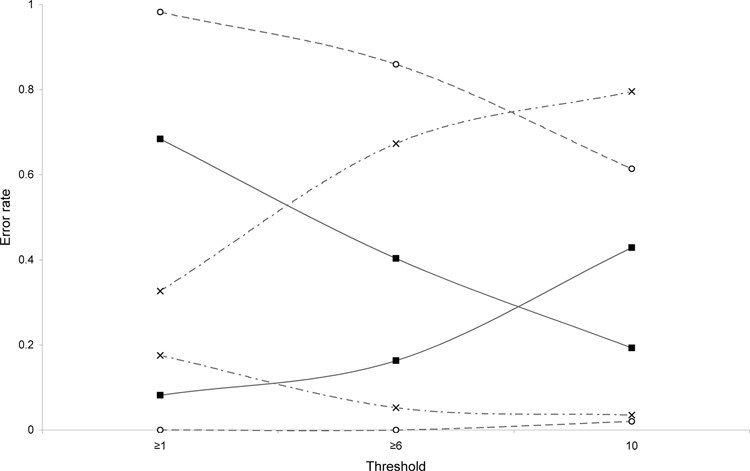
Evaluation of omission and commission error rates for three different models. Graph shows omission (false negatives) and commission (false positives) error rates for three models based on 7 high-incidence villages (crosses), 20 moderate-to-high villages (black squares), and 48 low-to-high incidence villages (open circles) evaluated under three different thresholds: ≥ 1 of 10 best subset models predicting presence, ≥ 6 of 10 best subset models predicting presence, and 10 of 10 best subset models predicting presence.
Relative contributions of the various environmental datasets were assessed using another jackknife manipulation. All variables contributed importantly to the best model, and elevation and mean EVIs in the first dry and first rainy period were the most important variables (Figure 3). Plague is most common between 1,200 and 2,000 m and in areas characterized by mean EVI values between 3,500 and 5,800 in the first rainy period and between 2,000 and 4,000 in the first dry period. Mean EVI across the year (2,700–5,000) and EVI seasonality (800–2,800) turned out to also be important predictors.
Figure 3.
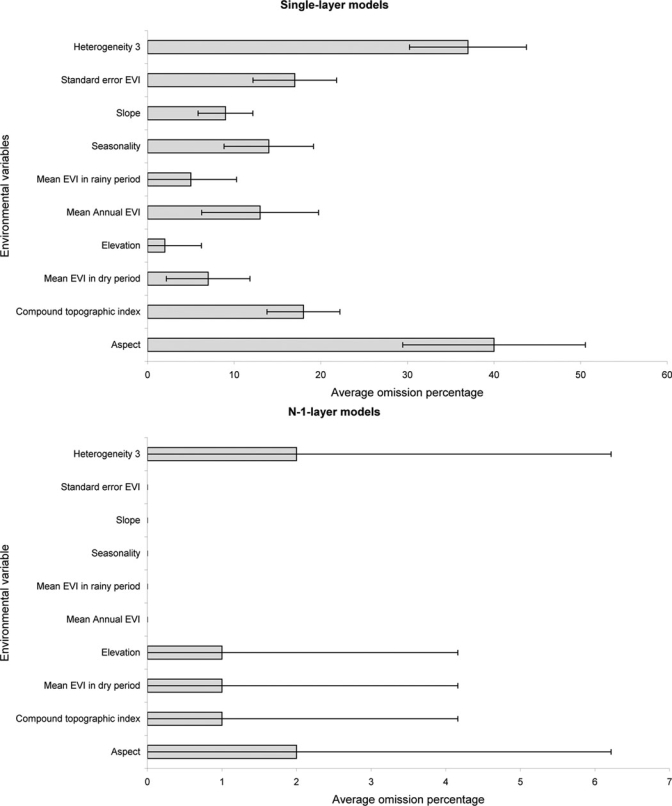
Relative contributions of environmental variables. Summary of results of single-layer (upper graph) and N-1-layer (lower graph) model analyses indicating mean omission percentages (and standard deviations) calculated based on predictions of 10 best subset models and independent testing points. Note that in single-layer models (upper graph), a positive contribution by a variable is indicated by low values, whereas in N-1-layer models, a positive contribution by a variable is indicated by a high value.
To test model transferability, we projected the ENM based on moderate-to-high incidence villages onto a broader region and predicted 15.5% (under majority-rule threshold T6) or 31,997 km2 of East Africa as suitable for plague transmission (Figure 4). We evaluated it using occurrences from southern Kenya, Tanzania, Uganda, and DRC. The six historical georeferenced plague locations from Tanzania and DRC were predicted with high probability by the model under T1–T9 (P < 0.01); only under the very restrictive threshold T10 were results not significant (P = 0.25; Figure 4).
Figure 4.
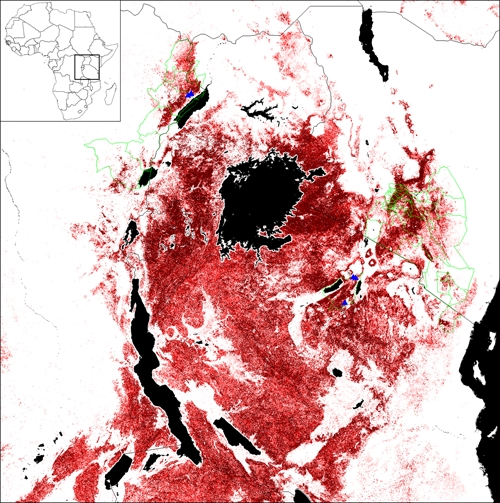
Geographic plague distribution in East Africa as predicted by the best Lushoto model. This map shows the potential geographic distribution of plague in East Africa, as predicted by the best niche model, using 20 moderate-to-high incidence Lushoto villages. Darker shades indicate areas with greater model agreement in predicting areas suitable to plague. Districts where plague has been reported since 1970 are indicated in green, including Ituri and Nord Kivu districts in the DRC, Okoro, Padyere, and Vurra counties in Uganda, Kajido, Kitui, Kiambu, and Taita districts and Nairobi and Machakos areas in Kenya, and Lushoto and Mbulu districts in Tanzania. This figure appears in color at www.ajtmh.org.
To visualize Lushoto plague niches in ecological dimensions, we integrated the three ENMs with the base environmental coverages. Figure 5 presents two examples (mean EVI in first rainy season versus mean EVI in first dry period and elevation versus mean annual EVI). The ecological niche defined by models based on high-incidence villages is nested within the niche defined by models based on moderate-to-high incidence villages, which in turn nests within that based on all presence points, consistent with the geographic predictions based on different subsets of occurrence points (Figure 1). More generally, plague in Lushoto only occurs at 1,200–2,000 m elevation and with moderate EVIs.
Figure 5.
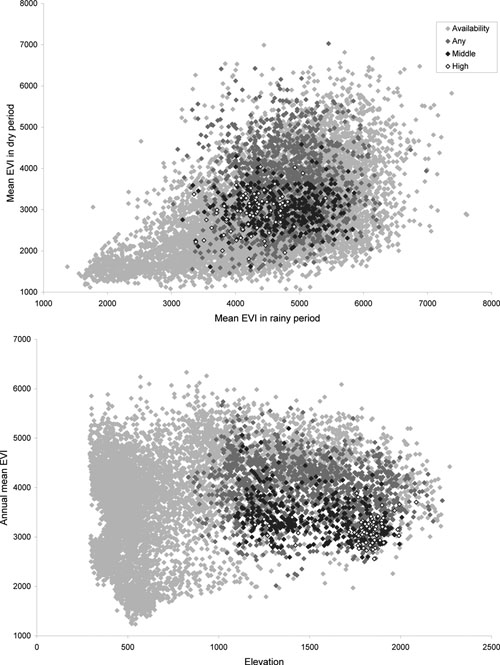
Visualizations of plague ecological-niche conditions in Lushoto. Two examples of visualizations of plague ecological-niche conditions shown in two-dimensional spaces. Displayed are all available habitat in the study area (light grey diamonds), predictions based on 48 low-to-high incidence villages (dark grey diamonds), predictions based on 20 moderate-to-high incidence villages (black diamonds), and predictions based on 7 high-incidence villages (white diamonds).
Finally, we produced a risk map of plague for East Africa based on moderate-to-high incidence villages that was combined with observed plague localities in surrounding regions (Figure 6). Areas around Lake Victoria and mountain areas of Kenya, Tanzania, DRC, and Uganda were predicted as most important plague-risk areas. Under T6, 45,745 km2 of the study region in East Africa (22.2%) was predicted as potential risk area for plague.
Figure 6.
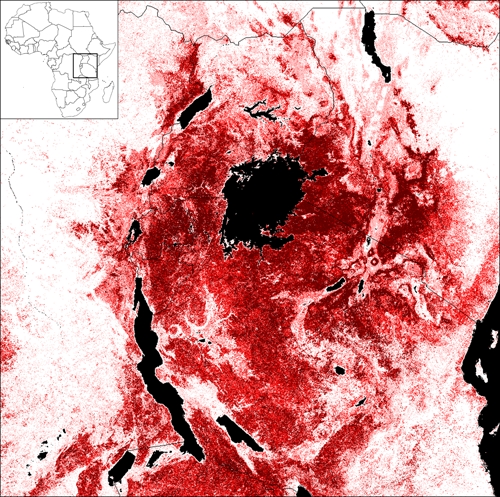
Risk map of plague for East Africa. The risk map of plague for East Africa is developed based on 10 environmental variables and 26 occurrence points (20 moderate-to-high villages from Lushoto, 3 locations in Tanzania, and 3 locations in DRC). Darker shades indicate areas with greater model agreement in predicting areas as suitable to plague. This figure appears in color at www.ajtmh.org.
Discussion
In our analyses, all environmental variables contributed importantly to the models. Plague areas are predicted to be focused at higher elevations (1,200–2,000 m), which coincides with previous studies in East Africa where plague was apparently known in the highlands even before the Third Pandemic.34,35 Plague occurs mainly in highlands in DRC7 and Uganda, where plague cases were more common at > 1,300 m.13,36 In Madagascar, plague-endemic areas focus on the high plateau of the island.37 In the western United States, a geographical information system (GIS)-based model indicated that suitability for plague increases for elevations up to 2,129 m but declines at higher elevations.38 Besides elevation, plague occurrences seem to be linked to seasonal vegetation changes, resulting from rainfall variability through the year, and to moderate EVI values and reasonably high seasonality, suggesting that forests are not suitable for plague; however, because people do not frequently live in the forests, this conclusion could be misleading.
We asked why we could predict plague occurrence in East Africa using 250-m resolution satellite imagery, which in the end provided us with a very real feasibility of risk mapping. Why are the conditions described above appropriate for plague transmission to humans? In other words, what is the basis for this predictability? Has it to do with host distributions or vector ecology, or are certain environmental conditions necessary for survival or transmission of Y. pestis? The plague cycle involves hosts (small mammals), vectors (fleas), and accidental hosts (susceptible rodents, domestic animals, and humans) as well as the bacterium itself. Climate may potentially act on all components of the system but particularly, on rodent- and flea-population dynamics and behavior. Principal climatic factors influencing plague actors and their ecology are temperature (related to elevation), rainfall (also possibly related to elevation), and humidity.39 The effects on the plague cycle can be direct: soil humidity and relative humidity may impact flea survival,40 and mild temperatures may favor rodent-population growth.41 However, climate and more particularly, rainfall can also indirectly synchronize rodent populations42 or act on food availability for rodents,43 reflecting general influences of climatic factors.35,44
With the relatively fine-resolution satellite imagery used here (250 m spatial resolution), occurrence data must be georeferenced accurately, because conditions within small areas define areas of suitability. The location of a residence or village may be a poor guide to sites of infection.45,46 Our focus on villages with moderate-to-high plague-incidence rates as input probably helps alleviate these problems. Nevertheless, instead of using village centers as occurrence points, it would be more accurate to use hamlet centers (hamlets are subdivisions of villages) with high incidence as plague-occurrence input data, but these data are not available. Better still would be to use animal-occurrence locations instead of human-plague incidence data, but long-term research in the area has not yielded useful occurrence data.
The different models we constructed can all be useful, despite the fact that we considered the model based on moderate-to-high villages as best for estimating the true potential distribution of plague in Lushoto. We defined this best model based on a balance between omission and commission error rates. Our model based on only high-incidence villages, predicting only a limited area as present, underestimates real plague distributional areas, but it may delineate areas of highest probability so that plague research could be focused there, reasoning that chances of finding plague are greatest. Contrarily, the model based on all plague-positive villages clearly overestimates potential plague habitat. The broad-extent risk map that was created based on our explorations may also overpredict the potential plague region; however, this possibility cannot be tested, because no data exist to establish that plague absence is real. Nevertheless, we believe that, even if part of the predicted potential plague region is overprediction, resources for surveillance and remediation can be spent better in the areas predicted than if they were spread randomly over the landscape. These areas are demonstrably higher in probability of plague presence than other areas.
We chose our best model based on a balance between omission and commission rates. We were interested in predicting plague presence versus plague absence, but we needed to adjust the threshold of the prediction simply to yield the desired level of sensitivity. More specifically, we developed models based on different subsets of occurrence points, from high-incidence models to low-to-high incidence models, seeking to predict where plague will occur, and we tuned our models accordingly. Certain error combinations were prioritized. For instance, a village predicted as plague present but with no actual plague records may be a suitable plague habitat, but the disease might not (yet) have reached or infected the place; infected plague hosts/vectors may be present in a village, but people might not have been infected. No transmission of plague occurred to humans for socio-cultural or other reasons, or human cases might have occurred but were not reported. A possible local explanation is that many local people in Lushoto often link plague incidence to witchcraft, so family members may hide sick persons. However, some false negatives may be accepted, because the possibility exists that villagers living in low-incidence villages may have been infected elsewhere but returned home with the infection. Indeed, for the model based on moderate-to-high incidence villages under T6, all eight false-negative–predicted villages had low mean plague-incidence rates with values between 0.02 and 0.86 per 1,000 inhabitants. Alternatively, misidentification of sick persons as plague cases, particularly in times of plague panic, may have caused overestimation of plague incidence. Moreover, the lack of reliable plague diagnostics at most rural clinics in plague-endemic areas of Africa also means that most cases are identified strictly on clinical grounds and lack laboratory confirmation, a situation that can lead to many cases of febrile illness being wrongly attributed to plague, especially after an outbreak or small cluster of actual plague cases.
In this study, an ENM approach was applied to the distribution of plague in the Lushoto District of northeastern Tanzania to test the potential of predicting its distributional area and identifying key environmental factors. Our main conclusion is that the typical focality of plague, observed in East Africa, can indeed be predicted using 250-m resolution satellite imagery. Known areas outside Lushoto District that are endemic to plague can be predicted well. Moreover, the link with environmental variables, such as elevation and EVI derivatives, makes it possible to develop a risk map for plague in East Africa, which is provided in this report.
Acknowledgments
We are grateful to the following individuals for generous provision of plague distribution data: Rhodes Makundi and Bukheti Kilonzo (Pest Management Centre, Sokoine University of Agriculture, Morogoro, Tanzania) and the District Health Officer of Lushoto, the late Mr. Msingwa. We also thank Monica Papeş (University of Kansas) for assistance with complex GIS manipulations. S.N. was supported by a PhD grant from the Institute for the Promotion of Innovation through Science and Technology in Flanders (IWT-Vlaanderen) and a travel assistance grant of the Research Foundation Flanders (FWO-Vlaanderen). We appreciate the support of the UK's Joint Environment and Human Health Program (funded by the Natural Environment Research Council (NERC); the Department for Environment, Food and Rural Affairs (DEFRA); the Environment Agency (EA); the Ministry of Defence (MOD); and the Medical Research Council (MRC)). A.T.P. was supported by a grant from the U.S. Centers for Disease Control and Prevention.
Footnotes
Authors' addresses: Simon Neerinckx and Herwig Leirs, Evolutionary Ecology Group, Department of Biology, Universiteit Antwerpen, Groenenborgerlaan 171, B-2020 Antwerp, Belgium, E-mail: simon.neerinckx@ua.ac.be. Simon Neerinckx, Hubert Gulinck, and Jozef Deckers, Department of Earth and Environmental Sciences, Katholieke Universiteit Leuven, Celestijnenlaan 200 E, B-3001 Heverlee, Belgium. A. Towsend Peterson, Natural History Museum and Biodiversity Research Center, University of Kansas, Lawrence, KS. Didas Kimaro, Department of Agricultural Engineering and Land Planning, Sokoine University of Agriculture, Morogoro, Tanzania. Herwig Leirs, Danish Pest Infestation Laboratory, University of Aarhus, Faculty of Agricultural Sciences, Department of Integrated Pest Management, Skovbrynet 14, DK-2800 Kongens Lyngby, Denmark.
References
- 1.Gage KL, Kosoy MY. Natural history of plague: perspectives from more than a century of research. Annu Rev Entomol. 2005;50:505–528. doi: 10.1146/annurev.ento.50.071803.130337. [DOI] [PubMed] [Google Scholar]
- 2.Dennis DT, Gage KL, Gratz NG, Poland JD, Tikhomirov E. Plague Manual: Epidemiology, Distribution, Surveillance and Control. Geneva: World Health Organization; 1999. [Google Scholar]
- 3.Stenseth NC, Atshabar BB, Begon M, Belmain SR, Bertherat E, Carniel E, Gage KL, Leirs H, Rahalison L. Plague: past, present, and future. PLoS Med. 2008;5:e3. doi: 10.1371/journal.pmed.0050003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Neerinckx S, Bertherat E, Leirs H. Historical plague occurrences in Africa–an overview from 1877 to 2008. Trans R Soc Trop Med Hyg. 2010;104:97–103. doi: 10.1016/j.trstmh.2009.07.028. [DOI] [PubMed] [Google Scholar]
- 5.WHO . Epidemic and pandemic alert and response. Proceedings of the Interregional Meeting on Prevention and Control of Plague. Antananarivo: Madagascar; 2008. April 7–11, 2006. [Google Scholar]
- 6.Laudisoit A, Leirs H, Makundi RH, Van Dongen S, Davis S, Neerinckx S, Deckers J, Libois R. Plague and the human flea, Tanzania. Emerg Infect Dis. 2007;13:687–693. doi: 10.3201/eid1305.061084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Misonne X. Les rongeurs des foyers de peste congolais. Ann Soc Belg Med Trop. 1920;39:437–493. [PubMed] [Google Scholar]
- 8.Kilonzo BS, Mvena ZS, Machangu RS, Mbise TJ. Preliminary observations on factors responsible for long persistence and continued outbreaks of plague in Lushoto district, Tanzania. Acta Trop. 1997;68:215–227. doi: 10.1016/s0001-706x(97)00096-x. [DOI] [PubMed] [Google Scholar]
- 9.Nakazawa Y, Williams R, Peterson AT, Mead P, Staples JE, Gage K. Climate change effects on plague and tularemia in the United States. Vector Borne Zoonotic Dis. 2007;7:529–540. doi: 10.1089/vbz.2007.0125. [DOI] [PubMed] [Google Scholar]
- 10.Ben Ari T, Gershunov A, Gage KL, Snall T, Ettestad P, Kausrud KL, Stenseth NC. Human plague in the USA: the importance of regional and local climate. Biol Lett. 2008;4:737–740. doi: 10.1098/rsbl.2008.0363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eisen RJ, Reynolds PJ, Ettestad P, Brown T, Enscore RE, Biggerstaff BJ, Cheek J, Bueno R, Targhetta J, Montenieri JA, Gage KL. Residence-linked human plague in New Mexico: a habitat-suitability model. Am J Trop Med Hyg. 2007;77:121–125. [PubMed] [Google Scholar]
- 12.Neerinckx SB, Peterson AT, Gulinck H, Deckers J, Leirs H. Geographic distribution and ecological niche of plague in sub-Saharan Africa. Int J Health Geogr. 2008;7:54. doi: 10.1186/1476-072X-7-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Winters AM, Staples JE, Ogen-Odoi A, Mead PS, Griffith K, Owor N, Babi N, Enscore RE, Eisen L, Gage KL, Eisen RJ. Spatial risk models for human plague in the West Nile region of Uganda. Am J Trop Med Hyg. 2009;80:1014–1022. [PubMed] [Google Scholar]
- 14.Kilonzo BS, Mhina JI. The first outbreak of human plague in Lushoto district, north-east Tanzania. Trans R Soc Trop Med Hyg. 1982;76:172–177. doi: 10.1016/0035-9203(82)90269-3. [DOI] [PubMed] [Google Scholar]
- 15.Davis S, Makundi RH, Machang'u RS, Leirs H. Demographic and spatio-temporal variation in human plague at a persistent focus in Tanzania. Acta Trop. 2006;100:133–141. doi: 10.1016/j.actatropica.2006.10.006. [DOI] [PubMed] [Google Scholar]
- 16.Laudisoit A, Neerinckx S, Makundi RH, Leirs H, Krasnov B. Are local plague endemicity and ecological characteristics of vectors and reservoirs related? A case study in north-east Tanzania. Curr Zool. 2009;55:199–211. [Google Scholar]
- 17.Makundi RH, Kilonzo BS, Massawe AW. Interaction between rodent species in agro-forestry habitats in the western Usambara Mountains, north-eastern Tanzania, and its potential for plague transmission to humans. Singleton GR, ed. Rats, Mice and People: Rodent Biology and Management. Canberra: Australian Centre for International Agricultural Research. 2003:20–24. [Google Scholar]
- 18.Lyamuya EF, Nyanda P, Mohammedali H, Mhalu FS. Laboratory studies on Yersinia pestis during the 1991 outbreak of plague in Lushoto, Tanzania. J Trop Med Hyg. 1992;95:335–338. [PubMed] [Google Scholar]
- 19.Kilonzo BS. Demonstration of Yersinia Pestis DNA and Antibodies in Humans and Animals in Lushoto District: Need to Improve Diagnostic Services. Moshi, Tanzania: Tanzania Public Health Association Publications; 2000. pp. 172–176. Proceedings of the 19th Annual Scientific Conference of the Tanzania Public Health Association, November 20–24, 2000. [Google Scholar]
- 20.SMRT 90 m digital elevation data. 2008. http://srtm.csi.cgiar.org/ Available at.
- 21.Soberón J, Peterson AT. Interpretation of models of fundamental ecological niches and species' distributional areas. Biodivers Inf. 2005;2:1–10. [Google Scholar]
- 22.Peterson AT. Biogeography of diseases: a framework for analysis. Naturwissenschaften. 2008;95:483–491. doi: 10.1007/s00114-008-0352-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stockwell D, Peters D. The GARP modeling system: problems and solutions to automated spatial prediction. Int J Geogr Inf Sci. 1999;13:143–158. [Google Scholar]
- 24.Elith J, Graham CH, Anderson RP, Dudik M, Ferrier S, Guisan A, Hijmans RJ, Huettmann F, Leathwick JR, Lehmann A, Li J, Lohmann LG, Loiselle BA, Manion G, Moritz C, Nakamura M, Nakazawa Y, Overton JM, Peterson AT, Phillips SJ, Richardson K, Scachetti-Pereira R, Schapire RE, Soberon J, Williams S, Wisz MS, Zimmermann NE. Novel methods improve prediction of species' distributions from occurrence data. Ecography. 2006;29:129–151. [Google Scholar]
- 25.Peterson AT, Papes M, Eaton M. Transferability and model evaluation in ecological niche modeling: a comparison of GARP and Maxent. Ecography. 2007;30:550–560. [Google Scholar]
- 26.Peterson AT, Papes M, Soberon J. Rethinking receiver operating characteristic analysis applications in ecological niche modeling. Ecol Modell. 2008;213:63–72. [Google Scholar]
- 27.Peterson AT, Bauer JT, Mills JN. Ecologic and geographic distribution of filovirus disease. Emerg Infect Dis. 2004;10:40–47. doi: 10.3201/eid1001.030125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Peterson AT, Sánchez-Cordero V, Beard CB, Ramsey JM. Ecologic niche modeling and potential reservoirs for Chagas disease, Mexico. Emerg Infect Dis. 2002;8:662–667. doi: 10.3201/eid0807.010454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Peterson AT, Shaw J. Lutzomyia vectors for cutaneous leishmaniasis in Southern Brazil: ecological niche models, predicted geographic distributions, and climate change effects. Int J Parasitol. 2003;33:919–931. doi: 10.1016/s0020-7519(03)00094-8. [DOI] [PubMed] [Google Scholar]
- 30.Anderson RP, Lew D, Peterson AT. Evaluating predictive models of species' distributions: criteria for selecting optimal models. Ecol Modell. 2003;162:211–232. [Google Scholar]
- 31.Pearson RG, Raxworthy CJ, Nakamura M, Peterson AT. Predicting species distributions from small numbers of occurrence records: a test case using cryptic geckos in Madagascar. J Biogeogr. 2007;34:102–117. [Google Scholar]
- 32.Peterson AT, Cohoon KP. Sensitivity of distributional prediction algorithms to geographic data completeness. Ecol Modell. 1999;117:159–164. [Google Scholar]
- 33.Phillips SJ, Anderson RP, Schapire RE. Maximum entropy modeling of species geographic distributions. Ecol Modell. 2006;190:231–259. [Google Scholar]
- 34.Roberts JI. The endemicity of plague in East Africa. East Afr Med J. 1935;12:200–219. [Google Scholar]
- 35.Davis DH. Plague in Africa from 1935 to 1949; a survey of wild rodents in African territories. Bull World Health Organ. 1953;9:665–700. [PMC free article] [PubMed] [Google Scholar]
- 36.Orochi Orach S. Plague Outbreaks: the Gender and Age Perspective in Okoro County, Nebbi District, Uganda. Nebbe. Uganda: Agency for Accelerated Regional Development; 2002. [Google Scholar]
- 37.Migliani R, Chanteau S, Rahalison L, Ratsitorahina M, Boutin JP, Ratsifasoamanana L, Roux J. Epidemiological trends for human plague in Madagascar during the second half of the 20th century: a survey of 20,900 notified cases. Trop Med Int Health. 2006;11:1228–1237. doi: 10.1111/j.1365-3156.2006.01677.x. [DOI] [PubMed] [Google Scholar]
- 38.Eisen RJ, Enscore RE, Biggerstaff BJ, Reynolds PJ, Ettestad P, Brown T, Pape J, Tanda D, Levy CE, Engelthaler DM, Cheek J, Bueno R, Targhetta J, Montenieri JA, Gage KL. Human plague in the southwestern United States, 1957–2004: spatial models of elevated risk of human exposure to Yersinia pestis. J Med Entomol. 2007;44:530–537. doi: 10.1603/0022-2585(2007)44[530:hpitsu]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 39.Gage KL, Burkot TR, Eisen RJ, Hayes EB. Climate and vectorborne diseases. Am J Prev Med. 2008;35:436–450. doi: 10.1016/j.amepre.2008.08.030. [DOI] [PubMed] [Google Scholar]
- 40.Krasnov BR, Khokhlova IS, Fielden LJ, Burdelova NV. Effect of air temperature and humidity on the survival of pre-imaginal stages of two flea species (Siphonaptera: Pulicidae) J Med Entomol. 2001;38:629–637. doi: 10.1603/0022-2585-38.5.629. [DOI] [PubMed] [Google Scholar]
- 41.Enscore RE, Biggerstaff BJ, Brown TL, Fulgham RF, Reynolds PJ, Engelthaler DM, Levy CE, Parmenter RR, Montenieri JA, Cheek JE, Grinnell RK, Ettestad PJ, Gage KL. Modeling relationships between climate and the frequency of human plague cases in the southwestern United States, 1960–1997. Am J Trop Med Hyg. 2002;66:186–196. doi: 10.4269/ajtmh.2002.66.186. [DOI] [PubMed] [Google Scholar]
- 42.Kausrud KL, Viljugrein H, Frigessi A, Begon M, Davis S, Leirs H, Dubyanskiy V, Stenseth NC. Climatically driven synchrony of gerbil populations allows large-scale plague outbreaks. Proc R Soc Lond B Biol Sci. 2007;274:1963–1969. doi: 10.1098/rspb.2007.0568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Parmenter RR, Yadav EP, Parmenter CA, Ettestad P, Gage KL. Incidence of plague associated with increased winter-spring precipitation in New Mexico. Am J Trop Med Hyg. 1999;61:814–821. doi: 10.4269/ajtmh.1999.61.814. [DOI] [PubMed] [Google Scholar]
- 44.Stenseth NC, Samia NI, Viljugrein H, Kausrud KL, Begon M, Davis S, Leirs H, Dubyanskiy VM, Esper J, Ageyev VS, Klassovskiy NL, Pole SB, Chan KS. Plague dynamics are driven by climate variation. Proc Natl Acad Sci USA. 2006;103:13110–13115. doi: 10.1073/pnas.0602447103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Peterson AT. Improving methods for reporting spatial epidemiologic data. Emerg Infect Dis. 2008;14:1335–1336. doi: 10.3201/eid1408.080145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Eisen L, Eisen RJ. Need for improved methods to collect and present spatial epidemiologic data for vectorborne diseases. Emerg Infect Dis. 2007;13:1816–1820. doi: 10.3201/eid1312.070211. [DOI] [PMC free article] [PubMed] [Google Scholar]


