Abstract
Monocots are known to respond differently to auxinic herbicides; hence, certain herbicides kill broadleaf (i.e., dicot) weeds while leaving lawns (i.e., monocot grasses) intact. In addition, the characters that distinguish monocots from dicots involve structures whose development is controlled by auxin. However, the molecular mechanisms controlling auxin biosynthesis, homeostasis, transport, and signal transduction appear, so far, to be conserved between monocots and dicots, although there are differences in gene copy number and expression leading to diversification in function. This article provides an update on the conservation and diversification of the roles of genes controlling auxin biosynthesis, transport, and signal transduction in root, shoot, and reproductive development in rice and maize.
Monocots and dicots share similar mechanisms for auxin biosynthesis, transport, and signaling, despite displaying big differences in the structures controlled by this phytohormone.
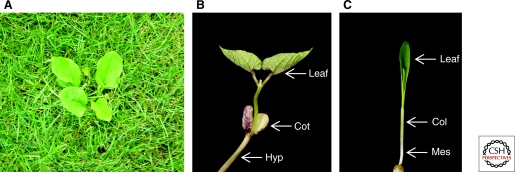
Auxinic herbicides have been used for decades to control dicot weeds in domestic lawns (Fig. 1A), commercial golf courses, and acres of corn, wheat, and barley, yet it is not understand how auxinic herbicides selectively kill dicots and spare monocots (Grossmann 2000; Kelley and Reichers 2007). Monocots, in particular grasses, must perceive or respond differently to exogenous synthetic auxin than dicots. It has been proposed that this selectivity is because of either limited translocation or rapid degradation of exogenous auxin (Gauvrit and Gaillardon 1991; Monaco et al. 2002), altered vascular anatomy (Monaco et al. 2002), or altered perception of auxin in monocots (Kelley and Reichers 2007). To explain these differences, there is a need to further understand the molecular basis of auxin metabolism, transport, and signaling in monocots.
Figure 1.
Differences between monocots and dicots. (A) A dicot weed in a lawn of grasses. Note the difference in morphology of the leaves. (B) Germinating dicot (bean) seedling. Dicots have two cotyledons (cot). Reticulate venation is apparent in the leaves. The stem below the cotyledons is called the hypocotyl (hyp). (C) Germinating monocot (maize) seedling. Monocots have a single cotyledon called the coleoptile (col) in grasses. Parallel venation is apparent in the leaves. The stem below the coleoptile is called the mesocotyl (mes).
Auxin, as we have seen in previous articles, plays a major role in vegetative, reproductive, and root development in the model dicot, Arabidopsis. However, monocots have a very different anatomy from dicots (Raven et al. 2005). Many of the characters that distinguish monocots and dicots involve structures whose development is controlled by auxin: (1) As the name implies, monocots have single cotyledons, whereas dicots have two cotyledons (Fig. 1B,C). Auxin transport during embryogenesis may play a role in this difference as cotyledon number defects are often seen in auxin transport mutants (reviewed in Chandler 2008). (2) The vasculature in leaves of dicots is reticulate, whereas the vasculature in monocots is parallel (Fig. 1). Auxin functions in vascular development because many mutants defective in auxin transport, biosynthesis, or signaling have vasculature defects (Scarpella and Meijer 2004). (3) Dicots often produce a primary tap root that produces lateral roots, whereas, in monocots, especially grasses, shoot-borne adventitious roots are the most prominent component of the root system leading to the characteristic fibrous root system (Fig. 2). Auxin induces lateral-root formation in dicots and adventitious root formation in grasses (Hochholdinger and Zimmermann 2008).
Figure 2.
The root system in monocots. (A) Maize seedling showing the primary root (1yR), which has many lateral roots (LR). The seminal roots (SR) are a type of adventitious root produced during embryonic development. Crown roots (CR) are produced from stem tissue. (B) The base of a maize plant showing prop roots (PR), which are adventitious roots produced from basal nodes of the stem later in development.
It is not yet clear if auxin controls the differences in morphology seen in dicots versus monocots. However, both conservation and diversification of mechanisms of auxin biosynthesis, homeostasis, transport, and signal transduction have been discovered so far. This article highlights the similarities and the differences in the role of auxin in monocots compared with dicots. First, the genes in each of the pathways are introduced (Part I, Table I) and then the function of these genes in development is discussed with examples from the monocot grasses, maize, and rice (Part II).
PART I—GENES CONTROLLING AUXIN BIOSYNTHESIS, TRANSPORT, AND SIGNAL TRANSDUCTION IN GRASSES
Auxin Biosynthesis
Maize has long been a model system for studies on the biochemistry of auxin biosynthesis (Kriechbaumer et al. 2006). Genes encoding enzymes required for the synthesis of auxin have been identified in maize, rice, and Arabidopsis (Table 1). Multiple pathways have been proposed, but the enzymes, genes that encode them, and intermediates are not fully known (Bartel 1997; Ljung et al. 2002; Cohen et al. 2003; Woodward and Bartel 2005). Recent advances have identified mutants and corresponding genes, which has led to revised pathways that still need to be defined further (Zhao et al. 2001; Zhao et al. 2002; Stepanova et al. 2008; Tao et al. 2008; Sugawara et al. 2009). Four tryptophan-dependent and one tryptophan-independent auxin biosynthetic pathways have been proposed. The revised tryptophan-dependent pathways proposed by Sugawara et al. (2009) will be used as a framework to introduce what is known of the genes controlling the pathway in the monocots, maize, and rice.
Table 1.
Arabidopsis genes required for auxin biosynthesis, transport, and signal transduction, and their homologs that have been functionally characterized in maize and rice
| Protein | Arabidopsis | Maize | Rice | Maize/Rice References |
|---|---|---|---|---|
| Biosynthesis | ||||
| Tryptophan synthase β | TRP2 | ORP1,2 | Wright et al. 1991 | |
| Amidase | AMI1 | |||
| Trytophan aminotransferase | TAA1 TAR1, 2 | VT2 | K. Phillips and P. McSteen, unpublished | |
| Aldehyde oxidase | AAO1 | ZmAO1 | OsAO1 | Sekimoto et al. 1997 |
| Flavin monoxygenase | YUC1-11 | SPI1 | OsYUC1 COW1/NAL7 | Fujino et al. 2007; Woo et al. 2007; Yamamoto et al. 2007; Gallavotti et al. 2008a |
| Cytochrome P450 | CYP79B2/3 | |||
| Nitrilase | NIT1-4 | ZmNIT1,2 | OsNIT | Park et al. 2003; Kriechbaumer et al. 2007 |
| Transport | ||||
| Auxin influx transporter | AUX1 | ZmAUX1 | Hochholdinger et al. 2000; Brooks et al. 2009 | |
| Auxin efflux carrier | PIN1 |
ZmPIN1a ZmPIN1b ZmPIN1c |
OsPIN1 | Xu et al. 2005; Carraro et al. 2006; Gallavotti et al. 2008b |
| Serine threonine kinase | PID | BIF2 | OsPID/OsBIF2 | McSteen et al. 2007; Morita and Kyozuka 2007 |
| ABC transporter | ABCB1,19 | BR2 | Multani et al. 2003 | |
| RCN1 | Yasuno et al. 2009 | |||
| ARF-GAP | VAN3/SFC | ZmSFC | OsAGAP | Zhuang et al. 2006; Zhang et al. 2007 |
| Signal transduction | ||||
| Aux/IAA transcription factor | IAA1-25 | RUM1 |
IAA1 IAA1/3 IAA3/31 |
Thakur et al. 2001; Nakamura et al. 2006; Song et al. 2009b |
| Auxin response factor | ARF1-23 |
ZmARF1 ZmARF2 ZmMP |
OsARF1 | Waller et al. 2002; Attia et al. 2009; Brooks et al. 2009 |
| F box | TIR1, AFB | ZmTIR1 | Zhang et al. 2007 | |
| Other | ||||
| Auxin binding protein | AtABP1 | ZmABP1,4 | Im et al. 2000 | |
| SAUR | SAUR | ZmSAUR2 | OsSAUR | Knauss et al. 2003; Jain et al. 2006c |
| LOB transcription factor | LBD16/29 | RTCS | ARL1/CRL1 | Inukai et al. 2005; Liu et al. 2005; Taramino et al. 2007 |
| BHLH transcription factor | BA1 | LAX1 | Komatsu et al. 2003; Gallavotti et al. 2004 |
Tryptophan-dependent Auxin Biosynthesis: (1) IAM Pathway
Genes controlling the conversion of tryptophan (TRP) to indole-3-acetamide (IAM) have not been detected in plants, although in bacteria the iaaM gene catalyzes this reaction (Comai and Kosuge 1982). The conversion of IAM to indole-3-acetic acid (IAA) is proposed to occur through the action of the amidase gene, AMI1, in Arabidopsis (Pollmann et al. 2003). Amidase genes have not yet been reported in monocots, though an IAM hydrolase activity has been detected from rice callus, so it is likely that this pathway is used in grasses (Arai et al. 2004).
(2). IPA Pathway
There is evidence that the indole-3-pyruvic acid (IPA) pathway is important in both monocot and dicot development. The conversion of TRP to IPA occurs through the action of the tryptophan aminotransferase genes TAA1, TAR1, and TAR2, which play a role in embryogenesis, root, vascular, and inflorescence development in Arabidopsis (Stepanova et al. 2008; Tao et al. 2008). TAA1-like genes from monocots have not yet been reported but the cloning of the vanishing tassel2 (vt2) mutant in maize provides evidence that this enzyme also functions in vegetative and inflorescence development in monocots (K. Phillips and P. McSteen, unpubl.). It has been proposed that IPA is converted to indole-3-acetaldehyde (IAAld) by an IPA decarboxylase activity although the gene encoding this enzyme has not been definitely identified in dicots or monocots (Woodward and Bartel 2005). The conversion of IAAld to IAA has been proposed to be catalyzed by an aldehyde oxidase encoded by the AAO1 gene of Arabidopsis (Sekimoto et al. 1998). A rice homolog, OsAO1 (Yamamoto et al. 2007) and maize homologs, ZmAO-1 and ZmAO-2 (Sekimoto et al. 1997), have been reported but not functionally characterized. However, from the phenotype of the taa1 and vt2 mutants, it is apparent that this pathway significantly contributes to vegetative and reproductive development in both dicots and monocots.
(3). TAM Pathway
There is good evidence that the tryptamine (TAM) pathway plays a significant role in development of both monocots and dicots. It is not known which genes convert TRP to TAM although it is thought to occur by a tryptamine decarboxylase activity (Woodward and Bartel 2005). The YUCCA (YUC) genes catalyze the rate-limiting step in the pathway by the conversion of TAM to N-hydroxyl tryptamine (HTAM) (Zhao et al. 2001). It is not known how HTAM is converted to IAA, though a recent report provides evidence that this does not occur through an indole-3-acetaldoximine (IAOx) intermediate (Sugawara et al. 2009).
There are a total of 10 YUC-like genes in Arabidopsis (Zhao et al. 2001). Each of the genes is expressed in a specific pattern and is proposed to function redundantly to produce auxin in a localized manner during embryogenesis, leaf, vascular, and inflorescence development (Zhao et al. 2001; Cheng et al. 2006; Cheng et al. 2007a) There are 13 YUC-like genes in rice, although functional information has been reported for only two of them. Seven OsYUC genes with similarity to AtYUC1-8 were reported by (Yamamoto et al. 2007) and another gene, OsYUC8/COW1/NAL7, was subsequently reported (Fujino et al. 2007; Woo et al. 2007). Five additional genes that are more closely related to AtYUC10 and 11 were included in a phylogenetic analysis reported in Gallavotti et al. 2008a. There is proposed to have been expansion, subfunctionalization, and diversification of this gene family in monocots and dicots (Gallavotti et al. 2008a).
The rice gene most closely related to AtYUC1 and 4 is OsYUC1 (Yamamoto et al. 2007). Overexpression and antisense inhibition of OsYUC1 causes opposite defects in root development (described in Part II) (Yamamoto et al. 2007). The ortholog of OsYUC1 based on sequence, phylogenetic analysis, and synteny is the SPARSE INFLORESCENCE1 (SPI1) gene of maize, yet loss of function of this gene causes defects in vegetative and reproductive development (Gallavotti et al. 2008a), indicating that there has been diversification of gene function even within the grasses. OsYUC1 and SPI1 belong to a monocot-specific rather than grass-specific clade as evidenced by the isolation of a SPI1 homolog from the monocot Joinvillea (Gallavotti et al. 2008a). A very similar phenotype to spi1 is seen in Arabidopsis plants with loss of function of four YUC genes, indicating that the YUC family is more redundant in Arabidopsis than in maize (Cheng et al. 2006). Although the basic biochemical function of the YUC genes may be conserved, the biological function of these genes appears to have diversified due to changes in copy number and expression pattern (Gallavotti et al. 2008a).
A gene more distantly related to AtYUC1 and 4 has been functionally characterized in rice. OsYUC8/COW1/NAL7 is sister to the Arabidopsis genes in clade AtYUC1-9 and clade AtYUC10-11 (Gallavotti et al. 2008a). constitutively wilted1 (cow1) mutants have defects in root mass, leading to leaf rolling and the wilty phenotype after which the mutant is named (Woo et al. 2007). The same gene was identified as a natural variant, narrow leaf7 (nal7), with narrow leaves (Fujino et al. 2007). More research is needed to uncover the functions of the other monocot YUC-like genes and to determine if they have additional functions masked by redundancy as seen for the Arabidopsis yuc mutants.
(4). IAOX Pathway
The indole-3-acetaldoximine (IAOx) pathway has been proposed to occur only in cruciferous species such as Arabidopsis, in which glucosinolate secondary metabolism occurs (Sugawara et al. 2009). However, there is conflicting evidence suggesting that this pathway may also occur in monocots (Kriechbaumer et al. 2006; Sugawara et al. 2009). TRP is converted to IAOx by genes encoding the cytochrome P450 enzymes CYP79B2 and CYP79B3 in Arabidopsis (Zhao et al. 2002). However, no orthologs of CYP79B2 and CYP79B3 have been found in noncruciferous species, including maize and rice, raising doubts as to whether this pathway occurs in other species. In support of this, Sugawara et al. (2009) were unable to detect IAOx in seedlings of maize, rice, or tobacco. In the next step of the proposed pathway, enzymes catalyzing the conversion of IAOx to indole-3-acetonitrile (IAN) are not known, but conversion of IAN to IAA occurs through the action of nitrilase genes in Arabidopsis (Pollmann et al. 2006). Nitrilase genes have also been well characterized in maize. For example, ZmNIT2 hydrolyses IAN to IAA in maize kernels and seedlings (Park et al. 2003; Kriechbaumer et al. 2007). Genes similar to ZmNIT2 have been reported but not characterized in rice (Yamamoto et al. 2007).
These results lead to the question of how IAN is produced in maize if there are no CYP79B2/3 genes and no detectable IAOx. On the other hand, enzyme activity that converts TRP to IAOx and converts IAOx to IAN and IAAld has been detected in maize tissues (reviewed in Kriechbaumer et al. 2006). Therefore, IAOx must be made by a different pathway in maize, although genes catalyzing this process have yet to be identified. Hence, although CYP79B2/3 genes are not present in monocots, a pathway related to the IAOx pathway may occur.
Tryptophan-independent Auxin Biosynthesis
Studies of the orange pericarp mutant, which fails to make tryptophan (because of a deficiency in tryptophan synthase β, which converts indole to tryptophan) but still makes auxin, provided the first evidence that tryptophan-independent biosynthesis occurs in maize (Wright et al. 1991). Studies of analogous mutants provided evidence that tryptophan-independent auxin biosynthesis also occurs in Arabidopsis (Normanly et al. 1993). When auxin is not made from tryptophan it is thought to be made via indole-3-glycerol phosphate or indole, although genes in the pathway have not been identified from monocots or dicots (Woodward and Bartel 2005).
In the future, it will be of interest not just to identity the genes in the entire biosynthetic pathway but also to understand why there are multiple pathways for auxin biosynthesis. Are different pathways used in different cells at different times in response to different conditions? Are different pathways used more in some species that others?
Auxin Homeostasis
Much of the auxin found in the plant is stored as conjugates to amino acids or sugars (Woodward and Bartel 2005). Auxin can also be produced by β oxidation of indole-3-butyric acid (IBA). These processes are thought to occur in monocots and some genes controlling the process have been identified. For example, ZmIAGLU, which conjugates IAA to glucose, has been characterized in maize (Ludwig-Muller et al. 2005). OsGH3-like genes, which conjugate IAA to amino acids, have been discovered in rice (Jain et al. 2006b). Furthermore, IAA has been found to be converted to IBA in maize (Ludwig-Muller 2000). Thus, it appears that there are conserved mechanisms of auxin homeostasis in monocots. However, more research needs to be done on the molecular basis for homeostasis. As the synthetic auxin herbicide 2,4,D is rapidly degraded in maize, auxin homeostasis has been proposed to be one of the mechanisms of selectivity of auxinic herbicides (Gauvrit and Gaillardon 1991; Monaco et al. 2002).
Auxin Transport
Evidence of the role of auxin transport in monocot development is illustrated by experiments in which monocots (in particular grasses) are treated with polar auxin transport inhibitors. Treatment with N-1-naphthylphthalamic acid (NPA) causes defects in embryogenesis (Fischer and Neuhaus 1996), leaf initiation and growth (Tsiantis et al. 1999; Scanlon 2003), root development (Morita and Kyozuka 2007), and reproductive development (Wu and McSteen 2007). Many of these effects are similar to the effects when dicots are treated with auxin transport inhibitors except for differences in morphology and in sensitivity to certain types of auxin transport inhibitors (Wu and McSteen 2007). The role of auxin transport in development is further discussed in Part II. In this section, genes controlling auxin transport in maize and rice are described.
AUX1
A homolog of the auxin influx carrier AUX1 has been identified in maize, although no functional evidence has been reported (Hochholdinger et al. 2000). ZmAUX1 is expressed in roots at the tip of all types of root and in the endodermis, pericycle, and epidermis, indicating it may play a role similar to Arabidopsis in roots. A recent analysis using laser capture microscopy and microarray analysis showed that ZmAUX1 is up-regulated at the site of leaf primordia initiation and down-regulated on treatment with NPA, indicating that ZmAUX1 may also function in shoots (Brooks et al. 2009).
PIN1
Three homologs of the PIN1 auxin efflux carrier have been reported in maize and rice. ZmPIN1a is expressed in a manner similar to AtPIN1 as it is up-regulated at the site of initiation of all primordia during shoot and reproductive development (Carraro et al. 2006; Gallavotti et al. 2008b; Brooks et al. 2009; Lee et al. 2009) and is induced by auxin (Lee et al. 2009). ZmPIN1a can rescue the Arabidopsis pin1 mutant, indicating that it may play a similar role in maize as in Arabidopsis (Gallavotti et al. 2008b). Knockdown of a homolog in rice, OsPIN1, does not produce the characteristic pin phenotype seen in Arabidopsis, perhaps because of redundancy with the three rice PIN1-like genes (Xu et al. 2005). However, knockdown of OsPIN1 does exhibit a phenotype somewhat similar to treatment of rice plants with NPA as there is a decrease in the number of crown roots. Furthermore, there is an increase in the number of tillers and an increase in tiller angle, phenotypes that differ from pin1 mutants. Therefore, there may be some differences in function that need to be further analyzed.
PID
Maize and rice orthologs of PID have been reported. Analysis of function shows that there are similarities and differences. OsPID is auxin induced and is expressed in lateral organs and axillary meristems (McSteen et al. 2007; Morita and Kyozuka 2007). No loss-of-function OsPID phenotype has been reported, but overexpression gives a root phenotype similar to treatment of plants with NPA (Morita and Kyozuka 2007).
One of the orthologs of PID in maize is BARREN INFLORESCENCE2 (BIF2) (McSteen et al. 2007). bif2 mutants have phenotypes very similar to pid mutants, indicating that function is conserved. Furthermore, BIF2 phosphorylates ZmPIN1a in vitro and affects ZmPIN1a localization in vivo similar to the function of PID (Skirpan et al. 2009). On the other hand, there may also be differences in function, as BIF2 has been reported to be nuclear localized and to phosphorylate a nuclear localized bHLH transcription factor, BARRENSTALK1 (BA1) (Skirpan et al. 2008). In the future, it would be interesting to determine the subcellular localization of OsPID and to determine whether other nuclear-localized PID-like kinases have taken over the role of BIF2 in Arabidopsis.
ABC Transporters
The adenosine triphosphate (ATP)–binding cassette (ABC) transporters, ABCB19/PGP19/MDR1, ABCB1/PGP1, and ABCB4/PGP4, play important roles in auxin transport in Arabidopsis (Bandyopadhyay et al. 2007). An ABC transporter also functions in auxin transport in maize and sorghum (Multani et al. 2003). The mutations, brachytic2 (br2) in maize and dwarf3 (d3) in sorghum, are caused by loss of function of an ABC transporter with high homology to ABCB1/PGP1 (Multani et al. 2003). br2 plants have short internodes because of reduced cell elongation and have defects in auxin transport (Multani et al. 2003). In contrast, knockout of ABCB1/PGP1 does not have a significant phenotype in Arabidopsis but double mutants with ABCB19/PGP19/MDR1 are short and have additional defects in vegetative and reproductive development. Further phylogenetic analysis is required to determine if there is less redundancy in the ABCB gene family in maize.
An ABC transporter of a different class, ABCG, has been identified in rice through the cloning of the reduced culm number1 (rcn1) mutant (Yasuno et al. 2009). rcn1 mutants, as the name implies, have fewer tillers (Takamure and Kinoshita 1985). The defect is caused by a defect in tiller outgrowth rather than tiller initiation. rcn1 is induced by auxin, indicating that it may play a role in auxin-mediated development.
ARF GAP
The ADP-ribosylation factor–GTPase-activating protein (ARF-GAP), VAN3/SCARFACE, functions in auxin-mediated vascular development in cotyledons, leaves, and roots of Arabidopsis (Koizumi et al. 2005; Sieburth et al. 2006). A maize VAN3/SFC homolog is reported to be expressed in leaf primordia (Zhang et al. 2007). Overexpression of OsAGAP, a rice ARF-GAP, in Arabidopsis and rice leads to defects in root development (Zhuang et al. 2005; Zhuang et al. 2006). The OsAGAP transgenics have defects in auxin transport, proposed to be caused by defects in auxin influx (Zhuang et al. 2005; Zhuang et al. 2006).
Homologs of other genes involved in auxin transport such as the guanine-nucleotide exchange factors for ADP-ribosylation factor GTPases (ARF-GEF) GNOM, the immunophilin-like TWISTED DWARF1 (TWD1), and the calossin-like BIG, have not yet been functionally characterized in monocots. However, from what has been discovered so far, there is conservation in the molecular mechanisms controlling auxin transport in monocots. Monocot-specific functions of auxin transport in controlling crown-root initiation and tiller angle is discussed in Part II.
Auxin Signal Transduction
Aux/IAA
Although there are 25 Aux/IAA genes in Arabidopsis, there are 31 members in the Aux/IAA gene family in rice (Jain et al. 2006a), 24 of which are regulated by auxin (Song et al. 2009a) and three of which have been functionally characterized. The first, Aux/IAA reported from rice was called OsIAA1 but is now renamed OsIAA3. OsIAA1/3 transcript is induced by auxin and suppressed by light (Thakur et al. 2001) and the protein has a short half life and is degraded by the proteosome (Thakur et al. 2005) similar to the stability of AUX/IAA proteins in Arabidopsis.
A very important finding in this area is the seminal paper by Nakamura et al. (2006), reporting the production of auxin-insensitive rice by expressing a degradation-resistant version of OsIAA3, now called OsIAA31. OsIAA3/31 transgenic plants have diverse phenotypes reminiscent of auxin-insensitive mutants in Arabidopsis, such as defects in shoot and root gravitropism, root length, and lateral-root initiation (Nakamura et al. 2006). This provided the first functional evidence that auxin signaling may occur similarly in rice as in Arabidopsis.
Much work remains to be done to characterize the specific functions of other Aux/IAA genes. Overexpression of OsIAA1 causes defects in root development and an additional defect of enlarged leaf angle (Song et al. 2009b). A transposon insertion into OsIAA25 is reported to be “dwarf with reduced fertility” but the phenotype has not been characterized in detail (Jain et al. 2006a). In maize, there has been very little characterization of auxin signal transduction. The rum1 gene of maize, which is a mutant with very specific defects in root development (discussed in Part II), has recently been reported in a patent application to be an Aux/IAA gene (Taramino et al. 2008). Additional auxin-induced genes such as homologs of the SAUR gene family have been identified in rice and maize (Knauss et al. 2003; Jain et al. 2006c). Therefore, it is likely that there is conservation in signal transduction of auxin in both maize and rice.
ARF
A gene family of AUXIN RESPONSE FACTOR (ARF) transcription factors has been described in rice with 25 OsARF genes compared with 23 ARFs in Arabidopsis (Sato et al. 2001; Wang et al. 2007). OsARF genes have diverse expression patterns but significant functional analysis has only been reported for OsARF1 (Waller et al. 2002; Attia et al. 2009), which is closely related to AtARF1 and AtARF2 in Arabidopsis.
OsARF1 is auxin induced and is expressed in coleoptiles, callus, and young panicles with weaker expression in leaves and roots (Waller et al. 2002; Attia et al. 2009). Antisense knockdown of OsARF1 in rice has defects in vegetative development, producing dwarf plants with small curled leaves and defects in reproductive development, as the plants either fail to flower or flower late and are sterile. Therefore, there are similarities and differences with arf1;arf2 double mutants in Arabidopsis, which have delayed flowering, sterility, and delayed senescence (Ellis et al. 2005). Transposon insertions in OsARF5, 11, and 12 are reported to have low fertility, whereas insertions in OsARF11, 19, and 24 are reported to be dwarf but the phenotypes have not been characterized in detail (Wang et al. 2007). Maize homologs of AtARF1, 2, and 5 are reported to be expressed at a higher level in leaf primordia than in the meristem (Brooks et al. 2009).
TIR1
The F box gene family, including homologs of the TRANSPORT INHIBITOR RESPONSE1 (TIR1) auxin receptor gene family have been catalogued in rice (Jain et al. 2007). A maize homolog ZmTIR1 is expressed in young leaf primordia (Zhang et al. 2007). However, no TIR1-like genes have been functionally characterized in maize and rice. It has been proposed that differences in selectivity to herbicides are caused by differences in the functions of TIR1 auxin receptors because different TIR1-like proteins have different affinities for different auxins in Arabidopsis (Walsh et al. 2006; Kelley and Reichers 2007).
ABP1
Another auxin-binding protein, AUXIN BINDING PROTEIN1, ABP1, was initially discovered in maize and subsequently in Arabidopsis (reviewed in Timpte 2001; Christian et al. 2006). In maize, knockouts of ABP1 and 4 did not have a phenotype, whereas Arabidopsis knockouts of ABP1 were embryo lethal, indicating that there may be greater redundancy in the gene family in maize (Im et al. 2000; Chen et al. 2001). Alternately, there could be differences in the role of ABP1 in maize, which has also been proposed as a possible mechanism for herbicide selectivity (Kelley and Reichers 2007).
PART II—ROLE OF AUXIN IN MONOCOT DEVELOPMENT
Root Development
The root systems of monocots and dicots differ in architecture, yet recent work suggests that auxin plays an equivalent role in maize and rice as in Arabidopsis (Hochholdinger and Zimmermann 2008). In most dicots, including Arabidopsis, there is a central primary root and the root system branches through the production of lateral roots. In grasses, there is often a fibrous root system such that much of the branching occurs through adventitious roots called crown roots (Raven et al. 2005). In maize and in rice, there is an embryonic and postembryonic root system (Hochholdinger et al. 2004). The embryonic root system consists of a short-lived primary root called a primary root in maize and a seminal root in rice, and embryonic adventitious roots called seminal roots in maize and crown roots in rice (Fig. 2A). The postembryonic root system consists of shoot-borne adventitious roots and lateral roots, which arise on all root types. Maize has three types of adventitious roots—seminal roots, which are embryonic adventitious roots, crown roots, which arise from stem tissue underground, and prop roots, which arise from stem tissue above ground (Fig. 2B). Rice also has the same three types of adventitious root but they are all called crown roots. For the sake of simplicity, all types of adventitious roots in maize and rice are referred to as crown roots in this section. From the analysis of mutants defective in auxin biosynthesis, transport, or signaling, auxin is required to inhibit root elongation and promote lateral-root and crown-root initiation in grasses. The role of auxin in each of these processes will be considered separately.
Crown-root Initiation
Crown-root initiation is of tremendous interest as adventitious roots are much more important for root architecture in grasses than in dicots. It appears that crown-root initiation is controlled somewhat similarly to lateral-root initiation even though these two root types develop from shoot and root tissues, respectively.
Evidence that auxin biosynthesis is required for crown-root formation comes from rice plants overexpressing OsYUC1, which have increased crown-root number (Yamamoto et al. 2007). Furthermore, application of exogenous IAA increases crown-root number (Inukai et al. 2005; Xu et al. 2005). Auxin transport is important as inhibition of auxin transport through either treatment with NPA, antisense inhibition of OsPIN1, or overexpression of OsPID reduces the number of crown roots (Inukai et al. 2005; Xu et al. 2005; Morita and Kyozuka 2007). In addition, auxin signal transduction plays a role because overexpression of OsIAA1 or degradation resistant OsIAA3/OsIAA31 abolishes crown-root initiation (Nakamura et al. 2006; Song et al. 2009b). Therefore, auxin biosynthesis, transport, and signaling are required for crown-root initiation.
Mutants that lack crown roots have been used to identify genes functioning in crown-root initiation. adventitious rootless1 (arl1)/crown rootless1 (crl1) mutants in rice lack crown roots, have fewer lateral roots, and have altered root gravitropsism, though they have no defects in the embryonic root system (Inukai et al. 2005; Liu et al. 2005). The ARL1/CRL1 gene encodes an auxin inducible LATERAL ORGAN BOUNDARY (LOB) domain containing transcription factor expressed in lateral- and crown-root primordia as well as floral tissue (Inukai et al. 2005; Liu et al. 2005). In maize, the ortholog of ARL1/CRL1 is ROOTLESS CONCERNING CROWN AND SEMINAL ROOTS (RTCS) (Taramino et al. 2007). rtcs mutants do not produce crown roots or seminal adventitious roots but can still produce an embryonic primary root with lateral roots (Hetz et al. 1996; Hochholdinger et al. 2004).
In rice, CRL1 has been shown to be an in vitro target of an OsARF1-like transcription factor (Inukai et al. 2005). However, OsARF1 is predicted to be a repressor ARF and overexpression does not affect root development, so the functional significance of this is unknown (Inukai et al. 2005). In Arabidopsis, the LOB domain, containing transcription factors LBD16 and LBD29 have been shown to be bona fide downstream targets of ARF7 and ARF19 (Okushima et al. 2005a; Okushima et al. 2007). arf7;arf19 double mutants fail to initiate lateral roots and lbd16 mutants have a mild defect in lateral-root initiation, whereas lbd29 knockouts have not been identified (Okushima et al. 2005b). ARF7 and ARF19 are activating ARFs, so perhaps homologs of these ARFs induce CRL1 in rice.
Recently, another transcription factor was found to play a role in crown-root formation in rice (Zhao et al. 2009). The WOX11 gene is a WUSCHEL-like homeodomain-containing transcription factor that is auxin and cytokinin inducible. Loss-of-function mutations have fewer crown roots, whereas overexpression causes an increase in the number of crown roots. WOX-like genes also play a role in lateral-root initiation in Arabidopsis, indicating that gene function is conserved (Deveaux et al. 2008).
Lateral-root Initiation
Auxin has been shown to play a role in lateral-root initiation in maize and rice as in Arabidopsis. However, mutant analysis has found that the processes of lateral-root and crown-root formation can be genetically separable.
Exogenous application of auxin promotes lateral-root initiation (Xu et al. 2005). Moreover, defects in auxin transport affect lateral-root initiation, for example, overexpression of OsPIN1 increases the number of lateral roots, whereas overexpression of OsAGAP decreases the number of lateral roots (Xu et al. 2005; Zhuang et al. 2006). Auxin signaling is also required because overexpression of OsIAA1 decreases the number of lateral roots (Song et al. 2009b).
Mutants that affect crown-root initiation can also affect lateral-root initiation. For example, crl1/arl1 have defects in both crown-root and lateral-root initiation and the gene is expressed in the primordia of both types (Inukai et al. 2005). On the other hand, the rootless with undetectable meristems 1 (rum1) mutant of maize does not initiate lateral roots or embryonic adventitious roots but crown roots initiate as normal, indicating that these two processes are separable (Woll et al. 2005). Hence, this mutant has been used for analysis of the proteome of different root types (Liu et al. 2006; Saleem et al. 2009). Analysis of the transcriptome indicates no change in the levels of ZmPIN1 or ZmAUX1 transcripts (Woll et al. 2005). However, there is reduced auxin transport in mutant roots. rum1 is reported to be caused by a mutation in an Aux/IAA gene (Taramino et al. 2008). This indicates conservation in auxin signal transduction mechanisms in both monocots and dicots, and indicates conservation in function during root development as several dominant mutants in Arabidopsis Aux/IAA genes produce no lateral roots (Fukaki et al. 2005; Uehara et al. 2008).
Root Length
Defects in auxin biosynthesis, transport, or signaling also cause defects in root length.
Exogenous auxin inhibits root elongation in monocots and dicots (Zhuang et al. 2006). Decreased auxin biosynthesis caused by cow1 mutants or antisense OsYUC1 in rice and ZmNIT2 mutants in maize have short roots (Kriechbaumer et al. 2007; Woo et al. 2007; Yamamoto et al. 2007). Changes in auxin transport, such as overexpression of OsPIN1, increased root length, whereas overexpression of OsAGAP decreased root length (Xu et al. 2005; Zhuang et al. 2006). In addition, the degradation-resistant OsIAA3/31 lines have longer roots on auxin (Nakamura et al. 2006).
In conclusion, despite the differences in root morphology, with Arabidopsis having a tap-root system and maize and rice having a fibrous-root system, auxin inhibits root length and promotes lateral-root formation in Arabidopsis and inhibits root length and promotes lateral-root and crown-root formation in maize and rice.
Vegetative Development
Stem Development
Dwarfism is a common defect in mutants defective in auxin transport in maize and rice as in Arabidopsis. The br2 mutant in maize has reduced auxin transport and short internodes (Multani et al. 2003). Another mutant in maize that has reduced auxin transport and short internodes is the semaphore (sem) mutant (Scanlon et al. 2002). The mutant has not yet been cloned but it has additional defects such as defects in vasculature in leaves and a reduction in the number of lateral roots, which implies it plays a role in an auxin-regulated process. The Developmental disaster (Dvd1) mutant of maize has short internodes without affecting leaf number (Phillips et al. 2009). Dvd1 mutants also have a pin inflorescence phenotype, indicating that Dvd1 may control an auxin-regulated process. In rice, narrow leaf1 (nal1) mutants have reduced auxin transport and short stems (Qi et al. 2008). Hence, auxin is required for stem elongation in monocots as well as in dicots.
Leaf Development
Genes required for auxin biosynthesis and transport also play a role in leaf initiation. spi1, bif2, and Bif1 mutants have a mild reduction in the number of leaves as single mutants (Barazesh and McSteen 2008a; Gallavotti et al. 2008a). However, in double-mutant combinations spi1;bif2 or Bif1;bif2, there is a synergistic effect such that about half the number of leaves as normal are initiated (Barazesh and McSteen 2008a; Gallavotti et al. 2008a). Leaf number defects are also seen in pin1 mutants in Arabidopsis (Bennett et al. 1995). Therefore, auxin biosynthesis and transport play a role in leaf initiation.
Mutants defective in auxin biosynthesis or transport sometimes have narrow leaves, indicating that auxin is required to promote leaf expansion. nal7 mutants have narrow leaves (Fujino et al. 2007; Woo et al. 2007), and overexpression causes the production of wider leaves (Fujino et al. 2007). nal1 mutants have narrow leaves as well as short stems and have vasculature defects in the stem and leaves (Qi et al. 2008). NAL1 encodes a protein with unknown biochemical function with homologs in maize and Arabidopsis.
Tiller Angle
A unique role of auxin in rice is in the control of tiller angle. This is a form of stem gravitropism and is critical for the ability of rice to grow at high density. In fact, through breeding programs, rice has been selected for increased tiller angle (Wang and Li 2008). Wild rice has a prostrate growth habit, whereas cultivated rice has a more upright growth habit.
The gravity sensing tissue in grass stems is the pulvinus region at the base of the stem. Leaves also sense gravity independently in a region known as the leaf sheath pulvinus. In fact, in the tiller-angle mutants, it is difficult to separate leaf gravitropism from stem gravitropism as the leaves ensheath the stem. It is likely that auxin functions in gravity sensing in the pulvinus as auxin differentially accumulates in the pulvinus on gravity stimulation (Long et al. 2002)
The first indication of a role for auxin transport in controlling tiller angle came from RNAi of OsPIN1, which increased tiller angle (Xu et al. 2005). The next evidence came from the cloning of LAZY1 (LA1) of rice, which has defects in auxin transport and tiller angle (Li et al. 2007; Yoshihara and Iino 2007). la1 mutants have defects in stem, leaf, and coleoptile gravitropism, but do not affect root gravitropism. LA1 encodes a novel gene that appears to be grass specific, as there is no clear homolog in Arabidopsis (Li et al. 2007; Yoshihara and Iino 2007). LA1 is expressed in the leaf sheath pulvinus, the coleoptile, and adjacent to vascular bundles in the stem and leaf (Li et al. 2007; Yoshihara and Iino 2007). la1 mutants have altered auxin distribution in the stem on gravity stimulation and have increased auxin transport in the coleoptile, indicating that LA1 is a negative regulator of auxin transport (Li et al. 2007).
It is not clear from this analysis of LA1 and OsPIN1 if auxin transport promotes or inhibits the prostrate growth habit. LA1 inhibits auxin transport and mutants have increased tiller angle, whereas OsPIN1 promotes auxin transport and loss of function causes increased tiller angle (Xu et al. 2005; Li et al. 2007). It should be noted that auxin transport was tested in la1 coleoptiles rather than tillers, so more research is needed to clarify this. What is clear though is that differential auxin distribution is important for stem and leaf gravitropism.
Two additional genes, TILLER ANGLE CONTROL1 (TAC1) and PROSTRATE GROWTH1 (PROG1), that have opposite effects on tiller angle have been identified as QTL. It is not known if these genes have an auxin connection, but it is clear that they have played a critical role in rice domestication. TAC1 encodes an unknown protein that is present in single copy in the rice genome and appears to be present only in grasses (Yu et al. 2007). Loss of function of TAC1 leads to upright tillers, whereas increased expression leads to wider tiller angle. Sequence analysis indicates that the tac1 mutation is associated with upright tiller angle in japonica rice varieties, whereas wild rice and indica varieties have wild-type TAC1 and spread out tiller angle. Similar to LA1, TAC1 is expressed at the base of tillers and in the leaf sheath pulvinus.
PROG1, which is responsible for prostrate growth in the wild rice Oryza rufipogon, was identified independently by two groups (Jin et al. 2008; Tan et al. 2008). Loss of function of PROG1 in cultivated lines led to more erect growth habit and reduced tiller number. In addition, the mutants also have increased panicle branch number and yield. PROG1 encodes a novel CYS2 HIS2 zinc finger in the EBF class. PROG1 is expressed at the base of tillers in the vascular bundles of the leaf sheath pulvinus and in the lamina joint of the leaf. Whether the absence of LA1, TAC1, or PROG1 homologs in Arabidopsis indicates a lack of function of auxin in controlling branch angle in Arabidopsis requires further analysis.
Reproductive Development
Auxin plays a fundamental role in axillary meristem initiation in the inflorescence (Cheng and Zhao 2007; Barazesh and McSteen 2008b). Axillary meristems give rise to flowers in Arabidopsis and branches and spikelets (branches containing the florets) in grasses (McSteen et al. 2000; Barazesh and McSteen 2008b). Defects in auxin biosynthesis, transport, or signal transduction cause defects in flower initiation characterized by the pin inflorescence phenotype in Arabidopsis (Bennett et al. 1995; Przemeck et al. 1996; Cheng et al. 2006; Cheng et al. 2007b). The equivalent phenotype in maize is the barren inflorescence phenotype seen in bif2 and spi1 mutants, which have defects in auxin transport and biosynthesis (McSteen and Hake 2001; Ritter et al. 2002; Gallavotti et al. 2008a). Bif1 and Dvd1 are semidominant mutants that are proposed to have auxin-related defects (Barazesh and McSteen 2008a; Phillips et al. 2009). An additional eight mutants in maize have similar phenotypes but have not been characterized in detail (www.AuxinEvoDevo.org). Furthermore, DR5 and PIN marker lines show an auxin maximum as lateral primordia and axillary meristems are initiated in maize (Gallavotti et al. 2008b). Thus, despite the differences in morphology between Arabidopsis and grasses, auxin still plays a critical role in inflorescence development.
A bHLH transcription factor that functions in axillary meristem formation has been discovered in maize and rice but its role in Arabidopsis has not yet been reported (Komatsu et al. 2003; Gallavotti et al. 2004). ba1 mutants in maize and lax1 mutants in rice do not produce axillary meristems during inflorescence development but the bract leaves that subtend axillary meristems are unaffected (Ritter et al. 2002; Komatsu et al. 2003). Recent analysis has shown that lax1 mutants have defects in axillary meristem outgrowth rather than initiation (Oikawa and Kyozuka 2009). BA1 has been proposed to function either upstream or downstream of auxin (discussed in Barazesh and McSteen 2008b).
An exciting recent development was the discovery that the LAX1 protein may traffic between cells (Oikawa and Kyozuka 2009). As BA1/LAX1 RNA expression is adjacent to rather than in axillary meristems, BA1/LAX1 was proposed to control the movement of a signal to promote axillary-meristem development (Komatsu et al. 2003; Gallavotti et al. 2004). It turns out that the signal may be the LAX1 protein itself because the protein is present throughout the axillary meristem (Oikawa and Kyozuka 2009). This discovery leads to the question of what controls the movement of LAX1. This will be an exciting area of future research considering how little is known about how transcription factors move between cells.
CONCLUDING REMARKS
The research described here illustrates the power of the comparative approach. In many cases, genes controlling auxin biosynthesis, transport, and signaling were cloned first in Arabidopsis and then used to identify homologs in maize and rice. However, in some cases, for example ABP1, genes were isolated first in maize and then homologs identified in Arabidopsis. Some gene families, for example PIN1, are more redundant in maize and rice, whereas others, for example YUC, are more redundant in Arabidopsis. in some cases, genes such as CYP79B2/3 are found only in species related to Arabidopsis, whereas, in other cases, genes such as LA1 are grass specific. In general, the mechanisms of auxin biosynthesis, transport, and signal transduction are conserved in monocots and dicots, emphasizing the fundamental importance of auxin in development. In the future, it will be interesting to discover how the genes mediating the effects of auxin have diversified, which eventually may lead to an understanding of what makes monocots different from dicots.
ACKNOWLEDGMENTS
Research in the author’s laboratory is funded by the National Science Foundation and the United States Department of Agriculture. I thank Amy Monko for providing the photograph in Figure 2A and Kim Phillips for the photograph in Figure 2B.
Footnotes
Editors: Mark Estelle, Dolf Weijers, Ottoline Leyser, and Karin Ljung
Additional Perspectives on Auxin Signaling available at www.cshperspectives.org
REFERENCES
- Arai Y, Kawaguchi M, Syono K, Ikuta A 2004. Partial purification of an enzyme hydrolyzing indole-3-acetamide from rice cells. J Plant Res 117:191–198 [DOI] [PubMed] [Google Scholar]
- Attia KA, Abdelkhalik AF, Ammar MH, Wei C, Yang J, Lightfoot DA, El-Sayed WM, El-Shemy HA 2009. Antisense phenotypes reveal a functional expression of OsARF1, an auxin response factor, in transgenic rice. Current Issues Mol Biol 11:I29–I34 [PubMed] [Google Scholar]
- Bandyopadhyay A, Blakeslee JJ, Lee OR, Mravec J, Sauer M, Titapiwatanakun B, Makam SN, Bouchard R, Geisler M, Martinoia E, et al. 2007. Interactions of PIN and PGP auxin transport mechanisms. Biochem Soc Transactions 35:137–141 [DOI] [PubMed] [Google Scholar]
- Barazesh S, McSteen P 2008a. Barren inflorescence1 functions in organogenesis during vegetative and inflorescence development in maize. Genetics 179:389–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barazesh S, McSteen P 2008b. Hormonal control of grass inflorescence development. Trends Plant Sci 13:656–662 [DOI] [PubMed] [Google Scholar]
- Bartel B 1997. Auxin biosynthesis. Ann Rev Plant Physiol Plant Mol Biol 48:49–64 [DOI] [PubMed] [Google Scholar]
- Bennett SRM, Alvarez J, Bossinger G, Smyth DR 1995. Morphogenesis in pinoid mutants of Arabidopsis thaliana. Plant J 8:505–520 [Google Scholar]
- Brooks L, Strable J, Zhang XL, Ohtsu K, Zhou RL, Sarkar A, Hargreaves S, Elshire RJ, Eudy D, Pawlowska T, et al. 2009. Microdissection of shoot meristem functional domains. PLOS Gen 5: e1000476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carraro N, Forestan C, Canova S, Traas J, Varotto S 2006. ZmPIN1a and ZmPIN1b encode two novel putative candidates for polar auxin transport and plant architecture determination of maize. Plant Physiol 142:254–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler JW 2008. Cotyledon organogenesis. J Exp Bot 59:2917–2931 [DOI] [PubMed] [Google Scholar]
- Chen JG, Ullah H, Young JC, Sussman MR, Jones AM 2001. ABP1 is required for organized cell elongation and division in Arabidopsis embryogenesis. Genes Develop 15:902–911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng YF, Zhao YD 2007. A role for auxin in flower development. J Integ Plant Biol 49:99–104 [Google Scholar]
- Cheng YF, Dai XH, Zhao YD 2006. Auxin biosynthesis by the YUCCA flavin monooxygenases controls the formation of floral organs and vascular tissues in Arabidopsis. Genes Develop 20:1790–1799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y, Dai X, Zhao Y 2007a. Auxin synthesized by the YUCCA flavin monooxygenases is essential for embryogenesis and leaf formation in Arabidopsis. Plant Cell 19:2430–2439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y, Qin G, Dai X, Zhao Y 2007b. NPY1, a BTB-NPH3-like protein, plays a critical role in auxin-regulated organogenesis in Arabidopsis. Proc Natl Acad Sci 104:18825–18829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christian M, Steffens B, Schenck D, Burmester S, Bottger M, Luthen H 2006. How does auxin enhance cell elongation? Roles of auxin-binding proteins and potassium channels in growth control. Plant Biol 8:346–352 [DOI] [PubMed] [Google Scholar]
- Cohen JD, Slovin JP, Hendrickson AM 2003. Two genetically discrete pathways convert tryptophan to auxin: More redundancy in auxin biosynthesis. Trends Plant Sci 8:197–199 [DOI] [PubMed] [Google Scholar]
- Comai L, Kosuge T 1982. Cloning and characterization of IAAM, a virulence determinant of Pseudomonas-Savastanoi. J Bacteriol 149:40–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deveaux Y, Toffano-Nioche C, Claisse G, Thareau V, Morin H, Laufs P, Moreau H, Kreis M, Lecharny A 2008. Genes of the most conserved WOX clade in plants affect root and flower development in Arabidopsis. BMC Evolutionary Biol 8:291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis CM, Nagpal P, Young JC, Hagen G, Guilfoyle TJ, Reed JW 2005. AUXIN RESPONSE FACTOR1 and AUXIN RESPONSE FACTOR2 regulate senescence and floral organ abscission in Arabidopsis thaliana. Development 132:4563–4574 [DOI] [PubMed] [Google Scholar]
- Fischer C, Neuhaus G 1996. Influence of auxin on the establishment of bilateral symmetry in monocots. Plant J 9:659–669 [Google Scholar]
- Fujino K, Matsuda Y, Ozawa K, Nishimura T, Koshiba T, Fraaije MW, Sekiguchi H 2007. NARROW LEAF 7 controls leaf shape mediated by auxin in rice. Mol Gen Genom 279:499–507 [DOI] [PubMed] [Google Scholar]
- Fukaki H, Nakao Y, Okushima Y, Theologis A, Tasaka M 2005. Tissue-specific expression of stabilized SOLITARY-ROOT/IAA14 alters lateral root development in Arabidopsis. Plant J 44:382–395 [DOI] [PubMed] [Google Scholar]
- Gallavotti A, Barazesh S, Malcomber S, Hall D, Jackson D, Schmidt RJ, McSteen P 2008a. sparse inflorescence1 encodes a monocot-specific YUCCA-like gene required for vegetative and reproductive development in maize. Proc Natl Acad Sci 105:15196–15201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallavotti A, Yang Y, Schmidt RJ, Jackson D 2008b. The relationship between auxin transport and maize branching. Plant Physiol 147:1913–1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallavotti A, Zhao Q, Kyozuka J, Meeley RB, Ritter M, Doebley JF, Pe ME, Schmidt RJ 2004. The role of barren stalk1 in the architecture of maize. Nature 432:630–635 [DOI] [PubMed] [Google Scholar]
- Gauvrit C, Gaillardon P 1991. Effect of low temperatures on 2,4-D behaviour in maize plants. Weed Res 31:135–142 [Google Scholar]
- Grossmann K 2000. Mode of action of auxinic herbicides: A new ending to a long, drawn out story. Trends Plant Sci 5:506–508 [DOI] [PubMed] [Google Scholar]
- Hetz W, Hochholdinger F, Schwall M, Feix G 1996. Isolation and characterization of rtcs, a maize mutant deficient in the formation of nodal roots. Plant J 10:845–857 [Google Scholar]
- Hochholdinger F, Woll K, Sauer M, Dembinsky D 2004. Genetic dissection of root formation in maize (Zea mays) reveals root-type specific developmental programmes. Ann Bot 93:359–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochholdinger F, Wulff D, Reuter K, Park WJ, Feix G 2000. Tissue-specific expression of AUX1 in maize roots. J Plant Physiol 157:315–319 [Google Scholar]
- Hochholdinger F, Zimmermann R 2008. Conserved and diverse mechanisms in root development. Curr Opinion Plant Biol 11:70–74 [DOI] [PubMed] [Google Scholar]
- Im KH, Chen JG, Meeley RB, Jones AM 2000. Auxin-binding protein mutants in maize. Maydica 45:319–325 [Google Scholar]
- Inukai Y, Sakamoto T, Ueguchi-Tanaka M, Shibata Y, Gomi K, Umemura I, Hasegawa Y, Ashikari M, Kitano H, Matsuoka M 2005. CROWN ROOTLESS1, which is essential for crown root formation in rice, is a target of an AUXIN RESPONSE FACTOR in auxin signaling. Plant Cell 17:1387–1396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain M, Tyagi AK, Khurana JP 2006c. Genome-wide analysis, evolutionary expansion, and expression of early auxin-responsive SAUR gene family in rice (Oryza sativa). Genomics 88:360–371 [DOI] [PubMed] [Google Scholar]
- Jain M, Kaur N, Garg R, Thakur J, Tyagi A, Khurana J 2006a. Structure and expression analysis of early auxin-responsive Aux/IAA gene family in rice (Oryza sativa ). Funct Integ Genom 6:47–59 [DOI] [PubMed] [Google Scholar]
- Jain M, Kaur N, Tyagi A, Khurana J 2006b. The auxin-responsive GH3 gene family in rice ( Oryza sativa ). Funct Integ Genom 6:36–46 [DOI] [PubMed] [Google Scholar]
- Jain M, Nijhawan A, Arora R, Agarwal P, Ray S, Sharma P, Kapoor S, Tyagi AK, Khurana JP 2007. F-box proteins in rice. Genome-wide analysis, classification, temporal and spatial gene expression during panicle and seed development, and regulation by light and abiotic stress. Plant Physiol 143:1467–1483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin J, Huang W, Gao JP, Yang J, Shi M, Zhu MZ, Luo D, Lin HX 2008. Genetic control of rice plant architecture under domestication. Nat Gen 40:1365–1369 [DOI] [PubMed] [Google Scholar]
- Kelley KB, Reichers DE 2007. Recent developments in auxin biology and new opportunities for auxinic herbicide research. Pesticide Biochem Physiol 89:1–11 [Google Scholar]
- Knauss S, Rohrmeier T, Lehle L 2003. The auxin-induced maize gene ZmSAUR2 encodes a short-lived nuclear protein expressed in elongating tissues. J Biol Chem 278:23936–23943 [DOI] [PubMed] [Google Scholar]
- Koizumi K, Naramoto S, Sawa S, Yahara N, Ueda T, Nakano A, Sugiyama M, Fukuda H 2005. VAN3 ARF-GAP-mediated vesicle transport is involved in leaf vascular network formation. Development 132:1699–1711 [DOI] [PubMed] [Google Scholar]
- Komatsu K, Maekawa M, Ujiie S, Satake Y, Furutani I, Okamoto H, Shimamoto K, Kyozuka J 2003. LAX and SPA: Major regulators of shoot branching in rice. Proc Natl Acad Sci 100:11765–11770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriechbaumer V, Park WJ, Gierl A, Glawischnig E 2006. Auxin biosynthesis in maize. Plant Biol 8:334–339 [DOI] [PubMed] [Google Scholar]
- Kriechbaumer V, Park WJ, Piotrowski M, Meeley RB, Gierl A, Glawischnig E 2007. Maize nitrilases have a dual role in auxin homeostasis and β-cyanoalanine hydrolysis. J Exp Bot 58:4225–4233 [DOI] [PubMed] [Google Scholar]
- Lee BH, Johnston R, Yang Y, Gallavotti A, Kojima M, Travencolo BAN, Costa LD, Sakakibara H, Jackson D 2009. Studies of aberrant phyllotaxy1 mutants of maize indicate complex interactions between auxin and cytokinin signaling in the shoot apical meristem. Plant Physiol 150:205–216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li PJ, Wang YH, Qian Q, Fu ZM, Wang M, Zeng DL, Li BH, Wang XJ, Li JY 2007. LAZY1 controls rice shoot gravitropism through regulating polar auxin transport. Cell Res 17:402–410 [DOI] [PubMed] [Google Scholar]
- Liu Y, Lamkemeyer T, Jakob A, Mi GH, Zhang FS, Nordheim A, Hochholdinger F 2006. Comparative proteome analyses of maize (Zea mays L.) primary roots prior to lateral root initiation reveal differential protein expression in the lateral root initiation mutant rum1. Proteomics 6:4300–4308 [DOI] [PubMed] [Google Scholar]
- Liu HJ, Wang SF, Yu XB, Yu J, He XW, Zhang SL, Shou HX, Wu P 2005. ARL1, a LOB-domain protein required for adventitious root formation in rice. Plant J 43:47–56 [DOI] [PubMed] [Google Scholar]
- Ljung K, Hull AK, Kowalczyk M, Marchant A, Celenza J, Cohen JD, Sandberg G 2002. Biosynthesis, conjugation, catabolism and homeostasis of indole-3-acetic acid in Arabidopsis thaliana. Plant Mol Biol 50:309–332 [DOI] [PubMed] [Google Scholar]
- Long JC, Zhao W, Rashotte AM, Muday GK, Huber SC 2002. Gravity-stimulated changes in auxin and invertase gene expression in maize pulvinal cells. Plant Physiol 128:591–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig-Muller J 2000. Indole-3-butyric acid in plant growth and development. Plant Growth Regulation 32:219–230 [Google Scholar]
- Ludwig-Muller J, Walz A, Slovin JP, Epstein E, Cohen JD, Dong WQ, Town CD 2005. Overexpression of maize IAGLU in Arabidopsis thaliana alters plant growth and sensitivity to IAA but not IBA and 2,4-D. J Plant Growth Regulation 24:127–141 [Google Scholar]
- McSteen P, Hake S 2001. Barren inflorescence2 regulates axillary meristem development in the maize inflorescence. Development 128:2881–2891 [DOI] [PubMed] [Google Scholar]
- McSteen P, Laudencia-Chingcuanco D, Colasanti J 2000. A floret by any other name: Control of meristem identity in maize. Trends Plant Sci 5:61–66 [DOI] [PubMed] [Google Scholar]
- McSteen P, Malcomber S, Skirpan A, Lunde C, Wu X, Kellogg E, Hake S 2007. Barren inflorescence2 encodes a co-ortholog of the PINOID serine/threonine kinase and is required for organogenesis during inflorescence and vegetative development in maize. Plant Physiol 144:1000–1011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monaco TJ, Weller SC, Ashton FM 2002. Weed Science: Principles and Practices Wiley-Blackwell, New York [Google Scholar]
- Morita Y, Kyozuka J 2007. Characterization of OsPID, the rice ortholog of PINOID, and its possible involvement in the control of polar auxin transport. Plant Cell Physiol 48:540–549 [DOI] [PubMed] [Google Scholar]
- Multani DS, Briggs SP, Chamberlin MA, Blakeslee JJ, Murphy AS, Johal GS 2003. Loss of an MDR transporter in compact stalks of maize br2 and sorghum dw3 mutants. Science 302:81–84 [DOI] [PubMed] [Google Scholar]
- Nakamura A, Umemura I, Gomi K, Hasegawa Y, Kitano H, Sazuka T, Matsuoka M 2006. Production and characterization of auxin-insensitive rice by overexpression of a mutagenized rice IAA protein. Plant J 46:297–306 [DOI] [PubMed] [Google Scholar]
- Normanly J, Cohen JD, Fink GR 1993. Arabidopsis thaliana auxotrophs reveal a tryptophan-independent biosynthetic pathway for indole-3-acetic-acid. Proc Natl Acad Sci 90:10355–10359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oikawa T, Kyozuka J 2009. Two-step regulation of LAX PANICLE1 protein accumulation in axillary meristem formation in rice. Plant Cell 21:1095–1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okushima Y, Fukaki H, Onoda M, Theologis A, Tasaka M 2007. ARF7 and ARF19 regulate lateral root formation via direct activation of LBD/ASL genes in Arabidopsis. Plant Cell 19:118–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okushima Y, Fukaki H, Theologis A, Tasaka M 2005a. Analysis of ARF7- and ARF19-regulated genes in Arabidopsis lateral root formation. Plant Cell Physiol 46:S197–S197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okushima Y, Overvoorde PJ, Arima K, Alonso JM, Chan A, Chang C, Ecker JR, Hughes B, Lui A, Nguyen D, et al. 2005b. Functional genomic analysis of the AUXIN RESPONSE FACTOR gene family members in Arabidopsis thaliana: Unique and overlapping functions of ARF7 and ARF19. Plant Cell 17:444–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park WJ, Kriechbaumer V, Muller A, Piotrowski M, Meeley RB, Gierl A, Glawischnig E 2003. The nitrilase ZmNIT2 converts indole-3-acetonitrile to indole-3-acetic acid. Plant Physiol 133:794–802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips KA, Skirpan AL, Kaplinsky NJ, McSteen P 2009. Developmental disaster1 (Dvd1): A novel mutation causing defects during vegetative and inflorescence development in maize (Zea mays, Poaceae). Am J Bot 96:420–430 [DOI] [PubMed] [Google Scholar]
- Pollmann S, Muller A, Weiler EW 2006. Many roads lead to “auxin”: Of nitrilases, synthases, and amidases. Plant Biol 8:326–333 [DOI] [PubMed] [Google Scholar]
- Pollmann S, Neu D, Weiler EW 2003. Molecular cloning and characterization of an amidase from Arabidopsis thaliana capable of converting indole-3-acetamide into the plant growth hormone, indole-3-acetic acid. Phytochemistry 62:293–300 [DOI] [PubMed] [Google Scholar]
- Przemeck GKH, Mattsson J, Hardtke CS, Sung ZR, Berleth T 1996. Studies on the role of the Arabidopsis gene MONOPTEROS in vascular development and plant cell axialization. Planta 200:229–237 [DOI] [PubMed] [Google Scholar]
- Qi J, Qian Q, Bu QY, Li SY, Chen Q, Sun JQ, Liang WX, Zhou YH, Chu CC, Li XG, et al. 2008. Mutation of the rice NARROW LEAF1 gene, which encodes a novel protein, affects vein patterning and polar auxin transport. Plant Physiol 147:1947–1959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raven PH, Evert RF, Eichhorn SE 2005. The Biology of Plants W. H. Freeman, New York [Google Scholar]
- Ritter MK, Padilla CM, Schmidt RJ 2002. The maize mutant barren stalk1 is defective in axillary meristem development. Am J Bot 89:203–210 [DOI] [PubMed] [Google Scholar]
- Saleem M, Lamkemeyer T, Schutzenmeister A, Fladerer C, Piepho HP, Nordheim A, Hochholdinger F 2009. Tissue specific control of the maize (Zea mays L.) embryo, cortical parenchyma, and stele proteomes by RUM1 which regulates seminal and lateral root initiation. J Proteome Res 8:2285–2297 [DOI] [PubMed] [Google Scholar]
- Sato Y, Nishimura A, Ito M, Ashikari M, Hirano HY, Matsuoka M 2001. Auxin response factor family in rice. Genes Gen Sys 76:373–380 [DOI] [PubMed] [Google Scholar]
- Scanlon MJ 2003. The polar auxin transport inhibitor N-1-naphthylphthalamic acid disrupts leaf initiation, KNOX protein regulation, and formation of leaf margins in maize. Plant Physiol 133:597–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scanlon MJ, Henderson DC, Bernstein B 2002. SEMAPHORE1 functions during the regulation of ancestrally duplicated knox genes and polar auxin transport in maize. Development 129:2663–2673 [DOI] [PubMed] [Google Scholar]
- Scarpella E, Meijer AH 2004. Pattern formation in the vascular system of monocot and dicot plant species. New Phytol 164:209–242 [DOI] [PubMed] [Google Scholar]
- Sekimoto H, Seo M, Dohmae N, Takio K, Kamiya Y, Koshiba T 1997. Cloning and molecular characterization of plant aldehyde oxidase. J Biol Chem 272:15280–15285 [DOI] [PubMed] [Google Scholar]
- Sekimoto H, Seo M, Kawakami N, Komano T, Desloire S, Liotenberg S, Marion-Poll A, Caboche M, Kamiya Y, Koshiba T 1998. Molecular cloning and characterization of aldehyde oxidases in Arabidopsis thaliana. Plant Cell Physiol 39:433–442 [DOI] [PubMed] [Google Scholar]
- Sieburth LE, Muday GK, King EJ, Benton G, Kim S, Metcalf KE, Meyers L, Seamen E, Van Norman JM 2006. SCARFACE encodes an ARF-GAP that is required for normal auxin efflux and vein patterning in Arabidopsis. Plant Cell 18:1396–1411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skirpan A, Wu X, McSteen P 2008. Genetic and physical interaction suggest that BARREN STALK 1 is a target of BARREN INFLORESCENCE2 in maize inflorescence development. Plant J 55:787–797 [DOI] [PubMed] [Google Scholar]
- Skirpan A, Culler AH, Gallavotti A, Jackson D, Cohen JD, McSteen P 2009. BARREN INFLORESCENCE2 interaction with ZmPIN1a suggests a role in auxin transport during maize inflorescence development. Plant Cell Physiol 50:652–657 [DOI] [PubMed] [Google Scholar]
- Song YL, Wang L, Xiong LZ 2009a. Comprehensive expression profiling analysis of OsIAA gene family in developmental processes and in response to phytohormone and stress treatments. Planta 229:577–591 [DOI] [PubMed] [Google Scholar]
- Song YL, You J, Xiong LZ 2009b. Characterization of OsIAA1 gene, a member of rice Aux/IAA family involved in auxin and brassinosteroid hormone responses and plant morphogenesis. Plant Mol Biol 70:297–309 [DOI] [PubMed] [Google Scholar]
- Stepanova AN, Robertson-Hoyt J, Jeonga Y, Benavente LM, Xie DY, Dolezal K, Schlereth A, Jurgens G, Alonso JM 2008. TAA1- mediated auxin biosynthesis is essential for hormone crosstalk and plant development. Cell 133:177–191 [DOI] [PubMed] [Google Scholar]
- Sugawara S, Hishiyama S, Jikumaru Y, Hanada A, Nishimura T, Koshiba T, Zhao Y, Kamiya Y, Kasahara H 2009. Biochemical analyses of indole-3-acetaldoxime dependent auxin biosynthesis in Arabidopsis. Proc Natl Acad Sci 106:5430–5435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takamure I, Kinoshita T 1985. Inheritance and expression of reduced culm number character in rice. Jap JBreeding 35:17–24 [Google Scholar]
- Tan LB, Li XR, Liu FX, Sun XY, Li CG, Zhu ZF, Fu YC, Cai HW, Wang XK, Xie DX, et al. 2008. Control of a key transition from prostrate to erect growth in rice domestication. Nat Gen 40:1360–1364 [DOI] [PubMed] [Google Scholar]
- Tao Y, Ferrer JL, Ljung K, Pojer F, Hong FX, Long JA, Li L, Moreno JE, Bowman ME, Ivans LJ, et al. 2008. Rapid synthesis of auxin via a new tryptophan-dependent pathway is required for shade avoidance in plants. Cell 133:164–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taramino G, Sakai H, Komatsu K, Xiaomu N 2008. Plants with altered root architechture, involving the RUM1 gene, related constructs and methods. Patent application #20080201803 E.I. Dupont de Nemours and Company, USA [Google Scholar]
- Taramino G, Sauer M, Stauffer JL, Multani D, Niu X, Sakai H, Hochholdinger F 2007. The maize (Zea mays L.) RTCS gene encodes a LOB domain protein that is a key regulator of embryonic seminal and post-embryonic shoot-borne root initiation. Plant J 50:649–659 [DOI] [PubMed] [Google Scholar]
- Thakur JK, Tyagi AK, Khurana JP 2001. OsIAA1, an Aux/IAA cDNA from rice, and changes in its expression as influenced by auxin and light. DNA Res 8:193–203 [DOI] [PubMed] [Google Scholar]
- Thakur JK, Jain M, Tyagi AK, Khurana JP 2005. Exogenous auxin enhances the degradation of a light down-regulated and nuclear-localized OsIAA1, an Aux/IAA protein from rice, via proteasome. Biochim Biophys Acta 1730:196–205 [DOI] [PubMed] [Google Scholar]
- Timpte C 2001. Auxin binding protein: Curiouser and curiouser. Trends Plant Sci 6:586–590 [DOI] [PubMed] [Google Scholar]
- Tsiantis M, Brown MIN, Skibinski G, Langdale JA 1999. Disruption of auxin transport is associated with aberrant leaf development in maize. Plant Physiol 121:1163–1168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uehara T, Okushima Y, Mimura T, Tasaka M, Fukaki H 2008. Domain II mutations in CRANE/IAA18 suppress lateral root formation and affect shoot development in Arabidopsis thaliana. Plant Cell Physiol 49:1025–1038 [DOI] [PubMed] [Google Scholar]
- Waller F, Furuya M, Nick P 2002. OsARF1, an auxin response factor from rice, is auxin-regulated and classifies as a primary auxin responsive gene. Plant Mol Biol 50:415–425 [DOI] [PubMed] [Google Scholar]
- Walsh TA, Neal R, Merlo AO, Honma M, Hicks GR, Wolff K, Matsumura W, Davies JP 2006. Mutations in an auxin receptor homolog AFB5 and in SGT1b confer resistance to synthetic picolinate auxins and not to 2,4-dichlorophenoxyacetic acid or indole-3-acetic acid in Arabidopsis. Plant Physiol 142:542–552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YH, Li JY 2008. Rice, rising. Nat Gen 40:1273–1275 [DOI] [PubMed] [Google Scholar]
- Wang DK, Pei KM, Fu YP, Sun ZX, Li SJ, Liu HQ, Tang K, Han B, Tao YZ 2007. Genome-wide analysis of the auxin response factors (ARF) gene family in rice (Oryza sativa). Gene 394:13–24 [DOI] [PubMed] [Google Scholar]
- Woll K, Borsuk LA, Stransky H, Nettleton D, Schnable PS, Hochholdinger F 2005. Isolation, characterization, and pericycle-specific transcriptome analyses of the novel maize lateral and seminal root initiation mutant rum1. Plant Physiol 139:1255–1267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo YM, Park HJ, Su’udi M, Yang JI, Park JJ, Back K, Park YM, An G 2007. CONSTITUTIVELY WILTED1, a member of the rice YUCCA gene family, is required for maintaining water homeostasis and an appropriate root to shoot ratio. Plant Mol Biol 65:125–136 [DOI] [PubMed] [Google Scholar]
- Woodward AW, Bartel B 2005. Auxin: Regulation, action, and interaction. Ann Bot 95:707–735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright AD, Sampson MB, Neuffer MG, Michalczuk L, Slovin JP, Cohen JD 1991. Indole-3-acetic-acid biosynthesis in the mutant maize orange pericarp, a tryptophan auxotroph. Science 254:998–1000 [DOI] [PubMed] [Google Scholar]
- Wu X, McSteen P 2007. The role of auxin transport during inflorescence development in maize, Zea mays (Poaceae). Am J Bot 11:1745–1755 [DOI] [PubMed] [Google Scholar]
- Xu M, Zhu L, Shou HX, Wu P 2005. A PIN1 family gene, OsPIN1, involved in auxin-dependent adventitious root emergence and tillering in rice. Plant Cell Physiol 46:1674–1681 [DOI] [PubMed] [Google Scholar]
- Yamamoto Y, Kamiya N, Morinaka Y, Matsuoka M, Sazuka T 2007. Auxin biosynthesis by the YUCCA genes in rice. Plant Physiol 143:1362–1371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuno N, Takamure I, Kidou S, Tokuji Y, Ureshi A, Funabiki A, Ashikaga K, Yamanouchi U, Yano M, Kato K 2009. Rice shoot branching requires an ATP-binding cassette subfamily G protein. New Phytol 182:91–101 [DOI] [PubMed] [Google Scholar]
- Yoshihara T, Iino M 2007. Identification of the gravitropism-related rice gene LAZY1 and elucidation of LAZY1-dependent and -independent gravity signaling pathways. Plant Cell Physiol 48:678–688 [DOI] [PubMed] [Google Scholar]
- Yu BS, Lin ZW, Li HX, Li XJ, Li JY, Wang YH, Zhang X, Zhu ZF, Zhai WX, Wang XK, et al. 2007. TAC1, a major quantitative trait locus controlling tiller angle in rice. Plant J 52:891–898 [DOI] [PubMed] [Google Scholar]
- Zhang X, Madi S, Borsuk L, Nettleton D, Elshire RJ, Buckner B, Janick-Buckner D, Beck J, Timmermans M, Schnable PS, et al. 2007. Laser microdissection of narrow sheath mutant maize uncovers novel gene expression in the shoot apical meristem. PLOS Gen 3:1040–1052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao YD, Christensen SK, Fankhauser C, Cashman JR, Cohen JD, Weigel D, Chory J 2001. A role for flavin monooxygenase-like enzymes in auxin biosynthesis. Science 291:306–309 [DOI] [PubMed] [Google Scholar]
- Zhao Y, Hu YF, Dai MQ, Huang LM, Zhou DX 2009. The WUSCHEL-related homeobox gene WOX11 Is required to activate shoot-borne crown root development in rice. Plant Cell 21:736–748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao YD, Hull AK, Gupta NR, Goss KA, Alonso J, Ecker JR, Normanly J, Chory J, Celenza JL 2002. Trp-dependent auxin biosynthesis in Arabidopsis: Involvement of cytochrome P450s CYP79B2 and CYP79B3. Genes Develop 16:3100–3112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang XL, Jiang JF, Li JH, Ma QB, Xu YY, Xue YB, Xu ZH, Chong K 2006. Over-expression of OsAGAP, an ARF-GAP, interferes with auxin influx, vesicle trafficking and root development. Plant J 48:581–591 [DOI] [PubMed] [Google Scholar]
- Zhuang X, Xu Y, Chong K, Lan L, Xue Y, Xu Z 2005. OsAGAP, an ARF-GAP from rice, regulates root development mediated by auxin in Arabidopsis. Plant Cell Environ 28:147–156 [DOI] [PubMed] [Google Scholar]