Abstract
γ-Secretase, an integral membrane protein complex, catalyzes the intramembrane cleavage of the β-amyloid precursor protein (APP) during the neuronal production of the amyloid β-peptide (Aβ). As such, the protease has emerged as a key target for developing agents to treat and prevent Alzheimer's disease. Existing biochemical studies conflict on the oligomeric assembly state of the protease complex, and its detailed structure is not known. Here, we report that purified active human γ-secretase in digitonin has a total molecular mass of ~230 kDa when measured by scanning transmission electron microscopy. This result supports a complex that is monomeric for each of the four component proteins. We further report the 3-dimensional structure of the γ-secretase complex at 12 Å resolution, as obtained by cryo-EM and single particle image reconstruction. The structure reveals several domains on the extracellular side, three solvent-accessible low-density cavities and a potential substrate-binding surface groove in the transmembrane region of the complex.
Introduction
γ-Secretase is a membrane protein complex composed of Presenilin (PS), Nicastrin (NCT), Aph-1 and Pen-2 1; 2. The necessity and the sufficiency of these four integral membrane proteins for forming the active protease complex have been established by functional reconstitution of γ-secretase activity in S. cerevisiae, which lacks these proteins 3, and by high-grade purification of the proteolytically active human complex from over-expressing mammalian cells 4.
Three properties make γ-secretase a highly interesting target for investigation. First, γ-secretase is an unconventional aspartyl protease that resides and cleaves its substrates within the lipid bilayer. It belongs to a unique group of intramembrane-cleaving proteases (I-CliPs) that includes site 2 protease (S2P), Rhomboids, and signal peptide peptidase (SPP) 5. The I-CliPs appear to have their catalytic residues located inside the membrane. For γ-secretase, the two catalytic aspartic acids reside in adjacent transmembrane domains at the interface of the PS heterodimer and inside the membrane 6. Second, Alzheimer's disease (AD) is believed to be caused by the progressive cerebral accumulation of Aβ, and γ-secretase effects the final cleavage of APP to release Aβ 7. Therefore, partially inhibiting γ-secretase could slow, halt or prevent AD. Third, in addition to this pathogenic function, γ-secretase processes a wide range of other type I membrane proteins, such as the receptors Notch and Erb-B4, the cell adhesion molecules N-cadherin and E-cadherin, and the neurotrophin co-receptor p75 8.
The recent crystal structures of the bacterial rhomboid homolog GlpG and the archaeal membrane protease mjS2P have begun to shed light on how peptide bond hydrolysis may occur in a lipid environment. GlpG has a core domain comprised of 6 transmembrane helices with the enzyme's active site located 10 Å under the outer surface of the lipid bilayer 9; 10; 11; 12. The archaeal site 2 protease (S2P) also has a six-transmembrane helix core domain; the enzyme's active site coordinates a Zn atom and is located near the middle of the bilayer 13. Except for the fact that they both have 6 transmembrane helices, these two enzymes share no common structural features. Nevertheless, the overall architectures of the two proteases allow the binding and potential entry of single helical substrates and the access of water to the active sites 14. However, the bacterial and archaeal enzymes are unrelated to γ-secretase in both sequence and structure. The recent success in bacterial expression and purification of SPP 15, an aspartyl protease that is related to PS 16, raises the hope that the crystal structure of this protease might be solved soon. In contrast, only modest amounts of active γ-secretase complex can be purified, due to its complex maturation and assembly of multiple components having 19 transmembrane domains 17, its requirement for certain lipids for activity 18, and its sensitivity to detergent type 19. These constraints will likely hinder the achievement of an atomic resolution structure for γ-secretase.
We recently reported a low-resolution, 3D structure of γ-secretase purified from over-expressing CHO cells (the γ-30 line) that was reconstructed from the single particle EM images of uranyl acetate-stained complexes 20. The structure revealed an irregular low-density interior chamber and apical and basal pore-like openings that could allow the entry of water molecules and exit of products. However, the concentration and amount of sample we were able to prepare from the γ-30 cells was not adequate for performing cryo-EM. For structural analyses, cryo-EM is more desirable than negative stain EM because in cryo-EM, the image contrast arises from the protein itself rather than from a contrast agent which can distort the image obtained, as occurs with negative stain EM. We have recently generated a new CHO cell line (called S-20) that over-expresses about five times more γ-secretase than the γ-30 line 21. The increased amount of material, while still modest compared to what can be produced for many bacterial or archaeal membrane proteins, has nevertheless enabled us to measure the mass of the γ-secretase complex by scanning transmission electron microscopy (STEM) and to further determine its cryo-EM structure at 12 Å resolution.
Results and Discussion
The purified, proteolytically active γ-secretase is a monomeric complex
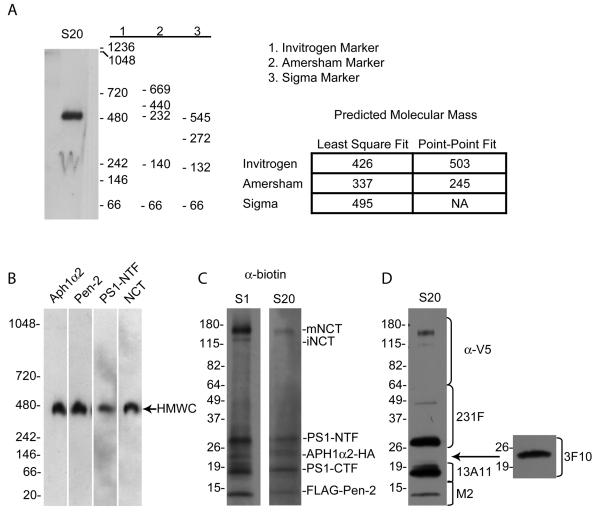
The oligomeric state (i.e., the stoichiometry) of the 4-component γ-secretase complex has been unclear. Earlier immunoprecipitation experiments raised the possibility that γ-secretase contained two copies of PS 22. This suggestion was supported by the reconstitution of γ-secretase activity with two inactive PS mutants that each had one of the two catalytic aspartates mutated to alanine 23. However, a recent biochemical study has suggested a 1:1:1:1 stoichiometry of the complex 24. In previous Blue Native (BN) gel analyses, γ-secretase was reported to have apparent masses varying from 250 19 to 500 22; 23; 24; 25 to 900 kDa 26. We attempted to clarify this discrepancy by running a BN gel of γ-secretase purified from the S-20 cell line side by side with the three most commonly used commercial molecular weight (MW) markers (Fig. 1A). We found that the predicted molecular mass of the same sample differed strikingly, from ~250 kDa to ~500 kDa, depending on the source of the MW markers as well as the type of line fit used to estimate the molecular mass. This observation demonstrates that caution must be used when interpreting sizing results of Blue Native gels and may explain in part the different molecular masses of γ-secretase reported in the literature heretofore.
Fig. 1. Molecular mass characterization of γ-secretase.
(A) Blue Native gel analysis of purified S-20 γ-secretase reveals different estimated masses of the complex dependent on source of the MW markers. The non-denatured, intact γ-secretase complex was probed with Ab14 to the presenilin-NTF. Molecular mass was estimated using three different commercial sources of MW markers as well as two different line fits in the AlphaEaseFC software. (B) Western blotting of a Blue Native PAGE gel of the S-20 γ-secretase complexes with antibodies to each known component confirms the presence of all four components in the purified complex (Invitrogen molecular weight markers used). (C) Western blotting of an SDS-PAGE gel of the biotinylated S-1 and S-20 γ-secretase with an anti-biotin antibody demonstrates the purity of the preparations. (D) Western blotting with antibodies to each of the known components establishes the identity of the observed bands in SDS-PAGE gel of the S-20 γ-secretase from (C).
To ascertain the mass and thus the assembly state of purified γ-secretase without relying on techniques (e.g., Blue Native gel electrophoresis, glycerol velocity gradients) that involve estimating protein mobility through a particular matrix, we used STEM, which is known to give accurate mass measurements of purified biological particles 27. The method is based on the principle that in a thin sample, the number of scattered electrons in the irradiated pixel is proportional to the number of atoms weighted by the atomic number. Since individual biological molecules are thin and composed mainly of light atoms, the STEM signal is directly proportional to the mass of the molecules 27.
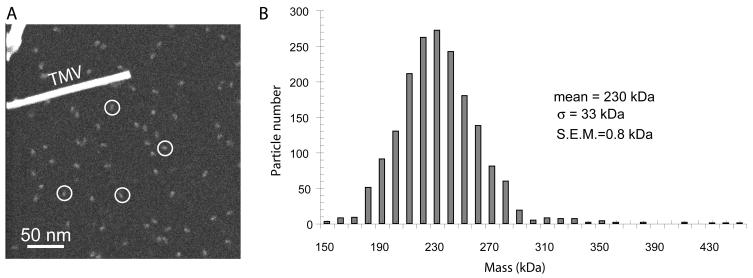
We used this technique to determine the molecular mass of γ-secretase purified from the S-20 cell line. Before performing STEM, we first confirmed that our purified γ-secretase preparation contained all four expected components, PS1, FLAG-Pen-2, Aph1α2-HA and NCT-V5/HIS, by Western blotting the sample with γ-secretase specific antibodies after Blue Native gel electrophoresis (Fig. 1B). Next, to address the purity of the S-1 and S-20 γ-secretase preparations, we biotinylated a small sample of the purified preparations, which resulted in specific biotin labeling of all of the γ-secretase components, as seen by Western blotting with an antibody to biotin (Fig. C). The α-biotin blots (Fig. 1C) revealed only the known proteins of the γ-secretase complex (Fig. 1D), confirming the purity of our γ-secretase preparations. Fig. 2A shows a cryo-STEM image of the γ-complexes purified from S-20 cells. Covering an area of 512 by 512 nm, the image contains a field of well-separated individual particles, each approximately 10 nm in size. We measured in unbiased fashion the masses of all particles that were separated from each other, obtaining a total of 1,800 particles. The particles were sorted according to their masses and binned at intervals of 10 kDa, and the numbers of particles in each bin were plotted against the mass (Fig. 2B). The mass exhibits a single modal distribution profile, with an average molecular mass of ~230 kDa, a standard deviation (SD or σ) of 33 kDa, and a standard error of the mean (SEM) of 0.8 kDa (33/√1800). The standard deviation, at 14% of the total mass, indicates a fairly homogeneous γ-secretase preparation, given the known heterogeneity in NCT glycosylation, and the relatively small size of the particle. For comparison, the standard deviation of STEM mass (56 kDa) of the small tetrameric Aquaporin is 36% of the total measured mass (157 kDa) 28. The homogeneity of that Aquaporin preparation was demonstrated by its ability to form 2D crystals. The calculated protein mass of the γ-secretase complex assuming a stoichiometry of one copy for each subunit is ~195 - 235 kDa (the estimated mass of the heterogeneously-glycosylated NCT is ~110 - 150 kDa 24; 29; 30, PS NTF + CTF 50 kDa, Aph1α2-HA ~23 kDa, Flaγ-Pen-2 ~12 kDa). Thus, these data are inconsistent with the dimeric γ-secretase hypothesis; they support the biochemical evidence for a 1:1:1:1 monomeric stoichiometry of the four component proteins in the over-expressed system 24. The difference between the STEM-measured mass and the calculated mass might be attributed to the digitonin detergent used during the purification and/or the presence of endogenous lipids. The purified γ-secretase complex we subjected to STEM and to cryo-EM lacks the exogenous lipids we routinely add for performing activity assays, and is not proteolytically active without lipid addition, supporting the notion that minimal endogenous lipids remain after the purification procedure, and may not have a significant impact on the STEM mass measurement 18.
Fig. 2. STEM mass measurement of γ-secretase from S-20 cells.
(A) A large-angle dark-field cryo-STEM image of purified γ-secretase particles. The image covers an area of 512 by 512 nm. The tobacco mosaic virus helical rod (TMV) served as a quality control during EM specimen preparation; the width of the TMV rod is 18 nm. White circles exemplify individual γ-secretase particles. (B) Mass distribution of a total of 1800 STEM-measured γ-secretase particles purified from the S-20 cell line.
2D classification of the GST-fused γ-secretase images
We previously probed the topological orientation of the isolated γ-secretase complex vis-à-vis the membrane by using Concanavalin A (Con A), a tetramer that selectively binds the mannose group of the glycosylated extracellular region of NCT (NCT is the only γ-component that has a glycosylated ectodomain) 20. Like other non-covalent recognition methods such as antibody labeling or functionalized heavy metal cluster labeling, the Con A labeling had a low occupancy problem, i.e. most of the γ-secretase particles did not bind the Con A. As such, image classification and averaging were not carried out due to the limited number of particles that had Con A bound, and the Con A density was localized only from the raw particle images. Although the raw-image based localization can be unambiguous in favorable cases where the protein particles are of distinctive shapes, the γ-secretase complex is near spherical and thus less distinctive. We previously utilized a maltose binding protein (MBP) fusion strategy coupled with 2D image classification to map the subunits of the eight-component yeast oligosaccharyl transferase membrane complex 31. We attempted this strategy on γ-secretase by stably transfecting our γ-30 cell line (which has no exogenous NCT) with NCT-MBP, containing the 44-kDa MBP on the cytosolic side of NCT. However, these stable cell lines lost expression of NCT-MBP over time. Purification of the MBP-containing complex via transient transfection on a large scale was carried out; however, this preparation was not homogeneous, and the MBP density could not be unambiguously identified in the averaged images of the negatively stained EM micrographs (data not shown).
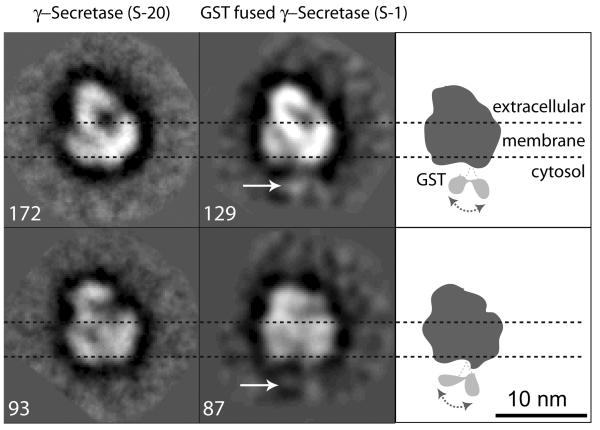
Next, we decided to use γ-secretase purified from our S-1 cell line that has a glutathione transferase (GST) tag fused to the carboxyl terminus of NCT 4. The GST-fused NCT migrated in SDS PAGE at higher molecular mass than the short V5/HIS peptide-taγed NCT from S-20 cells (data not shown). Consistent with this observation, we were able to detect densities that could be attributed to the relatively small GST protein tag (26 kDa) in the averaged negative stain EM images of the purified S-1 γ-secretase complexes (Fig. 3). Such densities were not detected in γ-secretase purified from S-20 cells (Fig. 3). We observed two weak densities at the bottom edge of each of two averaged views of the S-1 particles. Since there is only one NCT in γ-secretase, the two densities may correspond to two preferred positions of the GST protein on the complex, as these views are averages of 129 and 87 particles, respectively. The C-terminus of NCT is known to be cytosolic. Localization of the GST tag on the γ-secretase structure establishes this end as cytosolic and therefore the opposite end with larger density as extracellular. This membrane orientation agrees with our previous Con A labeling-based assignment of the NCT ectodomain 20.
Fig. 3. Localization of the GST tag of NCT-GST in the S-1 cell line helps orient the γ-secretase particle.
The first column shows two averaged side-view images of the purified, negatively stained S-20 γ-secretase containing the short V5/HIS-fused NCT. The second column shows two averaged side-view images of the purified, negatively stained S-1 γ-secretase containing the GST-fused NCT. Images in same row are in similar but not identical views. Numbers next to each image are raw particle images that were used for calculating the respective average. White arrows in second column point to the extra densities that are attributed to the GST tag; these were never observed in the images of the S-20 particles. The third column has cartoons indicating the apparent GST sites at the bottom of the S-1 γ-secretase particle. Dashed horizontal lines demarcate the estimated transmembrane region.
Three-dimensional reconstruction of cryo-EM images of the γ-secretase particles
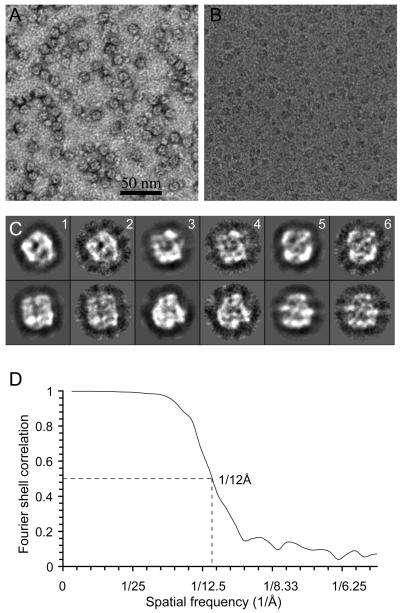
In order to obtain enough material for cryo-EM work, we used our new S-20 cell line that builds on the γ-30 line by also stably co-expressing a V5/HIS-tagged form of NCT. Purification from the S-20 cells yields substantially more γ-secretase protein and proportionally higher proteolytic activity in vitro than in the case of the γ-30 cells 21. Fig. 4A shows an electron micrograph of 5-times diluted and negatively stained γ-secretase particles from the S-20 cells. The image reveals highly homogeneous, round particles ~8-10 nm in diameter that are indistinguishable by negative stain EM from particles prepared similarly from the previous γ-30 cells 20. Fig. 4B displays a small portion of a micrograph of the purified S-20 γ-secretase sample embedded in vitreous ice. These cryo-images, recorded at under-focus values ranging from 1.2 to 3.5 μm at 200 kV, have good contrast, due to the selection of areas having very thin ice that we were able to observe only because we added a second layer of continuous, thin (20 nm) carbon film over the holey carbon film. We collected a total of ~110,000 γ-secretase particle images. We note that the stain-accessible central cavity (dark feature) in the uranyl acetate stained γ-secretase particles (Fig. 4A) is in agreement with the low-density feature (white feature) found at the center of most particles in the cryo-EM images (Fig 4B).
Fig. 4. Electron microscopy and 3D reconstruction of γ-secretase.
(A) A small area of a raw EM image of negatively stained γ-secretase particles purified from the S-20 cells. A dark spot in the middle of virtually every particle indicates a stain-accessible, low-density interior region of the complex. (B) A small area of a typical low-dose electron micrograph of S-20 γ-secretase particles embedded in vitreous ice. Consistent with the negative stained image (A), there is a white center (low density) in virtually every cryo-image of the γ-secretase particles, suggesting a low-density interior of the structure. (C) A comparison of the reprojections (columns 1, 3, 5) of the low-pass filtered cryo-EM map and the 2D class averages (columns 2, 4, 6) of the cryo-images of purified γ-secretase particles. (D) Resolution test of the cryo-EM map. The map has an estimated resolution of 12 Å, based on the threshold of 0.5 Å of the Fourier shell correlation calculated from two reconstructions from the two halves of the final data set.
We used three data sets (one negatively stained data set, one cryo-EM data set at high defocus, and a second cryo-EM dataset at lower defocus) and two complementary approaches (random conical tilt method and common line technique) to select a starting model (see Methods). The starting model we selected was calculated by the common-line technique with seven reference-free 2D class averages that were obtained from the high defocus cryo-EM dataset. We selected this starting model because it was similar to the cryo-EM reconstruction based on the random conical tilt model, and also because the reprojections of the model were most consistent with the reference-free 2D class averages calculated from the negative-stain data set or from the lower defocus cryo-EM data set. For 3D refinement, the contrast transfer function (CTF) phase-flipped images were used initially, but the CTF amplitude correction was applied at the final stages of refinement. Fig. 4C displays six pairs of refined class averages and their corresponding reprojections of the 3D model. The final 3D reconstruction has a resolution of 12Å, as estimated by Fourier shell correlation of two models c alculated from two halves of the data set (Fig. 4D). It is likely that heterogeneous glycosylation of NCT and the relatively small size of the particles have affected the image alignment accuracy, thus limiting the attainable resolution. Furthermore, due to use of the second layer of carbon film, some particles might have bound to the support film, causing slight non-isotropic orientation distribution, the effect of which is visible in Fig. 4C. However, the uneven angular distribution was not significant in the Eulerian angle plot (data not shown).
Structural features of the γ-secretase complex at 12 Å
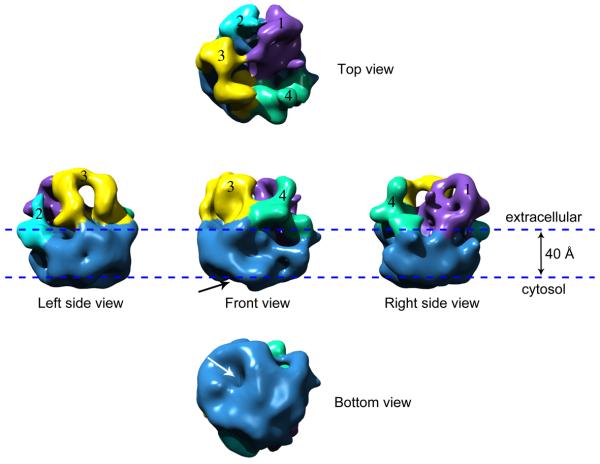
The 3D map is rendered as a surface representation at a threshold that encloses 100% of the expected protein mass of a monomeric γ-secretase complex (Fig. 5). Each view is labeled according to its orientation with respect to the membrane. The membrane orientation of the 3D cryo-EM map was established by first aligning the map with the negatively stained 3D map, and then by comparing the reprojections of the stained 3D map with 2D class averages of the stained GST-fused S-1 complex. The postulated lipid membrane is illustrated by a pair of horizontal lines 40 Å apart. Overall, the structure has a smooth cytosolic side and a larger, more irregular extracellular region. The structure has the belt-like feature that is typical of membrane protein complexes and represents the membrane-embedded region. In agreement with the previous negatively stained structure 20, the cryo-EM map reveals a globular structure of γ-secretase with dimensions of ~ 8 nm by 9 nm in the top (or extracellular) view and ~8.5 nm in height (Fig. 5). A central section in parallel to the membrane at the membrane embedded region has an area of ~8 × 9 nm2. This area is slightly larger than the required area for accommodating the predicted 19 transmembrane α-helices present in the 4-component purified complex. By comparison, the tetrameric aquaporin-1 water channel with 24 transmembrane α-helices occupies an area at the transmembrane region of 7 × 7 nm2 32; 33, and the P-glycoprotein with 12 transmembrane α-helices had a transmembrane area of 7 × 5 nm2 34. In addition, the overall size of the γ-secretase complex is slightly larger than a tightly packed soluble globular protein of this weight. As a result, the structure appears to be somewhat porous. For example, the extracellular region, instead of being a single compact domain, can be divided into two relatively large and two smaller domains that we have differentially colored and labeled as 1 through 4, for ease of visualization. At the cytosolic side, there is a sizable opening that reaches half way into the membrane region (Fig. 5 and Fig. 6A, arrows). Within the membrane region, there are two additional large cavities that are open towards the extracellular space, as seen in a vertically cut-open view of the structure (Fig. 6B, two dotted lines at top). We note that at this resolution (12Å), the low-density areas might not necessarily represent pores; they could also be unresolved loop structures that have lower density. Nevertheless, the presence of these low-density regions at both the extracellular and cytosolic surfaces and within the transmembrane region suggests that they may be required structural features for the enzyme to carry out intramembrane proteolysis of an embedded transmembrane helix of a type 1 substrate. These features appear to be consistent with biochemical studies demonstrating that the aspartate-containing active site, presumably located within the transmembrane domain, is accessible by chemical labels or inhibitors from both sides of the membrane 35; 36. Of particular interest is an almost continuous surface groove at the membrane region of the structure (as shown in Fig. 6A by a thick red curve) that could be the substrate entry site. The extracellular domain 4 located immediately above this groove might then serve as substrate receptor, i.e., the substrate-docking site previously suggested by biochemical studies 37.
Fig. 5. The 3D cryo-EM structure of the γ-secretase complex displayed in the top, bottom, and three side views.
These views are related by a 90° rotation around an axis either parallel or vertical to the membrane normal. The four densities on the extracellular surface are differentially colored and labeled as 1 through 4. The division into the four domains is based on the separation of the observed electron densities and is somewhat arbitrary and done for the convenience of structural description. The arrow indicates the apparent cavity on the cytoplasmic side.
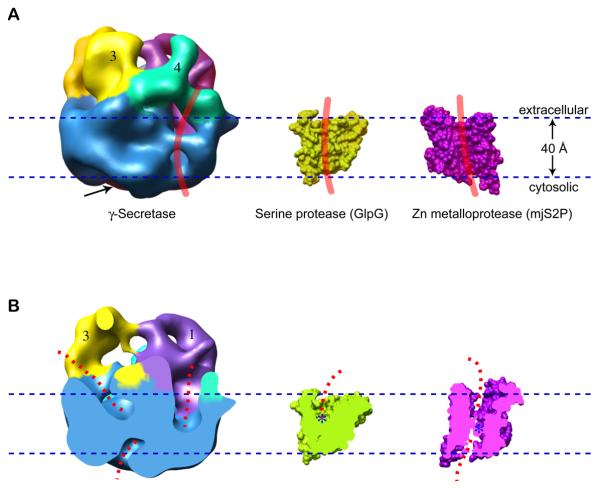
Fig. 6. A comparison of the γ-secretase cryo-EM structure with the crystal structures of the bacterial and archaeal intramembrane proteases.
(A) Surface-rendered side views of γ-secretase at the left, GlpG serine protease in the middle (PDB ID 2NRF), and the mjS2P Zn protease at the right (PDB ID 3B4R). A thick red curve is drawn at the potential membrane surface groove in each structure, representing the predicted position that an a-helical substrate might bind. The arrow indicates the apparent cytoplasmic cavity. (B) Vertically cut-open views of the three structures shown in (A). The dashed red lines indicate the potential water accessible space. The active sites in GlpG and mjS2P are indicated by blue asterisks. The exact location of active site in γ-secretase is currently unknown. The mjS2P structure is rotated 180° around an axis perpendicular to the membrane plane with respect to the view in (A).
A comparison of the cryo-EM map of γ-secretase with the crystal structures of the bacterial and archaeal intramembrane proteases
This is the first report that describes the structure of γ-secretase obtained by cryo-EM with single particle analysis, a method that avoids potential artifacts of negative stain EM and enables more detailed structural analysis. Although many of the structural features of the models obtained by negative stain and cryo-EM are consistent, some important structural features are resolved in the new cryo-EM structure. We now assign only the lower ~4 nm portion of the structure to the transmembrane region (previously assigned as ~6 nm in our negatively-stained structure 20), thus leaving a larger portion of the structure as extracellular (Fig. 5 and 6) More importantly, instead of an apparent single central chamber suggested by our negatively stained structure 20, we now observe multiple low-density cavities within the postulated transmembrane region (Fig. 6B).
Despite the improved resolution, our ability to accurately interpret the 12Å-resolution cryo-EM structure in terms of its function is limited by the lack of crystal structures of any of the γ-secretase subunits, particularly the catalytically active PS heterodimer. Nevertheless, we asked whether γ-secretase has architectural features similar to the bacterial or the archaeal intramembrane proteases 14. A side-by-side comparison reveals dramatic differences in size and shape between these structures (Fig. 6). Compared to the ~4 nm-sized, single-protein intramembrane proteases, GlpG 10 and mjS2P 13, the four-protein γ-secretase complex at ~8-9 nm in each dimension is significantly larger. In addition, whereas both GlpG and mjS2P have only small cap regions on their extracellular sides, the γ-secretase has a much larger multi-domain structure in this region (Fig. 6). Despite these important differences, we observed three aspects of structural similarity among these proteases: (1) all 3 enzymes have very small cytosolic regions; (2) there is a membrane-facing surface groove in each enzyme (as indicated by the red vertical lines in Fig. 6A) that may act as the docking and/or entry site of transmembrane substrates 14; and (3) water appears to be accessible deep into the membrane-embedded regions of the structures (illustrated by the dashed red curves in Fig. 6B), although in all three cases, the water access path does not appear to penetrate through the membrane. This latter feature might be important for helping to maintain the cellular membrane potential and chemical gradients.
In summary, we have extended the structural analysis of purified, active human γ-secretase by determining the structure at an improved resolution of 12 Å by cryo-EM and by showing that it has a mass of ~230 kDa by STEM measurement, consistent with a monomeric complex with a subunit stoichiometry of 1:1:1:1. Despite marked physical differences, we found that γ-secretase appears to share certain design principles with simpler intramembrane proteases in that it has a membrane-facing surface groove and routes for water access into the depth of the membrane-embedded region. Future work by cryo-EM of purified γ-secretase in complex with substrate plus a docking-site or a catalytic-site inhibitor will help shed light on how the substrates interact with the enzyme complex.
Materials and Methods
Cell line production and complex purification
The S-1 cell line stably expressing human PS1, FLAG-Pen-2, Aph1a2-HA and NCT-GST and the S-20 cell line stably expressing human PS1, FLAG-Pen-2, Aph1a2-HA and NCT-V5/HIS were cultured as previously described 4; 21. Additionally, the γ-30 cell line that stably overexpresses human PS1, FLAG-Pen-2 and Aph1a2-HA was transiently transfected with NCT-MBP in the pcDNA5.1 vector. γ-Secretase complexes were purified from these various cell lines as previously described 4; 20. The quality of the anti-FLAG affinity resin purchased from Sigma varied from batch to batch, often causing antibody contamination. The complex used for cryo-EM work was prepared using a batch with good quality such that the IgG contamination was insignificant.
Blue Native gel electrophoresis
One day prior to analysis, 5-13.5% polyacrylamide gels were poured as previously described 38; 39. For Blue Native analysis on these gels, the purified γ-secretase from S-20 cells was prepared in BN-PAGE lysis buffer 40. High molecular weight standards for native gel analysis were purchased from Invitrogen, Amersham, and Sigma. Gels were run as previously described 40 and Western blotted as described below. Molecular mass of the γ-secretase complex using various molecular weight standards was estimated using AlphaEaseFC software.
Western blot analysis
For Western blot analysis of PS1-NTF, PS1-CTF, Aph1a2-HA, FLAG-Pen-2, and NCT-V5/HIS, samples were run on a 4-20% Tris-glycine polyacrylamide gel, transferred onto a polyvinylidene difluoride (PVDF) membrane and probed with antibody AB14 (for PS1-NTF, 1:2000; a gift of S. Gandy), or 231F (for PS1-NTF, 1:1000, gift of Dr. B. Yanker), 13A11 (for PS1-CTF, 5 μg/ml; gift of Elan Pharmaceuticals), 3F10 (for A ph1a2-HA, 1:2000, Roche), anti-FLAG M2 (for FLAG-Pen-2, 1:1000, Sigma), or N1660 (for NCT, 1:1000, Sigma).
Biotinylation
Biotinylation of purified γ-secretase was described previously 18. Briefly, purified γ-secretase from the S-1 and S-20 cell line was biotinylated with EZ-Link Sulfo-NHS-LC-Biotin (Pierce) according to the manufacturer's protocol. Excess biotin was removed with a desalting column, and the biotinylated γ-secretase preparation was bound to M2 resin overnight, washed extensively with 0.1% digitonin in Tris-buffered saline, and eluted with 200 μg/ml FLAG peptide. The biotinylated protein preparation was run on a 4-20% Tris-glycine gel, transferred to a PVDF membrane and probed with an anti-biotin antibody (1:2000, Sigma).
STEM mass measurement
Scanning transmission EM (STEM) was carried out at the Brookhaven National Laboratory STEM user facility. Purified γ-secretase from S-20 cells was diluted in 0.1% digitonin-TBS in order to find the proper sample concentration that gave rise to images with appropriate particle distribution. Tobacco mosaic virus (TMV) was included during specimen preparation as an internal quality control. To avoid bias, we selected indiscriminately all 1,800 particles from 70 cryo-STEM images and measured their masses (after background subtraction) with the program PCMASS 27. The mean and standard deviation of the mass were then calculated. Details of the STEM operation and mass analysis have been described 27.
Cryo-EM
Cryo-EM grids were prepared using a Vitrobot (FEI, Hisbrone, CO) with the humidity of the specimen chamber set to 90% and the temperature set to 11 °C. A 4.5 μl undiluted sample purified from the S-20 cells was applied to a glow-discharged lacey carbon grid covered with an additional very thin layer of continuous carbon film (~20 nm thick). The grid was held inside the chamber by a pair of tweezers at the offset position −1 mm. After incubating the sample on the grid for 40 seconds, the grid was blotted for 4 seconds and then rapidly plunged into liquid ethane. Negative staining was carried out as described 20. The underfocus value used for image acquisition was in the ranges of 1.0–1.5 μm and 1.2–3.5 μm for the negative stained and cryo samples, respectively. Images of the negatively stained sample were recorded in a JEOL JEM 1200EX electron microscope operating at 120 kV with a magnification of 50,000. The frozen-hydrated γ-secretase samples were imaged under low-dose condition (10 e/Å2) in a JEOL 2010F electron microscope operating at 200 kV at the same magnification, while the grids were maintained at −170°C in a Gatan 626 cryo-specimen holder (Gatan, Pittsburg, PA). All electron micrographs were recorded on Kodak SO-163 negative film. The films were developed in Kodak D-19 solution and digitized using a Nikon Supercool scanner 8000ED at a step size of 12.7 μm, corresponding to a pixel size of 2.54 Å at the sample level.
Image processing and 3D reconstruction
The software packages EMAN 41 and SPIDER 42 were used for image processing and reconstruction of γ-secretase single particle images. We selected the individual γ-secretase particles with a square box of 80 pixels size in the semiautomatic mode of the Boxer in EMAN. The resulting particle images were inspected, and about 5% of the particles were manually rejected at this stage, based on the contrast and size. A total of 11,518 particles were included in the negative-stain data set of the GST-tagged γ-secretase purified from the S-1 cell line. A second negative stain data set containing 15,300 particles was collected with sample purified from the S-20 cells. We collected two cryo-EM datasets of the γ-secretase purified from S-20 cells, one at the higher defocus value of ~3.5 μm that contained 46,118 particles, and the other at the lower defocus value of ~1.8 μm that contained 61,635 particles. The parameters of the microscope contrast transfer function were estimated by manually fitting the averaged power spectrum of all particles belonging to the same micrograph with the computed CTF model. The contrast and the frequency-dependent phase reversal in cryo-EM images were corrected at this stage. Subsequently, 2D classification was carried out for these data sets. We found that various starting models calculated by the common-line technique with selected reference-free 2D averages from these datasets did not converge to essentially the same structures after multiple iterations of refinement. Thus, precaution was taken to ascertain the reliability of the reconstruction from the cryo-EM images of the small and asymmetrical γ-secretase complexes.
We first used our published negative-stain 3D map 20 as the starting model to refine the cryo-EM datasets. This starting model was produced by merging 4 random conical tilt (RCT)-generated models. Each of these RCT models was calculated from a distinctive view of the γ-secretase complex. Merging several RCT models minimizes the missing-cone caused structural distortion. Cryo-EM map refined from the cryo-EM datasets using this negatively stained starting model had features consistent with reference-free 2D class averages of the cryo-EM datasets. However, we were not certain if the negative-staining procedure itself introduced distortion in the starting model and if any distortion was propagated into the refined cryo-EM map. To alleviate this concern, we further calculated and refined an array of 3D models by common line technique from the 2D class averages of the high defocus cryo-EM dataset. These models shared certain features but could not be regarded as representing virtually the same structure. However, among these 3D models, we found one model that was most similar to the cryo-EM map that was refined from the RCT-based starting model. This new 3D model was then selected as the starting model for iterative refinement against the lower defocus cryo-EM dataset. About 20% of particles were rejected during refinement. The final cryo-EM map thus obtained was similar to the RCT-based cryo-EM map, but the new map was deemed better than the RCT-based map because the new map had a lower noise level at the same resolution and display threshold. The resolution of the final map was estimated by Fourier shell correlation of two maps calculated separately from two halves of the data set. The map was low-pass filtered to the estimated resolution (12Å), and viewed in UCSF Chimera 43. The crystal structures of GlpG and mjS2P had also been surface rendered and sectioned in Chimera 43.
Acknowledgements
The mass measurement was carried out at the BNL STEM facility, a user facility supported by the US Department of Energy. HL was partially supported by BNL LDRD grant 05-111 and by NIH R01 grant GM74985. MW and DJS were supported by NIH P01 grant AG15379. PO was supported by training grant TE AG00222-15. PCF was supported by the Swiss National Science Foundation grant 310000-116652/1 and by the NCCR “Neural Plasticity and Repair”.
References
- 1.Kimberly WT, LaVoie MJ, Ostaszewski BL, Ye W, Wolfe MS, Selkoe DJ. Gamma-secretase is a membrane protein complex comprised of presenilin, nicastrin, Aph-1, and Pen-2. Proc Natl Acad Sci U S A. 2003;100:6382–7. doi: 10.1073/pnas.1037392100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Takasugi N, Tomita T, Hayashi I, Tsuruoka M, Niimura M, Takahashi Y, Thinakaran G, Iwatsubo T. The role of presenilin cofactors in the gamma-secretase complex. Nature. 2003;422:438–41. doi: 10.1038/nature01506. [DOI] [PubMed] [Google Scholar]
- 3.Edbauer D, Winkler E, Regula JT, Pesold B, Steiner H, Haass C. Reconstitution of gamma-secretase activity. Nat Cell Biol. 2003;5:486–8. doi: 10.1038/ncb960. [DOI] [PubMed] [Google Scholar]
- 4.Fraering PC, Ye W, Strub JM, Dolios G, LaVoie MJ, Ostaszewski BL, van Dorsselaer A, Wang R, Selkoe DJ, Wolfe MS. Purification and characterization of the human gamma-secretase complex. Biochemistry. 2004;43:9774–89. doi: 10.1021/bi0494976. [DOI] [PubMed] [Google Scholar]
- 5.Wolfe MS, Kopan R. Intramembrane proteolysis: theme and variations. Science. 2004;305:1119–23. doi: 10.1126/science.1096187. [DOI] [PubMed] [Google Scholar]
- 6.Wolfe MS, Xia W, Ostaszewski BL, Diehl TS, Kimberly WT, Selkoe DJ. Two transmembrane aspartates in presenilin-1 required for presenilin endoproteolysis and gamma-secretase activity. Nature. 1999;398:513–7. doi: 10.1038/19077. [DOI] [PubMed] [Google Scholar]
- 7.Selkoe DJ. Cell biology of protein misfolding: the examples of Alzheimer's and Parkinson's diseases. Nat Cell Biol. 2004;6:1054–61. doi: 10.1038/ncb1104-1054. [DOI] [PubMed] [Google Scholar]
- 8.Selkoe DJ, Wolfe MS. Presenilin: running with scissors in the membrane. Cell. 2007;131:215–21. doi: 10.1016/j.cell.2007.10.012. [DOI] [PubMed] [Google Scholar]
- 9.Wang Y, Zhang Y, Ha Y. Crystal structure of a rhomboid family intramembrane protease. Nature. 2006;444:179–80. doi: 10.1038/nature05255. [DOI] [PubMed] [Google Scholar]
- 10.Wu Z, Yan N, Feng L, Oberstein A, Yan H, Baker RP, Gu L, Jeffrey PD, Urban S, Shi Y. Structural analysis of a rhomboid family intramembrane protease reveals a gating mechanism for substrate entry. Nat Struct Mol Biol. 2006;13:1084–91. doi: 10.1038/nsmb1179. [DOI] [PubMed] [Google Scholar]
- 11.Ben-Shem A, Fass D, Bibi E. Structural basis for intramembrane proteolysis by rhomboid serine proteases. Proc Natl Acad Sci U S A. 2007;104:462–6. doi: 10.1073/pnas.0609773104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lemieux MJ, Fischer SJ, Cherney MM, Bateman KS, James MN. The crystal structure of the rhomboid peptidase from Haemophilus influenzae provides insight into intramembrane proteolysis. Proc Natl Acad Sci U S A. 2007;104:750–4. doi: 10.1073/pnas.0609981104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Feng L, Yan H, Wu Z, Yan N, Wang Z, Jeffrey PD, Shi Y. Structure of a site-2 protease family intramembrane metalloprotease. Science. 2007;318:1608–12. doi: 10.1126/science.1150755. [DOI] [PubMed] [Google Scholar]
- 14.Urban S, Shi Y. Core principles of intramembrane proteolysis: comparison of rhomboid and site-2 family proteases. Curr Opin Struct Biol. 2008 doi: 10.1016/j.sbi.2008.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Narayanan S, Sato T, Wolfe MS. A C-terminal region of signal peptide peptidase defines a functional domain for intramembrane aspartic protease catalysis. J Biol Chem. 2007;282:20172–9. doi: 10.1074/jbc.M701536200. [DOI] [PubMed] [Google Scholar]
- 16.Martoglio B, Golde TE. Intramembrane-cleaving aspartic proteases and disease: presenilins, signal peptide peptidase and their homologs. Hum Mol Genet. 2003;12(Spec No 2):R201–6. doi: 10.1093/hmg/ddg303. [DOI] [PubMed] [Google Scholar]
- 17.Wolfe MS. The gamma-secretase complex: membrane-embedded proteolytic ensemble. Biochemistry. 2006;45:7931–9. doi: 10.1021/bi060799c. [DOI] [PubMed] [Google Scholar]
- 18.Osenkowski P, Ye W, Wang R, Wolfe MS, Selkoe DJ. Direct and potent regulation of gamma -secretase by its lipid microenvironment. J Biol Chem. 2008;283:22529–40. doi: 10.1074/jbc.M801925200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fraering PC, LaVoie MJ, Ye W, Ostaszewski BL, Kimberly WT, Selkoe DJ, Wolfe MS. Detergent-dependent dissociation of active gamma-secretase reveals an interaction between Pen-2 and PS1-NTF and offers a model for subunit organization within the complex. Biochemistry. 2004;43:323–33. doi: 10.1021/bi035748j. [DOI] [PubMed] [Google Scholar]
- 20.Lazarov VK, Fraering PC, Ye W, Wolfe MS, Selkoe DJ, Li H. Electron microscopic structure of purified, active {gamma}-secretase reveals an aqueous intramembrane chamber and two pores. Proc Natl Acad Sci U S A. 2006;103:6889–94. doi: 10.1073/pnas.0602321103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cacquevel M, Aeschbach L, Osenkowski P, Li D, Ye W, Wolfe MS, Li H, Selkoe DJ, Fraering PC. Rapid purification of active gamma-secretase, an intramembrane protease implicated in Alzheimer's disease. J Neurochem. 2008;104:210–20. doi: 10.1111/j.1471-4159.2007.05041.x. [DOI] [PubMed] [Google Scholar]
- 22.Schroeter EH, Ilagan MX, Brunkan AL, Hecimovic S, Li YM, Xu M, Lewis HD, Saxena MT, De Strooper B, Coonrod A, Tomita T, Iwatsubo T, Moore CL, Goate A, Wolfe MS, Shearman M, Kopan R. A presenilin dimer at the core of the gamma-secretase enzyme: insights from parallel analysis of Notch 1 and APP proteolysis. Proc Natl Acad Sci U S A. 2003;100:13075–80. doi: 10.1073/pnas.1735338100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cervantes S, Saura CA, Pomares E, Gonzalez-Duarte R, Marfany G. Functional implications of the presenilin dimerization: reconstitution of gamma-secretase activity by assembly of a catalytic site at the dimer interface of two catalytically inactive presenilins. J Biol Chem. 2004;279:36519–29. doi: 10.1074/jbc.M404832200. [DOI] [PubMed] [Google Scholar]
- 24.Sato T, Diehl TS, Narayanan S, Funamoto S, Ihara Y, De Strooper B, Steiner H, Haass C, Wolfe MS. Active gamma-secretase complexes contain only one of each component. J Biol Chem. 2007;282:33985–93. doi: 10.1074/jbc.M705248200. [DOI] [PubMed] [Google Scholar]
- 25.Edbauer D, Winkler E, Haass C, Steiner H. Presenilin and nicastrin regulate each other and determine amyloid beta-peptide production via complex formation. Proc Natl Acad Sci U S A. 2002;99:8666–71. doi: 10.1073/pnas.132277899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Evin G, Canterford LD, Hoke DE, Sharples RA, Culvenor JG, Masters CL. Transition-state analogue gamma-secretase inhibitors stabilize a 900 kDa presenilin/nicastrin complex. Biochemistry. 2005;44:4332–41. doi: 10.1021/bi0481702. [DOI] [PubMed] [Google Scholar]
- 27.Wall JS, Simon MN, Lin BY, Vinogradov SN. Mass mapping of large globin complexes by scanning transmission electron microscopy. Methods Enzymol. 2008;436:487–501. doi: 10.1016/S0076-6879(08)36027-3. [DOI] [PubMed] [Google Scholar]
- 28.Fotiadis D, Jeno P, Mini T, Wirtz S, Muller SA, Fraysse L, Kjellbom P, Engel A. Structural characterization of two aquaporins isolated from native spinach leaf plasma membranes. J Biol Chem. 2001;276:1707–14. doi: 10.1074/jbc.M009383200. [DOI] [PubMed] [Google Scholar]
- 29.Shirotani K, Edbauer D, Kostka M, Steiner H, Haass C. Immature nicastrin stabilizes APH-1 independent of PEN-2 and presenilin: identification of nicastrin mutants that selectively interact with APH-1. J Neurochem. 2004;89:1520–7. doi: 10.1111/j.1471-4159.2004.02447.x. [DOI] [PubMed] [Google Scholar]
- 30.Capell A, Kaether C, Edbauer D, Shirotani K, Merkl S, Steiner H, Haass C. Nicastrin interacts with gamma-secretase complex components via the N-terminal part of its transmembrane domain. J Biol Chem. 2003;278:52519–23. doi: 10.1074/jbc.C300435200. [DOI] [PubMed] [Google Scholar]
- 31.Li H, Chavan M, Schindelin H, Lennarz WJ, Li H. Structure of the oligosaccharyl transferase complex at 12 A resolution. Structure. 2008;16:432–40. doi: 10.1016/j.str.2007.12.013. [DOI] [PubMed] [Google Scholar]
- 32.Li H, Lee S, Jap BK. Molecular design of aquaporin-1 water channel as revealed by electron crystallography. Nat Struct Biol. 1997;4:263–5. doi: 10.1038/nsb0497-263. [DOI] [PubMed] [Google Scholar]
- 33.Sui H, Han BG, Lee JK, Walian P, Jap BK. Structural basis of water-specific transport through the AQP1 water channel. Nature. 2001;414:872–8. doi: 10.1038/414872a. [DOI] [PubMed] [Google Scholar]
- 34.Lee JY, Urbatsch IL, Senior AE, Wilkens S. Projection structure of Pglycoprotein by electron microscopy. Evidence for a closed conformation of the nucleotide binding domains. J Biol Chem. 2002;277:40125–31. doi: 10.1074/jbc.M206871200. [DOI] [PubMed] [Google Scholar]
- 35.Sato C, Morohashi Y, Tomita T, Iwatsubo T. Structure of the catalytic pore of gamma-secretase probed by the accessibility of substituted cysteines. J Neurosci. 2006;26:12081–8. doi: 10.1523/JNEUROSCI.3614-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tolia A, Horre K, De Strooper B. Transmembrane domain 9 of presenilin determines the dynamic conformation of the catalytic site of gamma -secretase. J Biol Chem. 2008 doi: 10.1074/jbc.M802461200. [DOI] [PubMed] [Google Scholar]
- 37.Esler WP, Kimberly WT, Ostaszewski BL, Ye W, Diehl TS, Selkoe DJ, Wolfe MS. Activity-dependent isolation of the presenilin- gamma -secretase complex reveals nicastrin and a gamma substrate. Proc Natl Acad Sci U S A. 2002;99:2720–5. doi: 10.1073/pnas.052436599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schagger H, Cramer WA, von Jagow G. Analysis of molecular masses and oligomeric states of protein complexes by blue native electrophoresis and isolation of membrane protein complexes by two-dimensional native electrophoresis. Anal Biochem. 1994;217:220–30. doi: 10.1006/abio.1994.1112. [DOI] [PubMed] [Google Scholar]
- 39.Schagger H, von Jagow G. Blue native electrophoresis for isolation of membrane protein complexes in enzymatically active form. Anal Biochem. 1991;199:223–31. doi: 10.1016/0003-2697(91)90094-a. [DOI] [PubMed] [Google Scholar]
- 40.LaVoie MJ, Fraering PC, Ostaszewski BL, Ye W, Kimberly WT, Wolfe MS, Selkoe DJ. Assembly of the gamma-secretase complex involves early formation of an intermediate subcomplex of Aph-1 and nicastrin. J Biol Chem. 2003;278:37213–22. doi: 10.1074/jbc.M303941200. [DOI] [PubMed] [Google Scholar]
- 41.Ludtke SJ, Baldwin PR, Chiu W. EMAN: semiautomated software for high-resolution single-particle reconstructions. J Struct Biol. 1999;128:82–97. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
- 42.Frank J, Radermacher M, Penczek P, Zhu J, Li Y, Ladjadj M, Leith A. SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J Struct Biol. 1996;116:190–9. doi: 10.1006/jsbi.1996.0030. [DOI] [PubMed] [Google Scholar]
- 43.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–12. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]