Abstract
The outermost layer of the vertebrate heart originates from migratory mesothelial cells (epicardium) that give rise to coronary vascular smooth muscles and fibroblasts. The role of the epicardium in myocardial morphogenesis and establishment of normal heart function is still largely unknown. Here, we use Drosophila to investigate non-autonomous influences of epicardial-like tissue surrounding the heart tube on the structural and functional integrity of the myocardium. It has previously been shown that during Drosophila heart formation, mesodermal expression of the homeobox transcription factor even-skipped (eve) is required for specification of a subset of non-myocardial progenitors in the precardiac mesoderm. These progenitors may share some similarities with the vertebrate epicardium. To investigate a non-autonomous epicardial-like influence on myocardial physiology, we studied the consequences of reduced mesodermal Eve expression and epi/pericardial cell numbers on the maturation of the myocardial heart tube, its contractility, and acquisition of a normal heart rhythm in the Drosophila model. Targeting the eve repressor ladybird early (lbe) with the minimal eve mesodermal enhancer efficiently eliminates the mesodermal Eve lineages. These flies exhibit defects in heart structure, including a reduction in systolic and diastolic diameter (akin to ‘restrictive cardiomyopathy’). They also exhibit an elevated incidence of arrhythmias and intermittent asystoles, as well as compromised performance under stress. These abnormalities are restored by eve reexpression or by lbe-RNAi co-overexpression. The data suggest that adult heart function in Drosophila is likely to be modulated non-autonomously, possibly by paracrine influences from neighboring cells, such as the epi/pericardium. Thus, Drosophila may serve as a model for finding genetic effectors of epicardial–myocardial interactions relevant to higher organisms.
Keywords: Myocardium, Epicardium, even-skipped, ladybird, Arrhythmia, Heart failure, Heart disease, Cardiomyopathy, Aging
Introduction
In vertebrates, the epicardium is increasingly recognized as playing a prominent role in heart morphogenesis and in the regulation of myocardial function, but the underlying mechanisms remain to be elucidated. The mammalian epicardium is the outermost layer of the heart; it derives from the so-called proepicardium, a transient structure comprised of mesothelial cells located in the septum transversum. These cells begin migrating towards the heart during the process of cardiac looping and, later on, cover the surface of the myocardium. The pro-epicardium also gives rise to cells that form the coronary vessels and cardiac fibroblasts (Manner et al., 2001; Mikawa and Gourdie, 1996; Muñoz-Chápuli et al., 2002). The epicardium is thought to secrete factors that signal to the developing muscle cells that form the myocardium (Chen et al., 2002; Lavine et al., 2005; Stuckmann et al., 2003). Many genes involved in heart specification have been elucidated, including some that act in the epicardium (Merki et al., 2005; Zamora et al., 2007; Winter et al., 2007; Cai et al., 2008; Zhou et al., 2008). Proepicardium, epicardium, and other epicardial derived cells are critical for proper heart development and they exert their effects by modulating the maturation of the myocardium (reviewed in Manner and Ruiz-Lozano, 2008). Yet, the mechanisms by which the epicardium communicates with the myocardium during development and in maintaining its functional integrity remain to be elucidated. Due to complexity of heart development in vertebrates, interactions between the epicardium and the myocardium may profit from studies in a simpler genetic model system, such as Drosophila, which also has epicardial-like cells apposed to the myocardium.
Contrary to the multichambered organ in vertebrates, the Drosophila heart is a relatively simple linear tube which is reminiscent of the early embryonic heart in vertebrates (Bodmer, 1995; reviewed in Bodmer et al., 2005; Qian et al., 2008). The fly heart originates from bilaterally symmetrical groups of mesodermal cells that meet at the dorsal midline beneath the epidermis to form a heart tube. This heart tube consists of inner contractile myocardial cells (myocardium), expressing muscle-specific proteins (Bour et al., 1995; Lilly et al.,1995; Nguyen et al., 1994; Zhang and Bernstein, 2001), and an outer layer of non-myogenic pericardial cells, referred to here as the epi/pericardium. A subset of these epi/pericardial cells in the fly has previously been implicated in contributing heart function (Fujioka et al., 2005). In this study, it was found that adult hearts with fewer epi/pericardial cells beat at lower rates and may have a propensity for stress induced heart failure. How exactly the heart’s structure and function is modulated by paracrine signaling is not yet known. Interestingly, a non-autonomous influence of the epicardium on the function of the vertebrate heart has also been postulated recently (see Manner and Ruiz-Lozano, 2008).
During dorsal closure of the embryonic epidermis, the bilateral cardioblast rows fuse and become aligned in a highly ordered fashion underneath the dorsal epidermal midline to form a rhythmically contracting heart tube that circulates haemolymph through the larval and adult body cavities (Rizki and Rizki, 1978; Cammarato et al., 2008; Mery et al., 2008; Ocorr et al., 2007a,b; Taghli-Lamallem et al., 2008; Wessells and Bodmer, 2004; Wessells et al., 2004). During metamorphosis, the heart undergoes morphological remodelling to form the adult heart. This process includes the recruitment of longitudinal muscle fibers ventral to the heart tube, but without the addition of new cells to the (tinman-expressing) myocardial tube itself (Curtis et al., 1999; Lo and Frasch, 2003; Molina and Cripps, 2001; Monier et al., 2005; Zeitouni et al., 2007).
The genetic hierarchy that specifies the fly heart has been elucidated in considerable detail (reviewed in Bodmer and Frasch, 1999; Cripps and Olson, 2002; Qian et al., 2008). The formation of the fly heart is specified by a cascade of cardiogenic transcription factors interacting in a conserved network. Genetic experiments suggested the involvement of the homeobox genes tinman (Azpiazu and Frasch, 1993; Bodmer, 1993; Zaffran et al., 2006), even-skipped (eve) (Frasch et al., 1987; Macdonald et al., 1986; Han et al., 2002), ladybird early (lbe) (Jagla et al., 1997, 2002), odd-skipped (odd) (Ward and Skeath, 2000), of T-box genes (Miskolczi-McCallum et al., 2005; Qian et al., 2005; Reim and Frasch, 2005; Reim et al., 2005), GATA factors (Gajewski et al., 2001; Klinedinst and Bodmer, 2003), and the COUP transcription factor seven-up (svp) (Mlodzik et al., 1990; Lo and Frasch, 2001). Interactions between these factors give rise to specific subpopulations of cardiac progenitors. Cardiac specification in both, flies and vertebrates, also involves complex inductive signaling across germ layers by decapentaplegic (dpp), wingless (wg) and hedgehog (hh). In response to combinatorial tin/dpp/wg/hh signaling, the mesodermal cells subdivide into small clusters of cells of cardiac progenitors (Frasch, 1995; Wu et al., 1995; Carmena et al., 1998; Halfon et al., 2000; Liu et al., 2006). The similar roles of these cardiogenic factors in vertebrates suggest strikingly conserved mechanisms in both the genetic and molecular regulation of cardiac development.
One gene that has been implicated in the interaction between the epi/pericardium and myocardium in Drosophila is eve (Su et al., 1999; Fujioka et al., 2005). The transcriptional regulation of mesodermal eve expression has been elucidated in detail (Fujioka et al., 1999; Halfon et al., 2000; Han et al., 2002; Knirr and Frasch, 2001; Jagla et al., 2002; Liu et al., 2008; Su et al., 1999). eve is first expressed in a subset of segmentally repeated clusters of three to four cells at the dorsal margin of the mesoderm giving rise to both a subset of pericardial cells (EPCs) and dorsal muscle progenitors by well-defined lineages (for details see Han and Bodmer, 2003). All the regulatory influences necessary for this highly restricted mesodermal pattern of eve expression are contained within a small enhancer element (‘eme’; Fujioka et al., 1999). Binding sites in eme have been identified for a plethora of cardiogenic transcription factors and cardiogenic signalling pathway effectors (Halfon et al., 2000; Han et al., 2002; Knirr and Frasch, 2001; Liu et al., 2008; Lo and Frasch, 2003).
Mesodermal eve repression, due to overexpression of the homeobox factor Lbe, has previously been shown to alter cell fates within the Eve lineage. In turn, ectopic expression of eve affects the Lbe lineage suggesting a mutual repression between these two homeobox factors (Jagla et al., 1997; 2002; Han et al., 2002). In addition, mutations in the Lbe site of eme render reporter gene expression insensitive to repression by lbe (Han et al., 2002; Liu et al., 2008). Further studies showed that elimination of the eme element in a genomic rescue construct (“eve meso-minus”) abolishes mesodermal eve expression, which results in viable flies with reduced numbers of pericardial cells and some indications of altered heart function (Fujioka et al., 2005), although the exact changes in heart physiology remained to be determined. Fujioka et al. (2005) showed that eve is an essential factor for the correct specification of cell types arising from the eve-expressing lineage. In the absence of mesodermal eve, the precursor cells in which eve would usually be expressed produce progenitors that are not localized in the EPC region, but assume other fates, some associated with the heart (Fujioka et al., 2005, see Figs. 1C–I therein). These cells are apparently naïve mesodermal cells as shown by a eme-driven lacZ reporter gene and do not seem to be a priori specified to a particular fate, since they can adopt a myocardial, other pericardial or a muscle fate with random segment to segment distribution (Fig. 1I in Fujioka et al., 2005). These findings would suggest that the epi/pericardium plays a non-autonomous role in modulating specific aspects of adult heart function, which we have further investigated in this study. Based on this non-autonomy, it may be possible to screen for myocardial-specific modifiers that can ameliorate the compromised heart function caused by a defective epi/pericardium.
Fig. 1.

Eve pericardial cells (EPCs) are reduced upon targeted lbe overexpression. (A–F) eme-lbe mediated repression of eve (SSlbe) in stage 14 embryos (dorsal view). (A) In each wildtype hemisegment, Eve labels the nuclei of two EPCs and those of the dorsal acute (DA1) muscle (indicated by arrow and asterisk, respectively). In the SSlbe lines, a transgene dose-dependent loss of both EPCs and the DA1 muscle was found. Note the virtually complete absence of Eve protein in the recombinant SSlbeAF line (B), a phenotype similar to eme-deficient eve (J43DR) rescue embryos (D). (C, E, F) Heterozygous SSlbeAF/+ or single transgenic lines show reduced but not abolished Eve staining. (G) Quantification of the extent of eve suppression was achieved by counting EPCs in each embryonic hemisegment (n=30; stage 12–16). A score of 2 corresponds to 50% repression of Eve protein, a score of 0 indicates the complete absence of detectable EPCs. Error bars represent standard deviation. (H–J) Posterior heart of late stage 16 embryos double-labeled for Eve (red) and Pericardin (PC, green). In wildtype flies, the two EPCs per hemisegment are situated within the Pericardin stained region of the epi/pericardium. In the SSlbe lines, Pericardin staining is reduced and more diffuse in appearance. Note the absence of EPCs in SSlbeAF (J).
Here, we explore how specific aspects of the heart’s structure and function are affected by the absence of eve in non-myocardial mesoderm, as a model for non-autonomous influences on cardiac physiology. The previously used eve meso-minus rescue flies are cumbersome to handle (in a screen, for example), since they combine multiple elements. Thus, we inhibited eve expression by driving its repressor lbe exclusively in the mesodermal eve lineage (eme-lbe). eme-lbe transgenic flies have reduced mesodermal eve expression, pericardial cell number, and lifespan as previously observed for eve meso-minus flies. Importantly, we found that eme-lbe strongly impacted multiple aspects of adult heart physiology including its contractility and the overall incidence of arrhythmias and asystoles. These data suggest an essential role for the eve-expressing non-myocardial cells in adult cardiac physiology. Additionally, we find that forced co-expression of eve with lbe or of lbe-RNAi in the mesodermal eve lineage is sufficient to restore proper heart structure, rhythm, and stress response. These findings suggest that the Eve-positive epi/pericardium is likely to play a critical, non-autonomous role in regulating myocardial structure and contractility and in the development of a regular heart beat. These results lay the foundation for future experiments designed to identify downstream genes in the myocardium that are capable of rescuing the structural and/or functional heart defects inflicted by an epi/pericardial deficit.
Materials and methods
Transgenic lines and genetic crosses
The transgenic lines named SSlbe carry a P-element insertion of the genomic eve mesodermal enhancer (eme) fragment (SphI +5.8 kb and StuI +6.6 kb, eme900), the HSP70 promoter and the lbe cDNA flanked by the 3′ and 5′ UTRs from eve, which have been shown to increase transgene activity driven by eme in the mesoderm (Fujioka et al., 1999; Han et al., 2002). lbe is ectopically expressed by eme exclusively in eve-expressing cells. The SSlbe A and F transgenic lines were selected by their ability to suppress mesodermal eve expression (Figs. 1A–F). yw was used for SSlbe microinjection, for outcrosses and as wildtype controls. The recombinant SSlbeAF with four functional copies of the SSlbe transgene was also generated and used for dose-dependent analysis. Overexpression was accomplished using the UAS-Gal4 system (Brand and Perrimon, 1993). The following fly stocks were used: eme-Gal4 (Han et al., 2002), UAS-lbe (Jagla et al., 1997), UASrpr;UAS-hid (Bloomington Drosophila stock center), UAS-lbeRNAi (VDRC, Vienna Drosophila RNAi Center), Cypher-GFP (gene trap line G189; Morin et al., 2001), J43DR, J49DR and EGN86T (Fujioka et al., 2005). J43DR, J49DR and EGN86T flies were described previously (Fujioka et al., 2005). J43DR and J49DR are also referred to as “eve meso-minus”.
Analysis of embryos with immunohistochemistry
Antibody staining and double-labeling was performed as described previously (Han et al., 2002; Qian et al., 2005). For anti-eve staining, the signal was amplified using the TSA system (Perkin-Elmer) and the Vectastain ABC Elite PK 6100 Standard system (Vector Laboratories). Antibody staining was visualized using 3,3′-diaminobenzidine (DAB). For fluorescent confocal microscopy, Cy3- or FITC-conjugated secondary antibodies were used (Jackson Laboratories). Embryos were mounted in VectaShield (Vector Laboratories) and analyzed using a Biorad MRC-1024MP confocal microscope. Primary antibodies used were: mouse anti-Eve used at 1:50; mouse anti-PC at 1:2 (Hybridoma Bank, University of Iowa), rabbit anti-Eve at 1:1000 (Frasch et al., 1987) and rabbit anti-Dmef2 at 1:1000 (Lilly et al., 1995). For expression analysis, 25–30 embryos were used as sample size.
Staining of adult hearts
Adult hearts were dissected as for physiological recordings (see below) and fixed in 4% paraformaldehyde for 10–15 min. Actin filaments were visualized using fluorescent conjugated phalloidin, 1:250 (Molecular Probes) and analyzed using a Biorad MRC-1024MP confocal microscope.
Adult heart physiology
For semi-intact heart preparations, adult flies were anesthetized with FlyNap™ (Carolina Biological, Burlington, NC, USA) and dissected at room temperature (22 °C) in oxygenated artificial adult hemolymph (AHL) containing 108 mM NaCl, 5 mK KCl, 2 mM CaCl2, 8 mM MgCl2, 10 mM sucrose, 5 mM trehalose, 5 mM HEPES (pH 7.1), 1 mM NaH2PO4 and 4 mM NaHCO3 (Singleton and Woodruff, 1994; Wang et al., 2003) to expose the heart (Ocorr et al., 2007a). Internal abdominal organs and fat were removed and the hearts were allowed to equilibrate under oxygenation for 20 min prior to recording. Adult hearts were observed under a direct immersion upright compound microscope (Leica DM LFSA) with a 10× objective and a highspeed camera (Hamamatsu EM-CCD). Simple PCI Software (Compix Inc., Cranberry Township, PA) was adapted to record the heart performance for a period of 60 s with a frame rate of 100–120 fps. A gradient filter of 45° and contrast enhancement were then applied to enhance the appearance of the heart edges. Recordings were taken from abdominal segments A3–A4 and analyzed as in Ocorr et al. (2007a).
Results
Lbe-mediated suppression of eve in the dorsal mesoderm
Previous experiments deleting the mesodermal enhancer of eve suggested that the Eve-expressing pericardial cells (EPCs) may have an influence on heart function (Fujioka et al., 2005). Here, we examined the precise non-autonomous impact of reducing eve expression in non-myocardial cells associated with the heart on specific aspects of cardiac physiology. First, we generated additional tools to reduce mesodermal eve expression, with the added benefit of that these flies may be suitable for genetic screening. For this purpose, we made transgenic flies that directly target lbe to the mesodermal Eve progenitors and thereby inhibit eve expression (using the eme enhancer in SSlbeA and F, see Materials and methods).
We assessed the efficiency of eme-lbe in reducing mesodermal eve expression (Figs. 1A–G). The homozygous combination of two independent transgene insertions of eme-lbe, SSlbeAF, shows a complete absence of Eve protein in the dorsal mesoderm (Figs. 1A, B), similar to the eve meso-minus embryos that lack eme (J43DR; Fig. 1D; Fujioka et al., 2005). Heterozygous SSlbeAF/+ or single transgenic lines show reduced Eve staining (Figs. 1C, E, F). Thus, eme-lbe efficiently suppresses eve in the mesodermal Eve lineages, which results in a reduced number of pericardial cells at larval stages (Suppl. Fig. S2). This phenotype is rescued by overexpression of wildtype eve cDNA using eme-Gal4, which suggests that the epi/pericardial eve is required for formation of the larval pericardial cells.
Reduced mesodermal eve also affects some aspects of cardiac morphogenesis. For example, we observed a significant reduction and a more diffuse localization of the pan-pericardial marker Pericardin (Figs. 1H–J; Suppl. Fig. S1), a collagen IV component of the epi/pericardial extracellular matrix (Chartier et al., 2002). This phenotype, which is similar to a mild ‘broken heart’ phenotype (Yi et al., 2006), is rescued by eme-mediated overexpression of eve in SSlbe flies or in complete eve rescue flies (Suppl. Fig. S1K–N). In addition to the epi/pericardium, morphogenesis of the myocardium is also moderately affected by the lack of mesodermal eve. When we examined the alignment of the bilateral rows of myocardial cells at the dorsal midline of the embryo (Dmef2 staining; Bour et al., 1995; Lilly et al., 1995), we observed slight myocardial misalignments in the regions where the pericardium is noticeably affected (Suppl. Fig. S1), and these myocardial misalignments were corrected by co-overexpression of eve. We also observed disorganization and loss of dorsal skeletal muscles. Taken together, a lack of eve in the dorsal mesoderm causes a non-autonomous effect on the myocardium causing moderate cellular misalignments of the forming heart tube. This suggests a crosstalk between eve-expressing epi/pericardium (and/or the dorsal somatic muscles) and the myocardium that does not express eve.
eve-deficiency in the epi/pericardium affects adult myocardial morphology
All tinman-expressing adult cardiomyocytes in the working myocardium are of embryonic origin and further proliferation does not seem to occur during metamorphosis (Monier et al., 2005). We analyzed the myocardial structure of 1-week old adults in order to explore the role of mesodermal eve expression on the adult heart. We observed a significant reduction in the heart diameter in SSlbeAF as well as in eve meso-minus J43DR flies (Figs. 2A, B, E). These phenotypes were reversed by overexpression of eve or co-overexpression of lbe-RNAi in a SSlbe AF background (Figs. 2C, D) or in complete eve mutant flies (EGN86T; Fig. 2F). To determine whether it is the lack of eve or simply the absence of mesodermal Eve cell specification we over-expressed the death genes reaper and hid using eme-Gal4 to eliminate the mesodermal Eve progenitors (Suppl. Fig. S6). The observed constricted heart phenotype is similar to preventing mesodermal eve expression (Fig. 2), which is in support of the notion that mesodermal Eve cells exert a non-autonomous influence on the heart.
Fig. 2.

The adult myocardial heart tube is affected in eve-deficient hearts. (A–F) Phalloidin staining of dissected 1-week old adults showing the heart in abdominal segments A2–A5. (A) Labeled myofibrils of a wildtype heart (yw). (B) In SSlbeAF flies, heart morphology is primarily affected in the A3/A4 region (indicated by asterisks), similar to the phenotype observed in eme-deficient eve rescue flies (J43DR, in E). Note the overall reduction in heart size and a significant heart thinning in B and E compared to wildtype (A). The heart thinning phenotype was reversed by overexpression of wildtype eve cDNA or of lbe RNAi in a SSlbeAF background (C, D), or in complete eve mutants rescue flies (EGN86T, in F). (G, H) Diameter measurement in live movie recordings of the myocardium in both segments (A3 in grey, A4 in white bars) in relaxed (diastolic) and contracted (systolic) stage. For SSlbeAF, the diastolic diameter (DD) is 55 μm and the systolic diameter (SD) 33 μm in segment A4, which is a significant reduction compared to 75 μm and 45 μm for yw, respectively (Student’s t-test, p<0.001, n=19). SSlbeAF hearts with eme-Gal4-driven eve cDNA show rescue to 70 μm (DD) and 42 μm (SD). Similar to SSlbe AF, both parameters were significantly reduced in J43DR hearts and were reversed in complete eve mutants rescue EGN86T flies. Student’s t-test, *p<0.01, **p<0.001; 15–19 hearts per data point.
In addition to fixed tissue measurements, we also took movies from live hearts with these genetic combinations and measured diastolic (DD) and systolic diameters (SD) of the beating hearts (Figs. 2G, H; as in Ocorr et al., 2007a). Compared to the wildtype (yw) diameters of 75 μm (DD) and 45 μm (SD) in A4 abdominal segments, both parameters were significantly reduced in SSlbeAF (DD: 55 μm and SD: 33 μm). These cardiac contractility phenotypes in SSlbeAF flies could be partially rescued by eme-Gal4-driven lbe-RNAi or wildtype eve co-expression (DD: 70 μm and SD: 42 μm). In A3 segments the effect was less pronounced but the tendency was similar. The anteriorly located conical chamber (in A2) was little changed in all genotypes examined (data not shown). Although we do not know the reason for this differential effect on heart tube diameter along the anterior–posterior axis we have seen similar effects in myosin mutants, which exhibit dilated or restrictive cardiomyopathy preferentially in the A3/4 region of the heart (Cammarato et al., 2008).
We also examined the myofibrillar fine-structure of hearts using a Z-line-specific marker (Cypher-GFP gene trap; see Mery et al., 2008). We observed a regular arrangement of the transverse myofibrils within the tinman-expressing myocardium (Molina and Cripps, 2001), which appeared normal (Figs. 3A, C). In contrast, the spacing of the Z-lines and the parallel arrangement of the myofibrils were significantly defective in the longitudinal heart-associated muscles of SSlbeAF (Figs. 3B, D). These defects in myofibrillar structure were ameliorated by overexpressing eve or lbe-RNAi in SSlbeAF flies (data not shown). This suggests that the observed morphological changes of the heart are likely a consequence of the loss of eve in the mesodermal Eve lineages. Since the EPCs of the epi/pericardium as well as the DA1/DO2 somatic muscle founders normally express eve, we formally cannot distinguish whether the observed effects are due to a lack of Eve in the DA1/DO2, the EPCs, or via a reduction in the overall number of epi/pericardial cells. Based on the close proximity of the epi/pericardial cells to the myocardium, and because other genetic manipulations of the epi/pericardium also result in myocardial contractility defects (H.-Y. Lim and R.B., unpubl.), we favor the idea that it is Eve activity in the epi/pericardium that is critical.
Fig. 3.

In vivo imaging of the myocardial microstructure. Organization of myofibrillar structures by in vivo imaging of a Z-line-specific marker (cypher-GFP gene trap). 60× magnification of the fourth abdominal segment is shown. (A, C) Regular arrangement of the transverse myofibrils is observed within the tinman-expressing myocardium, in both wildtype and mutant. (B, D) Significant defects in spacing of the Z-lines and the parallel arrangement of the myofibrils are observed in the longitudinal, heart-associated muscles of SSlbeAF flies compared to wildtype (yw).
To further explore the effect of mesodermal eve repression on other parameters of myocardial function, including aggravation of an age-dependent susceptibility to cardiac stress, we subjected the SSlbe flies to a regime of external electrical pacing of the heart that is designed to stress the heart by driving it at a high rate (Wessells et al., 2004). Under this pacing stress SSlbe flies exhibited an increased age-dependent susceptibility to heart failure (Suppl. Fig. S3A). Electrical stimulation often triggered a fibrillation-like rhythm even in young SSlbe flies, which was rarely observed in wildtype control flies. Thus, eme-lbe repression of eve causes an increase in cardiac stress susceptibility that is elevated with age. In addition, both methods of inducing mesodermal eve deficiency caused a reduction in longevity (Suppl. Fig. S3B). A lifespan shortening of about 30% was observed in the SSlbe and J43DR flies compared to the mean lifespan of wildtype flies. The reduction in lifespan is more pronounced in females than in males (data not shown).
Repression of eve in the epi/pericardium causes severe heart rhythm disturbances
A sensitive method of monitoring the fly’s cardiac physiology, and thus the functional consequences of mesodermal eve repression, is the analysis of high-speed digital movie clips in semi-intact heart preparations of adult flies (Ocorr et al., 2007a). In these myogenic heart preparations, young wildtype flies exhibit a regular heart rhythm (Figs. 4A, and 5B), but age and mutations can cause severe arrhythmias, as manifested by variable systolic and/or diastolic intervals, asystolic events as well as other defects in cardiac dynamics (Ocorr et al., 2007a). Compared to wildtype and control flies, SSlbe and eme deleted J43DR flies exhibited an increase in cardiac arrhythmias as shown by the broadening of the heart period distribution (Figs. 4B, C, E) and by the significantly increased Arrhythmia Index (Fig. 5B). In these mesodermal eve-deficient flies, we also observed rhythm disturbances along the anterior–posterior axis, in that the two ends of the heart were beating with different frequencies (akin to a 2:1 block; data not shown). This finding may suggest also a disturbance of the coordination of the two cardiac pacemakers at either end of the abdominal heart tube (Dulcis and Levine, 2003).
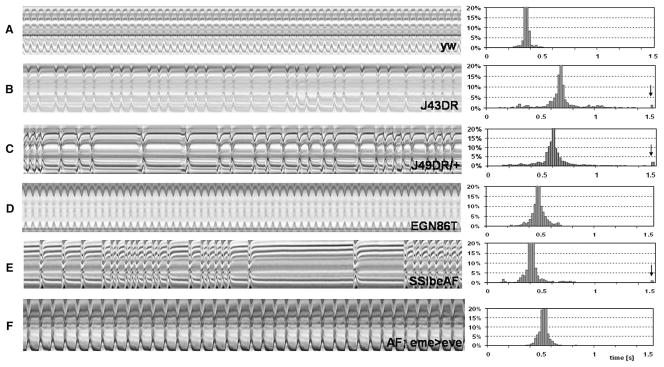
Fig. 4.
Movement analysis of adult heartbeats in semi-intact fly preparations. (A–F) Left panel: representative 20 s M-Mode traces from high speed movies. M-modes are made by electronically excising a one-pixel wide region that spans the upper and lower edges of the heart from the same position in every movie frame. The one-pixel wide strips are aligned horizontally to provide a trace of the heart edge movement in the y axis over time (x axis). Right panel: histograms of all the heart period lengths for the specified genotypes plotted as percentage of total beats (n=18–20 flies per genotype). Prolonged heart periods (>1.5 s) are binned together (indicated by arrows). (A) Wildtype (yw) M-modes show little variation of the heart period resulting in a narrow distribution around the median heart period. (B, C) Arrhythmic heart beats are evident in mesodermal eve-deficient flies. Asystolic events indicative of prolonged diastolic intervals result in longer and more variable heart periods, as illustrated by the broad distribution around the mean heart period for J43DR. Note the high percentage of periods longer than 1.5 s (indicated with arrows in the right panel) resulting in a significantly lower heart rate. (D) Few arrhythmias and asystolic events were observed in the complete eve rescue line EGN86T, with quite narrowly distributed heart periods of the individual flies. (E, F) SSlbeAF line shows a high incidence of arrhythmias and asystoles, which is reversed in SSlbeAF rescue flies, although the mean heart period remains longer than for wildtype.
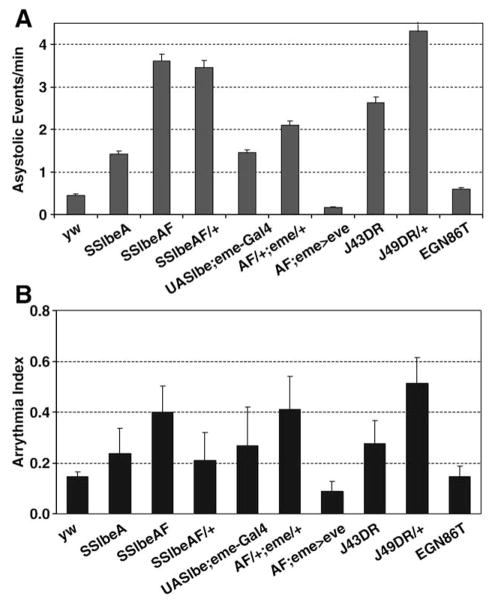
Fig. 5.
Quantification of asystolic events and arrhythmias. (A) Relative measurement of asystolic events using twice the median heart period of every individual fly for detecting prolonged diastolic intervals, normalized by the total time of the movies. Both, SSlbe flies and J43DR mutants exhibit a significant increase of asystoles relative to yw and the rescue controls. (B) Arrhythmia index is defined as the normalized standard deviation of the heart period. SSlbe and J43DR lines and UASlbe;eme-Gal4 all show an increased incidence in arrhythmias compared to wildtype (yw) and rescue controls (AF;eme>eve). The outcross of SSlbeAF;eme-Gal4 (AF/+;eme/+) is included as a control for the rescue SSlbeAF;eme-Gal4;UAS-eve (AF;eme>eve). (n is 18–20 hearts per data point).
Representative M-Mode examples for J43DR and SSlbeAF flies are shown in Fig. 4. The following cardiac phenotypes were observed: (1) non-contractile asystolic heart periods or temporary cardiac arrest (Fig. 5A), (2) double or multiple beats within otherwise regular periods, and (3) varying duration of diastolic intervals (DI) resulting in arrhythmias. The non-contractive asystolic heart periods were dramatically increased in SSlbeAF and eme deletion flies (arrows in histograms of Figs. 4B, C, E), and are quantitatively represented in Fig. 5A. The quantification of asystoles is based on detection in individual records of the diastolic interval lengths that are twice the median heart period, thus intervals larger than 0.7 s for yw, 0.8 s for SSlbeA and SSlbeAF, and 1.3 s for J43DR are recorded as asystolic events (Fig. 5A). The arrhythmia index, defined as the normalized standard deviation of the heart period, is also consistently higher in these eve repressed or mutant flies compared to the controls (Fig. 5B). Taken together, these observations suggest that the mesodermal eve expression is necessary for a normal set of adult epi/pericardial cells and in a yet to be determined non-autonomous fashion for a fully functional heart with a robust and regular myogenic heart rhythm.
Mesodermal eve repression in eme-lbe transgenic flies reproduces the cardiac abnormalities observed in eve meso-minus mutants. In addition, this genetic configuration is more amenable to genetic screening for modifiers capable of ameliorating heart function defects infiicted non-autonomously by the lack of eve expression in neighboring epi/pericardial cells or dorsal muscles. In order to test the feasibility of conducting such a screen, we examined SSlbeAF/+ flies crossed to a pilot set of heterozygous deficiencies to screen for second site modifiers of the SSlbeAF phenotype. We asked whether the myocardial dysfunction, observed in response to reduced mesodermal eve expression, could be rescued. In this pilot screen, most deficiencies aggravated or did not change the SSlbeAF phenotype (Suppl. Fig. S4, top 2 traces), except one that showed significant amelioration (Suppl. Fig. S4, bottom traces). Although more experiments are needed to confirm this result, these preliminary data suggest that efficient F1 screening with heterozygous deficiencies or with myocardial-specific expression of EP or RNAi lines (not shown) is indeed feasible and should provide clues as to the mechanisms of non-autonomous epi-myocardial interactions in normal heart function.
Discussion
A new tool for studying the role of non-myocardial eve expression in myocardial differentiation and establishment of heart function
During heart morphogenesis in Drosophila, mesodermal eve expression is required in a subset of cardiac progenitors that give rise to a subpopulation of epi/pericardial cells in the heart as well as a subset of dorsal somatic muscles. In the absence of mesodermal eve using eme-deficient rescue flies, the eve-dependent subset dorsal of muscles do not form and the number or epi/pericardial cells at larval and adult stages are reduced, which in turn seems to affect adult heart function (Fujioka et al., 2005). In that previous study, it was shown that heart rate and pacing-induced rate of heart failure was affected in eme-deficient rescue flies. Here, we have studied in detail the non-autonomous role of mesodermal eve on cardiac physiology (contractility, rhythmicity, etc.), and have generated new tools that can be used to investigate the genetic basis of epi/myocardial interactions. Since lbe represses mesodermal eve via the well-defined eme element (Han et al., 2002; Jagla et al., 2002), we directly targeted lbe expression to the mesodermal eve domains using this element (eme-lbe) resulting in phenotypes that were similar if not identical to that observed for eve meso-minus flies. Thus, it is likely that the main effect of eme-lbe is due to repression of mesodermal eve since both manipulations cause similar structural and functional defects. This is consistent with the notion that the lack of mesodermal eve prevents the formation of the derivatives of these Eve lineages, including a subset of the epi/pericardium.
It should be noted that the lineages of mesodermal eve-expressing progenitor cells (see Han and Bodmer, 2003) do not give rise to myocardial cells during development, nor are there new cells added to the inner tinman-expressing myocardial tube during metamorphosis (Molina and Cripps, 2001; Monier et al., 2005) and there is no evidence that the epi/pericardium contributes a cellular component to the myocardium at any time. Thus, the observed adult heart phenotype of the SSlbe lines, or of mesodermal eve deficiency lines, is likely due to an entirely non-autonomous effect of a compromised epi/pericardium (and missing dorsal muscles) on the establishment and functional integrity of the myocardium.
An important question is whether eve is required during development or (also) at adult stages to ensure normal heart function. Eve is present in a subset of epi/pericardial nuclei in the embryo (Fig. 1), but in the adult, Eve is present in the nucleolus of all pericardial cells (Suppl. Fig. S5; Frasch et al., 1987; Das et al., 2008a). Interestingly, eme-mediated lbe expression abolishes mesodermal eve only in the embryo but not in the adult (Fig. 1 and Suppl. Fig. S5), yet the adult heart structure and function are dramatically affected. Apparently, eme does not seem to mediate adult epi/pericardial eve expression (Suppl. Fig. S5, data not shown). Thus, we favor the interpretation that eve is required during development for the establishment of normal cardiac physiology in the adult. One aspect of heart function (overall heart rate in early pupae) does not seem to be altered by ablation of pericardial cells at larval stages (Das et al., 2008b). It will be interesting to see how other more subtle and specific aspects of the adult heart may be influenced by the ablation of all adult pericardial cells. Another question is whether the lack of mesodermal eve expression is equivalent to ablating the progenitors that normally give rise to Eve-pericardial and -muscle founder cells. To address this, we over-expressed the death genes reaper and hid using eme-Gal4. Remarkably, we observed the same constricted heart phenotype as with eliminating mesodermal eve expression by other means (Suppl. Fig. S6, compared to Fig. 2), suggesting that elimination of eve-expressing mesodermal cells abolishes the non-autonomous influences normally exerted of these cells onto the myocardium.
Eight Eve-positive cells anterior to the heart-associated epi/pericardial progenitors also express pericardial markers and they have recently been shown to give rise to the so-called “wing hearts”, two bilateral muscular organs in the thorax that pump hemolymph into the wings (Tögel et al., 2008). In the absence of eve expression in these progenitors, the flies are wing-heart-less and thus flightless, suggesting a role of eve in the formation of these pumps as well.
Mesodermal eve deficiency in the embryo causes a significantly reduced diameter of the adult heart in the abdominal A3 and A4 regions that is reminiscent of a ‘restrictive cardiomyopathy’-like phenotype also observed in certain myosin mutants (Cammarato et al., 2008). As the phenotypes could be reversed by co-over-expression of eve, we propose that the alterations in myocardial structure and function are likely due to the loss of mesodermal eve as opposed to a side effect of lbe overexpression. This conclusion is consistent with the rescue phenotype, accomplished by down-regulation of lbe in the eme-lbe background using a hairpin dsRNA construct (Fig. 2). As eve itself acts as a mesodermal repressor, loss of its repressive function is likely to cause the observed cardiac phenotype in the SSlbe lines. It will be interesting to study how repression by eve in the epi/pericardium influences the structure and function of the myocardium and whether this relationship is conserved in mammals. The eme-lbe flies provide a unique tool that allows us to address this question via transgenic manipulations. Genome-wide screens for non-autonomous modifiers can be conducted by driving any transgene in the myocardium of compromised eme-lbe lines. In a pilot experiment, we have demonstrated the feasibility of this approach (Suppl. Fig. S4).
Repression of mesodermal eve compromises cardiac performance
We further assessed the importance of mesodermal eve and an intact epi/pericardium on adult heart function. eme-lbe induced eve repression was found to trigger distinct physiological consequences, including a higher heart failure rate in response to cardiac pacing. In addition, we observe increased arrhythmias and incidences of asystoles suggesting that the non-autonomous influence of eve on the heart affects several distinct parameters of cardiac performance. Both the lack of eve in the developing epi/pericardium as well as the subsequent alterations in adult myocardial morphology may contribute to the observed physiological phenotypes. Although, we cannot formally exclude the possibility that the ‘unspecified’ EPC lineage without eve expression is entirely naïve as proposed by Fujioka et al. (2005), the finding that ablation of the mesodermal Eve progenitors results in a similarly constricted heart phenotype (Suppl. Fig. S6) further underlines a likely indispensible and non-autonomous contribution of the epi/pericardium to the establishment of a normal adult heart.
In vertebrates, signaling between the epicardium and the developing myocardium has been demonstrated (Chen et al., 2002; Lavine et al., 2005; Stuckmann et al., 2003) and this signaling likely plays a role in myocardial maturation and function (reviewed in Manner and Ruiz-Lozano, 2008). In order to test our hypothesis that the epi/pericardium regulates myocardial function in the fly, we analyzed in detail the heart contractions of mesodermal eve mutant flies. Indeed, we demonstrate that embryonic loss of mesodermal eve and reduced epi/pericardium provokes adult heart function defects. In addition, eve mutant adult flies exhibit compromised heart performance under stress conditions that may in part be due to the observed morphological changes of the myocardium. Based on these observations, we propose an essential, non-autonomous role for the epi/pericardium in developing a rhythmic heart beat and normal contractility.
Possible similarities between Drosophila Eve and vertebrate EVX2 function
It is possible that the epi/pericardial cell population in Drosophila shares some characteristics of the vertebrate epicardium (see also Fujioka et al., 2005). In vertebrates, there is also evidence for epicardial influences on the developing myocardium, although the physiology of the heart is affected is not known. Knockouts of epicardial-specific genes, such as the transcription factor Wilm’s tumor suppressor (WT-1) or α4-integrin, showed severe cardiac defects, most notably impaired epicardial development and myocardial thinning (Kreidberg et al., 1993; Yang et al., 1995; Merki et al., 2005; Zamora et al., 2007; Winter et al., 2007; Cai et al., 2008; Zhou et al., 2008). The expression patterns of these genes are largely restricted to the epicardium, suggesting that myocardium formation is dependent on signaling from the epicardium. More recently, epicardial RXR or β-catenin knockouts also cause a myocardial thinning phenotype (Merki et al., 2005; Zamora et al., 2007). Since the eve homolog Evx2 is expressed in the mouse epicardium (Fujioka et al., 2005), it is possible that Evx2 functions in a similar fashion in mammals during cardiogenesis and establishment of cardiac function, as eve does in flies.
Recently, signaling via pathways such as the retinoic acid (RA) or fibroblast growth factor (FGF) has been implicated in myocardial growth. One of the RA components, retinaldehyde dehydrogenase 2 (RALDH-2) is highly expressed in the epicardium, but not in the myocardium (Moss et al., 1998; Xavier-Neto et al., 2000) consistent with a role of this agent via the epicardium. While myocardial-specific knockouts of the retinoid receptor RXRα showed no abnormal myocardial phenotype, blocking of RA signaling from the epicardium in epicardial conditional RXRα mutants impairs myocyte proliferation (Chen et al., 1998; Merki et al., 2005; Stuckmann et al., 2003). These data suggest that the signaling from the epicardium plays a significant role in heart formation. Since eve is required in the epi/pericardium for myocardial maturation in flies, and its homolog Evx2 is expressed in the mammalian epicardium, the postulated epicardial-to-myocardial signaling might involve Evx2. Use of the Drosophila heart model should help identify additional genes that may be important in epicardial–myocardial interactions in vertebrates.
Acknowledgments
We are grateful to Barry Dickson and Georg Dietzl (Institute of Molecular Pathology, Vienna, Austria) for the gift of the UAS-lbe-RNAi Drosophila line. This work was supported by grants from the National Institutes of Health (NHLBI) to R.B and P.R.-L.
Appendix A. Supplementary data
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.ydbio.2009.02.013.
References
- Azpiazu N, Frasch M. tinman and bagpipe: two homeo box genes that determine cell fates in the dorsal mesoderm of Drosophila. Genes Dev. 1993;7:1325–1340. doi: 10.1101/gad.7.7b.1325. [DOI] [PubMed] [Google Scholar]
- Bodmer R. The gene tinman is required for specification of the heart and visceral muscles in Drosophila. Development. 1993;118:719–729. doi: 10.1242/dev.118.3.719. [DOI] [PubMed] [Google Scholar]
- Bodmer R. Heart development in Drosophila and its relationship to vertebrate systems. Trends Cardiovasc Med. 1995;5:21–28. doi: 10.1016/1050-1738(94)00032-Q. [DOI] [PubMed] [Google Scholar]
- Bodmer R, Frasch M. Genetic determination of Drosophila heart development. In: Rosenthal N, Harvey R, editors. Heart Development. Academic Press; 1999. pp. 65–99. [Google Scholar]
- Bodmer R, Wessells RJ, Johnson EC, Dowse H. Heart development and function. In: Gilbert LI, Iatrou K, Gill S, editors. Comprehensive Molecular Insect Science (volumes 1–7) Vol. 2. Elsevier; 2005. pp. 199–250. [Google Scholar]
- Bour BA, O’Brien MA, Lockwood WL, Goldstein ES, Bodmer R, Taghert PH, Abmayr SM, Nguyen HT. Drosophila MEF2, a transcription factor that is essential for myogenesis. Genes Dev. 1995;9:730–741. doi: 10.1101/gad.9.6.730. [DOI] [PubMed] [Google Scholar]
- Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- Cai CL, Martin JC, Sun Y, Cui L, Wang L, Ouyang K, Yang L, Bu L, Liang X, Zhang X, Stallcup WB, Denton CP, McCulloch A, Chen J, Evans SM. A myocardial lineage derives from Tbx18 epicardial cells. Nature. 2008;454:104–108. doi: 10.1038/nature06969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cammarato A, Dambacher CM, Knowles AF, Kronert WA, Bodmer R, Ocorr K, Bernstein SI. Myosin transducer mutations differentially affect motor function, myofibril structure, and the performance of skeletal and cardiac muscles. Mol Biol Cell. 2008;19:553–562. doi: 10.1091/mbc.E07-09-0890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmena A, Gisselbrecht S, Harrison J, Jiménez F, Michelson AM. Combinatorial signaling codes for the progressive determination of cell fates in the Drosophila embryonic mesoderm. Genes Dev. 1998;12:3910–3922. doi: 10.1101/gad.12.24.3910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chartier A, Zaffran S, Astier M, Sémériva M, Gratecos D. Pericardin, a Drosophila type IV collagen-like protein is involved in the morphogenesis and maintenance of the heart epithelium during dorsal ectoderm closure. Development. 2002;129:3241–3253. doi: 10.1242/dev.129.13.3241. [DOI] [PubMed] [Google Scholar]
- Chen J, Kubalak SW, Chien KR. Ventricular muscle-restricted targeting of the RXRalpha gene reveals a non-cell-autonomous requirement in cardiac chamber morphogenesis. Development. 1998;125:1943–1949. doi: 10.1242/dev.125.10.1943. [DOI] [PubMed] [Google Scholar]
- Chen T, Chang T, Kang J, Choudhary B, Makita T, Tran CM, Burch J, Eid H, Sucov HM. Epicardial induction of fetal cardiomyocyte proliferation via a retinoic acid-inducible trophic factor. Dev Biol. 2002;250:198–207. doi: 10.1006/dbio.2002.0796. [DOI] [PubMed] [Google Scholar]
- Cripps RM, Olson EN. Control of cardiac development by an evolutionarily conserved transcriptional network. Dev Biol. 2002;246:14–28. doi: 10.1006/dbio.2002.0666. [DOI] [PubMed] [Google Scholar]
- Curtis NJ, Ringo JM, Dowse HB. Morphology of the pupal heart, adult heart, and associated tissues in the fruit fly, Drosophila melanogaster. J Morphol. 1999;240:225–235. doi: 10.1002/(SICI)1097-4687(199906)240:3<225::AID-JMOR2>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- Das D, Ashoka D, Aradhya R, Inamdar M. Gene expression analysis in post-embryonic pericardial cells of Drosophila. Gene Expr Patterns. 2008a;8:199–205. doi: 10.1016/j.gep.2007.10.008. [DOI] [PubMed] [Google Scholar]
- Das D, Aradhya R, Ashoka D, Inamdar M. Post-embryonic pericardial cells of Drosophila are required for overcoming toxic stress but not for cardiac function or adult development. Cell Tissue Res. 2008b;331:565–570. doi: 10.1007/s00441-007-0518-z. [DOI] [PubMed] [Google Scholar]
- Dulcis D, Levine RB. Innervation of the heart of the adult fruit fly, Drosophila melanogaster. J Comp Neurol. 2003;465:560–578. doi: 10.1002/cne.10869. [DOI] [PubMed] [Google Scholar]
- Frasch M. Induction of visceral and cardiac mesoderm by ectodermal Dpp in the early Drosophila embryo. Nature. 1995;374:464–467. doi: 10.1038/374464a0. [DOI] [PubMed] [Google Scholar]
- Frasch M, Hoey T, Rushlow C, Doyle H, Levine M. Characterization and localization of the even-skipped protein of Drosophila. EMBO J. 1987;6:749–759. doi: 10.1002/j.1460-2075.1987.tb04817.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujioka M, Emi-Sarker Y, Yusibova GL, Goto T, Jaynes JB. Analysis of an even-skipped rescue transgene reveals both composite and discrete neuronal and early blastoderm enhancers, and multi-stripe positioning by gap gene repressor gradients. Development. 1999;126:2527–2538. doi: 10.1242/dev.126.11.2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujioka M, Wessells RJ, Han Z, Liu J, Fitzgerald K, Yusibova GL, Zamora M, Ruiz-Lozano P, Bodmer R, Jaynes JB. Embryonic even skipped-dependent muscle and heart cell fates are required for normal adult activity, heart function, and lifespan. Circ Res. 2005;97:1108–1114. doi: 10.1161/01.RES.0000191546.08532.B2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gajewski K, Zhang Q, Choi CY, Fossett N, Dang A, Kim YH, Kim Y, Schulz RA. Pannier is a transcriptional target and partner of Tinman during Drosophila cardiogenesis. Dev Biol. 2001;233:425–436. doi: 10.1006/dbio.2001.0220. [DOI] [PubMed] [Google Scholar]
- Halfon MS, Carmena A, Gisselbrecht S, Sackerson CM, Jiménez F, Baylies MK, Michelson AM. Ras pathway specificity is determined by the integration of multiple signal-activated and tissue-restricted transcription factors. Cell. 2000;103:63–74. doi: 10.1016/s0092-8674(00)00105-7. [DOI] [PubMed] [Google Scholar]
- Han Z, Bodmer R. Myogenic cells fates are antagonized by Notch only in asymmetric lineages of the Drosophila heart, with or without cell division. Development. 2003;130:3039–3051. doi: 10.1242/dev.00484. [DOI] [PubMed] [Google Scholar]
- Han Z, Fujioka M, Su MT, Liu M, Jaynes JB, Bodmer R. Transcriptional integration of competence modulated by mutual repression generates cell-type specificity within the cardiogenic mesoderm. Dev Biol. 2002;252:225–240. doi: 10.1006/dbio.2002.0846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jagla K, Frasch M, Jagla T, Dretzen G, Bellard F, Bellard M. ladybird, a new component of the cardiogenic pathway in Drosophila required for diversification of heart precursors. Development. 1997;124:3471–3479. doi: 10.1242/dev.124.18.3471. [DOI] [PubMed] [Google Scholar]
- Jagla T, Bidet Y, Ponte JP, Dastugue B, Jagla K. Cross-repressive interactions of identity genes are essential for proper specification of cardiac and muscular fates in Drosophila. Development. 2002;129:1037–1047. doi: 10.1242/dev.129.4.1037. [DOI] [PubMed] [Google Scholar]
- Klinedinst SL, Bodmer R. Gata factor Pannier is required to establish competence for heart progenitor formation. Development. 2003;130:3027–3038. doi: 10.1242/dev.00517. [DOI] [PubMed] [Google Scholar]
- Knirr S, Frasch M. Molecular integration of inductive and mesoderm-intrinsic inputs governs even-skipped enhancer activity in a subset of pericardial and dorsal muscle progenitors. Dev Biol. 2001;238:13–26. doi: 10.1006/dbio.2001.0397. [DOI] [PubMed] [Google Scholar]
- Kreidberg JA, Sariola H, Loring JM, Maeda M, Pelletier J, Housman D, Jaenisch R. WT-1 is required for early kidney development. Cell. 1993;74:679–691. doi: 10.1016/0092-8674(93)90515-r. [DOI] [PubMed] [Google Scholar]
- Lavine KJ, Yu K, White AC, Zhang X, Smith C, Partanen J, Ornitz DM. Endocardial and epicardial derived FGF signals regulate myocardial proliferation and differentiation in vivo. Dev Cell. 2005;8:85–95. doi: 10.1016/j.devcel.2004.12.002. [DOI] [PubMed] [Google Scholar]
- Lilly B, Zhao B, Ranganayakulu G, Paterson BM, Schulz RA, Olson EN. Requirement of MADS domain transcription factor D-MEF2 for muscle formation in Drosophila. Science. 1995;267:688–693. doi: 10.1126/science.7839146. [DOI] [PubMed] [Google Scholar]
- Liu J, Qian L, Han Z, Wu X, Bodmer R. Spatial specificity of mesodermal even-skipped expression relies on multiple repressor sites. Dev Biol. 2008;313:876–886. doi: 10.1016/j.ydbio.2007.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M, Su M, Lyons GE, Bodmer R. Functional conservation of zinc-finger homeodomain gene zfh1/SIP1 in Drosophila heart development. Dev Genes Evol. 2006;216:683–693. doi: 10.1007/s00427-006-0096-1. [DOI] [PubMed] [Google Scholar]
- Lo PC, Frasch M. A role for the COUP-TF-related gene seven-up in the diversification of cardioblast identities in the dorsal vessel of Drosophila. Mech Dev. 2001;104:49–60. doi: 10.1016/s0925-4773(01)00361-6. [DOI] [PubMed] [Google Scholar]
- Lo PC, Frasch M. Establishing A–P polarity in the embryonic heart tube: a conserved function of Hox genes in Drosophila and vertebrates? Trends Cardiovasc Med. 2003;13:182–187. doi: 10.1016/s1050-1738(03)00074-4. [DOI] [PubMed] [Google Scholar]
- Macdonald PM, Ingham P, Struhl G. Isolation, structure, and expression of even-skipped: a second pair-rule gene of Drosophila containing a homeo box. Cell. 1986;47:721–734. doi: 10.1016/0092-8674(86)90515-5. [DOI] [PubMed] [Google Scholar]
- Manner J, Ruiz-Lozano P. Development and function of the epicardium. In: Bodmer R, editor. Advances in Developmental Biology. Vol. 18. Elsevier; 2008. pp. 334–357. [Google Scholar]
- Manner J, Perez-Pomares JM, Macias D, Munoz-Chapuli R. The origin, formation and developmental significance of the epicardium: a review. Cells Tissues Organs. 2001;169:89–103. doi: 10.1159/000047867. [DOI] [PubMed] [Google Scholar]
- Merki E, Zamora M, Raya A, Kawakami Y, Wang J, Zhang X, Burch J, Kubalak SW, Kaliman P, Belmonte JC, Chien KR, Ruiz-Lozano P. Epicardial retinoid X receptor alpha is required for myocardial growth and coronary artery formation. Proc Natl Acad Sci U S A. 2005;102:18455–18460. doi: 10.1073/pnas.0504343102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mery A, Taghli-Lamallem O, Clark KA, Beckerle MC, Wu X, Ocorr K, Bodmer R. The Drosophila muscle LIM protein, Mlp84B, is essential for cardiac function. J Exp Biol. 2008;211:15–23. doi: 10.1242/jeb.012435. [DOI] [PubMed] [Google Scholar]
- Mikawa T, Gourdie RG. Pericardial mesoderm generates a population of coronary smooth muscle cells migrating into the heart along with ingrowth of the epicardial organ. Dev Biol. 1996;174:221–232. doi: 10.1006/dbio.1996.0068. [DOI] [PubMed] [Google Scholar]
- Miskolczi-McCallum CM, Scavetta RJ, Svendsen PC, Soanes KH, Brook WJ. The Drosophila melanogaster T-box genes midline and H15 are conserved regulators of heart development. Dev Biol. 2005;278:459–472. doi: 10.1016/j.ydbio.2004.11.026. [DOI] [PubMed] [Google Scholar]
- Mlodzik M, Hiromi Y, Weber U, Goodman CS, Rubin GM. The Drosophila seven-up gene, a member of the steroid receptor gene superfamily, controls photoreceptor cell fates. Cell. 1990;60:211–224. doi: 10.1016/0092-8674(90)90737-y. [DOI] [PubMed] [Google Scholar]
- Molina MR, Cripps RM. Ostia, the inflow tracts of the Drosophila heart, develop from a genetically distinct subset of cardial cells. Mech Dev. 2001;109:51–59. doi: 10.1016/s0925-4773(01)00509-3. [DOI] [PubMed] [Google Scholar]
- Monier B, Astier M, Sémériva M, Perrin L. Steroid-dependent modification of Hox function drives myocyte reprogramming in the Drosophila heart. Development. 2005;132:5283–5293. doi: 10.1242/dev.02091. [DOI] [PubMed] [Google Scholar]
- Morin X, Daneman R, Zavortink M, Chia W. A protein trap strategy to detect GFP-tagged proteins expressed from their endogenous loci in Drosophila. Proc Natl Acad Sci U S A. 2001;98:15050–15055. doi: 10.1073/pnas.261408198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss JB, Xavier-Neto J, Shapiro MD, Nayeem SM, McCaffery P, Dräger UC, Rosenthal N. Dynamic patterns of retinoic acid synthesis and response in the developing mammalian heart. Dev Biol. 1998;199:55–71. doi: 10.1006/dbio.1998.8911. [DOI] [PubMed] [Google Scholar]
- Muñoz-Chápuli R, Macías D, González-Iriarte M, Carmona R, Atencia G, Pérez-Pomares JM. [The epicardium and epicardial-derived cells: multiple functions in cardiac development. Rev Esp Cardiol. 2002;55:1070–1082. doi: 10.1016/s0300-8932(02)76758-4. [DOI] [PubMed] [Google Scholar]
- Nguyen HT, Bodmer R, Abmayr SM, McDermott JC, Spoerel NA. D-mef2: a Drosophila mesoderm-specific MADS box-containing gene with a biphasic expression profile during embryogenesis. Proc Natl Acad Sci U S A. 1994;91:7520–7524. doi: 10.1073/pnas.91.16.7520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ocorr K, Reeves NL, Wessells RJ, Fink M, Chen HS, Akasaka T, Yasuda S, Metzger JM, Giles W, Posakony JW, Bodmer R. KCNQ potassium channel mutations cause cardiac arrhythmias in Drosophila that mimic the effects of aging. Proc Natl Acad Sci U S A. 2007a;104:3943–3948. doi: 10.1073/pnas.0609278104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ocorr KA, Crawley T, Gibson G, Bodmer R. Genetic variation for cardiac dysfunction in Drosophila. PLoS ONE. 2007b;2:e601. doi: 10.1371/journal.pone.0000601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian L, Liu J, Bodmer R. Neuromancer Tbx20-related genes (H15/midline) promote cell fate specification and morphogenesis of the Drosophila heart. Dev Biol. 2005;279:509–524. doi: 10.1016/j.ydbio.2005.01.013. [DOI] [PubMed] [Google Scholar]
- Qian L, Liu J, Bodmer R. Heart development in Drosophila. In: Bodmer R, editor. Advances in Developmental Biology: Cardiac Development. Elsevier Publishing; 2008. pp. 1–29. [Google Scholar]
- Reim I, Frasch M. The Dorsocross T-box genes are key components of the regulatory network controlling early cardiogenesis in Drosophila. Development. 2005;132:4911–4925. doi: 10.1242/dev.02077. [DOI] [PubMed] [Google Scholar]
- Reim I, Mohler JP, Frasch M. Tbx20-related genes, mid and H15, are required for tinman expression, proper patterning, and normal differentiation of cardioblasts in Drosophila. Mech Dev. 2005;122:1056–1069. doi: 10.1016/j.mod.2005.04.006. [DOI] [PubMed] [Google Scholar]
- Rizki TM, Rizki RM. Larval adipose tissue of homoeotic bithorax mutants of Drosophila. Dev Biol. 1978;65:476–482. doi: 10.1016/0012-1606(78)90042-8. [DOI] [PubMed] [Google Scholar]
- Singleton K, Woodruff RI. The osmolarity of adult Drosophila hemolymph and its effect on oocyte-nurse cell electrical polarity. Dev Biol. 1994;161:154–167. doi: 10.1006/dbio.1994.1017. [DOI] [PubMed] [Google Scholar]
- Stuckmann I, Evans S, Lassar AB. Erythropoietin and retinoic acid, secreted from the epicardium, are required for cardiac myocyte proliferation. Dev Biol. 2003;255:334–349. doi: 10.1016/s0012-1606(02)00078-7. [DOI] [PubMed] [Google Scholar]
- Su MT, Fujioka M, Goto T, Bodmer R. The Drosophila homeobox genes zfh-1 and even-skipped are required for cardiac-specific differentiation of a numb-dependent lineage decision. Development. 1999;126:3241–3251. doi: 10.1242/dev.126.14.3241. [DOI] [PubMed] [Google Scholar]
- Taghli-Lamallem O, Akasaka T, Hogg G, Nudel U, Yaffe D, Chamberlain JS, Ocorr K, Bodmer R. Dystrophin deficiency in Drosophila reduces lifespan and causes a dilated cardiomyopathy phenotype. Aging Cell. 2008;7:237–249. doi: 10.1111/j.1474-9726.2008.00367.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tögel M, Pass G, Paululat A. The Drosophila wing hearts originate from pericardial cells and are essential for wing maturation. Dev Biol. 2008;318:29–37. doi: 10.1016/j.ydbio.2008.02.043. [DOI] [PubMed] [Google Scholar]
- Wang JW, Wong AM, Flores J, Vosshall LB, Axel R. Two-photon calcium imaging reveals an odor-evoked map of activity in the fly brain. Cell. 2003;112:271–282. doi: 10.1016/s0092-8674(03)00004-7. [DOI] [PubMed] [Google Scholar]
- Ward EJ, Skeath JB. Characterization of a novel subset of cardiac cells and their progenitors in the Drosophila embryo. Development. 2000;127:4959–4969. doi: 10.1242/dev.127.22.4959. [DOI] [PubMed] [Google Scholar]
- Wessells RJ, Bodmer R. Screening assays for heart function mutants in Drosophila. Biotechniques. 2004;37:58–60. doi: 10.2144/04371ST01. [DOI] [PubMed] [Google Scholar]
- Wessells RJ, Fitzgerald E, Cypser JR, Tatar M, Bodmer R. Insulin regulation of heart function in aging fruit flies. Nat Genet. 2004;36:1275–1281. doi: 10.1038/ng1476. [DOI] [PubMed] [Google Scholar]
- Winter EM, Grauss RW, Hogers B, van Tuyn J, van der Geest R, Lie-Venema H, Steijn RV, Maas S, DeRuiter MC, deVries AA, Steendijk P, Doevendans PA, van der Laarse A, Poelmann RE, Schalij MJ, Atsma DE, Gittenberger-de Groot AC. Preservation of left ventricular function and attenuation of remodeling after transplantation of human epicardium-derived cells into the infarcted mouse heart. Circulation. 2007;116:917–927. doi: 10.1161/CIRCULATIONAHA.106.668178. [DOI] [PubMed] [Google Scholar]
- Wu X, Golden K, Bodmer R. Heart development in Drosophila requires the segment polarity gene wingless. Dev Biol. 1995;169:619–628. doi: 10.1006/dbio.1995.1174. [DOI] [PubMed] [Google Scholar]
- Xavier-Neto J, Shapiro MD, Houghton L, Rosenthal N. Sequential programs of retinoic acid synthesis in the myocardial and epicardial layers of the developing avian heart. Dev Biol. 2000;219:129–141. doi: 10.1006/dbio.1999.9588. [DOI] [PubMed] [Google Scholar]
- Yang JT, Rayburn H, Hynes RO. Cell adhesion events mediated by alpha 4 integrins are essential in placental and cardiac development. Development. 1995;121:549–560. doi: 10.1242/dev.121.2.549. [DOI] [PubMed] [Google Scholar]
- Yi P, Han Z, Li X, Olson EN. The mevalonate pathway controls heart formation in Drosophila by isoprenylation of Ggamma1. Science. 2006;313:1301–1303. doi: 10.1126/science.1127704. [DOI] [PubMed] [Google Scholar]
- Zaffran S, Reim I, Qian L, Lo PC, Bodmer R, Frasch M. Cardioblast-intrinsic Tinman activity controls proper diversification and differentiation of myocardial cells in Drosophila. Development. 2006;133:4073–4083. doi: 10.1242/dev.02586. [DOI] [PubMed] [Google Scholar]
- Zamora M, Männer J, Ruiz-Lozano P. Epicardium-derived progenitor cells require beta-catenin for coronary artery formation. Proc Natl Acad Sci U S A. 2007;104:18109–18114. doi: 10.1073/pnas.0702415104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeitouni B, Senatore S, Severac D, Aknin C, Semeriva M, Perrin L. Signalling pathways involved in adult heart formation revealed by gene expression profiling in Drosophila. PLoS Genet. 2007;3:1907–1921. doi: 10.1371/journal.pgen.0030174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Bernstein SI. Spatially and temporally regulated expression of myosin heavy chain alternative exons during Drosophila embryogenesis. Mech Dev. 2001;101:35–45. doi: 10.1016/s0925-4773(00)00549-9. [DOI] [PubMed] [Google Scholar]
- Zhou B, Ma Q, Rajagopal S, Wu SM, Domian I, Rivera-Feliciano J, Jiang D, von Gise A, Ikeda S, Chien KR, Pu WT. Epicardial progenitors contribute to the cardiomyocyte lineage in the developing heart. Nature. 2008;454:109–113. doi: 10.1038/nature07060. [DOI] [PMC free article] [PubMed] [Google Scholar]