Summary
Many bacteria form Gln-tRNAGln and Asn-tRNAAsn by conversion of the misacylated Glu-tRNAGln and Asp-tRNAAsn species catalyzed by the GatCAB amidotransferase in the presence of ATP and an amide donor (glutamine or asparagine). Here we report the crystal structures of GatCAB from the hyperthermophilic bacterium Aquifex aeolicus complexed with glutamine, asparagine, aspartate, ADP, or ATP. In contrast to the Staphylococcus aureus GatCAB, the A. aeolicus enzyme formed acyl-enzyme intermediates with either glutamine or asparagine in line with the equally facile use by the amidotransferase of these amino acids as amide donors in the transamidation reaction. A water-filled ammonia channel is open throughout the length of the A. aeolicus GatCAB from the GatA active site to the kinase catalytic pocket in the B-subunit. A non-catalytic Zn2+ site in the A. aeolicus GatB stabilizes subunit contacts and the ammonia channel. Judged from sequence conservation in the known GatCAB sequences, the Zn2+ binding motif was likely present in the primordial GatB/E, but became lost in certain lineages (e.g. S. aureus GatB). Two divalent metal binding sites, one permanent and the other transient, are present in the catalytic pocket of the A. aeolicus GatB. The two sites enable GatCAB to first phosphorylate the misacylated tRNA substrate and then amidate the activated intermediate to form the cognate products, Gln-tRNAGln or Asn-tRNAAsn.1
Keywords: tRNA-dependent amidotransferase, GatCAB, crystal structure, amidase superfamily, amidotransferase evolution
Introduction
The correct pairing of an amino acid with its cognate tRNA is essential for the fidelity of protein synthesis. In general, aminoacyl-tRNAs (aa-tRNAs) are synthesized by a set of aminoacyl-tRNA synthetases (aaRSs) in the cell, each enzyme specific for one amino acid/tRNA cognate pair.1 However in the majority of bacteria and all known archaea, glutaminyl-tRNA synthetase (GlnRS) for Gln-tRNAGln synthesis is not encoded.2 Similarly, in most prokaryotes asparaginyl-tRNA synthetase (AsnRS) is absent.2 In these organisms, Gln-tRNAGln or Asn-tRNAAsn is formed by indirect routes in which an incorrectly acylated tRNA is converted to the cognate one. This involves a non-discriminating aaRS (ND-aaRS) possessing relaxed tRNA specificity, and a tRNA-dependent amidotransferase (AdT).2 For Gln-tRNAGln biosynthesis, a non-discriminating glutamyl-tRNA synthetase (ND-GluRS) first glutamylates tRNAGln to form Glu-tRNAGln.3 This misacylated Glu-tRNAGln is subsequently converted to Gln-tRNAGln by a glutamyl-tRNAGln amidotransferase (Glu-AdT).4 Analogously, Asn-tRNAAsn is formed by the concerted actions of a non-discriminating aspartyl-tRNA synthetase (ND-AspRS)5 and an Asp-tRNAAsn amidotransferase (Asp-AdT).6
Two AdTs are found in nature, a heterotrimeric GatCAB7 and a heterodimeric GatDE enzyme.8 The latter is specific for Glu-tRNAGln and found only in archaea. GatCAB is encoded in most bacteria as well as in all known archaea lacking an AsnRS.2 In vitro, bacterial GatCAB can act as both a Glu-AdT and an Asp-AdT to form either Gln-tRNAGln or Asn-tRNAAsn,2 while the Methanothermobacter thermautotrophicus GatCAB prefers Asp-tRNAAsn over homologous Glu-tRNAGln.9 The in vivo role of GatCAB in bacteria is determined by which ND-aaRS is encoded (ND-GluRS and/or ND-AspRS).2 In organisms that possess both ND-aaRSs (e.g. Helicobacter pylori and Aquifex aeolicus), GatCAB is employed for both Gln-tRNAGln and Asn-tRNAAsn biosynthesis.10
A similar mechanism for tRNA-dependent transamidation has been proposed for both AdTs: phosphorylation of the misacylated tRNA substrate using ATP to form an activated intermediate, hydrolysis of an amide donor (Gln or Asn) to liberate ammonia, and amidation of the activated intermediate using the liberated ammonia.11; 12 The B- and E-subunits are homologs8; 13 and serve as the synthetase domains of their respective AdTs, catalyzing the formation of the activated intermediate as well as the amidation step.12; 14; 15 GatA and GatD are functionally equivalent, serving as the glutaminase subunits to liberate ammonia from an amide donor.12; 16 However, GatA belongs to the amidase family of enzymes7; 16 while GatD is a homolog of L-asparaginases.12; 17 The small C-subunit of the heterotrimeric AdT is thought to play a stabilizing role in the holoenzyme.7; 14
The crystal structure of the S. aureus GatCAB complexed with either Gln or Asn suggested that Gln would serve as a better amide donor than Asn for transamidation.14 In agreement with the structural data, biochemical studies of other bacterial GatCAB enzymes have shown them to be significantly more active using Gln than Asn as the donor.10; 11; 18; 19; 20 Here we report the crystal structures of GatCAB from the hyperthermophilic bacterium A. aeolicus in the presence of Asn, Gln, Asp, and unreacted ATP with Mn2+ ions. In contrast to the properties of S. aureus GatCAB, both amide donors (Asn and Gln) formed covalent intermediates with the A. aeolicus GatCAB in agreement with our biochemical data showing that the enzyme uses the two amide donors with similar efficiencies. In addition, the A. aeolicus GatCAB structure revealed in the B-subunit a Zn2+ binding motif conserved in many other bacterial and all known archaeal GatB proteins, and in most GatE sequences. Such conservation is indicative that the ancestral GatB/E also had such a Zn2+ binding site. Free Asp, a mimic of the substrate Asp-tRNAAsn, bound to one of two Mn2+ ions in the GatB active site. Based on the complex structure of GatCAB/ATP/Asp, we propose a model for the phosphoryl- and ammonia-transfer reactions catalyzed by GatB, in which Mg2+ helps position the charged tRNA substrate in both reactions.
Results
A. aeolicus GatCAB structure
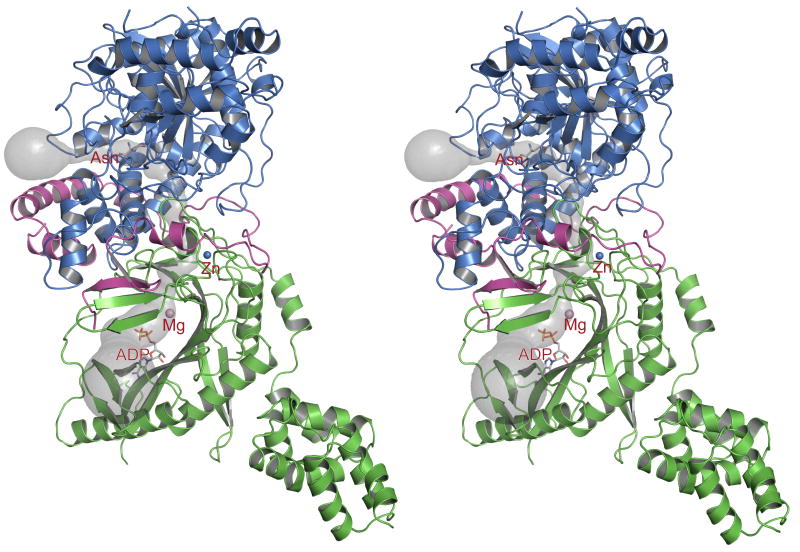
The crystal structure of A. aeolicus GatCAB-Asn/ADP complex (Fig. 1) was solved with a combination of experimental and molecular replacement phasing, and refined to 2.3 Å (R factor =23.8%, Rfree = 28.3%, Table 1). The asymmetric unit contained eight GatCAB molecules.
Figure 1.
Overall structure of the A. aeolicus GatCAB-Asn/ADP complex. Each subunit is colored differently in the stereo ribbon diagram: blue for the A-subunit, green for the B-subunit, and magenta for the C-subunit. The A-subunit contains the acyl enzyme from substrate Asn and the B-subunit contains ADP, Mg2+ and Zn2+. Ligands are drawn in stick form with atomic coloring: gray C, red O, blue N, orange P, pink Mg and blue Zn. A tunnel linking the amidase active site in the A-subunit with the synthetase active site in the B-subunit is shown as a transparent surface.
Table 1. Data Collection and Refinement Statistics.
| Hg | SeMet | Asn, ADP | Gln | Asn, ATP, Asp | |
|---|---|---|---|---|---|
| Crystallization/soaking ligands | 10 mM Asn | 10 mM Gln | 10 mM Asn | ||
| 10 mM Asp | |||||
| 0.6 mM ATP | 10 mM ATP | ||||
| 10 mM Mg2+ | 10 mM Mg2+ | 10 mM Mn2+ | |||
| Data Collection | |||||
| Wavelength (Å) | 1.007 | 0.9804 | 0.97934 | 1.0274 | 0.97934 |
| dmin (Å) | 3.2 | 3.2 | 2.3 | 2.8 | 3.0 |
| Space group | P21 | P21 | P1 | P1 | P1 |
| Cell dimensions a, b, c, (Å) α, β, γ (°) |
127.03 | 127.28 | 127.48 | 127.55 | 127.68 |
| 128.97 | 129.28 | 131.01 | 129.64 | 130.25 | |
| 154.68 | 155.17 | 154.67 | 154.33 | 153.76 | |
| 90.0 | 90.0 | 90.02 | 90.08 | 89.79 | |
| 90.0 | 90.3 | 90.00 | 90.27 | 90.25 | |
| 90.0 | 90.0 | 89.91 | 89.98 | 90.00 | |
| Unique reflections | 161,236 | 163,171 | 425,169 | 242,562 | 182,013 |
| Completeness (%) | 99.7(99.0)a | 99.9(100) | 96.3(90.2) | 98.6(97.2) | 91.2(84.5) |
| Rmergeb | 0.119(0.37) | 0.113(0.38) | 0.060(0.347) | 0.063(0.474) | 0.118(0.715) |
| Avg I/σI | 11(4) | 15(5) | 12.4(1.7) | 14.1(1.7) | 5.3(1.3) |
| Avg redundancy | 5 | 6.6 | 1.9 | 2.0 | 1.7 |
| Crystallographic Refinement | |||||
| No. atoms (protein/waters) | 62,952/983 | 62,952 | 62,952 | ||
| Rwork/Rfreec | 0.238/0.283 | 0.255/0.303 | 0.252/0.309 | ||
| RMSD - bond lengths (Å) | 0.010 | 0.013 | 0.014 | ||
| RMSD - bond angles (°) | 1.419 | 1.385 | 1.482 | ||
| Ramachandran plotd (favored/allowed/disallowed, %) | 97.0/3.0/0 | 95.6/4.4/0 | 95.1/4.9/0 | ||
| Avg. B factor (protein/ligand, Å2) | 50.3/53.7 | 59.2/58.6 | 60.5/49.9 | ||
| PDB code | 3H0L | 3H0M | 3H0R | ||
| Modeled Ligands | |||||
| 8 Asn | 8 Gln | 8 Asn | |||
| 8 ADP | 6 ATP, 2 ADP | ||||
| 2 Asp | |||||
| 8 Mg2+ | 6 Mg2+ | 16 Mn2+ | |||
| 8 Zn2+ | 8 Zn2+ | 8 Zn2+ | |||
Values in parenthesis are for the highest resolution bin.
Rmerge = ΣhΣj | <I>h - Ih,j | /ΣhΣjIh,j, where <I>h is the mean intensity of symmetry-equivalent reflections.
Rwork = Σ | Fobs - Fcal | /ΣFobs, where Fobs and Fcal are observed and calculated structure factor amplitudes. Rfree was calculated for a 5% subset of reflections excluded from refinement.
Ramachandran plot was calculated by MolProbity44.
The A. aeolicus A-subunit, which includes all 478 amino acids residues, is similar to the A-subunits of S. aureus (RMSD of 0.68 Å for 478 residues of PDB code 2G5H)14 and of Thermotoga maritima (RMSD of 0.71 Å for 475 residues of PDB 2GI3). Briefly, the A-subunit contains a conserved amidase signature (AS) sequence (62-192) that forms the enzymatic core comprised of an 11-stranded β sheet surrounded by twelve α-helices.
The B-subunit (478 amino acids) comprises an N-terminal “cradle” domain (3-293), similar to the domain in the B-subunit of S. aureus (RMSD of 1.1 Å)14, and the E. subunits of M. thermautotrophicus15 and P. abyssi17 (RMSD of 1.7 Å and 1.5 Å, respectively), followed sequentially by a helical domain (294-412). The two N-terminal residues of the A. aeolicus B-subunit were not visible in the electron density map, nor was the C-terminal YqeY-like tail (413-478) of the subunit implicated in tRNA binding,13; 14; 15; 21 as was the case with the S. aureus GatCAB structure.14
The A-B subunit interface buries a total of 2292 Å2 of molecular surface. Like in the S. aureus GatCAB,14 the A. aeolicus C-subunit (94 aa long) wraps around the interface of the A- and B-subunits. All residues of the C-subunit were well defined by electron density except residues 1, 93 and 94. The conserved side chains of Arg61 and Asp63 in an extended loop region of the C-subunit are hydrogen bonded with the main chain of Thr21 and the conserved side chain of Asn54 of the B-subunit. The main chain carbonyl of C-subunit Leu13 is hydrogen bonded with the side chain of conserved Arg271 of the B-subunit.
Substrate binding and catalysis in the A-subunit amidase active site
Covalent intermediate in the amidase active site
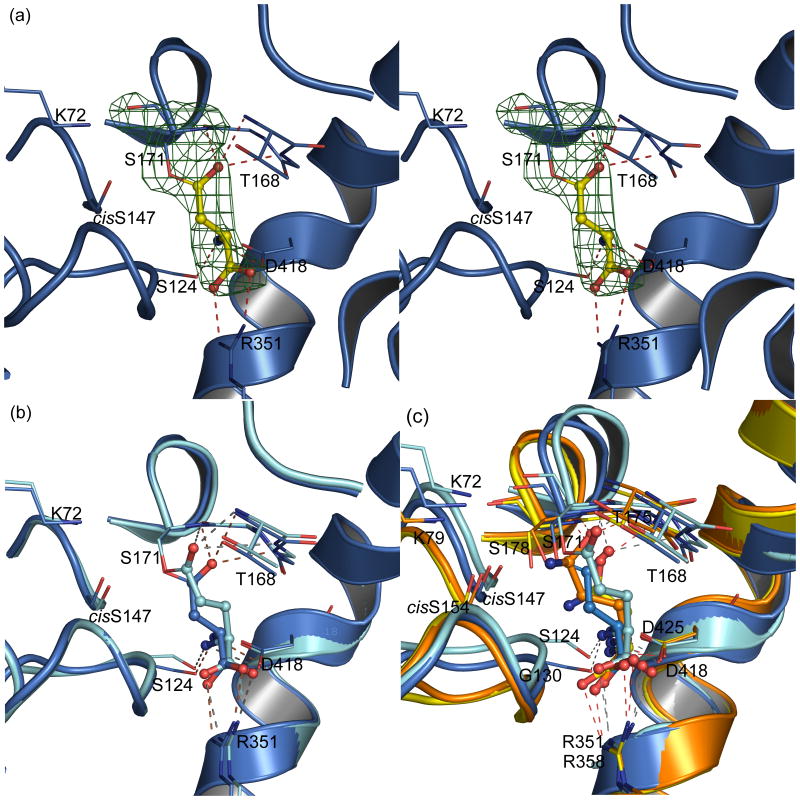
The A-subunit of GatCAB functions to liberate ammonia from an amide donor and belongs to the amidase family of enzymes.10; 14; 16 Amidases are characterized by Gly/Ser-rich sequence motif, which folds to assemble a Ser-cisSer-Lys “catalytic scissors” (Ser171, cisSer147, and Lys72 in the A. aeolicus A-subunit).22; 23 Co-crystals with Gln or with Asn resulted in electron density consistent with formation of the acyl intermediate at nucleophilic Ser171 (Fig. 2a,b). The carbonyl oxygen of the acyl group is stabilized in an oxyanion hole formed by the main chain amides of Thr168, Gly169, and Ser171 (Fig. 2a,b). The α-amino group of Gln/Asn is recognized through hydrogen bonding to the side chains of Asp418 and Ser124. The substrate α-carboxyl group is coordinated by a bidentate salt bridge to Arg351. Together these three residues anchor the substrate in the active site, placing the substrate's amide group in position for nucleophilic attack by Ser171.
Figure 2.
Acyl enzyme in the GatA amidase active site. (a). Stereo view of the A-subunit active site with acyl enzyme from substrate Asn. Critical residues in the active site are shown as thin sticks, and the substrate Asn in ball-and-stick form with atomic coloring: yellow C, blue N, red O. The omit |Fo-Fc| electron density map (contoured at 4σ, green mesh, with Asn and Ser171 omitted) demonstrates formation of the acyl-enzyme intermediate of the Asn with Ser171. Hydrogen bonds are shown as dashed lines. (b). Superposition of the amidase active site in complexes with Asn (blue) and Gln (cyan). The covalent acyl-enzyme intermediate is formed with both substrates. (c). Combined view of Asn and Gln complexes of the A. aeolicus Gat A (blue Asn complex; cyan Gln complex) and the S. aureus GatA (yellow Asn complex; orange Gln complex). The superposition is based on the common anchor residues (Arg351 and Asp418 in A. aeolicus GatA and Arg258 and Asp425 in S. aureus GatA) to illustrate the difference in distance between the anchor and the nucleophilic Ser. In both enzymes, the side chains of Asp418 and Arg351 anchor the α-amino and α-carboxyl groups. With both nitrogen donors, the acyl enzyme forms in the A. aeolicus A-subunit. In the S. aureus A-subunit, Asn does not reach the Ser178 nucleophile from its anchor.
Substrate selectivity in the amidase active site
The intriguing observation that the A-subunit formed an acyl-enzyme intermediate with both Gln and Asn suggested the A. aeolicus GatCAB would be able to efficiently use both amino acids as amide donors. Consistent with the structural results, we found the enzyme readily used both Gln and Asn as donors for transamidation (Table 2). The A. aeolicus GatCAB with Gln as the amide donor had a kcat about 2.6-fold greater than when Asn was the donor (Table 2). In contrast, the enzyme had about a 2.4-fold lower KM for Asn than Gln (11.2 μM and 26.4 μM, respectively). The net effect is that the A. aeolicus GatCAB was equally efficient using Asn as an amide donor as Gln (kcat/KM of 9.7 s-1/mM and 11.1 s-1/mM, respectively).
Table 2. Kinetic data for A. aeolicus GatCAB – amidotransferase activity*.
| Substratea | KM(μM) | kcat (s-1) | kcat/KM(s-1/μM) (x 10-3) |
|---|---|---|---|
| Glu-tRNAGln b | 1.71 ± 0.29 | 0.43 ± 0.03 | 242 ± 51.6 |
| Asp-tRNAAsn b | 1.68 ± 0.27 | 0.30 ± 0.01 | 180 ± 29.7 |
| Glnc | 26.4 ± 4.9 | 0.29 ± 0.01 | 11.1 ± 2.1 |
| Asnc | 11.2 ± 5.3 | 0.11 ± 0.01 | 9.7 ± 4.7 |
Steady-state kinetics of the A. aeolicus GatCAB amidotransferase activity, see Materials and Methods for details. Measurements were done three to four times. Standard deviations are reported.
KM was determined by varying the concentration of the substrate listed while adding the other two substrates required for transamidation in excess with 20 nM GatCAB.
4 mM Gln and 4 mM ATP were added in excess.
10-11 μM Asp-tRNAAsn and 4 mM ATP were added.
These results are in contrast to previous studies of bacterial GatCAB enzymes in which a preference was detected for either Gln or Asn.10; 11; 14; 18; 19; 20 For example, co-crystal structures of S. aureus GatCAB with Asn and Gln revealed binding of both substrates but acyl-enzyme formation only with Gln.14 The A-subunits of both A. aeolicus and S. aureus GatCAB recognize both Gln and Asn in an identical manner using two invariant “anchor” side chains: an Arg for the substrate α-carboxyl group and an Asp for the α-amino group (Fig. 2c). The ability to form an acyl-enzyme intermediate at the nucleophilic serine depends on the distance of the Ser-cisSer-Lys catalytic scissors from the Arg-Asp anchor residues. Asn binds in a more extended conformation and Gln in a less extended conformation in the active site of the A. aeolicus A-subunit than is seen in the S. aureus GatCAB structures.14 Superposition of the critical anchor residues (A. aeolicus GatA Arg351 and Asp418, S. aureus GatA Arg358 and Asp425) reveals the anchor is 1.3 Å closer to the Ser-cisSer-Lys catalytic scissors in the A. aeolicus A-subunit than in the S. aureus A-subunit (Fig. 2c). This shift, although small, is responsible for the acceptance of both Gln and Asn substrates by A. aeolicus GatCAB.
Structure alignment of the A. aeolicus and S. aureus A-subunits together with sequence alignment of the A-subunits of GatCAB enzymes with known preference for an amide donor (Supplemental Figure 1) does not reveal a clear pattern to predict whether a GatCAB enzyme will or will not prefer Gln. For example, a hydrogen bond between A. aeolicus GatA Ser124 and the substrate α-amino group is an obvious candidate in enabling the enzyme to use Asn as well as Gln because the analogous residue is a Gly (131) in the S. aureus A-subunit. However, the Bacillus subtilis, H. pylori, and Neisseria meningitidis GatCAB enzymes, which all prefer Gln over Asn (10-fold, 130-fold, and 830-fold respectively),10; 18; 20 have a Ser at the equivalent position in their A-subunits like the A. aeolicus AdT (Supplemental Figure 1). The critical difference in distance between catalytic scissors and anchor residues cannot be attributed to a few amino acid differences and likely results from many subtle changes due to sequence differences outside the active site pocket and perhaps outside the amidase core.
Comparison of GatA active site with other amidases
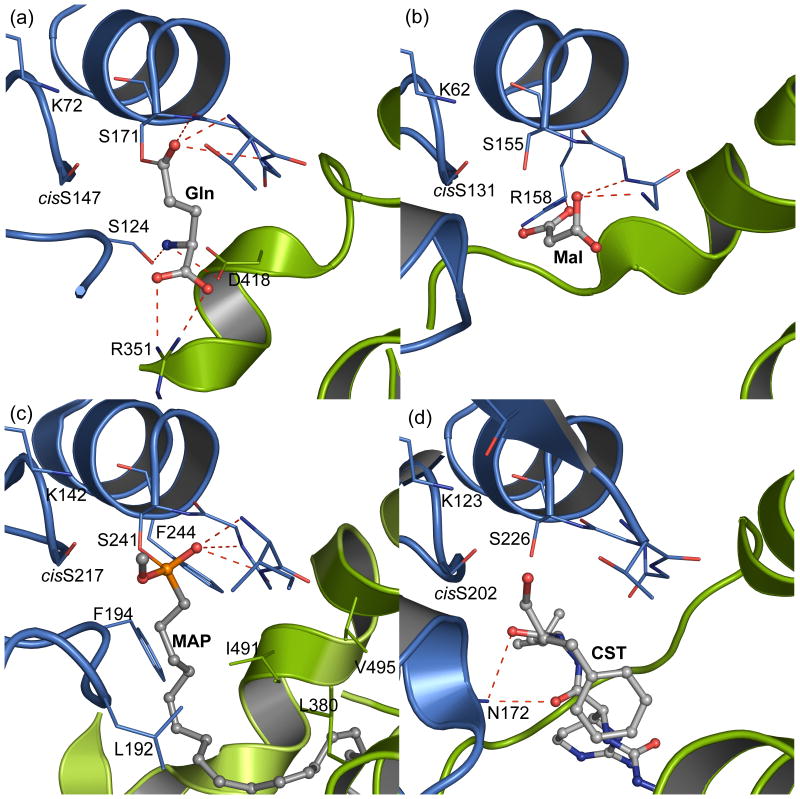
For insights to the structural basis of substrate specificity in the amidase family, we compared the active-site structures of the GatCAB A-subunit and other amidases (malonamidase E2 (MAE2),22 peptide amidase (PAM),24 and fatty acid amide hydrolase (FAAH)25). Whereas the detailed structures of the Ser-cisSer-Lys catalytic scissors and of the residues that stabilize the oxyanion intermediate are highly similar in the four amidases, substrate recognition differs in the four enzymes (Fig. 3a-d). In PAM, MAE2, and FAAH, substrate recognition is achieved through residues in the core region. While regions outside the core interact with substrates in all four amidases, critical substrate-recognition residues are located outside the core only in GatA (Arg351 and Asp418, Fig. 2).
Figure 3.
Comparison of amidase active sites. (a). A-subunit of A. aeolicus GatCAB, showing the acyl enzyme intermediate of substrate Gln with Ser171. (b). Active site of malonamidase (MAE2) in complex with product malonate.22 Arg158 in the amidase core region interacts with a carboxyl group of malonate (Mal). (c). Active site of fatty acid amide hydrolase (FAAH) with the inactivator methoxy arachidonyl phosphonate (MAP).25 The phosphonate of the covalent adduct at nucleophilic Ser241 mimics the tetrahedral intermediate of the hydrolytic reaction. Aromatic and aliphatic residues in the substrate binding pocket are indicated. (d). Active site of peptide amidase (PAM) in complex with the inhibitor chymostatin (CST)24. For each enzyme, the amidase core region (residues 62-192 of GatA, 52-176 of MAE2, 132-262 of FAAH and 113-246 of PAM) is colored blue, and residues outside the core region are colored green. Residues in the Ser-cisSer-Lys catalytic scissors of each enzyme and those interacting with ligands are shown as thin sticks; adducts and ligands are shown in ball-and-stick form with atomic coloring: gray C, red O, blue N, and orange P. Hydrogen bonds are shown as dashed lines.
Substrate and metal binding in the B-subunit synthetase active site
ADP binding
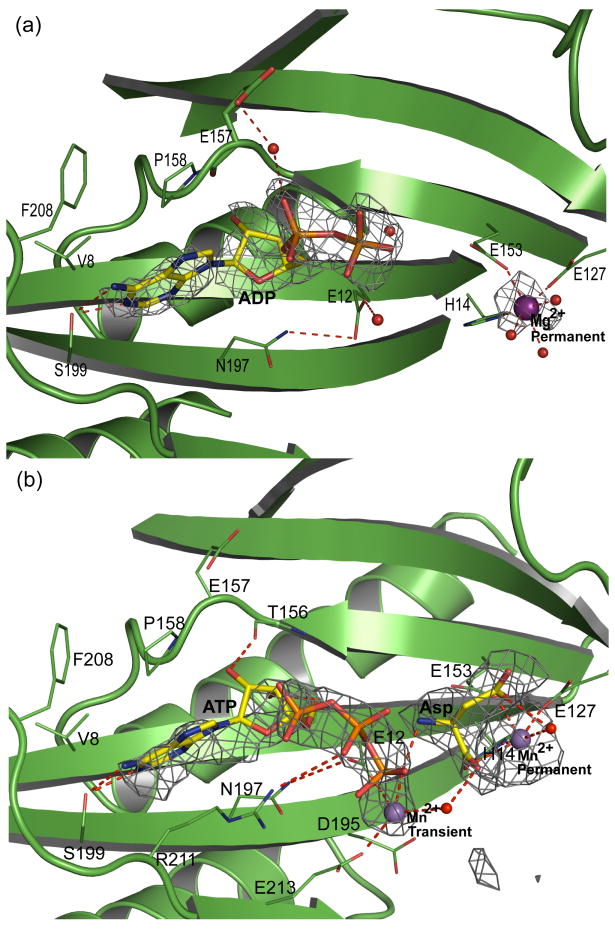
The ATP-dependent activation of substrate aa-tRNA (Glu-tRNAGln or Asp-tRNAAsn) leads to the formation of a phosphoryl-aa-tRNA intermediate that is then amidated with ammonia liberated by the A-subunit.11; 12; 26 To investigate the mechanism of activation, we prepared GatCAB crystals with ATP, divalent metal ions, and Asp as a mimic of Asp-tRNAAsn. The 2.3-Å electron density maps from crystals grown in presence of ATP and Mg2+ clearly indicated that the hydrolysis product ADP was bound in the B-subunit active site (Fig. 4a). Similar to ADP binding to the S. aureus B-subunit,14 the adenine base fits into a hydrophobic pocket formed by Val8, Phe208, and Pro158 with hydrogen bonds from N1 and N6 of the adenine to the conserved Ser199 side chain. The ADP α- and β-phosphates interact with bound water molecules. No bound metal ion was associated with the ADP.
Figure 4.
Synthetase active site in the B-subunit. (a). ADP/Mg2+ complex. (b). ATP/Mn2+ complex. The environment of the nucleotide is shown together with omit |Fo-Fc| electron density (3σ, grey mesh). Residues in contact with nucleotide, Asp, Mg2+ and Mn2+ are represented as thin sticks, ligands as thick sticks, metal ions as large spheres, and water molecules as small spheres in atomic coloring: yellow C, red O, blue N, orange P, purple Mg2+, gray Mn2+.
ATP and transient metal binding
To trap an ATP complex, we crystallized the protein without nucleotide, soaked crystals briefly in a solution containing ATP, Mn2+ and Asp, and trapped the resulting complex by quick freezing in liquid N2. In the 3.0-Å electron-density map from these crystals, density is present for ATP in 6 of 8 B-subunits (Fig. 4b). Additional new density adjacent to the ATP γ-phosphate was assigned to Mn2+, which is coordinated by the γ-phosphate of ATP, by the conserved side chains of Glu12 and Glu213, and by a water molecule. We designate this as the “transient” metal site and infer it assists phosphoryl transfer. Consistent with this proposed critical role of the transient Mg2+ site, when we mutated Glu11 to Ala in the B-subunit of the H. pylori GatCAB (equivalent to A. aeolicus Glu12), the AdT when expressed with Deinococcus radiodurans ND-AspRS was no longer able to rescue the Asn auxotrophy of the E. coli JF448 strain27 (Supplemental Figure 2). Asn prototrophy of E. coli JF448 requires expression of both a functional ND-AspRS and GatCAB.28
Permanent metal binding site
In addition to the transient metal site associated with ATP binding, a “permanent” site was occupied in all electron density maps from all crystals examined. Metal in this site is coordinated by His14, Glu127 and Glu153. The “permanent” metal is too far from the nucleotide (>9 Å) to participate directly in the phosphoryl transfer reaction. However, its three protein ligands are invariant in all GatB and GatE sequences. Furthermore, we find electron density consistent with metal binding to the permanent site in all deposited structures of GatCAB14 and GatDE15; 17 regardless of whether the deposited structure includes a metal, water or no ligand at the permanent site.
Intriguingly, mutation of His13 to Ala in the B-subunit of the H. pylori GatCAB (equivalent to A. aeolicus His14) resulted in a mutant AdT that was no longer able to rescue the Asn auxotrophy of the E. coli JF448 strain when co-expressed with ND-AspRS (Supplemental Figure 2). This result and the fact that mutations to the equivalent residues (A. aeolicus His14, Glu127, and Glu153) in the permanent metal site of M. thermautotrophicus GatDE resulted in phosphorylation- and transamidase-inactive mutant enzymes15 suggests a critical role for the site in the GatB/E protein family. In the nucleotide-free and ADP complexes, three water molecules together with the invariant protein side chains form an octahedral coordination sphere for the permanent metal. The permanent and transient metal sites are 8 Å apart, and both are occupied in the ATP complex.
Asp binding
The GatCAB crystal with ATP was also soaked with 10 mM Asp as a mimic of the aminoacyl end of Asp-tRNAAsn. New electron density adjacent to the permanent metal appeared in 2 of 8 copies of the B-subunit and was assigned to bound Asp (Fig. 4b). One of the Asp carboxyl groups coordinated the metal ion in the permanent site, demonstrating that this site has the capacity to coordinate another negative charge in addition to the two Glu side chains from the protein. As bound to the permanent metal, Asp faces the ATP γ-phosphate (Fig. 4b).
Channel connects active sites
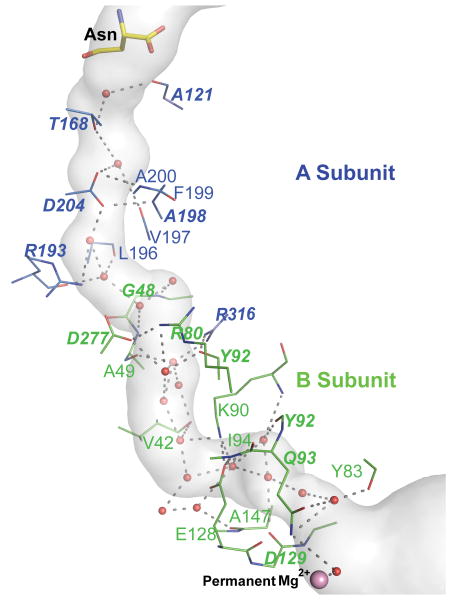
AdT catalyzed aa-tRNA activation and amide donor hydrolysis take place in active sites separated by more than 30 Å. Consequently ammonia is channeled from the A-subunit active site to the B-subunit active site. Like the S. aurues GatCAB,14 a continuous 35-Å long hydrophilic tunnel links the active sites in A. aeolicus GatCAB (Fig. 5). The tunnel in A. aeolicus GatCAB is open throughout its length (Fig. 5) and filled with 18 water molecules. These results differ from the interpretation of the crystal structure of S. aureus GatCAB,14 where it was reported that the Glu125 side chain blocks the channel via a salt bridge with Lys88. Both side chains are invariant in sequences of GatB and GatE. In A. aeolicus GatCAB, the analogous Glu128 forms a salt bridge with Lys90, but the channel is not blocked. We find the channel to be open in all A. aeolicus GatCAB structures and in all deposited structures of S. aureus GatCAB,14 based on accessibility calculations with the programs CAVER29 and castP30 using a probe radius ranging from 1.4-1.7 Å.
Figure 5.
Tunnel connecting the amidase (A-subunit) and synthetase (B-subunit) active sites. The putative ammonia channel (gray surface) in the GatCAB/Asn/ADP complex was calculated using the program CAVER29 with water molecules omitted. The 35-Å long channel is filled with 18 water molecules (red spheres), which interact with conserved polar residues (sticks with C atoms colored by subunit: A-subunit blue, B-subunit green). The acyl enzyme of Asn in the amidase active site is shown at the channel entrance, and Mg2+ in the permanent metal site (light purple sphere) in the synthetase active site is at the channel exit. Residues forming hydrogen bonds (dashed lines) with waters are shown in stick form. Conserved residues are labeled in bold type.
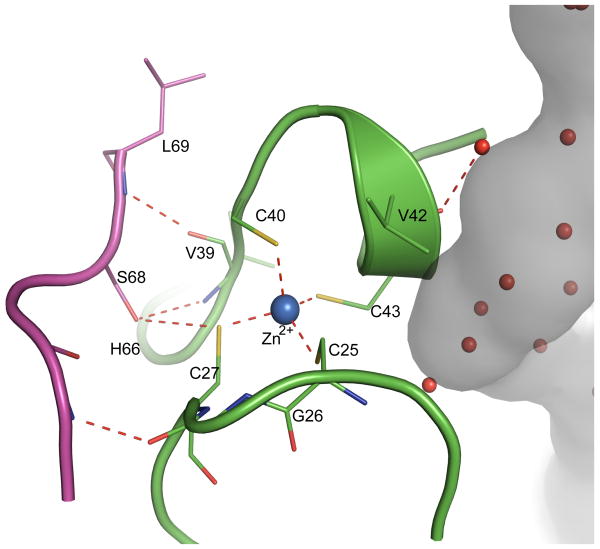
A. aeolicus GatCAB binds zinc
The initial electron density of GatCAB suggested a metal binding site with tetrahedral coordination by Cys25, Cys27, Cys40 and Cys43 in the cradle domain of the B-subunit. Subsequent data collection at an X-ray energy of 9.75 keV (λ = 1.27163 Å), just above the Zn K-edge (8.9 keV), yielded an anomalous difference electron density map clearly showing a Zn2+ in this site. The Zn2+ site sits squarely between the NH3 channel and the C-subunit. The Zn(Cys)4 stabilizes a B-subunit loop (residues 25 to 40) that forms several hydrogen bonds with the C-subunit, including a direct hydrogen bond from Ser68 in the C-subunit to the Cys27 Zn2+ ligand (Fig. 6a). In addition, residues Val42, Cys43 and Leu44 form part of the wall of the NH3 channel. Thus the Zn(Cys)4 is an important motif for maintenance of the channel and for binding the C-subunit. Sequence alignments reveal this motif comprised of four Cys residues is conserved in a number of bacterial and all archaeal B-subunits (Supplemental Figure 3). Among species whose B-subunits have the Cys motif, residue Ser68 is also conserved in the C-subunit. The S. aureus B-subunit lacks the Cys motif and no Zn2+ was found in the structure.14 The sequence alignment also suggests the Zn2+ motif is conserved with an Asp replacing the final Cys residue in all archaeal E-subunits except those from the Thermococci (Supplemental Figure 3). In agreement with the alignments, Zn2+ is coordinated by Cys26, Cys28, Cys79 and Asp82 in the M. thermautotrophicus E-subunit15 and no equivalent Zn2+ was found in the GatDE structure from the thermococcus P. abyssi.17 The path of the polypeptide is virtually identical in B-subunits and E-subunits whether or not they bind Zn, demonstrating the importance of this region to the subunit interface.
Figure 6.
Zn binding in the B-subunit. The Zn(Cys)4 center is bound in the B-subunit (green) between the ammonia channel (grey) and the C-subunit (magenta). C-subunit Ser68 is conserved in GatCABs having a Zn(Cys)4 motif in the B-subunit, and the hydrogen bond of Ser68 to Cys27 also likely conserved.
Discussion
Evolutionary perspective
The fact that A. aeolicus GatCAB can use the amide donors Gln and Asn about equally well means that its A-subunit is biochemically more similar to the M. thermautotrophicus enzyme9 than to the other bacterial homologs previously assayed.10; 11; 18; 19; 20 However, phylogenetic analysis indicates the A. aeolicus GatA is of the bacterial and not archaeal type.13 The lack of sequence conservation correlating with a preference, or lack thereof, for Gln over Asn may be indicative of sequence drift at permissible sites in the A-subunit. It should be noted while A. aeolicus GatCAB can use both amide donors, in vivo Gln is the likely donor. The organism cannot synthesize free Asn as its genome does not encode either of the known Asn synthetases.10 Thus, the A. aeolicus GatCAB equal use of Gln and Asn as amide donors in contrast to the S. aureus GatCAB preference for Gln14 may be due to chance (i.e. drift alone) and not selective pressure. The lack of clear sequence elements responsible for substrate specificity makes it difficult to predict whether the ancestral GatCAB preferred Gln or not.
In contrast, the conservation of the Zn2+ binding motif across the GatB/E family suggests that it was present in the ancestral GatB/E because the split of GatB and GatE occurred prior to the bacterial-archaeal phylogenetic divide.13 Whether the motif in the ancestral GatB/E was composed of four Cys residues or three Cys and one Asp residue is not clear, as the latter is found only in GatE sequences while the former only in GatB polypeptides. Why the Thermococci E-subunits and certain bacterial B-subunits such as S. aureus GatCAB now lack this motif is unclear. In bacteria, this motif appears to be lineage specific. For example, the B-subunits from Thermus thermophilus and the proteobacterium H. pylori retain the motif while the enzymes from Deinococcus geothermalis (evolutionarily related to T. thermophilus) and the proteobacterium N. meningitidis do not (Supplemental Figure 3). Thus loss of the Zn site appears to have occurred multiple times over the course of evolution.
GatB synthetase active site
The structures presented here extend our picture of catalysis in the synthetase active sites of both GatCAB and GatDE. The complex with ATP for the first time localizes the γ-phosphate within the active site. The new structures, together with previously determined ones14; 15; 17 and mutational analyses of H. pylori GatCAB and M. thermautotrophicus GatDE,15 highlight the importance of two divalent metal ions bound at “permanent” and “transient” sites. Based on the position of the ATP γ-phosphate, the metal in the transient site should directly assist in the activation reaction in which the γ-phosphate is transferred to the carboxyl group of misacylated substrate (Asp-tRNAAsn or Glu-tRNAGln). Consistent with this function, metal binding in the transient site is correlated with ATP binding in our structures.
In contrast, the metal ion in the permanent site is too far away to participate in the phosphoryl transfer reaction by direct interaction with the γ-phosphate. The three protein ligands of the permanent metal (His14, Glu127 and Glu153 of the B-subunit) are conserved across the GatB/GatE family, suggesting Mg2+ binds at this site in all the enzymes. However, not all deposited structures of GatCAB and GatDE have a metal in this site. By examining the deposited diffraction data, we found evidence of metal in the permanent site in all structures, regardless of the nucleotide-binding state. Therefore, the permanent Mg2+ site is likely occupied throughout the catalytic cycle. Moreover, the metal in this site is in a fixed position because its three protein ligands are part of secondary structures in the core of the AdT synthetase subunit.
In density maps of sufficient resolution, octahedral coordination of the metal is clear, with three water ligands in addition to the protein ligands. During the catalytic cycle, the water ligands in the active site cavity could be displaced by substrates or reaction intermediates. The permanent metal occupies a critical location in the synthetase active site (Fig. 7a). It faces the ATP γ-phosphate. It is adjacent to the mouth of the ammonia channel from the A-subunit and near the binding site for the tRNA aminoacyl 3′-adenosine. This prime location of the permanent metal site, its conservation, and the mutational analyses of H. pylori GatCAB (Supplemental Fig. 2) and M. thermautotrophicus GatDE15 argue for a critical function.
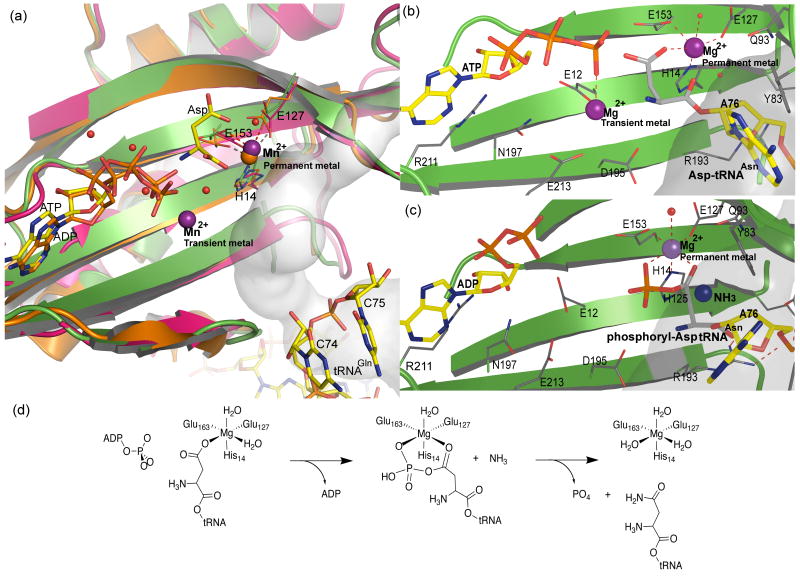
Figure 7.
Proposed reactions at the synthetase active site. (a). Superposition of synthetase active sites from crystal structures with informative ligands. The A. aeolicus GatCAB with ATP, Mn2+ and Asp (this work) is shown in green with gray C atoms for the ligands, the S. aureus GatCAB with ADP and Mg2+ 14 in orange with orange C for ligands, and the M. thermautotrophicus GatDE with tRNA 15 in magenta with yellow C for tRNA. Mn2+ ions are shown as purple spheres, Mg2+ in orange and waters as red spheres. Substrates ATP, ADP, Asp and the 3′-CCA of tRNAGln are represented as sticks. Residues interacting with substrates are represented by thick lines. The ammonia channel (grey surface) enters the synthetase active site from the right and is continuous with the tRNA binding site. (b). Model for the activation reaction. ATP is positioned as in the structure reported here. The terminus of Asp-tRNAAsn was modeled based on the GatDE-tRNA complex in which the 3′-terminal A was disordered. The Asp carboxyl group is coordinated by the metal in the permanent site, as in the Asp complex. (c). Model for the amidation complex. The activated substrate, phosphoryl-Asp-tRNAAsn, is shifted so that both phosphate and Oδ coordinate the permanent metal, thereby positioning the Asp Cγ atom at the exit of the ammonia tunnel, ready to receive ammonia from the amidase active site. (d). Schematic diagram of the reaction steps depicted in b and c.
The critical role of the permanent Mg2+ may be to position the substrate aa-tRNA for the activation and amidation reactions. A model for the two reactions (Fig. 7) includes essential elements of the crystal structures: ATP γ-phosphate coordination by the transient metal, permanent-metal coordination of an anionic substrate, and mobility of the tRNA aminoacyl 3′-adenosine (Fig. 7a). A precise location for the tRNA 3′-adenosine is unknown because the 3′-terminal nucleotide is disordered in the GatDE-tRNA co-crystal structure.15 However, a mimic of the aminoacyl-moiety of the substrate tRNA, Asp, bound to the permanent Mg2+ (Fig. 4b). Thus, we anticipate that the carboxyl group of Asp-tRNAAsn (or Glu-tRNAGln) coordinates the permanent metal (Fig. 7a), positioning the carboxyl group for nucleophilic attack on the ATP γ-phosphate (Fig. 7b). The newly phosphorylated Asp-tRNAAsn then rearranges to use the phosphate group of the activated intermediate to maintain coordination of the permanent Mg2+ by an anion (Fig. 7c). This small rearrangement shifts the phosphoester carbon atom towards the ammonia channel, in position for attack by ammonia emerging from the channel near Tyr83 and Gln93 (Fig. 7c). The amidation reaction releases the phosphate. The amidated aa-tRNA would be unable to coordinate to the permanent Mg2+, likely facilitating full product release.
Undoubtedly the substrate and intermediate motions proposed for the catalytic cycle are accompanied by changes in protein conformation. These may include motions of loops surrounding the tRNA docking site or associated with the ammonia channel, or small hinging motions of domains. Such motions are critical to the function of AdTs. An important signal that the synthetase active site is ready to receive ammonia must be transmitted to the partner catalytic subunit (A or D) in order to initiate ammonia production or release into the channel. For example, GatDE hydrolyzed glutamine only in presence of the misacylated tRNA substrate, and more efficiently when ATP was included.20 The signal could be sent upon binding of both synthetase substrates – tRNA and ATP – or upon formation of the activated intermediate. The proposed rearrangement of the activated intermediate in the synthetase active site could provide the requisite conformational signal to the A-subunit or D-subunit to send ammonia through the channel. The structures presented here provide additional data on the interconnection of individual steps in the formation of Asn-tRNAAsn and Gln-tRNAGln by the indirect pathways.
Materials and Methods
General
The Promega Wizard DNA purification kit was utilized for plasmid purification. The E. coli cells, XL1-Blue and BL21 (DE3) codon plus, were obtained from Stratagene Cloning System (La Jolla, CA). Restriction enzymes and T4 DNA ligase were purchased from New England Biolabs. DNA sequencing and primer syntheses were performed by the University of Michigan Biomedical Resources Core Facility and the Keck Foundation Biotechnology Research Laboratory at Yale University The QFF column, Phenyl Superose (HR10/10), and Superdex 200 (Hiload 16/60) chromatography columns, and [α-32P] ATP (10 mmol/μCi) were from Amersham Biosciences (GE Healthcare). Nickel-nitrilotriacetic acid-agarose (Ni-NTA) was from QIAGEN (Chatsworth, CA). Bio-Spin 30 columns were from Bio-Rad (Hercules, CA). High purity cold L-Glu, L-Asp, L-Gln and L-Asn were from Fluka (Deisenhofen, Germany). Phenol was from American Bioanalytical (Natick, MA). Protein concentrations were determined using the Bio-Rad Protein Assay Reagent, with bovine serum albumin (Sigma) serving as the standard.
Cloning of A. aeolicus gatCA and gatB
In order to obtain non-His-tagged GatCAB complex, gatCA and gatB were subcloned from pET16b-CA and pET16b-B, respectively, into the pET17b vector (Novagen). To construct plasmid pET17b-CA for expression of GatC and GatA, pET16b-CA was digested with NdeI and BamHI, and ligated into the similarly restricted expression vector, pET17b, which had been treated with calf intestinal alkaline phosphatase. The ligation mixture was used to transform E. coli XL1-Blue cells. The presence of plasmids containing the desired gene from several transformants was verified by restriction analysis, and the gene sequence was confirmed by DNA sequencing. Similar methodology was used to construct the plasmid pET17b-B, which expresses A. aeolicus gatB. In order to overcome the codon bias due to heterologous expression of A. aeolicus proteins in E. coli, transformation was performed with chemically competent E. coli Rosetta (DE3) cells for both pET17b-CA and pET17b-B.
Overproduction and purification of GatCAB
The E. coli Rosetta (DE3) cells harboring pET17-CA or pET17-B were grown in LB medium containing ampicillin (100 mg/L) and chloramphenicol (35 mg/mL) at 37 °C with shaking (280 rpm). Isopropyl-D-thiogalactoside was added to a final concentration of 0.4 mM when the culture reached an absorbance of 1.5. The cells were harvested 10 h post induction by centrifugation (29,000 × g, 20 min, 4 °C). The cell pellets of the two cultures were combined and suspended in buffer A (20 mM Tris-HCl, pH 7.4). The suspension was subjected to sonication on ice (five 30-s pulses with a 2-min rest between pulses). The lysate was centrifuged to remove cell debris (40,000 × g, 45 min, 4 °C).
Solid NaCl was added to the supernatant to a final concentration of 0.1 M and the solution was heated at 100 °C for 1.5 min and then at 80 °C for 10 min with gentle continuous hand swirling. The suspension was allowed to cool to 25 °C, placed on ice for 15 min and the precipitated protein was removed by centrifugation (29,000 × g, 20 min, 4 °C). The supernatant was dialyzed overnight against one liter of buffer A at 4 °C and then applied to a QFF column pre-equilibrated with buffer A. The column was developed at a flow rate of 1.0 mL/min using a linear gradient from 0 to 1 M KCl in the same buffer over 100 min. The fractions containing GatCAB as determined by SDS-PAGE were pooled. Solid (NH4)2SO4 was added slowly with stirring to the pooled fractions to a final concentration of 20% (w/v). The sample was filtered (0.22 μm) and loaded onto a Phenyl Superose column equilibrated with 20% (NH4)2SO4 in buffer A. A reverse gradient from 20% to 0% (NH4)2SO4 in buffer A was applied at a flow rate of 1.0 mL/min over 60 min and elution was continued with buffer A for 20 min. The fractions containing GatCAB were pooled and concentrated. The concentrated sample was loaded onto a Superdex 200 column pre-equilibrated with buffer B (10 mM HEPES pH 7.4). The column was developed at a flow rate of 1.0 mL/min in buffer B and the eluate was pooled and concentrated to 6 mg/mL. The final preparation was homogeneous as determined by SDS-PAGE. The purified enzyme was aliquoted and stored at -80 °C.
For the production of the selenomethionine (SeMet) derivative, cells were grown in SeMet minimal medium31 containing 25 mg/L L-SeMet. The purification procedure for the SeMet derivative was the same as that for native enzyme.
[32P]tRNA/nuclease P1 amidotransferase assay
H. pylori GluRS was over-produced and purified as previously described.32 H. pylori tRNAGln was over-expressed in E. coli XL1-Blue strain and purified as described.10 C. trachomatis tRNAAsn isoacceptor was in vitro transcribed and purified as described previously.33 The 3′ termini of the tRNA isoacceptors were 32P-labeled as previously described.34 H. pylori Glu-tRNAGln and C. trachomatis Asp-tRNAAsn were prepared as described.9; 10 The Glu-AdT and Asp-AdT activities of the A. aeolicus GatCAB were assayed as previously described in detail34 with slight modification. Briefly, amidotransferase reaction mixtures included A. aeolicus AdT buffer (50 mM Hepes-KOH, pH 7.2, 15 mM MgCl2, 25 mM KCl and 1 mM DTT) and unless otherwise noted 4 mM ATP, 9 to 11 μM 32P-labeled misacylated tRNA (Asp-tRNAAsn or Glu-tRNAGln) and 4 mM amide donor (Gln or Asn) and were carried out at 37°C. It should be noted that the optimal growth temperature for A. aeolicus is approximately 95 °C while these experiments were carried out at 37 °C in order to minimize deacylation of the aa-tRNA species, which occurred at elevated reaction temperatures. It is speculated that in vivo very little aa-tRNA is unbound as the tRNA substrate is channeled from the ND-aaRS to the AdT, protecting the aa-tRNA from deacylation.35; 36 For determination of the kinetic parameters, initial velocities were measured while varying the concentration of one substrate and saturating with the other two. For KM determinations of the two amide donors, substrate concentrations were varied between 3.25 μM to 4000 μM. For KM determinations of the misacylated tRNA substrate, concentrations varied from approximately 150 nM up to 12 μM. Reaction mixes were pre-incubated at 37 °C and started by addition of enzyme to a final concentration of 20 nM. Aliquots (2 μL) of the reaction mixes were quenched, digested and processed as previously described.
Complementation of E. coli JF448 with the D. radiodurans aspS2/H. pylori gatCAB artificial operon for asparagine synthesis
An artificial operon of D. radiodurans aspS2 and H. pylori gatCAB was made via overlap PCR similar to the previous artificial operon28 except a PacI restriction site was placed in front of gatB. The artificial operon was subcloned into pCBS2 between the NdeI and BamHI resistriction sites. Truncated H. pylori gatB mutants were made via PCR. Point mutations of H. pylori gatB were made by overlap PCR. The mutant gatB genes were subcloned into the pCBS2-operon vector between the PacI and BamHI restriction sites, replacing the wild-type gatB. The vectors were then transformed into E. coli JF448 cells and grown on M9 minimal media agar plates with or without Asn as previously described.28
Crystallization and data collection
Crystallization was carried out at 20 °C by hanging-drop vapor diffusion from a 1:1 mixture of protein stock (6-9 mg/mL GatCAB, 10mM HEPES pH 7.5, 50 μM Zn acetate) and well solution (10-12% (v/v) of PEG3350, 10 mM Mg formate). Crystals of the SeMet derivative were obtained under similar conditions. To prepare the heavy atom derivative, the crystals were soaked 3 hr in well solution containing 0.1 mM HgCl2. Crystals of the ligand complexes were obtained by co-crystallization or soaking. All crystals were dipped quickly into well solution containing 13% PEG3350 with 25% (v/v) glycerol for cryoprotection.
The best diffraction data (dmin = 2.3 Å) were recorded from a crystal of the GatCAB-Asn/ADP complex, which was grown in the presence of 0.6 mM ATP and 10 mM Asn. Data for the Gln complex (2.8 Å) were obtained from a crystal grown in the presence of 10 mM Gln. Data for the ATP-Asp-Asn complex (3.0 Å) came from a crystal grown with 10 mM Asn and soaked 5 hr in well solution containing 10 mM MnCl2, 10 mM Asp, and 10 mM ATP. All data were collected at GM/CA-CAT beamline 23ID-B at the Advanced Photon Source, and processed using HKL2000.37 GatCAB crystallized readily in apparently two crystal forms that are variants of a single lattice and have nearly identical cell constants (space group P21 with β ≈ 90° and four heterotrimers per asymmetric unit, and space group P1 with α ≈ β ≈ γ ≈ 90° and eight per asymmetric unit). A summary of the data collection statistics is given in Table 1. Crystallographic calculations were done using the CCP4 program suite38 unless otherwise noted.
Structure Determination and Refinement
The structure was solved in space group P21 by a combination of Hg-SAS, Se-SAD and molecular replacement. Twelve Hg sites were identified by visual inspection of anomalous difference Patterson maps. The native Patterson map contained a strong peak with height one-third of the origin peak at approximately (0.45, 0.5, 0), indicating pseudo C-centering in the primitive lattice. Initial Hg-SAD phases were applied to the Se Bijvoet data to locate 56 of 64 Se sites using the program SHARP/AUTOSHARP39, which subsequently yielded a 3.2-Å electron density map. The pseudo-translational symmetry complicated SIR and SAD phasing and also molecular replacement. Experimental phases were combined with molecular-replacement phases based on structures of T. maritima GatA (PDB 2GI3) and S. aureus GatB14 (PDB 2G5H). Approximate molecular replacement solutions were obtained with PHASER.40 The combined phases were refined and extended to 2.3 Å by density averaging the four copies of GatCAB using the program dm.41 The atomic structure was constructed using the program Coot.42 The initial model of GatCAB-Asn/ADP was refined against the 2.3 Å data with the program REFMAC.43 A bulk solvent correction and non-crystallographic symmetry restraints were applied throughout the refinement procedure. The refined model of GatCAB-Asn/ADP was used as a starting point for refinement of the other GatCAB complexes. After rigid-body refinement, difference Fourier maps showed clear density for Gln and for ATP, Asp and Mn2+ ions. Some crystals had nearly perfect monoclinic symmetry in data scaling, but in all cases, refinement was successful only in the lower-symmetry space group P1. The refinement statistics are summarized in Table 1.
Structure Analysis
The refined models were validated using the program Molprobity.44 Sequence alignment was done using ClustalW45. All figures were prepared with PyMOL [DeLano, W. L. The PyMOL Molecular Graphics System (2002)]†.
Supplementary Material
Acknowledgments
We are indebted to the staff of the GM/CA beamlines (supported by the NIGMS and NCI, National Institutes of Health) at the Advanced Photon Source (supported by the United States Department of Energy). This work was supported by NIH grants GM22854 to DS and DK42303 to JLS.
Footnotes
Abbreviations used: aa-tRNAs, aminoacyl-tRNAs; aaRSs, aminoacyl-tRNA synthetases; GlnRS, glutaminyl-tRNA synthetase; AsnRS, asparaginyl-tRNA synthetase; ND-aaRS, non-discriminating aaRS; AdT, tRNA-dependent amidotransferase; ND-GluRS, non-discriminating glutamyl-tRNA synthetase; Glu-AdT, glutamyl-tRNAGln amidotransferase; ND-AspRS, non-discriminating aspartyl-tRNA synthetase; Asp-AdT, Asp-tRNAAsn amidotransferase; RMSD, root mean square deviation; aa, amino acid; MAE2, malonamidase E2; PAM, peptide amidase; FAAH, fatty acid amide hydrolase; HEPES, 4-(2-hydroxyethyl)piperazine-1-ethanesulfonic acid; PEG, poly(ethylene glycol); SDS, sodium dodecyl sulfate; PAGE, polyacrylamide gel electrophoresis.
Accession Codes: The crystal structures and diffraction data have been deposited in the Protein Data Bank with accession codes 3H0L for the ADP complex and Asn adduct, 3H0M for the Gln adduct, and 3H0R for the ATP/aspartate complex and Asn adduct.
Supplementary Data: Supplementary data associated with the article can be found, in the online version, at doi:########.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Ibba M, Söll D. Aminoacyl-tRNA synthesis. Annu Rev Biochem. 2000;69:617–650. doi: 10.1146/annurev.biochem.69.1.617. [DOI] [PubMed] [Google Scholar]
- 2.Sheppard K, Yuan J, Hohn MJ, Jester B, Devine KM, Söll D. From one amino acid to another: tRNA-dependent amino acid biosynthesis. Nucleic Acids Res. 2008;36:1813–1825. doi: 10.1093/nar/gkn015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lapointe J, Duplain L, Proulx M. A single glutamyl-tRNA synthetase aminoacylates tRNAGlu and tRNAGln in Bacillus subtilis and efficiently misacylates Escherichia coli tRNAGln1in vitro. J Bacteriol. 1986;165:88–93. doi: 10.1128/jb.165.1.88-93.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wilcox M, Nirenberg M. Transfer RNA as a cofactor coupling amino acid synthesis with that of protein. Proc Natl Acad Sci U S A. 1968;61:229–236. doi: 10.1073/pnas.61.1.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Becker HD, Reinbolt J, Kreutzer R, Giegé R, Kern D. Existence of two distinct aspartyl-tRNA synthetases in Thermus thermophilus. Structural and biochemical properties of the two enzymes. Biochemistry. 1997;36:8785–8797. doi: 10.1021/bi970392v. [DOI] [PubMed] [Google Scholar]
- 6.Curnow AW, Ibba M, Söll D. tRNA-dependent asparagine formation. Nature. 1996;382:589–590. doi: 10.1038/382589b0. [DOI] [PubMed] [Google Scholar]
- 7.Curnow AW, Hong K, Yuan R, Kim S, Martins O, Winkler W, Henkin TM, Söll D. Glu-tRNAGln amidotransferase: a novel heterotrimeric enzyme required for correct decoding of glutamine codons during translation. Proc Natl Acad Sci U S A. 1997;94:11819–11826. doi: 10.1073/pnas.94.22.11819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tumbula DL, Becker HD, Chang WZ, Söll D. Domain-specific recruitment of amide amino acids for protein synthesis. Nature. 2000;407:106–110. doi: 10.1038/35024120. [DOI] [PubMed] [Google Scholar]
- 9.Sheppard K, Sherrer RL, Söll D. Methanothermobacter thermautotrophicus tRNAGln confines the amidotransferase GatCAB to asparaginyl-tRNAAsn formation. J Mol Biol. 2008;377:845–853. doi: 10.1016/j.jmb.2008.01.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sheppard K, Akochy PM, Salazar JC, Söll D. The Helicobacter pylori amidotransferase GatCAB is equally efficient in glutamine-dependent transamidation of Asp-tRNAAsn and Glu-tRNAGln. J Biol Chem. 2007 doi: 10.1074/jbc.M700398200. [DOI] [PubMed] [Google Scholar]
- 11.Wilcox M. Gamma-glutamyl phosphate attached to glutamine-specific tRNA. A precursor of glutaminyl-tRNA in Bacillus subtilis. Eur J Biochem. 1969;11:405–412. doi: 10.1111/j.1432-1033.1969.tb00788.x. [DOI] [PubMed] [Google Scholar]
- 12.Feng L, Sheppard K, Tumbula-Hansen D, Söll D. Gln-tRNAGln formation from Glu-tRNAGln requires cooperation of an asparaginase and a Glu-tRNAGln kinase. J Biol Chem. 2005;280:8150–8155. doi: 10.1074/jbc.M411098200. [DOI] [PubMed] [Google Scholar]
- 13.Sheppard K, Söll D. On the evolution of the tRNA-dependent amidotransferases, GatCAB and GatDE. Journal of Molecular Biology. 2008;377:831–844. doi: 10.1016/j.jmb.2008.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nakamura A, Yao M, Chimnaronk S, Sakai N, Tanaka I. Ammonia channel couples glutaminase with transamidase reactions in GatCAB. Science. 2006;312:1954–1958. doi: 10.1126/science.1127156. [DOI] [PubMed] [Google Scholar]
- 15.Oshikane H, Sheppard K, Fukai S, Nakamura Y, Ishitani R, Numata T, Sherrer RL, Feng L, Schmitt E, Panvert M, Blanquet S, Mechulam Y, Söll D, Nureki O. Structural basis of RNA-dependent recruitment of glutamine to the genetic code. Science. 2006;312:1950–1954. doi: 10.1126/science.1128470. [DOI] [PubMed] [Google Scholar]
- 16.Harpel MR, Horiuchi KY, Luo Y, Shen L, Jiang W, Nelson DJ, Rogers KC, Decicco CP, Copeland RA. Mutagenesis and mechanism-based inhibition of Streptococcus pyogenes Glu-tRNAGln amidotransferase implicate a serine-based glutaminase site. Biochemistry. 2002;41:6398–6407. doi: 10.1021/bi012126u. [DOI] [PubMed] [Google Scholar]
- 17.Schmitt E, Panvert M, Blanquet S, Mechulam Y. Structural basis for tRNA-dependent amidotransferase function. Structure. 2005;13:1421–1433. doi: 10.1016/j.str.2005.06.016. [DOI] [PubMed] [Google Scholar]
- 18.Strauch MA, Zalkin H, Aronson AI. Characterization of the glutamyl-tRNAGln-to-glutaminyl-tRNAGln amidotransferase reaction of Bacillus subtilis. J Bacteriol. 1988;170:916–920. doi: 10.1128/jb.170.2.916-920.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jahn D, Kim YC, Ishino Y, Chen MW, Söll D. Purification and functional characterization of the Glu-tRNAGln amidotransferase from Chlamydomonas reinhardtii. J Biol Chem. 1990;265:8059–8064. [PubMed] [Google Scholar]
- 20.Bailly M, Blaise M, Roy H, Deniziak M, Lorber B, Birck C, Becker HD, Kern D. tRNA-dependent asparagine formation in prokaryotes: characterization, isolation and structural and functional analysis of a ribonucleoprotein particle generating Asn-tRNAAsn. Methods. 2008;44:146–163. doi: 10.1016/j.ymeth.2007.11.012. [DOI] [PubMed] [Google Scholar]
- 21.Deniziak M, Sauter C, Becker HD, Paulus CA, Giegé R, Kern D. Deinococcus glutaminyl-tRNA synthetase is a chimer between proteins from an ancient and the modern pathways of aminoacyl-tRNA formation. Nucleic Acids Res. 2007;35:1421–1431. doi: 10.1093/nar/gkl1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shin S, Lee TH, Ha NC, Koo HM, Kim SY, Lee HS, Kim YS, Oh BH. Structure of malonamidase E2 reveals a novel Ser-cisSer-Lys catalytic triad in a new serine hydrolase fold that is prevalent in nature. EMBO J. 2002;21:2509–2516. doi: 10.1093/emboj/21.11.2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shin S, Yun YS, Koo HM, Kim YS, Choi KY, Oh BH. Characterization of a novel Ser-cisSer-Lys catalytic triad in comparison with the classical Ser-His-Asp triad. J Biol Chem. 2003;278:24937–24943. doi: 10.1074/jbc.M302156200. [DOI] [PubMed] [Google Scholar]
- 24.Labahn J, Neumann S, Buldt G, Kula MR, Granzin J. An alternative mechanism for amidase signature enzymes. J Mol Biol. 2002;322:1053–1064. doi: 10.1016/s0022-2836(02)00886-0. [DOI] [PubMed] [Google Scholar]
- 25.Bracey MH, Hanson MA, Masuda KR, Stevens RC, Cravatt BF. Structural adaptations in a membrane enzyme that terminates endocannabinoid signaling. Science. 2002;298:1793–1796. doi: 10.1126/science.1076535. [DOI] [PubMed] [Google Scholar]
- 26.Horiuchi KY, Harpel MR, Shen L, Luo Y, Rogers KC, Copeland RA. Mechanistic studies of reaction coupling in Glu-tRNAGln amidotransferase. Biochemistry. 2001;40:6450–6457. doi: 10.1021/bi002599l. [DOI] [PubMed] [Google Scholar]
- 27.Felton J, Michaelis S, Wright A. Mutations in two unlinked genes are required to produce asparagine auxotrophy in Escherichia coli. J Bacteriol. 1980;142:221–228. doi: 10.1128/jb.142.1.221-228.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Min B, Pelaschier JT, Graham DE, Tumbula-Hansen D, Söll D. Transfer RNA-dependent amino acid biosynthesis: an essential route to asparagine formation. Proc Natl Acad Sci U S A. 2002;99:2678–2683. doi: 10.1073/pnas.012027399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Petrek M, Otyepka M, Banas P, Kosinova P, Koca J, Damborsky J. CAVER:a new tool to explore routes from protein clefts, pockets and cavities. BMC Bioinformatics. 2006;7:316. doi: 10.1186/1471-2105-7-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dundas J, Ouyang Z, Tseng J, Binkowski A, Turpaz Y, L J. CASTp: computed atlas of surface topography of proteins with structrual and topographical mapping of funcationally annotated residues. Nucleic Acids Research. 2006;34:W116–118. doi: 10.1093/nar/gkl282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guerrero SA, Hecht HJ, Hofmann B, Biebl H, Singh M. Production of selenomethionine-labelled proteins using simplified culture conditions and generally applicable host/vector systems. Appl Microbiol Biotechnol. 2001;56:718–723. doi: 10.1007/s002530100690. [DOI] [PubMed] [Google Scholar]
- 32.Salazar JC, Ahel I, Orellana O, Tumbula-Hansen D, Krieger R, Daniels L, Söll D. Coevolution of an aminoacyl-tRNA synthetase with its tRNA substrates. Proc Natl Acad Sci U S A. 2003;100:13863–13868. doi: 10.1073/pnas.1936123100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fechter P, Rudinger J, Giegé R, Theobald-Dietrich A. Ribozyme processed tRNA transcripts with unfriendly internal promoter for T7 RNA polymerase: production and activity. FEBS Lett. 1998;436:99–103. doi: 10.1016/s0014-5793(98)01096-5. [DOI] [PubMed] [Google Scholar]
- 34.Sheppard K, Akochy PM, Söll D. Assays for transfer RNA-dependent amino acid biosynthesis. Methods. 2008;44:139–145. doi: 10.1016/j.ymeth.2007.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bailly M, Blaise M, Lorber B, Becker HD, Kern D. The transamidosome: a dynamic ribonucleoprotein particle dedicated to prokaryotic tRNA-dependent asparagine biosynthesis. Molecular Cell. 2007;28:228–239. doi: 10.1016/j.molcel.2007.08.017. [DOI] [PubMed] [Google Scholar]
- 36.Huot JL, Balg C, Jahn D, Moser J, Emond A, Blais SP, Chenevert R, Lapointe J. Mechanism of a GatCAB amidotransferase: Aspartyl-tRNA synthetase increases its affinity for Asp-tRNAAsn and novel aminoacyl-tRNA analogues are competitive inhibitors. Biochemistry. 2007;46:13190–13198. doi: 10.1021/bi700602n. [DOI] [PubMed] [Google Scholar]
- 37.Otwinowski Z, Minor W. Processing of X-Ray diffraction data collected in oscillation mode. Methods in Enzymology. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 38.Collaborative Computational Project, Number 4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 39.Vonrhein C, Blanc E, Roversi P, Bricogne G. Automated structure solution with autoSHARP. Methods Mol Biol. 2007;364:215–230. doi: 10.1385/1-59745-266-1:215. [DOI] [PubMed] [Google Scholar]
- 40.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Cryst. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cowtan K. ‘dm’: An automated procedure for phase improvement by density modification. Joint CCP4 and ESF-EACBM Newsletter on Protein Crystallography. 1994:34–38. [Google Scholar]
- 42.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 43.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 44.Lovell SC, Davis IW, Arendall WB, de Bakker PI, 3rd, Word JM, Prisant MG, Richardson JS, Richardson DC. Structure validation by Cα geometry: φ,ψ and Cβ deviation. Proteins. 2003;50:437–450. doi: 10.1002/prot.10286. [DOI] [PubMed] [Google Scholar]
- 45.Thompson JD, Gibson TJ, Higgins DG. Multiple sequence alignment using ClustalW and ClustalX. Curr Protoc Bioinformatics. 2002;2:3.1–3.22. doi: 10.1002/0471250953.bi0203s00. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.