SUMMARY
Oligodendrocytes form an insulating multilamellar structure of compact myelin around axons, which allows efficient and rapid propagation of action potentials. However, little is known about the molecular mechanisms operating at the onset of myelination and during maintenance of the myelin sheath in the adult. Here we use a genetic cell ablation approach combined with Affymetrix GeneChip microarrays to identify a number of oligodendrocyte-enriched genes that may play a key role in myelination. One of the “oligogenes” we cloned using this approach is Tmem10/Opalin, which encodes for a novel transmembrane glycoprotein. In situ hybridization and RT-PCR analysis revealed that Tmem10 is selectively expressed by oligodendrocytes and that its expression is induced during their differentiation. Developmental immunofluorescence analysis demonstrated that Tmem10 starts to be expressed in the white matter tracks of the cerebellum and the corpus callosum at the onset of myelination after the appearance of other myelin genes such as MBP. In contrast to the spinal cord and brain, Tmem10 was not detected in myelinating Schwann cells, indicating that it is a CNS-specific myelin protein. In mature oligodendrocytes, Tmem10 was present at the cell soma and processes, as well as along myelinated internodes, where it was occasionally concentrated at the paranodes. In myelinating spinal cord cultures, Tmem10 was detected in MBP-positive cellular processes that were aligned with underlying axons before myelination commenced. These results suggest a possible role of Tmem10 in oligodendrocyte differentiation and CNS myelination.
Keywords: Myelin, Axon-glial interaction, cell adhesion molecule, Paranodes, CNS-specific
INTRODUCTION
Oligodendrocytes are specialized cells that produce myelin in the central nervous system (CNS) (Baumann and Pham-Dinh 2001; Pfeiffer et al. 1993). They extend cellular processes that wrap spirally around axons, ultimately forming insulating myelin sheaths, which allow efficient and rapid propagation of action potentials. Oligodendrocytes consist of a heterogeneous population of cells with a complex morphology. They are divided into several groups based on a number of criteria such as the number and characteristics of their cellular processes, tissue distribution and the manner in which they associate with axons (Butt and Berry 2000; Szuchet 1995).
The formation of the CNS myelin sheaths is the outcome of a sequence of complex developmental steps that require close and reciprocal communication between oligodendrocytes and the axons they contact. Little is known about the proteins that mediate axon-oligodendrocyte recognition at the onset of myelination (Rosenberg et al. 2007). Oligodendrocytes express a number of cell surface proteins that could mediate their association with axons. It has been suggested that integrins play a key role during myelination (Colognato et al. 2002; Relvas et al. 2001), but a recent study demonstrated that although they are important for oligodendrocyte survival, they are unnecessary for axonal ensheathment and myelination (Benninger et al. 2006). Another transmembrane protein linking cell recognition with Rho GTPases is Lingo-1, which is expressed both by oligodendrocytes and neurons (Lee et al. 2007; Mi et al. 2005). Lingo-1 is a negative regulator of CNS myelination, and may control the timing of this process during development. An inhibitory role in myelination was also attributed to polysialated NCAM (PSA-NCAM) and L1, which are two axonal members of the immunoglobulin superfamily of cell-adhesion molecules (IgCAMs) whose expression is regulated by electrical activity (Itoh et al. 1995; Kiss et al. 1994). The disappearance of PSA-NCAM from the axon was found to be a prerequisite for myelination to begin, while enzymatic cleavage of the sialylated determinants resulted in increased myelination (Charles et al. 2000). Axonal L1 was found to be strongly downregulated during myelination, which suggested that it plays a role during the initial alignment of glial processes with the underlying axon (Barbin et al. 2004). A similar role was proposed for CNR/protocadherin alpha family protein, whose expression is downregulated locally in myelinating internodes (Morishita et al. 2004). However, analysis of the CNS mice lacking L1 or NCAM revealed no evidence for a role of these molecules in myelination (Bartsch 2003), further suggesting the involvement of other proteins in the recognition of axons by myelinating glia. Oligodendrocytes also express several members of the immunoglobulin superfamily (IgCAMs) - Neurofascin 155 (NF155) (Tait et al. 2000), TAG-1 (Traka et al. 2002) and Contactin (Koch et al. 1997), all of which are important for oligodendrocyte-axon interactions at and around the nodes of Ranvier, but appear to be dispensable for myelination (Boyle et al. 2001; Poliak et al. 2003; Sherman et al. 2005). Myelin-associated glycoprotein (MAG) is another IgCAM that was originally suggested to mediate glia-axon attachments (Trapp 1990). However, further evidence from gene targeting studies demonstrated that MAG is required for myelin maintenance rather than for the initial association between oligodendrocyte and the axon (Li et al. 1994; Montag et al. 1994). Hence, despite the growing number of oligodendrocyte genes being discovered (Dugas et al. 2006; Nielsen et al. 2006), the identity of the cell surface molecules that mediate oligodendrocyte-axon contact during myelination still remains elusive.
To identify novel proteins important for the function of myelinating oligodendrocytes, we have made use of an animal model in which oligodendrocytes were eliminated using a binary genetic system. We used a mouse line (R26;lacZbpA(flox)DTA), in which a loxP-conditional diphtheria toxin allele (DTA) was placed by a knock-in strategy under the control of the ubiquitously active ROSA26 promoter (Brockschnieder et al. 2006a; Brockschnieder et al. 2006b). Crossing this strain to the CNPase-Cre line in which Cre recombinase (Cre) is expressed under control of the oligodendrocyte specific CNPase (2′3′-cyclo-nucleotide 3′-phosphodiesterase) promoter (Lappe-Siefke et al. 2003), resulted in the expression of the toxin and the ablation of all oligodendrocytes in the offspring (Brockschnieder et al., 2006b). By comparing the expression profile of brain stems isolated from wild type and oligodendrocytes-depleted mice, we have identified several genes that encode for membrane proteins that are enriched in oligodendrocytes. We further show that one of the identified proteins, transmembrane protein 10 (Tmem10; also known as Opalin), is specifically expressed by myelinating oligodendrocytes, suggesting that it may play a role in CNS myelination.
MATERIAL AND METHODS
Microarray, plasmids and antibodies
Isolation of mRNA from wild type and OL-ablated brain stems, as well as microarray analysis was done as previously described (Brockschnieder et al. 2006b). The Myc-tagged Tmem10 expression construct was generated by PCR cloning of Tmem10 open reading frame (ORF) from rat brain cDNA into the pcDNA3.1myc vector (Invitrogen). Mutations of the N-glycosylation sites of Tmem10 (Asn to Ala mutants) were done with specific primer pairs using Phusion high-fidelity DNA polymerase (Finnzyme). Polyclonal antibodies to Tmem10 were generated by immunizing rabbits with a GST-fusion protein containing amino acids 65–142 of rat Tmem10.
RT-PCR and in situ hybridization
Total RNA was isolated from freshly dissected rat brains of the indicated age using TRI-reagent (Sigma). cDNAs were obtained with Super Script reverse transcriptase II (Invitrogen) and were normalized among different samples with actin-specific primers. Synthetic digoxigenin-labeled riboprobes (cRNA) were produced based on pGEM-T-easy vector (Promega) containing 1000–1500 base pairs of the indicated mRNA. In vitro transcription was done from both sides with either SP6 or T7 RNA polymerase, generating anti-sense or sense (control) cRNA probes. Probes were alkaline hydrolyzed to an average length of 200–400 bases. In situ hybridization was performed on cryosections of freshly frozen mouse brain as previously described (Spiegel et al. 2002).
Cell-culture methods
Cell-lines were grown in Dulbecco’s Modified Eagle’s Medium (DMEM; Gibco) supplemented with 10% Fetal Bovine Serum (FBS; Gibco). Dissociated spinal cord cells were prepared from mice embryos at 13.5 day of gestation using a modified procedure originally described for explant cultures (Thomson et al. 2006). Meninges-free spinal cord tissue was mechanically minced, incubated with trypsin and collagenase at 37°C, and the reaction was terminated with soybean trypsin inhibitor solution (Leibovitz’s L15 medium with 0.52mg/ml soybean trypsin inhibitor, 0.04mg/ml bovine pancreas DNAse, 3mg/ml bovine serum albumin). Cells were gently triturated, collected by centrifugation in plating medium (PM; 50% DMEM, 25% horse serum, 25% Hank’s balanced salt solution, 2mM glutamine), resuspended in fresh PM and seeded on Poly-L-Lysine (PLL) coated glass slides. After two hours incubation at 37°C, cells were supplemented with an equal volume of differentiation medium (DfM)+Insulin (DMEM with 4.5mg/ml glucose, 10ng/ml biotin, 50nM hydrocortisone, 5μg/ml apo-transferrin, 100μM putrescine, 20nM progesterone, 30nM selenium, 10μg/ml Insulin) and grown in 37°C with 5% CO2. After two weeks in cultures slides were grown in DfM without Insulin until used.
Immunofluorescence
Brain, spinal cord, and optic nerves were obtained from 4% PFA perfused mice. The tissues were cryoprotected in 30% sucrose in PBS, for 12 hours at 4°C, embedded in OCT (Tissue-Tek) and snap frozen in liquid nitrogen. 12 micron-thick sections were prepared and permeabilized with ice-cold methanol for 5 minutes. After consecutive PBS washes, the slides were incubated with blocking buffer (PBS with 0.5% Triton X-100 and 5% normal goat serum) for 1 hour at room temperature. Primary antibodies were diluted in blocking buffer and added to the slides for several hours at 4°C in a humid chamber. Slides were then washed with PBS, incubated with Cy3-, Cy5- or Alexa488-coupled secondary antibodies (Jackson Laboratories and Molecular Probes), rinsed again in PBS and mounted with elvanol. Staining of cultures was done as previously described (Brockschnieder et al. 2006b). Fluorescence images were obtained using an Axioskop 2 microscope equipped with Apotom imaging system (Carl Zeiss), or a Nikon eclipse E1000 microscope fitted with a Hamamatsu ORCA-ER CCD camera. Composition of Z-stack images was done using AxioVision 4.4 (Zeiss) and Photoshop (Adobe).
Immunoblotting, immunoprecipitation and cell surface labeling
Freshly dissected tissues and cultured cells were homogenized in RIPA buffer (50mM Tris-HCl pH=7.4, 1% NP-40, 0.25% Sodium-deoxycholate, 150mM NaCl, 1mM EDTA, 1mM PMSF, 10μg/ml Aprotinin and Leupeptin), incubated on ice for 30–60 minutes, centrifuged at 10,000 g for 30 minutes and supernatant was collected for further use. SDS-PAGE sample buffer was added to a 30 μg total protein lysate and resolved in Tris-Acetate 7% or gradient Bis-Tris 4%–12% acryl-amide gels (Invitrogen). Cell surface biotinylation, immunoprecipitation and immunoblotting were done as previously described (Gollan et al. 2002).
Immunoelectron Microscopy
Colloidal gold conjugate to protein A was obtained from the Cell Microscopy Center, Department of Cell Biology, University Medical Center Utrecht, The Netherlands. Sample preparation and immunolabeling was done as described previously (Werner et al. 2007). Mice were fixed by transcardial perfusion with 4% formaldehyde and 0.2% glutaraldehyde in 0.1 M phosphate buffer containing 0.5% NaCl. Dissected optic nerves were infiltrated in 2.3 M sucrose in 0.1M phosphate buffer over night, mounted onto aluminum pins for ultramicrotomy and frozen in liquid nitrogen. Ultrathin cryosections were picked in a 1:1 mixture of 2% methylcellulose and 2.3M sucrose. Sections were incubated with antibodies specific for Tmem10 followed by protein A-gold (10 nm). Sections were analyzed with a LEO EM912 Omega (Zeiss, Oberkochen) and digital micrographs were obtained with an on-axis 2048×2048-CCD camera (Proscan, Scheuring).
RESULTS
Identification of oligogenes by microarray-coupled genetic cell ablation
To identify novel proteins that are preferentially expressed in oligodendrocytes, we have compared gene expression profiles of the brain stem of wild type and oligodendrocyte-ablated mice by Affymetrix microarray (Brockschnieder et al. 2006b). We expected mRNAs level of genes that were uniquely expressed in oligodendrocytes to be dramatically reduced in brains missing this cell type. As summarized in Table 1, we noted a 30–200-fold reduction in the expression of known myelin genes (Mbp, Mag, Mal, Plp, Mog, Mobp, Ermin) in the oligodendrocytes-ablated brains, further validating our approach. A lesser, but significant reduction was also detected in the expression of genes encoding for myelin lipids enzymes such as UDP-galactosyltransferase (Ugt8a), fatty acid 2-Hydroxylase (Fa2h) and fatty acid desaturase (Elovl1), as well as in the expression of genes encoding for other known myelin proteins, CNPase (Cnp1) and Plasmolipin (Pllp). The fact that a very low level of expression was still detected for some of the genes in oligodendrocyte-ablated brains, suggest the existence of a minor population of oligodendrocytes that escaped the Cre-mediated genetic ablation. Further analysis of the genes whose expression was reduced in oligodendrocytes-ablated mice, revealed that a number of them encode for cell adhesion molecules (CAMs), channels, receptors and ligands (Table 2). The group of CAMs identified includes several tetraspanins (Cldn11, Cldn14, Tspan2, Tspan15, Tmem125), members of the immunoglobulin superfamily (Nfasc, Jam3, Mcam), protocadherins (Pcdh17), the ADAM like metalloproteases (Adamts4), as well as several novel CAMs encoded by Tmem10, Tmem2, Tjp4 and Tmcc3 genes. Only three of the identified CAMs, i.e., Claudin 11, Tetraspanin 2 and Neurofascin, were previously reported to be expressed by myelinating oligodendrocytes (Birling et al. 1999; Gow et al. 1999; Morita et al. 1999; Tait et al. 2000). Examination of the Allen Brain Atlas (http://brain-map.org/welcome.do), as well as in situ hybridization analysis done using specific probes to Tspan2, Jam3, Tmcc3 and Cldn11 (Supplementary Fig. 1), further confirm the presence of these genes in the mouse white matter.
Table 1.
Expression of myelin genes after oligodendrocyte ablation.
| UniGene | Gene | Description | WT | OL | Fold |
|---|---|---|---|---|---|
| Mm.40461 | Mobp | Myelin-Associated Oligodendrocytic Basic Protein | 1744 | 8 | 218 |
| Mm.210857 | Mog | Myelin Oligodendrocyte Glycoprotein | 1351 | 8 | 169 |
| Mm.241355 | Mag | Myelin-Associated Glycoprotein | 2985 | 43 | 69 |
| Mm.39040 | Mal | Myelin And Lymphocyte Protein | 2711 | 50 | 54 |
| Mm.1268 | Plp1 | Proteolipid Protein (Myelin) 1 | 5394 | 148 | 36 |
| Mm.252063 | Mbp | Myelin Basic Protein | 8149 | 225 | 36 |
| Mm.91712 | A330104H05Rik | Ermin | 402 | 12 | 35 |
| Mm.462581 | Cnp1 | Cyclic Nucleotide Phosphodiesterase 1 | 2394 | 158 | 15 |
| Mm.306021 | Ugt8a | UDP Galactosyltransferase 8A | 3431 | 253 | 13 |
| Mm.41083 | Fa2h | Fatty Acid 2-Hydroxylase | 1329 | 154 | 8 |
| Mm.282096 | Elovl1 | Elongation Of Very Long Chain Fatty Acids | 505 | 122 | 4 |
| Mm.279977 | Pllp | Plasma membrane proteolipid (Plasmolipin) | 1241 | 340 | 3.6 |
Numbers indicate relative levels of expression of the indicated genes in the brain stem of 12-day-old wild type (WT) and oligodendrocytes-ablated (OL) mice as determined by microarray analysis. Fold indicates fold reduction in the expression of each gene between WT and OL mice.
Table 2.
Oligogenes encoding for transmembrane proteins.
| UniGene | Gene | Description | WT | OL | Fold | |
|---|---|---|---|---|---|---|
| Cell Adhesion | Mm.23639 | Tmem125 | Transmembrane protein 125 | 471 | 3 | 188.2 |
| Mm.23156 | Adamts4 | Disintegrin-like and metalloprotease | 731 | 22 | 33.0 | |
| Mm.54119 | Tmem10 | Transmembrane protein 10 | 344 | 14 | 23.7 | |
| Mm.4425 | Cldn11 | Claudin 11 | 1383 | 76 | 18.2 | |
| Mm.153643 | Pcdh17 | Protocadherin 17 | 794 | 93 | 8.5 | |
| Mm.329776 | Tmem2 | Transmembrane protein 2 | 730 | 96 | 7.6 | |
| Mm.27469 | Tspan2 | Tetraspan 2 | 1977 | 402 | 4.9 | |
| Mm.416781 | Tspan15 | Tetraspanin 15 | 236 | 62 | 3.8 | |
| Mm.328716 | Cldn14 | Claudin 14 | 108 | 29 | 3.7 | |
| Mm.380307 | Nfasc | Neurofascin | 562 | 159 | 3.5 | |
| Mm.28770 | Jam3 | Junction Adhesion Molecule 3 | 259 | 75 | 3.4 | |
| Mm.275003 | Mcam | Melanoma cell adhesion molecule | 187 | 75 | 2.5 | |
| Mm.23047 | Tmcc3 | Transmembrane & coiled coil domains 3 | 228 | 113 | 2.0 | |
| Channels | Mm.21198 | Gjb1 | Gap junction channel protein b 1 (Cx32) | 189 | 4 | 48.0 |
| Mm.40016 | Gja12 | Gap junction channel protein a 12 (Cx47) | 230 | 9 | 24.9 | |
| Mm.332771 | Gje1 | Gap junction channel protein e (Cx29) | 94 | 24 | 4.0 | |
| Mm.270088 | Slc44a1 | Solute carrier family 44, member 1 | 911 | 365 | 2.5 | |
| Receptors and Ligands | AF44069 | Trf | Transferrin | 2513 | 75 | 33.7 |
| NM_031252 | Il23a | Interleukin 23, alpha subunit p19 | 84 | 6 | 14.6 | |
| Mm.190619 | Edg8 | G-protein-coupled receptor | 191 | 17 | 11.2 | |
| Mm.373043 | Erbb3 | Neuregulin receptor | 313 | 33 | 9.6 | |
| Mm.4772 | Edg2 | G-Protein-Coupled Receptor | 1160 | 122 | 9.4 | |
| Mm.326268 | Gpr62 | G Protein-coupled receptor 62 | 228 | 29 | 7.7 | |
| Mm.40909 | Sema6a | Semaphorin 6A | 153 | 34 | 4.5 | |
| Mm.275600 | Plexin B3 | Plexin B3 | 1064 | 243 | 4.3 | |
| Mm.290433 | Unc5b | Netrin receptor | 105 | 27 | 3.8 | |
| Mm.409670 | Gpr37 | G Protein-Coupled Receptor 37 | 661 | 199 | 3.3 | |
| Mm.245154 | Tmeff2 | TM with EGF & follistatin-like domains 2 | 1966 | 607 | 3.2 | |
| Mm.5021 | Ddr1 | Discoidin domain receptor | 462 | 170 | 2.7 | |
| Mm.2901 | Tyro3 | TYRO3 Protein Tyrosine Kinase 3 | 234 | 90 | 2.6 | |
| Mm.3213 | Ldlr | Low density lipoprotein receptor | 397 | 156 | 2.5 | |
| Mm.33903 | Sema4d | Semaphorin 4D | 552 | 222 | 2.4 | |
| Mm.192580 | Rtn4 | Reticulon 4 (Nogo) | 296 | 140 | 2.1 | |
| Mm.261182 | Tmbim1 | Transmembrane BAX Inhibitor | 359 | 130 | 2.7 | |
| Mm.27064 | March8 | Membrane-Associated Ring Finger 8 | 345 | 131 | 2.6 |
Numbers indicate relative levels of expression of the indicated genes in the brain stem of 12-day-old wild type (WT) and oligodendrocytes-ablated (OL) mice as determined by microarray analysis. Fold reduction in the expression of each gene after oligodendrocyte ablation is shown on the right. Three functional groups containing cell adhesion molecules, channels and transporters, and various receptors and ligands are indicated.
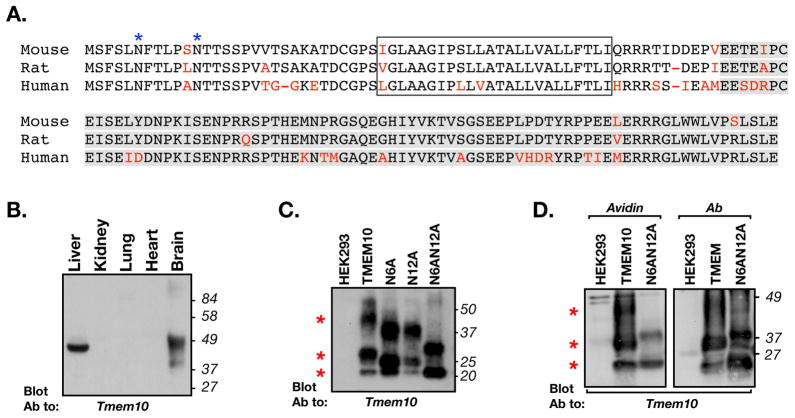
Tmem10 encodes for a cell surface protein
The expression of one of the novel genes, Tmem10, was reduced more than 23 fold in the dysmyelinated brain stem, suggesting that it is highly enriched in oligodendrocytes. Sequencing of Tmem10 cDNA cloned from mouse brain predicted that it encodes for a 143 amino acid-long transmembrane protein (Fig. 1A). The amino acid sequence of mouse Tmem10 share 95% and 85% similarity to the rat and human proteins, respectively. Tmem10 contains a predicted short (30 aa) extracellular region, containing two N-glycosylation sites (NetNGlyc 1.0; http://www.cbs.dtu.dk/services/NetNGlyc/), nine mucin-type O-glycosylation sites (NetOGlyc 3.1; http://www.cbs.dtu.dk/services/NetOGlyc/;(Julenius et al. 2005), a single transmembrane domain (Phobius; http://phobius.sbc.su.se/;(Kall et al. 2007), and an 89 amino acid-long cytoplasmic tail that shows no recognizable motifs. Western blot analysis using an antibody that recognizes the cytoplasmic domain of Tmem10, showed that it could be detected in extracts from liver and brain, but not from the kidney, lung, or heart (Fig. 1B). In the brain, Tmem10 appeared as a broad band centered at 40 kDa, indicating that it is indeed heavily glycosylated. This was even more apparent in lysates of transfected HEK293 cells, where Tmem10 was detected as three main bands of 22kDa, 30kDa and 45kDa (Fig. 1C). Mutating the two asparagine residues in the extracellular domain to alanine, reduced the apparent molecular mass of Tmem10, indicating that it undergoes N-linked glycosylation. Notably, even in the absence of the two N-linked glycosylation sites (N6AN12A), Tmem10 still appeared as two broad bands, suggesting that it undergoes additional posttranslational modifications such as O-glycosylation as predicted from its amino acid sequence. To determine whether Tmem10 is expressed on the cell surface, we labeled transfected HEK293 with a membrane impermeable biotinylation reagent, precipitated the biotinylated proteins with agarose-immobilized avidin and immunoblotted the resulting complexes with an antibody to Tmem10 (Fig. 1D). This analysis revealed that all the bands that were detected after immunoprecipitating Tmem10 with a specific antibody (Fig. 1D right panel), were also detected using the avidin-beads (Fig. 1D left panel), demonstrating that Tmem10 is a cell surface protein. The Tmem10-(N6AN12A) mutant was also detected on the cell surface although in a reduced manner, suggesting that full glycosylation of the protein is required for its stable appearance on the plasma membrane. The conclusion that Tmem10 is a cell surface protein was also supported by immunolabeling experiments, showing that in transfected epithelial cells (COS7 and MDCK) it was distributed along the cell membrane and was often concentrated at cell-cell contact sites (data not shown).
Figure 1. Tmem10 encodes for a transmembrane glycoprotein.
A. Alignment of the amino acid sequence of mouse, rat and human Tmem10 (different amino acids are shown in red. The transmembrane region is boxed and the region used to generate the antibody is highlighted in gray. The two asparagine-linked glycosylation sites found at the extracellular region of Tmem10 are labeled with blue asterisks. B. Tissue distribution. Immunoblot analysis of protein extracts prepared from the indicated tissues with an antibody to Tmem10. The location of molecular mass markers is shown on the right in kDa. C. Immunoblot analysis of glycosylation-site mutants of Tmem10. HEK293 cells transfected with an empty vector (HEK293) or cells expressing a myc-tagged Tmem10 (TMEM10), a mutant Tmem10 in which asparagine at position 6 was replaced with alanine (N6A), a mutant Tmem10 in which asparagine at position 12 was replaced with alanine (N12A), or a Tmem10 in which both asparagines were mutated to alanine (N6AN12A), were immunoprecipitated with an anti-myc antibody followed by immunoblotting with an antibody to Tmem10. Red asterisks indicate the locations of the three main bands of Tmem10. D. Cell surface biotinylation. HEK293 cells transfected with an empty vector or cells expressing a myc-tagged Tmem10 (TMEM10), or a mutant lacking its glycosylation sites (N6AN12A), were incubated with membrane impermeable NHS-biotin. Cell lysates were mixed with avidin-agarose (Avidin) or were immunoprecipitated with an antibody to myc (Ab), followed by immunoblotting with an antibody to Tmem10. Both wild type and the glycosylation mutant of Tmem10 reach the cell surface.
Tmem10 is expressed by myelinating oligodendrocytes
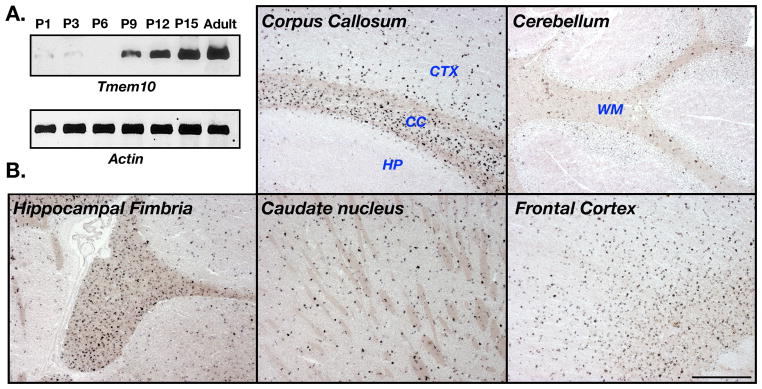
To study the expression of Tmem10, we first performed RT-PCR analysis on brain mRNAs isolated from rat at different ages (Fig. 2A). As expected from a myelin gene, Tmem10 transcript was detected from postnatal day 9 and its level increased thereafter until it reached its maximal level in adults. In situ hybridization analysis of adult mice brain using a Tmem10 antisense probe showed an intense labeling in the corpus callosum, hippocampal fimbria, caudate nucleus, putamen and the cerebral cortex (Fig. 2B). A weaker, scattered signal was detected in the white matter of the cerebellum. At large, and with the exception of the cerebellum, the signal obtained with the Tmem10 probe was very similar to that obtained with myelin genes such as MAG and MAL (data not shown).
Figure 2. Tmem10 transcript is found in CNS white matter.
A. Developmental expression of Tmem10 mRNA. RT-PCR analysis of brain RNA isolated from rat at the indicated postnatal days. B. In situ hybridization. Hybridization of the indicated adult mouse brain tissue was performed using an anti-sense probe containing the entire Tmem10 coding region. CTX, cortex; CC, corpus callosum; HP, hippocampus. Scale bar, 500 mm.
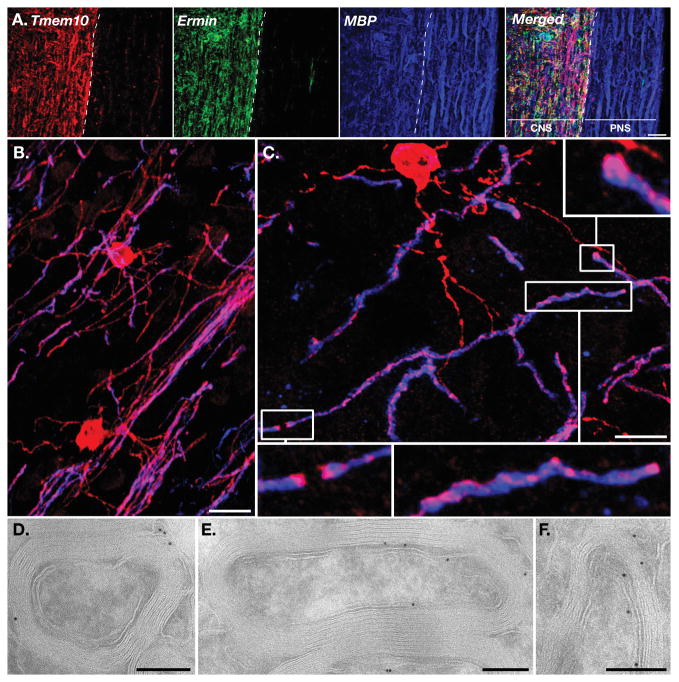
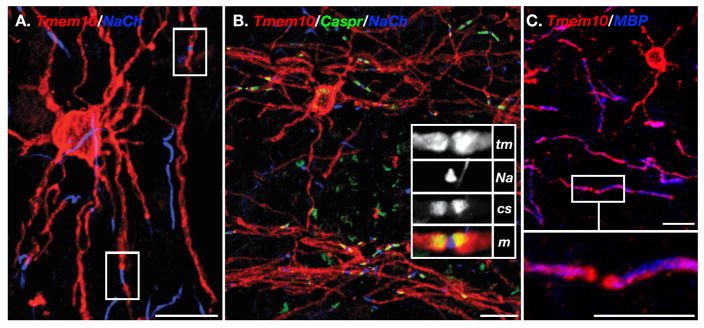
We next performed immunofluorescence staining of mouse brain and spinal cord sections. Triple labeling of mouse spinal cord with antibodies to Tmem10, the myelin marker MBP, and the CNS-specific myelin marker Ermin (Brockschnieder et al. 2006b), revealed that Tmem10 is strongly expressed in the white matter of the spinal cord (Fig. 3A). In contrast, Tmem10 immunoreactivity was completely absent in the myelinated ventral roots, demonstrating that it is specifically present in CNS but not in PNS myelin. No Tmem10 immunoreactivity was detected in GFAP-labeled astrocytes (data not shown). Double labeling of mouse frontal cortex with antibodies to Tmem10 and MBP further showed that Tmem10 is an oligodendrocyte-specific protein (Fig. 3B): Tmem10 was detected in the oligodendrocyte soma, myelin internodes and cellular processes, including very fine extensions that are negative for MBP. Notably, Tmem10 immunoreactivity was stronger in the perimeter of the cell body, which is consistent with a plasma membrane protein (see also Fig. 6A–B). Along the internodes, it was present in a line that circled around the compact myelin (Fig. 3C). Immunoelectron microscopy analysis of optic nerves and the corpus callosum revealed that along the internodes, Tmem10 was present at the outer (abaxonal) and inner (adaxonal) membranes, but was absent from the compact myelin (Fig. 3D–F). Tmem10 was also enriched in the paranodal region (Fig. 3C), but was completely absent from the nodes as observed by multiple immunolabeling with antibodies to Na+ channels and Caspr (Fig. 4A–B). This pattern was unchanged inparanodal junction mutants such as Caspr (Fig. 4C). We thus concluded that in myelinating oligodendrocytes, Tmem10 is found in the cell body, cell processes and in non-compact myelin along the internodes and the paranodal regions.
Figure 3. Tmem10 is expressed by myelinating glia in the CNS.
A. Immunolabeling of longitudinal spinal cord sections containing the ventral roots for Tmem10 (red), Ermin (green) and MBP (blue). The merged image is shown on the right panel. The border between the CNS and PNS is marked with a broken white line. Note that in contrast to MBP, both Tmem10 and Ermin are absent from the peripheral nerve. B–C. Immunolabeling of mouse frontal cortex for Tmem10 (red) and MBP (blue). Tmem10 immunoreactivity was detected in the cell bodies and processes. Higher magnifications of the boxed regions in panel C show focal concentration of Tmem10 at the paranodal loops and as a thread that circles around the MBP-labeled internodes. D–F. Immunoelectron microscopy analysis of rat optic nerve (D) or corpus callosum (E–F) using antibodies to for Tmem10. Scale bar: A, 20 μm; B–C, 10μm; D–F 200nm.
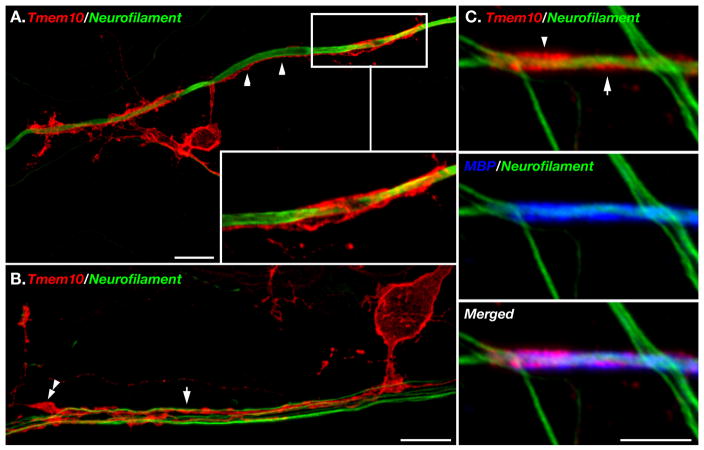
Figure 6. Localization of Tmem10 in myelinated cultures.
A–B. Spinal cord cultures were labeled with an antibody to Tmem10 (red) and neurofilament (green). Inset in A shows a higher magnification of an oligodendrocyte process that is aligned with the axon (arrowhead) and forms a tube that surrounds it. In B, Tmem10 is found along oligodendrocyte processes that are sorting a bundle of axons (arrow). One process ends in a filopodial protrusion (double arrowhead). C. Immunolabeling of myelin segments with antibodies to Tmem10 (red), MBP (blue) and neurofilament (green). Tmem10 is circled around the internodes (arrow) and accumulated at the paranodes (arrowhead). Scale bar: A–B, 10 μm; C, 5 μm.
Figure 4. Enrichment of Tmem10 at the paranodes.
A. Double immunolabeling of mouse frontal cortex with an antibody to Tmem10 (red), and Na+ channels (blue). The location of two nodes is boxed. B. Mouse frontal cortex tissue was immunolabeled with antibodies to Tmem10 (red), Caspr (green) and Na+ channels (blue). Higher magnification of a nodal area is shown in the inset (tm, Tmem10; Na, Na+ channels; cs, Caspr; m, merged). C. Immunolabeling of Caspr−/− mouse cortex with antibodies to Tmem10 (red) and MBP (blue). Higher magnification of the boxed area is shown in the lower panel. Scale bars: A–B, 20 μm; C, 10 μm.
Developmental expression of Tmem10 during myelination
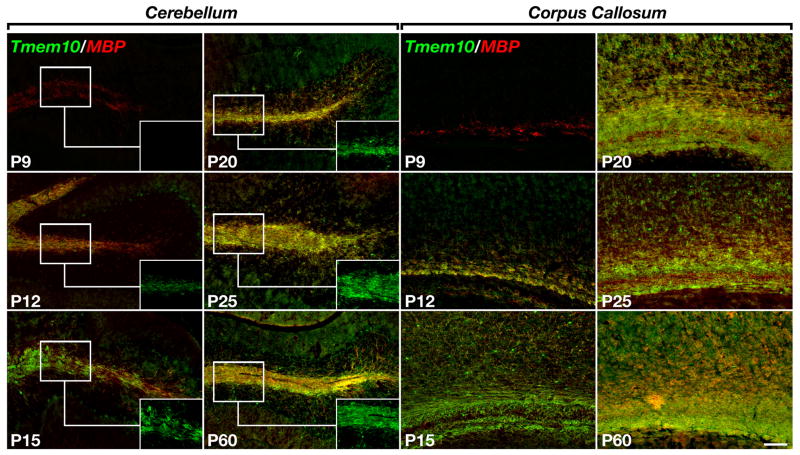
Next, we set out to determine when Tmem10 is expressed during myelination. To this end, we immunolabeled sections of mouse cerebellum and corpus callosum obtained at different postnatal ages (i.e. P7–P60), with antibodies to Tmem10 and MBP (Fig. 5.). We found that Tmem10 expression was restricted to MBP-labeled oligodendrocytes but lagged behind MBP by 3–4 days. At P7, no Tmem10-positive oligodendrocytes were found in the corpus callosum or cerebellum while a few dozen of Tmem10-labeled cells were present in the medulla (data not shown). At this stage, MBP-positive oligodendrocytes were located at the middle and front portion of the corpus callosum, whereas only a few MBP-positive cells were detected in some of the cerebellar lobes. At P9, a limited number of Tmem10-positive oligodendrocytes were present in the middle portion of the corpus callosum but not in the cerebellum. At P12, Tmem10-labeled MBP-positive oligodendrocytes were present throughout the corpus callosum, as well as in the white matter tracks of the cerebellum and hippocampal fimbria. The expression of Tmem10 increased thereafter and could be detected in all MBP-labeled oligodendrocytes from P15 to adult (Fig. 5). This analysis demonstrated that during myelination, the expression of Tmem10 by oligodendrocytes followed that of MBP. To further determine whether Tmem10 could play a role during myelination, we made use of myelinating spinal cord cultures. As depicted in Figure 6A–B, Tmem10 was present in cellular processes that contacted and ensheathed the axon prior to myelination. Similar to the situation in vivo, Tmem10 was always expressed by MBP-positive oligodendrocytes (data not shown). In myelinated segments, Tmem10 was found along the myelin internodes and was, to some extent, concentrated at paranodes (Fig. 6C).
Figure 5. Expression of Tmem10 during myelination.
A. Expression of Tmem10 in mouse brain. Tissue sections containing the cerebellum and corpus callosum obtained from mice at the indicated postnatal days (P9–P60), were double labeled with antibodies to Tmem10 (green) and MBP (red). Insets show high magnification of Tmem10 labeling in the cerebellar white matter. Scale bar: 100 μm.
DISCUSSION
In this work we combined gene expression profiling with genetic cell ablation to search for genes that encode for membrane proteins enriched in oligodendrocytes. The genes identified using this approach encode for cell adhesion molecules (CAMs), connexins, receptor tyrosine kinases, repellent molecules and their receptors, and G protein-coupled receptors (Table 2). The identified CAMs include members of the tetraspanin (Tspan2, Tspan15, Tmem125), claudin (Cldn11, Cldn14) and immunoglobulin (Nfasc, Jam3, Mcam) subfamilies. Claudin 11 is expressed by myelinating oligodendrocytes and is required for generating the intramembranous junctions within CNS myelin sheaths known as the radial component oligodendrocytes (Birling et al. 1999; Gow et al. 1999; Morita et al. 1999; Tait et al. 2000). It also generates a complex with other tetraspanins and integrins that regulate oligodendrocytes proliferation and migration (Tiwari-Woodruff et al. 2001). Similarly, Tspan2 was suggested to regulate extracellular signaling during the early stages of oligodendrocytes differentiation, as well as to function in stabilizing the mature sheath (Birling et al. 1999). No data is currently available on the expression or possible function of the other two tetraspanin proteins, Tspan15 and Tmem125 in myelinating glia. The expression of Tmem125 was dramatically (188 fold) reduced after oligodendrocyte depletion, suggesting that it is restricted to this cell type. Furthermore, although Tmem125 was not detected by gene expression profiling of differentiating oligodendrocytes in vitro (Dugas et al. 2006), it was detected in FACS-purified O4+ oligodendrocytes from rat brains (Nielsen et al. 2006). The difference between these results may indicate that its expression is tightly regulated in the presence of axons, a hypothesis that is worth further investigation. We also identified three members of the immunoglobulin superfamily – Neurofascin, Junction adhesion molecule 3 (Jam3) and Melanoma cell adhesion molecule (Mcam, also known as Gicerin, MUC18 and CD146). Neurofascin is expressed by myelinating oligodendrocytes (Birling et al. 1999; Gow et al. 1999; Morita et al. 1999; Tait et al. 2000), and is required for the establishment of the specialized axoglial junction that is formed between the paranodal loops and the axon (Boyle et al. 2001; Poliak et al. 2003; Sherman et al. 2005). A role for Jam3 and Mcam in CNS myelin is yet to be revealed, although interestingly, in endothelial cells Mcam triggers tyrosine phosphorylation of FYN (Anfosso et al. 1998), an intracellular tyrosine kinase that is required for myelination (Sperber et al. 2001). The expression and a possible role of Adamts4, Protocadherin 17, Tmem2, and Tmcc3 in oligodendrocytes are yet to be reported.
We have focused our present study on Tmem10, a novel oligodendrocyte transmembrane protein that may play a role in CNS myelination. Tmem10 contains a single transmembrane domain and a long cytoplasmic region, which show no apparent sequence homology to any other protein in the database. Its short, 30 amino acids- long extracellular region contains two N-linked and nine O-linked mucin type glycosylation sites, which render the protein highly glycosylated. Surface biotinylation experiments done by using avidin beads to precipitate labeled proteins followed by immunoblotting with anti-Tmem10 antibody (Fig. 1D), or by immunoprecipitating Tmem10 first followed by blotting with streptavidin HRP (data not shown), demonstrated that, although Tmem10 lacks a signal sequence, it is present on the cell surface. This conclusion was also supported by the observations that Tmem10 immunoreactivity appeared at the circumference of the oligodendrocyte cell body (Fig. 6) and of transfected epithelial cells (data not shown). Given the conflicting adhesive and repulsive roles of glycans in neural cell interactions (Kleene and Schachner 2004), the expression of heavily-glycosylated protein on the surface of oligodendrocytes during myelination may provide an anti-adhesive force important for the progression of the myelin membrane during wrapping. In line with this idea, we found that prior to myelination, Tmem10 was found in cellular processes of oligodendrocytes that were aligned along axons and begun had to ensheath them.
Developing oligodendrocytes undergo dramatic changes in shape, from bipolar precursors to multipolar myelinating cells. In culture, oligodendrocytes differentiate along a defined pathway, marked by a typical change in cell morphology and the expression of myelin-specific lipids and proteins (Pfeiffer et al. 1993). In the absence of axons, these differentiated oligodendrocytes form a myelin-like membrane (Pfeiffer et al. 1993), and express a similar repertoire of genes that is found in mature myelinated oligodendrocytes in vivo (Dugas et al. 2006). In the adult brain, Tmem10 was found along the myelin internodes, at the plasma membrane of the cell body, as well as on the thin cytoplasmic processes that bridge between the cell body and the myelin sheath. This localization was in marked contrast with MBP, which was found at the cell body early during myelination but was absent from this site in mature myelinating oligodendrocyte. Developmental immunofluorescence analysis of mouse brain revealed that Tmem10 appeared after MBP but before Ermin, which is the latest marker of oligodendrocyte differentiation (Fig. 5 and data not shown). Similarly, we also found that during the differentiation of oligodendrocytes precursors in culture, Tmem10 was present in O4+ cells that already expressed MBP and CNPase (data not shown). In agreement with our results, Tmem10 was found to be upregulated in differentiating oligodendrocytes after five days in vitro (Dugas et al. 2006), as well as during the transition between A2B5+ oligodendrocyte progenitors and O4+ oligodendrocytes in vivo (Nielsen et al. 2006). An insight into the molecular mechanism controlling Tmem10 expression was recently provided by Furuichi and colleagues (Aruga et al. 2007), who elegantly showed the presence of a short 490 bp oligodendrocyte-specific enhancer in the first intron of this gene (termed Opalin by this group). This enhancer sequence was sufficient to direct the expression of a LacZ reporter in oligodendrocytes when placed upstream of a housekeeping promoter in transgenic animals (Aruga et al. 2007). Furthermore, treatment of a oligodendroglial cell line with medium conditioned from neurons, led to a strong upragulation of Tmem10 transcript, demonstrating that its expression is tightly regulated by neurons (Mikael Simons, personal communication).
Immunolabeling of mouse spinal cord sections containing the attached ventral roots demonstrated that similarly to the CNS-specific cytoskeletal protein Ermin, Tmem10 is completely absent from white matter in the PNS. This result agrees with a previous microarray analysis, detecting Tmem10 in genes expressed by the lumbar spinal cord but not in dorsal root ganglion (DRG) (LeDoux et al. 2006). Thus, Tmem10 belongs to a small group of myelin proteins that includes Claudin 11 (Morita et al. 1999), MOBP (Montague et al. 2005), Connexin 47 (Kleopa et al. 2004), GPR62 (LeDoux et al. 2006), and Ermin (Brockschnieder et al. 2006b), which are expressed by mature oligodendrocytes and not by myelinating Schwann cells. Interestingly, all of these genes were identified after oligodendrocytes ablation (Tables 1–2), suggesting that the approach used here is useful to identify genes that are specifically expressed by myelinating glia in the CNS vs. the PNS. Further experiments using Schwann cell ablation will be required to test this suggestion, as well as to determine whether cell type specific genes serve key functions.
Supplementary Material
Acknowledgments
We would like to thank Klaus Armin-Nave for the CNP-Cre mice, and Mikael Simons and Teiichi Furuichi for discussions and sharing unpublished information. This work was supported by Shapell Family Biomedical Research Foundation and the Kekst Family Center for Medical Genetics at the Weizmann Institute, the Israel Science Foundation, the NIH (NINDS NS50220), and the Wolgin Prize for Scientific Excellence.
References
- Anfosso F, Bardin N, Frances V, Vivier E, Camoin-Jau L, Sampol J, Dignat-George F. Activation of human endothelial cells via S-endo-1 antigen (CD146) stimulates the tyrosine phosphorylation of focal adhesion kinase p125(FAK) J Biol Chem. 1998;273(41):26852–6. doi: 10.1074/jbc.273.41.26852. [DOI] [PubMed] [Google Scholar]
- Aruga J, Yoshikawa F, Nozaki Y, Sakaki Y, Toyoda A, Furuichi T. An oligodendrocyte enhancer in a phylogenetically conserved intron region of the mammalian myelin gene Opalin. J Neurochem. 2007;102(5):1533–47. doi: 10.1111/j.1471-4159.2007.04583.x. [DOI] [PubMed] [Google Scholar]
- Barbin G, Aigrot MS, Charles P, Foucher A, Schachner M, Zalc B, Lubetzki C. L1 adhesion molecule and CNS myelination. Neuron Glia Biol. 2004;1:65–72. doi: 10.1017/S1740925X04000092. [DOI] [PubMed] [Google Scholar]
- Bartsch U. Neural CAMS and their role in the development and organization of myelin sheaths. Front Biosci. 2003;8:d477–90. doi: 10.2741/1028. [DOI] [PubMed] [Google Scholar]
- Baumann N, Pham-Dinh D. Biology of oligodendrocyte and myelin in the mammalian central nervous system. Physiol Rev. 2001;81(2):871–927. doi: 10.1152/physrev.2001.81.2.871. [DOI] [PubMed] [Google Scholar]
- Benninger Y, Colognato H, Thurnherr T, Franklin RJ, Leone DP, Atanasoski S, Nave KA, Ffrench-Constant C, Suter U, Relvas JB. Beta1-integrin signaling mediates premyelinating oligodendrocyte survival but is not required for CNS myelination and remyelination. J Neurosci. 2006;26(29):7665–73. doi: 10.1523/JNEUROSCI.0444-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birling MC, Tait S, Hardy RJ, Brophy PJ. A novel rat tetraspan protein in cells of the oligodendrocyte lineage. J Neurochem. 1999;73(6):2600–8. doi: 10.1046/j.1471-4159.1999.0732600.x. [DOI] [PubMed] [Google Scholar]
- Boyle ME, Berglund EO, Murai KK, Weber L, Peles E, Ranscht B. Contactin orchestrates assembly of the septate-like junctions at the paranode in myelinated peripheral nerve. Neuron. 2001;30(2):385–97. doi: 10.1016/s0896-6273(01)00296-3. [DOI] [PubMed] [Google Scholar]
- Brockschnieder D, Pechmann Y, Sonnenberg-Riethmacher E, Riethmacher D. An improved mouse line for Cre-induced cell ablation due to diphtheria toxin A, expressed from the Rosa26 locus. Genesis. 2006a;44(7):322–7. doi: 10.1002/dvg.20218. [DOI] [PubMed] [Google Scholar]
- Brockschnieder D, Sabanay H, Riethmacher D, Peles E. Ermin, a myelinating oligodendrocyte-specific protein that regulates cell morphology. J Neurosci. 2006b;26(3):757–62. doi: 10.1523/JNEUROSCI.4317-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butt AM, Berry M. Oligodendrocytes and the control of myelination in vivo: new insights from the rat anterior medullary velum. J Neurosci Res. 2000;59(4):477–88. doi: 10.1002/(SICI)1097-4547(20000215)59:4<477::AID-JNR2>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- Charles P, Hernandez MP, Stankoff B, Aigrot MS, Colin C, Rougon G, Zalc B, Lubetzki C. Negative regulation of central nervous system myelination by polysialylated-neural cell adhesion molecule. Proc Natl Acad Sci U S A. 2000;97(13):7585–90. doi: 10.1073/pnas.100076197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colognato H, Baron W, Avellana-Adalid V, Relvas JB, Baron-Van Evercooren A, Georges-Labouesse E, ffrench-Constant C. CNS integrins switch growth factor signalling to promote target-dependent survival. Nat Cell Biol. 2002;4(11):833–41. doi: 10.1038/ncb865. [DOI] [PubMed] [Google Scholar]
- Dugas JC, Tai YC, Speed TP, Ngai J, Barres BA. Functional genomic analysis of oligodendrocyte differentiation. J Neurosci. 2006;26(43):10967–83. doi: 10.1523/JNEUROSCI.2572-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gollan L, Sabanay H, Poliak S, Berglund EO, Ranscht B, Peles E. Retention of a cell adhesion complex at the paranodal junction requires the cytoplasmic region of Caspr. J Cell Biol. 2002;157(7):1247–56. doi: 10.1083/jcb.200203050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gow A, Southwood CM, Li JS, Pariali M, Riordan GP, Brodie SE, Danias J, Bronstein JM, Kachar B, Lazzarini RA. CNS myelin and sertoli cell tight junction strands are absent in Osp/claudin-11 null mice. Cell. 1999;99(6):649–59. doi: 10.1016/s0092-8674(00)81553-6. [DOI] [PubMed] [Google Scholar]
- Itoh K, Stevens B, Schachner M, Fields RD. Regulated expression of the neural cell adhesion molecule L1 by specific patterns of neural impulses. Science. 1995;270(5240):1369–72. doi: 10.1126/science.270.5240.1369. [DOI] [PubMed] [Google Scholar]
- Julenius K, Molgaard A, Gupta R, Brunak S. Prediction, conservation analysis, and structural characterization of mammalian mucin-type O-glycosylation sites. Glycobiology. 2005;15(2):153–64. doi: 10.1093/glycob/cwh151. [DOI] [PubMed] [Google Scholar]
- Kall L, Krogh A, Sonnhammer EL. Advantages of combined transmembrane topology and signal peptide prediction--the Phobius web server. Nucleic Acids Res. 2007;35(Web Server issue):W429–32. doi: 10.1093/nar/gkm256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss JZ, Wang C, Olive S, Rougon G, Lang J, Baetens D, Harry D, Pralong WF. Activity-dependent mobilization of the adhesion molecule polysialic NCAM to the cell surface of neurons and endocrine cells. Embo J. 1994;13(22):5284–92. doi: 10.1002/j.1460-2075.1994.tb06862.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleene R, Schachner M. Glycans and neural cell interactions. Nat Rev Neurosci. 2004;5(3):195–208. doi: 10.1038/nrn1349. [DOI] [PubMed] [Google Scholar]
- Kleopa KA, Orthmann JL, Enriquez A, Paul DL, Scherer SS. Unique distributions of the gap junction proteins connexin29, connexin32, and connexin47 in oligodendrocytes. Glia. 2004;47(4):346–57. doi: 10.1002/glia.20043. [DOI] [PubMed] [Google Scholar]
- Koch T, Brugger T, Bach A, Gennarini G, Trotter J. Expression of the immunoglobulin superfamily cell adhesion molecule F3 by oligodendrocyte-lineage cells. Glia. 1997;19(3):199–212. doi: 10.1002/(sici)1098-1136(199703)19:3<199::aid-glia3>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- Lappe-Siefke C, Goebbels S, Gravel M, Nicksch E, Lee J, Braun PE, Griffiths IR, Nave KA. Disruption of Cnp1 uncouples oligodendroglial functions in axonal support and myelination. Nat Genet. 2003;33(3):366–74. doi: 10.1038/ng1095. [DOI] [PubMed] [Google Scholar]
- LeDoux MS, Xu L, Xiao J, Ferrell B, Menkes DL, Homayouni R. Murine central and peripheral nervous system transcriptomes: comparative gene expression. Brain Res. 2006;1107(1):24–41. doi: 10.1016/j.brainres.2006.05.101. [DOI] [PubMed] [Google Scholar]
- Lee X, Yang Z, Shao Z, Rosenberg SS, Levesque M, Pepinsky RB, Qiu M, Miller RH, Chan JR, Mi S. NGF regulates the expression of axonal LINGO-1 to inhibit oligodendrocyte differentiation and myelination. J Neurosci. 2007;27(1):220–5. doi: 10.1523/JNEUROSCI.4175-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Tropak MB, Gerlai R, Clapoff S, Abramow-Newerly W, Trapp B, Peterson A, Roder J. Myelination in the absence of myelin-associated glycoprotein. Nature. 1994;369(6483):747–50. doi: 10.1038/369747a0. [DOI] [PubMed] [Google Scholar]
- Mi S, Miller RH, Lee X, Scott ML, Shulag-Morskaya S, Shao Z, Chang J, Thill G, Levesque M, Zhang M, et al. LINGO-1 negatively regulates myelination by oligodendrocytes. Nat Neurosci. 2005;8(6):745–51. doi: 10.1038/nn1460. [DOI] [PubMed] [Google Scholar]
- Montag D, Giese KP, Bartsch U, Martini R, Lang Y, Bluthmann H, Karthigasan J, Kirschner DA, Wintergerst ES, Nave KA, et al. Mice deficient for the myelin-associated glycoprotein show subtle abnormalities in myelin. Neuron. 1994;13(1):229–46. doi: 10.1016/0896-6273(94)90472-3. [DOI] [PubMed] [Google Scholar]
- Montague P, McCallion AS, Barrie JE, Edgar JM, McLaughlin M, Davies RW, Griffiths IR. Characterization of the murine splice variant Mobp155: developmental CNS expression pattern and subcellular localization of epitope-tagged protein. Glia. 2005;50(1):80–5. doi: 10.1002/glia.20155. [DOI] [PubMed] [Google Scholar]
- Morishita H, Kawaguchi M, Murata Y, Seiwa C, Hamada S, Asou H, Yagi T. Myelination triggers local loss of axonal CNR/protocadherin alpha family protein expression. Eur J Neurosci. 2004;20(11):2843–7. doi: 10.1111/j.1460-9568.2004.03803.x. [DOI] [PubMed] [Google Scholar]
- Morita K, Sasaki H, Fujimoto K, Furuse M, Tsukita S. Claudin-11/OSP-based tight junctions of myelin sheaths in brain and Sertoli cells in testis. J Cell Biol. 1999;145:579–588. doi: 10.1083/jcb.145.3.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen JA, Maric D, Lau P, Barker JL, Hudson LD. Identification of a novel oligodendrocyte cell adhesion protein using gene expression profiling. J Neurosci. 2006;26(39):9881–91. doi: 10.1523/JNEUROSCI.2246-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer SE, Warrington AE, Bansal R. The oligodendrocyte and its many cellular processes. Trends Cell Biol. 1993;3(6):191–7. doi: 10.1016/0962-8924(93)90213-k. [DOI] [PubMed] [Google Scholar]
- Poliak S, Salomon D, Elhanany H, Sabanay H, Kiernan B, Pevny L, Stewart CL, Xu X, Chiu SY, Shrager P, et al. Juxtaparanodal clustering of Shaker-like K+ channels in myelinated axons depends on Caspr2 and TAG-1. J Cell Biol. 2003;162(6):1149–60. doi: 10.1083/jcb.200305018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Relvas JB, Setzu A, Baron W, Buttery PC, LaFlamme SE, Franklin RJ, ffrench-Constant C. Expression of dominant-negative and chimeric subunits reveals an essential role for beta1 integrin during myelination. Curr Biol. 2001;11(13):1039–43. doi: 10.1016/s0960-9822(01)00292-5. [DOI] [PubMed] [Google Scholar]
- Rosenberg SS, Powell BL, Chan JR. Receiving mixed signals: uncoupling oligodendrocyte differentiation and myelination. Cell Mol Life Sci. 2007 doi: 10.1007/s00018-007-7265-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman DL, Tait S, Melrose S, Johnson R, Zonta B, Court FA, Macklin WB, Meek S, Smith AJ, Cottrell DF, et al. Neurofascins are required to establish axonal domains for saltatory conduction. Neuron. 2005;48(5):737–42. doi: 10.1016/j.neuron.2005.10.019. [DOI] [PubMed] [Google Scholar]
- Sperber BR, Boyle-Walsh EA, Engleka MJ, Gadue P, Peterson AC, Stein PL, Scherer SS, McMorris FA. A unique role for Fyn in CNS myelination. J Neurosci. 2001;21(6):2039–47. doi: 10.1523/JNEUROSCI.21-06-02039.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiegel I, Salomon D, Erne B, Schaeren-Wiemers N, Peles E. Caspr3 and caspr4, two novel members of the caspr family are expressed in the nervous system and interact with PDZ domains. Mol Cell Neurosci. 2002;20(2):283–97. doi: 10.1006/mcne.2002.1110. [DOI] [PubMed] [Google Scholar]
- Szuchet S. The morphology and ultrastructure of oligodendrocytes and their functional implications. In: Kettenmann H, Ransom BR, editors. Neuroglia. New York: Oxford University Press; 1995. pp. 23–43. [Google Scholar]
- Tait S, Gunn-Moore F, Collinson JM, Huang J, Lubetzki C, Pedraza L, Sherman DL, Colman DR, Brophy PJ. An oligodendrocyte cell adhesion molecule at the site of assembly of the paranodal axo-glial junction. J Cell Biol. 2000;150(3):657–66. doi: 10.1083/jcb.150.3.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson CE, Hunter AM, Griffiths IR, Edgar JM, McCulloch MC. Murine spinal cord explants: A model for evaluating axonal growth and myelination in vitro. J Neurosci Res. 2006;84(8):1703–15. doi: 10.1002/jnr.21084. [DOI] [PubMed] [Google Scholar]
- Tiwari-Woodruff SK, Buznikov AG, Vu TQ, Micevych PE, Chen K, Kornblum HI, Bronstein JM. OSP/claudin-11 forms a complex with a novel member of the tetraspanin super family and beta1 integrin and regulates proliferation and migration of oligodendrocytes. J Cell Biol. 2001;153(2):295–305. doi: 10.1083/jcb.153.2.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traka M, Dupree JL, Popko B, Karagogeos D. The neuronal adhesion protein TAG-1 is expressed by Schwann cells and oligodendrocytes and is localized to the juxtaparanodal region of myelinated fibers. J Neurosci. 2002;22(8):3016–24. doi: 10.1523/JNEUROSCI.22-08-03016.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapp BD. Myelin-associated glycoprotein. Location and potential functions. Ann N Y Acad Sci. 1990;605:29–43. doi: 10.1111/j.1749-6632.1990.tb42378.x. [DOI] [PubMed] [Google Scholar]
- Werner HB, Kuhlmann K, Shen S, Uecker M, Schardt A, Dimova K, Orfaniotou F, Dhaunchak A, Brinkmann BG, Möbius W, Guarente L, Casaccia-Bonnefil P, Jahn O, Nave KA. Proteolipid protein is required for transport of sirtuin 2 into CNS myelin. J Neurosci. 2007;27(29):7717–30. doi: 10.1523/JNEUROSCI.1254-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.