Abstract
This study addresses the extent and consequences of gene exchange between populations of Darwin's finches. Four species of ground finches (Geospiza) inhabit the small island of Daphne Major in the centre of the Galápagos archipelago. We undertook a study of microsatellite DNA variation at 16 loci in order to quantify gene flow within species owing to immigration and between species owing to hybridization. A combination of pedigrees of observed breeders and assignments of individuals to populations by the program Structure enabled us to determine the frequency of gene exchange and the island of origin of immigrants in some cases. The relatively large populations of Geospiza fortis and G. scandens receive conspecific immigrants at a rate of less than one per generation. They exchange genes more frequently by rare but repeated hybridization. Effects of heterospecific gene flow from hybridization are not counteracted by lower fitness of the offspring. As a result, the standing genetic variation of the two main resident populations on Daphne Major is enhanced to a greater extent by introgressive hybridization than through breeding with conspecific immigrants. Immigrant G. fuliginosa also breeds with G. fortis. Conspecific immigration was highest in the fourth species, G. magnirostris. This species is much larger than the other three and perhaps for this reason it has not bred with any of them. The source island of most immigrants is probably the neighbouring island of Santa Cruz. Evolutionary change may be inhibited in G. magnirostris by continuing gene flow, but enhanced in G. fortis and G. scandens by introgressive hybridization.
Keywords: immigration, introgression, microsatellites, pedigrees, selection
1. Introduction
A well-known result of population genetics theory is that one breeding immigrant per generation (Nm) is sufficient to counteract the loss of genetic variation owing to drift (Crow & Kimura 1970; Miles & Allendorf 1996). Recent interest has focused on the question of how immigration affects the genetic structure of populations and, in particular, the outcome of selection and local adaptation in the face of immigration, referred to as gene flow (Slatkin 1985, 1987a). In the absence of selection, the effect of migration on the frequency of an allele at a locus is proportional to the difference in frequency between donor (d) and recipient (r) populations. The effect of selection on the frequency of an allele (a) at the locus is proportional to its relative fitness, that is the difference in fitness (W) between the donated allele and other alleles at that locus. Thus, the total effect of immigration (Hedrick 2000) is a change in allele frequency (Δp) as a function of the migration rate (m), the difference in allele frequency (pd−pr), and the relative fitness of the introduced allele 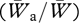
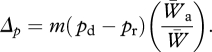 |
Immigrants (Hendry 2004; Bolnick & Nosil 2007; Harper & Pfennig 2008) or their descendants (Verhulst & van Eck 1996; Marr 2006) might be at a selective disadvantage, but if common enough they could counteract the effects of selection and local adaptation (García-Ramos & Kirkpatrick 1997; Kirkpatrick & Barton 1997; Hedrick 2000; Lenormand 2002; Hendry & Taylor 2004; Moore et al. 2007). Alternatively, they could introduce advantageous genes and thereby enhance local adaptation (Ebert et al. 2002; Saccheri & Brakefield 2002). A steady state may be reached at a point of balance between tendencies to diverge as a result of selection and drift (fission) and converge (fusion) as a result of immigration (Slatkin 1985, 1987b; Hendry et al. 2002; Cheviron & Brumfield 2009; reviewed in Bolnick et al. 2008).
This framework for understanding the dynamics of populations open to immigration should be extended to include interspecific hybridization and introgression. There is a rapidly increasing body of evidence that introgressive hybridization is widespread in a variety of animal taxa (Mallet 2005; Arnold 2008; Schwenk et al. 2008) and plant taxa (Arnold 1997, 2006; Rieseberg 1997), not to mention prokaryotes and their tendencies to exchange genes by horizontal transfer. Introgression is particularly prevalent in the early stages of adaptive radiations (Grant & Grant 2008a), as exemplified by cichlid fish of the African Great Lakes (Seehausen 2004, 2006) and the silversword alliance of plants (Compositae) in Hawaii (Barrier et al. 1999). Introgressive hybridization can have positive or negative effects upon the recipient population. It can inhibit divergence, but, on the other hand, it has the potential to increase standing genetic variation and to introduce new, selectively advantageous, alleles to a greater extent than is possible with conspecific gene flow. As such it may be especially important in evolution by creating selectively advantageous combinations of genes (Lewontin & Birch 1966; Svärdson 1970; Grant & Grant 1989, 2008b).
Attempts have rarely been made to quantify the effects of both introgression and immigration in the same study. Three components could affect the outcome in contrasting ways. First, conspecific gene flow might occur more often than heterospecific gene flow, even when conspecific and heterospecific encounter rates are equal, because responses to conspecific mating signals are likely to be stronger than responses to heterospecific ones: mc > mh, where subscripts c and h refer to conspecific and heterospecific, respectively. Second, on the other hand, the difference in allele frequencies between donors and recipients should be greater when the populations are heterospecific than when they are conspecific: (pcd−pcr) < (phd−phr). Third, genetic effects of heterospecific alleles may be beneficial with weakly differentiated species, yet disadvantageous with more strongly differentiated species. At equilibrium:
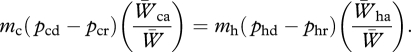 |
Departures from equilibrium, and the relative contribution of the three components on each side of the equation, have rarely been investigated. A few studies have found greater conspecific than heterospecific effects on allele frequencies (Alexandrino et al. 2006; Lorenzen et al. 2006; Harper et al. 2007), but they have relied on indirect estimates of gene flow from genetic data, and these are well known to be imprecise (Wilson & Rannala 2003; Faubert et al. 2007; Peery et al. 2008). In the study reported here, we use more direct methods involving genetic assignments of individuals to populations in order to estimate the two forms of gene flow. Estimating gene flow by direct observations of breeders is difficult but can be done with small populations of closely related species living in archipelagos (Keller et al. 2001b) or archipelago-like situations, such as clusters of ponds (Ebert et al. 2002) or fragmented patches of terrestrial habitat (Hanski et al. 1994).
This paper describes a study of Darwin's finch populations on Daphne Major Island designed in part to elucidate the role of introgression in young adaptive radiations. There are several advantages of studying this system. First, the island is only moderately isolated, by 8 km from the nearest large islands of Santa Cruz, Baltra and Seymour (Grant 1999), and immigration is known to occur (Grant et al. 2001). Second, finches are observable, parentage can be determined by genotyping and fitness can be quantified in small populations because the survival and reproductive fates of offspring can be documented. Finally, introgression between sympatric species occurs in the entirely natural environment of Daphne Major (Grant 1993) and neither is restricted to a hybrid zone as in many other taxa (Barton & Hewitt 1985; Harrison 1993; Barton 2001), nor is it restricted to sister species. This paper provides a new assessment of introgression on Daphne from genetic analysis of an expanded dataset, in conjunction with the first direct estimate of conspecific gene flow that results from immigration and breeding in the same species that hybridize. We provide additional information on relative fitness and genetic differences at microsatellite loci to assess the relative magnitude of conspecific and heterospecific gene exchange.
2. Finches and hybridization
Daphne Major is a small island, approximately 34 ha in area, near the centre of the Galápagos archipelago. Four species of ground finches breed on the island. Geospiza fortis (approx. 17 g), the medium ground finch, is a granivorous bird with a short and blunt beak; G. scandens (approx. 21 g), the cactus finch, which feeds on Opuntia cactus seeds, pollen and nectar in the dry season, has a long pointed beak; G. magnirostris (approx. 30 g), the large ground finch, feeds on large and hard seeds; and G. fuliginosa (approx. 12 g), the small ground finch, feeds on small seeds. Depending on environmental conditions, the population of G. fortis ranges from well over 1500 to less than 100 individuals, whereas the G. scandens population ranges from approximately 600 to less than 60 individuals. Geospiza magnirostris established a breeding population on Daphne in 1982–1983 and its numbers gradually increased to a maximum of approximately 350 in 2003, then fell during a severe drought (Grant & Grant 2006) and increased afterwards. Geospiza fuliginosa is a frequent immigrant that occasionally breeds on the island in numbers of less than 10.
Geospiza fortis occasionally breeds with G. scandens and G. fuliginosa (Grant 1993). The latter two species have not been observed to breed with each other, and none of the species has bred with G. magnirostris on Daphne. Geospiza fortis hybridizes at a low frequency in each year of full breeding (1–3% of pairs are mixed), more frequently with G. fuliginosa than with G. scandens. Interbreeding results in introgression because hybrids are viable and fertile (Grant & Grant 1992a; Grant et al. 2004). The direction of introgression is determined by the mating pattern of the F1 offspring: offspring choose mates on the basis of paternal song, with very few exceptions (Grant & Grant 2008a). The relative fitness of hybrids depends upon the availability of an appropriate food (seed) supply (Grant & Grant 1993). Seed composition varies according to environmental conditions that fluctuate from droughts to extremely wet conditions associated with periodic El Niño events (Gibbs & Grant 1987; Grant & Grant 2002).
A pair of fourth-generation offspring of an immigrant G. fortis bred on Daphne in 2005, and their offspring bred with each other (Grant & Grant 2008c, 2009). This endogamous group has not been included in the analyses reported here because they did not breed with residents.
3. Material and Methods
Beginning in 1973, we captured finches in mist nets, measured them and gave them a unique combination of coloured leg bands and a numbered metal band before releasing them. Six body size and beak traits were measured, described in Boag & Grant (1984) and illustrated in Grant & Grant (2008a). From 1988 onwards, we took a small drop of blood from the brachial vein, transferred it to EDTA-soaked filter paper and stored it in Drierite for later analysis of allelic variation at 16 microsatellite loci (Petren et al. 1999). This method bypassed the need for a buffer without sacrifice of DNA quality. For pedigree analysis when genotypes were not available, we identified social parents at nests. An attempt was made to find most nests in 1976 and 1992–1997, and all nests on the island in the years 1978–1991 and 1998. Nestlings (n = 7496) were banded at day 8; 802 of them were hybrids. From 1990 onwards, a drop of blood was taken from them at this time. Many were captured in nets as adults and measured. Genotyping was performed by Petren and colleagues (Grant et al. 2004), and by Ecogenics GmbH, Switzerland, and results were standardized (Grant & Grant 2008c). For an assessment of parentage, we allowed 2bp differences at two loci to be within the range of scoring variation and declared a mismatch when more differences or a single difference of at least 4bp were found (Keller et al. 2001a). Almost all offspring matched both parents at all loci. Extra-pair paternity was found in approximately 10 per cent of the 1794 offspring checked. The biological father was identified in 60–80% of the cases, depending on the year, where the social father was excluded as the biological father. To examine how well the hybrids and conspecific immigrants survived in comparison with the pure species hatched at the same time and living under the same conditions, we followed the survival of banded nestlings of the seven largest cohorts produced in the years of average or abundant rainfall (1978, 1981, 1983, 1984, 1987, 1991 and 1998).
Birds that lacked leg bands when captured in nets, and therefore not known to have hatched on the island, could be immigrants or residents. Prior to 1988, we used beak measurements to determine their identity from reference samples of non-overlapping distributions of measurements of the species at Borrero Bay, Santa Cruz Island (Grant 1993). From 1988 onwards, we used genotypic information from blood samples to identify hybrids and backcrosses, and island of origin of suspected immigrants, with v. 2.2 (Pritchard et al. 2007) of the program Structure (Pritchard et al. 2000; Falush et al. 2003). An attempt to use the alternative program NewHybrids (Anderson & Thompson 2002; Anderson 2008) was abandoned because it failed to identify known F1 hybrids from the pedigrees.
Structure employs a Bayesian analysis to assign individuals to specified groups with a probability estimated from frequencies of microsatellite alleles. We applied the majority rule (p > 0.500) to assign individuals to groups. The immigration problem involves each species, whereas hybridization involves G. fuliginosa, G. fortis and G. scandens but not G. magnirostris. Following the authors' recommendations, we used a burn-in of 50 000 iterations and a run length of 100 000. For each new analysis, we repeated the procedure once to make sure results were consistent. We used the Popinfo option to select a no-admixture model and chose the correlated allele option. For the immigration problem, the birds to be identified that lacked bands were given a value of zero in the Popflag column, and all individuals from the candidate source islands were given a value of unity. This allowed a repeated updating of allele frequencies of all groups except the target Daphne group. The number of previous generations was set at zero.
For the hybridization problem, an ancestry model with prior generations is appropriate. We set the number of previous generations at two. In this analysis, a given individual may be genetically identified with an estimated probability of belonging to another species (generation 0), having a parent (generation 1) or having a grandparent (generation 2) from another species. These last two are equivalent to F1 and first-generation backcross (B1) classes in most circumstances. Analyses with two prior generations performed better than those with either one or three prior generations. Those with one prior generation yielded fewer identified hybrids, and those with three yielded no more hybrids than did the two prior generation analyses, and typically at lower probabilities. Admixture and non-admixture models generally give similar results (Pritchard et al. 2007). We found the same, but when tested against pedigree information admixture models performed somewhat less well than no-admixture models; results were sometimes unrealistic and are not reported here.
We split the birds into an early (before 1998) and a late groups (1999–2008) for two reasons: (i) pedigree information was available up to 1998 but not afterwards and (ii) allele frequencies of the species changed as a result of introgressive hybridization. Hybrids in the early part of the study were detected in relation to contemporary allele frequencies of the species better than in relation to the total sample for the species.
To provide a methodological check on the ability of Structure to assign hybrids and backcrosses correctly, we constructed 25 artificial interspecific pairs from contemporaneous G. fortis and G. scandens individuals with assignment probabilities greater than 0.99. The male was a G. fortis individual in 13 pairs and a G. scandens in 12 pairs. To generate artificial offspring (n = 50), two per pair, we randomly drew alleles from the parents. The F1 hybrids were then backcrossed to each parental species, 15 families per species, to generate the B1 generation (n = 60) by the same procedure. F1s and B1s were assigned to one species or the other based on the father because paternal song determines mate choice (Grant & Grant 1997a,b). We ran the no-admixture model with the second-generation back option. Forty-eight of the 50 F1 assignments (0.96) were significantly different from the parental species, but only 24 (0.48) were correctly assigned to the F1 category. For the backcrosses, 46/60 B1 assignments (0.77) were significantly different from the parental species, but only 41 (0.68) were correctly assigned to the B1 category. The overall success rate is higher for identifying hybrids (0.85) than for the particular class of hybrids (0.68). Therefore, in reporting the results, we attach greater confidence to the identification of hybrids than to the particular class of hybrids. Oliveira et al. (2008) had a higher rate of success in a similar simulation with fewer microsatellite loci (12) but a larger sample of pairs of parents (40) and offspring (100).
Species are identified by song and morphology (Grant 1993, 1999). Gene exchange between populations is defined as the breeding of a member of one population with a member of the other. The F1 offspring in the pedigree were assigned to species according to the song sung by their father (Grant & Grant 2008a); paternal song indicates the direction of gene flow through backcrossing.
4. Intraspecific gene flow
(a). G. fortis
Sixty-seven genotyped birds lacking bands when captured in the years 1981–1998 were possible immigrants. They were identified as G. fortis by their measurements. Structure was run in order to assign them to the following defined populations: Santa Cruz (n = 39 genotyped individuals), Santiago (n = 9), Rábida (n = 3), Marchena (n = 17), San Cristóbal (n = 4), Pinta (n = 12), Isabela (n = 11) and Daphne (n = 969). The sample of 67 birds to be assigned was not defined. The defined Daphne population comprised all contemporaneous G. fortis known to have hatched on the island during the same period, and no hybrids. The small samples from most islands were not well characterized genetically, so the program was rerun with only the sample from the neighbouring Santa Cruz Island as a potential source. All birds lacking bands when captured on Daphne were assigned to the Daphne population except for four. These were assigned to Santa Cruz with high probabilities (p > 0.900), and two of them bred with resident G. fortis. Given an average generation length of 4.5 years for G. fortis on Daphne (Grant & Grant 1992b), two conspecific immigrants (Nc) in 18 years represent 0.50 per generation. They produced 15 and 10 fledglings (corrected for extra-pair young) and contributed five and one recruit to the next generation, respectively. One additional male that bred may have been an immigrant. It had unusually large measurements but was not genotyped. If it is included the number of breeding immigrants becomes three or 0.75 per generation.
(b). G. scandens
Sixty-four birds identified by measurements as G. scandens and lacking bands when captured in the years 1976–1998 were possible immigrants. Structure was run in order to assign them to the following defined populations: Santa Cruz (n = 23), Santiago (n = 4), Rábida (n = 12), Marchena and Pinta combined (n = 6), San Cristóbal (n = 6) and Daphne (n = 403). The defined Daphne population comprised all contemporaneous G. scandens known to have hatched on the island. All 64 candidate immigrants were assigned to the Daphne population except for three: one male was assigned to Santa Cruz (p = 0.734) and the other two were not assigned to any one population by the majority rule. Results remained unchanged when islands with the lowest assignment probabilities to any island were serially deleted until only Santa Cruz remained. The mean generation length of G. scandens is 5.5 years (Grant & Grant 1992b). One immigrant (Nc) in 24 years represents 0.23 per generation. It bred with a resident G. scandens female (F1 hybrid) in 1997 and produced at least three fledglings (all confirmed within-pair young), one of which bred successfully the following year.
(c). G. fuliginosa
At least two, and a maximum of 16, pairs of G. fuliginosa bred on the island, but only in the years 1976–1984. Most individuals lacked leg bands when captured, and therefore their identity and origin (Daphne resident or immigrant) could not be confirmed. Immigration is strongly suspected. Since identification poses a special problem, they are considered together with hybrids in §5a(iii). Assuming the two breeding pairs were immigrants, and assuming a generation length of 4.5 years like G. fortis, the number of immigrants is 0.78 when calculated over 23 years (1976–1998) or 2.0 over the period 1976–1984.
(d). G. magnirostris
The following populations were included in an analysis of 117 birds captured on Daphne without bands in the years 1988–1998 and treated as an undefined population: Santa Cruz (n = 12), Santiago (n = 10), Genovesa (n = 32), Rábida (n = 5), Marchena (n = 10), Pinta (n = 7), Fernandina (n = 9) and Isabela (n = 6). We serially deleted potential source populations with the lowest set of probability values without changing the results. Three islands were identified as sources of birds on Daphne: Santa Cruz (85), Santiago (7) and Pinta (3). None of the remaining 22 birds were assigned to an island by the majority rule. Twenty-two of the 117 bred on Daphne. Eighteen of them were identified by their assignments as coming from Santa Cruz, one was from Santiago and three were unassigned. These results differ from a previous analysis (Grant et al. 2001) in which Rábida and Marchena were identified as the most frequent sources of immigrants. The previous analysis lacked samples from Santiago and Pinta, however.
The analysis was repeated with 159 birds captured after 1998, the same non-Daphne populations as before, and a user-defined Daphne population comprising 11 residents that bred on Daphne in 1998 and their offspring (n = 100). Assignments of these were as follows: 117 to Daphne, 20 to Santa Cruz, three to Santiago and three to Pinta. Sixteen were unassigned.
Up to 1998, 22/117 = 0.118 bred. If the unknown generation length of G. magnirostris is the same as that of G. fortis, namely 4.5 years, the number of immigrants is 9.0 per generation. In the 10 years after 1998, a minimal estimate of the proportion of immigrants is 26/159 = 0.163 in 10 years. At least two immigrants bred, but the total number of breeders is not known. If all of them bred, which is highly unlikely, the number of immigrants is 7.3 per generation. If only two bred, the number of immigrants is 0.9 per generation. In both cases, it is lower than the earlier rate.
5. Interspecific gene flow
(a). G. fortis: interbreeding with G. fuliginosa immigrants
(i). Identification of G. fuliginosa and hybrids
Geospiza fortis and G. fuliginosa hybridize on Daphne (Grant & Price 1981; Boag & Grant 1984). Estimating the frequency is difficult because some G. fuliginosa individuals are morphologically indistinguishable from hybrids produced by G. fuliginosa × G. fortis pairs. In an earlier study (Grant 1993), we used measurements of G. fuliginosa from Borrero Bay on the north shore of the neighbouring island of Santa Cruz to define limits to the species. This is possible to do because at that locality there is a gap of 1.0 mm between the largest beak depth of G. fuliginosa (small ground finch: n = 109) and the smallest G. fortis (medium ground finch: n = 137), and a gap of 0.6 mm in beak width between the species. There are no such gaps within the frequency distributions of either species.
A total of 302 finches lacking leg bands when captured in mist nets on Daphne were classified as G. fuliginosa by their beak measurements. For the small sample genotyped (n = 39), we ran Structure twice, separately for the early and for the late samples. Two populations were specified: G. fuliginosa (Santa Cruz only) and G. fortis (G. fortis and G. fuliginosa on Daphne combined). The second-generation back option was chosen to allow identification of hybrid categories.
Nineteen of the 29 birds we originally identified as G. fuliginosa in the early sample on Daphne were assigned to G. fuliginosa (Santa Cruz) by applying the majority rule. Of the remaining 10, two were assigned to G. fortis, one to the F1 category, three to the first-generation backcross category, and four were not assigned to any category by the rule. Of the late sample of seven G. fuliginosa, three were assigned to G. fuliginosa (all p = 1), two were assigned to backcrosses (p = 0.620, 0.821), and two were assigned to G. fortis (p = 0.971, 0.988). Altogether 22/36 were confirmed genetically as G. fuliginosa. These results were not altered when the sample of G. fuliginosa from Santiago, the next closest island to Daphne, was substituted for the Santa Cruz sample in the analysis.
For the remaining birds on Daphne without genotypic or pedigree information we used morphological measurements to identify G. fuliginosa as follows. The smallest Daphne hybrid has beak depth and width measurements of 6.9 mm. It was assigned to the first-generation backcross class. All birds with smaller beak dimensions are considered to be unambiguously G. fuliginosa. The largest G. fuliginosa at Borrero Bay had a beak depth of 7.8 mm, and the largest beak width in the sample was 7.4 mm. Identities of birds on Daphne with measurements in the intervening range between these two limits of 6.9/6.9 and 7.8/7.4 (n = 116) are ambiguous. Thirteen are known hybrids (10 F1s, and three backcrosses: two B1 and one B2). The proportion of Borrero Bay G. fuliginosa in this ambiguous range of measurements is 36/109 = 0.33. The proportion of Daphne birds smaller than this maximum size and not known to be hybrids (n = 285) that are in this range is almost the same, 0.36. On these grounds, almost all birds classified as G. fuliginosa on Daphne are indeed likely to be G. fuliginosa.
(ii). Source of immigrants
We ran Structure to assign G. fuliginosa on Daphne (undefined) to the following user-defined populations: Santa Cruz (n = 24), Santiago (n = 19), Rábida (n = 10), Española (n = 10), Floreana (n = 10), San Cristóbal (n = 21), Pinta (n = 10) and Isabela (n = 13). None of the Daphne birds was assigned to a population with a probability exceeding 0.500. All were assigned to Santa Cruz and Santiago with about equal probabilities (0.337–0.478), and also when only Santa Cruz and Santiago populations were included as possible source populations. Therefore, Santa Cruz and Santiago G. fuliginosa are not different enough genetically at the 16 loci to make possible the identification of the source island of birds that immigrated to Daphne. On geographical grounds, Santa Cruz is the more likely, but both may have contributed immigrants to Daphne. Morphologically, the populations on Santa Cruz and Santiago are almost identical (Lack 1947; Grant et al. 1985).
(iii). Breeders
Most immigrant G. fuliginosa died without breeding or emigrated. To estimate the numbers that bred on Daphne, we applied criteria for inclusion at three levels of strictness. The strictest method requires genotype and/or unambiguous measurements. By this method, the total is 10: three males and seven females. Two of them were genotyped, six were measured and two more, although not measured, were the parents of an offspring with very small beak measurements (6.4 and 6.3 mm). When birds with measurements in the ambiguous zone are included the total rises to 15: five males and 10 females. When birds identified by observation alone are included, the total rises yet further. In the years 1976–1997, 44 breeding birds were identified by observation as G. fuliginosa. As noted above, two were the parents of a phenotypically confirmed G. fuliginosa and are already included in the estimates. The number remaining, 42, should be reduced to about half (21) to allow for errors in classification revealed by genotyping (above). In addition, three genotyped G. fuliginosa were suspected of breeding in 2002. When all these are included (the least strict criteria) roughly 40 are identified as breeding immigrant G. fuliginosa.
(iv). Number of immigrants
Four G. fuliginosa bred intraspecifically. More were observed but not confirmed because they were not captured. A maximum of 14 pairs of G. fuliginosa lacking leg bands bred in the years 1976–1984. The remainder of the known immigrants and at least two of the G. fuliginosa hatched on the island bred with G. fortis, as did all of the F1 offspring that survived to breed, i.e. they backcrossed to G. fortis. Depending on the criteria adopted for inclusion, the number of immigrants that bred with G. fortis is six, 11 or 18. In terms of G. fortis generation lengths (4.5 years on average: Grant & Grant 1992b) Nh, the number of migrants (heterospecific) per generation over 36 years is 0.75, 1.375 or 2.25. We believe the middle estimate is the most realistic.
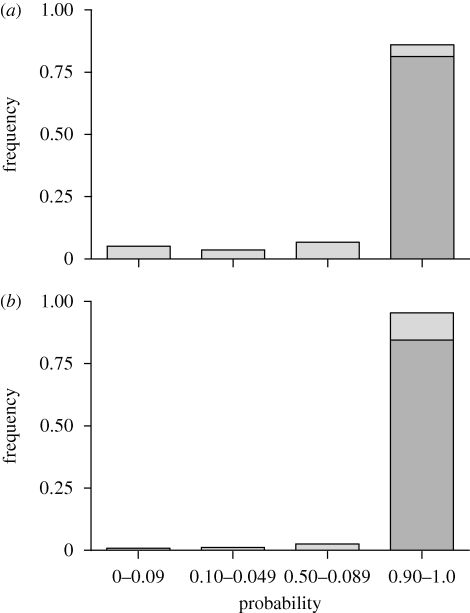
The estimate of the magnitude of migration is an average over eight generations. Immigration was not uniform across time, however, but was at an apparent maximum at the beginning of the study, declined markedly after 1986 according to observations and mist-net captures (Grant & Grant 1995) and remained low thereafter. For the last 6 years of the study, there was only one confirmed G. fuliginosa breeding (with G. fortis) on the island. The decline in immigration is reflected in the change in proportion of G. fortis identified as hybrids with G. fuliginosa by assignment tests (figure 1), from 14.0 per cent in 1981–1998 to 4.7 per cent in 1999–2008.
Figure 1.
Probabilities of assignment of G. fortis individuals in (a) early (1976–98; n = 1382) and (b) late (1998–2008; n = 344) samples. Probabilities of less than 0.900 indicate hybrids and backcrosses with mixtures of G. fortis and G. fuliginosa genes. Probabilities of greater than 0.99 are indicated in grey. Geospiza fortis × G. scandens hybrids and backcrosses are not included.
(b). G. fortis: interbreeding with G. scandens residents
(i). Detection by genotypes
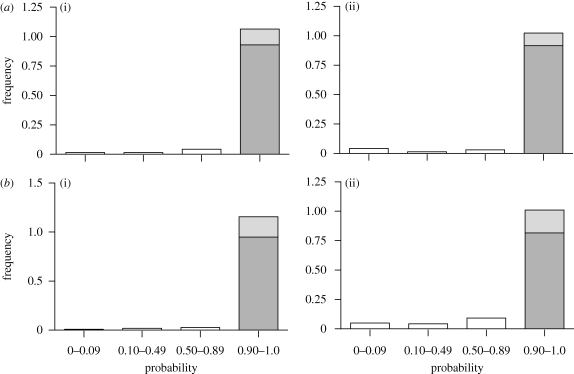
After excluding hybrids between G. fortis and G. fuliginosa (F1 + B1), we ran Structure with two prior generations to assign individuals in two defined populations, G. fortis and G. scandens, separately in early and late samples. Hybrids were found to be more common in the G. scandens than in the G. fortis samples, and more common in the late than in the early samples. By the majority rule, 40 (2.81%) of the early G. fortis sample (n = 1423) and nine (2.58%) of the late G. fortis sample (n = 349) were assigned to hybrids. Twenty-six (5.16%) of the early G. scandens sample (n = 504) and 17 (9.29%) of the late G. scandens sample (n = 183) were assigned to hybrids. The pattern is consistent with asymmetric gene exchange between the species, as reported before (Grant et al. 2004; Grant & Grant 2006), and a recent intensification of the asymmetry. The increase in gene flow is reflected in the change in the distribution of assignment probabilities from early to late samples (figure 2). Twenty-seven of the 92 hybrids in total (29.3%) were identified as F1s: note the percentage is subject to error (see §3).
Figure 2.
Probabilities of assignment of individuals to (i) G. fortis (n = 1642) or (ii) G. scandens (n = 689) in (a) early (1976–1998) and (b) late (1998–2008) samples. Probabilities of less than 0.900 reflect mixtures of G. fortis and G. scandens genes. Probabilities of greater than 0.99 are indicated in grey. Geospiza fortis × G. fuliginosa hybrids and backcrosses are not included.
(ii). Detection by pedigrees
In the 21 years from 1978 to 1998, there were 13 observed cases of interbreeding. Some of the offspring were observed to breed with either G. fortis or G. scandens. Others were assigned to species according to the song sung by their social father. Combining observed and potential breeding of the F1 offspring, we identified three as members of the G. fortis population and six as members of the G. scandens population. Two more did not breed and their fathers were not known. One was morphologically more like G. fortis than G. scandens, and therefore considered part of the G. fortis population, while the other more closely resembled G. scandens and was added to that population. Thus, the G. fortis population received genes from four G. scandens individuals in 21 years, or 0.86 per generation. Geospiza scandens received genes from at least seven G. fortis individuals, or 2.19 individuals per generation. Offspring without genotypes or measurements of the remaining two pairs have not been included. If they were included, G. scandens received genes from nine G. fortis individuals, or 2.36 individuals per generation.
6. Gene exchange through hybridization
Immigrant G. fuliginosa that bred with G. fortis brought to the island 21 alleles at the 16 microsatellite loci not detected in the G. fortis population at the time of their arrival. Three of the 21 alleles (14.7%) appeared in later samples of G. fortis or hybrids, presumably as a result of introgression. This indicates a slow addition of new alleles. Most introduced alleles gave rise to minor alterations in the frequencies of pre-existing alleles. Similarly, the interbreeding populations of G. fortis and G. scandens gained alleles from each other. Although most remained at low frequencies as with introgression of fuliginosa alleles into the G. fortis population, two increased substantially in the G. scandens population (Grant et al. 2004).
7. Genetic effects of gene flow
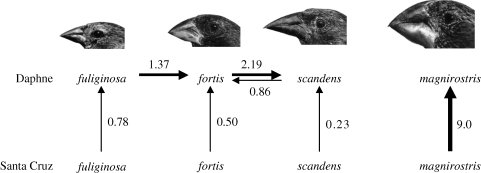
The flow of genes into the two main study populations on Daphne from conspecific and heterospecific sources is rare. Although the numbers of migrants per generation are low and are point estimates without confidence limits, they permit the conclusion that interbreeding is less frequent with conspecific immigrants (Nc = 0.50, 0.23) than with heterospecific immigrants (G. fortis × G. fuliginosa; Nh = 1.37) or residents (G. fortis × G. scandens; Nh = 0.86, 2.19). These contrasts are summarized in figure 3.
Figure 3.
Summary of gene flow through immigration and introgression on Daphne Major Island. Numbers refer to immigrants or hybridizing individuals per generation (see §9 for migration rates). Genes flow from G. fortis to G. scandens when the father of an F1 hybrid sings the G. scandens song, and vice versa for gene flow from G. scandens to G. fortis. The species hybridize on Santa Cruz Island to an unknown extent. Geospiza magnirostris hybridizes with G. fortis on Santa Cruz but not on Daphne.
The frequency of migrants is not a reliable index to the genetic effects of interbreeding when the source of the migrants, as here, is heterogeneous. For a given number of migrants per generation, the genetic effect of interbreeding is proportional to the mean of the absolute differences in allele frequencies between donor (Daphne or Santa Cruz) and recipient populations. A useful quantitative index for comparative purposes is the product of the number of migrants and the mean difference in allele frequencies (Nei's d) between the interbreeding populations, either conspecific or heterospecific. Summing intraspecific (Ncdc) and interspecific (Nhdh) gene inputs gives a value for this index of 1.6807 for G. fortis and 2.0456 for G. scandens. The interspecific contribution to total genetic input is 77.3 per cent for G. fortis and 95.9 per cent for G. scandens. Heterospecific sources do not contribute equally. The genetic effect on G. fortis of breeding with G. scandens (59.2%) is greater than the effect of breeding with G. fuliginosa (40.8%). All these refer to calculations based on data up to 1998. The contribution made by G. fuliginosa must have declined after this time because immigration declined.
8. Relative fitness
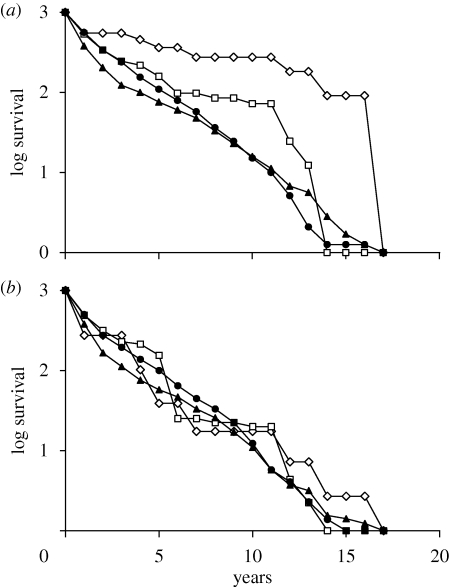
The greater genetic effect of heterospecific gene flow on Daphne populations is not counterbalanced by low relative fitness. On the contrary, F1 hybrid and conspecific resident individuals survived at least as well as the parental species on average, if not better (figure 4), over the same environmental conditions. Approximately half of the individuals in the seven major cohorts died in their first year. Mean survival over the first year was almost the same among G. fortis × G. fuliginosa F1s (0.52 ± 0.059 s.e.; n = 83) and G. fortis (0.51 ± 0.048; n = 3860) and was higher among the G. fortis × G. scandens F1s (0.679 ± 0.161; n = 10) than among G. scandens (0.404 ± 0.054; n = 1771) and G. fortis.
Figure 4.
Composite survival curves of seven cohorts of finches, hatched in the years 1978, 1981, 1983, 1984, 1987, 1991 and 1998. (a) Numbers were summed across years, standardized to 1000 at fledging and converted to logs. (b) Numbers in each cohort were first standardized to 1000 at fledging and converted to logs, then summed across years and averaged. For variation among three of the cohorts see Grant & Grant (2008a,b). Filled circle, G. fortis; solid triangles, G. scandens; open squares, G. fortis × G. fuliginosa; open diamond, G. fortis × G. scandens.
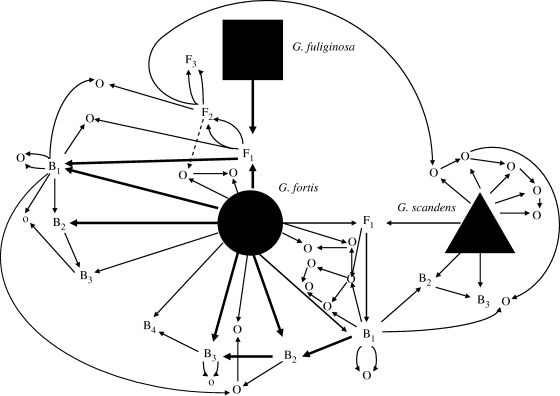
Hybrids do not experience a loss of fitness in acquiring mates (Grant & Grant 1997a,b) or in reproductive success (Grant & Grant 1992a). The same applies to conspecific immigrants that bred, although the numbers are too few for analysis. Therefore, overall, 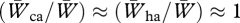 . As a consequence, backcrossing of F1 hybrids and successive generations of offspring has given rise to a complex network of genetic relationships among the species (figure 5).
. As a consequence, backcrossing of F1 hybrids and successive generations of offspring has given rise to a complex network of genetic relationships among the species (figure 5).
Figure 5.
Gene flow network reflecting known and quantified introgressive hybridization. All three species are connected by exchanging genes, but the relatively rare G. fuliginosa has not hybridized with G. scandens, and G. fortis × G. fuliginosa hybrids have not backcrossed to G. fuliginosa. Thick lines indicate the primary pathways of genes from one population to another.
9. Selection and migration
Selection, to counteract the effect of migration, must exceed the migration rate (m), i.e. the proportion of the breeding population that are migrants. Over the relevant time periods, the average sizes of the breeding populations were approximately 80 G. scandens and 230 G. fortis individuals. Using numbers in figure 3, we calculate m from the combined heterospecific and conspecific sources to be 0.030 for G. scandens and 0.018 for G. fortis. Natural selection has been far stronger at times, on both species (Grant & Grant 2002, 2006), with selection coefficients as high as 0.5–1 standard deviations. Therefore, migration has been insufficient to counteract local adaptation. In contrast, natural selection has been scarcely detectable in G. magnirostris (Grant et al. 2001), whose average size of the breeding population was 25 individuals in 1988–1998. Migration rate, estimated at 0.360 and one order of magnitude greater than in G. scandens and G. fortis, has been more than sufficient to counteract local adaptation.
10. Conclusions
The two main study populations of Darwin's finches on Daphne Major Island receive genes by breeding with allopatric conspecific individuals that have immigrated, and from heterospecific individuals, both allopatric immigrants (G. fuliginosa) and sympatric residents (G. fortis and G. scandens). The flow of genes from conspecific and heterospecific sources is rare and unequal. Genes flow at a faster rate from heterospecific sources than from conspecific sources and have stronger effects because species differ genetically more than do populations of the same species. The effects of heterospecific gene flow are not counteracted by lower fitness of the offspring. As a result, the standing genetic variation of the two main resident populations on Daphne Major is enhanced to a greater extent by introgressive hybridization than through interbreeding with rare immigrants from another island. The situation may be exceptional because most species do not hybridize, but where hybridization does occur, as in the coexistence of closely related species, it can have a greater effect than conspecific gene flow upon gene dynamics. Approximately 10 per cent of all bird species are known to hybridize (Grant & Grant 1992a), which is not unusual among animal taxa but is low compared with plants (Mallet 2005).
The study illustrates the dynamic nature of intraspecific and interspecific interactions in three ways. First, immigration (arrival) of G. fuliginosa declined after the mid-1980s. The unknown cause probably lies in the source islands (Grant & Grant 1995). Introgressive hybridization with G. fortis therefore declined, yet G. fuliginosa alleles persisted in the G. fortis population as a result of repeated backcrossing. The backcrossing was complex, resulting in a few cases in a combination of genes from three species in single individuals (figure 5). Second, introgressive hybridization between the resident species G. fortis and G. scandens increased at the same time as it decreased between G. fuliginosa and G. fortis. Here, the cause has been identified as an enduring transformation in the food supply resulting from a major, archipelago-wide, El Niño event in 1982–1983 (Grant & Grant 1996). Thus, the situation on Daphne is not equilibrial and is currently leading towards the fusion of G. fortis and G. scandens into a single panmictic population. Selection on G. scandens has not overridden effects of introgression on beak shape; instead selection may have augmented introgression (Grant et al. 2004). Nevertheless, the direction of change may be reversed if the climatic and floristic environment changes (Grant & Grant 2008b). Third, the El Niño event facilitated the establishment of a breeding population of G. magnirostris on Daphne in 1982–1983. The rate of immigration after the initial colonization was far higher than immigration of the other species, but showed signs of a density-related decline as population size increased. Immigration of this species is sufficiently frequent that it could overwhelm evolutionary change through natural selection on Daphne, but alternatively it might facilitate evolutionary change, as postulated for introgressive hybridization, by providing new genetic variation from as many as three source islands.
We conclude that conspecific gene flow as a result of immigration is insufficient to negate the strong effects of both hybridization and local selection on Daphne (Grant et al. 2004) and that conspecific and heterospecific gene flow in combination are sufficient to counteract random genetic drift. The chief implication of these findings is that gene exchange between populations is complex, heterogeneous and varies in time measured in decades. This dynamic perspective provides insight into population genetic structure, which is often used to infer average rates of gene flow at assumed steady state (Petren et al. 2005).
Additional tests of the relative importance of migration rate, genetic difference and relative fitness in hybridizing species could be conducted in hybrid zones in continental regions. Several avian hybrid zones are known to be moving northwards in the Northern Hemisphere (Cook 1975; Gill 1980, 2004; Rowher et al. 2001; Reudink et al. 2007), possibly influenced by climate warming (Cook 1975; Berthold et al. 1992). Their movement implies changing local dynamics of conspecific and heterospecific gene exchange. The local dynamics of the two sources of gene exchange are likely to vary wherever there are gradients in hybridization, as occurs in a variety of organisms, for example, cichlid fish (Seehausen & Magalhaes in press), Daphnia (Petrusek et al. 2008), Heliconius butterflies (Kronforst et al. 2006) and Triturus salamanders (Arntzen et al. 2009).
Acknowledgements
We thank the Galápagos National Parks Service and Charles Darwin Research Station for permission to carry out the fieldwork and for logistical support. We thank Arkhat Abzhanov for the invitation to contribute to this theme issue and many assistants over the years for help in the field. We thank Lukas Keller for help with assignment tests. We are especially grateful to Ken Petren for initial genotyping, analysis and supplying us with blood samples from Santiago and Isabela. Two reviewers provided important clarification of some issues. The research has been supported by grants from the National Science and Engineering Research Council (Canada) and the National Science Foundation (USA).
Footnotes
One contribution of 13 to a Theme Issue ‘Darwin's Galápagos finches in modern evolutionary biology’.
References
- Alexandrino P., Faria R., Linares D., Castro F., de la Corre M., Sabatié R., Bagliniere J. L., Weiss S.2006Interspecific differentiation and intraspecific substructure in two closely related clupeids with extensive hybridization, Alosa alosa and Alosa fallax. J. Fish. Biol. 69(Suppl.), 242–259 (doi:10.1111/j.1095-8649.2006.01289.x) [Google Scholar]
- Anderson E. C.2008Bayesian inference of species hybrids using multilocus dominant genetic markers. Phil. Trans. R. Soc. B 363, 2841–2850 (doi:10.1098/rstb.2008.0043) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson E. C., Thompson E. A.2002A model-based method for identifying species hybrids using multilocus genetic data. Genetics 160, 1217–1229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold M. L.1997Natural hybridization and evolution. Oxford, UK: Oxford University Press [Google Scholar]
- Arnold M. L.2006Evolution through genetic exchange. Oxford, UK: Oxford University Press [Google Scholar]
- Arnold M. L.2008Reticulate evolution and humans. Oxford, UK: Oxford University Press [Google Scholar]
- Arntzen J. W., Jehle R., Bardekci F., Burke T., Wallis G. P.2009Asymmetric viability of reciprocal cross-hybrids between crested and marbled newts (Triturus cristatus and T. marmoratus). Evolution 63, 1191–1202 (doi:10.1111/j.1558-5646.2009.00611.x) [DOI] [PubMed] [Google Scholar]
- Barrier M., Baldwin B. G., Robichaux R. H., Purugganan M.1999Interspecific hybrid ancestry of a plant adaptive radiation: allopolyploidy of the Hawaiian silversword alliance (Asteraceae) inferred from floral homeotic gene duplications. Mol. Biol. Evol 16, 1105–1113 [DOI] [PubMed] [Google Scholar]
- Barton N. H.2001The role of hybridization in evolution. Mol. Ecol. 10, 551–568 (doi:10.1046/j.1365-294x.2001.01216.x) [DOI] [PubMed] [Google Scholar]
- Barton N. H., Hewitt G. M.1985Adaptation, speciation, and hybrid zones. Annu. Rev. Ecol. Syst. 16, 113–148 (doi:10.1146/annurev.es.16.110185.000553) [Google Scholar]
- Berthold P., Helbig A. J., Mohr G., Querner U.1992Rapid microevolution of migratory behaviour in a wild bird species. Nature 360, 668–670 (doi:10.1038/360668a0) [Google Scholar]
- Boag P. T., Grant P. R.1984The classical case of character release: Darwin's finches (Geospiza) on Isla Daphne Major, Galápagos. Biol. J. Linn. Soc. 22, 243–287 (doi:10.1111/j.1095-8312.1984.tb01679.x) [Google Scholar]
- Bolnick D. I., Nosil P.2007Natural selection in populations subject to a migration load. Evolution 61, 2229–2243 (doi:10.1111/j.1558-5646.2007.00179.x) [DOI] [PubMed] [Google Scholar]
- Bolnick D. I., Caldera E. J., Matthews B.2008Evidence for asymmetric migration load in a pair of ecologically divergent stickleback populations. Biol. J. Linn. Soc. 94, 273–287 (doi:10.1111/j.1095-8312.2008.00978.x) [Google Scholar]
- Cheviron Z. A., Brumfield R. T.2009Migration-selection balance and local adaptation of mitochondrial haplotypes in rufous-collared sparrows (Zonotrichia capensis) along an elevational gradient. Evolution 63, 1593–1605 (doi:10.1111/j.1558-5646.2009.00644.x) [DOI] [PubMed] [Google Scholar]
- Cook A.1975Changes in a carrion-hooded crow hybrid zone and possible importance of climate. Bird Stud. 22, 165–168 (doi:10.1080/00063657509476460) [Google Scholar]
- Crow J. F., Kimura M.1970An introduction to population genetics theory. New York, NY: Harper & Row [Google Scholar]
- Ebert D., Haag C., Kirkpatrick M., Riek M., Hottinger J. W., Pajunen V. I.2002A selective advantage to immigrant genes in a Daphnia metapopulation. Science 295, 485–488 (doi:10.1126/science.1067485) [DOI] [PubMed] [Google Scholar]
- Falush D., Stephens M., Pritchard J. K.2003Inference of population structure: extensions to linked loci and correlated allele frequencies. Genetics 164, 1567–1587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faubert P., Waples R. S., Gaggliotti O. E.2007Evaluating the performance of a multilocus Bayesian method for an estimation of migration rates. Mol. Ecol. 16, 1149–1166 (doi:10.1111/j.1365-294X.2007.03218.x) [DOI] [PubMed] [Google Scholar]
- García-Ramos G., Kirkpatrick M.1997Genetic models of adaptation and gene flow in peripheral populations. Evolution 51, 21–28 (doi:10.2307/2410956) [DOI] [PubMed] [Google Scholar]
- Gibbs H. L., Grant P. R.1987Oscillating selection on Darwin's finches. Nature 327, 511–513 (doi:10.1038/327511a0) [Google Scholar]
- Gill F. B.1980Historical aspects of hybridization between blue-winged and golden-winged warblers. Auk 97, 1–18 [Google Scholar]
- Gill F. B.2004Blue-winged (Vermivora pinus) versus golden-winged warblers (V. chrysoptera). Auk 121, 1014–1018 (doi:10.1642/0004-8038(2004)121[1014:BWVPVG]2.0.CO;2) [Google Scholar]
- Grant P. R.1993Hybridization of Darwin's finches on Isla Daphne Major, Galápagos. Phil. Trans. R. Soc. Lond. B 340, 127–139 (doi:10.1098/rstb.1993.0052) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant P. R.1999Ecology and evolution of Darwin's finches, 2nd edn.Princeton, NJ: Princeton University Press [Google Scholar]
- Grant B. R., Grant P. R.1989Evolutionary dynamics of a natural population. The large cactus finch of the Galápagos. Chicago, IL: University of Chicago Press; [DOI] [PubMed] [Google Scholar]
- Grant P. R., Grant B. R.1992aHybridization of bird species. Science 256, 193–197 (doi:10.1126/science.256.5054.193) [DOI] [PubMed] [Google Scholar]
- Grant P. R., Grant B. R.1992bDemography and the genetically effective sizes of two populations of Darwin's finches. Ecology 73, 766–784 (doi:10.2307/1940156) [Google Scholar]
- Grant B. R., Grant P. R.1993Evolution of Darwin's finches caused by a rare climatic event. Proc. R. Soc. Lond. B 251, 111–117 (doi:10.1098/rspb.1993.0016) [Google Scholar]
- Grant P. R., Grant B. R.1995The founding of a new population of Darwin's finches. Evolution 49, 229–240 (doi:10.2307/2410333) [DOI] [PubMed] [Google Scholar]
- Grant B. R., Grant P. R.1996High survival of Darwin's finch hybrids: effects of beak morphology and diets. Ecology 77, 500–509 (doi:10.2307/2265625) [Google Scholar]
- Grant P. R., Grant B. R.1997aHybridization, sexual imprinting and mate choice. Am. Nat. 149, 1–28 (doi:10.1086/285976) [Google Scholar]
- Grant P. R., Grant B. R.1997bMating patterns of Darwin's finch hybrids determined by song and morphology. Biol. J. Linn. Soc. 60, 317–343 (doi:10.1111/j.1095-8312.1997.tb01499.x) [Google Scholar]
- Grant P. R., Grant B. R.2002Unpredictable evolution in a 30-year study of Darwin's finches. Science 296, 707–711 (doi:10.1126/science.1070315) [DOI] [PubMed] [Google Scholar]
- Grant P. R., Grant B. R.2006Evolution of character displacement in Darwin's finches. Science 313, 224–226 (doi:10.1126/science.1128374) [DOI] [PubMed] [Google Scholar]
- Grant P. R., Grant B. R.2008aHow and why species multiply: the radiation of Darwin's finches. Princeton, NJ: Princeton University Press [Google Scholar]
- Grant B. R., Grant P. R.2008bFission and fusion of Darwin's finches populations. Phil. Trans. R. Soc. B 363, 2821–2829 (doi:10.1098/rstb.2008.0051) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant P. R., Grant B. R.2008cPedigrees, assortative mating, and speciation in Darwin's finches. Proc. R. Soc. B 275, 661–668 (doi:10.1098/rspb.2007.0898) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant P. R., Grant B. R.2009The secondary contact phase of allopatric speciation in Darwin's finches. Proc. Natl Acad. Sci. USA 106, 20 141–20 148 (doi:10.1073/pnas.0911761106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant P. R., Price T. D.1981Population variation in continuously varying traits as an ecological genetics problem. Am. Zool. 21, 795–811 [Google Scholar]
- Grant P. R., Abbott I., Schluter D., Curry R. L., Abbott L. K.1985Variation in the size and shape of Darwin's finches. Biol. J. Linn. Soc. 25, 1–39 (doi:10.1111/j.1095-8312.1985.tb00384.x) [Google Scholar]
- Grant P. R., Grant B. R., Petren K.2001A population founded by a single pair of individuals: establishment, expansion, and evolution. Genetica 112/113, 359–382 (doi:10.1023/A:1013363032724) [PubMed] [Google Scholar]
- Grant P. R., Grant B. R., Markert J. A., Keller L. F., Petren K.2004Convergent evolution of Darwin's finches caused by introgressive hybridization and selection. Evolution 58, 1588–1599 [DOI] [PubMed] [Google Scholar]
- Hanski I., Kuussaari M., Nieminen M.1994Metapopulation structure and migration in the butterfly Melitaea cinxia. Ecology 75, 747–762 (doi:10.2307/1941732) [Google Scholar]
- Harper G. R., Jr, Pfennig D. W.2008Selection overrides gene flow to break down maladaptive mimicry. Nature 451, 1103–1106 (doi:10.1038/nature06532) [DOI] [PubMed] [Google Scholar]
- Harper F. M., Addison J. A., Hart M. W.2007Introgression versus immigration in hybridizing high dispersal echinoderms. Evolution 61, 2410–2418 (doi:10.1111/j.1558-5646.2007.00200.x) [DOI] [PubMed] [Google Scholar]
- Harrison R. G. (ed.) 1993Hybrid zones and the evolutionary process. New York, NY: Oxford University Press [Google Scholar]
- Hedrick P.2000Genetics of populations, 2nd edn.Sudbury, MA: Jones & Bartlett [Google Scholar]
- Hendry A. P.2004Selection against migrants contributes to the rapid evolution of ecologically-dependent reproductive isolation. Evol. Ecol. Res. 6, 1219–1236 [Google Scholar]
- Hendry A. P., Taylor E. B.2004How much of the variation in adaptive divergence can be explained by gene flow: an evaluation using lake-stream stickleback pairs. Evolution 58, 2319–2331 [DOI] [PubMed] [Google Scholar]
- Hendry A. P., Taylor E. B., McPhail J. D.2002Adaptive divergence and the balance between selection and gene flow in lake and stream sticklebacks in the Misty system. Evolution 56, 1199–1216 [DOI] [PubMed] [Google Scholar]
- Keller L. F., Grant P. R., Grant B. R., Petren K.2001aHeritability of morphological traits in Darwin's finches: misidentified paternity and maternal effects. Heredity 87, 325–336 (doi:10.1046/j.1365-2540.2001.00900.x) [DOI] [PubMed] [Google Scholar]
- Keller L. F., Jeffrey K. J., Arcese P., Beaumont M. A., Hochachka W. M., Smith J. N. M., Bruford M. W.2001bImmigration and the ephemerality of a natural population bottleneck: evidence from molecular markers. Proc. R. Soc. Lond. B 268, 1387–1394 (doi:10.1098/rspb.2001.1607) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkpatrick M., Barton N. H.1997Evolution of a species' range. Am. Nat. 150, 1–23 (doi:10.1086/286054) [DOI] [PubMed] [Google Scholar]
- Kronforst M. R., Young L. G., Blume L. M., Gilbert L. E.2006Multilocus analyses of admixture and introgression among hybridizing Heliconius butterflies. Evolution 60, 1254–1268 [PubMed] [Google Scholar]
- Lack D.1947Darwin's finches. Cambridge, UK: Cambridge University Press [Google Scholar]
- Lenormand T.2002Gene flow and the limits to natural selection. Trends Ecol. Evol. 17, 183–189 (doi:10.1016/S0169-5347(02)02497-7) [Google Scholar]
- Lewontin R. C., Birch L. C.1966Hybridization as a source of variation for adaptation to new environments. Evolution 20, 315–336 (doi:10.2307/3406633) [DOI] [PubMed] [Google Scholar]
- Lorenzen E. D., Simonsen B. T., Kat P. W., Arctander P., Siegesmund H. R.2006Hybridization between subspecies of waterbuck (Kobus ellipsiprymnus) in zones of overlap with limited introgression. Mol. Ecol. 15, 3787–3799 (doi:10.1111/j.1365-294X.2006.03059.x) [DOI] [PubMed] [Google Scholar]
- Mallet J.2005Hybridization as an invasion of the genome. Trends Ecol. Evol. 20, 229–237 (doi:10.1016/j.tree.2005.02.010) [DOI] [PubMed] [Google Scholar]
- Marr A. B.2006Immigrants and gene flow in small populations. In Conservation and biology of small populations (eds Smith J. N. M., Keller L. F., Marr A. B., Arcese P.), pp. 139–153 New York, NY: Oxford University Press [Google Scholar]
- Miles L. S., Allendorf F. W.1996The one-migrant-per-generation rule in conservation and management. Conserv. Biol. 10, 1509–1518 (doi:10.1046/j.1523-1739.1996.10061509.x) [Google Scholar]
- Moore J.-S., Gow J. L., Taylor E. B., Hendry A. P.2007Quantifying the constraining influence of gene flow on adaptive divergence in the lake-stream threespine stickleback system. Evolution 61, 2015–2026 (doi:10.1111/j.1558-5646.2007.00168.x) [DOI] [PubMed] [Google Scholar]
- Oliveira R., Godinho R., Randi E., Alves P. C.2008Hybridization versus conservation: are domestic cats threatening the genetic integrity of wildcats (Felis silvestris silvestris) in Iberian Peninsula? Phil. Trans. R. Soc. B 363, 2953–2961 (doi:10.1098/rstb.2008.0052) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peery M. Z., Beissinger S. R., House R. F., Bérubé M., Hall L. A., Sellas A., Palsbøll P. J.2008Characterizing source–sink dynamics with genetic parentage assignments. Ecology 89, 2746–2759 (doi:10.1890/07-2026.1) [DOI] [PubMed] [Google Scholar]
- Petren K., Grant B. R., Grant P. R.1999Extrapair paternity in the cactus finch, Geospiza scandens. Auk 116, 252–256 [Google Scholar]
- Petren K., Grant P. R., Grant B. R., Keller L. F.2005Comparative landscape genetics and the adaptive radiation of Darwin's finches: the role of peripheral isolation. Mol. Ecol. 14, 2943–2957 (doi:10.1111/j.1365-294X.2005.02632.x) [DOI] [PubMed] [Google Scholar]
- Petrusek A., Seda J., Machácek J., Ruthová S., Smilauer P.2008Daphnia hybridization along ecological gradients in pelagic environments: the potential for the presence of hybrid zones in plankton. Phil. Trans. R. Soc. B 363, 2931–2941 (doi:10.1098/rstb.2008.0026) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard J. K., Stephens M., Donnelly P.2000Inference of population structure using multilocus genotype data. Genetics 155, 945–959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard J. K., Wen X., Falush D. 2007. Documentation for structure software: Version 2.2. See http://pritch.bsd.uchicago.edu/software . [Google Scholar]
- Reudink M. W., Mech S. G., Mullen S. P., Curry R. L.2007Structure and dynamics of the hybrid zone between black-capped chickadee (Poecile atricapillus) and Carolina chickadee (P. carolinensis) in southeastern Pennsylvania. Auk 124, 463–478 (doi:10.1642/0004-8038(2007)124[463:SADOTH]2.0.CO;2) [Google Scholar]
- Rieseberg L. H.1997Hybrid origins of plant species. Annu. Rev. Ecol. Syst. 28, 359–389 (doi:10.1146/annurev.ecolsys.28.359) [Google Scholar]
- Rowher S., Bermingham E., Wood C.2001Plumage and mitochondrial DNA haplotype variation across a moving hybrid zone. Evolution 55, 405–422 [DOI] [PubMed] [Google Scholar]
- Saccheri I. J., Brakefield P. M.2002Rapid spread of immigrant genomes into inbred populations. Proc. R. Soc. Lond. B 269, 1073–1078 (doi:10.1098/rspb.2002.1963) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwenk K., Brede N., Streit B.2008Extent, processes and evolutionary impact of interspecific hybridization in animals. Phil. Trans. R. Soc. B 363, 2805–2811 (doi:10.1098/rstb.2008.0055) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seehausen O.2004Hybridization and adaptive radiation. Trends Ecol. Evol. 19, 198–207 (doi:10.1016/j.tree.2004.01.03) [DOI] [PubMed] [Google Scholar]
- Seehausen O.2006African cichlid fish: a model system in adaptive radiation research. Proc. R. Soc. B 273, 1987–1998 (doi:10.1098/rspb.2006.3539) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seehausen O., Magalhaes I. S.In press Geographical mode and evolutionary mechanism of ecological speciation in cichlid fish. In From field observations to mechanisms. A program in evolutionary biology (eds Grant P. R., Grant B. R.). Princeton, NJ: Princeton University Press [Google Scholar]
- Slatkin M.1985Gene flow in natural populations. Annu. Rev. Ecol. Syst. 16, 393–340 (doi:10.1146/annurev.ecolsys.16.1.393) [Google Scholar]
- Slatkin M.1987aRare alleles as indicators of gene flow. Evolution 39, 53–65 (doi:10.2307/2408516) [DOI] [PubMed] [Google Scholar]
- Slatkin M.1987bGene flow and the geographic structure of natural populations. Science 236, 787–792 (doi:10.1126/science.3576198) [DOI] [PubMed] [Google Scholar]
- Svärdson G.1970Significance of introgression in coregonid evolution. In Biology of coregonid fishes (eds Lindsey C. C., Woods C. S.), pp. 33–59 Winnipeg, MB: University of Manitoba Press [Google Scholar]
- Verhulst S., van Eck H. M.1996Gene flow and immigration rate in an island population of great tits. J. Evol. Biol. 9, 771–782 (doi:10.1046/j.1420-9101.1996.9060771.x) [Google Scholar]
- Wilson G. A., Rannala B.2003Bayesian inference of recent migration rates using multilocus genotypes. Genetics 163, 1177–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]