Abstract
Single-molecule imaging enables biophysical measurements devoid of ensemble averaging, gives enhanced spatial resolution beyond the diffraction limit, and permits superresolution reconstructions. Here, single-molecule and superresolution imaging are applied to the study of proteins in live Caulobacter crescentus cells to illustrate the power of these methods in bacterial imaging. Based on these techniques, the diffusion coefficient and dynamics of the histidine protein kinase PleC, the localization behavior of the polar protein PopZ, and the treadmilling behavior and protein superstructure of the structural protein MreB are investigated with sub-40-nm spatial resolution, all in live cells.
Single-molecule imaging techniques now allow us to visualize the structure and behavior of signaling molecules, polarity proteins, and cytoskeletal components in live bacteria.
Since its advent 20 years ago, single-molecule fluorescence imaging has given rise to a host of exciting experiments (Ambrose and Moerner 1991). Beyond enabling fundamental investigations of the physics of emissive molecules, one main advantage of this technique is its use in biologically relevant, live-cell experiments. Optical fluorescence microscopy is an important instrument for cell biology, as light can be used to noninvasively probe a sample with relatively small perturbation of the specimen, enabling dynamical observation of the motions of internal structures in living cells. Single-molecule epifluorescence microscopy extends these capabilities by achieving nanometer-scale resolution, taking advantage of the fact that one can precisely characterize the point spread function (PSF) of a microscope, allowing the center of a distribution, and thus the exact position of an emitter, to be localized with accuracy much better than the diffraction limit itself. This localization accuracy improves beyond the diffraction limit roughly as one over the square root of the number of detected photons (Thompson et al. 2002). Detecting 100 photons from a single, isolated molecule can therefore improve the resolution of an optical measurement from the ∼250-nm diffraction limit down to 25 nm.
Single-molecule imaging has been used in the investigation of a number of live-cell samples. In 2000, the lateral heterogeneity of the plasma membrane was investigated by tracing the motion of single dye-labeled lipids in native human airway smooth muscle (HASM) cells (Schütz et al. 2000), and epidermal growth factor (EGF) receptor signaling was explored with a fluorescent protein fusion and a labeled ligand (Sako et al. 2000). Single fluorophore-labeled molecules have subsequently been used in many ways (Moerner 2003), for instance to investigate the effect of varying cholesterol concentration on the mobility of proteins in the plasma membrane of Chinese hamster ovary (CHO) cells (Vrljic et al. 2002; Vrljic et al. 2005) and to explore the real-time dynamic behavior of cell-penetrating-peptide (CPP) molecular transporters on the plasma membrane of CHO cells (Lee et al. 2008). Furthermore, in 2001, Harms et al. characterized the emission of fluorescent proteins in biocompatible environments and noted that the yellow fluorescent protein EYFP was well-suited to single-molecule imaging in cells (Harms et al. 2001). Such fluorescent proteins can be genetically encoded as tags for native proteins in cells; these fusions have been used in many live-cell single-molecule experiments.
More recently, single-molecule epifluorescence microscopy has been used to probe the inner workings of live bacteria. The small size of prokaryotic cells makes the optical diffraction limit particularly noticeable, which has stimulated the push toward superlocalization and superresolution to overcome this obstacle. As a result, the nascent field of bacterial structural biology has benefited greatly from single-molecule investigations of proteins in live cells. The overall shapes of such cells can be seen in a standard light microscope, but those interested in probing subcellular details, such as protein structure and localization, have typically had to resort to in vitro characterization combined with extrapolation to the cellular environment, as well as to indirect methods such as biochemical assays. Although cryo-electron microscopy can provide extremely high spatial resolution, fixation or plunge-freezing is essential, and methods for identifying specific proteins out of many are still lacking. As a consequence, bacterial cell biology is an area of study ripe for investigation with direct, noninvasive optical methods of probing position, coupling and structure, with resolution below the standard diffraction limit.
Several groups have extended single-molecule imaging techniques to live bacterial samples. In 2004, single PleC proteins were visualized in Caulobacter crescentus cells (Deich et al. 2004), and the behavior of this system is described in more detail later. More recently, Xie and coauthors have used single-molecule fluorescence techniques to study DNA-binding proteins, mRNA, and membrane proteins to provide much insight into the mechanisms of bacterial gene expression; these efforts have been documented in a recent review (Xie et al. 2008). As well, Conley et al. used covalently linked Cy3-Cy5-thiol switchable fluorophores to illuminate the stalks of C. crescentus cells with high resolution (Conley et al. 2008). In this article, we focus on the application of single-molecule imaging and single-molecule-based superresolution imaging to investigate the localization, movement, and structure of three important proteins, PleC, PopZ, and MreB, in live C. crescentus cells.
METHODS FOR LIVE-CELL SINGLE-MOLECULE IMAGING
Sample Preparation
Single-molecule fluorescence imaging of proteins in living cells can be accomplished via a genetically encoded fusion of the protein of interest to a photostable fluorescent protein (FP) emitting at long enough wavelengths to avoid cellular autofluorescence. However, single-molecule imaging requires a low copy number of fluorophores in the cell at any one time, so that the ∼250-nm diameter image spots from single molecules do not overlap. This low concentration can be achieved in several ways. First, the protein-FP fusion can be integrated into the chromosome in lieu of the native protein copy. In such strains, the initial FP concentration is generally too high for single-molecule imaging, so the emitting FP population needs to be reduced via photobleaching before measurements are made (Deich et al. 2004). Second, the protein-FP fusion can be integrated into the chromosome under the control of an inducible promoter while maintaining the untagged wild-type protein in the strain. This has the advantage of allowing the experimenter to control the level of tagged protein, while maintaining a significant background of the native protein to minimize any physiological effects of the tagging (Kim et al. 2006; Bowman et al. 2008). Finally, a large number of protein-FP fusions can be expressed in the cell, and the number of emitting FPs in any imaging frame can be subject to optical control, for instance by bleaching all of the FPs and then photo-reactivating a sparse subpopulation (Biteen et al. 2008).
To resolve the emission from single FPs above background, cell and sample autofluorescence must be minimized. In Caulobacter crescentus, this is accomplished by growing cells in minimal M2G medium for >24 hours, and by avoiding blue and green GFP mutants. The experiments discussed later all use the enhanced yellow fluorescent protein EYFP (S65G, V68L, S72A, T203Y), which can be excited at 514 nm. The high resolution provided by single-molecule imaging also requires the cells to lie still on a substrate. This can be accomplished by preparing a pad of 1.5%–2% agarose in M2G onto which the cells are deposited for imaging. The sample of cells on agarose must also contain fiduciary markers that can quantify stage drift over the imaging time. Fiducials can be any bright, nonbleaching emitters, for example quantum dots (Kim et al. 2006) or fluorescent beads (Biteen et al. 2008).
Imaging
Single-molecule imaging of bacterial cells can be accomplished in a standard widefield epifluorescence microscopy configuration, and the general imaging considerations have been described in detail (Moerner and Fromm 2003). Confocal or total-internal-reflection methods are often not necessary because the entire cell itself is mostly within the depth of focus of a high numerical aperture (NA) microscope (∼300–700 nm). Because the photon emission rate from a single molecule is typically 10 orders of magnitude smaller than the number of photons per second irradiating each pixel of the detector, appropriate filtering is necessary to reject scattered laser light. To optimize the number of detected photons, the single-molecule fluorescence microscope should incorporate a high-NA oil-immersion objective, sharp and carefully chosen filters, and a sensitive detector capable of detecting single photons. In modern experiments, this detector is generally an electron-multiplying charge-coupled detector (EMCCD) (Kim et al. 2006; Biteen et al. 2008; Bowman et al. 2008), though single-molecule imaging has also been accomplished in live cells with an intensified CCD (Deich et al. 2004).
For imaging of EYFP-protein fusions, the sample is excited with a 514-nm Ar+ ion or solid state laser. The excitation beam is converted from linear to circular polarization with a λ/4 waveplate to avoid the preferential detection of certain chromophore orientations (which often cannot be directly correlated with the orientation of the protein under investigation because of the FP fusion). In the detection pathway, a 525-nm long-pass dichroic mirror and a 530-nm long-pass filter are used to reject scattered laser light with an optical density of 108–1010. In experiments in which photoswitching is useful, photoreactivation of EYFP to control the emitting fluorophore density is performed with a 407-nm laser. This is also circularly polarized, and is coupled into the excitation pathway with a dichroic mirror. For example, superresolution imaging based on single-molecule fluorescence and photoreactivation is accomplished by capturing different sparse subsets of emitters in each imaging frame. On photobleaching of a sparse subset, a new subset is generated by photoreactivation and the imaging cycle continues. The final image is generated based on the sum of many single-molecule imaging frames (Betzig et al. 2006; Hess et al. 2006; Rust et al. 2006; Biteen et al. 2008).
EMCCD cameras can operate at rates as fast as ∼50 frames/second for a fairly large field, and even faster for subimages. The integration time is chosen based on the experimental system. A longer integration time increases the signal-to-noise when detecting quasi-stationary or slow-moving FPs. On the other hand, a shorter integration time is needed to resolve freely or quickly diffusing FPs. Typical integra-tion times used in C. crescentus are 15.4–32.2 ms for the quickly diffusing unpolymerized proteins PopZ and MreB (Kim et al. 2006; Bowman et al. 2008), 100 ms for the slowly diffusing mobile protein PleC (Deich et al. 2004), and 100 ms for MreB monomers incorporated into filaments (Kim et al. 2006; Biteen et al. 2008).
Data Processing
Single molecules of the protein-FP fusion are identified by several criteria, including digital (one-frame) photobleaching, appropriate number of detected photons, and a diffraction-limited emission pattern (point-spread function, PSF). The position of a nanometer-sized isolated emitter is at the center of the PSF; this center can be identified by eye as the brightest pixel in the emission spot (Deich et al. 2004; Bowman et al. 2008), or the molecule can be more accurately localized by fitting the PSF to a 2D, symmetric Gaussian function (Kim et al. 2006; Biteen et al. 2008). High-density super-resolution images based on multiple cycles of single-molecule imaging frames require the fitting software to be automated (Biteen et al. 2008), and it is essential to carefully analyze the data in terms of numbers of photons detected and localization precision to represent the position determinations in an unbiased manner.
Absolute protein positions and direction of movement are determined relative to fiducials and to white-light or dark-field images of the C. crescentus cells. The midpoint and endpoints of each cell are observable, the stalked end is recognized by its stalk, and the cell cycle stage can be identified from the cell morphology and the presence or absence of a stalk. It would also be possible to use differential interference contrast (DIC) images, but the changes in microscope optics compared with those for single-molecule fluorescence imaging would have to be carefully calibrated. The techniques described in this article measure images in only two dimensions, but 2D measurements of diffusion on the surface of the bacterial cell membrane can be corrected appropriately by simulating 3D diffusional movement along an appropriately curved membrane (Deich et al. 2004; Kim et al. 2006).
LOCALIZATION MECHANISM OF THE HISTIDINE PROTEIN KINASE PleC
C. crescentus is characterized by an asymmetric cell division process. Each round of dimorphic C. crescentus cell division gives rise to two distinct daughter cells: A motile, nonreplicating swarmer cell with a polar flagellum and a replicating stalked cell with an adhesive stalk. The localization of the histidine kinase PleC regulates polar organelle formation, motility, and asymmetric cell division by localizing to the flagellar pole at specific points of the cell cycle. PleC is localized to the flagellar pole of swarmer cells. The swarmer cell must differentiate into a stalked cell, shedding its flagellum and building a stalk, to replicate and divide. During this differentiation, PleC becomes delocalized. As the stalked cell progresses into a predivisional cell, a new flagellum is formed opposite the stalk, and PleC is localized to the new flagellar pole (Wheeler and Shapiro 1999). Three different mechanisms have been postulated for the crucial process of PleC localization: free diffusion of PleC through the inner membrane followed by capture and immobilization at a binding site (Rudner et al. 2002; Shapiro et al. 2002), directed insertion of PleC into the inner membrane at the flagellar pole at the time of translation (Steinhauer et al. 1999; Robbins et al. 2001), and active transport of PleC to the pole by a motor protein. Deich et al. used single-molecule microscopy to distinguish between these putative mechanisms by measuring the diffusion coefficients and movement directionality of PleC molecules in C. crescentus cells as a function of cell type and position within the cell (Deich et al. 2004).
Strain EJ148 of C. crescentus was created with PleC-EYFP fusions replacing the wild-type PleC on the chromosome and expressed under control of the PleC promoter. The EYFP tags were exposed to 514-nm illumination until all but a sparse subset remained in the emissive state. These isolated single-molecule emitters were imaged and their positions recorded as a function of time to measure the movement of PleC molecules in the cells. Figure 1 shows representative fluorescence images of this system. The first image in each of the series A–C is a darkfield image illustrating the position, size, and orientation of the cell under investigation. The other images in each series represent successive 100-ms imaging frames of the same cells in which each punctate spot is the fluorescence image of a single PleC-EYFP molecule. Figure 1A shows images acquired uninterruptedly (every 100 ms). Because of the relatively slow dynamics of the membrane-bound PleC, such continuous imaging bleached the molecules before significant motion could be observed. Indeed, the lower molecule in Figure 1A is bleached during the last frame, after only 600 ms of imaging. Time-lapse imaging was therefore used to increase the information attained before photobleaching. Figures 1B and C show two different cells in which a 100-ms imaging frame is acquired every 1 second. In this way, more movement is observed before photobleaching.
Figure 1.
Dynamics of single PleC histidine kinase molecules. (A–C) Leftmost image: Darkfield image of the cell under investigation. Columns 2–7: Consecutive fluorescence images of single PleC-EYFP molecules spaced by different time intervals: (A) images acquired every 100 ms, (B–C) images acquired every 1 s. (D) Measured mean-square displacement (triangles) and geometry-corrected mean-square displacement (circles) versus time lag. The corrected data slope of 0.049 µm2/s corresponds to a diffusion coefficient, D= 12 ±2 µm2/s. (E) Direction of motion in stalked cells without localized PleC (black bars) and swarmer and predivisional cells with localized PleC (gray bars), in which −s indicates motion toward the flagellar pole. Each bin represents one tenth of the cell length, with the polar bins excluded. (Reprinted, with permission, from Deich et al. 2004 [National Academy of Sciences].)
The single-molecule investigation of PleC in C. crescentus showed several key aspects of PleC dynamics. Bulk microscopy indicates the presence of a bright fixed PleC locus at the flagellar pole of swarmer and predivisional cells, but by resolving PleC-EYFP on a single-molecule level, a population of mobile PleC molecules could be identified in these cells. Mobile PleC molecules were also seen in the stalked cells, which show no evidence of localization in bulk microscopy. The trajectories of 400 such mobile molecules were further investigated by tracking the PleC-EYFP molecules, and the measured mean-square displacement (MSD) of the molecules at different time lags is shown in the triangles in Figure 1D. Because the observed motion is a 2D projection of 3D motion along a membrane surface, the actual distance moved by the molecule is larger than the observed movement. By simulating diffusion on a cell surface, modeled as a cylinder with spherical ends, the measured MSD is converted to a geometry-corrected MSD (circles in Fig. 1D). The geometry-corrected MSD is linear with the time lag, Δt, consistent with Brownian diffusion with a diffusion coefficient, D = MSD/(4Δt) of 12 ±2 µm2/s.
A normalized coordinate system was created in which the variable s (0 < s < 1) indicates the position of a molecule along the long axis of a C. crescentus cell. The position s = 0 represents the flagellar pole, and s = 1 the stalked pole. The geometry-corrected coefficient D for the mobile PleC-EYFP molecules were classified based on the cell type in which they were imaged (stalked, swarmer, or predivisional) and their position in the cell along the s axis. No measurable difference in D was observed for the different cell types. The geometry-corrected coefficient D was also not found to vary significantly as a function of s, except for a small reduction in D at the cell flagellar pole of swarmer and predivisional cells, perhaps attributable to immobilization at the binding site during the course of observation. Finally, Figure 1E shows the direction of PleC-EYFP motion along the s axis, in which −s indicates motion toward the flagellar pole, as a function of position and cell type. Here, stalked cells, which do not have localized PleC, are in black, and swarmer and predivisional cells, which have localized PleC in addition to the mobile PleC under consideration, are in gray, and the cell is divided into ten equal bins, so length bin 2 includes data for molecules starting with 0.1 < s < 0.2. All molecules were equally likely to move in the +s or −s direction, regardless of cell type or PleC position.
The motion of PleC in the membrane appears to be governed by Brownian diffusion, and is the same at all times in the cell cycle and all positions in the cell. In addition, the absence of directed motion argues against the existence of an active transport mechanism. Overall, the results of single-molecule investigations of PleC in C. crescentus were consistent with the diffusion and capture model of PleC localization at the flagellar pole.
DIFFUSION DYNAMICS OF THE POLAR PROTEIN PopZ
The ability of single-molecule microscopy to track individual intracellular proteins was subsequently applied to the proline-rich protein PopZ in live C. crescentus cells. PopZ is required to anchor the C. crescentus chromosome origins to the cell poles via the DNA-binding protein ParB, an anchoring that is critical for the control of temporal and spatial localization in the cell cycle. PopZ is localized to the flagellar end of nonreplicating swarmer cells. On differentiation into a stalked cell, a second concentration of PopZ begins to accumulate at the opposite pole (distal from the stalk) with an intensity that increases as the cell cycle progresses. The mechanism of PopZ localization was investigated on a single-molecule level (Bowman et al. 2008).
The merodiploid strain GB175 of C. crescentus was created with an unperturbed popZ locus and popZ-eyfp under control of the vanillate promoter at the vanA chromosomal locus. Leakage of the vanillate promoter ensured the expression of 1–10 copies of PopZ-EYFP per cell in a background of untagged PopZ molecules. These isolated single-molecule emitters were imaged and their positions recorded as a function of time, and two subpopulations of PopZ were identified: pole-localized molecules that remained fixed within the measurement localization accuracy (60 nm), and mobile molecules that were resolved with fast (32.2-ms) imaging frames. Figures 2A and D show the motion of two molecules tracked in one cell under investigation. In Figure 2A, the tracks are overlaid on a transmitted white-light image of the cell with the cell outline in black, whereas in Figure 2D, the longitudinal positions of the molecules are recorded as a function of imaging time. In Figures 2A and D, the red traces show the position of a polar-localized molecule, and the green traces show the trajectory of a mobile molecule that explores a large portion of the cell interior. Figures 2B–F show the trajectories of 12 fixed (red) and mobile (green) PopZ-EYFP molecules in five cells.
Figure 2.
Visualization of single PopZ-YFP molecules in live cells. (A) Positions of two PopZ-EYFP molecules within a cell, with colored lines tracking the distance moved between 32.2-ms frames. One molecule (red) remains localized to the pole, and the other (green) has increased mobility. The tracks are overlaid on a white-light image of the cell, which is outlined in black. (B–F) Time-dependent behavior of 12 single molecules in five cells. Red and green lines follow stationary and mobile molecules, respectively. The colored lines are dotted during dark (blinking-off) periods. Black horizontal dotted lines in (B) and (D) mark the positions of the poles opposite to the pole at which stationary molecules are localized. The trajectories in (D) correspond to the longitudinal position of the molecules tracked in (A) as a function of time. (Reprinted, with permission, from Bowman et al. 2008 [© Elsevier].)
The mobile PopZ-EYFP molecules showed random, diffusive movement in the cytoplasm during most of the time that they were tracked, though on some occasions, they remained fixed at a pole (e.g., Fig. 2D at 3–4 s). This movement is consistent with a diffusion and capture mechanism for PopZ localization at the cell pole. The spatial dynamics of single PopZ molecules would not have been observable without single-molecule imaging.
MOVEMENT OF THE STRUCTURAL PROTEIN MreB
The structural protein MreB mediates polarity, chromosome segregation, and cell shape in C. crescentus (Gitai et al. 2004; Dye et al. 2005; Gitai et al. 2005). Bulk-level imaging and biochemical studies of the cytoplasmic MreB protein are consistent with the formation of a dynamic superstructure made up of actin-like filaments. This structure provides the ma-chinery for key cellular processes, and the dynamics of MreB molecules were investigated on a single-molecule level (Kim et al. 2006).
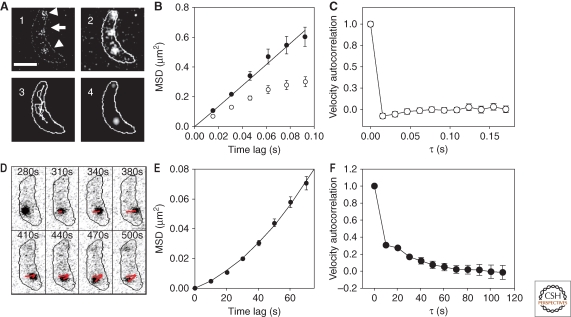
The merodiploid strain LS3813 of C. crescentus was generated by integrating a single Pxyl::eyfp-mreB construct into the xylX locus in a C. crescentus chromosome already containing a wild-type, unlabeled copy of MreB under its endogenous promoter (Gitai et al. 2004). Cells with only a small number of isolated EYFP-MreB molecules were prepared by incubating the cells in the M2G minimal medium with 0.0006%–0.003% xylose for 4 hours. The cells were imaged with 15.4-ms imaging frames to resolve the tagged MreB molecules. Figure 3A1 shows one such frame, in which three single molecules were captured. The image is smoothed by applying a low-pass filter (3×3 kernels of 0.0625, 0.125, 0.0625, 0.125, 0.25, 0.125, 0.0625, 0.125, and 0.0625) in Figure 3A2 for enhanced visibility. In this cell, two subpopulations of MreB were distinguished from one another: slow-moving molecules and fast-moving molecules (arrowheads and arrow, respectively, in Fig. 3A1). The trajectory of the fast-moving molecule in Figure 3A1, obtained by fitting the fluorescence image in every frame to a 2D Gaussian function, is plotted in Figure 3A3. This molecule diffused rapidly and explores a large portion of the cell. 450 successive frames were summed to form the fluorescence image in Figure 3A4. After this 7-second integration time, the fluorescence from the two slow-moving molecules was still evident, but emission from the fast-moving molecule was smeared out over many pixels and did not appear. The white line in Figure 3A shows the outline of the C. crescentus cell.
Figure 3.
Analysis of motion of EYFP-MreB. (A) 15.4-ms integration time fluorescence images of single EYFP-MreB molecules in a C. crescentus cell. White line shows the cell outline. (A1) 15.4-ms epifluorescence image showing three single EYFP-MreB molecules. The top and bottom molecules (arrowheads) are stationary on this timescale, the middle molecule (arrow) is mobile. (A2) Smoothed image of (A1) obtained by applying a low-pass filter. (A3) Recorded trajectory of the mobile (middle) molecule in (A1). (A4) Summed image of 450 sequential imaging frames. The fluorescence from the two stationary molecules is still evident, whereas the middle molecule does not appear on this 6.93-s integration time scale. (Scale bar, 1µm.) (B) Measured (open circles) and geometry-corrected (filled circles) MSD of fast-moving MreB molecules. The solid line is a linear fit of the corrected data. (C) Velocity autocorrelation of fast-moving molecules; this quantity drops to zero at the very first time lag. (D) Inverted-contrast, 100-ms integration time fluorescence images of single slow-moving EYFP-MreB molecules in a C. crescentus cell at 30-s intervals. The red arrows illustrate the directional movement of the molecules and the black line shows the cell outline. (E) Measured MSD versus time lag for slow-moving MreB molecules. The solid line is a quadratic fit to the data, indicative of directional motion. (F) Velocity autocorrelation of slow-moving MreB; this quantity remains positive over at least 80 s. (Reprinted, with permission, from Kim et al. 2006 [National Academy of Sciences].)
Because the dynamics of these molecules were unchanged in the presence of the MreB-depolymerizing drug A22, the fast-moving EYFP-MreB molecules were ascribed to a free, unpolymerized population. The behavior of the unpolymerized single molecules was further characterized based on 111 trajectories. The observed mean-square displacement (MSD) of the fast-moving molecules is plotted as a function of time lag (Δt) in the open circles of Figure 3B. Based on the first four points, a diffusion coefficient of D= MSD/(4Δt) of 1.11 ±0.18 µm2/s was extracted. This value of D is smaller than expected for a free, cytoplasmic protein, but is consistent with the motion of a membrane-associated protein. Because the observed motion represents a 2D projection, simulations were again performed to correct for the three-dimensionality of the cell, and the geometry-corrected MSD of the molecules is plotted in the filled circles of Figure 3B. The geometry-corrected MSD is linear with the time lag, Δt, consistent with diffusion along the cell membrane with D= MSD/(4Δt) of 1.75 ±0.17 µm2/s. The velocity autocorrelation function, CV(τ), was also calculated for these molecules. As shown in Figure 3C, for the fast-moving molecules, CV(τ) dropped to zero at the very first time step, indicating a nondirected random walk.
Further experiments addressed the behavior of the slow-moving MreB. Because slow-moving EYFP-MreB molecules were not observed in the presence of the MreB-depolymerizing drug A22, the slow-moving molecules represent polymerized MreB. The dynamics of 120 single members of this subpopulation were carefully examined. Because the molecules were stationary within the 30-nm resolution of the measurement over the course of the 7-second integration time shown in Figure 3A4, time-lapse imaging was used. Specifically, 100-ms imaging frames were separated by 9.9 seconds. In this way, the slow movement of polymerized MreB molecules could be followed over a longer time period before photobleaching. Figure 3D shows 8 such fluorescence images in reverse contrast, in which a single molecule is tracked for 220 seconds. The molecule moves from left to right, then turns and moves right to left at a downward angle, shown by the red line. The observed MSD of the slow-moving MreB molecules is plotted as a function of Δt in Figure 3E. Here, the MSD was characterized by a quadratic dependence on time lag, as is typical of directed motion. Also consistent with directed motion is the computed velocity autocorrelation function, CV(τ), shown in Figure 3F, in which CV(τ) remains positive until τ = 80 s.
The slow, directed motion of polymerized MreB was further explored by analyzing the trajectories of each individual molecule in the context of two putative models for the motion of an MreB molecule in an MreB filament: (1) the filaments into which the monomers are incorporated were moving, or (2) the monomers themselves were treadmilling though largely stationary filaments, analogous to the motion of actin. If the motion of polymerized MreB is because of whole-filament movement, then the observation time for the slow-moving single molecules should be limited by the photobleaching time of the EYFP, independent of experiment timescale. In the inset to Figure 4A, the total emission time of EYFP-MreB before photobleaching under continuous emission was measured, with an average on-time of 4.6 seconds. However, the true irradiation time before photobleaching of the molecules imaged with 9.9-second time lapses, shown in Figure 4A, was found to be only 0.8 seconds. Most of the fluorescence from the polymerized MreB molecules therefore disappeared before photobleaching occurred, likely as a result of dissociation from the end of a filament and the onset of fast diffusive motion not resolvable with the 100-ms imaging frames. This result indicates that MreB molecules treadmill through filaments with fixed ends.
Figure 4.
Slow movement of MreB molecules through filaments. (A) Distribution of observed true irradiation time of slow-moving MreB molecules with time-lapse imaging (average = 0.8 s) and (inset) average emission time under continuous illumination (average = 4.6 s). (B) Speed distribution of slow-moving MreB (average = 6.0 ±0.2 nm/s). (C) Distribution of end-to-end contour lengths measured from the movement of single slow-moving MreB molecules (average = 332 nm). (D) Representative trajectories of MreB filaments in a stalked (ST) cell and in a predivisional cell, plotted on normalized cell. Representative global direction assignments are given by “+” (toward the swarmer [SW] pole) or “−” [toward the ST pole]). (Reprinted, with permission, from Kim et al. 2006 [National Academy of Sciences].)
Given that the polymerized MreB molecules showed directed motion, a speed value was extracted from each single-molecule trajectory (Fig. 4B). The average speed was 6.0 ± 0.2 nm/s. From its crystal structure, the length of an MreB monomer is 5.4 nm (van den Ent et al. 2001); the average speed therefore corresponded to 1.2 monomer additions per second in steady-state fixed MreB filaments. The average length traveled by a polymerized MreB molecule before dissociation, which corresponds to the filament length, was also measured by considering only MreB molecules that became polymerized after the start of imaging and that dissociated before the end of imaging. The MreB filament lengths of 128 trajectories are shown in Figure 4C, in which the average filament length was 392 ±23 nm—quite small relative to the average cell length of 3.5 µm. A representative subset of these trajectories is plotted on normalized cell shapes in Figure 4D. Of note, most of the trajectories moved perpendicular to the cell long axis. A smaller number of trajectories were oblique. These nonperpendicular trajectories were characterized as moving toward the swarmer pole (+) or toward the stalked pole (−). No preferred orientation was observed.
CELL-CYCLE-DEPENDENT SUPERRESOLUTION STRUCTURE OF MreB
By fitting the point-spread function (PSF) of isolated nanoscale emitters with 30-nm localization accuracy, and by following these superlocalized molecules over time, the previous experiment attained superresolution via single-molecule tracking, as displayed in Figure 4D. This approach was, however, limited by the isolated fluorophore requirement to ∼1–3 tagged MreB molecules, and therefore ∼1–3 trajectories per cell. To address this, photoactivated localization microscopy (PALM) was used to extend the investigation of EYFP-MreB in C. crescentus cells to cells expressing high concentrations of these molecules, and thus to visualize the superstructure formed by a collection of MreB filaments in these cells (Biteen et al. 2008). In particular, bulk visualization of MreB in C. crescentus indicates that MreB forms a cell-cycle-dependent structure (Gitai et al. 2004; Dye et al. 2005), and single-molecule-based superresolution imaging was applied to measure this structure.
C. crescentus cells expressing 100–1000 copies of EYFP-MreB were created by incubating the strain LS3813, described above, in the M2G minimal medium with 0.006%–0.075% xylose for 4 hours. The cells were imaged with 100-ms imaging frames to resolve only the slow-moving, polymerized MreB molecules. The initial density of fluorescent tags in these cells was initially too great for isolated, single-molecule imaging, but this could be addressed based on the fact that apparently bleached EYFP molecules can be reactivated with violet light (Dickson et al. 1997). Therefore, in live C. crescentus cells, after the bleaching of all emissive EYFP-MreB molecules, a sparse subset could be reactivated with 407-nm irradiation, in which the reactivation intensity of 103–104 W/cm2 was chosen such that at most one EYFP molecule was reactivated in each diffraction-limited region. The positions of these isolated molecules were determined by PSF fitting, and the reactivation and imaging process was repeated many times until a superresolution image could be reconstructed from the sum of the localizations. To illustrate the photoreactivation probes, Figures 5A–F show the fluorescence emission from EYFP-MreB molecules in a single cell over time. Each punctate, white spot is the emission from a single molecule, and the fluorescence images (acquired with 100-ms integration times) are superimposed on a reversed-contrast white-light image of the cell. After the initial bleaching step, two single EYFP-MreB molecules are observed in the cell (Fig. 5A). Additional imaging with 514-nm light bleached all the fluorophores (Fig. 5B). A 2-second dose of 407-nm laser illumination returned some EYFP-MreB to an emissive state (Figs. 5C and E), though all fluorophores are apparently bleached prior to the 407-nm pulse (Figs. 5D and F).
Figure 5.
Superresolution imaging of MreB superstructure. (A–F) Reactivation of single EYFP-MreB fusions in the same live C. crescentus cell using 407 nm light. Fluorescence images of single EYFP-MreB molecules from 514 nm excitation are overlaid on a reversed-contrast white-light image of the cell. (A) Initial image showing two isolated emissive EYFP-MreB molecules. (B), (D), (F) After photobleaching by 514-nm irradiation, the cell contains no emissive EYFP-MreB. (C), (E) Reactivated EYFP-MreB molecules are observed following a short 407-nm reactivation pulse. (Scale bar, 1 µm.) (G–J) TL-PALM images of EYFP-MreB in C. crescentus cells created by fitting molecule positions in imaging frames such as those in (A), (C), and (E). (G), (H) Quasi-helical structure in a stalked cell. (I), (J) Midplane ring in a predivisional cell. Fluorescence PALM images are shown in (G) and (I). The PALM images in (H) and (J) are from the same cells as in (G) and (I), respectively, overlaid on a reversed-contrast white-light image of the cell. (Scale bars, 300 nm). (Reprinted, with permission, from Biteen et al. 2008 [Nature Publishing Group].)
The positions of these molecules, calculated from a fit of the fluorescence images to a 2D Gaussian function, were recorded for every such frame, and at the end of a 4-minute acquisition period, these localization events were summed. To form a final superresolution image, each localized molecule was depicted in the reconstruction as a unit-area Gaussian with fixed width equal to the localization accuracy of 30–40 nm. This technique, termed live-cell PALM, enabled the imaging of the EYFP-MreB superstructure in C. crescentus. In particular, it resolved several MreB bands spanning the length of stalked cells, and a tightly-focused midplane ring in predivisional cells. Importantly, understanding superresolution features derived from many image acquisitions requires careful consideration of the emitter photophysics and the dynamics, if any, of the underlying structure. Because, as described above, polymerized MreB molecules treadmill slowly along MreB filaments, time-lapse (TL) imaging, as described above, was incorporated into live-cell PALM to increase the number of localization events given the fact that the maximum labeling concentration in live-cell experiments is limited because of changes in the cell morphology at high concentrations of fusion protein. Specifically, a 900-ms delay was introduced between each 100-ms imaging frame, and the sample was only illuminated with 514-nm light during the short acquisition time. 407-nm reactivation pulses were again applied after photobleaching to generate novel sparse subsets of emissive EYFP-MreB.
Figures 5G–J present the results from TL-PALM imaging of EYFP-MreB in C. crescentus. Two distinct MreB superstructures were identified in the cells: a quasi-helical arrangement in a stalked cell (Figs. 5G,H) and a midplane ring in the predivisional cell (Figs. 5I,J). In Figures 5H and J, the false-color PALM reconstruction is superimposed on a reversed-contrast, white-light image of the cell in which the EYFP-MreB was visualized. The images obtained by TL-PALM were more continuous than those acquired without the introduction of dark periods. Furthermore, the use of TL increased the number of localization events (487 in Fig. 5G and 330 in Fig. 5J) to the point in which the Nyquist criterion for 40-nm resolution was satisfied (Shroff et al. 2008), giving rise to a true superresolution reconstruction of the MreB superstructure in C. crescentus.
CONCLUSIONS
Single-molecule imaging permits high-resolution measurements of fluorescent emitters in time and space. This noninvasive, nonperturbative technique is quite useful for the study of single protein molecules in live cells. By applying the techniques of single-molecule microscopy to EYFP-tagged proteins in live C. crescentus cells, the diffusion dynamics, localization patterns, and structure of three key proteins, PleC, PopZ, and MreB, could be explored. By providing a noninvasive tool for high-resolution imaging that is beginning to approach the spatial resolution of cryo-electron microscopy, single-molecule, superresolution imaging can enable investigations of many more intracellular processes and has become an important biophysical tool.
Because the single-molecule-based imaging techniques described here are based on widefield microscopy, they can further provide the basis for more sophisticated experiments. Specific challenges that are being addressed include improving the axial resolution of single-molecule microscopes (Huang et al. 2008; Juette et al. 2008; Pavani et al. 2009), incorporating multiple excitation and emission wavelengths (Bates et al. 2007; Shroff et al. 2007), and imaging thick samples (Vaziri et al. 2008).
ACKNOWLEDGMENTS
The authors warmly thank members of the Lucy Shapiro laboratory, specifically Ellen Judd, Harley McAdams, Grant Bowman, and Zemer Gitai, as well as members of the Moerner laboratory, specifically Jason Deich, Marcelle Koenig, So Yeon Kim, Anika Kinkhabwala, Michael Thompson, and Nicole Tselentis, for their contributions to this work. This research was supported in part by DARPA Grant MDA-972-00-1-0032, NSF Grant MCB-0212503, Department of Energy Grant DE-FG02-04ER63777, and NIH Grants P20-HG003638, R01-GM086196, and R01-GM085437.
Footnotes
Editors: Lucy Shapiro and Richard Losick
Additional Perspectives on Cell Biology of Bacteria available at www.cshperspectives.org
REFERENCES
- Ambrose WP, Moerner WE 1991. Fluorescence spectroscopy and spectral diffusion of single impurity molecules in a crystal. Nature 349:225–227 [Google Scholar]
- Bates M, Huang B, Dempsey GT, Zhuang X 2007. Multicolor super-resolution imaging with photo-switchable fluorescent probes. Science 317:1749–1753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betzig E, Patterson GH, Sougrat R, Lindwasser OW, Olenych S, Bonifacino JS, Davidson MW, Lippincott-Schwartz J, Hess HF 2006. Imaging intracellular fluorescent proteins at nanometer resolution. Science 313:1642–1645 [DOI] [PubMed] [Google Scholar]
- Biteen JS, Thompson MA, Tselentis NK, Bowman GR, Shapiro L, Moerner WE 2008. Super-resolution imaging in live Caulobacter crescentus cells using photoswitchable EYFP. Nat Meth 5:947–949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman GR, Comolli LR, Zhu J, Eckart M, Koenig M, Downing KH, Moerner WE, Earnest T, Shapiro L 2008. A polymeric protein anchors the chromosomal origin/ParB complex at a bacterial cell pole. Cell 134:945–955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conley NR, Biteen JS, Moerner WE 2008. Cy3-Cy5 covalent heterodimers for single-molecule photoswitching. J Phys Chem B 112:11878–11880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deich J, Judd EM, McAdams HH, Moerner WE 2004. Visualization of the movement of single histidine kinase molecules in live Caulobacter cells. Proc Nat Acad Sci 101:15921–15926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson RM, Cubitt AB, Tsien RY, Moerner WE 1997. On/Off blinking and switching behavior of single green fluorescent protein molecules. Nature 388:355–358 [DOI] [PubMed] [Google Scholar]
- Dye NA, Pincus Z, Theriot JA, Shapiro L, Gitai Z 2005. Two independent spiral structures control cell shape in Caulobacter. Proc Nat Acad Sci 102:18608–18613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gitai Z, Dye N, Shapiro L 2004. An actin-like gene can determine cell polarity in bacteria. Proc Nat Acad Sci 101:8643–8648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gitai Z, Dye NA, Reisenauer A, Wachi M, Shapiro L 2005. MreB actin-mediated segregation of a specific region of a bacterial chromosome. Cell 120:329–341 [DOI] [PubMed] [Google Scholar]
- Harms GS, Cognet L, Lommerse PHM, Blab GA, Schmidt T 2001. Autofluorescent proteins in single-molecule research: applications to live cell imaging microscopy. Biophys J 80:2396–2408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess ST, Girirajan TPK, Mason MD 2006. Ultra-high resolution imaging by fluorescence photoactivation localization microscopy. Biophys J 91:4258–4272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang B, Wang W, Bates M, Zhuang X 2008. Three-dimensional super-resolution imaging by stochastic optical reconstruction microscopy. Science 319:810–813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juette MF, Gould TJ, Lessard MD, Mlodzianoski MJ, Nagpure BS, Bennett BT, Hess ST, Bewersdorf J 2008. Three-dimensional sub-100 nm resolution fluorescence microscopy of thick samples. Nat Meth 5:527–529 [DOI] [PubMed] [Google Scholar]
- Kim SY, Gitai Z, Kinkhabwala A, Shapiro L, Moerner WE 2006. Single molecules of the bacterial actin MreB undergo directed treadmilling motion in Caulobacter crescentus. Proc Nat Acad Sci 103:10929–10934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H-D, Dubikovskaya EA, Hwang H, Semyonov AN, Wang H, Jones LR, Twieg RJ, Moerner WE, Wender PA 2008. Single-molecule motions of oligoarginine transporter conjugates on the plasma membrane of chinese hamster ovary cells. J Am Chem Soc 130:9364–9370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moerner WE 2003. Optical measurements of single molecules in cells. Trends Analyt Chem 22:544–548 [Google Scholar]
- Moerner WE, Fromm DP 2003. Methods of single-molecule fluorescence spectroscopy and microscopy. Rev Sci Instrum 74:3597–3619 [Google Scholar]
- Pavani SRP, Thompson MA, Biteen JS, Lord SJ, Liu N, Twieg RJ, Piestun R, Moerner WE 2009. Three-dimensional, single-molecule fluorescence imaging beyond the diffraction limit by using a double-helix point spread function. Proc Nat Acad Sci 106:2995–2999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbins JR, Monack D, McCallum SJ, Vegas A, Pham E, Goldberg MB, Theriot JA 2001. The making of a gradient: IcsA (VirG) polarity in Shigella flexneri. Mol Microbiol 41:861–872 [DOI] [PubMed] [Google Scholar]
- Rudner DZ, Pan Q, Losick RM 2002. Evidence that subcellular localization of a bacterial membrane protein is achieved by diffusion and capture. Proc Nat Acad Sci 99:8701–8706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rust MJ, Bates M, Zhuang X 2006. Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM). Nat Meth 3:793–795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sako Y, Minoghchi S, Yanagida T 2000. Single-molecule imaging of EGFR signalling on the surface of living cells. Nat Cell Biol 2:168–172 [DOI] [PubMed] [Google Scholar]
- Schütz GJ, Kada G, Pastushenko VP, Schindler H 2000. Properties of lipid microdomains in a muscle cell membrane visualized by single molecule microscopy. EMBO J 19:892–901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro L, McAdams H, Losick R 2002. Generating and exploiting polarity in bacteria. Science 298:1942–1946 [DOI] [PubMed] [Google Scholar]
- Shroff H, Galbraith CG, Galbraith JA, Betzig E 2008. Live-cell photoactivated localization microscopy of nanoscale adhesion dynamics. Nat Meth 5:417–423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shroff H, Galbraith CG, Galbraith JA, White H, Gillette J, Olenych S, Davidson MW, Betzig E 2007. Dual-color superresolution imaging of genetically expressed probes within individual adhesion complexes. Proc Nat Acad Sci 104:20308–20313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinhauer J, Agha R, Pham T, Varga AW, Goldberg MB 1999. The unipolar Shigella surface protein IcsA is targeted directly to the bacterial old pole: IcsP cleavage of IcsA occurs over the entire bacterial surface. Mol Microbiol 32:367–377 [DOI] [PubMed] [Google Scholar]
- Thompson RE, Larson DR, Webb WW 2002. Precise nanometer localization analysis for individual fluorescent probes. Biophys J 82:2775–2783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Ent E, Amos LA, Lowe J 2001. Prokaryotic origin of the actin cytoskelecton. Nature 413:39–44 [DOI] [PubMed] [Google Scholar]
- Vaziri A, Tang J, Shroff H, Shank CV 2008. Multilayer three-dimensional super resolution imaging of thick biological samples. Proc Nat Acad Sci 105:20221–20226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrljic M, Nishimura SY, Brasselet S, Moerner WE, McConnell HM 2002. Translational diffusion of individual class II MHC membrane proteins in cells. Biophys J 83:2681–2692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrljic M, Nishimura SY, Moerner WE, McConnell HM 2005. Cholesterol depletion suppresses the translational diffusion of class II major histocompatibility complex proteins in the plasma membrane. Biophys J 88:334–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler RT, Shapiro L 1999. Differential localization of two histidine kinases controlling bacterial cell differentiation. Mol Cell 4:683–694 [DOI] [PubMed] [Google Scholar]
- Xie XS, Choi PJ, Li GW, Lee NK, Lia G 2008. Single-molecule approach to molecular biology in living bacterial cells. Annu Rev Biophys 37:417–444 [DOI] [PubMed] [Google Scholar]