Abstract
The peptidoglycan glycosyltransferases (PGTs) catalyze the processive polymerization of a C55 lipid-linked disaccharide (Lipid II) to form peptidoglycan, the main component of the bacterial cell wall. Our ability to understand this reaction has been limited due to challenges identifying the appropriate substrate analogs to selectively interrogate the donor (the elongating strand) and acceptor (Lipid II) sites. To address this problem, we have developed an assay using synthetic substrates that can discriminate between the donor and acceptor sites of the PGTs. We have shown that each site has a distinct lipid length preference. We have also established that processive polymerization depends on the length of the lipid attached to the donor.
The peptidoglycan glycosyltransferases (PGTs) catalyze the polymerization of Lipid II (LP2, 1a) to form the bacterial cell wall. These enzymes are processive polymerases that catalyze multiple rounds of chain elongation without releasing the growing glycan strand, but the molecular basis for processivity is not understood.1,2a Because the function of these extracellular enzymes is unique and essential to bacteria, inhibitors of the PGTs could be developed into new antibiotics. Toward this end, crystal structures of these enzymes have been determined, both in the apo form and bound to moenomycin, a natural product inhibitor.2 Although these structures have provided information about the PGT’s interaction with the inhibitor, we know comparatively little about the enzyme’s interactions with its substrates, LP2 and the elongating polymer, due in part to challenges synthesizing mechanistically informative substrate analogs. Recently, we demonstrated that PGTs elongate the growing glycan polymer by addition to its reducing end.3 In this paper we have used differentiated substrates to reveal that the donor and acceptor sites have distinct lipid preferences and that substrate lipid length plays a critical role in processive polymerization.
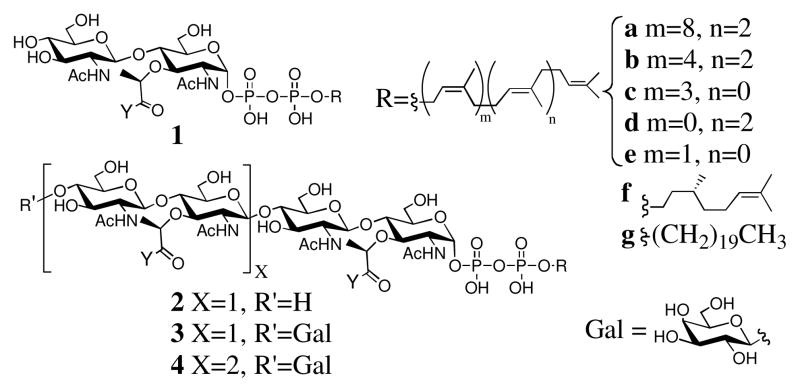
Like many other bacterial cell surface polymers, peptidoglycan precursors are assembled on undecaprenol, a 55 carbon lipid carrier (a, Figure 1). The membrane-associated enzymes involved in undecaprenyl-dependent pathways often accept much shorter (ten to fifteen carbon) lipid substituents in vitro.4 Thus, we were surprised when early efforts to define the PGT substrate requirements showed that these enzymes required at least a 35 carbon substituent (1b).5 We had no way to distinguish whether the observed requirement reflected the preferences of the donor site, of the acceptor site, or of both.
Figure 1.
Structures of LP2 (1), LP4 (2), blocked LP4 (GLP4, 3), blocked LP6 (GLP6, 4). Y = L-Ala–γ-D-Glu–L-Lys–D-Ala–D-Ala.
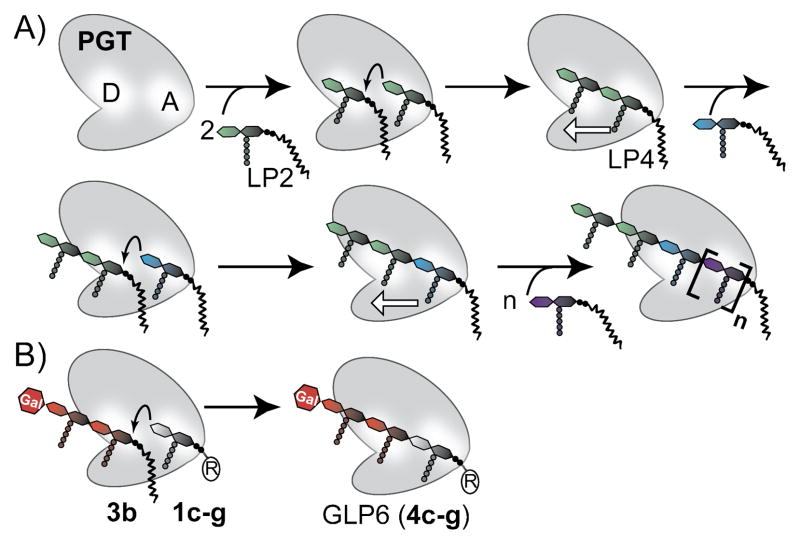
Distinguishing between the donor and acceptor sites of any polymerase that converts a single substrate into a polymeric product is a challenging task.6 To differentiate between the sites, we synthesized an analog of the elongating strand, Lipid IV (2b, LP4).7 LP4, however, was homopolymerized by several PGTs and therefore it could not be used to selectively probe the polymer (donor) site. To generate donor-only substrates, we developed a method to block the nonreducing end of PGT substrates via attachment of galactose.3 Access to a combination of blocked (e.g., GLP4, 3b) and unblocked PGT substrates containing different lipid lengths allowed us to directly probe the substrate lipid requirements in the acceptor site for the first time (Figure 2B).
Figure 2. The PGT homopolymerization (A) and acceptor site (B) assays.
A) PGTs contain two substrate binding sites; one for a glycosyl donor (D) and one for a glycosyl acceptor (A). During initiation, one LP2 serves as the donor and another as acceptor. Following the first turnover (second row), the elongating strand is the donor and LP2 is the acceptor. In this assay, LP2 must react from both donor and acceptor site to form product. B) For the acceptor site assay, GLP4 (3b) is the donor and the acceptor is varied. Formation of GLP6 demonstrates that the LP2 analog (1c–g) can be an acceptor.
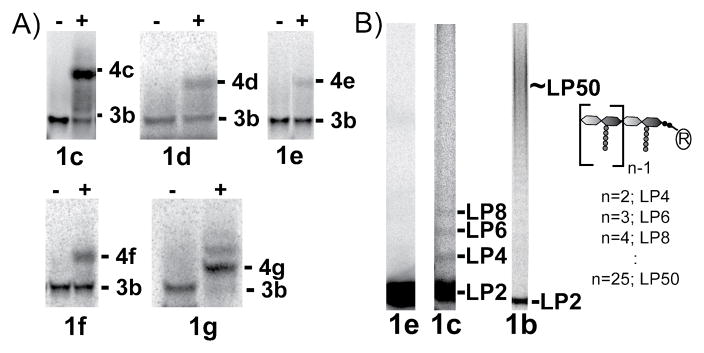
We have prepared a series of LP2 analogs (1c–g) and used a SDS-PAGE assay, which separates peptidoglycan chains to single disaccharide resolution,7b to monitor the ability of the substrates to react with the blocked Lipid IV donor substrate, GLP4 (3b, Figure 2B). The LP2 analogs investigated varied in both the length and geometry of the lipid. For example, 1c and 1e are shortened analogs of 1b (15 and 25 carbons shorter, respectively) and they maintain the cis-isoprene bonds of the natural lipid, while the lipid of 1d is shorter and contains all trans-isoprene units. The citronellyl derivative (f) and saturated C20 derivative (g) allowed us to probe the effects of unsaturation and lack of methyl branching. All the short chain lipid analogs 1c–1g reacted with radiolabeled GLP4 (3b*) in the presence of the PGT domain from Aquifex aeolicus PBP1A (AaPBP1A)2a to produce a single major product with migratory properties consistent with the formation of GLP6 (4, Figure 3A). To confirm this assignment, we analyzed the products formed by reaction of 3b and 1d by Escherichia coli PBP1A via high resolution mass spectrometry. A product with an exact mass identical to 4d was identified (Figure S3).
Figure 3. SDS-PAGE analysis of the acceptor site assay (A) and the homopolymerization assay (B).
A) [14C]-3b and [14C]-1c–g were incubated in the absence (−) or presence (+) of AaPBP1A.2a In all cases, predominantly one product was formed, GLP6 (4c–g). B) Comparison of the homopolymerization of 1e, 1c, and 1b by AaPBP1A. The C10 derivative (1e) cannot be homopolymerized, while the C20 (1c) and C35 (1b) derivatives are homopolymerized via a distributive and a processive mechanism, respectively.
In addition to providing a direct readout of acceptor substrate specificity, our assay also revealed information about the lipid requirements in the donor site. Previous work had demonstrated that lipid analogs 1c–1g do not react in a homopolymerization assay, which requires LP2 to act as both donor and acceptor (Figure 2A). Since we now know that all these analogs are substrates for the acceptor site,8 we have concluded that PGTs are promiscuous with respect to the acceptor lipid but have a stringent requirement for longer lipids in the donor site. This conclusion explains the observation that all of the short lipid substrates react to give one major product, GLP6.9 Since these products contain short lipids, they are poor donors and chain extension does not readily occur.
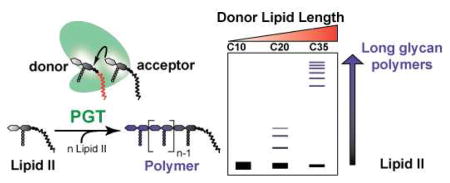
In the acceptor assay reactions with 1c, we noted the presence of a few faint bands in addition to the band for GLP6 (Figure S1). High resolution mass spectrometry was used to analyze reactions of GLP4 (3b) with 1c and revealed products that could only arise from reaction of the C20 derivative from the donor site (C20-GLP8; Figure S3). To further examine this reactivity, we compared the products produced via the homopolymerization of 1e, 1c, or 1b (containing 10, 20 and 35 carbon lipids, respectively) by AaPBP1A. With the shortest LP2 substrate, C10-LP2 (1e), we did not observe even a single turnover at high (10 μM) enzyme concentrations (Figure 3B), indicating that under no circumstances can it function as a glycosyl donor. In contrast, C20-LP2 (1c), which contains a lipid chain with two more isoprene units, did react, producing bands of decreasing electrophoretic mobility that represent products differing in size by a single disaccharide unit (Figure 3B, panel 2). The largest product observed had undergone three rounds of coupling. These results show that C20-LP2 is capable of serving as a glycosyl donor, but the product distribution is in striking contrast to that observed for C35-LP2 (1b), which reacts in a processive manner to give long polymers even at enzyme:substrate ratios of 1:1.1 Therefore, the length of the lipid in the donor site is a major determinant of the processivity of the enzymatic reaction.
Our results show that PGTs have different lipid length requirements in the donor and acceptor sites. Although the acceptor site can tolerate short lipid chains, the donor site requires a lipid longer than 20 carbons to undergo processive polymerization. Kiessling and coworkers have recently reported that the glycan polymerase GlfT2 requires a long lipid “tether” for processive polymerization.4h Since GlfT2 elongates its glycan polymer at its nonreducing end while PGTs elongate from the reducing end, the PGTs represent a different kind of lipid-assisted processivity. During each round of elongation, the polymer must translocate (white arrow, Figure 2A) to position the new donor substrate in the active site. It has been suggested that the primary role of the lipid in PG substrates is to anchor them to the membrane. Since these experiments were carried out in the absence of membranes, this lipid-dependent processivity is a function of intrinsic interactions between the donor lipid and the enzyme. Recent structures of several PGTs have revealed the presence of a large hydrophobic patch that funnels down towards the membrane.2 This hydrophobic patch may play a role in tethering the growing strand to the donor site during processive polymerization.
Supplementary Material
Acknowledgments
This work was supported by the NIH (GM076710 to SW and DK and a postdoctoral fellowship to DLP). Emma Doud was supported by an NSF Fellowship.
Footnotes
Supporting information available. Experimental procedures and mass spectrometry data are available.
References
- 1.Wang TA, Manning SA, Walker S, Kahne D. J Am Chem Soc. 2008;130:14068–9. doi: 10.1021/ja806016y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.(a) Yuan Y, Barrett D, Zhang Y, Kahne D, Sliz P, Walker S. Proc Natl Acad Sci USA. 2007;104:5348–53. doi: 10.1073/pnas.0701160104. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Lovering AL, de Castro LH, Lim D, Strynadka NC. Science. 2007;315:1402–5. doi: 10.1126/science.1136611. [DOI] [PubMed] [Google Scholar]; (c) Yuan Y, Fuse S, Ostash B, Sliz P, Kahne D, Walker S. ACS Chem Biol. 2008;3:429–36. doi: 10.1021/cb800078a. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Sung MT, Lai YT, Huang CY, Chou LY, Shih HW, Cheng WC, Wong CH, Ma C. Proc Natl Acad Sci USA. 2009;106:8824–9. doi: 10.1073/pnas.0904030106. [DOI] [PMC free article] [PubMed] [Google Scholar]; (e) Heaslet H, Shaw B, Mistry A, Miller AA. J Struct Biol. 2009;167:129–35. doi: 10.1016/j.jsb.2009.04.010. [DOI] [PubMed] [Google Scholar]
- 3.Perlstein DL, Zhang Y, Wang TA, Kahne DE, Walker S. J Am Chem Soc. 2007;129:12674–5. doi: 10.1021/ja075965y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.(a) Chen L, Men H, Ha S, Ye X, Brunner L, Hu Y, Walker S. Biochemistry. 2002;41:6824–33. doi: 10.1021/bi0256678. [DOI] [PubMed] [Google Scholar]; (b) Zhang YH, Ginsberg C, Yuan Y, Walker S. Biochemistry. 2006;45:10895–904. doi: 10.1021/bi060872z. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Ginsberg C, Zhang YH, Yuan Y, Walker S. ACS Chem Biol. 2006;1:25–8. doi: 10.1021/cb0500041. [DOI] [PubMed] [Google Scholar]; (d) Chen MM, Weerapana E, Ciepichal E, Stupak J, Reid CW, Swiezewska E, Imperiali B. Biochemistry. 2007;46:14342–8. doi: 10.1021/bi701956x. [DOI] [PMC free article] [PubMed] [Google Scholar]; (e) Faridmoayer A, Fentabil MA, Haurat MF, Yi W, Woodward R, Wang PG, Feldman MF. J Biol Chem. 2008;283:34596–604. doi: 10.1074/jbc.M807113200. [DOI] [PMC free article] [PubMed] [Google Scholar]; (f) Brown S, Zhang YH, Walker S. Chem Biol. 2008;15:12–21. doi: 10.1016/j.chembiol.2007.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]; (g) Sewell EW, Pereira MP, Brown ED. J Biol Chem. 2009;284:21132–8. doi: 10.1074/jbc.M109.010215. [DOI] [PMC free article] [PubMed] [Google Scholar]; (h) May JF, Splain RA, Brotschi C, Kiessling LL. Proc Natl Acad Sci USA. 2009;106:11851–6. doi: 10.1073/pnas.0901407106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ye XY, Lo MC, Brunner L, Walker D, Kahne D, Walker S. J Am Chem Soc. 2001;123:3155–6. doi: 10.1021/ja010028q. [DOI] [PubMed] [Google Scholar]
- 6.Stubbe J, Tian J, He A, Sinskey AJ, Lawrence AG, Liu P. Annu Rev Biochem. 2005;74:433–80. doi: 10.1146/annurev.biochem.74.082803.133013. [DOI] [PubMed] [Google Scholar]
- 7.(a) Zhang Y, Fechter EJ, Wang TSA, Barrett D, Walker S, Kahne DE. J Am Chem Soc. 2007;129:3080–1. doi: 10.1021/ja069060g. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Barrett D, Wang TS, Yuan Y, Zhang Y, Kahne D, Walker S. J Biol Chem. 2007;282:31964. doi: 10.1074/jbc.M705440200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Short lipid analogs are also acceptor substrates for PBP1A and PBP1B from E. coli (Figure S1).
- 9.The ratio of GLP4:L2 in these experiments was roughly 1:1, increasing the ratio to 1:5 in reactions of 3b with C10-LP2 (1f) does not produce products longer than GLP6 (4f) whereas reactions of 3b and 1b produce numerous higher molecular weight products (Figure S2).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.