Abstract
The neuronal repressor REST/NRSF is expressed at high levels in mouse embryonic stem (mES) cells1, but its role in these cells has not been understood. Here we show that REST maintains self-renewal and pluripotency in mES cells through suppression of microRNA (miR)-21. We found that, as with known self-renewal markers, REST expression is much higher in self-renewing mES cells than in differentiating mES (mEB) cells. The heterozygous deletion of REST (REST+/−) and its siRNA-mediated knockdown in mES cells causes a loss of self-renewal, even when these cells are grown under self-renewal conditions, and leads to the expression of markers specific for multiple lineages. Conversely, exogenously added REST maintains self-renewal in mEB cells. In addition, REST+/− mES cells cultured under self-renewal conditions express substantially reduced levels of several self-renewal regulators, including Oct4, Nanog, Sox2, and c-Myc, and exogenously added REST in mEB cells maintains the self-renewal phenotypes and expression of these self-renewal regulators. We further show that in mES cells, REST is bound to the gene chromatin of a set of miRs that potentially target self-renewal genes. Whereas mES cells and mEB cells containing exogenously added REST express lower levels of these miRs, mEB cells, REST+/− mES cells, and siREST-treated mES cells express higher levels of these miRs. At least one of these REST-regulated miRs, miR-21, specifically suppresses the self-renewal of mES cells, corresponding to the decreased expression of Oct4, Nanog, Sox2, and c-Myc. Thus, REST is a newly discovered element of the interconnected regulatory network that maintains the self-renewal and pluripotency of mES cells.
Repressor element-1 silencing transcription factor/neuron-restrictive silencer factor (REST/NRSF) is believed to be a major transcriptional repressor of neurogenesis2–5 and activation of REST target genes was found to be sufficient to convert neural stem/progenitor cells to neuronal phenotypes6, 7. However, REST activity appears to depend on the cellular context; for example, REST can show both an oncogenic8–10 and tumor suppressor function5 as well as involvement in hematopoietic and cardiac differentiation3–5. Embryonic stem (ES) cells are pluripotent cells that have the potential for both indefinite self-renewal and differentiation into all three germ layers of the body11. Here we provide evidence that REST has a unique role as a protector of self-renewal and pluripotency in mouse ES (mES) cells, corresponding to the expression of critical regulators such as Oct4, Nanog, Sox2, and c-Myc.
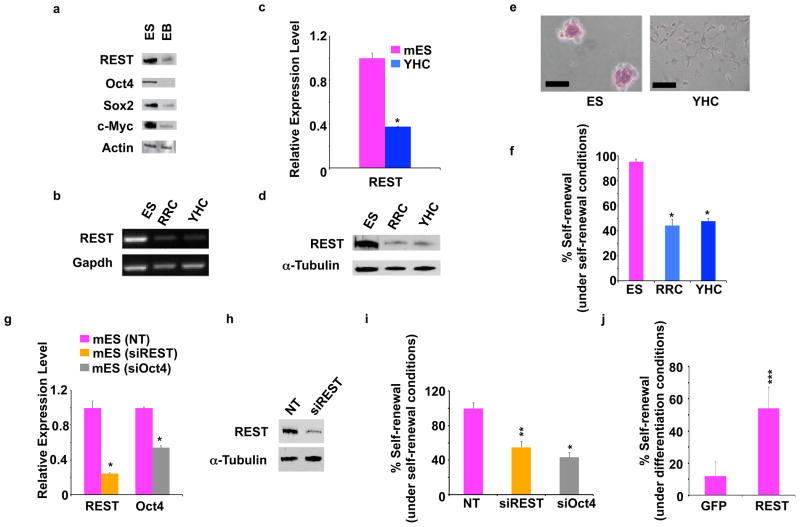
We began by assessing the levels of REST protein in mES cells growing under self-renewal conditions and differentiation conditions (Fig. 1a; ES and EB, respectively). As expected, Western blotting showed that the ES cells had higher expression of REST and the representative markers of the self-renewal proteins Oct4, Sox2, and c-Myc than did the EB cells, indicating that REST expression is associated with self-renewal. To determine whether REST regulates the self-renewal of mES cells, we took two approaches using two cell lines—YHC334 (YHC) and RRC160 (RRC) (see Supplementary Materials). In the first approach, we performed RT-PCR (Fig. 1b), quantitative RT-PCR (Q.RT-PCR; Fig. 1c) using different primer sets than those used for regular RT-PCR, and Western blotting (Fig. 1d), which showed that both of the REST+/− mES cell lines had substantially lower levels of REST transcript and protein than the parental mES cells had. Alkaline phosphatase assays12, performed to measure self-renewal within these cells, revealed that the levels of self-renewal in both of the REST+/− mES cell lines were less than half that in the parental mES cells (Fig. 1e and 1f).
Figure 1. REST regulates self-renewal in mouse ES cells.
a, EB cells (ES cells grown in the absence of LIF under nonadherent conditions for 4 d) showed reduced levels of self-renewal markers and REST compared with ES cells (grown in the presence of LIF under adherent conditions). Western blot analysis of whole-cell extracts prepared from ES cells and EB cells showed an association between expression of REST and self-renewal markers. Actin was used as a loading control. b–f, Mouse ESC lines with heterozygous deletion of REST (RRC and YHC) show loss of self-renewal. b, RT-PCR analysis and c, Q.RT-PCR analysis with different primer sets of total RNA isolated from ES, RRC, and YHC cells showed reduced REST transcripts in REST+/− cells. Gapdh was used as a loading control. * p < 0.0001. The values are represented as mean +/− SD (n=3). d, Western blot analysis of whole-cell extracts from ES, RRC, and YHC cells showed reduced REST protein in REST+/− cells. α-Tubulin was used as a loading control. e, Alkaline phosphatase staining of ES colonies showed loss of self-renewal in REST+/− cells as compared with wild-type ES cells f, Percentages of self-renewing colonies of ES, RRC, and YHC cells calculated after alkaline phosphatase assays when cultured under self-renewing conditions showed significant reductions in the self-renewal capacity of both REST+/− cell lines compared with ES. * p < 0.0001. The error bars correspond to three replicates (n=3). g–i, siRNA-mediated knockdown of REST caused loss of self-renewal in mES cells. g, Specific knockdown of targeted genes was achieved using siRNA. Q.RT-PCR of total RNA purified from mES cells treated with siREST or siOct4 showed knockdown of specific genes. Analysis was performed 5 d after transfection. * p < 0.0001. The values are represented as mean +/− SD (n=3). h, Western blotting showed reduced REST protein levels in siREST treated cells compared with control (NT siRNA). i, siREST- and siOct4-treated cells showed less self-renewal than NT-treated cells did. mES cell colonies were screened by alkaline phosphatase assays. * p < 0.0001; ** p < 0.001. The error bars correspond to three replicates (n=3). j, Exogenously added REST, but not GFP, maintained self-renewal in mES cells cultured under differentiation conditions. mES cells were transfected with plasmids encoding GFP or REST and grown in the absence of LIF. Percentages of self-renewing colonies from three independent experiments were averaged after alkaline phosphatase assay and are shown for each transfected gene. *** p < 0.01. The error bars correspond to three replicates (n=3).
In the second approach, done to rule out any adaptive response accumulated by REST+/− mES cell lines, we determined whether the decreased self-renewal seen in the REST+/− mES cells could also be seen in mES cells treated with small interfering RNA (siRNA) against REST (siREST). Q.RT-PCR assays done to confirm that siRNA-treated cells actually had decreased REST and Oct4 levels showed a 50% knockdown of REST and Oct4, respectively, in both siREST- and siOct4-treated cells (Fig. 1g). This was further confirmed by Western blotting (Fig. 1h). Self-renewal assays of these cells (Fig. 1i) then showed that whereas mES cells treated with NT siRNA showed almost no change in self-renewal (94.5%), mES cells treated with siREST showed about a 50% decrease in self-renewal (see Fig. S1 in Supplementary Materials for the effect of four different siREST constructs on the self-renewal capacity of mES cells). SiOct4-treated mES cells also showed a decrease in self-renewal (47%) consistent with the role of Oct4 in mES cell self-renewal13. Thus, siREST-mediated knockdown of REST transcripts, and not nonspecific siRNA activity, caused decreased self-renewal in mES cells, a result similar to that seen in the REST+/− mES cell lines.
Gain-of-function experiments (Fig. 1j), in which self-renewing mES cells were transfected with plasmids encoding a neomycin resistance gene and either green fluorescent protein (GFP) or REST12 showed that cells transfected with GFP showed very little maintenance of self-renewal, whereas cells transfected with REST showed approximately 55% maintenance of self-renewal. Taken together, these results further confirmed the role of REST in maintaining self-renewal of mES cells.
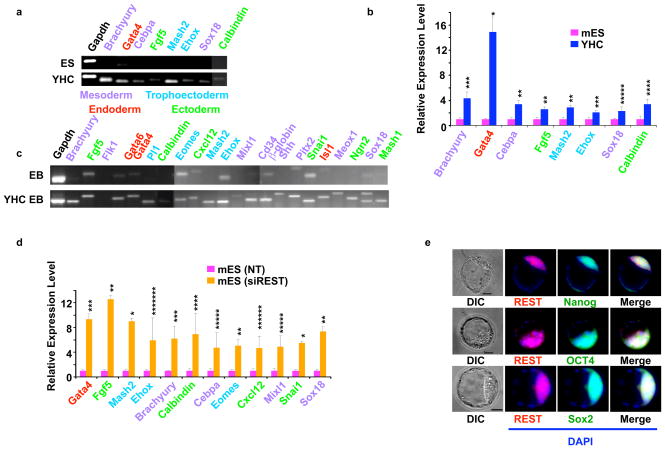
To ascertain the lineages into which REST+/− haploinsufficient mES cells differentiate on their exit from REST-mediated self-renewal, we performed RT-PCR (Fig. 2a) and Q.RT-PCR (Fig. 2b) assays of parental mES cells, YHC, and RRC cells growing under self-renewal conditions in the presence of LIF. REST+/− cells showed activation of markers specific for mesoderm, endoderm, ectoderm, and trophectoderm. Expression of all of these differentiation markers was absent or detectable at only very low levels in self-renewing parental mES cells. The results in RRC cells (data not shown) and YHC cells were very similar. Notably, while the neuronal marker Calbindin is a direct target of REST1, the other differentiation markers are not known to be direct REST targets, suggesting that REST can maintain the pluripotency of mES cells partly by directly repressing the expression of some differentiation genes and partly by an indirect mechanism in which the non-REST target differentiation genes are repressed as a consequence of the REST-mediated maintenance of self-renewal.
Figure 2. Mouse ES cells cells with heterozygous deletion of REST or mouse ES cells treated with siREST differentiate into multiple lineages.
a, RT-PCR analysis and b, Q.RT-PCR analysis of total RNA isolated from ES and YHC cells grown under self-renewing conditions were performed using primers specific for markers color-coded for ectoderm, mesoderm, endoderm, and trophectoderm. Gapdh was used as an internal control. * p = 0.001; ** p = 0.003; *** p = 0.006; **** p = 0.012. The values are represented as mean +/− SD (n=3). c, EB cells derived from YHC cells show lineage markers that are either absent or significantly reduced in EB cells derived from wild-type mES cells. RT-PCR analysis of total RNA isolated from wild-type EB cells and YHC EB cells was performed using specific primers for various differentiated lineage genes (shown above each lane). Gapdh was used as a loading control. d, Q.RT-PCR analysis of total RNA isolated from mES cells treated with either non-targeting siRNA (NT) or siREST was performed using primers specific for different lineage markers. Analysis was performed 5 d after transfection. * p < 0.0001; ** p = 0.001; *** p = 0.002; **** p = 0.004; **** p = 0.005; ****** p = 0.01. The values are represented as mean +/− SD (n=3). e, Inner cell mass of blastocysts showed coexpression of REST and self-renewal markers Oct4, Nanog, and Sox2. Blastocysts were stained for nuclei (DAPI, blue), REST (red), and self-renewal markers (green). Merged and differential interference contrast (DIC) images are also shown. Scale bars represent 25 μm.
The differentiation of REST+/− haploinsufficient cells into multiple lineages growing under self-renewal conditions prompted us to determine the differentiation characteristics of EB cells derived from wild-type as well as REST+/− ES cells. RT-PCR of REST+/− EB cells detected the expression of markers for all lineages, including ectoderm, mesoderm, endoderm, and trophectoderm (Fig. 2c). Some of these markers were either absent or markedly reduced in wild-type mES cell-derived EB cells. Thus, although both wild-type and REST+/− cells started expressing multiple differentiation markers in EB cells, the REST+/− cells expressed a greater number of lineage markers than did the wild-type mES cells, suggesting that REST deficiency predisposes mES cells to differentiation. To determine whether siREST-treated mES cells also differentiate into various lineages, we performed a Q.RT-PCR assay with these cells growing under self-renewal conditions (Fig. 2d). The results indicated that siREST indeed increased the expression of various lineage markers. Interestingly, the exact level of expression of each differentiation marker varied between siREST-treated mES and REST+/− cells (Fig. 2b), presumably because of the differences in the methods used to reduce the REST levels in these cells; that is, whereas REST+/− cells may show additional adaptive responses as a result of being cultured for longer than the siREST-treated mES cells, the latter show more immediate effects of REST knockdown. Nonetheless, the overall results indicated that upon exit of from REST-mediated self-renewal, both siREST-treated REST+/− cells and mES cells were able to differentiate into various lineages.
The inner cell mass (ICM) of the blastocyst has been shown to harbor self-renewing cells that express proteins such as Nanog, Oct4, and Sox2. To determine whether REST is expressed in blastocysts, we subjected mouse blastocysts to double-immunofluorescence analysis using anti-REST antibodies and antibodies against Nanog, Oct4, or Sox2. Consistent with its role in the self-renewal of mES cells, REST was co-expressed with these self-renewal regulators in the blastocyst ICM (Fig. 2e) (see Supplementary Materials for a discussion on the role of REST in the ICM cells).
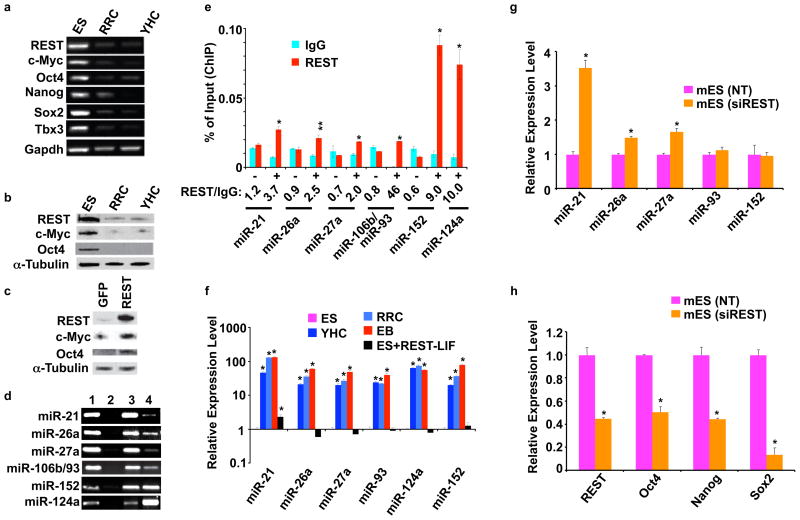
When we subjected the parental mES cells and REST+/− (YHC and RRC) haploinsufficient cells growing under self-renewal conditions to RT-PCR assays to examine the transcript levels of various self-renewal markers, we were surprised to find that the decreased REST levels in REST+/− cells corresponded with decreased transcript levels of several other self-renewal genes, such as Oct4, Nanog, Sox2, Tbx3, and c-myc (Fig. 3a), suggesting that REST protects the expression of these self-renewal genes. Western blot analysis of the same cells also indicated that REST+/− cells have substantially reduced levels of c-Myc and Oct4 proteins compared with the parental mES cells (Fig. 3b), indicating that the downregulation of REST negatively affects self-renewal signals. To further confirm the role of REST in maintaining the self-renewal signals, we performed the same experiment shown in Fig. 1j, in which exogenously added REST maintained the self-renewal of mES cells growing under differentiation conditions, and then analyzed the expression of c-Myc and Oct4 by Western blotting. As shown in Fig. 3c, the exogenously added REST, but not GFP, maintained c-Myc and Oct4 protein levels in mES cells growing under differentiation conditions. Taken together, these results indicate that REST protects the expression of several self-renewal genes.
Figure 3. REST maintains the expression of critical self-renewal regulators and represses expression of a set of miRNAs in mES cells.
a and b, Heterozygous deletion of REST results in decreased expression of self-renewal genes. a, RT-PCR analysis of total RNA from ES, RRC, and YHC cells. Genes are indicated on the left side of each panel. Gapdh was used as a loading control. b, Western blot analysis of whole-cell extracts from ES, RRC, and YHC cells. Antibodies specific for REST, c-Myc, and Oct4 proteins were used for the analysis. α-Tubulin was used as a loading control. c, Exogenously added REST, but not GFP, maintained the expression of c-Myc and Oct4 in ES cells growing without LIF. Western blot analysis of whole-cell extracts from mES cells transfected with plasmids encoding GFP or REST and then grown in the absence of LIF. Antibodies specific for REST, c-Myc, and Oct4 proteins were used for the analysis. α-Tubulin was used as a loading control. d–h, REST suppresses the expression of miRs, which are associated with the absence of self-renewal regulators. d, Chromatin immunoprecipitation assays and e, quantitative chromatin immunoprecipitation assays showed that REST binds to the gene chromatin of a set of miRs in ES cells (1=input; 2=IgG, negative control; 3=anti-H3 antibody, positive control; 4=anti-REST antibody; “+” and “−” represent predicted RE-1 binding sites occupied and unoccupied by REST, respectively). * p < 0.0001; ** p = 0.001. The values of three replicates are represented as mean +/− SD. f, Q.RT-PCR analysis of ES, YHC, RRC, EB, and EB+REST showed that expression of the miRs (shown in D and E) was lower in ES than in YHC, RRC, and EB cells. The higher expression of miRs in EB cells could be extinguished in a gain-of-function experiment in the presence of exogenously added REST. * p < 0.0001. The values are represented as mean +/− SD (n=3). g, The expression of the miRs shown in (D) was upregulated in a loss-of-function experiment when mES cells were treated with siREST but not when they are treated with non-targeting siRNA (NT). * p < 0.0001. The values are represented as mean +/− SD (n=3). Analysis was performed 3 d after transfection. h, siREST-mediated knockdown of REST that produced increased expression of the miRs in (G) corresponded to the decreased expression of REST and the known self-renewal genes Oct4, Nanog, and Sox2. Q.RT-PCR assay with mES cells treated with siREST and NT are shown. Analysis was performed 5 d after transfection. * p < 0.0001. The values are represented as mean +/− SD (n=3).
To determine how REST maintains the expression of multiple self-renewal genes, we examined the possible role of microRNAs (miRs) in REST-mediated mES cell self-renewal. We examined the miR profiles of wild-type and REST+/− mES cells growing under self-renewal conditions. The miRs from self-renewing wild-type mES cells fell into two major groups, representing expressed and repressed miRs (Supplementary Fig. S1 and Table 1). REST+/− YHC and RRC cell lines had similar miR profiles, which were strikingly opposite to those of wild-type mES cells. Also, miR-124a, which was recently shown to be a target of REST14, and miR-106a and miR-106b, whose predicted targets include REST, were up-regulated in REST+/− cells, suggesting that regulation involving double negative-feedback loops may exist to maintain homeostasis. Using miR databases (http://cbio.mskcc.org/mirnaviewer and http://pictar.bio.nyu.edu/), we found a set of miRs that can potentially target self-renewal genes such as Nanog, Sox2, Tbx3, and c-myc (Supplementary Materials, Table 2). These miRs were present only at low levels in self-renewing mES cells and were present at higher levels in both the REST+/− heterozygous cells and EB cells. To determine whether REST directly binds to the regulatory sequences of these miR-containing gene chromatin, we determined such potential REST binding sites (Supplementary Materials, Table: 3) using the MatInspector module of the Genomatix database15. We found several potential REST binding sites for each of these genes. Chromatin immunoprecipitation assays (Fig. 3d) and quantitative chromatin immunoprecipitation assays (Fig. 3e) in ES cells further revealed that REST was bound only to specific sites of the gene chromatin for each of these miRs. In a gain-of-function experiment similar to those shown in Figs. 1j and 3c, in which exogenously added REST restored the self-renewal function of mES cells growing under differentiation conditions, we analyzed the expression of the miRs in these cells by Q.RT-PCR. As shown in Fig. 3f, these miRs were expressed at lower levels in self-renewing mES cells, expressed at higher levels in both REST+/− heterozygous cells and EB cells, and expressed at lower levels in EB cells transfected with exogenous REST. To further confirm REST-mediated repression of the miRs, we performed a loss-of-function experiment, in which we treated mES cells with siREST and analyzed the expression of the miRs by Q.RT-PCR. As shown, siREST-mediated knockdown of REST resulted in increased levels of miR expression when compared with non-targeting siRNA (Fig. 3g). To confirm that the siREST-mediated knockdown of REST that increased the expression of the miRs also decreased the expression of the self-renewal genes, we performed a Q.RT-PCR assay. As shown in Fig. 3h, siREST-treated mES cells showed decreased levels of REST, Oct4, Nanog, and Sox2 expression. Thus, taken together, these results indicate that REST represses this set of miRs and strongly suggest that at least some of the miRs, in turn, interfere with the expression of critical self-renewal regulators such as Oct4, Nanog, and Sox2.
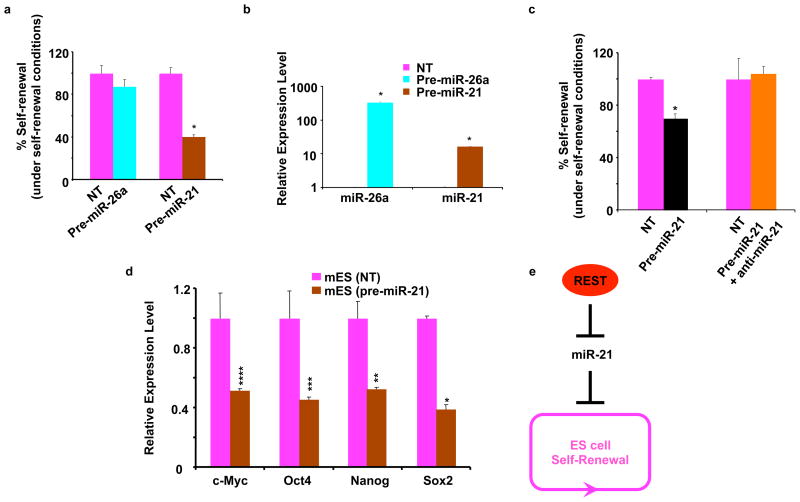
To determine whether the REST-regulated miRs are directly involved in maintaining self-renewal, we performed self-renewal assays in mES cells after transfecting individual precursors of these miRs. At least one of these precursor miRs, pre-miR-21, dramatically decreased the self-renewing capacity of mES cells by 60%, compared with that of a nontargeting control siRNA (NT). In contrast, pre-miR-26a did not effectively alter the self-renewal efficiency when compared with the same NT (Fig. 4a). To confirm that the decreased self-renewal seen in mES cells was actually due to the intracellular increase in the corresponding miRs, we measured the miR-21 and miR-26a levels by Q.RT-PCR. Indeed, the pre-miR-transfected cells showed more than 10-fold and 300-fold increases in miR-21 and miR-26a expression, respectively (Fig. 4b).
Figure 4. MiR-21 regulates self-renewal in mouse ES cells.
a, Addition of exogenous precursor-miR-21 (pre-miR-21), but not non-targeting siRNA (NT) or pre-miR-26a, caused a decrease of self-renewal in mES cells. * p < 0.0001. The error bars correspond to three replicates (n=3). b, Addition of exogenous pre-miR-21 and exogenous pre-miR-26a but not NT to mES cells resulted in increased levels of their corresponding miR expression levels. Results of Q.RT-PCR assays performed with NT-, pre-miR-26a- and pre-miR-21-transfected cells 1 d after transfection are shown. * p < 0.0001. The values are represented as mean +/− SD (n=3). c, The effect of miR-21 on loss of self-renewal was specific. Addition of exogenous pre-miR-21 along with anti-miR-21 rescued the loss of self renewal by pre-miR-21 alone. * p < 0.0001 (there was no significant difference between NT and Pre-miR-21 plus anti-miR-21 values). The error bars correspond to three replicates (n=3). d, Addition of exogenous pre-miR-21, but not NT, to mES cells resulted in loss of self-renewal markers Oct4, Nanog, Sox2, and c-Myc. Q.R-PCR after transfection of mES cells with pre-miR-21 and NT was performed 5 d after transfection to detect the expression levels of the self-renewal markers. * p < 0.0001; ** p = 0.002; *** p = 0.007; **** p = 0.008. The values are represented as mean +/− SD (n=3). e, Proposed model of REST-mediated maintenance of self-renewal through miR-21. REST maintains self-renewal and pluripotency of mES cells by suppressing miR-21 expression.
To confirm that the effect of pre-miR-21 on self-renewal was specific, we transfected mES cells with either NT or a mixture of pre-miR-21 plus either NT or anti-miR-21 and performed self-renewal assays. As shown in Fig. 4c, pre-miR-21 plus NT lowered the self-renewal capacity by 30% when compared with NT alone. This lowering of self-renewal by 50 nM pre-miR-21 was half of what we had observed with 100 nM pre-miR-21 (Fig. 4a), indicating that under these conditions, 50–100 nM pre-miR-21 was in the linear range of its activity in suppressing self-renewal in mES cells. Furthermore, the suppression of self-renewal by pre-miR-21 was rescued by anti-miR-21 but not by NT, indicating that the lowering of self-renewal was pre-miR-21-specific. We then performed Q.RT-PCR assays to determine the expression levels of the self-renewal markers Oct4, Nanog, Sox2, and c-Myc in pre-mir-21-treated cells. The results showed a corresponding decrease in the expression levels of these markers in pre-mir-21-treated cells (Fig. 4d). This is the first evidence of a single miR as a regulator of self-renewal in mES cells. Taken together, our results indicate that REST-repressed miR-21 regulates the self-renewal of mES cells. Thus, we report that REST has a newly discovered role in maintaining self-renewal and pluripotency of mES cells. Furthermore, while REST can perform this function partly by direct repression of some of its target differentiation genes, such as Calbindin, we provide evidence that REST functions in this process by repressing the transcription of a specific miR, miR-21, which suppresses self-renewal with a corresponding loss of expression of the critical self-renewing regulators Oct4, Nanog, Sox2, and c-Myc (Fig. 4e). Whether miR-21 directly suppresses the expression of these self-renewing regulators, thereby inhibiting self-renewal, or whether miR-21 inhibits self-renewal by a different mechanism, with consequent suppression of these self-renewal regulators, is unclear. It is, however, interesting to note that bioinformatics-predicted miR-21 binding sites are present in the mRNAs of Sox2 and Nanog but not those of Oct4 and c-Myc (Supplementary Materials, Table 2). If these sites truly function as miR-21 target sites and if miR-21 target sites are indeed not present in the mRNAs of Oct4 and c-Myc, then these observations indicate that the miR-21-mediated regulation of mES cell self-renewal involves a regulatory cascade initiated by the loss of Sox2 and Nanog that leads to the loss of Oct4 and c-Myc. It is noteworthy that Oct4, Nanog, and Sox2 were found to co-occupy the REST promoter/enhancer sequences apparently as an activator complex16. In addition, REST was found to be a part of the Nanog/Oct4 complex17 and was predicted to be a major component of an Oct4-Sox-Nanog network in ES cells18. Thus, taken together with our data presented here, these observations indicate that REST is a new element of the interconnected regulatory network required to maintain the self-renewal and pluripotency of cultured mES cells.
Methods summary
Embryonic stem cells (E14Tg2a) cells were cultured in presence of LIF on gelatin coated dishes. Embryoid bodies were grown on bacterial dishes in absence of LIF and β-mercaptoethanol. Alkaline phosphatase staining after loss- and gain- of function was carried out using a detection kit from Chemicon. ChIP assays were performed as described in the methods in full. For the miRNA microarrays, total RNA was prepared using TRIzol, and RNA samples were analyzed by LC Sciences on their microarray platform with a probe set based on Sanger version 9.0. Blastocysts were obtained from C57BL/6 NCr mice, fixed in 4% paraformaldehyde, permeabilized with 0.25% Triton X- 100, and were then double immunostained with antibodies against REST, Sox2, Nanog and Oct4. Primers used for RTPCR and ChIP are available in the methods in full (see Supplementary Methods).
Online Methods
ESC Lines and Culture Conditions
mESCs (E14Tg2A) obtained from Bay Genomics, were cultured under self-renewal conditions (in the presence of leukemia inhibitory factor, LIF, cultured on adherent tissue culture dishes) without feeder cells in 1000 units ml−1 LIF (ESGRO) on gelatin-coated tissue culture dishes19. Embryoid bodies in absence of LIF and b-mercaptoethanol on non-adherent tissue culture dishes were grown for 3 days in bacterial dishes as described1. The REST+/− cell lines YHC334 (YHC) and RRC160 (RRC) that have gene traps at REST exons 1 and 2, respectively were obtained from BayGenomics. Both of these lines were derived from parental E14Tg2A mES cells, the same cells we used for all of our wild-type mES cell experiments.
ChIP Assays
ChIP assays of cultured mESCs and EBs were performed as described 20. We used antibodies against mouse IgG (2 μl; cat. no. I 5381; Sigma), REST (2μl; cat. no. 07-579, Upstate) and histone H3 (2 μl; cat. no. ab1791; Abcam). One microgram of each antibody and approximately 15 μg of chromatin were added to each reaction mixture and incubated overnight at 4 °C. The mixtures were then incubated with 33 μl of protein A-agarose beads (Amersham Pharmacia Biotech AB; pre-equilibrated with 1 mg ml−1 bovine serum albumin and 0.3 mg ml−1 salm on sperm DNA).
The PCR primer sequences used are as follows:
miR-93/miR-106b Forward: cccccaaaaccagtatcctt
miR-93/miR-106b Reverse: tcgtacttcccggatcactc
miR-27a Forward: aaatccccagaagctggagt
miR-27a Reverse: agcacttgaaaggcaaagga
miR-21 Forward: agggcaggaagatgacacac
miR-21 Reverse: gggctgatgagcactaagga
miR-26a Forward: gcctaacccaagaagggaaa
miR-26a Reverse: tccctctcatctggacaacc
miR-152 Forward: tggtctctgtccagcacaac
miR-152 Reverse: caggtcacagctgcactcat
miR-124a Forward: ctctgcgtgttcacagcgg
miR-124a Reverse: ctcttggcattcaccgcgtg
Quantitative ChIP PCR was performed using the same extracts used above. Briefly, QPCR was performed and analyzed using real-time PCR (ABI prism 7500). Primers were designed by Primer Express 2.0 (Applied Biosystems) to amplify 60- to 150-bp amplicons. Amplicon was measured by SYBR Green fluorescence (SYBR Green Master mix, Applied Biosystems) in 10-μl reactions. Reactions were performed in triplicates. The amount of product was determined relative to a standard curve of input chromatin. Melting curves showed that PCRs yielded single product. Ct values for each sample were normalized against that of input DNA and percentage recovery is plotted. The primers used for QPCR will be available upon request.
Western Blotting, RT-PCR and miRNA Microarray
Whole-cell extracts were prepared, and approximately 10 μg of proteins were resolved on SDS-PAGE using antibodies against REST (#07-579, Upstate), c-myc (#sc-764, Santa Cruz Biotechnology), Oct4 (#ab19857, Abcam), Sox2 (#ab15830, Abcam), Actin (#sc-1616, Santa Cruz), and α-Tubulin (#MMS-407R, Covance) per the manufacturers’ recommendations. Total RNA was extracted using TRIzol (Invitrogen), and approximately 50 ng of total RNA was used as a template. RT-PCR was performed using a OneStep RT-PCR kit (Qiagen).
The following gene-specific primers were designed: REST forward, 5′-AGCGAGTACCACTGGAGGAA-3′; REST reverse, 5′-CTGAATGAGTCCGCATGTGT-3′; Calbindin forward, 5′-GCTTCTATCTGGCGGAAGG-3′; Calbindin reverse, 5′-TGTCATCTGGCTACCTTCCC-3′; Pitx2 forward, 5′-CGGCAGAGGACTCATTTCAC-3′; Pitx2 reverse, 5′-GTACGAATAGCCGGGGTACA-3′; Snai1 forward, 5′-CTTGTGTCTGCACGACCTGT-3′; Snai1 reverse, 5′-CTTCACATCCGAGTGGGTTT-3′, Sox18 forward, 5′-AACAAAATCCGGATCTGCAC-3′; Sox18 reverse, 5′-CGAGGCCGGTACTTGTAGTT-3′; Cebpa forward, 5′-CCGACTTCTACGAGGTGGAG-3′; Cebpa reverse, 5′-TGGCCTTCTCCTGCTGTC-3′. The following primers were based on published sequences: Gapdh, Oct4, Nanog, Gata6, Fgf5, brachyury, Flk1, and Pl121; Mash1, Mash2, β-globin, CD34, Cxcl12, Ehox, Eomes, Gata4, Isl1, Meox1, Mixl1, Ngn2, Shh, and Tbx322.
For quantitative RT PCR, first strand synthesis was performed using Applied Biosystems’ high capacity cDNA reverse transcription kit (cat. No. 4368814). Real time PCR was carried out using cDNAs with SYBR Green PCR master mix from Applied biosystems (cat. No. 4309155). Reactions were carried out in triplicates using ABI prism 7500 and the analysis was performed similar to that described for quantitative ChIP PCR above except that Delta;ct was obtained after normalization with ct of Gapdh for mouse specific primers and that of Actin for human specific primers. Mouse specific primers will be provided upon request and human specific primers are as published23.
For the miRNA microarrays, total RNA was prepared using TRIzol, and RNA samples were analyzed by LC Sciences on their microarray with a probe set based on Sanger version 9.0. Multiple sample analysis involves normalization, data adjustment, t-Test/ANOVA analysis, and clustering. Normalization is carried out using a cyclic LOWESS(Locally-weighted Regression) method. The normalization is to remove system related variations, such as sample amount variations, different labeling dyes, and signal gain differences of scanners so that biological variations can be faithfully revealed. Data adjustment includes data filtering, Log2 transformation, and gene centering and normalization. The data filtering removes genes (or miRNAs) with (normalized) intensity values below a threshold value of 32 across all samples. The Log2 transformation converts intensity values into Log2 scale. Gene centering and normalization transform the Log2 values using the mean and the standard deviation of individual genes across all samples using the following formula:
In certain cases, expression ratios of dual-sample arrays are used in clustering analysis. Gene normalization would then be performed using the following formula:
t-Test is performed between “control” and “test” sample groups with each group contains at least two samples. T-values are calculated for each miRNA, and p-values are computed from the theoretical t-distribution. miRNAs with p-values below a critical p-value (typically 0.01) are selected for cluster analysis. The clustering is done using hierarchical method and is performed with average linkage and Euclidean distance metric.
ANOVA (Analysis of Variance) is an extension of the t-test to more than two experimental conditions. It picks out miRNAs that have significant differences in means across three or more groups of samples, with each group containing at least two samples. In this analysis, p-values are computed from the F-distribution. In certain cases, such as time point experiments, in which only one sample is collected and assayed for each condition or time point, repeating sets of reporters (or probes) on each array may be used as a “group” in an ANOVA analysis.
All data processes, except clustering plot, are carried out using in-house developed computer programs. The clustering plot is generated using TIGR MeV (Multiple Experimental Viewer) software from The Institute for Genomic Research.
miRNA validation was carried out using Ambion’s mirVana™ qRT-PCR miRNA detection kit (Cat. # AM1558). mirVana qRT-PCR primer sets for the miRNA of interest were also obtained from Ambion (Cat. #30000–30999). Reactions were performed as per manufacturer’s recommendations. The values obtained were normalized with primer set corresponding to 5S rRNA (cat. # AM 30302).
RNAi-Mediated Knockdown, exogenous expression of miR-21, anti-miR-21 and Gain-of-Function Experiments
RNAi-mediated knockdown of REST and Oct4 were carried out per the manufacturer’s protocol (Dharmacon). siREST (60pmol), siOct4 (60 pmol), and nontargeting siRNA (60 pmol) were transfected using Amaxa nucleofection method. Colonies were assayed for alkaline phosphatase activity 3d after transfection.
Amaxa nucleofection was also used to deliver precursor miR-21 and anti-miR-21 molecules into mES cells. Transfection was carried out using 100 pmoles of precursor or 50 pmoles of miR-21 and anti-miR-21 each using 4×106 cells. Non-targeting siRNA was used as control. Delivery and mature miR-21 product was inspected in cells by quantitative RTPCR 1d after transfection as described above.
For the gain-of-function experiments, mESCs were transfected with 2 μg of plasmid DNA using Lipofectamine 2000. The Flag-REST construct was a generous gift from G. Mandel. The transfected cells were selected with G418 (125 μg ml−1) under self- renewing conditions in the presence of LIF for 3 d. After selection, we grew the cells under differentiation conditions in the absence of LIF for 4 days, and subjected them to self-renewal assays. An alkaline phosphatase assay was then performed using an alkaline phosphatase detection kit (Chemicon) after 4d. Percentages of self-renewing colonies from three independent experiments were plotted. Standard deviation was calculated and is depicted as error bars in the figures.
Immunofluorescence Microscopy of Blastocysts
Blastocysts from C57BL/6 NCr mice were fixed in 4% paraformaldehyde, permeabilized with 0.25% Triton X-100, and blocked with 1% goat serum in PBS24. The blastocysts were then double immunostained with mouse anti-REST antibody (raised against N-terminal portion of human REST, A372) in combination with antibodies against Sox2 (Abcam, cat # AB15830), Oct4 (Abcam, cat. # AB19857), Nanog (Abcam, Cat # AB21603) followed by Alexa Fluor 488-conjugated secondary antibody detecting mouse IgG and Fluor 555-conjugated secondary antibody detecting rabbit IgG (Molecular Probes, Carlsbad, CA). Images were captured with a Nikon microscope (Nikon Eclipse TE 2000U).
Supplementary Material
Acknowledgments
We would like to thank the National Institutes of Health for funding (grants CA97124 and CA81255 to S.M.). MNK and SKS were both recipients of the Dodie Hawn Fellowship in Cancer Genetics.
Footnotes
Author Contributions
SKS and MNK: all aspects including manuscript preparation; JP-T: blastocyst experiments; HA: immunofluorescence studies; SM: project planning and manuscript preparation.
References
- 1.Ballas N, Grunseich C, Lu DD, Speh JC, Mandel G. REST and its corepressors mediate plasticity of neuronal gene chromatin throughout neurogenesis. Cell. 2005;121:645–57. doi: 10.1016/j.cell.2005.03.013. [DOI] [PubMed] [Google Scholar]
- 2.Ballas N, Mandel G. The many faces of REST oversee epigenetic programming of neuronal genes. Curr Opin Neurobiol. 2005;15:500–6. doi: 10.1016/j.conb.2005.08.015. [DOI] [PubMed] [Google Scholar]
- 3.Ooi L, Wood IC. Chromatin crosstalk in development and disease: lessons from REST. Nat Rev Genet. 2007;8:544–54. doi: 10.1038/nrg2100. [DOI] [PubMed] [Google Scholar]
- 4.Coulson JM. Transcriptional regulation: cancer, neurons and the REST. Curr Biol. 2005;15:R665–8. doi: 10.1016/j.cub.2005.08.032. [DOI] [PubMed] [Google Scholar]
- 5.Majumder S. REST in good times and bad: roles in tumor suppressor and oncogenic activities. Cell Cycle. 2006;5:1929–35. doi: 10.4161/cc.5.17.2982. [DOI] [PubMed] [Google Scholar]
- 6.Su X, Kameoka S, Lentz S, Majumder S. Activation of REST/NRSF target genes in neural stem cells is sufficient to cause neuronal differentiation. Mol Cell Biol. 2004;24:8018–25. doi: 10.1128/MCB.24.18.8018-8025.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Watanabe Y, et al. Conversion of myoblasts to physiologically active neuronal phenotype. Genes Dev. 2004;18:889–900. doi: 10.1101/gad.1179004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lawinger P, et al. The neuronal repressor REST/NRSF is an essential regulator in medulloblastoma cells. Nat Med. 2000;6:826–31. doi: 10.1038/77565. [DOI] [PubMed] [Google Scholar]
- 9.Fuller GN, et al. Many human medulloblastoma tumors overexpress repressor element-1 silencing transcription (REST)/neuron-restrictive silencer factor, which can be functionally countered by REST-VP16. Mol Cancer Ther. 2005;4:343–9. doi: 10.1158/1535-7163.MCT-04-0228. [DOI] [PubMed] [Google Scholar]
- 10.Su X, et al. Abnormal expression of REST/NRSF and Myc in neural stem/progenitor cells causes cerebellar tumors by blocking neuronal differentiation. Mol Cell Biol. 2006;26:1666–78. doi: 10.1128/MCB.26.5.1666-1678.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pan G, Thomson JA. Nanog and transcriptional networks in embryonic stem cell pluripotency. Cell Res. 2007;17:42–9. doi: 10.1038/sj.cr.7310125. [DOI] [PubMed] [Google Scholar]
- 12.Pritsker M, Ford NR, Jenq HT, Lemischka IR. Genomewide gain-of-function genetic screen identifies functionally active genes in mouse embryonic stem cells. Proc Natl Acad Sci U S A. 2006;103:6946–51. doi: 10.1073/pnas.0509861103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Loh YH, et al. The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat Genet. 2006;38:431–40. doi: 10.1038/ng1760. [DOI] [PubMed] [Google Scholar]
- 14.Conaco C, Otto S, Han JJ, Mandel G. Reciprocal actions of REST and a microRNA promote neuronal identity. Proc Natl Acad Sci U S A. 2006;103:2422–7. doi: 10.1073/pnas.0511041103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cartharius K, et al. MatInspector and beyond: promoter analysis based on transcription factor binding sites. Bioinformatics. 2005;21:2933–42. doi: 10.1093/bioinformatics/bti473. [DOI] [PubMed] [Google Scholar]
- 16.Boyer LA, et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–56. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang J, et al. A protein interaction network for pluripotency of embryonic stem cells. Nature. 2006;444:364–368. doi: 10.1038/nature05284. [DOI] [PubMed] [Google Scholar]
- 18.Zhou Q, Chipperfield H, Melton DA, Wong WH. A gene regulatory network in mouse embryonic stem cells. Proc Natl Acad Sci U S A. 2007;104:16438–43. doi: 10.1073/pnas.0701014104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nichols J, Evans EP, Smith AG. Establishment of germ-line-competent embryonic stem (ES) cells using differentiation inhibiting activity. Development. 1990;110:1341–8. doi: 10.1242/dev.110.4.1341. [DOI] [PubMed] [Google Scholar]
- 20.Cui R, et al. Family members p53 and p73 act together in chromatin modification and direct repression of alpha-fetoprotein transcription. J Biol Chem. 2005;280:39152–60. doi: 10.1074/jbc.M504655200. [DOI] [PubMed] [Google Scholar]
- 21.Kaji K, et al. The NuRD component Mbd3 is required for pluripotency of embryonic stem cells. Nat Cell Biol. 2006;8:285–92. doi: 10.1038/ncb1372. [DOI] [PubMed] [Google Scholar]
- 22.Pritsker M, Ford NR, Jenq HT, Lemischka IR. Genomewide gain-of-function genetic screen identifies functionally active genes in mouse embryonic stem cells. Proc Natl Acad Sci U S A. 2006;103:6946–51. doi: 10.1073/pnas.0509861103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cai J, et al. Assessing self-renewal and differentiation in human embryonic stem cell lines. Stem Cells. 2006;24:516–30. doi: 10.1634/stemcells.2005-0143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang J, et al. Sall4 modulates embryonic stem cell pluripotency and early embryonic development by the transcriptional regulation of Pou5f1. Nat Cell Biol. 2006 doi: 10.1038/ncb1481. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.