Abstract
IL-23 plays an important role in autoimmune tissue inflammation and induces the generation of not fully characterized effector cells that mediate protection against pathogens. Here, we established the essential role of IL-23R in the host response against intracellular pathogens. IL-23 was critical for the expansion/maintenance of γδ and double negative (DN) αβ T cells. These cells were rapidly recruited to the site of infection, and produced large amounts of IL-17, IFN-γ and TNF-α. Notably, DN T cells transferred into L. monocytogenes-infected RAG2−/− mice prevented bacteria growth confirming their protective role against intracellular pathogens. Our results show that IL-23 regulates the function of IL-17-producing γδ and DN T cells, two essential components of the early protective immune response directed against intracellular pathogens.
IL-23 is secreted by macrophages and dendritic cells (DCs) in response to microbial products and inflammatory cytokines(1). IL-23 is composed of a specific p19 subunit and the p40 subunit that is shared with IL-12. Its effects are mediated by a receptor composed of the IL-12Rβ 1 and the specific IL-23R. Although we have witnessed an intense investigation on the involvement of IL-23 in the pathogenesis of many autoimmune diseases over the past few years, including experimental autoimmune encephalitis (EAE)(2–5), rheumatoid arthritis(6–8) and inflammatory bowel disease(9, 10), increasing evidence suggests that IL-23 is also critical in mediating protection against pathogens(11). In addition, it is now clear that IL-23 is necessary for the full differentiation and maintenance of CD4+ TH17 effector cells(4, 12). CD4+ TH17 cells along with CD8+, NK T, γδ-TCR+ and αβ–TCR+ CD4− CD8− (double negative (DN)) T cells(13–16) are collectively named neutrophil-regulatory “Tn” cells due to their high production of IL-17 which can induce G-CSF-dependent neutrophilia(17–19).
IL-17 is required to initiate the production of inflammatory cytokines important for granulopoiesis and neutrophil chemotaxis and is essential for the development of autoimmune diseases. On the other hand, it is crucial for protection against pathogens such as Klebsiella pneumoniae, Candida albicans, Toxoplasma gondii (20–25). However, it has been unclear which cells produce IL-17 at the early stages before IL-23R+ CD4+ TH17 cells develop later during infection. Similarly, IL-23 confers protective immune mechanisms against pathogens such as Salmonella enterica, Escherichia coli, K. pneumoniae or Citrobacter rodentium by inducing the production of IL-22 and activating cells that still have to be fully characterized(11, 26–28).
In this study we investigated the role of IL-23R in the host response to L. monocytogenes, a useful model for studying protective mechanisms against intracellular pathogens. The early response, which occurs during the first 48 h after the onset of primary infection, has been attributed to macrophages, DCs, NK cells and neutrophils that limit growth of the organism. The late listericidal response begins around day 4 and involves CD4+ TH1 and cytotoxic CD8+ T cells(29, 30). Since IL-23 is produced by macrophages and DCs very rapidly during infection, IL-23 might bind IL-23R on the surface of cells at early stages of inflammation in order to control the bacteria until the late adaptive immune response develops several days later. Thus, we propose to identify early IL-23R responding cells and elucidate their role in the control of bacterial infections.
Due to he low expression of IL-23R and the lack of appropriate reagents to track IL-23R bearing cells in vivo, it has been very difficult to identify IL-23R expressing and IL-23 responsive cells in vivo. To detect the cell types responding to IL-23 and their effector functions during infection, we used a knock-in “reporter” mouse. In heterozygous mice, IL-23R-expressing cells can be followed by their expression of GFP, and when bred as homozygotes, the deletion of the IL-23R abrogates their responsiveness to IL-23(5). Here, we report that IL-23R regulates the function of specific IL-17-producing-γδ-TCR+ and αβ-TCR+ CD4− CD8− DN T cells which contribute to cover the gap between the early and late immune responses in order to fight intracellular bacterial infections.
MATERIAL AND METHODS
Mice
C57BL/6 (WT), β2M−/−, RAG2−/− and IL6 −/− mice were obtained from Jackson Laboratories and IL-17F-CreEYFP were a gift from A. Waisman. Heterozygous IL-23R-GFP.KI and IL-23R−/− mice were generated as previously described(5). Mice were housed in a conventional, pathogen-free facility at 65 Landsdowne Street, Cambridge, MA and at the Harvard Institute of Medicine, 77 Ave Louis Pasteur, Boston, MA. For F. tularensis LVS or L. Monocytogenes experiments, mice were housed in the BSL2 animal facility at Harvard School of Public Health, 651 Huntington Ave, Boston, MA or 65 Landsdowne Street, Cambridge, MA, respectively. All experiments were performed in accordance with guidelines from the standing committee of animals at Harvard Medical School.
Microorganisms and bacterial infection
For aerosol infection, 1.5×109/ml F. tularensis LVS (New England Regional Center of Excellence for Biodefense and Emerging Infectious Disease) were diluted in 25 ml Mueller-Hilton broth, and were grown at 37°C for 5 h prior to infection. The log-phase bacteria were re-suspended in 20% glycerol at 8×108/ml concentration. Mice were exposed to the aerosol-containing bacteria using nose-only exposure unit (In-Tox Products, Albuquerque, NM) for 20 min using Lovelace nebulizer, followed by 5 minutes of air-only. 24 h later, lungs from 2 mice were homogenized and plated on Mueller-Hilton plates to determine the colony forming units (CFU) of bacteria recovered from the lung. Generally, 104 CFU were recovered from the lungs of infected mice using this protocol.
L. monocytogenes, strain EGD (ATCC) was inoculated in WT mice, fresh isolates were obtained from infected spleens, grown in brain heart infusion broth, resuspended in PBS and stored at −80°C in small aliquots. Mice were infected i.p. with 105 CFU of L. monocytogenes per mouse for RAG2−/− mice or 106 CFU for WT, IL-23RGFP.KI, IL-17F-CreEYFP and IL-23R−/− mice. For bacterial counts, mice were sacrificed on day 3 after infection and their livers were collected and homogenized in PBS and plated onto brain heart infusion agar.
MOG35-55/CFA immunization and in vivo BrdU incorporation
Mice were immunized subcutaneously with 100 μl of an emulsion containing 100 μg of MOG35-55 peptide (MEVGWYRSPFSRVVHLYRNGK) and CFA. 4 days after immunization cell suspension of LN cells were analyzed. For in vivo proliferation assay, unimmunized or MOG immunized mice obtained 2 mg/mouse of BrdU i.p. every other day. For the detection of BrdU incorporation, samples were permeabilized with Cytofix/Cytoperm Plus buffer (BD Pharmingen) and treated with 30 mg DNase for 60 min at 37°C to expose BrdU epitopes. After washing, cells were stained with anti-BrdU APC (BD Pharmingen) for 45 min at room temperature and then washed.
DN T cells activation and differentiation in vitro
Generation of double negative T cells was performed culturing LN cells from IL-23R-GFP.KI mice at a concentration of 4 × 106/ml in the presence of soluble anti-CD3 (1 μg/ml) with IL-23 (30 ng/ml) for 6 d.
RNA was extracted 48h after in vitro stimulation using RNAeasy columns (Qiagen, Valencia, CA) and subjected to quantitative RT-PCR according to the manufacturer’s instruction (Applied Biosystems). Primer/probe mixtures of mouse IL-17A, IL-23R, IFN-γ, T-bet, and ROR-γt, CD4 and CD8 were obtained from Applied Biosystems.
Cytokine analysis
Cytokines from culture supernatants were determined by either ELISA according to the manufacturer’s instructions (Biolegend) or bead array (BD Bioscience). For the measurement of cytokines in the peritoneal fluid, mice were infected i.p. with L. monocytogenes. After 24 h, we performed a peritoneal lavage with 2 ml of PBS and harvested after gentle massage. PEC were collected by centrifugation cultured overnight plus PMA (50 ng/ml) and Ionomycin 1μg/ml (both Sigma) at a concentration of 4.5 × 106/ml. Concentration of IL-17 and INF-γ was determined by ELISA.
For the intracellular cytokine staining, cells were re-stimulated with PMA 50 ng/ml, Ionomycin 1μg/ml (Sigma), and Golgi Stop (1μl/ml, BD Bioscience) at 37° C/10% CO2 for 4 h followed by surface and intracellular staining according to the manufacturer’s (BD Bioscience) instruction and cells were analyzed with a FACS Calibur (BD Biosciences).
Cellular phenotypic analysis by flow cytometry
PEC, splenocytes, LN cells, lung and liver mononuclear cells, Lamina propria lymphocytes (LPL)(5) were collected from uninfected and LVS or L. monocytogenes infected mice. Viable cells were stained with CD4, CD8, B220, NK1.1, CD11b, CD11c, δ-TCR and β-TCR antibodies (all purchased from BD Biosciences Pharmingen) or PBS57-CD1d loaded tetramers (NIH Tetramer Core Facility; Emory Vaccine Center at Yerkes, Atlanta, GA). The percentage of IL-23 (GFP)+ cells was determined, and the absolute numbers of IL-23R (GFP)+ cells was calculated using the following formula: % surface marker+GFP+(live cell gate) × total number of cells.
In vitro macrophage killing assays
WT and β2M−/− bone marrow cells were plated at 2×106/ml in a 48-well plate and grown in macrophage DMEM complete medium supplemented with 10% heat-inactivated FBS, 10% L-929 conditioned supernatant, 0.2 mM L-glutamine, 1 mM HEPES buffer and 0.1 mM non-essential amino acids. 7 d after in vitro differentiation, macrophages were infected with LVS at MOI 10 for 2 h at 37°C. Following infection macrophages were washed twice with warm media, and incubated with 50 μg/ml of gentamicin for 45 min at 37°C to kill extracellular bacteria. After extensive washing, LVS-infected macrophages were cultured with media only or in the presence of DN T cells isolated from the spleens of naïve or immune mice (isolated 1 month post-infection). DN cells were isolated after negative selection using magnetic beads against CD4, CD8, CD19, δ–TCR, NK1.1 and CD49b (clone DX5) purchased from Stem Cell Technologies). The negative fraction was washed and added to the infected macrophages at 1:3 ratio (1 DN: 3 MΦ). LVS-infected bone marrow derived macrophages (BMMΦ) were co-cultured with effector cells for 72 hours and the number of intracellular bacteria was determined after lysing infected macrophages in water for 3 minutes and plating on Mueller-Hilton plates.
Statistics
Histograms generally show the mean ± SD of results from at least 3 separate experiments. Significance of differences between 2 series of results was assessed using the two tail Student’s unpaired t test.
RESULTS
IL-23R expressing γδ-TCR+ and αβ-TCR+ CD4− CD8− (DN) T cells are recruited to the peritoneal cavity in response to L. monocytogenes infection
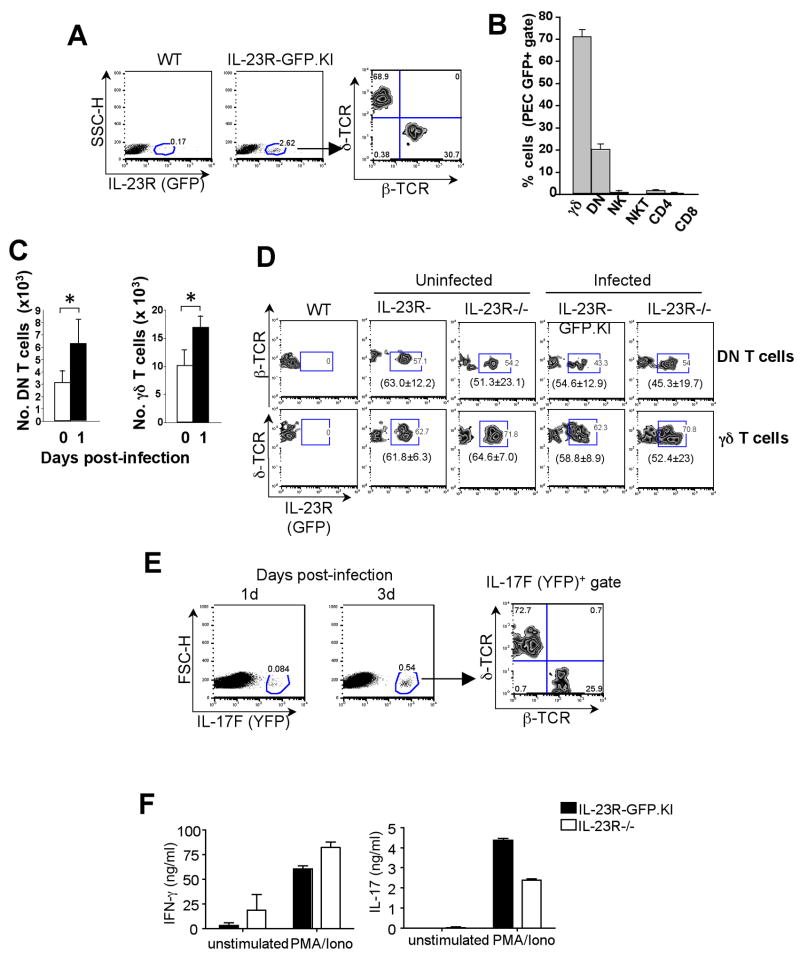
To investigate the local expression of IL-23R at the site of infection we examined the expression of IL-23R (GFP) in peritoneal exudate cells (PEC) of IL-23R-GFP.KI mice 1 d after intraperitoneally (i.p.) infection L. monocytogenes. A small percentage of these PEC expressed IL-23R (~2.6%) (Fig. 1A). The majority of these GFP+ cells (68.9%) expressed the γδ-TCR, while the remaining (30.7%) GFP+ cells were positive for αβ–TCR. Surprisingly most of the GFP+ αβ-TCR+ cells did not express CD4, CD8 co-receptors or any of the NK T cell markers (Fig. 1B). Since these cells lacked expression of either CD4, or CD8 or NK1.1 co-receptors, they were characterized as IL-23R+αβ–TCR+ double negative (DN) T cells (Fig. 1A). Furthermore, we observed that γδ and DN T cells were rapidly recruited to the peritoneal cavity of IL-23R-GFP.KI mice in response to infection (Fig. 1C). To determine whether this early recruitment of γδ and DN T cells to the site of infection is driven by IL-23R, we infected IL-23R-GFP.KI and IL-23R deficient mice with L. monocytogenes and assessed the number of PEC 24 h after infection. However, we did not find any significant difference in numbers of total or IL-23R (GFP) expressing γδ or DN T cells (Fig. 1D and data not shown). Additionally, similar numbers of neutrophils, macrophages or dendritic cells recruited to the peritoneal cavity were obtained from either IL-23R-GFP.KI or IL-23R−/− mice (data not shown). These experiments showed that the initial recruitment of immune cells to the peritoneum was independent of IL-23R in case of L. monocytogenes infection.
Figure 1. Expression of IL-23R in the peritoneal cavity of L. monocytogenes-infected mice.
Mice were infected via the intraperitoneal route with viable L. monocytogenes. (A, B, C, D and F) 24 h or 72 h (E) after infection. PEC were collected from WT, IL-23R-GFP.KI or IL-23R−/− (A, B, C, D and F) or IL-17F-CreEYFP (E) mice and β–TCR and δ–TCR surface stainings were performed. (A) IL-23R expression was analyzed in PEC of IL-23R-GFP.KI mice (B) Percentages of IL-23R (GFP)+ expressing cells were calculated within δ–TCR+, DN, CD4+, CD8+, NK1.1+, CD49b+ cell populations isolated from the peritoneal cavity of infected IL-23R-GFP.KI mice. The bars represent mean ± SD of 3 independent experiments (3 mice/group/experiment). (C) Histograms represent total numbers of γδ and DN collected from the peritoneum of IL-23R-GFP.KI mice 1d after infection. [*p<0.05 (two tail Student t-test)]. (D) The zebra plots represent the IL-23R (GFP) expression of peritoneal γδ or DN T cells from IL-23R-GFP.KI or IL-23R−/− mice 1 day after infection. In brackets, mean ± SD of 3 experiments with 3 mice/group is shown. (E) 1 d after infection, IL-23R-GFP.KI or IL-23R−/− PEC were isolated and stimulated or not with PMA/Ionomycin for an additional 18 h. Supernatants were collected and cytokine ELISA was performed for IL-17 and IFN-γ. Data shown are representative of 2 experiments with 3 mice/group. (F) 4× 106 PEC where cultured in the presence of PMA/Ionomicin. After 24h the supernatants were collected and ELISA for IL-17 and INF-γ were performed.
Identification of IL-17-producing cells in the peritoneum of L. monocytogenes-infected mice
IL-17 is required to initiate inflammation by inducing neutrophil maturation and recruitment from the bone marrow. To identify the specific source of IL-17 in vivo, we infected IL-17F-CreEYFP reporter mice(31, 32) with L. monocytogenes, and IL-17F (YFP) expression was monitored in PEC 1 or 3 d after infection. We found a small percentage (0.54%) of IL-17F (YFP) positive cells in the peritoneum 3 d after infection with L. monocytogenes. Interestingly, IL-17F (YFP) showed a similar distribution pattern as the IL-23R (GFP) expression since the vast majority (72.7%) of the IL-17F (YFP)+ cells corresponded to γδ–TCR+ cells and the remaining 25.9% were αβ–TCR+ cells (Fig. 1E). These data suggest that two distinct populations of IL-17-producing cells, γδ and DN T cells, are rapidly recruited to the site of infection and may initiate and control early events of inflammation in order to limit bacterial dissemination. Therefore, we sought to determine whether IL-17 produced by PEC upon infection was dependent on IL-23R. PEC from IL-23R-GFP.KI and IL-23R−/− mice were isolated 1 d post infection (d.p.i.) and cultured ex vivo for 18 h. In the presence of PMA and Ionomycin, IL-23R−/− PEC produced consistently lower amounts of IL-17 than those from IL-23RGFP.KI, while IFN-γ production was slightly higher in PEC harvested from IL-23R−/− compared to IL-23R-GFP.KI mice (Fig. 1F).
All together, our results suggest that γδ and DN T cells are rapidly recruited to the site of infection and produced large quantities of IL-17. However, only the latter seems to be IL-23 dependent.
IL-23R expression defines subsets of γδ T cells
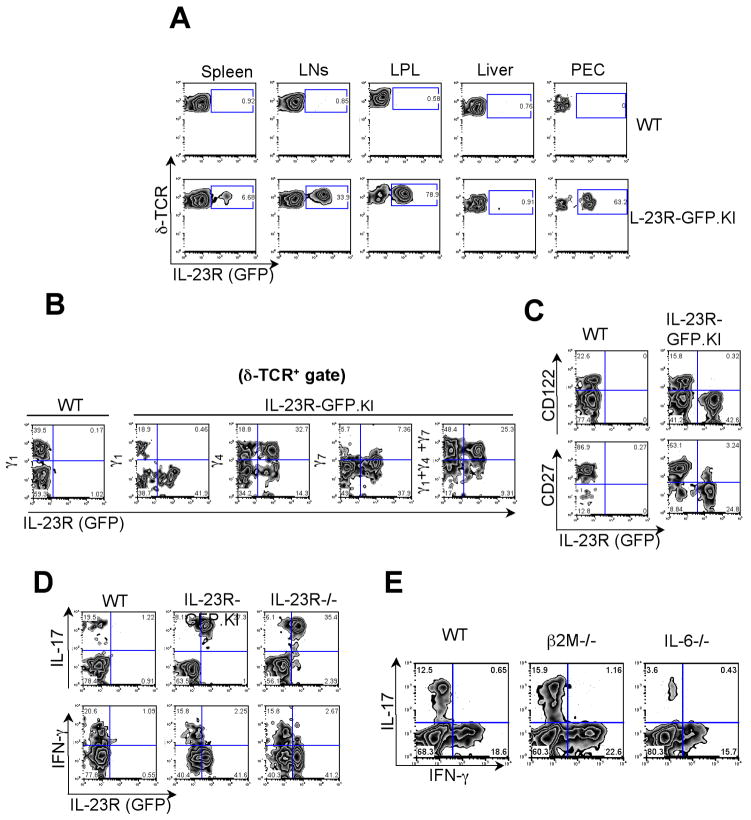
In the next step, we characterized the organ distribution of IL-23R expressing γδ T cells. We observed only very few IL-23R expressing γδ T cells in the spleen (approximately 6.7%). Interestingly, we found a higher frequency of γδ T cells expressing IL-23R in the peripheral lymph nodes (33.9%) and even a greater percentages at the mucosal surfaces, such as in the Lamina Propria (LP) (78.9%) and in the peritoneal cavity (63.2%). However, we could not detect any significant presence of IL-23R expressing γδ T cells in the liver of naïve mice (Fig. 2A).
Figure 2. Identification of IL-23R expressing γδ T cells.
(A) IL-23R (GFP) expression was analyzed on γδ T cells from LNs, spleen, Lamina Propria (LP), liver and Peritoneal Exudate Cells (PEC) in IL-23R-GFP.KI naïve mice. (B, C) LN cells from IL-23R-GFP.KI naïve mice were collected and analyzed for δ-TCR and Vγ1 Vγ4 and Vγ7 (B) or CD27 or CD122 (C) expression on γδ T cells. (D) LN cells collected from IL-23R-GFP.KI or IL-23R−/− were stimulated with PMA/Ionomycin and intracellular cytokine staining for IFN-γ and IL-17 was performed. The quadrants represent intracellular cytokine staining and IL-23R (GFP) expression on γδ T cells. (E) Single cell suspensions were prepared from LNs from WT, β2M−/− or IL-6−/− mice. Quadrants represent intracellular cytokine staining for IL-17 and IFN-γ on γδ T cells after PMA/Ionomycin stimulation. The experiment was repeated 2 times, 3 mice/group.
Subsequently, we sought to determine whether IL-23R expression on γδ T cells is associated with a specific Vγ TcR usage. Figure 2B shows a representative FACS staining of specific Vγ receptors of δ-TCR+ cells in the LNs of IL-23R-GFP.KI mice. IL-23R expression was not detected among Vγ1 and barely detected in the Vγ7 subsets of γδ T cells. In contrast, more than 60% of TCR Vγ4+γδ T cells were positive for IL-23R (GFP). Since Vγ5+γδ T cells are usually not present in the LNs, the remaining 27% IL-23R (GFP) positive γδ T cells are likely Vγ6 expressing γδ T cells. Therefore, expression of IL-23R seems to be mainly restricted to γδ T cells that utilize Vγ4 and Vγ6. It has recently been reported that most γδ T cells that produce IL-17 do not express CD122 or CD27(33, 34) and the use of these two markers could faithfully discriminate between IFN-γ–and IL-17-producing γδ T cells. However, the mechanisms leading to this biased functional segregation is still unclear. Here, we validated IL-23R as a marker that defines functional segregation of γδ T cell subsets. While IL-23R (GFP) expressing γδ T cells did not express CD122, we identified a small subset of IL-23R (GFP)+γδ T cells featuring low levels of CD27 (Fig. 2C).
Next, we sought to determine whether the IL-23R (GFP) expression could faithfully report IL-17 expression in γδ T cells. Thus, we analyzed IL-17 production by LN γδ T cells from WT, IL-23RGFP.KI and IL-23R−/− mice. We found that most of the IL-23R (GFP) expressing γδ T cells produced IL-17. However, 22% of the IL-17-producing γδ T cells expressed only very low levels of GFP (Fig. 2D). It is likely that the low GFP expression in these cells is due to some loss of the fluorescence that occurs during the fixation and permeabilization procedures performed prior intracellular cytokine stainings. Further analysis confirmed that IL-17 expression of γδ T cells was very similar in cells derived from either WT, IL-23R-GFP.KI or IL-23 −/− mice. This finding supports the notion that at the steady state, IL-17 production in γδ T cells, as well as in CD4+ TH17 cells, is completely independent of IL-23R expression (Fig. 2D).
In β2M−/− mice, γδ T cells exhibit low or no expression of CD122 characteristic for T cells that have not encountered antigen yet(33). Consistent with other studies, our results demonstrate that encountering ligand is not required for γδ T cells in order to produce effector cytokines since the percentages of IL-17+ or INF-γ+ γδ T cells found in β2M−/−were comparable to WT mice (Fig. 2E).
Since IL-6 is required for the generation of CD4+ TH17 cells, we sought to determine whether IL-6 is also required for IL-17 production by γδ T cells. As shown in Fig. 2E, only few IL-17- producing γδ T cells were detected in IL-6−/− mice while the percentages of IFN-γ+ γδ T cells were similar between WT and IL-6−/− Accordingly, both CD4+ TH17 cells and IL-17-producing γδ T cells are dependent on IL-6.
IL-23 controls the balance between IL-17- and IFN-γ-producing γδ T cells at early stages of inflammation
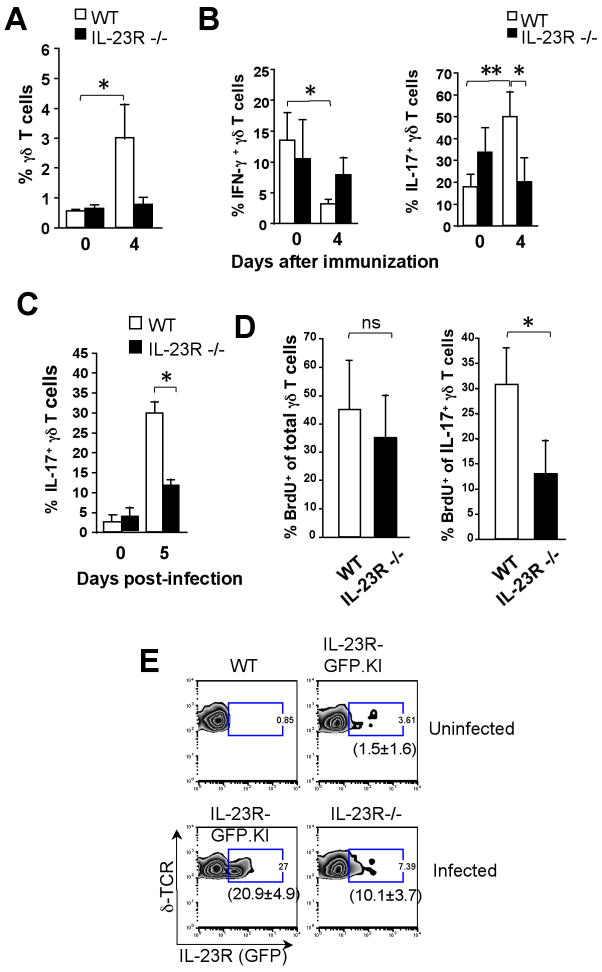
γδ T cells contribute to the host defense and are ideally equipped to provide IL-17 at early stages of an inflammatory response. Therefore, we wanted to explore the in vivo role of IL-23R for γδ T cells during this period(33). Thus, WT and IL-23R−/− mice were immunized with CFA, as a surrogate for the induction of inflammation, and draining LN cells were harvested 4 d later. We observed that the frequency of γδ T cells was significantly augmented in the draining LNs of WT but not in IL-23R−/− mice 4 d after immunization with of CFA/MOG35-55 compared to naïve mice (Fig. 3A). We also noticed a remarkable increase of IL-17-producing γδ T cells in WT but not in IL-23R−/− mice. However, WT but not IL-23−/− mice showed a significant decrease of IFN-γ–producingγδ T cells 4 d after immunization (Fig. 3B). Furthermore, IL-17 but not IFN-γ-producing γδ T cells showed 5′-Bromo-2-deoxyuridine (BrdU) incorporation after CFA/MOG35-55 immunization and such BrdU incorporation by IL-17+ γδ T cells was diminished in IL-23R−/− compared to WT mice (data not shown). Collectively, these results show that the capacity of IL-17+ γδ T cells to rapidly proliferate upon stimulation is largely dependent on IL-23R.
Figure 3. IL-17-producing γδ T cells require Il-23R for expansion/survival during L. monocytogenes infection.
(A, B) IL-23R-GFP.KI or WT mice were immunized with MOG35-55 emulsified in CFA. (A) Percentages of draining LN γδ T cells in naïve mice or 4 d after MOG35-55/CFA immunization of WT or IL23R−/− mice. The bars represent mean ± SD of 3 independent experiments [*p<0.05, **p<0.01 (two tail Student t-test)]. (B) Draining LN cells were stimulated with PMA/Ionomycin. Percentages of IFN-γ or IL-17+γδ T cells from naïve or MOG35-55/CFA-immunized mice after 4 d are represented. (C, D) Mice were i.p infected with viable L. monocytogenes. Mononuclear cells were isolated from the livers of IL- 23R−/− or WT mice and stimulated with PMA/Ionomycin. (C) Percentages of hepatic IL-17-producing γδ T cells from WT or IL-23R−/− during L. monocytogenes infection are represented. The bars represent mean ± SD of 3 independent experiments. (D) WT or IL23R−/− mice were infected with L. monocytogenes and received 2 mg of BrdU intraperitoneally on day 1 and 2 after infection. Intracellular staining of IL-17 and BrdU was performed in γδ T cells from livers of WT or IL-23R−/− mice 3 d after infection. The bars represent mean ± SD of 3 independent experiments [*p<0.05 (two tail Student t-test)]. (E). IL-23R (GFP) expression was assessed in intrahepatic γδ T cells from IL-23R-GFP.KI or IL-23R−/− mice during L. monocytogenes infection. In brackets, mean ± SD of 3 experiments with 3 mice/group is shown.
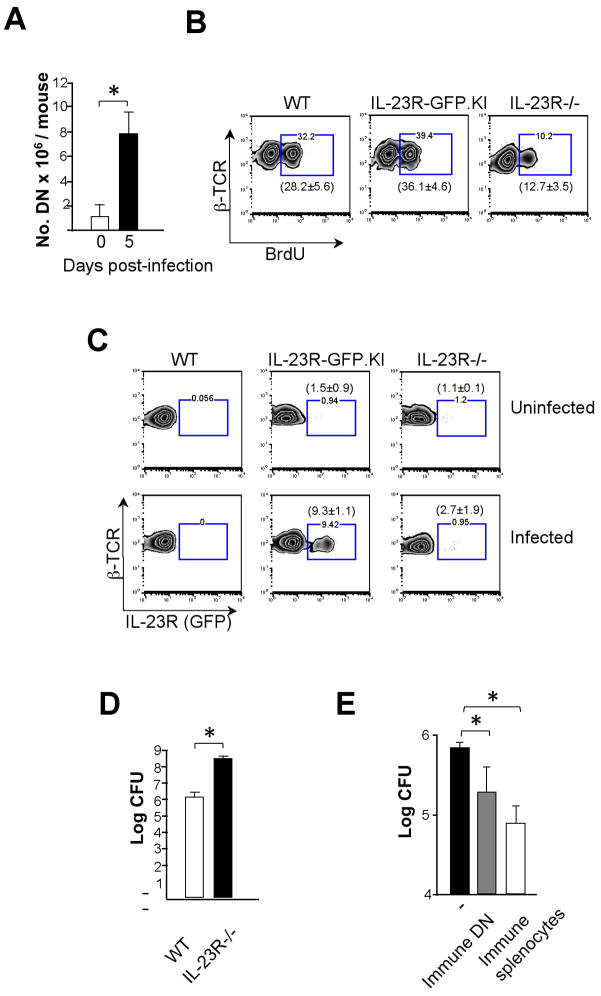
IL-23R controls the expansion of IL-17-producing γδ T cells during infection
After bacterial infections, γδ T cell deficient mice show an increased bacterial load with early bacterial dissemination and higher mortality rates than WT mice(35). On the other hand IL-17-producing γδ T cells are essential for protection against infection(15, 25, 36). Therefore, we sought to characterize the functional properties of IL-23R during L. monocytogenes infection in γδ T cells. Thus, we infected either WT or IL-23R−/− mice via the intraperitoneal route with viable L. monocytogenes and the livers were harvested after 3–5 d after infection. First, WT mice showed increased IL-17 expression 5 d.p.i. whereas IL-23R−/− mice did not (Fig. 3C). Second, we monitored γδ T cells turnover rates in vivo by measuring the amount of BrdU incorporation 3 d after L. monocytogenes infection. As shown in Fig. 3d, we found no significant differences in the frequency of total BrdU positive γδ T cells between WT and IL-23R−/− mice. However, whereas around 30% of IL-17-producing γδ T cells incorporated BrdU in WT mice, only 13% of IL-17-producingγδ T cells isolated from IL-23R−/− livers showed BrdU incorporation (Fig. 3D). Since IL-23R expression was required for the expansion of IL-17 producing γδ T cells during infection, we next wanted to analyze the IL-23R levels of γδ T cells in this situation. Thus, we infected IL-23R-GFP.KI and IL-23R−/− mice and monitored the IL-23R (GFP) expression 3 d after infection. Livers from IL-23R–GFP.KI mice showed an enrichment of IL-23R (GFP) bearing γδ T cells during infection (from 1.5 ± 1.6% to 20.9 ± 4.9%). In contrast, the percentage of IL-23R (GFP)+γδ T cells was significantly reduced (10.0±3.7%) in the liver of IL-23R−/− mice (Fig. 3E). These data suggest that IL-23R expression at early stages of infection is extremely important for maintenance/expansion of IL-17-producing γδ T cells in the liver during L.monocytogenes infection.
DN T cells in vitro expansion relies on IL-23R
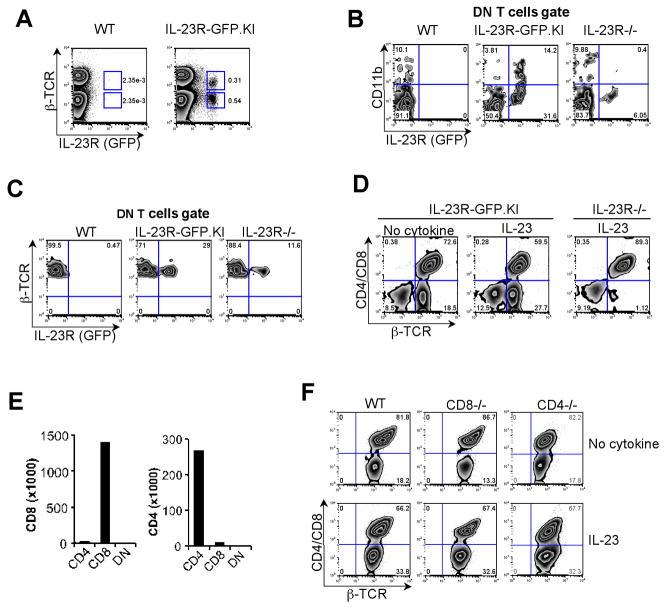
In previous studies, αβ-TCR+ CD4− CD8− DN cells appeared in the peritoneal cavity at an early stage of i.p. L. monocytogenes infection suggesting their role in combating this intracellular bacterium(37, 38). It is also known that DN T cells contribute to control of intracellular infections in vivo(39). However the control mechanisms exerted by DN T cells remain unknown. Thus, we sought to investigate the role of IL-23R in DN T cells during infections. In this regard, we started examining IL-23R (GFP) expression in the LNs. We observed that more than 50% of the IL-23R (GFP) bearing cells did not express the αβ-TCR and were mainly composed of γδ T cells, macrophages and DCs (Fig. 4A)(5). The remaining IL-23R (GFP) expressing cells consisted of DN T cells which surprisingly expressed the myeloid marker CD11b. The number of IL-23R (GFP) expressing DN T cells was significantly reduced in LNs of IL23R−/− (~10%), compare to IL-23R-GFP.KI (~18%) mice, and the remaining DN T cells were CD11b negative (Fig. 4B). Around 29% of these DN T cells expressed IL-23R (GFP) in the LN of IL-23R-GFP.KI mice, and this percentage was diminished to 11.6% in IL-23R−/− mice (Fig. 4C). In naïve mice, DN T cells represent about ~2% of all αβ-TCR+ cells (data not shown) and expanded in a IL-23R fashion after 6 d of activation in vitro in the presence of IL-23 (Fig. 4D). Furthermore, we found that the majority (~87%) of DN T cells that expressed IL-23R (GFP) were CD62Llow CD45RBlow, a surface phenotype commonly found on effector/memory cells (data not shown). Consistent with our observations that the expansion of DN T cells was completely dependent on IL-23R expression, other cytokines such as IL-12 and IL-27 could not substitute for IL-23 (Supplementary Fig. 1).
Figure 4. Expansion of DN T cells depends on IL-23R in vitro.
LNs were collected from naïve WT and IL-23R-GFP.KI mice. (A) Single cell suspensions were prepared for surface staining of β–TCR, CD4 and CD8. (B) Quadrants show CD11b and IL-23R (GFP) expression in β–TCR+ CD4− CD8− DN T cells from LNs collected from WT, IL-23R-GFP.KI and IL-23R−/− mice. (C) Single cell suspensions from LNs of naïve WT, IL-23R-GFP.KI and IL-23R−/−mice were prepared and stained for β–TCR, CD4 and CD8. IL-23R (GFP) expression in the DN T cell gate is shown. (D) LNs from IL-23R-GFP.KI or IL-23R−/− mice were cultured with anti-CD3 alone or with rIL-23 (30 ng/ml). On day 6, cultured cells were stained for CD4, CD8 and β–TCR. The numbers in the quadrants represent percentages of β–TCR+ CD4− CD8− DN and β–TCR+ CD4+/CD8+ T cells. (E) Real time PCR was performed for CD4 and CD8 genes on FACS sorted DN T cells. (F) LNs were collected from CD8−/−, CD4 −/− and WT mice. Cells were activated with either anti-CD3 alone or in the presence of rIL-23. On day 6, cells were stained for β–TCR, CD4 and CD8. Cells were analyzed for generation of DN T cells.
FACS sorted DN T cells did not show any expression of surface or intracellular staining of either CD4 or CD8 at protein or RNA level (Fig. 4e and Supplementary Fig. 2A). Neither CD4−/− nor CD8−/− mice showed any defect in the generation of DN T cells (Fig. 4F) confirming that the generation of DN T cells is not dependent on CD4+ and CD8+ T cells. Interestingly, complete absence of DN T cells in athymic nude mice confirmed their thymic origin (Supplementary Fig. 2B).
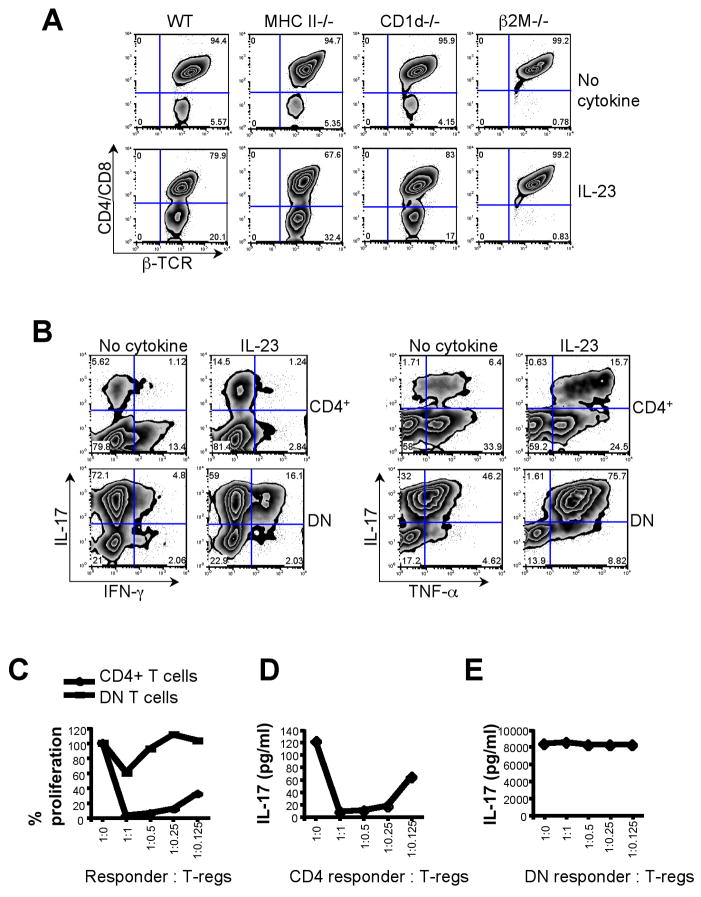
This subset of DN T cells expressed αβ-TCR but not NK T lineages markers such as NK1.1 and CD49b and showed very low levels of CD1d tetramer binding (Supplementary Fig. 3). Furthermore, the frequencies of DN T cells in WT and CD1d−/−mice were similar (around 5%) and could be expanded by IL-23 (Fig. 5A), suggesting that these cells are not of the NK T lineage.
Figure 5. Characterization of DN T cells.
(A) LNs were collected from MHCII−/−, CD1d−/−, β2M−/− and WT littermate control mice. LN cells were activated with either anti-CD3 alone or in the presence of rIL-23. On day 6, cells were stained for β–TCR, CD4 and CD8 and analyzed for generation of DN T cells. (B) On day 6, CD4+ and DN T cells were FACS sorted and then re-stimulated with PMA and Ionomycin and intracellular cytokine staining was performed for IL-17, IFN-γ and TNF-α. (C, D, E) CD4+ and DN T cells generated as described were FACS sorted. CD4+ and DN T cells were used as effector cells with indicated ratio of Foxp3+ Tregs in an in vitro suppression assay. (E) Cell proliferation was measured by thymidine incorporation. (D, E) Culture supernatant was collected at 48 h and cytokine analysis was performed for IL-17.
DN T cells hold a unique IL-17+ IFN-γ+TNF-α+ effector phenotype dependent on IL-23R
DN T cells do not express CD4 or CD8 co-receptors and the restriction elements used for the selection of these cells are not known. We first assessed whether the generation of DN T cells depended on MHC class II molecules. We did not find any significant difference in the generation/expansion of DN T cells in MHC class II or CD1d deficient mice. In contrast, DN T cells were completely absent in β2M−/− mice. Addition of IL-23 increased the fraction of DN T cells in WT mice (20.1%) but failed to do so in β2M−/−mice (Fig. 5A). This suggests that the expansion of DN T cells is dependent on MHC class I or Ib molecules which utilize β2M for complex formation.
Since IL-23R cross-linking expands IL-17 producing CD4+ T cells, we also determined the cytokines produced by DN T cells. While only ~7% of CD4+ T cells produced IL-17 after stimulation with anti-CD3 for 6 d, 77% of the DN T cells were positive for IL-17 expression. Addition of IL-23 to DN T cells did not further increase the frequency of IL-17+ cells, but a sizeable population of DN T cells (18%) began to co-produce IL-17 and IFN-γ. This is in contrast to CD4+ T cells where addition of IL-23 reduced the number of IFN-γ-expressing cells from ~14.5% to ~4% but enhanced IL-17 production (Fig. 5B left panel). The total frequency of TNF-α producers remained unchanged in CD4+ T cells upon exposure to IL-23. However, the fraction of IL-17/TNF-α double-expressors increased dramatically in DN T cells in the presence of IL-23 (Fig. 5B right panel). Compared with conventional CD4+ T cells, DN cells produced massive amounts of IFN-γ, TNF-α and IL-17 (Supplementary Fig. 4). Furthermore, DN T cells also expressed high mRNA levels of genes associated with the CD4+ TH17 lineage such as ROR-γt, IL-22 and IL-21 (data not shown).
Since IL-23R expressing DN T cells expanded upon IL-23 exposure and produced such high levels of pro-inflammatory cytokines, we next determined whether Foxp3+ regulatory T cells (Treg) could suppress these DN T cells in vitro. Thus, conventional CD4+ T cells and DN T cells were used as responder cells and cultured with Tregs in an in vitro suppression assay. Whereas Tregs significantly suppressed the proliferation of CD4+ T cells, DN T cells were refractory to suppression (Fig. 5C). Accordingly, the production of IL-17 by CD4+ T cells was dramatically inhibited by Tregs whereas IL-17 production by DN T cells was completely unaffected. Furthermore, DN T cells produced significantly larger amounts of IL-17 than CD4+ T cells (Figs. 5D, E).
Protective role of DN T cells against L. monocytogenes infection
It was recently reported that DN T cells mediated protective immunity against infection with F. tularensis LVS and Mycobacterium tuberculosis(39–41). This raised the question whether those IL-17-producing IL-23R+ DN T cells identified here might be involved in mediating resistance to infection.
DN T cells rapidly expanded in the spleens and livers of mice infected with L. monocytogenes (Fig. 6A). Consistently, proliferative ability measured by in vivo BrdU incorporation, was significantly diminished in DN T cells from livers of L. monocytogenes-infected IL-23R−/− compared to WT or IL-23R-GFP.KI mice (Figs. 6B). Furthermore, we observed a remarkable increase of IL-23R (GFP) expression on DN T cells only in IL-23R-GFP.KI but not in IL-23R−/− mice during infection (Fig. 6C). This data suggest that the DN T cells expansion during L. monocytogenes infection is controlled by IL-23R. We also observed a ~100-fold increase in the bacterial burden in infected livers of IL-23R−/− mice compared to WT ones 3 d.p.i. indicating impaired bacterial clearance in the absence of IL-23R signaling (Fig. 6D). Notably, adoptive transfer of DN T cells isolated from L. monocytogenes-infected mice (immune DN T cells) into RAG2−/− mice(42) significantly reduced the bacterial burden in the liver following L. monocytogenes challenge (Fig. 6E). Collectively, these results suggest that DN T cells are involved in a protective immune response against L. monocytogenes.
Figure 6. DN T cells are involved in resistance to L. monocytogenes infection.
(A) Absolute numbers of DN T cells from spleen and liver pooled cells before or 5 d after L. monocytogenes infection are shown. The bars represent mean ± SD of 3 independent experiments (5–10 mice pooled/group/experiment). (B, C) WT, IL-23R-GFP.KI or IL-23R−/− mice were infected with L. monocytogenes. After 3 d, BrdU incorporation (B) and IL-23R (GFP) expression (C) were analyzed in DN T cells. In brackets, mean ± SD of 2 experiments with 3 mice/group is shown. (D) After 3 d bacterial CFU counts were determined in livers of IL-23R−/− or WT mice infected with L. monocytogenes. (E) No cells, immune splenocytes or immune DNs isolated from WT mice 5 d after infection with L. monocytogenes were transferred intravenously into RAG2−/− mice. 24 h later mice were challenged i.p. with L. monocytogenes. After 3 d, the CFU in the liver was assessed to quantify bacterial burden. The bars represent mean ± SD of 3 independent experiments (5 mice pooled/group/experiment) [*p<0.05, **p<0.01 (two tail Student t-test)].
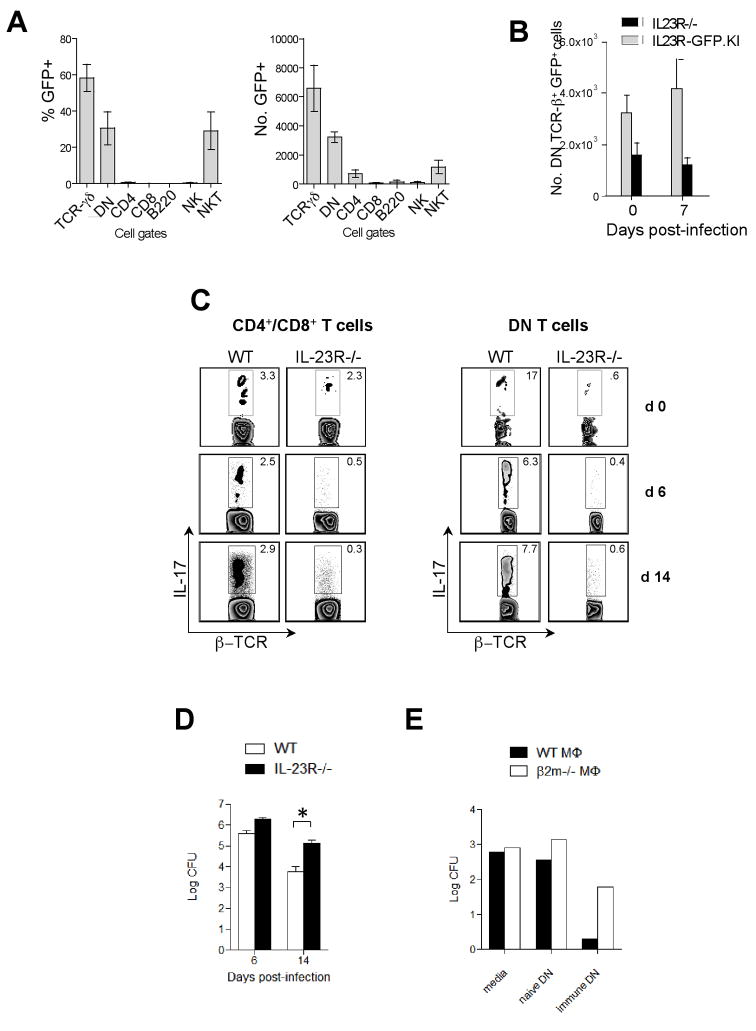
IL-23R governs effector features of DN T cells during F. tularensis LVS infection
To investigate whether DN T cells are involved in immune responses directed against other intracellular pathogens, we monitored the expansion of IL-23R (GFP)+ DN T cells in IL-23R-GFP.KI mice in response to F. tularensis LVS(40) infection using the IL-23R-GFP.KI strain. First, we determined IL-23R (GFP) expression in the lungs of uninfected mice. IL-23R was expressed on a number of different cell types with significant expression on γδ, DN, and NK T cells. Approximately 30% of pulmonary DN T cells expressed IL-23R (GFP) at baseline (Fig. 7A). While similar numbers of CD4+ and CD8+ T cells were present in IL-23R−/− and WT lungs during acute infection (data not shown), there was no expansion of IL-23R (GFP)+ DN T cells in IL-23R−/− mice (Fig. 7B). In fact, the number of IL-23R (GFP)+ DN cells in IL-23R−/− mice after infections was approximately one third of the number found in IL-23R-GFP.KI mice, suggesting that IL-23R is crucial for the expansion/survival of DN T cells during in vivo infection. The number of IL-17 producing CD4+, CD8+ and DN T cells was markedly reduced in IL-23R−/− mice in response to F. tularensis LVS infection (Fig. 7C). In contrast, the percentage of conventional and DN T cells producing IFN-γ was increased in IL-23R−/−compared to WT mice 14 d.p.i. (data not shown). Although there was no difference in the survival of IL-23R−/− and WT mice, we noted that IL-23R−/− but not WT mice infected with F. tularensis LVS showed weight loss and signs of respiratory distress. These signs were accompanied by a 10-fold increase in the bacterial burden in the lungs of infected IL-23R−/− mice 14 d.p.i. (Fig. 7D), indicating impaired bacterial clearance in the absence of IL-23R signaling.
Figure 7. Role of IL-23R in F. tularensis LVS infection.
(A) Percentages and absolute numbers of IL-23R (GFP) expressing cells within δ-TCR+, DN, CD4+, CD8+, B220+, NK1.1+, PBS57/CD1d tetramer+ cell populations isolated from the lungs of uninfected IL-23R-GFP.KI mice. The bars represent mean ± SEM of 3 independent experiments (3–5 mice pooled/group/experiment). (B) Absolute numbers of IL-23R (GFP) expressing DN T cells isolated from the lungs of uninfected and F. tularensis LVS-infected IL-23R−/− and IL-23R-GFP.KI mice. The bars represent mean ± SEM of 3 independent experiments (3–5 mice pooled/group/experiment). (C) The dot plots represent IL-17 production by β–TCR+ CD4+/CD8+ and β–TCR+ DN T cells isolated from uninfected and F. tularensis LVS infected lungs of IL-23R−/− and WT mice. The experiment was repeated twice with 3–4 mice/group pooled in each experiment. (D) Bacterial CFU was determined in the F. tularensis LVS infected lungs of IL-23R−/− and WT mice [*p<0.05, **p<0.01 (two tail Student t-test)]. (E) F. tularensis LVS infected β2M−/− and WT macrophages were incubated with DN T cells isolated from the spleens of naïve or immune mice and bacterial burden was determined 72 h after culture. The experiment was repeated 3 times.
To further evaluate the MHC restriction and antigen specificity of DN T cells in controlling intracellular F. tularensis LVS infection, we isolated DN T cells from the spleens of naïve and F. tularensis LVS-immune mice and incubated them with F. tularensis LVS-infected WT or β2M−/− macrophages. While DN T cells from naïve mice failed to restrict replication of intracellular bacteria, DN T cells obtained from immune mice inhibited intracellular growth of F. tularensis LVS (Fig. 7E). This inhibition was partly dependent on β2M expression since the protective effect of DN cells was diminished in infected β2M−/− macrophages (Fig. 7E).
DISCUSSION
It is now well established that IL-23 activates local resident CD4+ TH17 IL-23R-bearing cells to produce IL-17 and IL-22 in order to control extracellular bacteria dissemination such as K. pneumoniae or C. rodentium infection (18, 19, 43). However, very few reports have explored the importance of the IL-23/IL-17 axis in combating intracellular bacteria. Here, we have established the essential role of IL-23R in the host response against the intracellular pathogens L. monocytogenes and F. tularensis LVS. The experiments performed in L. monocytogenes-infected mice point out the importance of IL-23R regulation of γδ and DN T cells function in order to limit the growth of the organism at early stages until the immune adaptive response clears the infection days later.
At the first line of the host defense against L. monocytogenes we observe a recruitment of IL-23R+ IL-17-producing-γδ and DN T cells to the peritoneum(44). Although such initial recruitment is not mediated by IL-23R, IL-17 production is partially diminished in IL-23R−/− mice suggesting that other factors such as IL-6, IL-21 or TGF-β may also contribute to IL-17 production. Phagocytes or granulocytes do not express IL-23R nor their recruitment to the peritoneal cavity is dismissed in IL-23−/− mice, suggesting a modest role of IL-23R in fighting the pathogen at the very initial events of infection in the peritoneum. The latter observation contrasts which is observed in the liver 2 d later since IL-23R−/− mice are much more susceptible to L. monocytogenes infection compared to WT mice. At these early stages IL-17 produced by γδ T cells is important for protection against L. monocytogenes and the contribution of IL-23 in the induction of IL-17 expression by γδ T cells which has been proposed in L. monocytogenes-infected IL-12/23p40 deficient mice but not faithfully proven(25). However, we demonstrate that IL-23R cross-linking induces the enrichment of IL-17-producing γδ T cells in the livers of L. monocytogenes-infected mice. We propose that IL-23 may act as a growth factor for IL-17-producing IL-23R bearing γδ T cells at an initial stage of infection contributing to the expansion/maintenance of IL-17-producing γδ T cells and the contraction of the IFN-γ–producing counterparts. An alternative explanation could be that the IL-23R status could determine the trafficking behavior of specific γδ T cell subsets regulating the balance between IL-17-and IFN-γ–producing γδ T cells in different organs. Moreover, most of the IL-23R+γδ T cells, which utilize Vγ4 and Vγ6 and do not express CD122 or CD27, produce IL-17 and are found in the LNs. On the other hand, IL-23R− CD122+ CD27+γδ T cells which produce IFN-γ are located in the spleen(33, 34). Thus, IL-23R may represent one mechanism leading to this biased distribution of γδ T cells in different organs. It is also likely, as previously suggested, that IL-23R expression is imprinted during the development of γδ T cells, and that mechanism of imprinting is probably influenced by γδ-TCR, CD122 and CD27 agonists in the thymus (33, 34).
In previous studies, a population of αβ-TCR+ CD4− CD8− DN cells along with γδ T cells appeared in the peritoneal cavity at an early stage of i.p. L. monocytogenes infection suggesting their role in combating this intracellular bacterium(37, 38). This rare T cell subset expands after infection with F. tularensis LVS and M. tuberculosis controlling the dissemination of these organisms(39–41). Here, we confirm this protective role of DN T cells as adoptive transfer of DN T cells into RAG2−/− mice significantly reduced the bacterial burden compared with mice that have not received any cells. However, the mechanisms of this antibacterial effect exerted by DN T cells remain to be characterized. In this regard, we show that DN T cells recognize bacterial antigens in the context of MHC I class or class Ib. This is consistent with previous studies performed with human αβ-TCR+ CD4− CD8− DN T cell clones that recognized the MHC class Ib restricted antigens as well(45). Remarkably, DN T cells expand in the course of L. monocytogenes infection, which is controlled by IL-23R. This contrasts with previous studies that showed no DN T cell proliferation in the presence of L. monocytogenes-infected macrophages, however this could be due to the absence of growth factors in that in vitro setting(38). Our data elucidate that IL-23R cross-linking promotes differentiation into CD62Llow CD45RBlow CD11b+ effector DN T cells uniquely equipped in order to exert protection against intracellular bacterial infection. DN T cells may indirectly promote inflammation collaborating with the innate system by initiating neutrophil recruitment via IL-17 production or priming macrophages and neutrophils for bactericidal activity via INF-γ and TNF-α. DN T cells may constitute a good source of TNF-α which in turn is essential for CD8+ T cell mediated immunity to intracellular bacteria. Furthermore, addition of IL-23 increases perforin and granzyme-B levels. Hence DN T cells may mediate cytolysis of bacteria-infected cells. Our findings show that IL-23R+ DN T cells are induced during infection contributing to the control of F. tularensis LVS and L. monocytogenes dissemination suggesting that DN T cells might be involved in protective immune responses against a number of intracellular pathogens.
In summary, IL-23 is involved in expanding different types of IL-17-producing effector T cells. CD4+ MHC class II restricted TH17 cells mediate tissue inflammation and induce autoimmunity while β2M-dependent DN and γδ T cells are involved in clearing intracellular pathogens. These data also provide a means by which beneficial effects of IL-23 in protecting against infections and detrimental effects in inducing tissue inflammation and autoimmunity can be separated at the effector cell level.
Supplementary Material
Acknowledgments
We thank D. Kozoriz for cell sorting and technical assistance, G. Matsuzaki and Pablo Pereira for anti-Vγ4, Vγ5 and Vγ7 mAbs and T. Korn for critical reading of the manuscript.
Footnotes
This work was supported by grants from National Institutes of Health (R01AI073542-01 to M.O. and 1R01NS045937-01, 2R01NS35685-06, 2R37NS30843-11, 1R01A144880-03, 2P01A139671-07, 1P01NS38037-04 and 1R01NS046414 to V.K.K. and AI32412, P01 AI56296 to L.H.G.) and National Multiple Sclerosis Society (RG-2571-D-9 to V.K.K. and RG-3882-A-1 to M.O.), and the Juvenile Diabetes Research Foundation Center for Immunological Tolerance at Harvard Medical School. V.K.K. is a recipient of the Javits Neuroscience Investigator Award from the US National Institutes of Health. Ellison Medical Foundation to L.H.G. LR-B is supported by a post-doctoral fellowship from the Human Frontiers Science Program (HFSP). VL is supported by a fellowship from the Irvington Institute. AA is supported by a post-doctoral fellowship from the National Multiple Sclerosis Society (NMSS), New York. MM is supported by the Deutsche Forschungsgemeinschaft (DFG).
References
- 1.Langrish CL, McKenzie BS, Wilson NJ, de Waal Malefyt R, Kastelein RA, Cua DJ. IL-12 and IL-23: master regulators of innate and adaptive immunity. Immunol Rev. 2004;202:96–105. doi: 10.1111/j.0105-2896.2004.00214.x. [DOI] [PubMed] [Google Scholar]
- 2.Langrish CL, Chen Y, Blumenschein WM, Mattson J, Basham B, Sedgwick JD, McClanahan T, Kastelein RA, Cua DJ. IL-23 drives a pathogenic T cell population that induces autoimmune inflammation. J Exp Med. 2005;201:233–240. doi: 10.1084/jem.20041257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Park H, Li Z, Yang XO, Chang SH, Nurieva R, Wang YH, Wang Y, Hood L, Zhu Z, Tian Q, Dong C. A distinct lineage of CD4 T cells regulates tissue inflammation by producing interleukin 17. Nat Immunol. 2005;6:1133–1141. doi: 10.1038/ni1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McGeachy MJ, Chen Y, Tato CM, Laurence A, Joyce-Shaikh B, Blumenschein WM, McClanahan TK, O’Shea JJ, Cua DJ. The interleukin 23 receptor is essential for the terminal differentiation of interleukin 17-producing effector T helper cells in vivo. Nat Immunol. 2009;10:314–324. doi: 10.1038/ni.1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Awasthi A, Riol-Blanco L, Jager A, Korn T, Pot C, Galileos G, Bettelli E, Kuchroo VK, Oukka M. Cutting edge: IL-23 receptor gfp reporter mice reveal distinct populations of IL-17-producing cells. J Immunol. 2009;182:5904–5908. doi: 10.4049/jimmunol.0900732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nakae S, Saijo S, Horai R, Sudo K, Mori S, Iwakura Y. IL-17 production from activated T cells is required for the spontaneous development of destructive arthritis in mice deficient in IL-1 receptor antagonist. Proc Natl Acad Sci U S A. 2003;100:5986–5990. doi: 10.1073/pnas.1035999100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nakae S, Nambu A, Sudo K, Iwakura Y. Suppression of immune induction of collagen-induced arthritis in IL-17-deficient mice. J Immunol. 2003;171:6173–6177. doi: 10.4049/jimmunol.171.11.6173. [DOI] [PubMed] [Google Scholar]
- 8.Lubberts E, Schwarzenberger P, Huang W, Schurr JR, Peschon JJ, van den Berg WB, Kolls JK. Requirement of IL-17 receptor signaling in radiation-resistant cells in the joint for full progression of destructive synovitis. J Immunol. 2005;175:3360–3368. doi: 10.4049/jimmunol.175.5.3360. [DOI] [PubMed] [Google Scholar]
- 9.Fujino S, Andoh A, Bamba S, Ogawa A, Hata K, Araki Y, Bamba T, Fujiyama Y. Increased expression of interleukin 17 in inflammatory bowel disease. Gut. 2003;52:65–70. doi: 10.1136/gut.52.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yen D, Cheung J, Scheerens H, Poulet F, McClanahan T, McKenzie B, Kleinschek MA, Owyang A, Mattson J, Blumenschein W, Murphy E, Sathe M, Cua DJ, Kastelein RA, Rennick D. IL-23 is essential for T cell-mediated colitis and promotes inflammation via IL-17 and IL-6. J Clin Invest. 2006;116:1310–1316. doi: 10.1172/JCI21404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Khader SA, Bell GK, Pearl JE, Fountain JJ, Rangel-Moreno J, Cilley GE, Shen F, Eaton SM, Gaffen SL, Swain SL, Locksley RM, Haynes L, Randall TD, Cooper AM. IL-23 and IL-17 in the establishment of protective pulmonary CD4+ T cell responses after vaccination and during Mycobacterium tuberculosis challenge. Nat Immunol. 2007;8:369–377. doi: 10.1038/ni1449. [DOI] [PubMed] [Google Scholar]
- 12.Veldhoen M, Hocking RJ, Atkins CJ, Locksley RM, Stockinger B. TGFbeta in the context of an inflammatory cytokine milieu supports de novo differentiation of IL-17-producing T cells. Immunity. 2006;24:179–189. doi: 10.1016/j.immuni.2006.01.001. [DOI] [PubMed] [Google Scholar]
- 13.Huber M, Heink S, Grothe H, Guralnik A, Reinhard K, Elflein K, Hunig T, Mittrucker HW, Brustle A, Kamradt T, Lohoff M. A Th17-like developmental process leads to CD8(+) Tc17 cells with reduced cytotoxic activity. Eur J Immunol. 2009;39:1716–1725. doi: 10.1002/eji.200939412. [DOI] [PubMed] [Google Scholar]
- 14.Intlekofer AM, Banerjee A, Takemoto N, Gordon SM, Dejong CS, Shin H, Hunter CA, Wherry EJ, Lindsten T, Reiner SL. Anomalous type 17 response to viral infection by CD8+ T cells lacking T-bet and eomesodermin. Science. 2008;321:408–411. doi: 10.1126/science.1159806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Roark CL, Simonian PL, Fontenot AP, Born WK, O’Brien RL. gammadelta T cells: an important source of IL-17. Curr Opin Immunol. 2008;20:353–357. doi: 10.1016/j.coi.2008.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Michel ML, Keller AC, Paget C, Fujio M, Trottein F, Savage PB, Wong CH, Schneider E, Dy M, Leite-de-Moraes MC. Identification of an IL-17-producing NK1.1(neg) iNKT cell population involved in airway neutrophilia. J Exp Med. 2007;204:995–1001. doi: 10.1084/jem.20061551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stark MA, Huo Y, Burcin TL, Morris MA, Olson TS, Ley K. Phagocytosis of apoptotic neutrophils regulates granulopoiesis via IL-23 and IL-17. Immunity. 2005;22:285–294. doi: 10.1016/j.immuni.2005.01.011. [DOI] [PubMed] [Google Scholar]
- 18.Schwarzenberger P, La Russa V, Miller A, Ye P, Huang W, Zieske A, Nelson S, Bagby GJ, Stoltz D, Mynatt RL, Spriggs M, Kolls JK. IL-17 stimulates granulopoiesis in mice: use of an alternate, novel gene therapy-derived method for in vivo evaluation of cytokines. J Immunol. 1998;161:6383–6389. [PubMed] [Google Scholar]
- 19.Schwarzenberger P, Huang W, Ye P, Oliver P, Manuel M, Zhang Z, Bagby G, Nelson S, Kolls JK. Requirement of endogenous stem cell factor and granulocyte-colony-stimulating factor for IL-17-mediated granulopoiesis. J Immunol. 2000;164:4783–4789. doi: 10.4049/jimmunol.164.9.4783. [DOI] [PubMed] [Google Scholar]
- 20.Huang W, Na L, Fidel PL, Schwarzenberger P. Requirement of interleukin-17A for systemic anti-Candida albicans host defense in mice. J Infect Dis. 2004;190:624–631. doi: 10.1086/422329. [DOI] [PubMed] [Google Scholar]
- 21.Ye P, Rodriguez FH, Kanaly S, Stocking KL, Schurr J, Schwarzenberger P, Oliver P, Huang W, Zhang P, Zhang J, Shellito JE, Bagby GJ, Nelson S, Charrier K, Peschon JJ, Kolls JK. Requirement of interleukin 17 receptor signaling for lung CXC chemokine and granulocyte colony-stimulating factor expression, neutrophil recruitment, and host defense. J Exp Med. 2001;194:519–527. doi: 10.1084/jem.194.4.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kelly MN, Kolls JK, Happel K, Schwartzman JD, Schwarzenberger P, Combe C, Moretto M, Khan IA. Interleukin-17/interleukin-17 receptor-mediated signaling is important for generation of an optimal polymorphonuclear response against Toxoplasma gondii infection. Infect Immun. 2005;73:617–621. doi: 10.1128/IAI.73.1.617-621.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Umemura M, Yahagi A, Hamada S, Begum MD, Watanabe H, Kawakami K, Suda T, Sudo K, Nakae S, Iwakura Y, Matsuzaki G. IL-17-mediated regulation of innate and acquired immune response against pulmonary Mycobacterium bovis bacille Calmette-Guerin infection. J Immunol. 2007;178:3786–3796. doi: 10.4049/jimmunol.178.6.3786. [DOI] [PubMed] [Google Scholar]
- 24.Lockhart E, Green AM, Flynn JL. IL-17 production is dominated by gammadelta T cells rather than CD4 T cells during Mycobacterium tuberculosis infection. J Immunol. 2006;177:4662–4669. doi: 10.4049/jimmunol.177.7.4662. [DOI] [PubMed] [Google Scholar]
- 25.Hamada S, Umemura M, Shiono T, Tanaka K, Yahagi A, Begum MD, Oshiro K, Okamoto Y, Watanabe H, Kawakami K, Roark C, Born WK, O’Brien R, Ikuta K, Ishikawa H, Nakae S, Iwakura Y, Ohta T, Matsuzaki G. IL-17A produced by gammadelta T cells plays a critical role in innate immunity against listeria monocytogenes infection in the liver. J Immunol. 2008;181:3456–3463. doi: 10.4049/jimmunol.181.5.3456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Siegemund S, Schutze N, Schulz S, Wolk K, Nasilowska K, Straubinger RK, Sabat R, Alber G. Differential IL-23 requirement for IL-22 and IL-17A production during innate immunity against Salmonella enterica serovar Enteritidis. Int Immunol. 2009;21:555–565. doi: 10.1093/intimm/dxp025. [DOI] [PubMed] [Google Scholar]
- 27.Nakamura R, Shibata K, Yamada H, Shimoda K, Nakayama K, Yoshikai Y. Tyk2-signaling plays an important role in host defense against Escherichia coli through IL-23-induced IL-17 production by gammadelta T cells. J Immunol. 2008;181:2071–2075. doi: 10.4049/jimmunol.181.3.2071. [DOI] [PubMed] [Google Scholar]
- 28.Zheng Y, Valdez PA, Danilenko DM, Hu Y, Sa SM, Gong Q, Abbas AR, Modrusan Z, Ghilardi N, de Sauvage FJ, Ouyang W. Interleukin-22 mediates early host defense against attaching and effacing bacterial pathogens. Nat Med. 2008;14:282–289. doi: 10.1038/nm1720. [DOI] [PubMed] [Google Scholar]
- 29.Edelson BT, Unanue ER. Immunity to Listeria infection. Curr Opin Immunol. 2000;12:425–431. doi: 10.1016/s0952-7915(00)00112-6. [DOI] [PubMed] [Google Scholar]
- 30.Pamer EG. Immune responses to Listeria monocytogenes. Nat Rev Immunol. 2004;4:812–823. doi: 10.1038/nri1461. [DOI] [PubMed] [Google Scholar]
- 31.Croxford AL, Kurschus FC, Waisman A. Cutting edge: an IL-17F-CreEYFP reporter mouse allows fate mapping of Th17 cells. J Immunol. 2009;182:1237–1241. doi: 10.4049/jimmunol.182.3.1237. [DOI] [PubMed] [Google Scholar]
- 32.Haak S, Croxford AL, Kreymborg K, Heppner FL, Pouly S, Becher B, Waisman A. IL-17A and IL-17F do not contribute vitally to autoimmune neuro-inflammation in mice. J Clin Invest. 2009;119:61–69. doi: 10.1172/JCI35997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jensen KD, Su X, Shin S, Li L, Youssef S, Yamasaki S, Steinman L, Saito T, Locksley RM, Davis MM, Baumgarth N, Chien YH. Thymic selection determines gammadelta T cell effector fate: antigen-naive cells make interleukin-17 and antigen-experienced cells make interferon gamma. Immunity. 2008;29:90–100. doi: 10.1016/j.immuni.2008.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ribot JC, deBarros A, Pang DJ, Neves JF, Peperzak V, Roberts SJ, Girardi M, Borst J, Hayday AC, Pennington DJ, Silva-Santos B. CD27 is a thymic determinant of the balance between interferon-gamma- and interleukin 17-producing gammadelta T cell subsets. Nat Immunol. 2009;10:427–436. doi: 10.1038/ni.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Andrew EM, Newton DJ, Dalton JE, Egan CE, Goodwin SJ, Tramonti D, Scott P, Carding SR. Delineation of the function of a major gamma delta T cell subset during infection. J Immunol. 2005;175:1741–1750. doi: 10.4049/jimmunol.175.3.1741. [DOI] [PubMed] [Google Scholar]
- 36.Hamada S, Umemura M, Shiono T, Hara H, Kishihara K, Tanaka K, Mayuzumi H, Ohta T, Matsuzaki G. Importance of murine Vdelta1gammadelta T cells expressing interferon-gamma and interleukin-17A in innate protection against Listeria monocytogenes infection. Immunology. 2008;125:170–177. doi: 10.1111/j.1365-2567.2008.02841.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Matsuzaki G, Li XY, Kadena T, Song F, Hiromatsu K, Yoshida H, Nomoto K. Early appearance of T cell receptor alpha beta + CD4- CD8- T cells with a skewed variable region repertoire after infection with Listeria monocytogenes. Eur J Immunol. 1995;25:1985–1991. doi: 10.1002/eji.1830250728. [DOI] [PubMed] [Google Scholar]
- 38.Kadena T, Matsuzaki G, Fujise S, Kishihara K, Takimoto H, Sasaki M, Beppu M, Nakamura S, Nomoto K. TCR alpha beta+ CD4- CD8- T cells differentiate extrathymically in an lck-independent manner and participate in early response against Listeria monocytogenes infection through interferon-gamma production. Immunology. 1997;91:511–519. doi: 10.1046/j.1365-2567.1997.00303.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cowley SC, Hamilton E, Frelinger JA, Su J, Forman J, Elkins KL. CD4-CD8- T cells control intracellular bacterial infections both in vitro and in vivo. J Exp Med. 2005;202:309–319. doi: 10.1084/jem.20050569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cowley SC, Elkins KL. Multiple T cell subsets control Francisella tularensis LVS intracellular growth without stimulation through macrophage interferon gamma receptors. J Exp Med. 2003;198:379–389. doi: 10.1084/jem.20030687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yee D, Rhinehart-Jones TR, Elkins KL. Loss of either CD4+ or CD8+ T cells does not affect the magnitude of protective immunity to an intracellular pathogen, Francisella tularensis strain LVS. J Immunol. 1996;157:5042–5048. [PubMed] [Google Scholar]
- 42.Carrero JA, Calderon B, Unanue ER. Lymphocytes are detrimental during the early innate immune response against Listeria monocytogenes. J Exp Med. 2006;203:933–940. doi: 10.1084/jem.20060045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Iwakura Y, Ishigame H. The IL-23/IL-17 axis in inflammation. J Clin Invest. 2006;116:1218–1222. doi: 10.1172/JCI28508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ohga S, Yoshikai Y, Takeda Y, Hiromatsu K, Nomoto K. Sequential appearance of gamma/delta- and alpha/beta-bearing T cells in the peritoneal cavity during an i.p. infection with Listeria monocytogenes. Eur J Immunol. 1990;20:533–538. doi: 10.1002/eji.1830200311. [DOI] [PubMed] [Google Scholar]
- 45.Dellabona P, Casorati G, Friedli B, Angman L, Sallusto F, Tunnacliffe A, Roosneek E, Lanzavecchia A. In vivo persistence of expanded clones specific for bacterial antigens within the human T cell receptor alpha/beta CD4-8-subset. J Exp Med. 1993;177:1763–1771. doi: 10.1084/jem.177.6.1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.