Abstract
Genetic analysis of museum specimens offers a direct window into a past that can predate the loss of extinct forms. We genotyped 18 Galápagos finches collected by Charles Darwin and companions during the voyage of the Beagle in 1835, and 22 specimens collected in 1901. Our goals were to determine if significant genetic diversity has been lost since the Beagle voyage and to determine the genetic source of specimens for which the collection locale was not recorded. Using ‘ancient’ DNA techniques, we quantified variation at 14 autosomal microsatellite loci. Assignment tests showed several museum specimens genetically matched recently field-sampled birds from their island of origin. Some were misclassified or were difficult to classify. Darwin's exceptionally large ground finches (Geospiza magnirostris) from Floreana and San Cristóbal were genetically distinct from several other currently existing populations. Sharp-beaked ground finches (Geospiza difficilis) from Floreana and Isabela were also genetically distinct. These four populations are currently extinct, yet they were more genetically distinct from congeners than many other species of Darwin's finches are from each other. We conclude that a significant amount of the finch biodiversity observed and collected by Darwin has been lost since the voyage of the Beagle.
Keywords: ancient DNA, historical, microsatellite, NHC, population structure, SSR
1. Introduction
It is widely acknowledged that Charles Darwin was not impressed by the Galápagos finches during HMS Beagle's tour of the Galápagos in 1835. While he was marvelling at the differences among island forms of the mockingbirds, he overlooked the task of recording which islands most of his finch specimens came from (Sulloway 1982). Darwin thought the Galápagos finches were formed by perhaps five separate colonizations of very different looking mainland taxa. It was not until sometime later that John Gould informed him that all were finches and they were closely related to each other (Sulloway 1982). There are some indications that Darwin wanted to use the finches as an example in his Origin of species because he felt sure that they would support the pattern of different forms occurring on different islands, yet he was uncertain that his reconstructions of island origins were accurate, and the resulting patterns did not present a strong case for natural selection (Sulloway 1983a,b). Thus, the finches that bear his name went unmentioned in Darwin's major contribution to science and humanity (Darwin 1859).
The voyage of the Beagle occurred during a time when whaling ships frequented the Pacific. In the Galápagos, tortoises were harvested and goats and other domesticated animals were left to breed and then hunted on subsequent visits to provide fresh meat. However, the Galápagos were unusual in that no major or permanent human settlements were established until the very end of the 1800s (Lundh 2001). Population growth was limited until the latter part of the 1900s and remained small until the recent surge in residents, tourists and introduced species (Watkins & Cruz 2007). The delayed history of human settlement has left the Galápagos unusually intact, and it provides a valuable model for understanding how humans affect island ecosystems. Although no species of Darwin's finch is known to have become extinct, at least a dozen populations appear to have gone extinct, including several that were collected during the Beagle voyage (Grant 1999, p. 53; Grant et al. 2005a; unpublished field surveys). A full understanding of what has been lost since Darwin's time may provide insights about the current state and future of this valuable model system.
DNA sequence analysis of museum specimens has been used widely to understand the evolutionary history of extinct forms (Herrmann & Hummel 1994; Hedrick & Waits 2005). Historical specimens can also be used to assess trends in population growth and decline over time (Westemeier et al. 1998; Groombridge et al. 2000), which is an important consideration for conservation (Allendorf & Luikart 2007). Given the numbers of specimens available for cross-temporal comparisons, and the wide use of microsatellite length variation for demographic studies (Ellegren 2004), it is surprising that relatively few studies have used microsatellite length variation to assess trends in demography over time (Wandeler et al. 2007). The limited application of this approach probably arises from the generally poor quality of DNA obtained from older museum specimens along with the need to successfully amplify several loci and two alleles per locus. Even a large investment to circumvent these problems may not fully remove the uncertainty from individual genotypes. Studies that assess several different predicted patterns for larger numbers of specimens may therefore provide a valuable guide for how to extract information from multilocus genotypes from museum specimens (Gilbert et al. 2005; Wandeler et al. 2007).
Our main goal was to understand the degree to which genetic diversity in Darwin's finches has been lost since the voyage of the Beagle. We reconstructed multilocus genotypes from the specimens collected by Charles Darwin and his shipmates, along with relevant specimens that were collected on later expeditions. Using a stratified sampling strategy to address the challenges associated with PCR of poorly preserved specimens, we assessed whether the genetic information obtained appears to be robust, whether populations that are suspected of extinction ever existed in the first place and whether extinct populations were genetically different from currently existing populations. A further goal was to help resolve the issue of the island origin of several historically important specimens and to determine if there is support for the inferred island of origin in several instances.
This study is focused on populations of two species: the large ground finch (Geospiza magnirostris) and the sharp-beaked ground finch (Geospiza difficilis). These species occur as several distinct island populations (figure 1), many of which are morphologically different from other populations and many of which are now extinct (Grant 1999, ch. 4). Specimens from two populations of large ground finches on San Cristóbal and Floreana that were collected in 1835 have beak sizes that are larger than any other Darwin's finch, and among the largest known for seed-eating birds (Grant 1999, p. 88). By virtue of their morphological distinctness and the fact that these islands are visited frequently, these two populations are almost certainly extinct, yet their genetic distinctness remains unknown. We tested the hypothesis that their morphological distinctness reflects genetic distinctness and, for this purpose, included an analysis of populations collected during later expeditions in 1900–1901 from the remote islands of Darwin and Wolf that are currently of unknown status.
Figure 1.
Map of the Galápagos Islands including the route of the HMS Beagle in 1835 (Grant & Estes 2009) and the sites where Darwin landed (solid circles). Existing and presumed extinct (underlined) island breeding populations of G. magnirostris (asterisk) and G. difficilis (plus) are indicated. Scale bar, 50 km.
Several populations of the sharp-beaked ground finch are more genetically distinct from each other than many other species of Darwin's finches (Petren et al. 2005). Darwin is thought to have collected three specimens of G. difficilis: the only known specimen ever recorded from San Cristóbal, one of two specimens collected from Floreana (Habel collected one other in 1869) and one from a population that persists on Santiago. A later expedition in 1900–1901 collected the only specimen ever known from Isabela. We asked whether these single specimens are genetically distinct from other forms. We also asked whether individuals suspected as being from currently extant populations of G. difficilis and phenotypically similar Geospiza scandens show a genetic affinity to these groups.
2. Material and methods
Darwin collected 31 finch specimens while in Galápagos, of which 21 are available today in the British Museum of Natural History (BMNH, n = 19) and the University Museum of Zoology, Cambridge (UMZC, n = 2). Darwin's shipmates Fitzroy, Fuller and Covington collected 25–27 additional finch specimens from the same locations. A full account of these specimens, their beak measurements and uncertain history regarding the island of origin for many has been extensively documented by Sulloway (1982). We removed a small piece of skin (approx. 1 × 1 × 3 mm) from the pad of the large hind digit (Mundy & Woodruff 1997) from each of 15 ground finch specimens from the Beagle, including putative G. magnirostris, G. difficilis and G. scandens (tables 1 and 2). We also genotyped birds from the American Museum of Natural History (AMNH) collected in 1900–1901, including several G. magnirostris from Darwin and Wolf, and the putative G. difficilis specimen from Isabela.
Table 1.
Island populations of G. magnirostris genotyped from historical collections and compared with recently sampled existing (reference) populations from Fernandina, Isabela, Santiago, Santa Cruz, Rábida, Pinta, Marchena and Genovesa. Data for references are averages among reference populations. He, expected heterozygosity.
| island | date | museum number | average quality | He (%) | genetic distances DA (range) | genetic differentiation |
|---|---|---|---|---|---|---|
| references (n = 96) | 1997–2006 | — | 9.5 | 60–68 | 0.25 (0.09–0.39) | — |
| Floreana (n = 3) | 1835 | BNHM1837.2.21 (−0.398, −0.402, −403) | 6.3 | 65 | 0.63 (0.57–0.68) | highly distinct |
| San Cristóbal (n = 6) | 1835 | BNHM1855.12.19. (−80, −81, −83, −104, −113); 1837.2.21.396 | 5.4 | 68 | 0.67 (0.61–0.71) | highly distinct |
| Darwin (n = 6) | 1906 | AMNH516875-82 | 7.1 | 60 | 0.53 (0.34–0.70) | distinct |
| Wolf (n = 15) | 1906 | AMNH516976-90; 517047 | 7.6 | 60 | 0.41 (0.34–0.53) | distinct |
Table 2.
Historical specimens of individual birds, with current species and island affiliations, observed heterozygosity and genetic classification based on assignment tests. Specimen labels with ‘m’, ‘d’ or ‘s’ reflect current classifications as G. magnirostris, G. difficilis and G. scandens, respectively. Ho, observed heterozygosity.
| specimen | species | island | date | museum number | genotype quality | Ho (%) | genetic classification |
|---|---|---|---|---|---|---|---|
| 1m | G. magnirostris | Santiago | 1835 | UCZM27/Fri(E)/26/e/1 | 7.1 | 64 | Santiago |
| 2m | G. magnirostris | ? | 1835 | BNHM1885.15.14.281 | 6.9 | 71 | Floreana |
| 3m | G. magnirostris | ? | 1835 | BNHM1885.15.14.280 | 8.7 | 50 | (no match) |
| 4d | G. difficilis | San Cristóbal | 1835 | AMNH517815 | 6.6 | 50 | G. fuliginosa |
| 5d | G. difficilis | Floreana | 1835 | BNHM1837.2.21.400 | 4.8 | 71 | distinct: G. difficilis |
| 6d | G. difficilis | Isabela | 1901 | AMNH517793 | 8.4 | 57 | distinct: G. difficilis |
| 7d | G. difficilis | Santiago | 1835 | BNHM1855.12.19.20 | 5.4 | 43 | G. difficilis or G. fuliginosa |
| 8s | G. scandens | Santiago | 1835 | BNHM1925.12.22.62 | 6.9 | 64 | G. difficilis |
| 9s | G. scandens | Santiago | 1835 | BNHM1837.2.21.415 | 4.9 | 64 | G. fuliginosa or G. scandens |
| 10s | G. scandens | Santiago | 1835 | BNHM1855.12.19.15 | 5.7 | 64 | G. fuliginosa or G. scandens |
Genotypes from 26 reference populations (n = 423 birds) were available from previous studies of blood samples of birds collected in the field (Petren et al. 1999a, 2005) and a recent survey of G. scandens (n = 19) from Isabela in 2006. Reference populations include G. scandens, G. fuliginosa and G. fortis from each of the four islands visited by Darwin, G. magnirostris and G. difficilis from Santiago, seven other island populations of G. magnirostris and five other populations of G. difficilis.
For museum specimens, DNA was extracted in a facility where fresh finch tissue, DNA or PCR products have never been present. This dedicated room is periodically wiped with bleach and the hood and equipment used for DNA extraction was bathed in UV daily to render any trace DNA unamplifiable. All tissue was subdivided for DNA extraction using a glass micro-bead affinity method (GeneClean for Ancient DNA, BIO101). In our hands, this method outperforms filter cartridges (Qiagen), standard phenol–chloroform and other methods we have used previously (Petren et al. 2005). Final elution occurred in 30 µl 0.1X Tris and 1–3 µl were used in a 15 µl PCR reaction. An extraction blank and no-DNA control were included in every batch of PCR amplifications. Original published primer sequences yielded an excessive number of PCR failures (Petren 1998; Petren et al. 1999b), thus primers were redesigned to yield smaller PCR products (generally between 90 and 160 bp), which greatly improved genotyping success (see electronic supplementary material). PCR amplification was performed for 3–4 loci simultaneously using a multiplex Taq polymerase kit (Qiagen). PCR products were separated on an ABI 3730XL sequencer and allele sizes were scored relative to a size standard (LIZ labelled) included in each lane using GeneMapper software.
Each genotype was obtained by compiling a consensus from at least three repeated PCR reactions. A quality score for each locus × individual was computed, then averaged among all 15 loci. Quality scores were calculated by deducting (from a maximum score of 10) two points for evidence of allele dropout, three points for a failed amplification, four points for reciprocal dropout (one allele missing in one PCR and the other allele missing in another PCR) and five points for spurious alleles (greater than two alleles per run or greater than three alleles among runs). Individuals with quality scores less than 6 were subjected to additional rounds of genotyping, such that 25 per cent of all museum specimens were repeated fivefold and three samples were repeated sevenfold across all loci (Taberlet et al. 1996). A study of mangrove finch museum specimens from approximately 1900 showed that few additional dropout alleles are recovered after two repeated PCRs (Petren et al. in preparation). The final genotype was compiled by eliminating spurious alleles that appeared once (in fewer than 30% of replicates). Heterozygosity was assumed if less than 70 per cent of genotypes were homozygous (Arandjelovic et al. 2009), otherwise the homozygous genotype was used. Unresolvable genotypes (greater than 1 spurious allele) were coded as missing data.
Measures of genetic diversity, tests for equilibrium and genetic distances were computed in Genalex (Peakall & Smouse 2006). We use Nei's (1972) standard genetic distance (DA) because it captures genetic distinctness among Darwin's finches and remains more linear than FST and other distances over longer time scales (see fig. 5 in Petren et al. 2005). Principal coordinate axis (PCA) variation was used to produce a coarse graphical summary of genetic variation (Peakall & Smouse 2006). The 14 autosomal loci were subjected to assignment tests, first using the method of assigning individual likelihood scores (Paetkau et al. 2004; Piry et al. 2004), with zero frequencies set to 0.01, test samples unassigned but with all allele frequencies incorporated. This method allows for individuals to be assigned with low likelihood to all reference populations. Another method of genetic assignment (Pritchard et al. 2000) was used for fine-scaled assessment of genetic affinity and to test for admixture. We ran simulations for 7000 generations with a burn-in of 2000 and default parameters except where specified. We first used no population information to assess natural clustering, then used hypothesized population groupings and assessed admixture up to two generations back.
3. Results
Of the 600 possible individual × locus genotypes that could be obtained for the 40 museum specimens, 94 per cent were recovered. Allele dropout affected approximately 30 per cent of genotypes, which is higher than the number (approx. 20%) found in a separate study on Galápagos museum specimens from approximately 1900 (Petren et al. in preparation) and other studies (Bonin et al. 2004). Spurious allele sizes affected approximately 3 per cent of all alleles scored and approximately 20 per cent of specimens. Spurious alleles disproportionately affected one locus (Gf16), in which nearly half the genotypes were affected. The vast majority were accounted for by 2 bp shifts attributable to early PCR slippage or misscoring. Expected heterozygosities for populations (table 1) and observed heterozygosities for individual specimens (table 2) were similar to those from recent field-collected populations (Petren et al. 2005). Deviations from Hardy–Weinberg equilibrium (Peakall & Smouse 2006) occurred in five of 56 population/locus combinations before correction, a rate similar to previous studies (Petren et al. 2005).
(a). Genetic distinctness of G. magnirostris populations
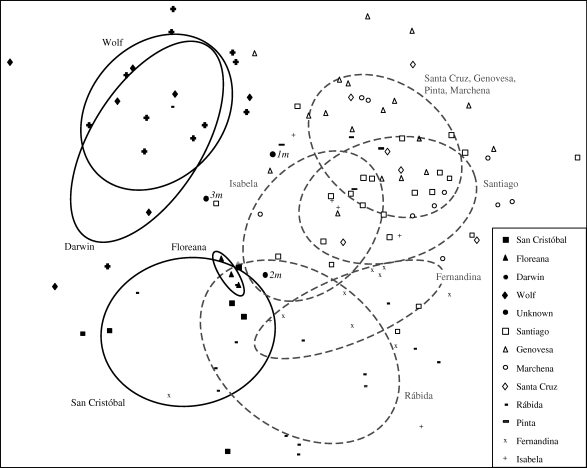
The four populations of large ground finches (G. magnirostris) from museum collections were genetically distinct from eight other field-sampled populations of G. magnirostris. Genetic distances among previously sampled reference populations were significantly lower than the distances between the four museum populations and the rest (table 1). The first two principal component axes depict Darwin and Wolf as more distinct than the San Cristóbal and Floreana populations (figure 2), but this is only part of the story. Nei's genetic distance captures the distinct nature of the giant forms from San Cristóbal and Floreana collected by Darwin (table 1). Large ground finches from the extremely remote (and neighbouring) islands of Darwin and Wolf had lower (and identical) expected heterozygosities than the Floreana and San Cristóbal populations, as would be expected owing to their small size and isolation from immigrant gene flow.
Figure 2.
Principal coordinate plot of genetic distinctness of historical populations collected during the Beagle voyage in 1835 (San Cristóbal and Floreana) and in 1900–1901 (Darwin, Wolf), in relation to eight currently existing G. magnirostris populations and three specimens of questionable origin (1m–3m). The first two axes account for 9.7% and 8.6% of the variation in the data, respectively. Specimen numbers are given in table 2. Density ellipses (50%) are provided as a heuristic guide.
(b). Genetic affinity of G. magnirostris individuals
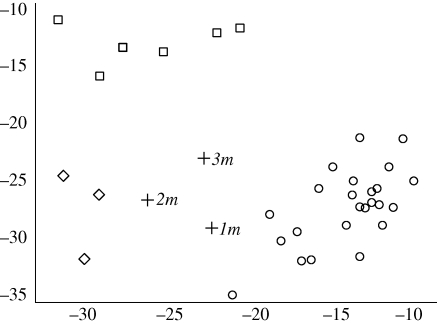
The 1835 specimens considered to be from San Cristóbal (n = 6) and Floreana (n = 3) appeared to form natural groupings. Likelihood scores for cross-membership in other populations were much lower than for the specified population (figure 3). One G. magnirostris of questionable origin (1m) appears to be from Santiago (table 2). This bird was earlier thought to be from San Cristóbal before Sulloway (1982) showed that it is more typical morphologically of the Santiago population. Another G. magnirostris of unknown origin appears to be from Floreana (3m), while a third (2m) could not be classified.
Figure 3.
Likelihood scores from assignment tests for historical G. magnirostris specimens of questionable origin (1, 2 and 3 m) in relation to scores from San Cristóbal (squares), Floreana (diamonds) and the recently sampled existing population from Santiago (circles). Axes reflect increasing likelihood of being from Santiago (along the x-axis) and San Cristóbal (along the y-axis). Specimen numbers are given in table 2.
(c). Genetic affinity of individual G. difficilis and G. scandens
For putative G. difficilis, there is a possibility that singleton samples from islands could be mistaken for either G. scandens or G. fuliginosa, or to a lesser extent G. fortis, or they may be hybrids of these species. On the coarse scale of variation captured by the first two PCAs, several of the putative G. difficilis (4d–7d) and G. scandens (8s–10s) appear to be genetically distinct from other Geospiza species, while several others appear to be clearly allied with other Geospiza species (figure 4).
Figure 4.
Principal coordinate plot of genetic differences among individual G. difficilis and G. scandens museum specimens in relation to extant, recently field-sampled G. scandens, G. fuliginosa and G. fortis genotypes from several islands and G. difficilis populations from the six islands indicated in the key. The first (x) and second (y) axes account for 27% and 17% of the variation in the data, respectively. Density ellipses (50%) are provided as a heuristic guide. Individual historical specimen numbers (specimens 4–9) are given in table 2.
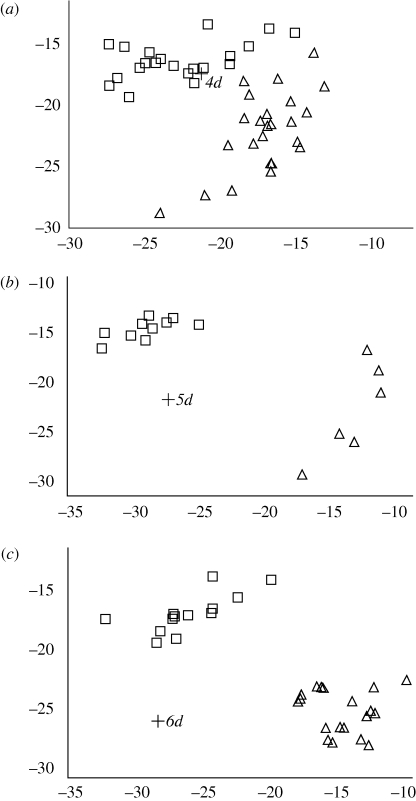
The putative G. difficilis from Floreana (5d) and Isabela (6d) were subjected to assignment tests and appear to be distinct from G. fuliginosa and G. scandens from those islands (figure 5). Similar analyses showed that these birds were not closely allied to G. fortis on these islands (results not shown). The putative G. difficilis from San Cristóbal (4d) could not be distinguished from G. fuliginosa on the same island (figure 5). The putative G. difficilis from Santiago (7d) showed high affinity to present day G. difficilis from Santiago, but it also showed high affinity to G. fuliginosa from the same island (figure 6).
Figure 5.
Likelihood scores from assignment tests for historical G. difficilis specimens from (a) San Cristóbal (4d), (b) Floreana (5d) and (c) Isabela (6d), in relation to recently field-sampled G. scandens (triangles) and G. fuliginosa (squares) from the same islands. Axes reflect increasing likelihood of being a G. scandens (along the x-axis) and G. fuliginosa (along the y-axis).
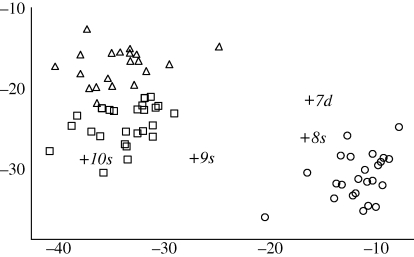
Figure 6.
Likelihood scores from assignment tests for Santiago birds, including historical G. difficilis (7d) and G. scandens (8s–10s), and recently field-sampled populations including G. difficilis (circles), G. scandens (triangles) and G. fuliginosa (squares). Axes are likelihoods of being a G. difficilis (x) and G. scandens (y). Specimen numbers are given in table 2.
Of the birds thought to be G. scandens, the first (8s) is genetically clearly a G. difficilis from the same island (figure 6). The last two of uncertain island origin (9s, 10s) do not show a high likelihood of belonging to any reference population. These birds did not show affinity to any populations on San Cristóbal or Floreana, nor to G. fortis from any of the islands (results not shown).
Fine-scaled population structure analysis of the four museum specimens from Santiago (7d, 8s–10s) supported the above results. Average likelihood values for the number of populations plateaued at k = 3 (structure, k/Ln: 3/−3500; 2/−3651; 4/−3496; 5/−3476). Without the use of prior population information, all but one of 24 reference G. difficilis individuals clustered together with p = 1, and all but one of the 19 G. scandens clustered together (overall p = 0.87). Reference G. fuliginosa did not form a strong natural cluster (overall, p = 0.42), and they showed extensive overlap with G. scandens (p = 0.58). One reference G. difficilis and museum specimens 4d, 9s and 10s all showed a higher probability of belonging to the G. fuliginosa cluster (p > 0.99) than most of the other reference G. fuliginosa specimens. When population information was used to define populations a priori, better resolution among species was obtained, but specimen 4d could be ‘forced’ (i.e. not be considered misassigned) to belong to either G. difficilis (p = 0.77) or G. fuliginosa (p > 0.99), and birds 9s and 10s could be ‘forced’ to belong to either G. scandens or G. fuliginosa (all p > 0.95). Admixture analysis did not reveal any evidence of hybridization over the last two generations.
4. Discussion
Museum specimens more than 170 years old yielded usable multilocus information that allowed us to verify species identifications, document demographic changes, such as extinction, and assess the loss of biodiversity over time. We first evaluate the methods employed and their limitations, then discuss the results in terms of the patterns of genetic and morphological diversity in Darwin's finches and finally consider the implications for understanding recent changes and future persistence of Darwin's finches and Galápagos biodiversity.
(a). Multilocus markers and ancient DNA
To avoid being misled by potential artefacts, multilocus studies of museum DNA should take opportunities to test the reliability of the data for internal consistency, beyond the specific question at hand (Gilbert et al. 2005; Wandeler et al. 2007). Our study produced several instances of good agreement between past and current genotypes when specimen origin was not in question, and historical specimens from the same population tended to cluster together. Methodological refinements such as designing shorter PCR target products (less than 160 bp), repeating all genotyping several times and repeating problem specimens as many as seven times appeared to improve results. For Darwin's finches, we were able to derive the most information from well-sampled populations. Although most individual specimens could be classified, and many were classified as expected, there is a clear limitation to the tests of genetic affinity that are confined to single museum specimens. The approximately 175-year-old Beagle specimens were generally more difficult to amplify than the approximately 100-year-old specimens, perhaps due to time and/or conditions on the Beagle. With the larger numbers of individuals available from collections around approximately 1900, our genotyping success has varied such that each specimen tends to be either reliable or problematic across most loci. The conditions after death, drying time and perhaps even the size of a bird's toe pad may all be important factors in later genotyping success.
Some populations contained birds that did not classify into species very well. Even data from recently field-sampled birds show that some species (e.g. G. fortis, G. fuliginosa and G. scandens) are not very distinct genetically on some islands. Their recent origin and occasional genetic exchange through hybridization likely play a role in limiting genetic differences (Grant et al. 2005b). Chance sharing of alleles present in both reference populations could also hinder classification of individual birds, in which case genotyping additional loci might improve resolution. Within-island population substructure or genetic changes over time could limit the ability to classify historical specimens using recently sampled DNA. Genetic change is a possibility given the dramatic episodes of natural selection (Grant 1999; Grant & Grant 2002) and population crashes (e.g. Grant & Grant 2006). We suspect that the instances of limited resolution we encountered were more a product of the natural system than the condition of the specimens.
(b). Individual historical specimens and the sharp-beaked ground finch
Following in the footsteps of Swarth (1931), Sulloway (1982) made a heroic effort to rectify a problem that has persisted since Darwin's visit: where were the Beagle specimens collected? Our results suggest that in most instances, Sulloway's reasoning was correct. Particularly impressive was the delineation of G. magnirostris specimens into three island populations. Miscategorizations were mostly attributable to original species identifications (or lack of resolution as discussed above), and in no case was Sulloway's island designation found to be in error.
The sharp-beaked ground finch (G. difficilis) is notable for several reasons. Lack (1947) could not discern where to place this species on an evolutionary tree, since island populations varied widely, and their traits were shared with differing subsets of other species. Based on their unique beak shape, G. difficilis populations form a natural group (Schluter et al. 1991; Grant 1999). Genetically, G. difficilis is highly structured, with genetic differences among populations that surpass genetic differences among many other species (Grant et al. 2000; Petren et al. 2005). The current study supports the notion that two additional populations of this highly divergent species existed on Floreana and Isabela. The genetic distinctness of the Floreana specimen supports Steadman's (1986) attribution of fossil remains found on this island to G. difficilis. The genetic distinctness of these birds from other G. difficilis and other species may be substantial if the single specimens are a reasonable representation of the sampled populations (figure 4). On San Cristóbal, recent field studies suggest that G. scandens and G. fuliginosa may be occasionally hybridizing in a part of the island that was visited by the Beagle. Thus, what we concluded is a misclassified G. difficilis (4d) may be a bird of hybrid ancestry and somewhat distinct from most G. fuliginosa and G. scandens in terms of its beak shape. It is tempting to interpret the decline of G. difficilis, especially on the smaller, more disturbed island of Floreana, as being due to human impact. However, there is a possibility that this species was already in decline owing to competition with ecologically similar species that may have evolved more recently, such as G. fuliginosa (Schluter & Grant 1982).
(c). Historical populations of the large ground finch
Our results suggest a considerable amount of genetic diversity in the large ground finch (G. magnirostris) has been lost since Darwin's time. The populations on the islands of Darwin and Wolf are very distinct genetically (mean DA = 0.53, 0.41, respectively) compared with other currently existing populations, which show much less differentiation (mean DA = 0.39). Their genetic affinity also lies closer to other G. magnirostris and not to G. conirostris (DA = 0.73) with which they share superficial similarities in beak shape (Grant 1999, p. 131). The current status of these remote populations is uncertain, but they are rare at best on both islands and potentially declining, according to field observations over the last 30 years. Populations on San Cristóbal and Floreana are even more genetically distinct (mean DA = 0.63, 0.67, respectively) and they are certainly extinct on these islands. To place these genetic distances into perspective, the average DA between all G. fortis and G. fuliginosa populations is 0.26 (0.13–0.50), between G. fortis and G. scandens is 0.34 (0.26–0.53) and between the small and large tree finches is 0.15 (0.08–0.30), but among populations of G. difficilis, the genetic distances are comparable (mean 0.73, 0.17–1.23).
We confirmed the prediction that greater morphological divergence of the exceptionally large ground finch populations should also indicate greater genetic divergence. A similar pattern has been shown in G. difficilis (Grant et al. 2000). Darwin was clearly inspired by the differences he observed among populations on neighbouring islands, most notably in the mockingbirds. If Darwin's efforts to reconstruct the island of origin of his finch specimens had succeeded, he might have been able to use the large ground finches as an example of recent evolution and adaptation to different conditions in his most influential work (Darwin 1859).
(d). Population divergence in Darwin's finches
A great deal of the genetic variation among Darwin's finches comes among populations that are currently (or were formerly) recognized as the same species (Petren et al. 2005; Tonnis et al. 2005). Differentiation among island populations may set the stage for speciation (Lack 1947; Grant 1999; Petren et al. 2005). Lack viewed interisland variation as adaptation to local conditions, which on rare occasions led to colonization, secondary contact of two slightly different forms and then further divergence in sympatry owing to ecological character displacement. He supposed that interisland movements must be rare to allow differences to accumulate. However, recent evidence suggests that interisland movements are not rare and typically result in interbreeding (Petren et al. 2005). It is probably only on rare occasions that circumstances permit a group of colonists to breed among themselves and not among residents of the same species, yet parts of this phase have recently been observed in the field (Grant & Grant 2009). If the two incipient species manage to persist in sympatry, the new species would probably spread rapidly across the archipelago. Consistent with this model is the observation that more recently evolved species (G. fuliginosa, G. fortis and the Camarhynchus tree finches) have less genetic structure among island populations, while older species such as Certhidea warbler finches (Tonnis et al. 2005), G. difficilis and G. magnirostris are more structured, presumably owing to the accumulation of local adaptations over time (Petren et al. 2005). Island populations of several species may therefore be critical evolutionary entities and worthy of consideration when assessing biodiversity loss and preservation.
(e). Species divergence in Darwin's finches
In spite of the pronounced morphological differences easily observed among species, the expected positive correlation of morphological and genetic divergence is absent across the group (Petren et al. 2005). Examples of the patterns that inspired Darwin, with different species occupying different islands, are rare in Darwin's finches (Tonnis et al. 2005). Most species occur together on the same islands. Furthermore, most recognized species of Darwin's finches are not diagnosably different according to mitochondrial DNA (mtDNA) sequence variation (Petren et al. 2005). This lack of mtDNA resolution may raise questions concerning whether these species are in fact different evolutionary entities. We favour the interpretation that mtDNA diversification simply cannot keep up with the pace of speciation in this group. Infrequent hybridization also likely affects mtDNA resolution, but genetic mixing is limited by restricted patterns of backcrossing (Grant & Grant 1997a,b; Grant et al. 2003, 2004, 2005b). On any island, all species are diagnosably different in their beak morphology, song and mating patterns (Grant 1999). Beak size and shape differences are heritable and are critical determinants of natural selection, and even the most closely related species respond differently to environmental changes (Grant & Grant 2002, 2006). So, although many species of Darwin's finches would escape detection by DNA barcoding and similar sequence-based approaches, there is little question that they are important evolutionary entities worthy of preservation.
(f). The decline of finch biodiversity over time
It is illuminating to consider the status of the finch populations sampled by Charles Darwin and his Beagle shipmates. A large proportion of the birds they collected were from populations that are now extinct. There is no indication that they paid any particular attention to sampling rare forms disproportionately, and the reasonably high genetic diversity of the now extinct populations does not suggest that decline was imminent. The decline to extinction must have occurred since the onset of major human occupation and disturbance. Floreana and San Cristóbal were among the earliest islands to be impacted by permanent human settlement, and they are foci of population extinction (Grant et al. 2005a). Although species extinctions are not known in Darwin's finches, the mangrove finch (Cactospiza heliobates) is critically endangered (Grant & Grant 1997a,b; Dvorak et al. 2004), and the medium tree finch (Camarhynchus pauper) on Floreana has recently been listed as vulnerable (IUCN 2009). The growing number of extinct populations, coupled with the fact that some were genetically distinct, is an indication that this natural engine of biodiversity is in danger of collapse. The primary threats are introduced disease (Wikelski et al. 2004; Thiel et al. 2005), parasites (e.g. Fessl et al. 2006), other introduced species and human disturbance. The question remains whether the Galápagos will suffer the fate of so many other island endemic communities, and whether a cascade of events can be prevented to ensure its survival. Note added in proof. Analysis of the putative G. difficilis from Floreana collected by Habel in 1868 (courtesy of the Swedish Museum of Natural History) suggests this bird is also distinct from extant species on this island and therefore likely represents a second remnant G. difficilis specimen (assignment likelihoods as in figure 5b: x = −31.9; y = −26.8).
Acknowledgements
We thank Robert Prys-Jones of the British Natural History Museum, Terry Chesser and Joel Cracraft of the American Museum of Natural History, and the University Museum of Zoology, Cambridge, for permission to obtain tissue from historical specimens. We thank the Galápagos National Parks for permission to conduct field studies, and the Charles Darwin Research Station for support in the field. We thank Jeffrey Markert and Heather Farrington for help developing methods to genotype museum specimens, Kristen Short, John Niedzwiecki and Elizabeth Ristagno for field assistance and two anonymous reviewers for helpful comments. This work was partially supported by the National Science Foundation (DEB-0317687 to K.P.).
Footnotes
One contribution of 13 to a Theme Issue ‘Darwin's Galápagos finches in modern evolutionary biology’.
References
- Allendorf F. W., Luikart G.2007Conservation and the genetics of populations. Malden, MA: Blackwell [Google Scholar]
- Arandjelovic M., Guschanski K., Schubert G., Harris T. R., Thalmann O., Siedel H., Vigilant L.2009Two-step multiplex PCR improves the speed and accuracy of genotyping using DNA from noninvasive and museum samples. Mol. Ecol. Resour. 9, 28–36 (doi:10.1111/j.1755-0998.2008.02387.x) [DOI] [PubMed] [Google Scholar]
- Bonin A., Bellemain E., Bronken E. P., Pompanon F., Brochmann C., Taberlet P.2004How to track and assess genotyping errors in population genetics studies. Mol. Ecol. 13, 3261–3273 (doi:10.1111/j.1365-294X.2004.02346.x) [DOI] [PubMed] [Google Scholar]
- Darwin C. R.1859On the origin of species by means of natural selection. London, UK: John Murray [Google Scholar]
- Dvorak M., Vargas H., Fessl B., Tebbich S.2004On the verge of extinction: a survey of the mangrove finch Cactospiza heliobates and its habitat on the Galápagos Islands. Oryx 38, 171–179 (doi:10.1017/S0030605304000316) [Google Scholar]
- Ellegren H.2004Microsatellites: simple sequences with complex evolution. Nat. Rev. Genet. 5, 435–445 (doi:10.1038/nrg1348) [DOI] [PubMed] [Google Scholar]
- Fessl B., Kleindorfer S., Tebbich S.2006An experimental study on the effects of an introduced parasite in Darwin's finches. Biol. Conserv. 127, 55–61 (doi:10.1016/j.biocon.2005.07.013) [Google Scholar]
- Gilbert M. T. P., Bandelt H.-J., Hofreiter M., Barnes I.2005Assessing ancient DNA studies. Trends Ecol. Evol. 20, 541–544 (doi:10.1016/j.tree.2005.07.005) [DOI] [PubMed] [Google Scholar]
- Grant P. R.1999Ecology and evolution in Darwin's finches. Princeton, NJ: Princeton University Press [Google Scholar]
- Grant K. T., Estes G. B.2009Darwin in Galápagos: footsteps to a New World. Princeton, NJ: Princeton University Press [Google Scholar]
- Grant P. R., Grant B. R.1997aThe rarest of Darwin's finches. Conserv. Biol. 11, 119–126 (doi:10.1046/j.1523-1739.1997.95399.x) [Google Scholar]
- Grant P. R., Grant B. R.1997bMating patterns of Darwin's finch hybrids determined by song and morphology. Biol. J. Linn. Soc. 60, 317–343 (doi:10.1111/j.1095-8312.1997.tb01499.x) [Google Scholar]
- Grant P. R., Grant B. R.2002Unpredictable evolution in a 30-year study of Darwin's finches. Science 296, 707–711 (doi:10.1126/science.1070315) [DOI] [PubMed] [Google Scholar]
- Grant P. R., Grant B. R.2006Evolution of character displacement in Darwin's finches. Science 313, 224–226 (doi:10.1126/science.1128374) [DOI] [PubMed] [Google Scholar]
- Grant P. R., Grant B. R.2009The secondary contact phase of allopatric speciation in Darwin's finches. Proc. Natl Acad. Sci. USA 106, 20 141–20 148(advanced online) (doi:10.1073/pnas.0911761106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant P. R., Grant B. R., Petren K.2000The allopatric phase of speciation: the sharp-beaked ground finch (Geospiza difficilis) on the Galápagos Islands. Biol. J. Linn. Soc. 69, 287–317 (doi:10.1111/j.1095-8312.2000.tb01207.x) [Google Scholar]
- Grant P. R., Grant B. R., Keller L. F., Markert J. A., Petren K.2003Inbreeding and interbreeding in Darwin's finches. Evolution 57, 2911–2916 [DOI] [PubMed] [Google Scholar]
- Grant P. R., Grant B. R., Markert J. A., Keller L. F., Petren K.2004Convergent evolution of Darwin's finches caused by introgressive hybridization and selection. Evolution 58, 1588–1599 [DOI] [PubMed] [Google Scholar]
- Grant P. R., Grant B. R., Petren K., Keller L. F.2005aExtinction behind our backs: the possible fate of one of the Darwin's finch species on Isla Floreana, Galápagos. Biol. Conserv. 122, 499–503 (doi:10.1016/j.biocon.2004.09.001) [Google Scholar]
- Grant P. R., Grant B. R., Petren K.2005bHybridization in the recent past. Am. Nat. 166, 56–67 (doi:10.1086/430331) [DOI] [PubMed] [Google Scholar]
- Groombridge J. J., Jones C. G., Bruford M. W., Nichols R. A.2000‘Ghost’ alleles of the Mauritius kestrel. Nature 403, 616 (doi:10.1038/35001148) [DOI] [PubMed] [Google Scholar]
- Hedrick P., Waits L.2005What ancient DNA tells us. Heredity 94, 463–464 (doi:10.1038/sj.hdy.6800647) [DOI] [PubMed] [Google Scholar]
- Herrmann B., Hummel S.1994Ancient DNA: recovery and analysis of genetic material from paleontological, archaeological, museum, medical and forensic specimens. New York, NY: Springer [Google Scholar]
- IUCN 2009Red list of threatened species. See http://www.iucnredlist.org [Google Scholar]
- Lack D.1947Darwin's finches. Cambridge, UK: Cambridge University Press [Google Scholar]
- Lundh J. P. Galápagos: a brief history. 2001 ISBN 82-92294-00-7. See http://www.lundh.no/jacob/Galapagos . [Google Scholar]
- Mundy N. I., Woodruff D. S.1997Skin from feet of museum specimens as a non-destructive source of DNA for avian genotyping. Auk 114, 126–129 [Google Scholar]
- Nei M.1972Genetic distance between populations. Am. Nat. 106, 283–292 (doi:10.1086/282771) [Google Scholar]
- Paetkau D., Slade R., Burdens M., Estoup A.2004Genetic assignment methods for the direct, real-time estimation of migration rate: a simulation based exploration of accuracy and power. Mol. Ecol. 13, 55–65 (doi:10.1046/j.1365-294X.2004.02008.x) [DOI] [PubMed] [Google Scholar]
- Peakall R., Smouse P. E.2006Population genetic software for teaching and research (Genalex 6.0). Mol. Ecol. Notes 6, 288–295 (doi:10.1111/j.1471-8286.2005.01155.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petren K.1998Microsatellite primers from Geospiza fortis and cross-species amplification in Darwin's finches. Mol. Ecol. 7, 1782–1784 [DOI] [PubMed] [Google Scholar]
- Petren K., Grant B. R., Grant P. R.1999aA phylogeny of Darwin's finches based on microsatellite DNA length variation. Proc. R Soc. Lond. B 266, 321–329 (doi:10.1098/rspb.1999.0641) [Google Scholar]
- Petren K., Grant B. R., Grant P. R.1999bLow extrapair paternity in the cactus finch (Geospiza scandens). Auk 116, 252–256 [Google Scholar]
- Petren K., Grant P. R., Grant B. R., Keller L. F.2005Comparative landscape genetics and the adaptive radiation of Darwin's finches: the role of peripheral isolation. Mol. Ecol. 14, 2943–2957 (doi:10.1111/j.1365-294X.2005.02632.x) [DOI] [PubMed] [Google Scholar]
- Petren K., Farrington H., Fessl B., Vargas H., Clack A. A.In preparation A genetic signature of impending extinction in Darwin's rarest finch. [Google Scholar]
- Piry S., Alapetite A., Cornuet J., Paetkau D., Baudouin L., Estoup A.2004Geneclass2: a software for genetic assignment and first generation migrant detection. J. Hered. 95, 536–539 (doi:10.1093/jhered/esh074) [DOI] [PubMed] [Google Scholar]
- Pritchard J. K., Stephens M., Donnelly P.2000Inference of population structure using multilocus genotype data. Genetics 155, 945–959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schluter D., Grant P. R.1982The distribution of G. difficilis in relation to G. fuliginosa in the Galápagos Islands: tests of three hypotheses. Evolution 36, 1213–1226 (doi:10.2307/2408154) [DOI] [PubMed] [Google Scholar]
- Schluter D., Ratcliffe L. M., Grant P. R.1991The taxonomic status of the small Genovesa ground-finch in the Galápagos. Auk 108, 201–204 [Google Scholar]
- Steadman D.1986Holocene vertebrate fossils from Isla Floreana, Galápagos. Smithsonian Contrib. Zool. 413, 1–104 [Google Scholar]
- Sulloway F. J.1982The Beagle collections of Darwin's finches [Geospizinae]. Bull. Br. Mus. [Nat. Hist.] Zool. Ser. 43, 49 [Google Scholar]
- Sulloway F. J.1983aDarwin and his finches: the evolution of a legend. J. Hist. Biol. 15, 1–53 (doi:10.1007/BF00132004) [Google Scholar]
- Sulloway F. J.1983bThe legend of Darwin's finches. Nature 303, 372 (doi:10.1038/303372a0) [Google Scholar]
- Swarth H. S.1931The avifauna of the Galápagos Islands. Occ. Pap. Cal. Acad. Sci. 18, 1–299 [Google Scholar]
- Taberlet P., Griffin S., Goossens B., Questiau S., Manceau V., Escaravage N., Waits L. P., Bouvet J.1996Reliable genotyping of samples with very low DNA quantities using PCR. Nucleic Acids Res. 24, 3189–3194 (doi:10.1093/nar/24.16.3189) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiel T., Whiteman N. K., Tirape A., Maquero M. I., Cedeno V., Walsh T., Jimenez G., Parker P. G.2005Characterization of Canarypox-like viruses infecting endemic birds in the Galápagos Islands. J. Wildl. Dis. 41, 342–353 [DOI] [PubMed] [Google Scholar]
- Tonnis B., Grant P. R., Grant B. R., Petren K.2005Habitat selection and ecological speciation in Galápagos warbler finches (Certhidea olivacea and Certhidea fusca). Proc. R. Soc. B 272, 819–826 (doi:10.1098/rspb.2004.3030) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wandeler P., Hoeck P. A. E., Keller L. F.2007Back to the future: museum specimens in population genetics. Trends Ecol. Evol. 22, 634–642 (doi:10.1016/j.tree.2007.08.017) [DOI] [PubMed] [Google Scholar]
- Watkins G., Cruz F.2007Galápagos at risk: a socioeconomic analysis of the situation in the archipelago. Puerto Ayora, Ecuador: Charles Darwin Foundation [Google Scholar]
- Westemeier R. L., Brawn J. D., Simpson S. A., Esker T. L., Jansen R. W., Walk J. W., Kershner E. L., Bouzat J. L., Paige K. N.1998Tracking the long-term decline and recovery of an isolated population. Science 282, 1695–1698 (doi:10.1126/science.282.5394.1695) [DOI] [PubMed] [Google Scholar]
- Wikelski M., Foufopoulos J., Vargas H., Snell H.2004Galápagos birds and diseases: invasive pathogens as threats for island species. Ecol. Soc. 9, 5 (http://www.ecologyandsociety.org/vol9/iss1/art5) [Google Scholar]