Abstract
A linear epitope on catfish IgM has been identified as the docking site for the catfish soluble FcμR (IpFcRI). Western blot analyses and latex bead binding assays identified the consensus octapeptide motif FxCxVxHE located at the second cysteine that forms the intrachain disulfide bond of the catfish Cμ3 and Cμ4 immunolglobulin (Ig) domains as the IpFcRI binding sites. Furthermore, molecular modeling of catfish Cμ3 and Cμ4 confirmed that the octapeptide in both of these domains is accessible for IpFcRI interactions. In addition, since this octapeptide motif is also found in other vertebrate Ig domains, IpFcRI binding to Ig heavy (H) and light (L) chains from rainbow trout, chicken, mouse, rabbit, and goat were examined by Western blot analyses and latex bead binding assays. IpFcRI readily bound reduced rainbow trout (Igμ), chicken (Igν), mouse (Igμ, Igγ1, Igγ2a, Igγ2b, and Igα), rabbit (Igμ and Igγ) and goat (Igγ) IgH chains, and mouse Igκ and Igλ, and chicken Igλ IgL chains. IpFcRI also bound mouse IgM, IgA and IgG subclasses when examined under native conditions.
Keywords: Channel catfish, soluble FcR, IgM, Comparative Immunology
1. Introduction
Teleost B cell responses share numerous functional similarities with those of mammals (Miller et al., 1998; Miller et al., 1994), including immunoglobulin (Ig) gene rearrangements, allelic exclusion and production of both secreted and membrane forms of IgM, and IgD (Bengten et al., 2006a; Bengten et al., 2006b; Bengten et al., 2002; Bengten et al., 2000; Wilson et al., 1997; Wilson et al., 1990). However, while much is known about teleost IgM structure and function, much less is known about the function of teleost IgD, and little is known about the receptors that bind teleost immunoglobulins (Coosemans and Hadji-Azimi, 1988; Hamuro et al., 2007; Haynes et al., 1988; O’Dowd et al., 1998; Rombout et al., 2008; Sekizawa et al., 1984; Shen et al., 2003). In mammals, receptors specific for the Fc portion of Ig, i.e. FcRs, participate in a multitude of immune functions, and for the most part are found as integral membrane proteins, expressed on immune effector cells, including B cells, monocytes, macrophages, neutrophils, NK cells, and mast cells (Daeron, 1997). The FcR mediated functions include phagocytosis, antibody-dependent cell mediated cytotoxicity (ADCC), lymphocyte proliferation, mast cell degranulation, release or secretion of cytokines and chemokines, antigen presentation, regulation of antibody production and clearance of immune complexes, as well as Ig transport (Daeron, 1997; Nimmerjahn and Ravetch, 2006; Ravetch and Kinet, 1991). Typically, Ig (or immune complex) binding to FcRs leads to receptor aggregation that activates or inhibits effector cell functions depending on the type(s) of FcR involved. Cellular activation or inhibition occurs through recruitment of kinases or phosphatases to the signaling motifs present within the FcR cytoplasmic tail (CYT) or the associated adaptor molecules (Daeron and Lesourne, 2006; Garcia-Garcia and Rosales, 2002). In addition to membrane bound FcRs, soluble forms of FcRs (sFcRs), are produced either by alternative splicing or proteolytic cleavage of the membrane-bound FcRs (Astier et al., 1994; Fridman et al., 1993; Sautes et al., 1992; Sautes et al., 1991). sFcRs have been described for IgG (CD32/FcγRII and CD16/FcγRIII; Fleit et al., 1992; Fridman et al., 1992; Gachet et al., 1995; Huizinga et al., 1994; Huizinga et al., 1988; Lanier et al., 1989; Middelhoven et al., 2001; Teillaud et al., 1992; Tosi and Zakem, 1992), IgA (CD89/FcαRI; van Zandbergen et al., 1999; Yodoi et al., 1987) and IgE (CD23/FcεRII; Letellier et al., 1989; Sarfati et al., 1996). The sFcγ-receptors are believed to function by competing with their membrane-bound counterparts for Ig (or immune complex) binding, which in turn down-regulates B cell proliferation and antibody production (Fernandez-Botran, 1991; Fridman et al., 1993). In comparison, the function of sFcαRI, which is found covalently linked with IgA in the serum of normal individuals and patients with IgA nephropathy, is not well understood (Launay et al., 2000; van der Boog et al., 2002). Finally, FcεRII, the low affinity receptor for IgE, exhibits significant homology to C-type lectins, and is not an Ig superfamily member (reviewed in Daeron, 1997). Soluble FcεRII is produced by endogenous proteases and has been shown to activate monocytes and resting T cells (Armant et al., 1995) and can serve as a positive as well as a negative regulator of IgE synthesis in human B cells (Delespesse et al., 1992; Gould et al., 2003; McCloskey et al., 2007).
The strongest evidence for FcR homologs in ectothermic vertebrates came from studies in the channel catfish, Ictalurus punctatus (Shen et al., 2004; Shen et al., 2003), which showed that catfish NK-like cells were armed with IgM and were positive for two Ig light (L) chain isotypes. Since these cells did not express message for either Ig heavy (H) or IgL chains, it was hypothesized that serum IgM is bound on their surface via a putative FcμR. Moreover, when the surface bound IgM was replaced by catfish anti-trinitrol-phenol (TNP) IgM antibodies, these specifically armed NK cells were able to kill TNP-labeled target cells by ADCC (Shen et al., 2003). More recently, we identified a catfish FcR homolog, termed IpFcRI, which represents the first FcR cloned from an ectothermic vertebrate (Stafford et al., 2006). The single copy IpFcRI gene encodes a protein of three constant (C)-2-like Ig domains and lacks a transmembrane (TM) and CYT. Notably, the encoded Ig domains are phylogenetically and structurally related to mammalian FcRs and the putative Fc-binding region appears to be conserved. In addition, IpFcRI-related genomic sequences were found in pufferfish and rainbow trout, indicating the likely presence of a soluble FcR in other fish species. However, while IpFcRI message was highly expressed in catfish lymphoid tissues and peripheral blood leukocytes (PBL) clonal leukocyte cell lines, including NK cells, expressed little (if any) message, which suggested that IpFcRI is not the putative FcμR observed in catfish NK cells. Nevertheless, IpFcRI was shown to bind catfish IgM as demonstrated by co-immunoprecipations and cell transfection studies, and native IpFcRI was detected in catfish plasma using a mouse polyclonal antiserum to IpFcRI (Stafford et al., 2006). Together, these observations suggest that the IpFcRI functions as a soluble IgM-binding FcR that may have immunoregulatory functions in vivo. Here, we further analyze the Ig binding properties of IpFcRI and determine that the IpFcRI docking site on catfish IgM is a conserved eight amino acid linear epitope involving the second cysteine that forms the intra-chain disulfide bond in Cμ3 and Cμ4 domains. Furthermore comparative mapping studies show that the epitope is conserved in several other vertebrate Ig C domains, including mouse, goat, rabbit, chicken and rainbow trout.
2. Material and Methods
2.1 Monoclonal antibodies, recombinant protein production, and catfish IgM affinity purification
Hybridoma culture supernatant was used directly as the monoclonal antibody (mAb) source for the anti-catfish reagents and all supernatants contained equivalent Ig concentrations as determined by ELISA (Southern Biotechnology, Birmingham, AL) and Western blot analysis. Anti-catfish IgM antibodies 9E1 (IgG1, κ) and 1H12 (IgG1, κ) react with catfish Igμ H chains (Lobb and Olson, 1988; Sizemore et al., 1984), and anti-trout IgM antibody 1.14 (IgG1, κ), an isotype control, reacts with trout Igμ H chains (DeLuca et al., 1983). The anti-epitope tag mAbs, anti-FLAG M2 (IgG1; Sigma-Aldrich) and anti-Xpress (IgG1; Invitrogen Life Technologies) were used at concentrations according to the manufacturer’s recommended protocol. All mAbs are of mouse origin, unless otherwise stated in the text.
Recombinant (r) IpFcRI proteins were produced using a eukaryotic expression system. Briefly, IpFcRI D1-D2-D3 domains and partial IpFcRI (two domain combinations of D1-D2 and D2-D3, as well as single domains D1, D2, and D3) were cloned into the p3×FLAG-CMV-9 (Sigma-Aldrich) using EcoRV and BamHI restriction sites. This vector introduces three adjacent FLAG epitopes at the N-terminus of the cloned sequence allowing for identification and purification of the FLAG tagged fusion proteins by anti-FLAG M2 mAb. The constructs were transfected into human embryonic kidney (HEK) 293T.17 cells using GeneJuice transfection reagent (Novagen EMD Biosciences) according to the manufacturer’s protocol. Briefly, 293T.17 cells were grown to 70–80% confluency in 6-well plates, and transfected with a mixture of 1 μg plasmid DNA in 4 μl GeneJuice (added per well). At 48 h post transfection, cells were harvested following trypsinization (Trypsin–Versene Mixture, Lonza Inc), washed twice with phosphate buffered saline (PBS; 137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2PO4, pH 7.4) and solublized with 1% IGEPAL CA-630 (Sigma-Aldrich) in lysis buffer (50 mM Tris-HCl, 150 mM NaCl, pH 7.8) containing a set of protease inhibitors (Complete, Mini; Roche Applied Science). The 293T.17 lysates were cleared of cellular debris by centrifugation at 1500 × g for 10 min and the supernatant was passed though an anti-FLAG M2 affinity gel column (Sigma-Aldrich). The FLAG-fusion proteins were eluted by 100 μg/ml of FLAG peptides (Sigma-Aldrich) according to manufacturer’s protocol and residual peptides were removed by passing the eluents through D-Salt Dextran desalting columns (Pierce). Purity of the FLAG-fusion proteins was verified by Western blotting with anti-FLAG M2-HRP mAb (Sigma-Aldrich).
Catfish rIgμ proteins spanning two Cμ domains (Cμ2-Cμ3 and Cμ3-Cμ4), single Cμ domains (Cμ1, Cμ2, Cμ3 and Cμ4) and partial (or split) Cμ3 and Cμ4 domain proteins were cloned into the Champion pET 100/D-TOPO vector (Invitrogen) and expressed in Escherichia coli BL21 star cells as previously described (Silva et al., 2007). This vector introduces a 6×His and an Xpress N-terminal epitope tag to the rCμ proteins for purification and detection. The polyhistidine tagged rCμ proteins were purified using MagneHis Ni-particles (Promega) following the manufacturer’s guidelines and their purity was verified by Western blotting using peroxidase conjugated anti-Xpress mAb (Invitrogen).
Catfish anti-TNP IgM was prepared from immunized catfish as previously described (Lobb, 1985; van Ginkel et al., 1992) and the affinity purified IgM served as the source of free (unbound) IgM used in latex bead and Western blot immunoassays.
2.2 Latex bead microsphere solid phase immunoassays
Five micron carboxyl (-COOH) modified latex bead microspheres (Bangs Laboratories) were covalently coupled with rIpFcRI (D1-D2-D3), affinity purified catfish IgM, different mouse Igs (IgM, IgG1, IgG2a, IgG2b, IgG3 or IgA; Southern Biotech), or with BSA as a negative control using a PolyLink protein coupling kit (Bangs Laboratories). This protocol conjugates proteins via their amino-terminus (NH2-) onto carboxylated microspheres using 1-Ethyl-3(3-dimethylaminopropyl) carbodiimide hydrochloride as a linker. Briefly, 10 mg of latex beads were coupled with 500 μg of protein for 2 h at room temperature following the supplier’s guidelines and the protein-coupled beads were maintained at a concentration of 10 μg/μl in PolyLink Wash/Storage buffer (Bangs Laboratories).
Flow cytometry analyses of IpFcRI-IgM binding was performed using rIpFcRI coupled latex beads (L-IpFcRI) incubated with catfish serum or catfish IgM coupled latex beads (L-IgM) incubated with rIpFcRI. Briefly, for assaying L-rIpFcRI, 10 μg of beads were mixed with either 2 μg of catfish affinity purified IgM or 0.5 μl of catfish serum and mixed end-over-end for 1 h at 24°C in PBS-blocking buffer (with 0.1% Tween-20 and 1% BSA; pH 7.4). The latex beads were then washed 3× in PBS-blocking buffer, centrifuged at 1200 × g for 10 min and stained with 20 μl of anti-catfish IgM (9E1) mAb that had been fluorescently labeled with Alexa Fluor-488 (AF-488) using a Zenon Mouse IgG1 labeling kit (Molecular Probes). After a final wash in PBS-blocking buffer, the beads were analyzed using a BD BioSciences FACScan. Ten thousand events per sample were analyzed at a rate of ≥ 1000 events per second. Conditions for assaying L-IgM were identical, except that the L-IgM coupled beads were mixed with 2 μg of rIpFcRI and stained with anti-FLAG M2 mAb fluorescently labeled with AF-488. This protocol was also used to assess IpFcRI binding to different mouse immunoglobulins and for these experiments, the latex beads were covalently coupled with mouse IgM, IgG1, IgG2a, IgG2b, IgG3 or IgA (Southern Biotech); mixed with rIpFcRI and stained with fluorescently labeled anti-FLAG M2 mAb as described above.
2.3 SDS-PAGE and Western blot analyses
IpFcRI binding to reduced Ig was examined by Western blot analyses using normal sera from catfish, rainbow trout, mouse and rabbit and various affinity purified Igs, including catfish IgM, chicken IgY (Aves Labs), rabbit IgG (Southern Biotech) and goat IgG (Southern Biotech). Briefly, 0.2 μl of sera or 1 μg of Ig were analyzed under reducing conditions in 10% SDS-PAGE using standard protocols (Sambrook and Russell, 2001). Proteins were transferred to Hybond-ECL nitrocellulose membranes (Amersham Biosciences) and incubated in Tris-buffered saline (TBS; 20 mM Tris, 150 mM NaCl, pH 7.4) containing 1.0% BSA and 0.1% Tween 20 (TTBS-BSA) overnight at 4°C. The membranes were incubated with rIpFcRI (1 μM in TTBS-BSA) for 1h at 24°C, followed by incubation with anti-FLAG M2-HRP (0.25 μg/ml in TTBS-BSA) for 1 h at 24°C. Blots were then washed 3× in TTBS and immunoreactive bands were visualized using Supersignal West Pico Chemiluminescent Substrate (Pierce Biotechnology, Rockford, IL). To insure complete reduction of catfish IgM, 2 μg of affinity purified IgM was incubated at 37°C overnight in the presence of 100 mM 2-β-mercaptoethanol, 500 mM Tris and 6 M guanidine-HCl. The reduced IgM was subsequently alkylated with a 3-fold molar excess of iodoacetamide (Sigma-Aldrich) at 4°C for 6 h, and dialyzed overnight in 2× in TBS, pH 7.4 before using (Clem and Small, 1967).
2.4 Mapping of IpFcRI binding
IpFcRI binding to catfish IgM was initially mapped using rCμ domain proteins. One μg of catfish recombinant proteins or 1 μg of affinity purified IgM were analyzed by SDS-PAGE and Western blot analyses as described above. Synthetic biotinylated peptides corresponding to the flanking sequence surrounding the second cysteine that forms the interchain disulfide bond of the different catfish Cμ domains array were synthesized (Mimotopes) and used in Western blot and flow cytometry analyses in order to map the IpFcRI binding site(s). Briefly, peptides (Pep) 1 and 2 were from Cμ1 and Cμ2, respectively, and served as negative binding controls. In contrast, a series of truncated peptides from Cμ3 (Pep3.1-Pep3.4) and Cμ4 (Pep4.1-4.5) were designed to determine the required length of the IpFcRI docking site. Peptide 5 (Pep5) was designed to be a mismatch of peptide 4.2 (Pep4.2), i.e. four amino acids (389, 390, 392, and 394) surrounding the cysteine391 residue were substituted based on residues found at corresponding positions in mouse and rabbit Igs, but not in catfish Ig (FSCLVYHE → YTCSVGHE). For the Western immunoblot analyses 2–2.5 μg of each peptide was spotted onto Hybond-ECL nitrocellulose membranes (Amersham Biosciences) and allowed to air dry. Blots were incubated and developed using rIpFcRI and anti-FLAG M2-HRP as described above. For flow cytometry analysis the ability of each peptide to bind rIpFcRI was examined. Briefly, 10 μg of L-IpFcRI beads was mixed with 2.5 μg of peptide in 50 μl of TBS-Tween (0.5%) and incubated at 24°C for 1 h. The beads were washed once with TBS-Tween before adding 10 μl of phycoerythrin (PE)-conjugated streptavidin (BD Biosciences). Following a 30 min incubation at 24°C, the beads were washed in TBS-Tween and analyzed as described above.
2.5 Sequence analyses and homology modeling
The putative binding motif in Cμ3 and Cμ4 was found by dot plot alignment using MegAlign software (DNASTAR Lasergene, Madison, WI) and the default settings 20% match and a window size of 30. The catfish IpFcRI binding site 3-D structure was predicted by amino acid homology modeling using the SWISS_MODEL alignment interface mode (http://swissmodel.expasyporg/) and the PHYRE protein fold recognition server (Kelley and Sternberg, 2009); version 0.2, http://www.sbg.bio.ic.ac.uk/phyre/). The catfish Cμ3-Cμ4 sequence was used to query the RCSB Protein Data Bank (PDB, http://www.rcsb.org/pdb/home/home.do) and the best match was IgG1 C region (entry 1L6X with a 29% amino acid identity, an E-value of 2.5e-28 and prediction confidence 100%), which was used to build a structural model of the catfish Cμ3-Cμ4 region. Molecular decorations and visualization was done through PyMOL version 1.1 (DeLano Scientific: http://www.pymol.org/).
3. Results
3.1 IpFcRI binds a conserved epitope found on catfish constant Cμ3 and Cμ4 domains
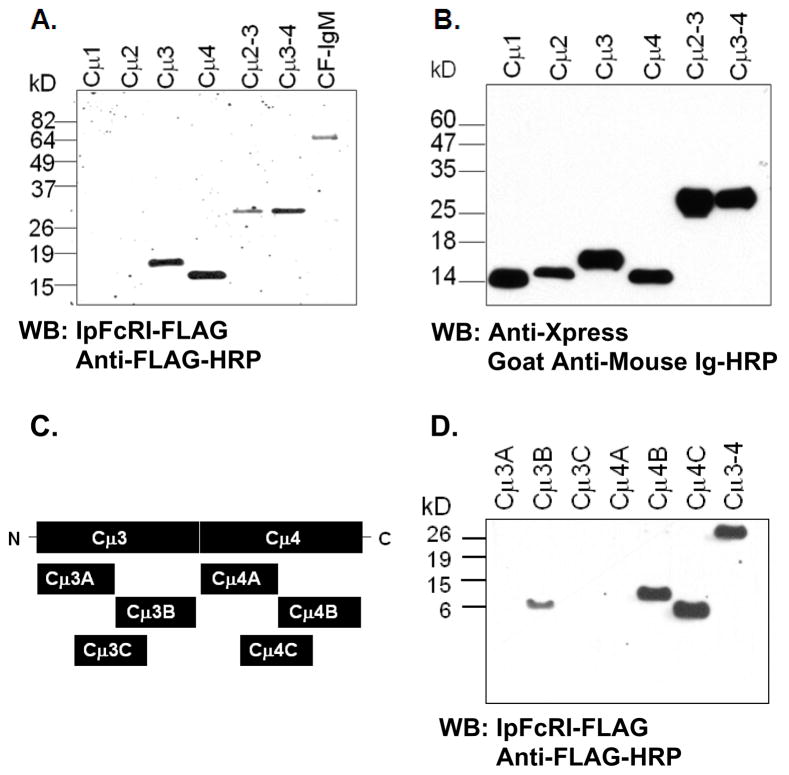
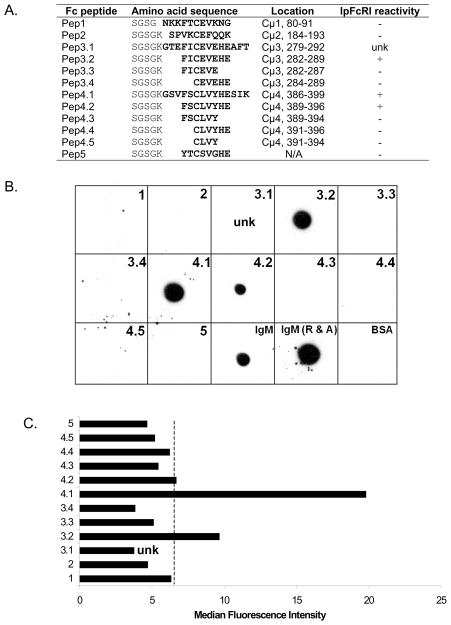
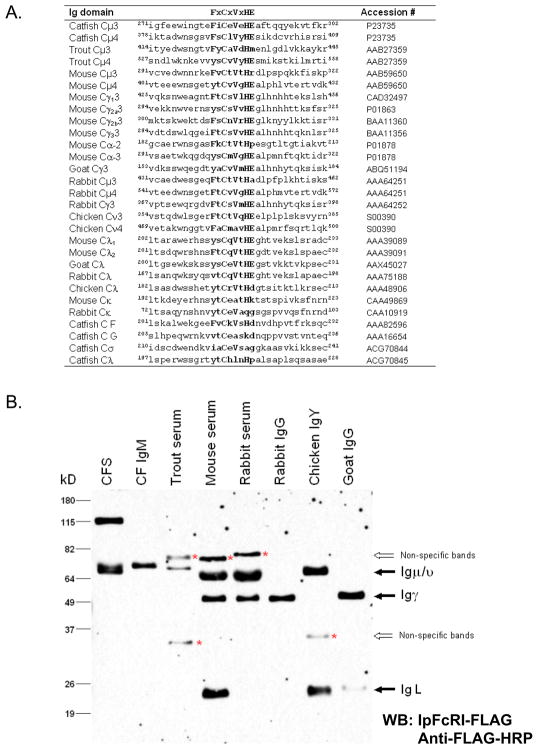
As an alternative to using surface expression on transfected cells as a means to assay the binding of soluble IpFcRI to catfish IgM, a solid phase latex bead assay was developed. Using flow cytometry, latex beads coated with rIpFcRI protein were shown to readily bind catfish serum IgM (Fig. 1) or purified catfish IgM (data not shown). Conversely, beads coated with affinity purified catfish IgM were shown to bind rIpFcRI protein, which confirms our previous studies demonstrating that IpFcRI binds catfish IgM as assessed by co-immunmunoprecipation and cell transfection studies (Stafford et al., 2006). Using this assay it was demonstrated that IpFcRI bound catfish IgM at room temperature but not at 4°C. Subsequently, it was also found that IpFcRI binds IgM in reduced form (Fig. 2A). Briefly, catfish serum and purified IgM were separated by 10% SDS-PAGE under reducing conditions (Sambrook and Russell, 2001) and subjected to Western blot analysis using either rIpFcRI or anti-catfish IgM 1H12 mAb (Lobb and Olson, 1988) as the primary binding reagent. Both reacted with the ~70 kD catfish Igμ chain, demonstrating that IpFcRI and 1H12 mAb react with denatured IgM, which implies that rIpFcRI binds to an exposed linear Igμ epitope. Here it should be noted that the anti-FLAG mAb by itself does not react with catfish IgM or any other catfish serum proteins (Fig. 2B). Therefore in order to map the IpFcRI binding site(s) on the catfish Igμ H chain, recombinant proteins of individual constant domains (Cμ) and pairs Cμ2-3 and Cμ3-4 were produced in E. coli and subjected to 10% SDS-PAGE under reducing conditions and analyzed by Western blot (Fig. 3A and 3B). Both rCμ2-Cμ3 and rCμ3-Cμ4 proteins were detected by rIpFcRI and of the four individual rCμ domain proteins only rCμ3 and rCμ4 were IpFcRI reactive. Subsequently, truncated amino- and carboxyl-terminal rCμ3 and rCμ4 proteins were produced and analyzed as described above, and were found to bind rIpFcRI (Figs. 3C and 3D). This finding that IpFcRI bound to noncontiguous linear epitopes present on Cμ3B, Cμ4B and Cμ4C suggested either the presence of 1) an independent binding site in each of the Cμ3 and Cμ4 domains or 2) a conserved epitope common to both Cμ3 and Cμ4, but not to Cμ1 and Cμ2. By comparing the four catfish Cμ domain sequences using pair-wise dot plot analysis a consensus sequence of eight amino acid residues (FxCxVxHE) including the second cysteine that forms the intrachain disulfide bond was identified and hypothesized to be a part of the putative IpFcRI binding site. These residues located on the F strand of the Ig domain are conserved in many different vertebrate Ig C domains and are structurally important for forming the Ig domain fold, e.g. the amino acids F, C, and V are located in domain core positions and participate in the interaction between the two β-sheets of the domain (Potapov et al., 2004). Moreover, the FxCxVxHE consensus was present in all three of the catfish recombinant proteins, Cμ3B, Cμ4B, and Cμ4C. To prove that the FxCxVxHE amino acid sequence was the site for IpFcRI binding and to further define the core binding region, custom peptides flanking the second cysteine residue in the different catfish Cμ domains were synthesized. The peptides ranged from four to 14 amino acids in length and each had a short amino-terminal linker sequence added to improve solubility (Fig. 4A). IpFcRI binding to these peptides was analyzed by dot-blot analyses and solid phase latex bead assay. By dot-blot, peptides Pep4.1, Pep4.2, and Pep3.2 containing the core epitope FxCxVxHE were positive for rIpFcRI binding, however in the latex bead assay only peptides Pep4.1 and Pep3.2 bound rIpFcRI strongly (Figs. 4B and 4C). In both assays IpFcRI failed to bind the Cμ3 and Cμ4 peptides that did not contain the full-length core consensus motif, and the Cμ1 and Cμ2 control peptides. However, an exception did occur with the Cμ3 peptide Pep3.1, which despite containing the core epitope was IpFcRI negative. This was likely due to Pep3.1’s insolubility since it formed aggregates in both H2O (polar) and 50% dimethyl sulfoxide (organic) solutions. In addition, the peptide Pep5 that contained a mismatched motif YtCsVgHE, based on amino acid residues occurring in rabbit and mouse IgH, was also negative for IpFcRI binding. Since F → Y is a conserved amino acid substitution this result was unexpected. Also, because the FxCxVxHE motif is conserved in vertebrate Ig domains and the F → Y substitution is a common occurrence in some of these domains (Fig. 5A), it seemed reasonable to determine if rIpFcRI could bind immunoglobulins from other species and to correlate binding with the presence or absence of the FxCxVxHE consensus motif. Here it should be stated that while we did not examine any human Ig isotypes for IpFcRI binding, the consensus octapeptide F/Y xCxVxHE motif is present in human Cμ4, Cδ3, and Cλ domains and in each of the IgG γ3 and IgA α3 domains (see sTable 1).
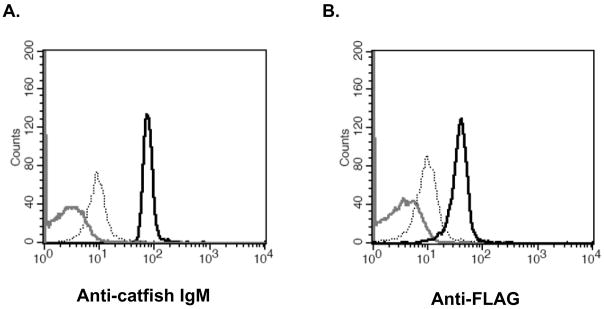
Figure 1.
IpFcRI binds catfish IgM. A. Latex beads coated with rIpFcRI (black) and beads coated with BSA (gray) were incubated in catfish serum, stained with anti-catfish IgM 9E1 mAb, and analyzed by flow cytometry. rIpFcRI coated beads not incubated in catfish serum (dotted), but stained with anti-catfish IgM 9E1 mAb served as background control. B. Latex beads coated with catfish IgM (black line) and beads coated with BSA (gray) were incubated with rIpFcRI, stained with anti-FLAG M2 mAb, and analyzed by flow cytometry. IgM coated beads not incubated with rIpFcRI (dotted), but stained with anti-FLAG mAb served as background control.
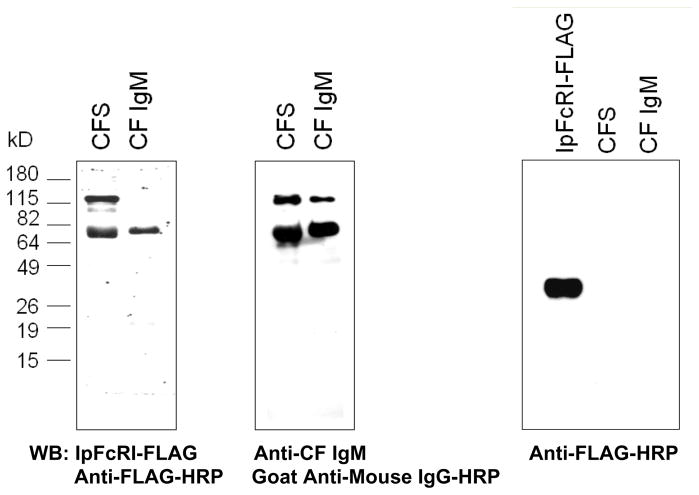
Figure 2.
IpFcRI binds reduced catfish IgM. A. Catfish serum and affinity purified catfish IgM were electrophoresed under reducing conditions by 10% SDS-PAGE and visualized by Western blot using rIpFcRI as the primary binding reagent followed by anti-FLAG M2-HRP (left panel) or anti-catfish IgM 1H12 mAb as the primary antibody followed by goat anti-mouse IgG-HRP (right panel). The observed ~120 kD molecular weight protein bands detected by IpFcRI and anti-catfish IgM are partially reduced catfish Igμ chains as verified by MS/MS sequencing. B. Catfish serum and affinity purified catfish IgM were analyzed as above using anti-FLAG M2-HRP. Recombinant IpFcRI-FLAG served as a positive control. Molecular weight size markers are at left.
Figure 3.
IpFcRI binds linear epitopes found on catfish Cμ3 and Cμ4 domains. A. Catfish Xpress-tagged-Cμ proteins were electrophoresed under reducing conditions by 10% SDS-PAGE and examined by Western blot. Recombinant IpFcRI was used as the primary binding reagent followed by anti-FLAG M2-HRP. Affinity purified catfish IgM served as positive control. B. Catfish rCμ proteins were analyzed as above using anti-Xpress mAb followed by goat anti-mouse Ig (H + L)-HRP. C. Schematic illustrates where Cμ3 and Cμ4 domains were split into segments, which were cloned and expressed as truncated proteins. D. Catfish rCμ proteins were analyzed using rIpFcRI as the primary binding reagent followed by anti-FLAG M2-HRP. Catfish rCμ3-4 served as the positive control for IpFcRI binding. Molecular weight size markers are at left.
Figure 4.
IpFcRI binds to catfish Cμ3 and Cμ4 linear epitopes. A. Synthetic peptides derived from amino acid sequences flanking the second cysteine of catfish Cμ domains are shown with their reactivity for rIpFcRI listed at right. An amino acid-linker consisting of SGSG or SGSGK was added at the N-terminus to improve peptide solubility, amino acid location numbers are shown for catfish IgM (GenBank accession number: P23735), “unk” indicates unknown reactivity due to peptide insolubility. B. Each peptide was spotted onto nitrocellulose and incubated with rIpFcRI as the primary binding reagent. The rIpFcRI reactive spots were developed using anti-FLAG M2-HRP. 2 μg of unreduced and reduced and alkylated affinity purified catfish IgM (IgM and IgM R&A, respectively) served as positive controls, BSA served as negative control. C. Biotinylated peptides were incubated with IpFcRI beads stained with streptavidin-PE and analyzed by flow cytometry. The dashed line indicates cutoff value for a positive reaction. Peptide names are shown on Y-axis and median fluorescence intensity on X-axis.
Figure 5.
IpFcRI binds Ig from different species. A. Potential core IpFcRI-binding motifs and their flanking sequences of different vertebrate Ig domains were compared to the catfish core motifs. Consensus amino acids are in upper case and the different core motifs are shown in bold. B. Serum from catfish (CFS), trout, mouse and rabbit; affinity purified catfish IgM, rabbit IgG, chicken IgY and goat IgG were electrophoresed under reducing conditions by 10% SDS-PAGE and examined by Western blot using rIpFcRI as described in Fig. 2. Labeled arrows mark the IgH and IgL chains, and non-specific protein bands that react with the anti-FLAG M2 mAb are marked (*; see sFig. 1). Molecular weight size markers are at left.
3.2 IpFcRI binds immunoglobulins from different species
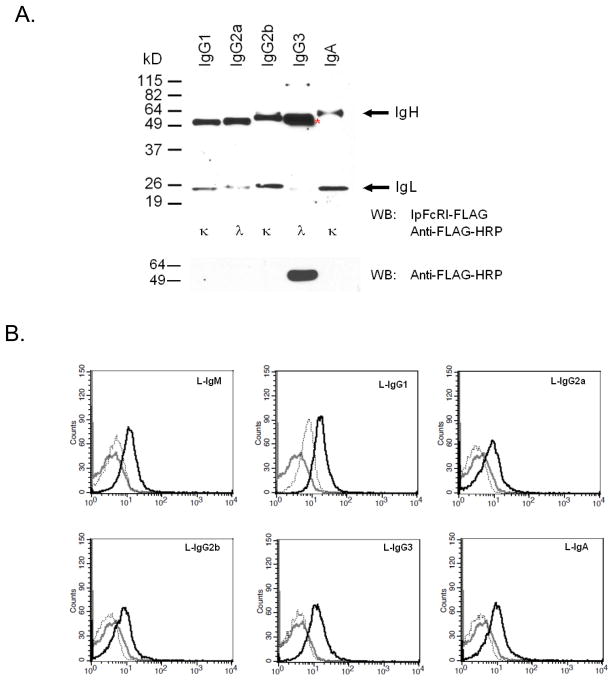
IpFcRI-reactive bands were readily detected in rainbow trout, mouse, and goat serum and with purified rabbit and goat IgG and chicken IgY (Fig. 5B). Briefly, serum, and purified total IgG and IgY were separated by 10% SDS-PAGE under reducing conditions and analyzed by Western blot using rIpFcRI as the primary binding reagent and anti-FLAG mAb as the secondary. Consistent with the presence of F/YxCxVxHE, IpFcRI binding was observed in rainbow trout, mouse, and rabbit Igμ chains, the three mouse Igγ chains, the single rabbit and goat Igγ chains, and chicken Igυ. Specifically, rIpFcRI recognized the ~70 kDa Igμ chains found in rainbow trout (verified by anti-trout 1.14 mAb, data not shown), as well as protein bands of the appropriate size, for mouse and rabbit Igμ and Igγ, chicken Igυ and goat Igγ. As expected catfish Igμ was also readily detected in serum and in affinity purified Ig. Here it should be noted that IpFcRI does not bind to catfish Igδ. In addition to IgH chains, IpFcRI also bound IgL chains present in mouse serum and the IgL chain associated with chicken Igυ and goat Igγ. The mouse Ig Cλ domain contains a consensus FtCqVtHE and the goat and chicken Ig Cλ domains have a F → Y substitution in their consensus motif (ysCeVtHE and ytCrVtHd), demonstrating that IpFcRI is also able to bind to the octapeptide motif if a Y is in that first position. While it seemed likely that rIpFcRI binds only to mouse Cλ, using this assay, it was not possible to exclude binding to mouse Cκ, which contains a different motif of ytCeatHk. Also, our assay could not determine if only one of the mouse, rabbit, and goat IgG subclasses or if all of their IgG classes could be bound by IpFcRI. However, by examining affinity purified mouse IgG mAbs of different subclasses individually the issue of IpFcRI binding specific Igγ, Igκ and/or Igλ chains, as well as mouse Igα chains, was addressed. As assessed by Western blot analysis and the latex bead immunoassay, rIpFcRI bound to IgG1, IgG2a, IgG2b H chains and to a lesser degree to IgA (Fig. 6A). Also both Igκ and Igλ light chains were detected by rIpFcRI, albeit Igκ chains appeared to be bound more readily. However, since the mouse IgG3 H chain contained a partial FLAG epitope (DYK), and was shown to react with the anti-FLAG M2 mAb in the absence of IpFcRI, it could not be determined whether the reduced form of IgG3 was recognized by rIpFcRI. When IpFcRI binding to native mouse Ig isotypes was examined in the flow cytometric solid phase latex bead assay, rIpFcRI was shown to bind all of the mouse Ig isotypes (Fig. 6B). However, the mean fluorescence intensities were approximately 5-fold lower than when the beads were coated with an equal amount of affinity purified catfish IgM (see Fig. 1B). Here it should be emphasized that our assay was designed to demonstrate IpFcRI binding and not meant to comprehensively map the Ig binding epitope, but as might be expected, sequence comparisons of the different Ig motifs combined with the binding data suggest that the overall amino acid composition of the eight amino acid F/YxCxVxHE motif is important for binding.
Figure 6.
IpFcRI binds to different murine Ig isotypes. A. Mouse IgG1, IgG2a, IgG2b, IgG3, and IgA were electrophoresed under reducing conditions by 10% SDS-PAGE and examined by Western blot using rIpFcRI as described in Fig. 2. Molecular weight size markers are at left, IgL chain isotypes are listed below each lane. That the anti-FLAG mAb directly reacts with mouse IgG3 is marked (*) and shown in the bottom panel. B. Latex beads coated with the different mouse Ig proteins (black) and beads coated with BSA (grey) were incubated with rIpFcRI, stained with anti-FLAG M2 mAb and analyzed by flow cytometry. Coated latex beads not incubated with rIpFcRI (dotted), but stained with anti-FLAG M2 mAb served as background staining controls.
In addition, since anti-catfish IgM mAb 1H12, reacts with both native and denatured Igμ chains, it was of interest to examine its reactivity with the different rCμ proteins and synthetic peptides. Similar to IpFcRI, mAb 1H12 was found to recognize rCμ3 and rCμ4 domains in Western blot (sFigs. 2A and 2B) and to react with the longest Cμ4 peptide Pep4.1 (385GSVFSCLVYHESIK399). Moreover, it was demonstrated that mAb 1H12 binding to rCμ3 and rCμ4 could be blocked by IpFcRI (sFig. 2C). Taken together, these findings suggest that anti-catfish IgM mAb 1H12 and IpFcRI bind to an overlapping epitope found at the second cysteine of Cμ4. However, because blocking by mAb 1H12 did not inhibit rIpFcRI binding, it appears that the binding sites either do not completely overlap or IpFcRI has a higher affinity for the binding site than mAb 1H12 (sFig. 2D).
3.3 IpFcRI structural modeling
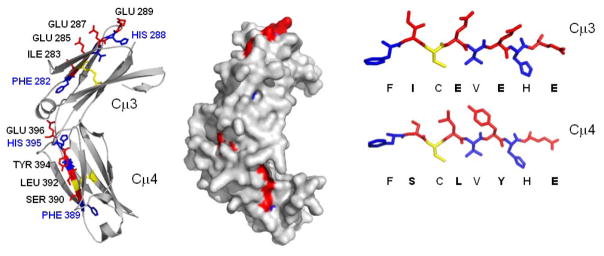
To determine the location of the IpFcRI binding linear epitope on catfish IgM, the Cμ3-Cμ4 region was subjected to homology modeling based on the crystal structure of human IgG1, the best match in the PDB database. A three dimensional model was built at an estimated precision of 100% and an E-value of 2.5e-28 (Fig. 7). The epitope FxCxVxHE, found on the F-strand of the beta-sheet sandwich in both the Cμ3 and Cμ4 domains, has the side chains of amino acids F, C, V, and H buried in the hydrophobic center of the domain. This finding, as expected, matches the previous accepted models of other Ig C domains (Edmundson et al., 1975; Potapov et al., 2004). Thus, it is likely that IpFcRI binding to the native protein is dependent on interactions with the side chains of the alternating exposed residues I283, E285, E287 and E289 of Cμ3 and S390, L392, Y394 and E396 of Cμ4 as illustrated by topology modeling. Also, it appears that the interactions between IpFcRI and Cμ3 involve ionic and hydrophobic bonds; in contrast the binding of Cμ4 depends on non-ionic interactions.
Figure 7.
Predicted 3-D structure of catfish Cμ3 and Cμ4 domains based on comparative homology modeling. The amino acid sequence of catfish Cμ3 and Cμ4 (aa 204A-434Y, Entrez Protein ID# P23735) was used to query the PDB database using the PHYRE server (Kelley and Sternberg, 2009). The human IgG1 C region PDB accession code 1L6X was used to build the 3D model. The ribbon diagram and space fill model were made in Pymol. The side chain of the amino acids in the FxCxVxHE motif are shown with the domain core residues in grey and putative IpFcRI interacting residues in black. The modeled core motifs in Cμ3 and Cμ4 are shown for comparison. For the web version of this article the side chain of the amino acids in the FxCxVxHE motif are shown with the domain core residues in blue, interchain disulfide bonds in yellow and putative IpFcRI interacting residues in red.
4. Discussion
In this study we used a latex bead assay to confirm our previous findings that the catfish soluble FcμR binds catfish IgM (Stafford et al., 2006), regardless of whether IgM or rIpFcRI was immobilized on the beads (Fig. 1). Since it had been demonstrated that the interaction between certain mammalian IgGs and their corresponding FcRs involves linear epitopes (Goldsmith et al., 1997; Kristoffersen et al., 1994; Radaev and Sun, 2001b; Zhang et al., 2006) it was of interest to examine whether IpFcRI would demonstrate any binding to reduced catfish IgM. Not only did rIpFcRI react with reduced purified catfish IgM, but it also bound reduced serum IgM and did not cross-react with any other serum proteins (Fig. 2). Consequently, based on the ability to bind reduced IgM, the IpFcRI docking site on IgM was mapped using recombinant Cμ proteins and a consensus octapeptide FxCxVxHE sequence present in Cμ3 and Cμ4 was hypothesized to be a part of the putative IpFcRI binding site (Fig. 3). The putative IpFcRI docking site was then mapped using custom made oligopeptides of varying lengths with N- and C-terminal deletions to define the minimum core epitope. Peptides 3.2, 4.1, and 4.2 each containing the FxCxVxHE motif were IpFcRI reactive (Fig. 4) and because none of the truncated peptides shorter than the octapeptide core were found to be recognized by IpFcRI, the octapeptide FxCxVxHE was designated to be the minimally required IpFcRI docking site. However, the binding to Pep4.2, which consists of only the core sequence, appeared to be weaker than the binding to Pep4.1 and Pep3.2. While both Pep4.1 and Pep3.2 contain the core octapeptide sequence, Pep4.1 is longer with three additional residues at both the N- and C- terminal ends and Pep3.2 has three glutamic acids at the variant positions. Such sequence differences suggest that residues additional to the octapeptide core also influence the binding of IpFcRI to catfish IgM. For example, the six extra amino acid residues present in the longer Cμ4 epitope represented by Pep4.1 may further stabilize IpFcRI binding, while the three negatively charged residues (E 285, E287 and E289) of Cμ3 represented by Pep3.2 likely influences IpFcRI binding by forming a negatively charged patch similar to that observed with the high affinity FcγRII binding site found on the mouse IgG2b (Kato et al., 2000). Conversely, the Cμ4 binding site motif represented by Pep4.1 and Pep4.2, includes only one negatively charged residue (E396) and may represent a hydrophobic patch. Nevertheless, the IpFcRI peptide binding studies show the octapetide core containing the second Ig intradomain-forming cysteine of Cμ3 and Cμ4 to be the IpFcRI binding site on reduced catfish Igμ. Also, here it is important to note that while the binding studies do not address whether IpFcRI uses one or both of these sites to bind native Igμ, the IgM binding studies performed using the anti-catfish Igμ mAb 1H12 strongly support that the FxCxVxHE motif is accessible for IpFcRI interaction in catfish native IgM. For example, mAb 1H12 binds native and reduced catfish Igμ (Lobb and Olson, 1988), and rCμ3 and rCμ4 proteins (sFig. 2A), and together such results strongly imply that mAb 1H12 recognizes a linear epitope accessible under native conformation. This notion is further supported by our peptide binding studies that show mAb 1H12 binds Pep 4.1 (sFig. 2B), which confirms there must be an overlap between mAb 1H12 and IpFcRI binding sites. However, because mAb 1H12 did not bind to the core octapeptide motif represented by Pep3.2 and Pep4.2, it may be that binding of Igμ by mAb 1H12 requires a few additional amino acids for interaction (sFig. 2B). In agreement with this, pre-incubating the reduced rCμ blots with rIpFcRI inhibited mAb 1H12 binding to rCμ3 and rCμ4 proteins (sFig. 2C). In contrast, blocking of replica rCμ blots with mAb 1H12 did not inhibit IpFcRI binding (sFig. 2D) and while implications for this observation are not clear-cut, it may be that the IpFcRI and mAb 1H12 binding sites do not completely overlap, or that IpFcRI has a higher affinity for the binding site than that of mAb 1H12. Alternatively, it may be possible that IpFcRI binds to the Igγ1 chain of mAb 1H12, which has bound catfish IgM. However, together the IpFcRI and mAb 1H12 binding studies support the hypothesis that the FxCxVxHE octapeptide motifs found on catfish IgM are accessible epitopes for catfish soluble FcR interaction.
The IpFcRI binding sites were also predicted to be accessible since structural modeling of the Cμ3 and Cμ4 domains show them to be localized on the F-strands of their respective Ig domains (Fig. 7). Also, the catfish IgM Cμ3-Cμ4 topography revealed the residues F, C, and V from the consensus octapeptide motif to be buried in the Ig domain beta sandwich fold. This result was expected, since these “domain core residues” are conserved in many Ig C domains and are known to provide structural support for the Ig domain (Edmundson et al., 1975; Potapov et al., 2004). In contrast, the side chains of the alternate variable amino acids that fill the positions between the “domain core residues” in catfish Cμ3 and Cμ4 (I283, E285, E287 and E289 of Cμ3 and S390, L392, Y394 and E396 of Cμ4) appear to be exposed. Hence, they are accessible to serve as contact points for IpFcRI interaction (see Fig. 7). Moreover, the E289 and E396at the 8th position of the core peptide of Cμ3 and Cμ4, respectively, are the most accessible and exposed residues of the motifs and may therefore have a greater role in IpFcRI recognition/binding as compared to other residues in the motif.
Consistent with the presence of the FxCxVxHE core sequence in IgH chains of trout (Igμ), chicken (Igν), mouse (Igμ, Igγ1, Igγ2a, Igγ2b, and Igα), rabbit (Igμ and Igγ) and goat (Igγ) these IgH chains were also readily bound by IpFcRI under reducing conditions (Fig. 5). In addition, IpFcRI bound mouse IgM, IgG and IgA when examined under native conditions. This broad-spectrum Ig binding across mammalian, avian, and teleost Ig isotypes is in contrast to the previously described mammalian FcR-Ig cross-reactivities that are frequently observed with only certain Ig isotype subclasses from closely related species (Canfield and Morrison, 1991; Lubeck et al., 1985; Nimmerjahn and Ravetch, 2005). Due to the presence of the core IpFcRI binding motif in both Cμ3 and Cμ4 domains from the different species, as well as in the mouse Cα2 and Cα3 domains, the contributions from the individual binding sites and also from the individual amino acids are difficult to discern. Even so, based on sequence comparisons of the octapeptide core motif present in the different mammalian Igγ and IgL chains and in chicken Igυ and Igλ chains that were positive for IpFcRI binding in Western blot analyses, a substitution of F (a less hydrophobic residue) for Y did not appear to significantly impact IpFcRI binding (see Figures 5 and 6). However, again because variable IpFcRI reactivity was readily apparent with the different IgL chains emphasizes that the contribution made by the amino acids (either individually or perhaps in combination) found in the variable positions of the octapeptide are important. Also, it seems likely that IpFcRI binding to various IgL chains merely reflects the conservation of the Ig domain fold and is not physiologically significant. Consistent with this notion, IpFcRI did not bind to catfish IgL chains and here it should be noted that four IgL isotypes have been identified in catfish (Edholm et al., 2009). Together the Igκ F and G isotypes represent nearly 90% of total serum IgL (Lobb et al., 1984) and along with Igσ, they are readily detectable in catfish serum by Western blot (Wilson unpublished). Currently, there are no available antibodies to catfish Igλ and its presence in serum cannot yet be measured. The lack of IpFcRI binding to proteins in the 20–27kD range in Western blot strongly argues against IpFcRI interaction with catfish IgL chains F, G and σ and although not formally proven we consider it highly unlikely that the IpFcRI would interact with catfish Cλ. All four of the catfish IgL chains contain one or more non-favorable residue(s) on the predicted F strand and three of them (IgL G, λ, and σ) have only the cysteine of the three conserved “domain core residues” found in the FxCxVxHE motif. IpFcRI also did not bind to rabbit or trout IgL chains or to Pep5 (YTCSVGHE), which was synthesized based on amino acid residues present in the IpFcRI binding motif of IgM and IgG in mouse and rabbit. This nonbinding may be due to the absence of charged or bulky side chain amino acids occupying the 2nd, 4th, and 6th positions of the octapeptide motif (Fig. 4 and sFig 3). The G at the position 6 may be a particularly unfavorable residue for binding because of its small size and non-chiral properties. Similarly, the absence of IpFcRI binding to catfish IgD domain 6, the only domain Igδ domain that contains a core binding motif, is likely due to the lack of charged or bulky residues at the same 2nd, 4th, and 6th positions (sTable 2 and sFig. 3).
In summary, since IpFcRI is the first FcR homolog identified in an ectothermic vertebrate and phylogenetic analyses shows it is more closely related to the classical FcRs and the FCR-like homologs than to the leukocyte receptor complex (LRC) encoded receptors (Stafford et al., 2006), it was of interest to determine whether IpFcRI bound to any of the two distinct locations targeted by other vertebrate FcRs. Briefly, mammalian classical FcRs, that are homologs of the human FcRs encoded on chromosome 1, i.e. IgG and IgE binding receptors FcγRs I, II, III and FcεRIα bind to similar sites in the lower hinge region located in homologous domains Cγ2 and Cε3, respectively (Canfield and Morrison, 1991; Garman et al., 2000; Radaev and Sun, 2001a; Sondermann et al., 2000; Wines et al., 2000). In contrast, the mammalian FcαRI (CD89) and the chicken IgY binding receptor, CHIR-AB1, are both encoded within their respective LRC and interact with homologous sites found at the Cα2-Cα3 and Cυ3-Cυ4 interface, respectively (Herr et al., 2003; Pleass et al., 1999; Purzel et al., 2009; Taylor et al., 2009). The data presented here demonstrates that the catfish soluble FcμR, IpFcRI, while more related to the classical FcRs does not bind to the N-terminal side of Cμ3, but interacts with a novel and conserved epitope present on the F-strand of catfish Cμ3 and Cμ4 domains. Interestingly, the Ig binding sites recognized by human FcαRI and chicken CHIR-AB1 both overlap with the IpFcRI binding site and a mutation of the conserved histidine of IgY that is homologous to the histidine at position 7 in the catfish octapeptide motif abolishes the CHIR-AB1 binding. At the very least, this overlap in binding sites support the conserved histidine residue to be highly accessible and phylogenetically important in Fc binding.
Overall, this study represents the first characterization of an FcR binding site on any teleost Ig and is an important step in understanding the evolution not only of FcμR receptors (Ghumra et al., 2009; Kinet and Launay, 2000), but of FcRs in general.
Supplementary Material
sFigure 1. Serum from catfish (CFS), trout, mouse and rabbit; affinity purified catfish IgM, rabbit IgG, chicken IgY and goat IgG were electrophoresed under reducing conditions by 10% SDS-PAGE and examined by Western blot using Anti-FLAG-HRP as in Fig 5. Protein bands that react with the anti-FLAG M2 mAb are marked (*). Molecular weight size markers are at left.
sFigure 2. The binding sites of IpFcRI and mAb 1H12 overlap. A. Catfish rCμ proteins or catfish IgM were electrophoresed under reducing conditions by 10% SDS-PAGE and examined by Western blot. Reactive proteins were visualized using anti-catfish IgM 1H12 mAb as the primary antibody followed by goat anti-mouse Ig (H + L)-HRP. B. Synthetic peptides as in Fig 4 were spotted onto a nitrocellulose membrane and incubated with mAb 1H12 followed by goat anti-mouse IgG-HRP. C. Catfish rCμ domain proteins or catfish IgM were electrophoresed as in A and examined by Western blot. Transferred proteins were first blocked by rIpFcRI then incubated with anti-catfish IgM 1H12 mAb and goat anti-mouse Ig (H + L)-HRP. The lack of reactive bands shows rIpFcRI prevents anti-catfish Igμ 1H12 mAb from binding to catfish IgM. D. Catfish rCμ domain proteins or catfish IgM were electrophoresed as in A and examined by Western blot. Transferred proteins were first blocked by mAb 1H12 then incubated with rIpFcRI as the primary binding reagent followed by anti-FLAG M2-HRP. Molecular weight size markers are at left.
sFigure 3. Comparisons of FxCxVxHE homologous regions in IpFcRI and non-binding sequences. The different sequences were modeled as in Fig 7 and only the carbon side chains of the core octapeptide sequence are shown. Domain core residues are in blue, cysteines involved in interchain disulfide bonds in yellow, and the variable alternating residues in red. IpFcRI interacting sequences chicken Igλ (AAA48906), goat Igλ (AAX45027) and mouse Igλ (X558411) were modeled on the Igλ chain of human IgG1 PDB accession number 2FB4 and mouse Igκ (CAA49869) was modeled on the mouse IgG1 Fab PBD accession number 2HMI. IpFcRI non-interacting sequences catfish IgL F (AAA82596), catfish IgL G (AAA16654), catfish Igσ (ACG70844) were modeled on the human surrogate IgL chain PDB accession number 2H3N; catfish Igλ (ACG70845) was modeled on the Igλ chain of human IgG1 PDB accession number 1MFB; rabbit Igλ (AAA75188) was modeled on 2FB4 and rabbit Igκ (CAA10919) and catfish IgD domain 6 (AF363450) were modeled on 2HMI.
Acknowledgments
We thank professor Steve Kaattari for the gift of trout serum. This work was supported by grants from the National Institutes of Health (RO1AI-19530), UMMC IRSPs (59917, 59908) awarded to E.B. and M.W., respectively.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Armant M, Rubio M, Delespesse G, Sarfati M. Soluble CD23 directly activates monocytes to contribute to the antigen-independent stimulation of resting T cells. J Immunol. 1995;155:4868–75. [PubMed] [Google Scholar]
- Astier A, de la Salle H, de la Salle C, Bieber T, Esposito-Farese ME, Freund M, Cazenave JP, Fridman WH, Teillaud JL, Hanau D. Human epidermal Langerhans cells secrete a soluble receptor for IgG (Fc gamma RII/CD32) that inhibits the binding of immune complexes to Fc gamma R+ cells. J Immunol. 1994;152:201–12. [PubMed] [Google Scholar]
- Bengten E, Clem LW, Miller NW, Warr GW, Wilson M. Channel catfish immunoglobulins: repertoire and expression. Dev Comp Immunol. 2006a;30:77–92. doi: 10.1016/j.dci.2005.06.016. [DOI] [PubMed] [Google Scholar]
- Bengten E, Quiniou S, Hikima J, Waldbieser G, Warr GW, Miller NW, Wilson M. Structure of the catfish IGH locus: analysis of the region including the single functional IGHM gene. Immunogenetics. 2006b;58:831–44. doi: 10.1007/s00251-006-0139-9. [DOI] [PubMed] [Google Scholar]
- Bengten E, Quiniou SM, Stuge TB, Katagiri T, Miller NW, Clem LW, Warr GW, Wilson M. The IgH locus of the channel catfish, Ictalurus punctatus, contains multiple constant region gene sequences: different genes encode heavy chains of membrane and secreted IgD. J Immunol. 2002;169:2488–97. doi: 10.4049/jimmunol.169.5.2488. [DOI] [PubMed] [Google Scholar]
- Bengten E, Wilson M, Miller N, Clem LW, Pilstrom L, Warr GW. Immunoglobulin isotypes: structure, function, and genetics. Curr Top Microbiol Immunol. 2000;248:189–219. doi: 10.1007/978-3-642-59674-2_9. [DOI] [PubMed] [Google Scholar]
- Canfield SM, Morrison SL. The binding affinity of human IgG for its high affinity Fc receptor is determined by multiple amino acids in the CH2 domain and is modulated by the hinge region. J Exp Med. 1991;173:1483–91. doi: 10.1084/jem.173.6.1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clem LW, Small PA., Jr Phylogeny of immunoglobulin structure and function. I. Immunoglobulins of the lemon shark. J Exp Med. 1967;125:893–920. doi: 10.1084/jem.125.5.893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coosemans V, Hadji-Azimi I. Immunoglobulin Fc receptor molecules on Xenopus laevis splenocytes. Immunology. 1988;65:641–5. [PMC free article] [PubMed] [Google Scholar]
- Daeron M. Fc receptor biology. Annu Rev Immunol. 1997;15:203–234. doi: 10.1146/annurev.immunol.15.1.203. [DOI] [PubMed] [Google Scholar]
- Daeron M, Lesourne R. Negative signaling in Fc receptor complexes. Adv Immunol. 2006;89:39–86. doi: 10.1016/S0065-2776(05)89002-9. [DOI] [PubMed] [Google Scholar]
- Delespesse G, Sarfati M, Wu CY, Fournier S, Letellier M. The low-affinity receptor for IgE. Immunol Rev. 1992;125:77–97. doi: 10.1111/j.1600-065x.1992.tb00626.x. [DOI] [PubMed] [Google Scholar]
- DeLuca D, Wilson M, Warr GW. Lymphocyte heterogeneity in the trout, Salmo gairdneri, defined with monoclonal antibodies to IgM. Eur J Immunol. 1983;13:546–51. doi: 10.1002/eji.1830130706. [DOI] [PubMed] [Google Scholar]
- Edholm ES, Wilson M, Sahoo M, Miller NW, Pilstrom L, Wermenstam NE, Bengten E. Identification of Igsigma and Iglambda in channel catfish, Ictalurus punctatus, and Iglambda in Atlantic cod, Gadus morhua. Immunogenetics. 2009;61:353–70. doi: 10.1007/s00251-009-0365-z. [DOI] [PubMed] [Google Scholar]
- Edmundson AB, Ely KR, Abola EE, Schiffer M, Panagiotopoulos N. Rotational allomerism and divergent evolution of domains in immunoglobulin light chains. Biochem. 1975;14:3953–61. [PubMed] [Google Scholar]
- Fernandez-Botran R. Soluble cytokine receptors: their role in immunoregulation. FASEB J. 1991;5:2567–74. doi: 10.1096/fasebj.5.11.1868981. [DOI] [PubMed] [Google Scholar]
- Fleit HB, Kobasiuk CD, Daly C, Furie R, Levy PC, Webster RO. A soluble form of Fc gamma RIII is present in human serum and other body fluids and is elevated at sites of inflammation. Blood. 1992;79:2721–8. [PubMed] [Google Scholar]
- Fridman WH, Bonnerot C, Daeron M, Amigorena S, Teillaud JL, Sautes C. Structural bases of Fc gamma receptor functions. Immunol Rev. 1992;125:49–76. doi: 10.1111/j.1600-065x.1992.tb00625.x. [DOI] [PubMed] [Google Scholar]
- Fridman WH, Teillaud JL, Bouchard C, Teillaud C, Astier A, Tartour E, Galon J, Mathiot C, Sautes C. Soluble Fc gamma receptors. J Leukoc Biol. 1993;54:504–12. [PubMed] [Google Scholar]
- Gachet C, Astier A, de la Salle H, de la Salle C, Fridman WH, Cazenave JP, Hanau D, Teillaud JL. Release of Fc gamma RIIa2 by activated platelets and inhibition of anti-CD9-mediated platelet aggregation by recombinant Fc gamma RIIa2. Blood. 1995;85:698–704. [PubMed] [Google Scholar]
- Garcia-Garcia E, Rosales C. Signal transduction during Fc receptor-mediated phagocytosis. J Leukoc Biol. 2002;72:1092–108. [PubMed] [Google Scholar]
- Garman SC, Wurzburg BA, Tarchevskaya SS, Kinet JP, Jardetzky TS. Structure of the Fc fragment of human IgE bound to its high-affinity receptor Fc epsilonRI alpha. Nature. 2000;406:259–66. doi: 10.1038/35018500. [DOI] [PubMed] [Google Scholar]
- Ghumra A, Shi J, McIntosh RS, Rasmussen IB, Braathen R, Johansen FE, Sandlie I, Mongini PK, Areschoug T, Lindahl G, Lewis MJ, Woof JM, Pleass RJ. Structural requirements for the interaction of human IgM and IgA with the human Fcalpha/mu receptor. Eur J Immunol. 2009;39:1147–56. doi: 10.1002/eji.200839184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldsmith EB, Erickson BW, Thompson NL. Synthetic peptides from mouse Fc receptor (MoFc gamma RII) that alter the binding of IgG to MoFc gamma RII. Biochemistry. 1997;36:952–9. doi: 10.1021/bi961564b. [DOI] [PubMed] [Google Scholar]
- Gould HJ, Sutton BJ, Beavil AJ, Beavil RL, McCloskey N, Coker HA, Fear D, Smurthwaite L. The biology of IGE and the basis of allergic disease. Annu Rev Immunol. 2003;21:579–628. doi: 10.1146/annurev.immunol.21.120601.141103. [DOI] [PubMed] [Google Scholar]
- Hamuro K, Suetake H, Saha NR, Kikuchi K, Suzuki Y. A teleost polymeric Ig receptor exhibiting two Ig-like domains transports tetrameric IgM into the skin. J Immunol. 2007;178:5682–9. doi: 10.4049/jimmunol.178.9.5682. [DOI] [PubMed] [Google Scholar]
- Haynes L, Fuller L, McKinney EC. Fc receptor for shark IgM. Dev Comp Immunol. 1988;12:561–71. doi: 10.1016/0145-305x(88)90072-9. [DOI] [PubMed] [Google Scholar]
- Herr AB, Ballister ER, Bjorkman PJ. Insights into IgA-mediated immune responses from the crystal structures of human FcalphaRI and its complex with IgA1-Fc. Nature. 2003;423:614–20. doi: 10.1038/nature01685. [DOI] [PubMed] [Google Scholar]
- Huizinga TW, de Haas M, van Oers MH, Kleijer M, Vile H, van der Wouw PA, Moulijn A, van Weezel H, Roos D, von dem Borne AE. The plasma concentration of soluble Fc-gamma RIII is related to production of neutrophils. Br J Haematol. 1994;87:459–63. doi: 10.1111/j.1365-2141.1994.tb08298.x. [DOI] [PubMed] [Google Scholar]
- Huizinga TW, van der Schoot CE, Jost C, Klaassen R, Kleijer M, von dem Borne AE, Roos D, Tetteroo PA. The PI-linked receptor FcRIII is released on stimulation of neutrophils. Nature. 1988;333:667–9. doi: 10.1038/333667a0. [DOI] [PubMed] [Google Scholar]
- Kato K, Sautes-Fridman C, Yamada W, Kobayashi K, Uchiyama S, Kim H, Enokizono J, Galinha A, Kobayashi Y, Fridman WH, Arata Y, Shimada I. Structural basis of the interaction between IgG and Fcgamma receptors. J Mol Biol. 2000;295:213–24. doi: 10.1006/jmbi.1999.3351. [DOI] [PubMed] [Google Scholar]
- Kelley LA, Sternberg MJ. Protein structure prediction on the Web: a case study using the Phyre server. Nat Protoc. 2009;4:363–71. doi: 10.1038/nprot.2009.2. [DOI] [PubMed] [Google Scholar]
- Kinet JP, Launay P. Fc alpha/microR: single member or first born in the family? Nat Immunol. 2000;1:371–2. doi: 10.1038/80805. [DOI] [PubMed] [Google Scholar]
- Kristoffersen EK, Matre R, Ulvestad E, Vedeler CA. A dot-immunobinding assay for the demonstration of soluble Fc gamma receptors. J Immunol Methods. 1994;167:15–9. doi: 10.1016/0022-1759(94)90070-1. [DOI] [PubMed] [Google Scholar]
- Lanier LL, Phillips JH, Testi R. Membrane anchoring and spontaneous release of CD16 (FcR III) by natural killer cells and granulocytes. Eur J Immunol. 1989;19:775–8. doi: 10.1002/eji.1830190431. [DOI] [PubMed] [Google Scholar]
- Launay P, Grossetete B, Arcos-Fajardo M, Gaudin E, Torres SP, Beaudoin L, Patey-Mariaud de Serre N, Lehuen A, Monteiro RC. Fcalpha receptor (CD89) mediates the development of immunoglobulin A (IgA) nephropathy (Berger’s disease). Evidence for pathogenic soluble receptor-Iga complexes in patients and CD89 transgenic mice. J Exp Med. 2000;191:1999–2009. doi: 10.1084/jem.191.11.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letellier M, Sarfati M, Delespesse G. Mechanisms of formation of IgE-binding factors (soluble CD23)--I. Fc epsilon R II bearing B cells generate IgE-binding factors of different molecular weights. Mol Immunol. 1989;26:1105–12. doi: 10.1016/0161-5890(89)90054-0. [DOI] [PubMed] [Google Scholar]
- Lobb CJ. Covalent structure and affinity of channel catfish anti-dinitrophenyl antibodies. Mol Immunol. 1985;22:993–9. doi: 10.1016/0161-5890(85)90087-2. [DOI] [PubMed] [Google Scholar]
- Lobb CJ, Olson MO. Immunoglobulin heavy H chain isotypes in a teleost fish. J Immunol. 1988;141:1236–45. [PubMed] [Google Scholar]
- Lobb CJ, Olson MO, Clem LW. Immunoglobulin light chain classes in a teleost fish. J Immunol. 1984;132:1917–23. [PubMed] [Google Scholar]
- Lubeck MD, Steplewski Z, Baglia F, Klein MH, Dorrington KJ, Koprowski H. The interaction of murine IgG subclass proteins with human monocyte Fc receptors. J Immunol. 1985;135:1299–304. [PubMed] [Google Scholar]
- McCloskey N, Hunt J, Beavil RL, Jutton MR, Grundy GJ, Girardi E, Fabiane SM, Fear DJ, Conrad DH, Sutton BJ, Gould HJ. Soluble CD23 monomers inhibit and oligomers stimulate IGE synthesis in human B cells. J Biol Chem. 2007;282:24083–91. doi: 10.1074/jbc.M703195200. [DOI] [PubMed] [Google Scholar]
- Middelhoven PJ, Van Buul JD, Hordijk PL, Roos D. Different proteolytic mechanisms involved in Fc gamma RIIIb shedding from human neutrophils. Clin Exp Immunol. 2001;125:169–75. doi: 10.1046/j.1365-2249.2001.01548.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller N, Wilson M, Bengten E, Stuge T, Warr G, Clem W. Functional and molecular characterization of teleost leukocytes. Immunol Rev. 1998;166:187–97. doi: 10.1111/j.1600-065x.1998.tb01263.x. [DOI] [PubMed] [Google Scholar]
- Miller NW, Rycyzyn MA, Wilson MR, Warr GW, Naftel JP, Clem LW. Development and characterization of channel catfish long term B cell lines. J Immunol. 1994;152:2180–9. [PubMed] [Google Scholar]
- Nimmerjahn F, Ravetch JV. Divergent immunoglobulin g subclass activity through selective Fc receptor binding. Science. 2005;310:1510–2. doi: 10.1126/science.1118948. [DOI] [PubMed] [Google Scholar]
- Nimmerjahn F, Ravetch JV. Fcgamma receptors: old friends and new family members. Immunity. 2006;24:19–28. doi: 10.1016/j.immuni.2005.11.010. [DOI] [PubMed] [Google Scholar]
- O’Dowd AM, Ellis AE, Secombes CJ. Binding of immune complexes to Atlantic salmon peripheral blood leucocytes. Dev Comp Immunol. 1998;22:439–48. doi: 10.1016/s0145-305x(98)00018-4. [DOI] [PubMed] [Google Scholar]
- Pleass RJ, Dunlop JI, Anderson CM, Woof JM. Identification of residues in the CH2/CH3 domain interface of IgA essential for interaction with the human fcalpha receptor (FcalphaR) CD89. J Biol Chem. 1999;274:23508–14. doi: 10.1074/jbc.274.33.23508. [DOI] [PubMed] [Google Scholar]
- Potapov V, Sobolev V, Edelman M, Kister A, Gelfand I. Protein--protein recognition: juxtaposition of domain and interface cores in immunoglobulins and other sandwich-like proteins. J Mol Biol. 2004;342:665–79. doi: 10.1016/j.jmb.2004.06.072. [DOI] [PubMed] [Google Scholar]
- Purzel J, Schmitt R, Viertlboeck BC, Gobel TW. Chicken IgY binds its receptor at the CH3/CH4 interface similarly as the human IgA: Fc(alpha)RI interaction. J Immunol. 2009;183:4554–9. doi: 10.4049/jimmunol.0901699. [DOI] [PubMed] [Google Scholar]
- Radaev S, Sun P. Recognition of immunoglobulins by Fcgamma receptors. Mol Immunol. 2001a;38:1073–83. doi: 10.1016/s0161-5890(02)00036-6. [DOI] [PubMed] [Google Scholar]
- Radaev S, Sun PD. Recognition of IgG by Fcgamma receptor. The role of Fc glycosylation and the binding of peptide inhibitors. J Biol Chem. 2001b;276:16478–83. doi: 10.1074/jbc.M100351200. [DOI] [PubMed] [Google Scholar]
- Ravetch JV, Kinet JP. Fc receptors. Annu Rev Immunol. 1991;9:457–92. doi: 10.1146/annurev.iy.09.040191.002325. [DOI] [PubMed] [Google Scholar]
- Rombout JH, van der Tuin SJ, Yang G, Schopman N, Mroczek A, Hermsen T, Taverne-Thiele JJ. Expression of the polymeric Immunoglobulin Receptor (pIgR) in mucosal tissues of common carp (Cyprinus carpio L.) Fish Shellfish Immunol. 2008;24:620–8. doi: 10.1016/j.fsi.2008.01.016. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Russell DW. Molecular cloning: a laboratory manual. 3 Vol. 1. Cold Spring Harbor Laboratory Press; New York: 2001. [Google Scholar]
- Sarfati M, Chevret S, Chastang C, Biron G, Stryckmans P, Delespesse G, Binet JL, Merle-Beral H, Bron D. Prognostic importance of serum soluble CD23 level in chronic lymphocytic leukemia. Blood. 1996;88:4259–64. [PubMed] [Google Scholar]
- Sautes C, Mazieres N, Galinha A, Tartour E, Bonnerot C, Amigorena S, Teillaud C, Spagnoli R, Fridman WH. Murine soluble Fc gamma receptors/IgG-binding factors (IgG-BF): analysis of the relation to Fc gamma RII and production of milligram quantities of biologically active recombinant IgG-BF. Immunol Res. 1992;11:181–90. doi: 10.1007/BF02919125. [DOI] [PubMed] [Google Scholar]
- Sautes C, Varin N, Teillaud C, Daeron M, Even J, Hogarth PM, Fridman WH. Soluble Fc gamma receptors II (Fc gamma RII) are generated by cleavage of membrane Fc gamma RII. Eur J Immunol. 1991;21:231–4. doi: 10.1002/eji.1830210135. [DOI] [PubMed] [Google Scholar]
- Sekizawa A, Fujii T, Tochinai S. Membrane receptors on Xenopus macrophages for two classes of immunoglobulins (IgM and IgY) and the third complement component (C3) J Immunol. 1984;133:1431–5. [PubMed] [Google Scholar]
- Shen L, Stuge TB, Bengten E, Wilson M, Chinchar VG, Naftel JP, Bernanke JM, Clem LW, Miller NW. Identification and characterization of clonal NK-like cells from channel catfish (Ictalurus punctatus) Dev Comp Immunol. 2004;28:139–52. doi: 10.1016/s0145-305x(03)00119-8. [DOI] [PubMed] [Google Scholar]
- Shen L, Stuge TB, Evenhuis JP, Bengten E, Wilson M, Chinchar VG, Clem LW, Miller NW. Channel catfish NK-like cells are armed with IgM via a putative FcmicroR. Dev Comp Immunol. 2003;27:699–714. doi: 10.1016/s0145-305x(03)00042-9. [DOI] [PubMed] [Google Scholar]
- Silva EF, Medeiros MA, McBride AJ, Matsunaga J, Esteves GS, Ramos JG, Santos CS, Croda J, Homma A, Dellagostin OA, Haake DA, Reis MG, Ko AI. The terminal portion of leptospiral immunoglobulin-like protein LigA confers protective immunity against lethal infection in the hamster model of leptospirosis. Vaccine. 2007;25:6277–86. doi: 10.1016/j.vaccine.2007.05.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sizemore RC, Miller NW, Cuchens MA, Lobb CJ, Clem LW. Phylogeny of lymphocyte heterogeneity: the cellular requirements for in vitro mitogenic responses of channel catfish leukocytes. J Immunol. 1984;133:2920–4. [PubMed] [Google Scholar]
- Sondermann P, Huber R, Oosthuizen V, Jacob U. The 3.2-A crystal structure of the human IgG1 Fc fragment-Fc gammaRIII complex. Nature. 2000;406:267–73. doi: 10.1038/35018508. [DOI] [PubMed] [Google Scholar]
- Stafford JL, Wilson M, Nayak D, Quiniou SM, Clem LW, Miller NW, Bengten E. Identification and characterization of a FcR homolog in an ectothermic vertebrate, the channel catfish (Ictalurus punctatus) J Immunol. 2006;177:2505–17. doi: 10.4049/jimmunol.177.4.2505. [DOI] [PubMed] [Google Scholar]
- Taylor AI, Sutton BJ, Calvert RA. Mutations in an avian IgY-Fc fragment reveal the locations of monocyte Fc receptor binding sites. Dev Comp Immunol. 2009 doi: 10.1016/j.dci.2009.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teillaud C, Fridman WH, Sautes C. Demonstration of soluble IgG-Fc type II receptors (or s-Fc-gamma-II-R) in human whole saliva. J Biol Buccale. 1992;20:3–10. [PubMed] [Google Scholar]
- Tosi MF, Zakem H. Surface expression of Fc gamma receptor III (CD16) on chemoattractant-stimulated neutrophils is determined by both surface shedding and translocation from intracellular storage compartments. J Clin Invest. 1992;90:462–70. doi: 10.1172/JCI115882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Boog PJ, van Zandbergen G, de Fijter JW, Klar-Mohamad N, van Seggelen A, Brandtzaeg P, Daha MR, van Kooten C. Fc alpha RI/CD89 circulates in human serum covalently linked to IgA in a polymeric state. J Immunol. 2002;168:1252–8. doi: 10.4049/jimmunol.168.3.1252. [DOI] [PubMed] [Google Scholar]
- van Ginkel FW, Miller NW, Lobb CJ, Clem LW. Characterization of anti-hapten antibodies generated in vitro by channel catfish peripheral blood lymphocytes. Dev Comp Immunol. 1992;16:139–51. doi: 10.1016/0145-305x(92)90014-4. [DOI] [PubMed] [Google Scholar]
- van Zandbergen G, Westerhuis R, Mohamad NK, van De Winkel JG, Daha MR, van Kooten C. Crosslinking of the human Fc receptor for IgA (FcalphaRI/CD89) triggers FcR gamma-chain-dependent shedding of soluble CD89. J Immunol. 1999;163:5806–12. [PubMed] [Google Scholar]
- Wilson M, Bengten E, Miller NW, Clem LW, Du Pasquier L, Warr GW. A novel chimeric Ig heavy chain from a teleost fish shares similarities to IgD. Proc Natl Acad Sci U S A. 1997;94:4593–7. doi: 10.1073/pnas.94.9.4593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson MR, Marcuz A, van Ginkel F, Miller NW, Clem LW, Middleton D, Warr GW. The immunoglobulin M heavy chain constant region gene of the channel catfish, Ictalurus punctatus: an unusual mRNA splice pattern produces the membrane form of the molecule. Nucleic Acids Res. 1990;18:5227–33. doi: 10.1093/nar/18.17.5227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wines BD, Powell MS, Parren PW, Barnes N, Hogarth PM. The IgG Fc contains distinct Fc receptor (FcR) binding sites: the leukocyte receptors Fc gamma RI and Fc gamma RIIa bind to a region in the Fc distinct from that recognized by neonatal FcR and protein A. J Immunol. 2000;164:5313–8. doi: 10.4049/jimmunol.164.10.5313. [DOI] [PubMed] [Google Scholar]
- Yodoi J, Adachi M, Noro N. IgA binding factors and Fc receptors for IgA: comparative studies between IgA and IgE Fc receptor systems. Int Rev Immunol. 1987;2:117–41. doi: 10.3109/08830188709044750. [DOI] [PubMed] [Google Scholar]
- Zhang G, Guo J, Zhou J, Wang X, Li Q, Yang Y, Shen H, Zhao D, Zhang H, Xi J, Wang L, Qiao S, Jin X. Identification of the linear epitope for Fc-binding on the bovine IgG2 Fc receptor (boFcgamma2R) using synthetic peptides. FEBS Lett. 2006;580:1383–90. doi: 10.1016/j.febslet.2006.01.060. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
sFigure 1. Serum from catfish (CFS), trout, mouse and rabbit; affinity purified catfish IgM, rabbit IgG, chicken IgY and goat IgG were electrophoresed under reducing conditions by 10% SDS-PAGE and examined by Western blot using Anti-FLAG-HRP as in Fig 5. Protein bands that react with the anti-FLAG M2 mAb are marked (*). Molecular weight size markers are at left.
sFigure 2. The binding sites of IpFcRI and mAb 1H12 overlap. A. Catfish rCμ proteins or catfish IgM were electrophoresed under reducing conditions by 10% SDS-PAGE and examined by Western blot. Reactive proteins were visualized using anti-catfish IgM 1H12 mAb as the primary antibody followed by goat anti-mouse Ig (H + L)-HRP. B. Synthetic peptides as in Fig 4 were spotted onto a nitrocellulose membrane and incubated with mAb 1H12 followed by goat anti-mouse IgG-HRP. C. Catfish rCμ domain proteins or catfish IgM were electrophoresed as in A and examined by Western blot. Transferred proteins were first blocked by rIpFcRI then incubated with anti-catfish IgM 1H12 mAb and goat anti-mouse Ig (H + L)-HRP. The lack of reactive bands shows rIpFcRI prevents anti-catfish Igμ 1H12 mAb from binding to catfish IgM. D. Catfish rCμ domain proteins or catfish IgM were electrophoresed as in A and examined by Western blot. Transferred proteins were first blocked by mAb 1H12 then incubated with rIpFcRI as the primary binding reagent followed by anti-FLAG M2-HRP. Molecular weight size markers are at left.
sFigure 3. Comparisons of FxCxVxHE homologous regions in IpFcRI and non-binding sequences. The different sequences were modeled as in Fig 7 and only the carbon side chains of the core octapeptide sequence are shown. Domain core residues are in blue, cysteines involved in interchain disulfide bonds in yellow, and the variable alternating residues in red. IpFcRI interacting sequences chicken Igλ (AAA48906), goat Igλ (AAX45027) and mouse Igλ (X558411) were modeled on the Igλ chain of human IgG1 PDB accession number 2FB4 and mouse Igκ (CAA49869) was modeled on the mouse IgG1 Fab PBD accession number 2HMI. IpFcRI non-interacting sequences catfish IgL F (AAA82596), catfish IgL G (AAA16654), catfish Igσ (ACG70844) were modeled on the human surrogate IgL chain PDB accession number 2H3N; catfish Igλ (ACG70845) was modeled on the Igλ chain of human IgG1 PDB accession number 1MFB; rabbit Igλ (AAA75188) was modeled on 2FB4 and rabbit Igκ (CAA10919) and catfish IgD domain 6 (AF363450) were modeled on 2HMI.