Abstract
Small and isolated island populations provide ideal systems to study the effects of limited population size, genetic drift and gene flow on genetic diversity. We assessed genetic diversity within and differentiation among 19 mockingbird populations on 15 Galápagos islands, covering all four endemic species, using 16 microsatellite loci. We tested for signs of drift and gene flow, and used historic specimens to assess genetic change over the last century and to estimate effective population sizes. Within-population genetic diversity and effective population sizes varied substantially among island populations and correlated strongly with island size, suggesting that island size serves as a good predictor for effective population size. Genetic differentiation among populations was pronounced and increased with geographical distance. A century of genetic drift did not change genetic diversity on an archipelago-wide scale, but genetic drift led to loss of genetic diversity in small populations, especially in one of the two remaining populations of the endangered Floreana mockingbird. Unlike in other Galápagos bird species such as the Darwin's finches, gene flow among mockingbird populations was low. The clear pattern of genetically distinct populations reflects the effects of genetic drift and suggests that Galápagos mockingbirds are evolving in relative isolation.
Keywords: Nesomimus, genetic drift, historic specimens, population differentiation
1. Introduction
Biologists have long recognized islands as ideal natural laboratories, and islands have thus contributed substantially to our understanding of evolution (Grant 1998). Two defining features of islands are their isolation and restricted land mass, both of which result in populations of limited size with clearly defined geographical boundaries. Genetic drift (the random changes of allele frequencies over generations) reduces genetic variation to an extent inversely proportional to population size (Crow & Kimura 1970); therefore, drift is expected to be pronounced in island systems (Barton 1998). Like mutation and selection, genetic drift leads to divergence among populations, whereas gene flow (migration) has a homogenizing effect (Slatkin 1985). Hence, low rates of gene flow and substantial genetic drift in perpetually small populations are the most probable explanations for the lower genetic diversity generally observed in island populations and species when compared with their mainland relatives (Frankham 1997). However, particularly in very vagile species such as birds, the effects of drift may be counteracted by gene flow. Darwin's finches in the Galápagos archipelago represent one well-known example where drift has been shown to be relatively weak due to the ubiquity of gene flow even over substantial distances (Petren et al. 2005). Here we examine genetic drift and diversification in another genus of birds in the Galápagos Islands, the mockingbirds, whose diversification differs substantially from that of Darwin's finches.
The mockingbirds of Galápagos played a key role in the development of Darwin's (1839) thinking on speciation. The phenotypic variation they exhibit across islands was one of Darwin's crucial observations that would ultimately lead him to propose his famous theory of evolution by natural selection (Darwin 1859). In contrast to the Darwin's finches, Galápagos mockingbird species do not occur in sympatry. Four endemic species are found in the Galápagos (Harris 1974), one of which, Mimus parvulus, is widespread in the archipelago occurring on most of the major islands except those occupied by the other species, whereas the other three species are very restricted in their geographical range: Mimus trifasciatus occurs on the islets Champion and Gardner-by-Floreana close to Floreana, Mimus macdonaldi on Española and Gardner-by-Española, and Mimus melanotis on San Cristóbal (figure 1). Unfortunately, the Floreana mockingbird (M. trifasciatus) today is classified as critically endangered with only 20–50 individuals left on Champion and approximately 300–500 on Gardner-by-Floreana (P. E. A. Hoeck & L. F. Keller 2006–2009, unpublished census data) after extinction of the main population on Floreana at around 1880. Galápagos mockingbirds are relatively sedentary birds and hypothesized to be weak fliers as they have rarely been observed flying over water or visiting islands where there is no established population (P. R. Grant & R. L. Curry 2005, personal communication). Because of this, migration rate is presumed to be low and genetic drift along with selection may have played a key role in the genetic structuring of mockingbird populations.
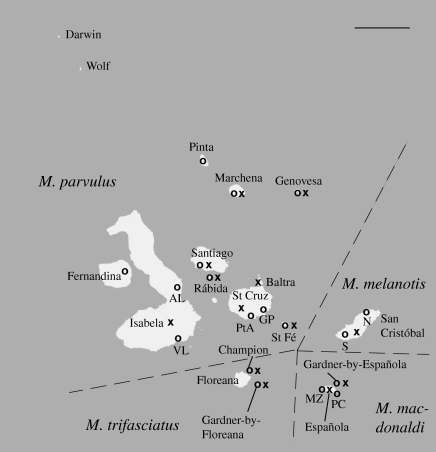
Figure 1.
Distribution of the four mockingbird species in Galápagos with island names and locations from which contemporary (o) and historic (x) samples were obtained. On Isabela (AL and VL), St Cruz (PtA and GP), San Cristóbal (N and S) and Española (MZ and PC) we collected samples in two different locations (Scale bar, 50 km).
In this study, we examined genetic diversity and differentiation within and among the four mockingbird species and their populations, covering nearly their entire range and testing for evidence of genetic drift and gene flow. Using microsatellite markers, we described genetic diversity present in contemporary populations and compared it with the genetic diversity of historic populations using samples collected in the early twentieth century. We tested the prediction that genetic diversity increases with population size using island size as a proxy for population size (Frankham 1996; Petren et al. 2005) and assessed the role of population size in maintaining genetic diversity over time. If gene flow is limited by distance, genetic and geographical distances should be positively correlated (Wright 1943). We therefore tested for an isolation-by-distance pattern across the archipelago. Furthermore, with the help of the temporal samples, we estimated effective population size (Crow & Kimura 1970). Effective population size (Ne) has become an important measure not only in evolution but also in conservation biology because of its role in maintaining adaptive genetic diversity and evolutionary potential (Palstra & Ruzzante 2008). Finally, we compared the genetic structure among populations and species from our nuclear markers with the results published in a study on the phylogeny of the Galápagos mockingbirds based on mitochondrial DNA (mtDNA) sequences (Arbogast et al. 2006). Our study provides an example of the usefulness of temporal samples to describe the change in the genetic structure between populations and species in order to disentangle temporal from spatial variation and learn more about the mechanisms underlying genetic diversification in small and isolated populations.
2. Material and methods
(a). Sample collection
(i). Contemporary samples
Blood samples from a total of 543 individuals from 14 islands in the Galápagos were collected between 2003 and 2008 (figure 1). We sampled on all islands inhabited by mockingbirds, except for two small, remote islands in the northwest of the archipelago (Darwin and Wolf). In order to estimate within-island differentiation, we obtained samples from two separate locations on three large islands (Isabela: AL and VL; St Cruz: GP and PtA; San Cristóbal: N and S) and one medium-sized island (Española: PC and MZ). On Champion we managed to sample the entire population except one individual. Sample sizes from the different locations varied between 10 and 69 individuals (mean: 30 individuals; table 1).
Table 1.
Populations studied and number of successfully genotyped samples from each population (n). Measures of genetic variation based on 16 microsatellite loci: Na: number of alleles, AR: average allelic richness, He: expected heterozygosity, Ho: observed heterozygosity, P: polymorphism. Island size is shown in hectares and island isolation was calculated as nearest shore-to-shore distances from all other islands (km). Island location was assigned as central (c) or peripheral (p) based on the position of the island within the species range of M. parvulus. Island age is shown in million years.
| n | total Na | mean Na | AR | He | Ho | P | island size | island size (ln) | island isolation | island location | island age | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Baltra-CAS | 18 | 60 | 3.75 | 3.25 | 0.560 | 0.488 | 0.94 | 2619.6 | 7.87 | 68.34 | n.a | 1.1 |
| Champion | 48 | 21 | 1.31 | 1.20 | 0.072 | 0.082 | 0.25 | 9.5 | 2.25 | 87.69 | n.a. | 1.5 |
| Champion-CAS | 11 | 20 | 1.25 | 1.25 | 0.118 | 0.084 | 0.25 | 9.5 | 2.25 | 87.69 | n.a. | 1.5 |
| Española, MZ | 29 | 35 | 2.19 | 1.86 | 0.225 | 0.220 | 0.50 | 6048.0 | 8.71 | 111.15 | n.a. | 3 |
| Española, PC | 58 | 40 | 2.50 | 1.81 | 0.211 | 0.222 | 0.44 | 6048.0 | 8.71 | 111.15 | n.a. | 3 |
| Española-CAS | 25 | 40 | 2.50 | 2.05 | 0.279 | 0.215 | 0.69 | 6048.0 | 8.71 | 111.15 | n.a. | 3 |
| Fernandina | 24 | 66 | 4.13 | 2.93 | 0.438 | 0.419 | 0.81 | 64248.0 | 11.07 | 122.84 | p | 0.035 |
| Gardner-by-Española | 10 | 27 | 1.69 | 1.59 | 0.167 | 0.194 | 0.44 | 58.0 | 4.06 | 116.47 | n.a. | 3 |
| Gardner-by-Española-CAS | 12 | 24 | 1.50 | 1.45 | 0.174 | 0.177 | 0.38 | 58.0 | 4.06 | 116.47 | n.a. | 3 |
| Gardner-by-Floreana | 69 | 34 | 2.13 | 1.77 | 0.261 | 0.250 | 0.56 | 81.2 | 4.40 | 93.59 | n.a. | 1.5 |
| Gardner-by-Floreana-CAS | 27 | 38 | 2.38 | 1.95 | 0.276 | 0.234 | 0.50 | 81.2 | 4.40 | 93.59 | n.a. | 1.5 |
| Genovesa | 37 | 37 | 2.31 | 1.95 | 0.290 | 0.296 | 0.63 | 1410.8 | 7.25 | 120.09 | p | 0.3 |
| Genovesa-CAS | 29 | 34 | 2.13 | 1.76 | 0.221 | 0.197 | 0.56 | 1410.8 | 7.25 | 120.09 | p | 0.3 |
| Isabela, AL | 32 | 72 | 4.50 | 2.97 | 0.430 | 0.436 | 0.81 | 458812.0 | 13.04 | 69.37 | c | 0.5 |
| Isabela, VL | 30 | 73 | 4.56 | 3.24 | 0.486 | 0.475 | 0.81 | 458812.0 | 13.04 | 69.37 | c | 0.5 |
| Isabela-CAS | 27 | 75 | 4.69 | 3.18 | 0.476 | 0.417 | 0.88 | 458812.0 | 13.04 | 69.37 | c | 0.5 |
| Marchena | 38 | 55 | 3.44 | 2.63 | 0.455 | 0.451 | 0.88 | 12996.0 | 9.47 | 107.75 | p | 0.6 |
| Marchena-CAS | 24 | 52 | 3.25 | 2.74 | 0.470 | 0.429 | 0.88 | 12996.0 | 9.47 | 107.75 | p | 0.6 |
| Pinta | 27 | 46 | 2.88 | 2.30 | 0.370 | 0.359 | 0.69 | 5940.0 | 8.69 | 134.41 | p | 0.7 |
| Rábida | 21 | 50 | 3.13 | 2.56 | 0.423 | 0.461 | 0.69 | 499.3 | 6.21 | 78.05 | c | 1.3 |
| Rábida-CAS | 27 | 49 | 3.06 | 2.45 | 0.410 | 0.402 | 0.75 | 499.3 | 6.21 | 78.05 | c | 1.3 |
| San Cristóbal, N | 17 | 46 | 2.88 | 2.46 | 0.353 | 0.353 | 0.63 | 55808.6 | 10.93 | 101.23 | n.a. | 2.4 |
| San Cristóbal, S | 20 | 54 | 3.38 | 2.67 | 0.378 | 0.381 | 0.63 | 55808.6 | 10.93 | 101.23 | n.a. | 2.4 |
| San Cristóbal-CAS | 27 | 57 | 3.56 | 2.70 | 0.390 | 0.348 | 0.63 | 55808.6 | 10.93 | 101.23 | n.a. | 2.4 |
| Santiago | 27 | 82 | 5.13 | 3.50 | 0.534 | 0.502 | 1.00 | 58465.0 | 10.98 | 69.29 | c | 0.8 |
| Santiago-CAS | 29 | 84 | 5.25 | 3.56 | 0.521 | 0.504 | 0.88 | 58465.0 | 10.98 | 69.29 | c | 0.8 |
| St Cruz, GP | 22 | 70 | 4.38 | 3.38 | 0.579 | 0.586 | 1.00 | 98555.0 | 11.50 | 53.24 | c | 1.1 |
| St Cruz, PtA | 13 | 66 | 4.13 | 3.46 | 0.594 | 0.582 | 0.94 | 98555.0 | 11.50 | 53.24 | c | 1.1 |
| St Cruz-CAS | 27 | 84 | 5.25 | 3.62 | 0.568 | 0.453 | 1.00 | 98555.0 | 11.50 | 53.24 | c | 1.1 |
| St Fé | 21 | 33 | 2.06 | 1.81 | 0.255 | 0.235 | 0.56 | 2413.0 | 7.79 | 73.64 | p | 2.9 |
| St Fé-CAS | 25 | 35 | 2.19 | 1.72 | 0.218 | 0.148 | 0.56 | 2413.0 | 7.79 | 73.64 | p | 2.9 |
| total/average | 851 | 143 | 3.18 | 2.47 | 0.366 | 0.348 | 0.69 |
(ii). Historic samples
Historic tissue samples from 349 specimens from 13 islands were obtained from the museum collection of the California Academy of Sciences (CAS). The majority of the specimens were collected during the CAS expedition to the Galápagos in the years 1905 and 1906 (called 1906 below), with a few samples collected in 1899. We were thus able to obtain both contemporary and historic samples from 12 islands, contemporary samples only from two (Fernandina and Pinta) and historic samples only from one island (Baltra; figure 1). The Baltra mockingbird population went extinct during or after World War II (Curry 1986); however, the historic samples are interesting to determine the former population's genetic relationship to the surrounding populations. Sample sizes for the historic populations (referred to as CAS-populations) ranged from 11 to 29 specimens per island (mean: 24; table 1).
(b). DNA extraction and microsatellite analysis
(i). Contemporary samples
Blood samples were collected on filter paper after a small puncture of the wing vein of live birds. Extraction and PCR were performed using previously published methods (Hoeck et al. 2009). Concentrations of DNA extracts were standardized at 20 ng µl−1 (Quant-iT PicoGreen dsDNA Quantitation, Invitrogen) and the following 17 microsatellite loci were amplified: MpAAT26, Nes01, Nes03, Nes04, Nes06, Nes10, Nes12, Nes13, Nes14, Nes15, Nes16, Nes17, Nes18, Nes19, Nes20, Nes22 and Nes23. Except for MpAAT26 which was developed in Mimus polyglottos by Hughes & Deloach (1997), all microsatellite loci were previously designed in our laboratory (Hoeck et al. 2009) with the aim of obtaining short microsatellite products (less than 200 bp) for amplification in highly fragmented, low quality DNA. Microsatellites were amplified in four independent multiplex reactions as described in Hoeck et al. (2009). MpAAT26 was amplified separately under the same conditions as markers in multiplex reactions B and C. Fragment analyses were performed on a 3730 DNA Analyser using Gene-Scan-500 LIZ size standard (ABI) and Genemapper v. 4 software (ABI) followed by manual proofreading of genotypes. To estimate the frequency of genotyping error rates, six per cent of the contemporary samples were amplified and genotyped a second time at each locus.
(ii). Historic samples
Small toe pad samples (approx. 4 mm2 in size) were collected from the historic specimens and half of each sample was used for DNA extraction using QIAamp DNA Micro kit (QIAGEN) following the manufacturer's tissue protocol. Negative controls were included and all work with historic samples was carried out in a dedicated historic DNA laboratory where no contemporary mockingbird DNA had ever been present. The laboratory had an independent air-handling system, was under positive air displacement and was irradiated with UV light to destroy DNA following each laboratory session. The DNA concentration in the historic samples was measured through quantitative PCR (QPCR) using SYBR Green I detection format (Roche Diagnostics, Switzerland) by amplifying part of the 7 intron of the fibrinogen gene β-subunit (Prychitko & Moore 1997). Using the FIB-BI7U and FIB-BI7L primers developed by Prychitko & Moore (1997), we sequenced M. trifasciatus DNA to design two new primers for QPCR, NesFib7F (5′-CTGGATGCAATAGTCAGAGACTG-3′) and NesFib7R (5′-CCTGCCTCTTTCTTCAGGAC-3′), in order to reduce the amplicon length to 104 bp. The ABI 7500 Fast Real-Time PCR System (Applied Biosystems) was used for QPCR amplification and detection. Negative controls were included in the experimental runs and 1–2 replicates were done for each historic sample. QPCR was prepared in a 20 µl reaction volume containing 10 µl of FastStart Universal SYBR Green Master (ROX), 300 nM of each primer and 2 µl template DNA following the operator's manual for PCR conditions. DNA concentrations were determined using a standard curve consisting of 11 dilutions (of modern M. trifasciatus DNA) ranging from 0.005 to 20 ng µl−1.
PCR amplification of the 17 microsatellites was carried out as described in Hoeck et al. (2009) with the exception that the total reaction volume of 5 µl contained 2.5 µl Multiplex PCR Master Mix (QIAGEN) and 2 µl of template historic DNA. Negative controls were included to monitor potential contamination. PCR conditions were changed slightly from the protocol described in Hoeck et al. (2009), with an initial denaturation step of only 12 min followed by 38 cycles of amplification at 59°C for all four panels. To assure reliable genotyping of the historic samples, PCR amplification was replicated four times for each sample at each locus. This should be sufficient as 2–3 replicates have previously been shown to accurately score the genotype in 99 per cent of sample- and locus-combinations in museum samples containing reasonable amounts of DNA (Sefc et al. 2003). Fragment analyses and genotyping were done as described above. The software Gimlet (Valiere 2002) was used to determine dropout and genotyping error rates per locus as well as consensus genotypes for each sample based on the four replicates.
(c). Diversity within populations
Deviations from Hardy–Weinberg equilibrium (HWE) for each locus were tested with allele randomizations within samples (1000 permutations per test) and overall samples (10 000 permutations) using FStat 2.9.3.1 package (Goudet 2001) and Bonferroni corrections. Genotypic equilibrium between all pairs of loci in each population was tested using G-statistics with Bonferroni corrections (FStat; 84 000 permutations). To describe within-population genetic diversity, we calculated standard parameters such as mean number of alleles (Na), allelic richness (AR, standardized to the smallest sample size), observed (Ho) and expected heterozygosity (He) and polymorphism (P) in FStat and Genetix v 4.05 (Belkhir et al. 2004).
(i). Contemporary populations
We tested for the predicted positive correlation between genetic diversity and population size using He, AR and P as estimators of genetic diversity. Because no empirical information on current population sizes was available except for the two M. trifasciatus populations, we used island size as a surrogate for population size (Frankham 1996). In a multiple regression analysis, we entered island size as an explanatory variable and the within-population indices of genetic diversity as dependent variables in separate analyses. As more isolated islands are less likely to receive gene flow than islands situated at the centre of the archipelago and older island populations might have lost more genetic diversity due to drift and reduced gene flow, we entered island isolation and island age as further explanatory variables. Average isolation for each island was calculated by adding up nearest shore-to-shore distances to all other islands and dividing the sum by the total number of islands minus one (Hamilton & Rubinoff 1967). For island age, we used the youngest age estimate for each island (D. Geist 2005–2008, unpublished data). The three explanatory variables (island size, isolation and age) were not correlated (all r2 < 0.18). Island size was ln-transformed, but all other variables and their residuals showed no significant deviation from normality.
(ii). Temporal change within populations
We performed a Wilcoxon signed-rank test with the variables He, AR and P to test whether these within-population indices of genetic diversity changed significantly over the last century. Based on the assumption that genetic drift is stronger in smaller and more isolated populations, we also investigated whether change in genetic diversity was dependent on island size or isolation. To this end, we performed a multiple regression analysis using size and isolation as explanatory variables and the relative change in He, AR and P between the historic and contemporary populations as dependent variables (i.e. 1−HeContemporary/HeCAS, etc). To quantify the change in gene frequencies within each island since 1906 (i.e. temporal differentiation), we calculated Weir & Cockerham's (1984) estimator τ for Wright's FST (Genepop on the web v. 3.4; Raymond & Rousset 1995) for each CAS-contemporary population pair (called ‘temporal FST’ below) and related it to island size or isolation, respectively, in a linear regression analysis. We chose FST to estimate temporal differentiation within islands because of the relatively small time scale involved (approx. 25 generations assuming a generation time of 4 years; Grant et al. 2000). Over such short time scales drift is the dominant process creating local differentiation and the effects of mutation are minimal (Slatkin 1995).
(iii). Effective population size
In the absence of migration, selection and mutation, effective population size can be estimated using temporal changes in allele frequencies (Wang 2001). We used our temporal dataset and the Bayesian coalescent-based method implemented in the program CoNe (Anderson 2005) to calculate the variance effective population size (Ne) for each island population, setting the time between the two sampling periods to 25 generations, the likelihood range for Ne between 2 and 20 000 in steps of 5, and using 1000 Monte Carlo replications.
(d). Differentiation among populations and species
(i). Pair-wise population differentiation
Differentiation over all loci for all contemporary population pairs was estimated using Nei's standard genetic distance Ds (Nei 1972) calculated in Populations v. 1.2.30 (Langella 2000). We chose Ds because it allows for mutation and increases more linearly with time than FST when considering large time scales and, hence, is a more accurate estimator when estimating evolutionary times (Takezaki & Nei 1996). Furthermore, Ds does not assume a specific mutation model and has been shown to perform well with microsatellite data (e.g. Takezaki & Nei 1996; Paetkau et al. 1997; Petren et al. 1999). FST (Weir & Cockerham 1984) was calculated for comparison. Including only populations of M. parvulus to avoid species bias, we also tested whether overall differentiation between peripheral islands (table 1) was higher than between centrally located islands in the archipelago. We tested for isolation-by-distance by contrasting geographical distances and multi-locus Ds-values between all contemporary population pairs of M. parvulus and, separately, also between all populations of all four species, using a series of Mantel tests (1000 permutations; Raymond & Rousset 1995). Geographical distance was measured as the logarithm of each island's nearest shore-to-shore distance from the other islands in the archipelago (Hamilton & Rubinoff 1967; Google Earth v 5.0, Google Inc.).
(ii). Genetic affinities among species and populations
Genetic affinities among contemporary species and populations were described with a factorial correspondence analysis (FCA) on multilocus genotypes using Genetix v 4.05 (Belkhir et al. 2004). FCA displays the genetic differences among populations in a two-dimensional graphical space. Genetic distances among populations were also assessed by building an evolutionary tree based on Nei's Ds using UPGMA and performing 1000 bootstrap resamplings among loci with Populations v. 1.2.30 (Langella 2000).
All statistical analyses were done using JMP v. 8 (SAS Institute Inc., Cary, NC).
3. Results
(a). Genotyping
Amplification and genotyping of the contemporary samples was very successful reaching nearly 100 per cent, with only one locus not amplifying in a single individual. Genotyping error and dropout rates in the contemporary samples were below 0.1 per cent. Not surprisingly, amplification success for the historic samples was lower, most probably due to the much lower DNA concentrations (0.01–254 pg µl−1 with an average of 28 pg µl−1) and lower DNA quality of historic samples in general (Wandeler et al. 2007). On average, 78 per cent of the PCR reactions with the historic samples resulted in successful amplification (across individuals and loci) and the combined allelic dropout and false allele rates of all four replicates of the historic samples were seven per cent (s.d. = 0.08) and 2.8 per cent (s.d. = 0.04), respectively. Consensus genotypes were cross-checked for reliability by hand. Additionally, blank negative controls confirmed that cross-contamination was negligible. Forty-two individuals from the CAS collection amplified successfully for less than 10 loci and were therefore excluded from all further analyses.
(b). Diversity within populations
Fourteen of 17 loci were in HWE in all populations, and no genotypic disequilibrium was detected for any pairs of loci in any population. Nes22 significantly deviated from HWE in various populations (San Cristóbal-CAS, Santiago-CAS, Marchena and Marchena-CAS) and was therefore excluded from all further analyses. Nes16 and Nes04 deviated significantly from HWE in a single population each (Española-CAS and Santiago, respectively) showing an excess of homozygotes. If null alleles were the cause of these deviations, we would expect to find other populations out of HWE for these loci as well. Therefore, these deviations most probably reflect substructure within the populations although an overall Wahlund effect is unlikely with only one locus out of HWE.
We identified a total of 143 alleles across all 16 remaining loci (table 1), with individual loci having between 3 and 19 alleles (average: 8.9 alleles) and individual populations having between 1 and 11 alleles per locus (average: 3.1 alleles). Genetic diversity, measured as mean number of alleles (Na), allelic richness (AR) and heterozygosities (He and Ho), varied greatly between different populations (table 1 and figure 2). Ho ranged from 0.08 to 0.59 (mean: 0.35) and correlated strongly (r2 = 0.95, p < 0.0001) with He (range: 0.07–0.59; mean: 0.37). Average AR ranged between 1.2 and 3.6 and P between 25 and 100 per cent. Na and AR correlated strongly (r2 = 0.95, p < 0.0001) indicating that both measures are equally suited to describe genetic diversity. Overall, Champion and Gardner-by-Española showed the lowest and Santiago and St Cruz the highest estimates of genetic diversity (table 1 and figure 2).
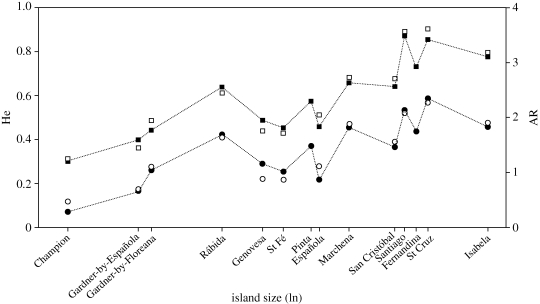
Figure 2.
Expected heterozygosity (He) and allelic richness (AR) of contemporary and historic populations as a function of the natural logarithm of island size (in ha). To improve visual representation, points that overlapped were slightly moved (filled circle, contemporary He; open circle, historic He; filled square, contemporary AR; open square, historic AR).
(i). Contemporary populations
All measures of genetic diversity were significantly related to island size (He: F1,14 = 15.9, b = 0.03 ± 0.01, p = 0.001; AR: F1,14 = 33.4, b = 0.14 ± 0.05, p < 0.0001; P: F1,14 = 12.1, b = 0.04 ± 0.01, p = 0.004; figure 2) and island age (He: F1,14 = 6.3, b = −0.05 ± 0.02, p = 0.025; AR: F1,14 = 8.7, b = −0.21 ± 0.07, p = 0.011; P: F1,14 = 6.4, b = −0.08 ± 0.03, p = 0.024) but not isolation (He: F1,14 = 3.1, b = −0.0014 ± 0.0008, p = 0.101; P: F1,14 = 1.7, b = −0.002 ± 0.001, p = 0.22) except for AR (F1,14 = 5.6, b = -0.007 ± 0.003, p = 0.033). The results showed an overall pattern of genetic diversity increasing significantly with island size and decreasing with island age and, at least for AR, also with isolation.
(ii). Temporal change within populations
Overall, within-population genetic diversity estimates of the CAS-populations were not significantly different from the contemporary populations (Wilcoxon: He, p = 0.67; AR, p = 0.42; P, p = 1.0), indicating that archipelago-wide genetic diversity did not change significantly since 1906 (figure 2). Also, we detected no significant relationship between island size or isolation and change in He, AR or P over the last century (all p-values above 0.34). However, when studying the genetic diversity estimates for individual populations, it becomes evident that changes did occur in some cases and some contemporary populations have individually lost or gained genetic diversity: the Champion population lost 39 per cent, Española 22 per cent and the two Gardners 4–5% of their expected heterozygosity during the last 100 years, whereas He for the populations on Genovesa and St Fé increased by 32 and 17 per cent, respectively (table 1 and figure 2).
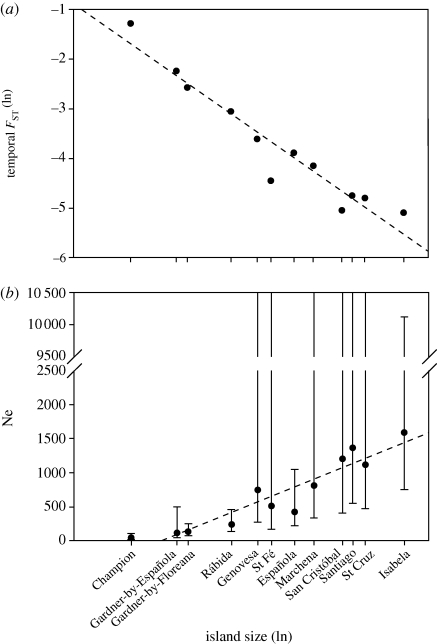
Using temporal FST, which quantifies the change in gene frequencies within each island since 1906, we found that the degree of genetic differentiation was significantly negatively correlated with island size (r2 = 0.93, p < 0.0001) but not island isolation (r2 = 0.14, p = 0.23). Genetic differentiation was stronger in smaller than in larger populations during the last 100 years as expected from genetic drift (figure 3a; see the electronic supplementary material).
Figure 3.
(a) Temporal FST between contemporary and historic populations as a function of island size (in ha; log scale). Differentiation clearly increased more strongly in smaller populations as shown by the dotted regression line (FST = −0.899–0.354 * log(island size)). Overlapping data points from islands with similar size were slightly modified to improve visual representation. (b) Estimates of effective population size (Ne) with lower and, where available, upper estimation limits plotted against island size (in ha; log scale). The dotted line shows the linear regression line (Ne = −507.07 + 148.94 * log(island size)). A strong positive linear correlation was also found when both Ne and island size were ln-transformed (r2 = 0.92, p < 0.0001).
(iii). Effective population size
We were able to estimate effective population size for all 12 populations for which we had temporal samples. The lowest maximum likelihood Ne estimate was 43 individuals for Champion, and the highest was 1591 individuals for Isabela, reflecting the smallest and largest islands investigated (see the electronic supplementary material). In six cases no upper confidence interval could be calculated, resulting in an infinite upper support limit. Ne estimates were strongly positively correlated with island size (r2 = 0.88, p < 0.0001; figure 3b).
(c). Differentiation among populations and species
(i). Pair-wise population differentiation
Pair-wise differentiation (Ds) between all contemporary populations and species from different islands ranged from 0.004 to 1.988 (see the electronic supplementary material). Pair-wise Ds and FST correlated strongly (r2 = 0.75, p < 0.0001; see the electronic supplementary material) and qualitatively provided the same results. In general, the highest values occurred between populations belonging to different mockingbird species (mean Ds = 0.86 ± s.d. 0.37). Ds-values were lower for within-species comparisons (mean Ds = 0.37 ± s.d. 0.19) and lowest for comparisons between localities within islands (mean Ds = 0.04 ± s.d. 0.03). However, differentiation between the Alcedo (AL) population on Isabela and the population on neighbouring Fernandina (Ds = 0.024) was lower than the differentiation between the two sites on Isabela (AL and VL, Ds = 0.042). Also, differentiation between Isabela VL and Fernandina (Ds = 0.037) was slightly lower than between Isabela AL and VL. We found that overall differentiation between M. parvulus populations was higher among peripheral than among centrally located islands (mean peripheral Ds = 0.54 ± s.d. 0.12 versus mean central Ds = 0.32 ± s.d. 0.17). The Mantel test showed a highly significant relationship between genetic differentiation and geographical distance for pairs of M. parvulus populations (r2 = 0.22, p = 0.001) and also across populations of all four species (r2 = 0.16, p = 0.001). Thus, isolation-by-distance, i.e. an increase in genetic differentiation with increasing between-island distances, was evident.
(ii). Genetic affinities among species and populations
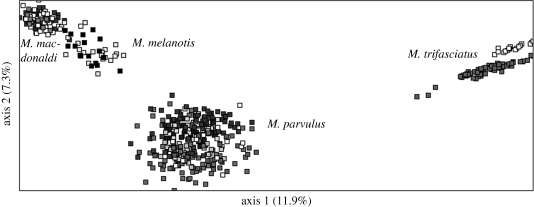
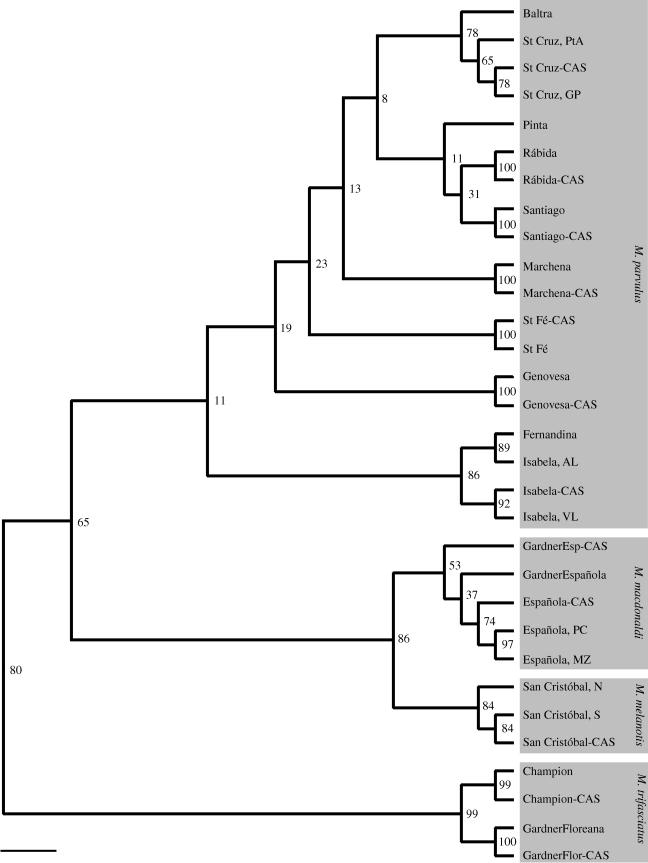
Genetic differences among contemporary populations based on an FCA analysis revealed three main clusters, with M. trifasciatus most clearly differentiated from the other species (figure 4), all populations of M. parvulus forming a second cluster and M. macdonaldi and M. melanotis together forming a third cluster. Similar population relationships were found in the UPGMA tree, which showed all populations of M. parvulus separated from the other three species, populations of M. macdonaldi most closely together with M. melanotis and M. trifasciatus forming a separate branch (figure 5). As we were unable to root our tree due to the lack of microsatellite data from a related species, we cannot show the evolutionary position of the clusters.
Figure 4.
A two-dimensional diagram representing the relationships between the four mockingbird species based on a factorial correspondence analysis on multilocus genotypes. Only the first two axes are represented with the percentage of variance explained by the axes in parentheses.
Figure 5.
Unrooted UPGMA tree based on Nei's Ds with bootstrap values over loci. The shaded areas behind the population names represent the four different Mimus species. Scale bar, 0.1.
4. Discussion
Our analyses revealed that genetic drift has strongly shaped the distribution of genetic variance within and between mockingbird populations and species. Furthermore, on an archipelago-wide scale genetic diversity did not change over the past 100 years, suggesting that overall the mockingbird populations in Galápagos are in or close to migration-drift equilibrium at neutral loci.
(a). Diversity within populations
Levels of genetic diversity varied greatly among mockingbird populations (table 1). Overall, the amount of genetic diversity was lower in M. trifasciatus and M. macdonaldi than in M. melanotis and, especially, M. parvulus (table 1 and figure 2). These results reflect the wide distribution of M. parvulus in the archipelago (figure 1) and are in line with the general finding of increased genetic diversity on larger islands. Additionally, the lower genetic diversity in the three range-restricted species might also reflect their occurrence on older and more isolated islands. Island age, and to a lesser degree, island isolation were also found to affect levels of genetic diversity. The positive relationship between genetic diversity and island size and the negative relationship with island isolation and age suggest that gene flow is generally limited. The pattern found here is unlikely due to the older age and smaller size of peripheral islands because we found no significant correlation between the explanatory variables island size, isolation and age. However, the effects of gene flow seem to depend on the neighbouring islands. Smaller islands that are adjacent to islands with high levels of genetic diversity harboured considerably higher levels of genetic diversity than expected for their size (e.g. Rábida and Baltra; figure 2 and table 1). On the other hand, although close to a larger island, Gardner-by-Española did not have elevated levels of genetic diversity presumably because Española itself showed low levels of genetic diversity.
Levels of genetic diversity in the mockingbirds were much lower than those found in other Galápagos bird species such as Darwin's finches (Petren et al. 2005) or the Galápagos dove (Santiago-Alarcon et al. 2006). This could be due to the mockingbirds’ relatively sedentary behaviour in comparison to the much more vagile finches and doves. In addition, mockingbirds may have smaller effective population sizes (see below). Comparing levels of genetic diversity at microsatellite loci across species is often hampered by ascertainment bias resulting from the selection for polymorphism during marker development (Brandstrom & Ellegren 2008). In cross-species comparisons, this ascertainment bias can lead to artifactual differences because the loci will be more polymorphic in the species in which the microsatellite loci were developed (e.g. Ellegren et al. 1995). In designing our microsatellite loci, we deliberately avoided creating such ascertainment bias among the four mockingbird species in Galápagos by including all loci that were polymorphic in at least one of the species in our panels (Hoeck et al. 2009). However, when comparing the mockingbirds to other bird species in Galápagos we cannot rule out that ascertainment bias affects our conclusions.
Overall there was little difference in estimates of genetic diversity between the CAS- and contemporary populations. However, change in diversity was considerable in a few cases (figure 2): the Champion and Española populations lost a substantial amount of He, clearly showing that these populations are individually not in drift–gene-flow equilibrium. Interestingly, levels of He were higher in the contemporary populations on Genovesa and St Fé than in the historic ones, a result that is less intuitive. We can rule out genotyping errors in the CAS samples as a major cause of these findings because estimates of allelic dropout rates among the historic samples on these two islands were very small. It seems more likely that these increases were due to biased sampling in the field, genetic drift or immigration. The occurrence of immigration is a possible explanation for the contrasting nuclear and mtDNA patterns found in the Genovesa population (see discussion below).
Overall, allele frequency distributions changed more in smaller than in larger populations: within-island genetic differentiation between 1906 and the present was much stronger on smaller islands (figure 3a), indicating more pronounced genetic drift in small populations. However, despite these clear signals of genetic drift, absolute levels of genetic diversity changed remarkably little on an archipelago-wide scale between 1906 and today. This is perhaps not so surprising since it simply implies that overall Galápagos mockingbird populations are in migration–drift equilibrium at neutral loci. Two other studies of undisturbed island populations (Taylor et al. 2007; Pertoldi et al. 2008) have similarly shown stability in genetic diversity over time, suggesting that our finding of migration–drift equilibrium at neutral loci may not be unusual for islands without major anthropogenic disturbances.
Estimates of effective population size (Ne) were strongly related to island size, suggesting that the latter is a reliable estimator of Ne (figure 3b). On the two islands where we had data on census sizes and estimates of Ne, the two were in reasonable agreement (Champion: Nc = 20–50, Ne = 43 (95% CI: 17–107), Gardner-by-Floreana: Nc ≈ 300–500, Ne = 133 (95% CI: 75–245)). The strong correlation between Ne and island size indicates that mockingbird habitats are quite equally distributed throughout the different islands despite the vast areas of lava occurring on some of the larger islands. The more humid transitional zones at higher altitudes that only occur on these larger islands and provide good mockingbird habitat might compensate for the lack of habitat in lava field areas. In addition, the fact that the relationship between island size and Ne was linear (figure 3b) suggests that subdivision within island was not very strong, a view supported by the significant but small degrees of differentiation between populations on the same island (see the electronic supplementary material).
Ne estimates found in this study ranged from 47 to 1591 with a mean of 692 (see the electronic supplementary material). Thus, all but five mockingbird populations in Galápagos have Ne values at levels currently thought to represent viable populations (Ne = 500–5000; e.g. Franklin & Frankham 1998). On smaller islands such as Rábida and Gardner-by-Española Ne estimates are lower, most probably due to restricted territorial space, but occasional gene flow from the adjacent larger populations probably contributes to maintaining genetic diversity. In contrast, since the extinction of the population on Floreana, this is no longer possible for the two M. trifasciatus populations. As a consequence, Champion in particular experienced low Ne and loss of genetic diversity over the past 100 years.
(b). Differentiation among populations and species
Genetic differentiation among contemporary populations was high for most population pairs (see the electronic supplementary material). As expected, we found differentiation to be strongest between populations belonging to different species and lowest between populations of the same species on adjacent islands. The low differentiation found between the two M. macdonaldi populations suggests gene flow between Española and its satellite population. Levels of differentiation between the two M. trifasciatus populations were about 10 times higher, of a magnitude comparable to those of between-species comparisons. This pattern can be explained by the very low population size on Champion resulting in strong genetic drift, and by a lack of gene flow between Gardner and Champion since the extinction of the population on Floreana (Hoeck et al. in press). In addition, the low percentage of polymorphic loci on Champion may also have contributed to the high FST estimates (Hedrick 1999). We also measured within-island differentiation on four islands and found that it was lower than between-island differentiation for three of them. The higher differentiation within Isabela than among Isabela and Fernandina (see the electronic supplementary material) suggests that mockingbird dispersal is affected more by geographical proximity than separation by water, a view also reflected in the general isolation-by-distance pattern detected in this study. However, the substantial within-island genetic diversity on Isabela and Fernandina could also contribute to the low Ds (FST) values (Hedrick 1999).
We found significant isolation-by-distance among populations of all four species and also within M. parvulus. Under isolation-by-distance, the most striking differences are expected to occur between peripheral populations, a pattern that was corroborated by the higher pair-wise Ds-values between peripheral mockingbird populations compared with central ones. Stronger genetic differentiation between peripheral and geographically more distant populations was also detected in cactus and ground finches (Petren et al. 2005), but not in warbler finches where dispersal was limited by habitat similarity (Tonnis et al. 2005).
As with genetic diversity, overall levels of genetic differentiation among mockingbird populations contrast with that found for other species in the Galápagos, which show much lower inter-population differentiation (e.g. Ciofi et al. 2002; Petren et al. 2005; Santiago-Alarcon et al. 2006). However, our findings are comparable to levels of differentiation found between Galápagos hawk (Bollmer et al. 2005) or land iguana populations (Tzika et al. 2008), for example, reflecting the effects of pronounced genetic drift and restricted gene flow.
Genetic affinities among species. Our multilocus microsatellite data revealed three major clusters, with M. macdonaldi and M. melanotis grouping closely together, all M. parvulus populations forming a second group, and M. trifasciatus, the most distant of the four species, forming a third group (figure 4). The UPGMA tree confirmed this pattern (figure 5). Phylogenetic analyses based on mtDNA identified four distinct clades which differ from current taxonomy (Arbogast et al. 2006). Mimus macdonaldi, M. melanotis and M. parvulus from Genovesa clustered on the same branch despite belonging to three different species and formed the most distant branch within the Galápagos mockingbird genus. All other M. parvulus grouped in a second branch except for individuals from Isabela which formed a third phylogenetically divergent class. Mimus trifasciatus formed a fourth branch. Our micorsatellite data confirm this pattern except that individuals from Genovesa did not group with M. macdonaldi and M. melanotis but instead with the other M. parvulus populations (figure 5). Differential introgression of mitochondrial and nuclear genes might be responsible for this pattern (Arbogast et al. 2006). Alternatively, the discrepancy between the nuclear and mitochondrial data could arise from the differing lineage sorting times of the two types of markers. Given the recent evolutionary history of the Galápagos bird fauna, contrasting nuclear and mtDNA patterns are not surprising and have indeed been found in other phylogenies of Galápagos birds (Petren et al. 1999, 2005; Bollmer et al. 2006).
Based on mtDNA data, Arbogast et al. (2006) suggested that Galápagos mockingbirds diverged approximately 1.6–5.5 Myr ago following a single colonization event, thus forming a monophyletic clade. Our data do not provide information about the colonization since we did not include taxa from outside Galápagos as outgroups in our study. However, we can compare the divergence patterns within Galápagos between the published mtDNA and our microsatellite data. The mtDNA data suggested that M. macdonaldi, M. melanotis and M. parvulus from Genovesa were the first to split from all others (Arbogast et al. 2006), while our microsatellite data suggest that M. trifasciatus diverged before M. macdonaldi and M. melanotis (figure 5). Both patterns of diversification generally match information about island age and the directionality of prevailing winds (Colinvaux 1984). Mimus trifasciatus, M. macdonaldi and M. melanotis inhabit the eastern-most and oldest islands of the Galápagos archipelago (figure 1 and table 1), likely to be colonized first. Within Galápagos, the phylogenetic pattern detected here is consistent with a model of wind-based dispersal following initial colonization, i.e. from the southeast to the northwest, as previously suggested by Arbogast et al. (2006). Further investigations with nuclear genetic markers will hopefully improve our understanding of the relationship between the Galápagos mockingbirds and their continental relatives, their time of divergence and rate of diversification, and resolve the contrasting pattern found between the mitochondrial and nuclear DNA.
(c). Conclusions
Using observations on a temporal and spatial scale, we have quantified the effects of small population size and drift on the genetic diversity and structuring at neutral loci of mockingbird populations in Galápagos. Temporally, we measured the change in genetic diversity over 100 years (approx. 25 generations) and spatially we compared many different-sized populations across the archipelago. Although archipelago-wide genetic diversity did not change significantly over the last century, genetic drift was pronounced in small populations where it led to substantial variation in allele frequencies over time and to loss of genetic diversity. This was particularly obvious in the tiny Champion population. The significant isolation-by-distance pattern implies that gene flow occurs but the high levels of interisland differentiation emphasize the existence of substantial barriers to gene flow between islands.
We conclude that for the mockingbird populations in Galápagos, genetic drift is strong and gene flow limited. Our results contrast with the findings in Darwin's finches where interisland migration is widespread, isolation-by-distance is weak and substantial genetic drift has only been found in populations of the warbler finch on small, peripheral islands (Petren et al. 2005; Tonnis et al. 2005). Future studies will have to show whether, in contrast to the situation in Darwin's finches, isolation and genetic drift may have contributed to the phenotypic divergence among mockingbird populations.
Acknowledgements
We thank the Galápagos National Park Service for permission to conduct this research, the Charles Darwin Research Station for logistical support and TAME airlines for providing scientists a reduced airfare. Thanks to the California Academy of Sciences to let us collect toe pad samples of the historic specimens. Our special thanks go to Felipe Cruz, Marilyn Cruz and Sonia Cisneros who provided invaluable support in Galápagos. Thanks to our field assistants Iris Biebach, Herbert Biebach, Wilson Cabrera, Stefan Henrich, María Fernanda Holguín, Michael Janssen, Karla Muñoz and Michèle Wegmann. Thomas Bucher, Peter Wandeler and Andrea Gubler provided invaluable help in the laboratory. Erik Postma, Iris Biebach, Sonia Angelone and Peter Wandeler gave very useful inputs on the analyses. Thanks to Peter and Rosemary Grant for helpful discussions and partial funding of our fieldwork. This research was funded by the Zoological Museum of the University of Zurich, the Forschungskredit of the University of Zurich, the Balzan Foundation (Balzan Prize 2005 awarded to Peter and Rosemary Grant) and the Basler Stiftung für biologische Forschung.
Footnotes
One contribution of 13 to a Theme Issue ‘Darwin's Galápagos finches in modern evolutionary biology’.
References
- Anderson E. C.2005An efficient Monte Carlo method for estimating N-e from temporally spaced samples using a coalescent-based likelihood. Genetics 170, 955–967 (doi:10.1534/genetics.104.038349) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arbogast B. S., Drovetski S. V., Curry R. L., Boag P. T., Seutin G., Grant P. R., Grant B. R., Anderson D. J.2006The origin and diversification of Galapagos mockingbirds. Evolution 60, 370–382 [PubMed] [Google Scholar]
- Barton N. H.1998Natural selection and random genetic drift as causes of evolution on islands. In Evolution on islands (ed. Grant P. R.). New York, NY: Oxford University Press [Google Scholar]
- Belkhir K., Borsa P., Chikhi L., Raufaste N., Bonhomme F.2004Genetix 4.05, logiciel sous Windows TM pour la génétique des populations. Montpellier, France: Université de Montpellier II [Google Scholar]
- Bollmer J. L., Whiteman N. K., Cannon M. D., Bednarz J. C., De Vries T., Parker P. G.2005Population genetics of the Galapagos hawk (Buteo galapagoensis): genetic monomorphism within isolated populations. The Auk 122, 1–15 [Google Scholar]
- Bollmer J. L., Kimball R. T., Whiteman N. K., Sarasola J. H., Parker P. G.2006Phylogeography of the Galapagos hawk (Buteo galapagoensis): a recent arrival to the Galapagos Islands. Mol. Phylogenet. Evol. 39, 237–247 (doi:10.1016/j.ympev.2005.11.014) [DOI] [PubMed] [Google Scholar]
- Brandstrom M., Ellegren H.2008Genome-wide analysis of microsatellite polymorphism in chicken circumventing the ascertainment bias. Genome Res. 18, 881–887 (doi:10.1101/gr.075242.107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciofi C., Milinkovitch M. C., Gibbs J. P., Caccone A., Powell J. R.2002Microsatellite analysis of genetic divergence among populations of giant Galapagos tortoises. Mol. Ecol. 11, 2265–2283 (doi:10.1046/j.1365-294X.2002.01617.x) [DOI] [PubMed] [Google Scholar]
- Colinvaux P. A.1984The Galapagos climate: past and present. In Galapagos (key environments) (ed. Perry R.). Oxford, UK: Pergamon [Google Scholar]
- Crow J. F., Kimura M.1970An introduction to population genetic theory New York, NY: Harper and Row Publishers [Google Scholar]
- Curry R. L.1986Whatever happened to the Floreana mockingbird? Noticias de Galapagos 43, 13–15 [Google Scholar]
- Darwin C.1839Voyage of the Beagle London, UK: Henry Colburn [Google Scholar]
- Darwin C. R.1859On the origin of species by means of natural selection, or the preservation of favoured races in the struggle for life London, UK: John Murray; [PMC free article] [PubMed] [Google Scholar]
- Ellegren H., Primmer C. R., Sheldon B. C.1995Microsatellite evolution—directionality or bias. Nat. Genet. 11, 360–362 (doi:10.1038/ng1295-360) [DOI] [PubMed] [Google Scholar]
- Frankham R.1996Relationship of genetic variation to population size in wildlife. Conserv. Biol. 10, 1500–1508 (doi:10.1046/j.1523-1739.1996.10061500.x) [Google Scholar]
- Frankham R.1997Do island populations have less genetic variation than mainland populations? Heredity 78, 311–327 (doi:10.1038/hdy.1997.46) [DOI] [PubMed] [Google Scholar]
- Franklin I. R., Frankham R.1998How large must populations be to retain evolutionary potential? Anim. Conserv. 1, 69–70 (doi:10.1111/j.1469-1795.1998.tb00228.x) [Google Scholar]
- Goudet J.2001FStat, a program to estimate and test gene diversities and fixation indices (version 2.9.3). Lausanne, Switzerland: Lausanne University [Google Scholar]
- Grant P. R.1998Evolution on islands New York, NY: Oxford University Press [Google Scholar]
- Grant P. R., Curry R. L., Grant B. R.2000A remnant population of the Floreana mockingbird on Champion island, Galapagos. Biol. Conserv. 92, 285–290 (doi:10.1016/S0006-3207(99)00092-0) [Google Scholar]
- Hamilton T. H., Rubinoff I.1967On predicting insular variation in endemism and sympatry for Darwin finches in Galapagos Archipelago. Am. Nat. 101, 161–171 [Google Scholar]
- Harris M. P.1974The birds of Galapagos New York, NY: Houghton-Mifflin [Google Scholar]
- Hedrick P. W.1999Perspective: highly variable loci and their interpretation in evolution and conservation. Evolution 53, 313–318 (doi:10.2307/2640768) [DOI] [PubMed] [Google Scholar]
- Hoeck P. E. A., Bucher T. B., Wandeler P., Keller L. F.2009Microsatellite primers for the four Galápagos mockingbird species (Mimus parvulus, Mimus macdonaldi, Mimus melanotis, and M. trifasciatus). Mol. Ecol. Resources 9, 1538–1541 (doi:10.1111/j.1755-0998.2009.02704.x) [DOI] [PubMed] [Google Scholar]
- Hoeck P. E. A., Beaumont M. A., James K. E., Grant B. R., Grant P. R., Keller L. F.In press Saving Darwin's muse: evolutionary genetics for the recovery of the Floreana mockingbird. Biol. Lett. (doi:10.1098/rsbl.2009.0778) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes C. R., Deloach D. M.1997Developing microsatellites when they are rare: trinucleotide repeat loci in the northern mockingbird Mimus polyglottos. Mol. Ecol. 6, 1099–1102 (doi:10.1046/j.1365-294X.1997.d01-109.x) [DOI] [PubMed] [Google Scholar]
- Langella O.2000Populations 1.2.30. Laboratoire Populations, Génétique et Evolution, Centre National de la Recherche Scientifique, CNRS UPR9034, Gif Sur Yvette, France [Google Scholar]
- Nei M.1972Genetic distance between populations. Am. Nat. 106, 283–292 [Google Scholar]
- Paetkau D., Waits L. P., Clarkson P. L., Craighead L., Strobeck C.1997An empirical evaluation of genetic distance statistics using microsatellite data from bear (Ursidae) populations. Genetics 147, 1943–1957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palstra F. P., Ruzzante D. E.2008Genetic estimates of contemporary effective population size: what can they tell us about the importance of genetic stochasticity for wild population persistence? Mol. Ecol. 17, 3428–3447 (doi:10.1111/j.1365-294X.2008.03842.x) [DOI] [PubMed] [Google Scholar]
- Pertoldi C., Barker S. F., Madsen A. B., Jorgensen H., Randi E., Munoz J., Baagoe H. J., Loeschcke V.2008Spatio-temporal population genetics of the Danish pine marten (Martes martes). Biol. J. Linn. Soc. 93, 457–464 (doi:10.1111/j.1095-8312.2007.00892.x) [Google Scholar]
- Petren K., Grant B. R., Grant P. R.1999A phylogeny of Darwin's finches based on microsatellite DNA length variation. Proc. R. Soc. Lond. B 266, 321–329 (doi:10.1098/rspb.1999.0641) [Google Scholar]
- Petren K., Grant P. R., Grant B. R., Keller L. F.2005Comparative landscape genetics and the adaptive radiation of Darwin's finches: the role of peripheral isolation. Mol. Ecol. 14, 2943–2957 (doi:10.1111/j.1365-294X.2005.02632.x) [DOI] [PubMed] [Google Scholar]
- Prychitko T. M., Moore W. S.1997The utility of DNA sequences of an intron from the beta-fibrinogen gene in phylogenetic analysis of woodpeckers (Aves: Picidae). Mol. Phylogenet. Evol. 8, 193–204 (doi:10.1006/mpev.1997.0420) [DOI] [PubMed] [Google Scholar]
- Raymond M., Rousset F.1995Genepop (v. 1.2)—population-genetics software for exact tests and ecumenicism. J. Heredity 86, 248–249 [Google Scholar]
- Santiago-Alarcon D., Tanksley S. M., Parker P. G.2006Morphological variation and genetic structure of Galapagos Dove (Zenaida galapagoensis) populations: issues in conservation for the Galapagos bird fauna. Wilson J. Ornithol. 118, 194–207 (doi:10.1676/05-010.1) [Google Scholar]
- Sefc K. M., Payne R. B., Sorenson M. D.2003Microsatellite amplification from museum feather samples: effects of fragment size and template concentration on genotyping errors. The Auk 120, 982–989 (doi:10.1642/0004-8038(2003)120[0982:MAFMFS]2.0.CO;2) [Google Scholar]
- Slatkin M.1985Gene flow in natural populations. Annu. Rev. Ecol. Syst. 16, 393–430 (doi:10.1146/annurev.ecolsys.16.1.393) [Google Scholar]
- Slatkin M.1995A measure of population subdivision based on microsatellite allele frequencies. Genetics 139, 457–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takezaki N., Nei M.1996Genetic distances and reconstruction of phylogenetic trees from microsatellite DNA. Genetics 144, 389–399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor S. S., Jamieson I. G., Wallis G. P.2007Historic and contemporary levels of genetic variation in two New Zealand passerines with different histories of decline. J. Evol. Biol. 20, 2035–2047 (doi:10.1111/j.1420-9101.2007.01362.x) [DOI] [PubMed] [Google Scholar]
- Tonnis B., Grant P. R., Grant B. R., Petren K.2005Habitat selection and ecological speciation in Galapagos warbler finches (Certhidea olivacea and Certhidea fusca). Proc. R. Soc. B 272, 819–826 (doi:10.1098/rspb.2004.3030) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzika A. C., et al. 2008Population genetics of Galapagos land iguana (genus Conolophus) remnant populations. Mol. Ecol. 17, 4943–4952 (doi:10.1111/j.1365-294X.2008.03967.x) [DOI] [PubMed] [Google Scholar]
- Valiere N.2002Gimlet: a computer program for analysing genetic individual identification data. Mol. Ecol. Notes 2, 377–379 (doi:10.1046/j.1471-8286.2002.00228.x) [Google Scholar]
- Wandeler P., Hoeck P. E. A., Keller L. F.2007Back to the future: museum specimens in population genetics. Trends Ecol. Evol. 22, 634–642 (doi:10.1016/j.tree.2007.08.017) [DOI] [PubMed] [Google Scholar]
- Wang J. L.2001A pseudo-likelihood method for estimating effective population size from temporally spaced samples. Genet. Res. 78, 243–257 (doi:10.1017/S0016672301005286) [DOI] [PubMed] [Google Scholar]
- Weir B. S., Cockerham C. C.1984Estimating F-statistics for the analysis of population structure. Evolution 38, 1358–1370 (doi:10.2307/2408641) [DOI] [PubMed] [Google Scholar]
- Wright S.1943Isolation by distance. Genetics 28, 114–138 [DOI] [PMC free article] [PubMed] [Google Scholar]