Abstract
Although DNA in eukaryotes is packaged in nucleosomes, it remains vulnerable to oxidative damage that can result from normal cellular metabolism, ionizing radiation, and various chemical agents. Oxidatively damaged DNA is repaired in a stepwise fashion via the base excision repair (BER) pathway, which begins with the excision of damaged bases by DNA glycosylases. We reported recently that the human DNA glycosylase hNTH1 (human Endonuclease III), a member of the HhH GpG superfamily of glycosylases, can excise thymine glycol lesions from nucleosomes without requiring or inducing nucleosome disruption; optimally oriented lesions are excised with an efficiency approaching that seen for naked DNA [1]. To determine if this property is shared by human DNA glycoylases in the Fpg/Nei family, we investigated the activity of NEIL1 on defined nucleosome substrates. We report here that the cellular concentrations and apparent kcat/KM ratios for hNTH1 and NEIL1 are similar. Additionally, after adjustment for non-specific DNA binding, hNTH1 and NEIL1 proved to have similar intrinsic activities towards nucleosome substrates. However, NEIL1 and hNTH1 differ in that NEIL1 binds undamaged DNA far more avidly than hNTH1. As a result, hNTH1 is able to excise both accessible and sterically occluded lesions from nucleosomes at physiological concentrations, while the high non-specific DNA affinity of NEIL1 would likely hinder its ability to process sterically occluded lesions in cells. These results suggest that, in vivo, NEIL1 functions either at nucleosome-free regions (such as those near replication forks) or with cofactors that limit its non-specific binding to DNA.
Keywords: Base excision repair, Chromatin, Nucleosome, DNA glycosylase, Human endonuclease III, Endonuclease VIII-like I, Mesothelioma
1. INTRODUCTION
Oxidative damage to DNA occurs as a consequence of normal cellular metabolism, ionizing radiation such as that used in cancer therapy, and various chemical agents [2–5]. Some of the resulting lesions are mutagenic, while others are cytotoxic. Most oxidative lesions are repaired via the base excision repair (BER) pathway which, in its simplest form (known as “short patch BER”), consists of four enzymatic steps (for reviews see [6–10]). The first step is the recognition and excision of a damaged base by either a mono- or bi-functional DNA glycosylase. This is followed by cleavage of the DNA backbone at the resulting apurinic site, either by the lyase activity associated with bifunctional DNA glycosylases or by AP Endonuclease (APE). Then, either APE or polynucleotide kinase (PNK) removes inhibitory moieties at the incision site, leaving a single base gap that is filled by Polymerase β and sealed by Ligase IIIα.
BER enzymes have been extensively characterized (for structure reviews, see [11, 12], see also [13]), and the entire BER pathway reconstituted in vitro with naked DNA substrates (e.g. [14, 15]). However, much less is known about how BER enzymes function in the context of the chromatin that packages DNA in eukaryotes. The basic subunit of chromatin is the nucleosome, which consists of 147 base pairs of DNA, wrapped in a left-handed toroidal helix around a histone octamer [16]. Histone octamers create a steric impediment to many of the factors and enzymes that act on DNA. The binding of such factors may also be influenced by the bending of DNA around the octamer, which alternately compresses and expands its major and minor grooves. Several groups have reported considerable variation in the capacity of selected BER enzymes to act on lesions in nucleosomes reconstituted in vitro [1, 17–19]. Some of this variation can be attributed to the position of the lesion relative to the underlying histone octamer. For example, the efficiency with which the human, bifunctional DNA glycosylase hNTH1 excises lesions from nucleosomes varies with the helical orientation of the lesion and its distance from the dyad axis (i.e. center) of the nucleosome [1]. The extent to which these structural variables affect lesion processing depends as well on enzyme concentration. Specifically, at high concentrations, hNTH1 is able to capture and process sterically-occluded lesions during episodes of transient, partial unwrapping of lesion-containing DNA from the histone octamer [1].
To further elucidate determinants that influence BER of oxidative lesions in chromatin, we have extended our studies to include the human DNA glycosylase NEIL1, a member of the Fpg/Nei family (hNTH1 is a member of the HhH GpG superfamily of glycosylases). While both NEIL1 and hNTH1 recognize and excise thymine glycol (Tg) from double-stranded DNA, several observations suggest that the two enzymes act in different cellular contexts. First, only NEIL1 is able to process lesions in single strand DNA and bubble substrates [20], a property that might make NEIL1 especially suited to removal of DNA polymerase blocking lesions (such as Tg) at replication forks [21–24]. Second, the inhibitory moiety left by hNTH1 is removed by APE1 while that left by NEIL1 is removed by PNK [15], and coimmunoprecipitation experiments indicate that NEIL1 interacts with PCNA and FEN1, factors that act in both DNA replication and long-patch BER [25, 26]. Thus, the two glycosylases channel their substrates into different BER sub-pathways. Third, the abundance of hNTH1 does not change during the cell cycle, while that of NEIL1 increases during S phase [27]. Taken together, these observations led to the hypothesis that NEIL1 acts on lesions at replication forks while hNTH1 acts more globally and independently of DNA replication [28].
The possibility that NEIL1 functions at replication forks, and with a different set of protein partners than does hNTH1, led us to ask if NEIL1 and hNTH1 differ in their capacity to process lesions in nucleosomes. We report here that cellular concentrations of hNTH1 and NEIL1 are similar, as are their catalytic rate constants. Additionally, the two enzymes exhibit similar intrinsic activity towards Tg lesions in nucleosomes and are similarly sensitive to the helical orientation of the lesion. However, the two enzymes differ in that NEIL1 binds undamaged double-stranded DNA far more tightly than does hNTH1. Our measurements of the in vivo abundance of NEIL1 and its relatively high affinity for lesion-free DNA suggest that NEIL1 would be severely restricted in its capacity to recognize and act on lesions in chromatin without the aid of protein partners that either recruit NEIL1 to sites of damage or reduce its non-specific interactions with DNA. By contrast, hNTH1 is more likely to be able to recognize lesions in chromatin without the aid of accessory factors.
2. EXPERIMENTAL PROCEDURES
2.1 In vivo concentrations of hNTH1 and NEIL1
The human mesothelial cell line LP9, and the human mesothelioma cell lines Gates, Mills, and Mont were seeded at 50,000 cells/ml in 60-mM dishes and grown overnight in CMRL 1066 medium with 10% fetal bovine serum (FBS; HyClone), 100 U of penicillin/ml, and 100 µg of streptomycin/ml at 37°C and in 5% CO2. At harvest, the cells were washed once with cold PBS, pH 7.4, and lysed by the addition of 100 µl lysis buffer (Laemmli buffer containing 100 mM DTT instead of β-mercaptoethanol, plus Roche Protease Inhibitor Cocktail) per plate. Lysates were collected by scraping with a rubber policeman, and the insoluble fraction was removed by centrifugation in a microfuge for 5 min. The protein concentration of the soluble fraction was determined using a Lowry protein assay [29]. Cell extracts and recombinant hNTH1 and NEIL1 were fractionated by 10% SDS polyacrylamide gel electrophoresis (SDS-PAGE), and transferred onto polyvinylidene fluoride (PVDF) membranes with the aid of a semi-dry electroblotting system (The WEP Company). Membranes were blocked with 1% bovine serum albumin (BSA) in TBS containing 0.1% Tween prior to addition of antibodies to either hNTH1 (Novus Biologicals NB100-302SS), NEIL1 (Abcam ab21337) or Actin (Chemicon MAB1501). Immunoreactive bands were visualized by using the appropriate secondary antibodies and an ECL detection kit (USB Rodeo ECL).
2.2 DNA and nucleosome substrates
For enzyme kinetic and non-specific DNA binding studies, a 35 nt DNA oligomer containing a single thymine glycol (Tg) residue (5′ -TGTCAATAGCAAGTgGGAGAAGTCAATCGTGAGTCT-3′), and a complementary oligomer containing dA opposite the Tg position, were purchased from Midland Certified Reagent Co. (Midland, Texas) and gel purified. Appropriate amounts of the lesion containing oligomer were end-labeled using [γ-32P]-ATP and T4 polynucleotide kinase (New England Biolabs), and annealed to equimolar amounts of the complementary oligomer in 10 mM Tris-HCl, pH 8.0, 50 mM NaCl and 1 mM EDTA. A 14 nt DNA oligomer containing a single guanidinohydantoin (Gh) residue (5′-GCGTCCAGhGTCTAC-3′, kindly provided by Cynthia Burrows (Univ. Utah; [30]), was end-labeled and annealed to a complementary oligomer containing dC opposite the Gh position in the same fashion. Tg-containing 184 bp DNA substrates and nucleosomes were prepared as previously described [1]. Briefly, DNA oligomers containing discretely positioned Tg residues (either 30 or 35 nucleotides from the 5′ end, for “Tg-out” and “Tg-in” containing nucleosomes, respectively), were end-labeled with [γ-32P] ATP, annealed to a single strand DNA template containing the Lytechinus variegatus 5S ribosomal DNA (rDNA) nucleosome positioning sequence, and extended with (exo-) Klenow enzyme (New England Biolabs). The resulting double-stranded 184 bp DNA was gel purified, quantified, and assembled into nucleosomes, using high salt mediated transfer of histone octamers from chicken erythrocyte donor chromatin prepared as described [1]. The efficiency of reconstitution (typically 85–95%) was assessed by electrophoresis of nucleosomes through a 5% polyacrylamide gel in 50 mM Tris base, 50 mM borate, and 1 mM H4EDTA.
2.3 Expression and purification of NEIL1 and hNTH1
To prepare NEIL1, BL21 cells were co-transformed with pRARE2 plasmid (Novagen) and a pET30a vector containing a C-terminal, His-tagged version of NEIL1. Cells were grown to saturation at 20°C for 48–60 hours in 1.5 L Terrific Broth (Fisher Scientific) containing 0.5% glycerol, 0.05% glucose, and 0.2% α-lactose monohydrate, to induce transcription by auto-induction [31]. Cells were harvested and lysed in buffer A (50 mM sodium phosphate buffer, pH 8.0, 10 mM imidazole, 300 mM NaCl, and 5 mM β-mercaptoethanol) containing freshly added 1 mM PMSF and 10 mM benzamidine. Cell debris was removed by centrifugation at 26,000 g for 15 min at 4°C. The supernatant was brought to 30% (NH4)2SO4 and centrifuged at 26,000 g for 20 min at 4°C. The supernatant was transferred to a clean tube, brought to 45% (NH4)2SO4 and centrifuged once again. The resulting NEIL1-containing pellet was suspended in buffer A containing fresh PMSF and benzamidine as before, and dialyzed against two changes of buffer A. The dialysate was loaded onto a 5 mL HiTrap Chelating HP column (GE Lifesciences) that had been charged with 2.5 mL of 100 mM NiSO4, and NEIL1 was eluted using a 100 mL linear gradient of 10 to 500 mM imidazole, pH 8.0 in buffer A. Fractions containing NEIL1 were identified by SDS-PAGE, pooled and dialyzed against buffer B (25 mM Tris-HCl, pH 7.5, 300 mM NaCl, 10% glycerol, and 5 mM β-mercaptoethanol). The dialysate was loaded onto a 1 mL HiTrap Heparin HP column (GE Lifesciences), and NEIL1 was eluted using a 20 mL linear gradient from 0.3 to 2 M NaCl in buffer B. Fractions containing NEIL1 were identified by SDS-PAGE, pooled, dialyzed back to 300 mM NaCl in buffer B, divided into aliquots, flash frozen in liquid nitrogen, and stored at −80° C until use. The 56 amino acid C-terminal NEIL1 truncation mutant was expressed and purified as previously described [32, 33].
To prepare hNTH1, an N-terminal GST-fusion construct was expressed in BL21 Star (DE3) cells using an auto-induction protocol identical to that described for NEIL1. Cells were lysed in buffer C500 (50 mM Tris-HCl, pH 8.0, and 5 mM β-mercaptoethanol containing 500 mM NaCl) containing freshly added 1 mM PMSF. Cell debris was removed by centrifugation at 26,000 g for 15 min, and the supernatant was loaded onto a 5 mL GSTrap FF column (GE Lifesciences), pre-equilibrated with buffer C500. Protein was eluted with a linear gradient of 0–10 mM reduced glutathione in buffer C500. Fractions containing hNTH1 were identified by SDS-PAGE, pooled and dialyzed against buffer C200. The GST moiety was then removed by the addition of 5 units Precision Protease (Amersham Pharmacia Biotech) for every mg of recombinant hNTH1. After an overnight incubation at 4° C, protein was loaded onto a second 5 mL GSTrap FF column (GE Lifesciences) as above, in order to separate the cleaved and uncleaved protein fractions. Flow-through fractions containing hNTH1 were pooled, dialyzed against buffer D (50 mM Tris-HCl, pH 8.0, 100 mM NaCl, 10% glycerol, and 5 mM β-mercaptoethanol), loaded onto a 5 mL SPFF column (GE Lifesciences) equilibrated in buffer D, and eluted using a linear gradient of 100–800 mM NaCl in buffer D. Fractions containing hNTH1 were identified by SDS-PAGE, pooled, dialyzed against 50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 1 mM DTT, 0.005% Triton-X, and 50% glycerol, and stored at −20°C. The 55 amino acid N-terminal hNTH1 truncation mutant was expressed and purified in the same manner.
2.4 Enzyme assays
All enzyme concentrations reported in the text and figures refer to active enzyme concentrations, determined as described by Blaisdell and Wallace (2007) [34]. Enzyme stocks were diluted into ice cold reaction buffer (25 mM NaHEPES NaOH, pH 8.0, 1 mM EDTA, 1 mM DTT, 100 mM NaCl) containing 0.1 mg/mL BSA (New England Biolabs) immediately prior to use. Nucleosome and nucleosome length DNA control reactions were conducted in reaction buffer containing 0.05% NP-40 and 0.02 mg/mL donor chromatin. Reactions were stopped by the addition of one volume formamide containing 0.1% bromophenol blue and 0.1% xylene cyanol. Samples were subsequently made 0.1 N in NaOH by addition of 1/10 volume 1 N NaOH, boiled for 3 min, and fractionated on 8% sequencing gels; reaction products from reactions with 14 and 35 bp substrates were fractionated on 12% sequencing gels. In all cases, reaction products were visualized and quantified by phosphorimagery. Glycosylase activity toward nucleosomes was determined as described in Prasad et al [1], which adjusts for small amounts of contaminating naked DNA substrate present in the nucleosome preparation.
2.5 Determination of non-specific binding affinity
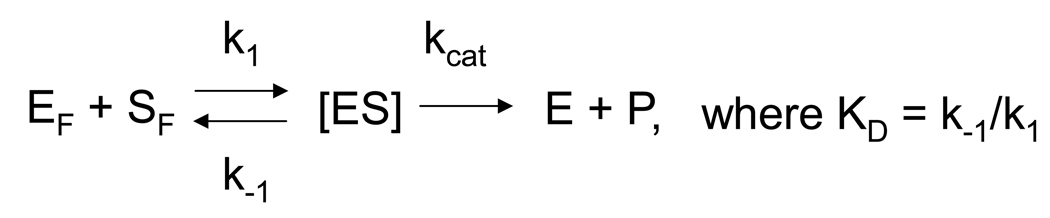
To compare NEIL1 and hNTH1 under conditions that were equivalent with respect to fraction of lesion bound by each enzyme, it was necessary to compensate for differences between the two enzymes in their affinity for Tg lesions and for non-specific binding to DNA. As diagrammed in Scheme 1 and Scheme 2, these DNA binding events can be represented as simple association reactions (where [S] and [NS] are the concentrations of specific and non-specific binding sites in DNA). In pseudo-steady-state conditions, the distribution of enzyme between specific and non-specific substrates will be dominated by k1 and k−1, allowing us to ignore kcat, and model both the specific and non-specific DNA binding of the two enzymes as one would for DNA sequence specific binding proteins [35], [36]. This enabled us to derive Equation 1, which describes the effect of both specific and non-specific binding constants (KD and KNS, respectively) on fraction of lesion in an enzyme-lesion complex (i.e. ES/ST) under a specified set of reaction conditions.
| (1) |
Scheme 1.
Scheme 2.
To solve Equation 1 for NEIL1, we first measured its affinity for Tg (KD) through a series of single-turnover reactions containing a fixed amount of a 32P end-labeled, Tg-containing DNA substrate (42 pM) and varying amounts of NEIL1. Reaction rates were determined by fitting data to a one-phase association using GraphPad Prism 5.0. The reaction rates thus obtained were plotted against the NEIL1 concentration and fitted to a one site binding (hyperbola) equation. kcat and KM values were obtained from the resulting curves as described [37]. To measure the non-specific DNA binding affinity of NEIL1 (KNS) we conducted a series of reactions in which a fixed amount of Tg- or Gh-containing oligomeric substrate (25 nM) was added to a fixed amount of enzyme (25 nM NEIL1, 17 nM hNTH1) that had been pre-incubated with varying amounts of non-specific competitor (either donor chromatin or DNA purified from the donor chromatin). The fraction of specific complexes formed in each reaction was inferred from the relative fraction of Tg-containing DNA cleaved in 45 seconds. The data were fit using a one-site competitive inhibition model on GraphPad Prism 5.0, which enabled us to extract KNS. Parallel measurements indicated that the non-specific DNA binding constant KNS for hNTH1 is much greater than the concentration of non-specific binding sites in our standard assay (~61 µM). Under these conditions, Equation 1 simplifies to Equation 2, which allowed us to estimate ES/ST for hNTH1 knowing just its KD for Tg.
| (2) |
hNTH1 undergoes a monomer to dimer transition that affects its activity [38], making it difficult to obtain kinetic constants as precise as those for NEIL1. Therefore, in separate studies, we measured approximate KD values for interactions between selected lesions and a hNTH1 N-terminal truncation mutant. This mutation does not detectably affect the DNA binding or catalytic activity of hNTH1 but reduced dimer formation enough for us to estimate the KD for hNTH1 binding to Tg as 6.7 +/− 2.1 nM (Table 1; Prasad, Barbour, Wallace and Pederson, submitted).
Table 1.
| kcat (sec−1) | KM (nM) | kcat/KM (sec−1 nM−1) | KNS (µM) | |
|---|---|---|---|---|
| NEIL1 | 0.13 | 0.25 | 0.52 | 0.029 |
| hNTH1 | 0.073 | 6.7 | 0.01 | 30 |
3. RESULTS
3.1 Cellular abundance of NEIL1 and hNTH1
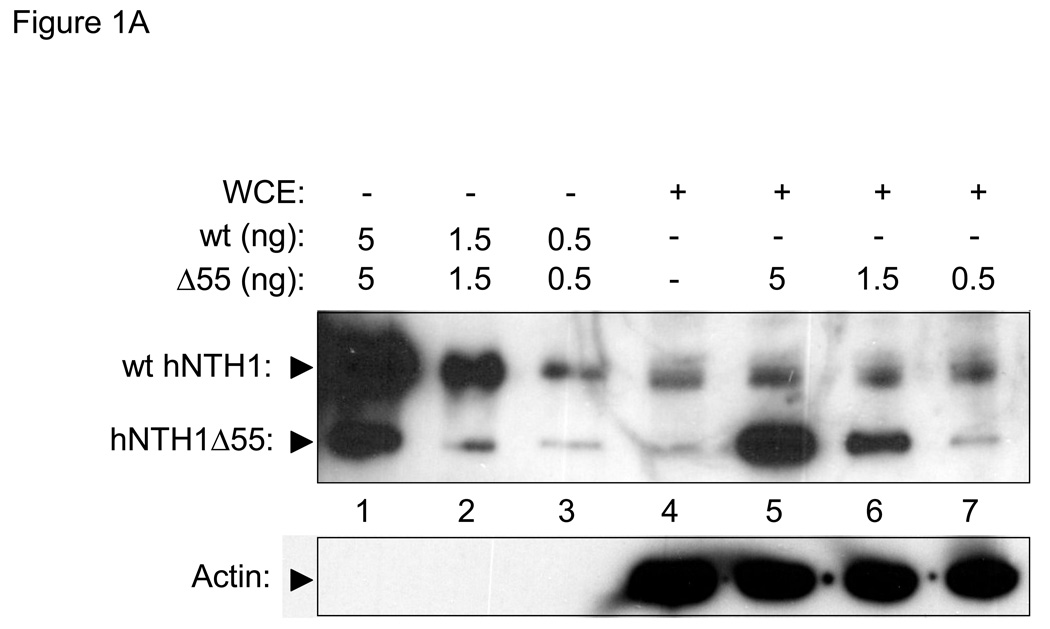
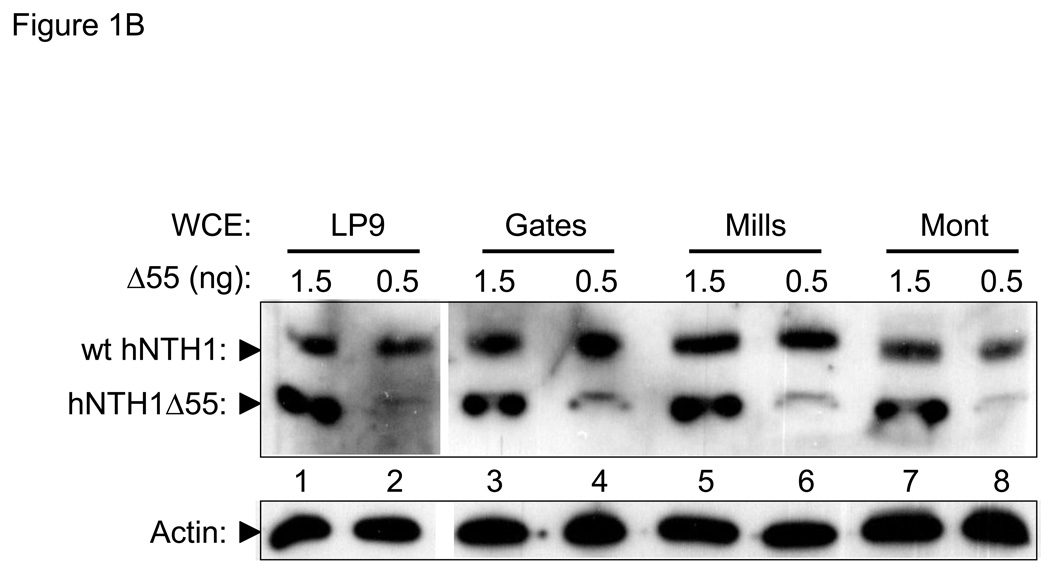
To compare NEIL1 and hNTH1 at physiologically meaningful concentrations, we first measured the amount of each enzyme in a mesothelial cell line (LP9) that is normal in most respects except for having been immortalized by constitutive expression of the telomerase catalytic subunit, hTERT [39]. Lanes 1–3 in Figure 1A show the antibody signal from serially diluted, recombinant full-length hNTH1 as well as that from a serially diluted, recombinant hNTH1 truncation mutant. The intensity of the antibody signal from hNTH1 in 40 µg of whole cell extract (lane 4 in Figure 1A) is similar to that generated by 0.5 ng of the full-length recombinant protein. In lanes 5–7, serial dilutions of the recombinant hNTH1 truncation protein were mixed with whole cell extract prior to electrophoresis. The intensity of the antibody signals from the full-length endogenous protein lies between those corresponding to 0.5 and 1.5 ng of the truncated enzymes. Since the antibody signal generated by full-length enzyme is 2–3 fold higher than the signal from the truncated enzyme (c.f. lanes 1–3), these results also indicate that cells contain about 0.5 ng of hNTH1 per 40 µg total protein, or roughly 0.5 fg per cell. Virtually all of the cellular hNTH1 is confined to the nucleus [40], which we estimated has a volume between one and three fl. Knowing that hNTH1 has a molecular weight of 34 kDa, we calculated that the nuclear concentration of hNTH1 lies between 250 and 800 nM.
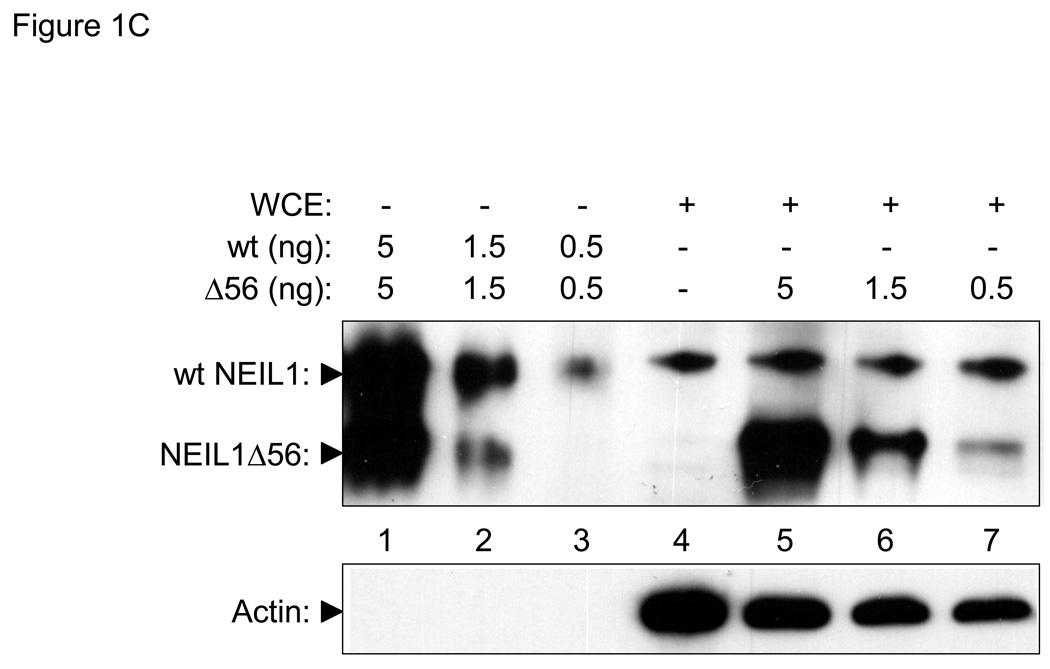
Figure 1. In vivo concentrations of hNTH1 and NEIL1.
(A) Upper panel: hNTH1 western blot showing, in lanes 1–3, serially diluted, full length, recombinant hNTH1 (“wt”) and a 55 amino acid N-terminal truncation mutant (hNTH1Δ55). In lanes 4–7, 0, 5, 1.5, and 0.5 ng of hNTH1Δ55 were added to 40 µg of LP9 whole cell extract prior to electrophoresis. Lower panel: hNTH1 blots were stripped and incubated with an anti-actin antibody as a loading control. (B) Upper panel: western blot of hNTH1 amounts in LP9 whole cell extracts compared to those in three mesothelioma cell lines (Gates, Mills, and Mont). Extracts were mixed with 1.5 or 0.5 ng of hNTH1Δ55 prior to electrophoresis, to facilitate quantification. Lower panel: actin control as above. (C) Upper panel: NEIL1 western blot showing, in lanes 1–3, serially diluted, recombinant full length NEIL1 (“wt”) and a 56 amino acid C-terminal truncation mutant of NEIL1 (NEIL1Δ56). In lanes 4–7, 0, 5, 1.5, and 0.5 ng of NEIL1Δ56 were added to 40 µg LP9 whole cell extract prior to electrophoresis. Lower panel: actin control as above.
The hNTH1 concentration calculated above is far lower than that reported earlier, in a study using HeLa cell nuclei [38]. This led us to ask if the abundance of hNTH1 in oncogenically transformed cells is higher than normal. Figure 1B shows that the hNTH1 abundance in three independently derived mesothelioma cell lines is similar to those in the relatively normal LP9 cell line.
To measure the cellular concentration of NEIL1, we conducted western blot studies similar to those described above for hNTH1. Lanes 1–3 of Figure 1C show the antibody signal from serially diluted, recombinant full-length NEIL1 as well as a serially diluted, recombinant NEIL1 truncation mutant. The intensity of the antibody signal from 40 µg of whole cell extract (lane 4 in Figure 1C) lies between the signals from 0.5 and 1.5 ng of the full-length recombinant protein. In lanes 5–7, serial dilutions of the recombinant NEIL1 truncation protein were mixed with 40 µg whole cell extract as described for hNTH1. The antibody signal from the full-length endogenous protein lies between that of 0.5 and 1.5 ng of the truncated enzyme. These results indicated that cells contain about 0.7 ng NEIL1 per 40 µg total protein, or 0.7 fg per cell. In situ localization studies and the phenotype of the NEIL1 knockout mouse suggest that NEIL1 is present in mitochondria as well as nuclei [41, 42]. However, given that most of the DNA in cells is nuclear, we expect that most of the NEIL1 enzyme is nuclear. Given this assumption, and that NEIL1 has a molecular weight of 44 kDa, we estimate that its nuclear concentrations ranges between 250 and 800 nM, approximately the same as that measured for hNTH1.
3.2 Impact of non-specific DNA binding on the capacity of hNTH1 and NEIL1 to excise oxidative lesions from naked DNA
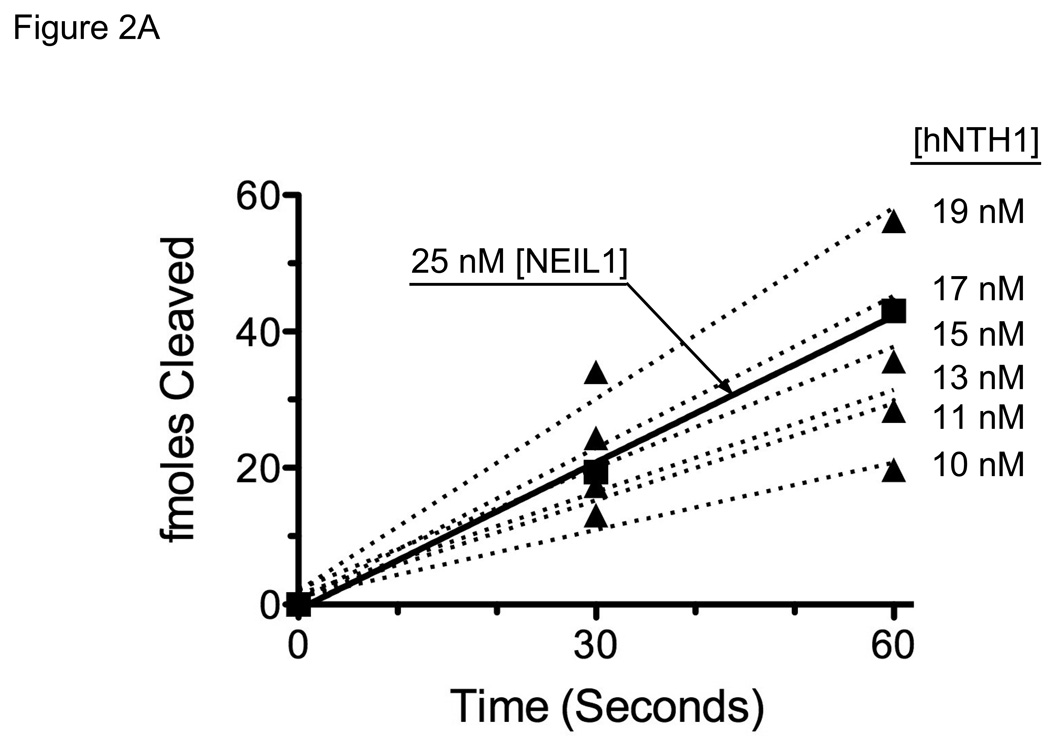
Based on the cellular abundance of hNTH1 and NEIL1, we decided initially to compare the activity of hNTH1 on selected substrates with that of 25 nM NEIL1. To compensate for kcat/KM differences between the two enzymes, we first determined (empirically) the hNTH1 concentration that would give an initial reaction velocity on naked DNA equivalent to that produced by 25 nM NEIL1. Figure 2A shows initial reaction velocities for 25 nM NEIL1 and an array of hNTH1 concentrations. Inspection of these data indicated that, for Tg-containing substrates, 17 nM hNTH1 best matched that of 25 nM NEIL1. Thus, hNTH1 and NEIL1 are not only similar in cellular abundance but they also exhibit similar activity toward Tg lesions in naked double-stranded DNA.
Figure 2. Adjustment of hNTH1 and NEIL1 concentrations to compensate for differences in enzyme efficiency and non-specific DNA binding affinity.
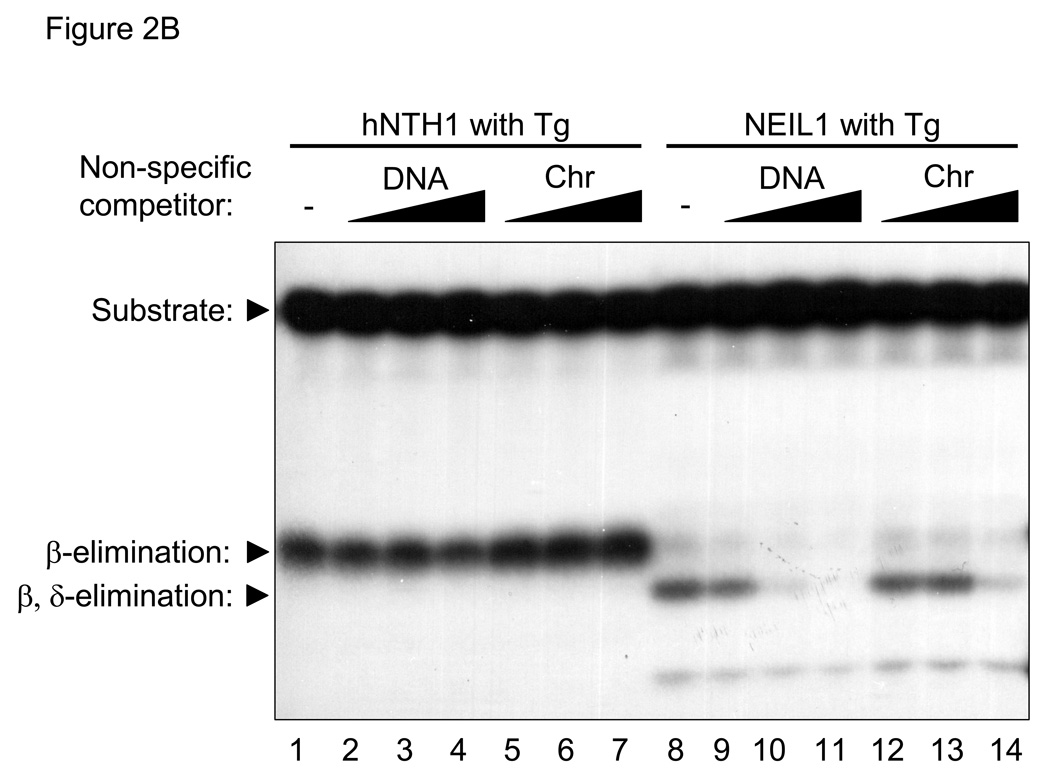
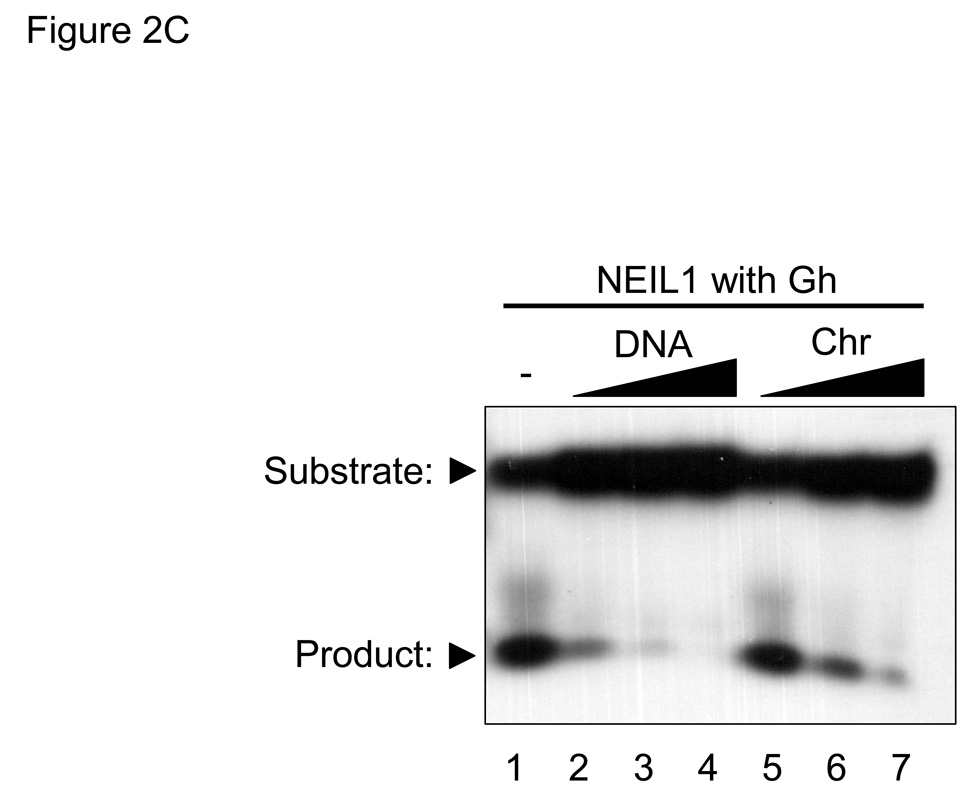
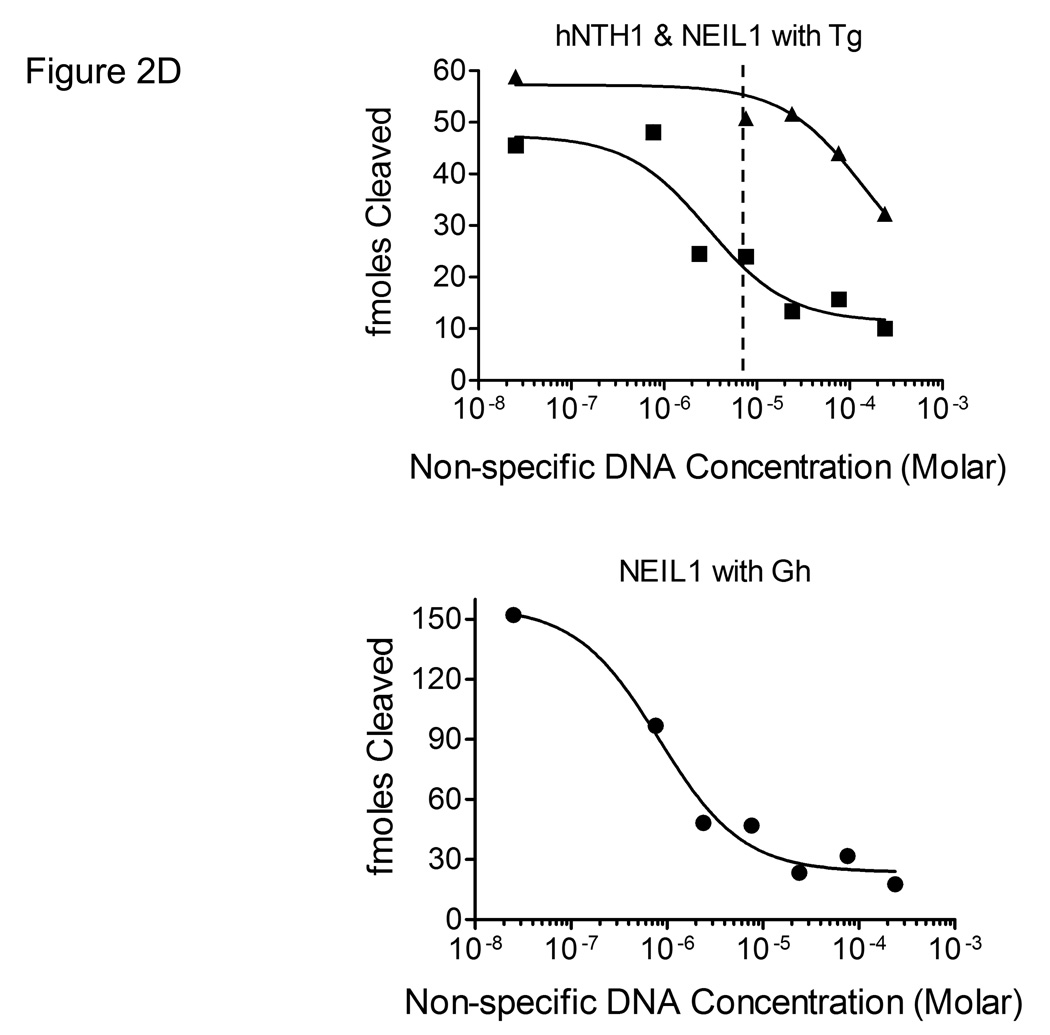
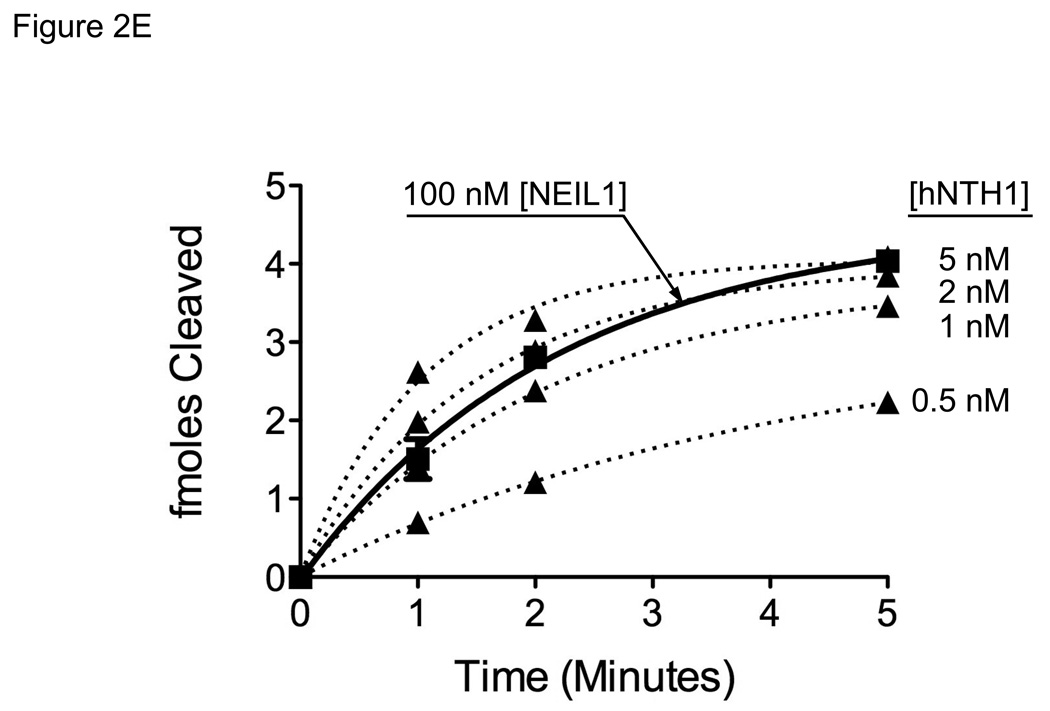
(A) Graph showing initial rates of excision of Tg lesions by 25 nM NEIL1 (■, solid line) and varying concentrations of hNTH1 (▲, dotted lines for reactions containing 10, 11, 13, 15, 17 and 19 nM hNTH1), assayed as described in the Methods. Data from the single-turnover portion of each reaction curve were fit by linear regression. Note that the rate of excision by 17 nM hNTH1 closely matches that observed with 25 nM NEIL1. (B) Non-specific DNA binding assay. A 35 bp, end-labeled, Tg-containing DNA substrate was added to reaction tubes containing varying amounts of undamaged competitor DNA or chromatin and either 17 nM hNTH1 (Lanes 1–7) or 25 nM NEIL1 (lanes 8–14). Reaction extents were determined after 45 seconds at 37°C. Lanes 1 and 8, 2 and 9, 3 and 10 and 4 and 11 contained, respectively, 0, 0.76, 7.6 and 76 µM DNA. Lanes 5 and 12, 6 and 13, and 7 and 14 contained, respectively, 0.76, 7.6 and 76 µM chromatin. (C) 25 nM NEIL1 was incubated with a 14 bp double-stranded oligomer containing Gh. Lanes 1, 2, 3, and 4 contained, respectively, 0, 2.4, 24, and 240 µM undamaged DNA and lanes 5, 6, and 7 contained, respectively, 2.4, 24, and 240 µM chromatin. (D) The amounts of substrate cleaved in the reactions shown in figures 2B and 2C, were quantified by phosphorimagery. These and data from additional, independent competition experiments were plotted against the logarithm of the concentration of lesion-free DNA. The upper panel depicts the activities of NEIL1 (■) and hNTH1 (▲) towards Tg when incubated with 25 nM substrate alone, or together with the indicated amounts of lesion-free DNA. The lower panel depicts the activity of NEIL1 (●) towards Gh when incubated with the same concentrations of lesion-free DNA competitor. The vertical dashed line in the upper panel indicates the concentration of lesion-free chromatin in our standard chromatin assay (61 µM). (E) Graph showing reaction curves for 100 nM NEIL1 in the presence of 61 µM chromatin (■, solid line) and varying concentrations of hNTH1 (▲, dotted lines for reactions containing 0.5, 1, 2 and 5 nM hNTH1). Note that, in the presence of 61 µM chromatin, the rate of excision by 2 nM hNTH1 most closely matches that observed with 100 nM NEIL1.
The concentration of undamaged DNA in our standard chromatin repair assay is approximately 61 µM, about 35-fold higher than in the above-described reactions with double-stranded DNA oligomers. This made it important to determine if non-specific binding of NEIL1 and hNTH1 to DNA or chromatin would affect their relative activity. We therefore conducted a series of reactions with the same amount of enzyme as before (i.e. 25 nM NEIL1 and 17 nM hNTH1) but in the presence of increasing amounts of either soluble, lesion-free chromatin or naked DNA isolated from the same preparation of chromatin. Apart from the non-specific competitor, reaction conditions were identical to those in Figure 2A, where the rate of product formation by either hNTH1 or NEIL1 was approximately constant during the first 60 sec. Therefore, we reasoned that the amount of product generated during the first 45 sec in the competition reactions would appropriately reflect the fraction of total substrate bound by enzyme. Lanes 1–7 in Figure 2B show that the activity of hNTH1 was relatively unaffected by addition of either a ~30, ~300, or ~3000 fold molar excess of undamaged DNA or chromatin. Subsequent experiments revealed moderate inhibition of hNTH1 when the fold-excess of non-specific competitor DNA was increased to ~10,000 (Figure 2D and data not shown).
In stark contrast, inhibition of the activity of NEIL1 toward Tg was evident in reactions with as little as a ~100-fold molar excess of non-specific DNA competitor (Figure 2D and lanes 8–14 in Figure 2B). Although NEIL1 excises Tg lesions with reasonably high efficiency, it is far more active toward hydantoin lesions that form as oxidation products of 7,8-dihydro-8-oxoguanine (8-oxoG) [43]. If the higher activity of NEIL1 toward lesions such as Gh (guanidinohydantoin) were due to more avid binding, we would expect that a non-specific DNA competitor would not have as great an impact as it did in reactions with Tg substrates. On the other hand, if the higher activity of NEIL1 toward Gh were due to a higher rate of catalysis, we would expect that a non-specific DNA competitor would suppress the activity of NEIL1 toward Gh to the same degree as it did for Tg. To test these predictions, we conducted competition experiments with Gh-containing substrates. The results in Figure 2C support the latter prediction, and are consistent with the recent finding that the catalytic constants for NEIL1 acting on hydantoin lesions are far higher than for Tg lesions [43]. The results in Figure 2C also support a qualitative observation by the same authors, that addition of a small quantity of lesion-free DNA (25 nM) helped stabilize hNEIL1 during stopped-flow reaction conditions but that larger amounts suppressed the activity of hNEIL1.
The median length of the DNA competitor used in these studies was no more than ~400 bp, making it important to rule out the possible inhibition of NEIL1 due to non-specific binding to DNA ends. We therefore repeated the above study using full-length phage lambda DNA as a non-specific DNA competitor. At equivalent concentrations, the ~48,000 bp phage DNA proved to be an equally or slightly more effective competitor than DNA isolated from soluble chromatin, despite a >100-fold lower concentration of DNA ends (data not shown). Thus, binding to DNA ends has little or no impact on the non-specific affinity of NEIL1 for DNA. These results collectively indicated that the specific-to-nonspecific DNA binding ratio for hNTH1 is far higher than that for NEIL1, and that it would be necessary to further adjust the relative concentrations of hNTH1 and NEIL1 to ensure equivalent amounts of free enzyme in reactions with chromatin substrates.
To estimate the relative amounts of NEIL1 and hNTH1 needed to mitigate the differential impact of non-specific DNA binding, we first quantified the non-specific DNA binding data, as shown in Figure 2D. We next determined the equilibrium dissociation constants (KD) for NEIL1 and hNTH1 binding to Tg, as outlined in the Methods and tabulated in Table 1. We then were able to calculate non-specific DNA binding affinities (KNS) of approximately 29 nM for NEIL1 and 30 µM for hNTH1. The non-specific affinity of NEIL1 for lambda DNA was slightly higher (KNS ~9 nM), although the likelihood that this difference is statistically significant is only about ~85%. As a non-specific competitor, undamaged chromatin was not as effective as DNA isolated from the chromatin (KNS ~930 nM). Indeed, the difference between the naked DNA and chromatin competitors is statistically significant and consistent with the idea that, through steric occlusion, nucleosomes reduce the impact of non-specific DNA binding on the activity of certain enzymes in chromatin.
Having measured the non-specific affinity of NEIL1 for DNA, we were able to calculate (using the equations described in the Methods) that 100 nM NEIL1 in our standard chromatin assay would exhibit about the same activity on naked DNA as would hNTH1 in the range of 1–5 nM. To further refine this estimate, we compared the activity of 100 nM NEIL1 in a reaction with a Tg-containing naked DNA substrate and 61 µM non-lesion containing chromatin to results from a series of parallel reactions containing 0.5 to 5.0 nM hNTH1. Figure 2E shows the rate of Tg excision by 100 nM NEIL1 was similar to that exhibited in reactions with 1 to 2 nM hNTH1.
3.3 Comparison of hNTH1 and NEIL1 on nucleosome substrates
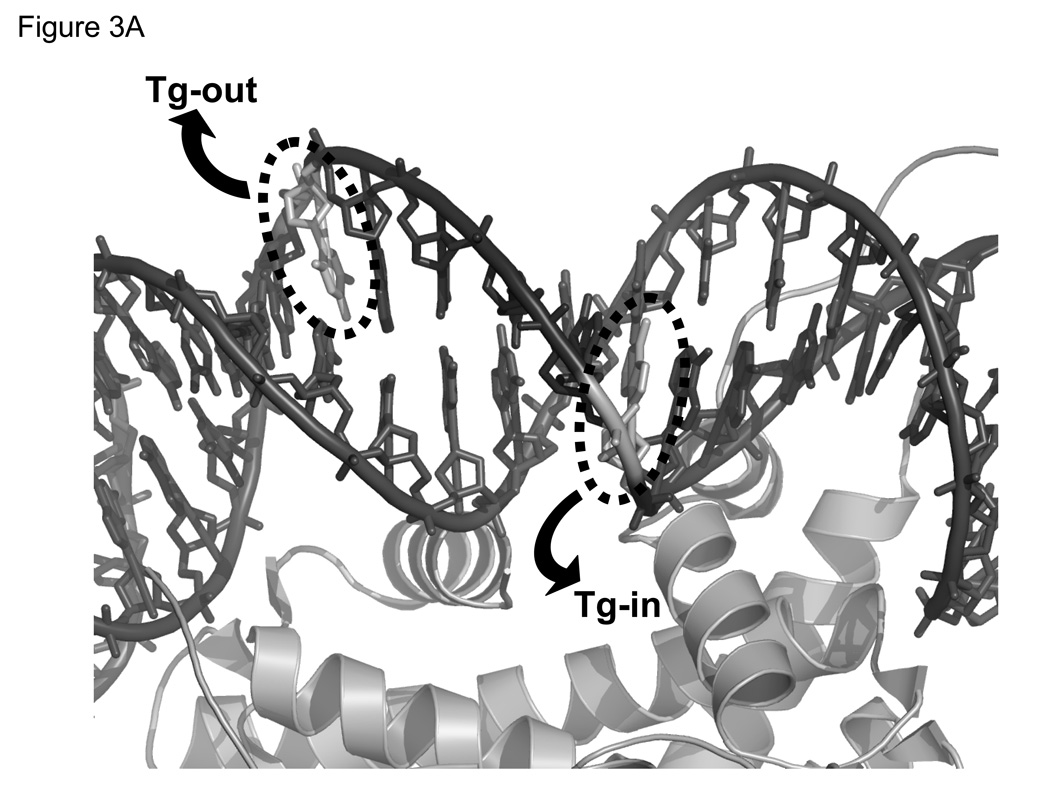
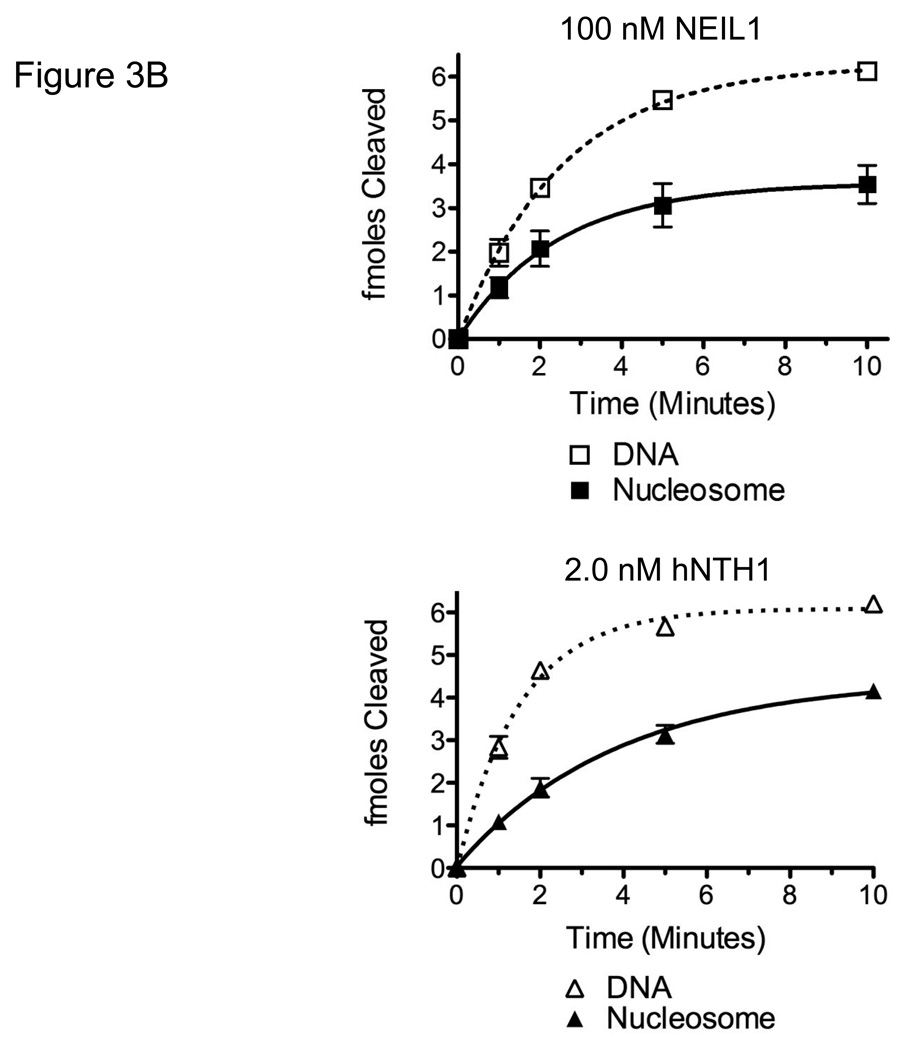
Having determined conditions that would compensate for differences between hNTH1 and NEIL1 in their non-specific DNA binding, we next measured the relative activity of the two enzymes toward Tg-containing nucleosomes. Both hNTH1 and NEIL1 require the oxidized base to flip out of the DNA helix via the minor groove [44, 45]. Hence, a Tg positioned such that the minor groove faces away from the histone octamer (Tg-out) is more accessible than a Tg residue at which the minor groove faces toward the histone octamer (Tg-in), as illustrated in Figure 3A [1, 19]. As shown in Figure 3B, both 2 nM hNTH1 and 100 nM NEIL1 were able to excise lesions from Tg-out nucleosomes with a relatively high efficiency. For hNTH1, this result is in accord with earlier studies [1] and indicates that hNTH1 can efficiently process sterically accessible lesions in nucleosomes at enzyme concentrations equivalent to or below those in cells (though somewhat higher than those required for naked DNA substrates). For NEIL1, efficient excision of thymine glycol residues occurred only when NEIL1 was used at concentrations equal to or higher than those in cells.
Figure 3. NEIL1 and hNTH1 exhibit similar intrinsic activities toward nucleosomes with ‘outward-facing’ Tg lesions but the capacity of NEIL1 to excise lesions from sterically occluded sites in nucleosomes is limited by its high non-specific DNA binding.
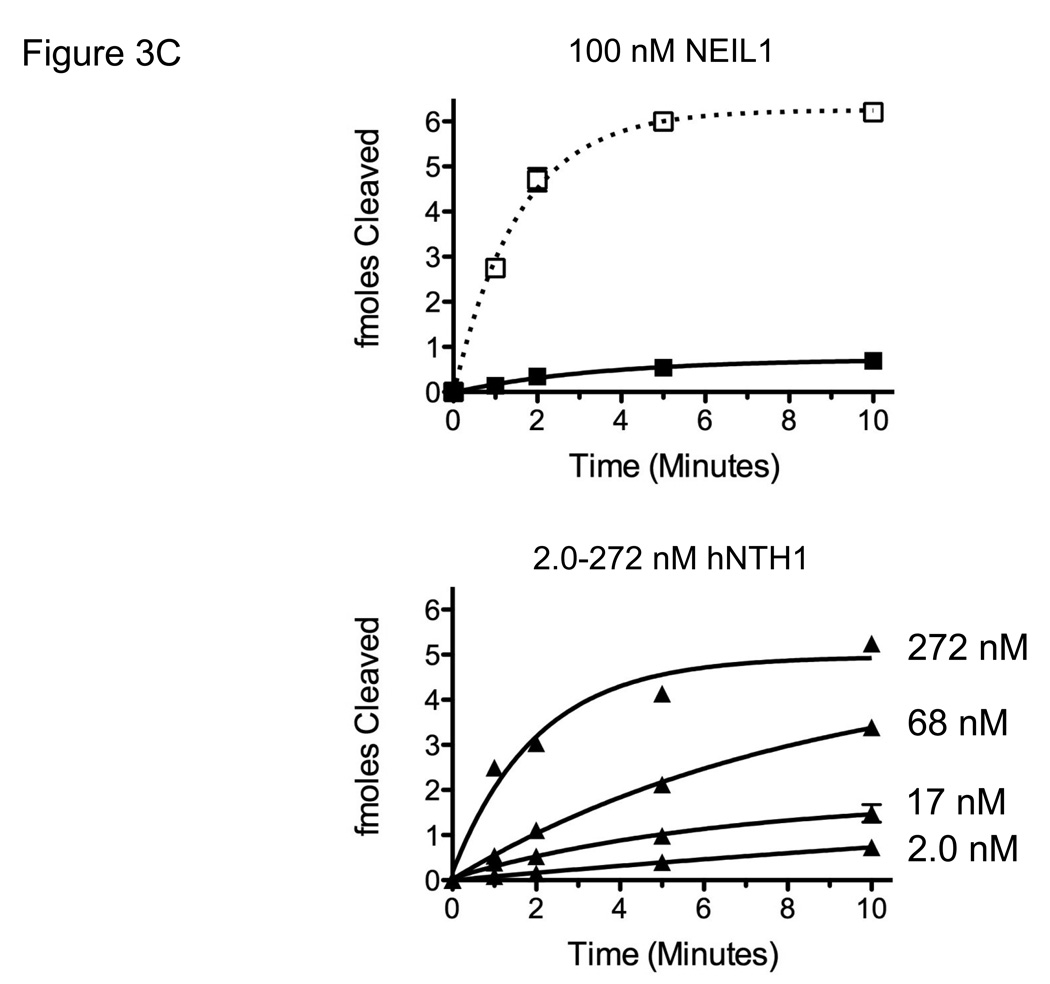
(A) Depiction of the helical orientation of thymine glycol lesions in Tg-in and Tg-out nucleosomes, based on a 1.9 Å resolution nucleosome crystal structure (PDB 1KX5 [52]). Arrows indicate direction of Tg flipping needed to enter the active site of either hNTH1 or NEIL1. (B) Reaction curves for 100 nM NEIL1 and 2 nM hNTH1 with Tg-out containing nucleosomes (solid lines) and naked DNA (dashed lines). (C) Reaction curves obtained with 100 nM NEIL1 and varying concentrations of hNTH1 with Tg-in containing nucleosomes (solid lines) and naked DNA controls (dotted lines). Note that 100 nM NEIL1 matches or slightly exceeds its estimated concentration in vivo. The activity of NEIL1 at concentrations above 100 nM (active enzyme) was similar or only slightly higher than the activity shown for 100 nM (not shown). Note that while the histone octamer substantially inhibits hNTH1 at low concentrations, hNTH1 is increasingly able to process the sterically occluded lesion in Tg-in nucleosomes when its concentration is increased from 2 nM to 17, 68, and 272 nM hNTH1. The 68 nM reaction curve closely matches the estimated in vivo concentration of hNTH1.
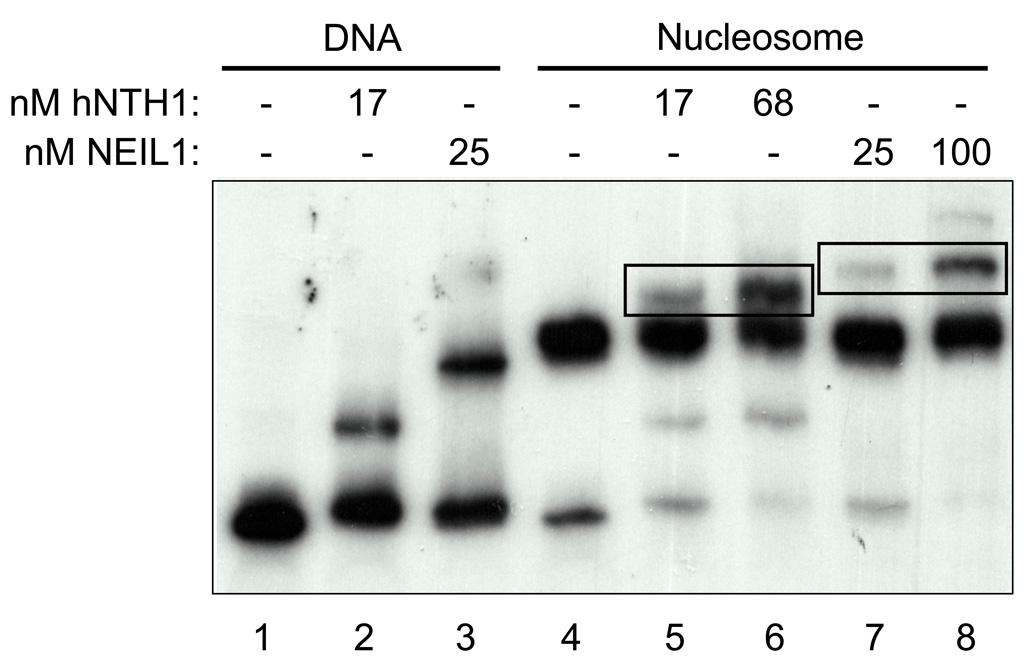
Previous studies had indicated that hNTH1 forms a ternary complex with lesion-containing nucleosomes and can remove damaged bases without inducing or requiring nucleosome disruption [1]. To determine if this was the case for NEIL1 as well, and to rule out the possibility that NEIL1 disrupts lesion-containing nucleosomes, we gel-fractionated Tg-out nucleosomes in the absence and presence of hNTH1 and NEIL1. Lanes 1–3 in Figure 4 show that both hNTH1 and NEIL1 alter the migration of naked, nucleosome-length Tg-containing DNA. Lanes 5–8 in Figure 4 show the effect of both low and high concentrations of hNTH1 and NEIL1 on the mobility of lesion-containing nucleosomes. The failure of hNTH1 and NEIL to increase the amount of naked DNA in lanes 5–8 indicated that neither enzyme disrupted nucleosomes. Lanes 5–8 in Figure 4 also show that both enzymes form ternary complexes with Tg-containing nucleosomes.
Figure 4. Both hNTH1 and NEIL1 remove sterically accessible lesions from nucleosomes without requiring or inducing nucleosome disruption.
Electrophoretic mobility shift assay of hNTH1 and NEIL1 bound to Tg-containing naked DNA and Tg-out nucleosomes. Note that both hNTH1 and NEIL1 form ternary complexes with nucleosomes, indicated by rectangles in lanes 5 and 6 and 7 and 8. Note that neither enzyme increases the amount of material that co-migrates with naked DNA, indicating that neither enzyme irreversibly disrupts the nucleosome during lesion processing.
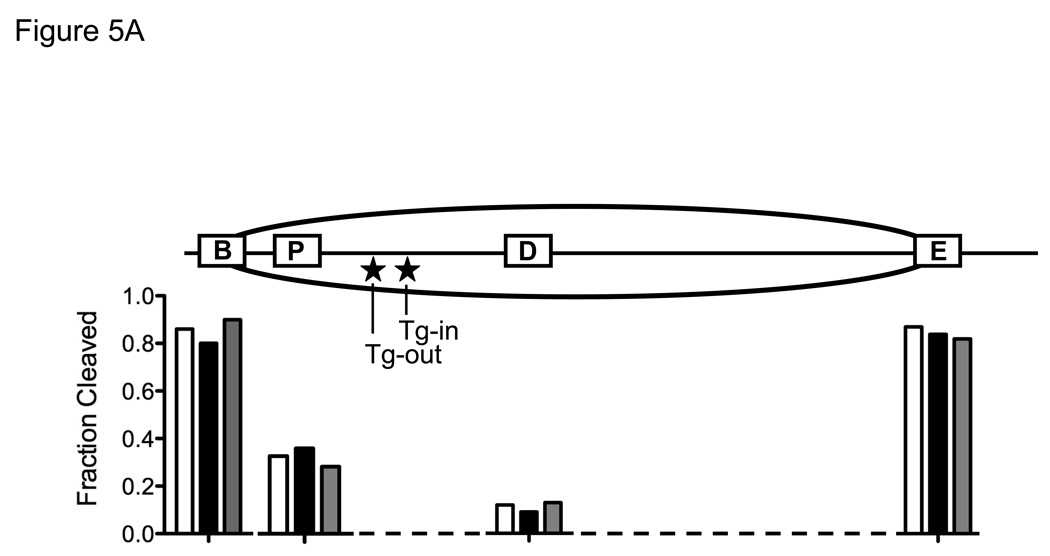
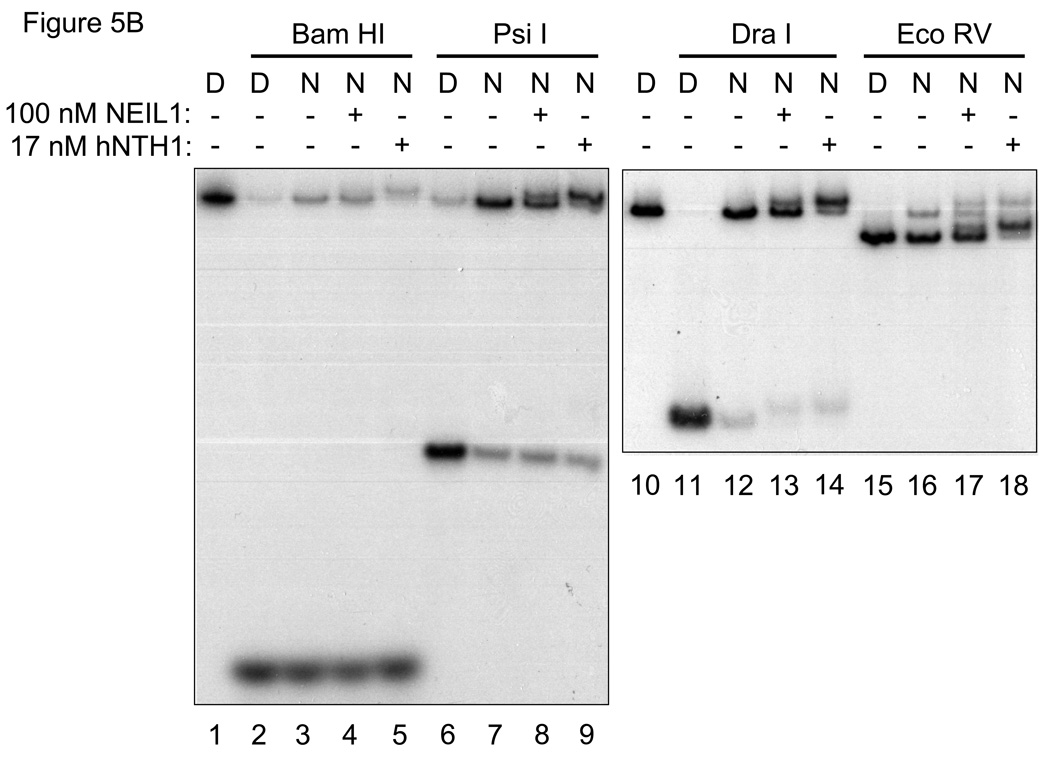
The above described mobility gel shift study indicated that NEIL1 can excise lesions from Tg-out nucleosomes without inducing nucleosome disruption, but it was necessary as well to rule out the possibility that NEIL1 had altered the translational position of DNA relative to the underlying octamer, thereby moving the Tg lesion into a nucleosome-free segment of DNA. To map the predominant translational position of Tg-out nucleosomes, we conducted quantitative restriction enzyme cleavage analyses in the absence and presence of hNTH1 and NEIL1. Nucleosomes and naked DNA controls were incubated with either Bam HI or Eco RV, which cleave at sites immediately adjacent to either edge of the nucleosome in its dominant translational position, or with Psi I or Dra I, whose cognate sites lie within the nucleosome (c.f. Figure 5A). Figures 5A and 5B show patterns of restriction site exposure and protection for lesion-containing nucleosomes identical to those described previously [1], indicating that most of the histone octamers resided at a single common position. Minor positional variants do exist but, as described previously [1], do not appear to expose the Tg lesion. Figures 5A and 5B also show that neither NEIL1 nor hNTH1 altered the restriction enzyme cleavage pattern. Thus, as with hNTH1, NEIL1 can excise sterically accessible lesions from nucleosomes without altering nucleosome positions.
Figure 5. Both hNTH1 and NEIL1 remove sterically accessible lesions from nucleosomes without detectably altering nucleosome position.
(A) Diagram that depicts the predominant translational setting of the model, Tg-containing nucleosomes used in this study, and the restriction enzyme sites used to monitor nucleosome position. The extent of cleavage of restriction sites in nucleosomes, assessed as described below in Figure 5B, is expressed in the histogram as a fraction of the cleavage obtained at the corresponding sites in naked DNA. The open, solid, and grey bars indicate, respectively, the relative double-stranded DNA cleavage by the indicated restriction enzyme when incubated alone, with 100 nM NEIL1, and with 17 nM hNTH1. Note that both the Bam H1 and Eco RV sites, located immediately adjacent to either side of the nucleosome, remain accessible following the addition of either hNTH1 or NEIL1, suggesting that neither enzyme alters the dominant translational position of the nucleosome. (B) Restriction enzyme cleavage analyses of Tg-containing nucleosomes in the absence and presence of either 100 nM NEIL1 or 17 nM hNTH1. Naked DNA (“D”) and nucleosomes (“N”) were incubated with a 50-fold unit excess of the indicated restriction enzyme for 10 min at 37°C. The resulting DNA products were separated on non-denaturing gels, and quantified by phosphorimagery to assess the extent of cleavage at selected restriction sites. Note that single strand DNA cleavage by either glycosylase slightly retards the migration of the double-stranded DNA, which accounts for the minor bands, such as those seen in lanes 13 and 14. Lanes 1 and 10 contain uncleaved naked DNA controls.
Earlier studies indicated that the histone octamer substantially blocks access to Tg lesions at sites where the minor groove of the lesion faces toward the histone octamer. However, at high concentrations hNTH1 was able to process such lesions in a relatively efficient manner, probably because spontaneous, transient partial unwrapping of DNA from the histone octamer enables hNTH1 to capture lesions in the unwrapped DNA population [1]. Figure 3C shows that at an active enzyme concentration of 100 nM, NEIL1 was unable to excise more than 10% of the lesions from Tg-in nucleosomes in a 10 min interval. At an active enzyme concentration of 2 nM, the activity of hNTH1 on Tg-in nucleosomes was equally poor. The key difference, however, is that 100 nM NEIL1 equals or slightly exceeds our estimate of its concentration in vivo while the 2 nM hNTH1 used for comparative purposes is far below its in vivo concentration. In reactions conducted with hNTH1 amounts comparable to those in vivo, the efficiency with which hNTH1 excises occluded lesions rises dramatically, as shown in Figure 3C. (Because of aggregation of NEIL1 at high concentrations it was not possible to examine its activity at very high concentrations.) In summary, hNTH1 is able to function efficiently towards both accessible and occluded lesions in chromatin due to its high substrate specificity and low non-specific DNA binding. On the other hand, NEIL1 (which has an even higher affinity toward Tg lesions in naked DNA) may be unable to efficiently process lesions in canonical nucleosomes without the aid other proteins that either suppress its non-specific DNA binding or actively recruit it to sites of oxidative damage.
4. DISCUSSION
In this study we have determined that the in vivo concentrations of the DNA glycosylases hNTH1 and NEIL1 are similar to one another, and that both enzymes are able to excise thymine glycol lesions from nucleosomes without inducing nucleosome movement or disruption. Previous studies that compared the capacity of selected DNA glycosylases to function on chromatin substrates have not (explicitly) accounted for enzymatic variables that could influence results, such as differences in kcat or KM or in non-specific DNA binding affinity. By adjusting enzyme concentrations to compensate for such variables, this study provides a standard template for future comparisons of enzyme action on chromatin substrates.
The key difference between NEIL1 and hNTH1, which bears on their likely roles in vivo, is that NEIL1 has a much higher affinity for undamaged DNA than hNTH1. The likely in vivo concentration of hNTH1 (~250 to ~800 nM) is sufficiently high to imagine that the enzyme finds oxidative lesions in two steps, beginning with a diffusion-driven, three-dimensional search for DNA, that results in non-specific DNA binding. Once bound to DNA the enzyme would engage in a highly efficient ‘one-dimensional’ search along the DNA for oxidative lesions. This model is predicated on classic studies of lac repressor (notably by Riggs and von Hippell and their colleagues [35, 46]). In the case of hNTH1, the efficiency of lesion discovery may be further enhanced through a charge-transport mechanism proposed by Barton and colleagues [47, 48]. Specifically, hNTH1, like its prokaryotic counterpart, contains an iron-sulfur cluster that may become oxidized upon DNA binding [49]. If hNTH1 binds more avidly to DNA in its oxidized [4Fe4S]+3 state, its dissociation rate would be enhanced by reduction of the iron-sulfur cluster, which could occur by electron transport from an electron donor through the DNA base pair stack. DNA lesions that interfere with base stacking would likely interfere with this charge transport and thereby increase the dwell-time of hNTH1 near sites of DNA damage. Thus, a combination of diffusion-driven and electron transport mediated search mechanisms may permit highly efficient lesion discovery.
In sharp contrast to hNTH1, NEIL1 exhibited a relatively high non-specific DNA binding affinity. This would make it virtually impossible for NEIL1 to efficiently discover oxidative lesions in nuclear DNA via the mechanism suggested for hNTH1 (assuming that NEIL1 is homogenously distributed within the nucleus at a concentration ranging between ~250 to ~800 nM). There are several potential evolutionary solutions to this conundrum that are consistent with known properties of NEIL1. Specifically, NEIL1 is found in both mitochondria and nuclei, and its abundance increases during S phase [27, 41, 50, 51]. Given the relatively small size of the mitochondrial genome, NEIL1 might be able to discover oxidative lesions in mitochondria via the simple diffusion-driven search mechanism like that described for hNTH1. In nuclei, an increase in the concentration of NEIL1 during S phase would increase the efficiency of lesion discovery. Additionally, our non-specific competition studies indicate that NEIL1 has a higher affinity towards undamaged naked DNA than nucleosomal DNA. Thus, the packaging of DNA into chromatin likely reduces the fraction of NEIL1 that is lost to non-specific interactions with DNA. It is unlikely, however, that these factors, even in combination, would enable NEIL1 to discover lesions at an adequate rate without the additional involvement of proteins that either recruit NEIL1 to sites of oxidative damage or bind NEIL1 in such a way as to reduce its non-specific DNA affinity. Relevant to this point, Mitra and colleagues have reported that NEIL1 interacts with such DNA replication and repair factors as PCNA and FEN-1, which prompted them to hypothesize that the primary role of NEIL1 is in replication associated repair [20, 25, 26, 28]. It remains to be determined if these or other NEIL1-interacting proteins influence its non-specific interactions with DNA.
ACKNOWLEDGEMENTS
We thank Dr. Cynthia J. Burrows for supplying the Gh-containing oligomers, Lauren Harvery and April Averill for the expression and purification of enzymes used in this study and Dr. Viswanath Bandaru for providing the GST-hNTH1 expression vector. We thank Dr. Amalthiya Prasad and Joy-El Barbour for guidance in the preparation and assay of lesion-containing nucleosomes, and for sharing kinetic data on hNTH1Δ55. We thank Drs. Scott Morrical, Jeffrey Bond, and Susan Robey-Bond for helpful discussions of enzyme kinetics. The research was supported in part by an NSF grant MCB-0821941 to D.S.P., and NIH grant P01-CA098993 awarded by the National Cancer Institute.
ABBREVIATIONS
- APE
AP endonuclease
- BER
base excision repair
- BSA
bovine serum albumin
- FBS
fetal bovine serum
- Fpg
formamidopyrimidine DNA glycosylase
- Gh
guanidinohydantoin
- Nei
endonuclease VIII
- PBS
phosphate buffered saline
- PNK
Polynucleotide kinase
- PVDF
polyvinylidene fluoride
- rDNA
ribosomal DNA
- SDS-PAGE
SDS polyacrylamide gel electrophoresis
- Tg
thymine glycol
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
CONFLICT OF INTEREST STATEMENT.
The above authors have no financial interests.
REFERENCES
- 1.Prasad A, Wallace SS, Pederson DS. Initiation of base excision repair of oxidative lesions in nucleosomes by the human, bifunctional DNA glycosylase NTH1. Molecular and cellular biology. 2007;27:8442–8453. doi: 10.1128/MCB.00791-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lindahl T, Wood RD. Quality control by DNA repair. Science (New York, N.Y. 1999;286:1897–1905. doi: 10.1126/science.286.5446.1897. [DOI] [PubMed] [Google Scholar]
- 3.Klaunig JE, Kamendulis LM. The role of oxidative stress in carcinogenesis. Annual review of pharmacology and toxicology. 2004;44:239–267. doi: 10.1146/annurev.pharmtox.44.101802.121851. [DOI] [PubMed] [Google Scholar]
- 4.Engel RH, Evens AM. Oxidative stress and apoptosis: a new treatment paradigm in cancer. Front Biosci. 2006;11:300–312. doi: 10.2741/1798. [DOI] [PubMed] [Google Scholar]
- 5.Hada M, Georgakilas AG. Formation of clustered DNA damage after high-LET irradiation: a review. Journal of radiation research. 2008;49:203–210. doi: 10.1269/jrr.07123. [DOI] [PubMed] [Google Scholar]
- 6.Almeida KH, Sobol RW. A unified view of base excision repair: lesion-dependent protein complexes regulated by post-translational modification. DNA repair. 2007;6:695–711. doi: 10.1016/j.dnarep.2007.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hegde ML, Hazra TK, Mitra S. Early steps in the DNA base excision/single-strand interruption repair pathway in mammalian cells. Cell research. 2008;18:27–47. doi: 10.1038/cr.2008.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.David SS, O'Shea VL, Kundu S. Base-excision repair of oxidative DNA damage. Nature. 2007;447:941–950. doi: 10.1038/nature05978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wallace SS, Bandaru V, Kathe SD, Bond JP. The enigma of endonuclease VIII. DNA repair. 2003;2:441–453. doi: 10.1016/s1568-7864(02)00182-9. [DOI] [PubMed] [Google Scholar]
- 10.Wilson DM, 3rd, Sofinowski TM, McNeill DR. Repair mechanisms for oxidative DNA damage. Front Biosci. 2003;8:d963–d981. doi: 10.2741/1109. [DOI] [PubMed] [Google Scholar]
- 11.Fromme JC, Banerjee A, Verdine GL. DNA glycosylase recognition and catalysis. Current opinion in structural biology. 2004;14:43–49. doi: 10.1016/j.sbi.2004.01.003. [DOI] [PubMed] [Google Scholar]
- 12.Hitomi K, Iwai S, Tainer JA. The intricate structural chemistry of base excision repair machinery: implications for DNA damage recognition, removal, and repair. DNA repair. 2007;6:410–428. doi: 10.1016/j.dnarep.2006.10.004. [DOI] [PubMed] [Google Scholar]
- 13.Imamura K, Wallace SS, Doublie S. Structural characterization of a viral NEIL1 ortholog unliganded and bound to abasic site-containing DNA. The Journal of biological chemistry. 2009 doi: 10.1074/jbc.M109.021907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harrison L, Hatahet Z, Wallace SS. In vitro repair of synthetic ionizing radiation-induced multiply damaged DNA sites. Journal of molecular biology. 1999;290:667–684. doi: 10.1006/jmbi.1999.2892. [DOI] [PubMed] [Google Scholar]
- 15.Wiederhold L, Leppard JB, Kedar P, Karimi-Busheri F, Rasouli-Nia A, Weinfeld M, Tomkinson AE, Izumi T, Prasad R, Wilson SH, Mitra S, Hazra TK. AP endonuclease-independent DNA base excision repair in human cells. Molecular cell. 2004;15:209–220. doi: 10.1016/j.molcel.2004.06.003. [DOI] [PubMed] [Google Scholar]
- 16.Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 17.Menoni H, Gasparutto D, Hamiche A, Cadet J, Dimitrov S, Bouvet P, Angelov D. ATP-dependent chromatin remodeling is required for base excision repair in conventional but not in variant H2A.Bbd nucleosomes. Molecular and cellular biology. 2007;27:5949–5956. doi: 10.1128/MCB.00376-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nilsen H, Lindahl T, Verreault A. DNA base excision repair of uracil residues in reconstituted nucleosome core particles. The EMBO journal. 2002;21:5943–5952. doi: 10.1093/emboj/cdf581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Beard BC, Wilson SH, Smerdon MJ. Suppressed catalytic activity of base excision repair enzymes on rotationally positioned uracil in nucleosomes. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:7465–7470. doi: 10.1073/pnas.1330328100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dou H, Mitra S, Hazra TK. Repair of oxidized bases in DNA bubble structures by human DNA glycosylases NEIL1 and NEIL2. The Journal of biological chemistry. 2003;278:49679–49684. doi: 10.1074/jbc.M308658200. [DOI] [PubMed] [Google Scholar]
- 21.Clark JM, Beardsley GP. Thymine glycol lesions terminate chain elongation by DNA polymerase I in vitro. Nucleic acids research. 1986;14:737–749. doi: 10.1093/nar/14.2.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hayes RC, LeClerc JE. Sequence dependence for bypass of thymine glycols in DNA by DNA polymerase I. Nucleic acids research. 1986;14:1045–1061. doi: 10.1093/nar/14.2.1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ide H, Kow YW, Wallace SS. Thymine glycols and urea residues in M13 DNA constitute replicative blocks in vitro. Nucleic acids research. 1985;13:8035–8052. doi: 10.1093/nar/13.22.8035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McNulty JM, Jerkovic B, Bolton PH, Basu AK. Replication inhibition and miscoding properties of DNA templates containing a site-specific cis-thymine glycol or urea residue. Chemical research in toxicology. 1998;11:666–673. doi: 10.1021/tx970225w. [DOI] [PubMed] [Google Scholar]
- 25.Hegde ML, Theriot CA, Das A, Hegde PM, Guo Z, Gary RK, Hazra TK, Shen B, Mitra S. Physical and functional interaction between human oxidized base-specific DNA glycosylase NEIL1 and flap endonuclease 1. The Journal of biological chemistry. 2008;283:27028–27037. doi: 10.1074/jbc.M802712200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dou H, Theriot CA, Das A, Hegde ML, Matsumoto Y, Boldogh I, Hazra TK, Bhakat KK, Mitra S. Interaction of the human DNA glycosylase NEIL1 with proliferating cell nuclear antigen. The potential for replication-associated repair of oxidized bases in mammalian genomes. The Journal of biological chemistry. 2008;283:3130–3140. doi: 10.1074/jbc.M709186200. [DOI] [PubMed] [Google Scholar]
- 27.Hazra TK, Izumi T, Boldogh I, Imhoff B, Kow YW, Jaruga P, Dizdaroglu M, Mitra S. Identification and characterization of a human DNA glycosylase for repair of modified bases in oxidatively damaged DNA. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:3523–3528. doi: 10.1073/pnas.062053799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hazra TK, Das A, Das S, Choudhury S, Kow YW, Roy R. Oxidative DNA damage repair in mammalian cells: a new perspective. DNA repair. 2007;6:470–480. doi: 10.1016/j.dnarep.2006.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lowry OH, Rosebrough NJ, Farr AL, Randall RJ. Protein measurement with the Folin phenol reagent. The Journal of biological chemistry. 1951;193:265–275. [PubMed] [Google Scholar]
- 30.Kornyushyna O, Berges AM, Muller JG, Burrows CJ. In vitro nucleotide misinsertion opposite the oxidized guanosine lesions spiroiminodihydantoin and guanidinohydantoin and DNA synthesis past the lesions using Escherichia coli DNA polymerase I (Klenow fragment) Biochemistry. 2002;41:15304–15314. doi: 10.1021/bi0264925. [DOI] [PubMed] [Google Scholar]
- 31.Studier FW. Protein production by auto-induction in high density shaking cultures. Protein expression and purification. 2005;41:207–234. doi: 10.1016/j.pep.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 32.Bandaru V, Cooper W, Wallace SS, Doublie S. Overproduction, crystallization and preliminary crystallographic analysis of a novel human DNA-repair enzyme that recognizes oxidative DNA damage. Acta crystallographica. 2004;60:1142–1144. doi: 10.1107/S0907444904007929. [DOI] [PubMed] [Google Scholar]
- 33.Doublie S, Bandaru V, Bond JP, Wallace SS. The crystal structure of human endonuclease VIII-like 1 (NEIL1) reveals a zincless finger motif required for glycosylase activity. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:10284–10289. doi: 10.1073/pnas.0402051101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Blaisdell JO, Wallace SS. Rapid determination of the active fraction of DNA repair glycosylases: a novel fluorescence assay for trapped intermediates. Nucleic acids research. 2007;35:1601–1611. doi: 10.1093/nar/gkm021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.von Hippel PH, Revzin A, Gross CA, Wang AC. Non-specific DNA binding of genome regulating proteins as a biological control mechanism: I. The lac operon: equilibrium aspects. Proceedings of the National Academy of Sciences of the United States of America. 1974;71:4808–4812. doi: 10.1073/pnas.71.12.4808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ptashne M. A genetic switch : phage lambda revisited. 3rd ed. Cold Spring Harbor, N.Y.: Cold Spring Harbor Laboratory Press; 2004. [Google Scholar]
- 37.Porello SL, Leyes AE, David SS. Single-turnover and pre-steady-state kinetics of the reaction of the adenine glycosylase MutY with mismatch-containing DNA substrates. Biochemistry. 1998;37:14756–14764. doi: 10.1021/bi981594+. [DOI] [PubMed] [Google Scholar]
- 38.Liu X, Choudhury S, Roy R. In vitro and in vivo dimerization of human endonuclease III stimulates its activity. The Journal of biological chemistry. 2003;278:50061–50069. doi: 10.1074/jbc.M309997200. [DOI] [PubMed] [Google Scholar]
- 39.Dickson MA, Hahn WC, Ino Y, Ronfard V, Wu JY, Weinberg RA, Louis DN, Li FP, Rheinwald JG. Human keratinocytes that express hTERT and also bypass a p16(INK4a)-enforced mechanism that limits life span become immortal yet retain normal growth and differentiation characteristics. Molecular and cellular biology. 2000;20:1436–1447. doi: 10.1128/mcb.20.4.1436-1447.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ikeda S, Kohmoto T, Tabata R, Seki Y. Differential intracellular localization of the human and mouse endonuclease III homologs and analysis of the sorting signals. DNA repair. 2002;1:847–854. doi: 10.1016/s1568-7864(02)00145-3. [DOI] [PubMed] [Google Scholar]
- 41.Shinmura K, Tao H, Goto M, Igarashi H, Taniguchi T, Maekawa M, Takezaki T, Sugimura H. Inactivating mutations of the human base excision repair gene NEIL1 in gastric cancer. Carcinogenesis. 2004;25:2311–2317. doi: 10.1093/carcin/bgh267. [DOI] [PubMed] [Google Scholar]
- 42.Vartanian V, Lowell B, Minko IG, Wood TG, Ceci JD, George S, Ballinger SW, Corless CL, McCullough AK, Lloyd RS. The metabolic syndrome resulting from a knockout of the NEIL1 DNA glycosylase. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:1864–1869. doi: 10.1073/pnas.0507444103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Krishnamurthy N, Zhao X, Burrows CJ, David SS. Superior removal of hydantoin lesions relative to other oxidized bases by the human DNA glycosylase hNEIL1. Biochemistry. 2008;47:7137–7146. doi: 10.1021/bi800160s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hollis T, Ichikawa Y, Ellenberger T. DNA bending and a flip-out mechanism for base excision by the helix-hairpin-helix DNA glycosylase, Escherichia coli AlkA. The EMBO journal. 2000;19:758–766. doi: 10.1093/emboj/19.4.758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zharkov DO, Golan G, Gilboa R, Fernandes AS, Gerchman SE, Kycia JH, Rieger RA, Grollman AP, Shoham G. Structural analysis of an Escherichia coli endonuclease VIII covalent reaction intermediate. The EMBO journal. 2002;21:789–800. doi: 10.1093/emboj/21.4.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Riggs AD, Bourgeois S, Cohn M. The lac repressor-operator interaction. 3. Kinetic studies. Journal of molecular biology. 1970;53:401–417. doi: 10.1016/0022-2836(70)90074-4. [DOI] [PubMed] [Google Scholar]
- 47.Boal AK, Genereux JC, Sontz PA, Gralnick JA, Newman DK, Barton JK. Redox signaling between DNA repair proteins for efficient lesion detection. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:15237–15242. doi: 10.1073/pnas.0908059106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yavin E, Stemp ED, O'Shea V L, David SS, Barton JK. Electron trap for DNA-bound repair enzymes: a strategy for DNA-mediated signaling. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:3610–3614. doi: 10.1073/pnas.0600239103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gorodetsky AA, Boal AK, Barton JK. Direct electrochemistry of endonuclease III in the presence and absence of DNA. Journal of the American Chemical Society. 2006;128:12082–12083. doi: 10.1021/ja064784d. [DOI] [PubMed] [Google Scholar]
- 50.Hu J, de Souza-Pinto NC, Haraguchi K, Hogue BA, Jaruga P, Greenberg MM, Dizdaroglu M, Bohr VA. Repair of formamidopyrimidines in DNA involves different glycosylases: role of the OGG1, NTH1, and NEIL1 enzymes. The Journal of biological chemistry. 2005;280:40544–40551. doi: 10.1074/jbc.M508772200. [DOI] [PubMed] [Google Scholar]
- 51.Takao M, Kanno S, Kobayashi K, Zhang QM, Yonei S, van der Horst GT, Yasui A. A back-up glycosylase in Nth1 knock-out mice is a functional Nei (endonuclease VIII) homologue. The Journal of biological chemistry. 2002;277:42205–42213. doi: 10.1074/jbc.M206884200. [DOI] [PubMed] [Google Scholar]
- 52.Davey CA, Sargent DF, Luger K, Maeder AW, Richmond TJ. Solvent mediated interactions in the structure of the nucleosome core particle at 1.9 a resolution. Journal of molecular biology. 2002;319:1097–1113. doi: 10.1016/S0022-2836(02)00386-8. [DOI] [PubMed] [Google Scholar]