Abstract
Gastrointestinal stromal tumors (GISTs) generally harbor activating mutations in KIT or PDGFRA. Mutations in these receptor tyrosine kinases lead to dysregulation of downstream signaling pathways that contribute to GIST pathogenesis. GISTs with KIT or PDGFRA mutations also undergo secondary cytogenetic alterations that may indicate the involvement of additional genes important in tumor progression. Approximately 10–15% of adult and 85% of pediatric GISTs do not have mutations in KIT or in PDGFRA. Most mutant adult GISTs display large-scale genomic alterations, but little is know about the mutation-negative tumors. Using genome-wide DNA arrays, we investigated genomic imbalances in a set of 31 GISTs, including 10 KIT/PDGFRA mutation-negative tumors from 9 adults and 1 pediatric case and 21 mutant tumors. While all 21 mutant GISTs exhibited multiple copy number aberrations, notably losses, 8 of the 10 KIT/PDGFRA mutation-negative GISTs exhibited few or no genomic alterations. One KIT/PDGFRA mutation-negative tumor exhibiting numerous genomic changes was found to harbor an alternate activating mutation, in the serine-threonine kinase BRAF. The only other mutation-negative GIST with significant chromosomal imbalances was a recurrent metastatic tumor found to harbor a homozygous deletion in chromosome 9p. Similar findings in several KIT-mutant GISTs identified a minimal overlapping region of deletion of ~0.28 Mbp in 9p21.3 that includes only the CDKN2A/2B genes, which encode inhibitors of cell-cycle kinases. These results suggest that GISTs without activating kinase mutations, whether pediatric or adult, generally exhibit a much lower level of cytogenetic progression than that observed in mutant GISTs.
Keywords: GISTs, KIT, PDGFRA, BRAF, imatinib mesylate, DNA copy number analysis
Introduction
Gastrointestinal stromal tumors (GISTs) are the most common mesenchymal tumors of the gastrointestinal tract, with an estimated annual incidence of 5,000–6,000 cases in the United States (Fletcher, et al. 2002). GISTs are thought to arise from the interstitial cells of Cajal (ICC) (Sircar, et al. 1999), or from a common progenitor cell (Kindblom, et al. 1998), and like the ICC are defined by immunohistochemical expression of the type III receptor tyrosine kinase (RTK) KIT (CD117 antigen), the product of the KIT (v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog) gene. Approximately 70–75% of GISTs harbor gain-of-function mutations in the KIT gene, while mutations in the related RTK gene PDGFRA (platelet-derived growth factor receptor, alpha polypeptide) have been identified in another ~10% of GISTs (Corless, et al. 2002; Heinrich, et al. 2003b; Hirota, et al. 1998; Rubin, et al. 2001; Tarn, et al. 2005). Approximately 10–15% of GISTs from adult patients lack mutations in these genes (Corless, et al. 2004; Corless, et al. 2002; Tarn, et al. 2005). Recently, activating mutations in the serine/threonine protein kinase BRAF were identified in 3/41 (~7%) of adult GISTs that otherwise lacked RTK-mutations (Agaram, et al. 2008b). The discovery of oncogenic mutations in KIT and PDGFRA has led to the clinical use of the RTK-inhibitor imatinib mesylate (IM or Gleevec, Novartis, Switzerland) for advanced or metastatic GIST. Tumor response to IM correlates with KIT or PDGFRA mutational status (Corless and Heinrich 2007; Debiec-Rychter, et al. 2004; Heinrich, et al. 2003a; van Oosterom, et al. 2001). Approximately 70% of GISTs with KIT exon 11 (juxtamembrane domain) mutations have been shown to respond to IM therapy, while ~38% of GISTs with KIT exon 9 (extracellular dimerization domain) mutations and only ~28% of GISTs that lack RTK mutations show an objective response to IM therapy (Heinrich, et al. 2003a). Duration of response as indicated by progression-free and overall survival is also dependent on mutational status (Heinrich, et al. 2003a).
Although mutational activation of KIT or PDGFRA and potentially BRAF is essential to the oncogenesis of the great majority of GISTs, other genetic and cytogenetic changes have been implicated in the progression to malignancy and metastasis. Specific genomic imbalances, such as losses in chromosomes 1p, 14, and 22, are commonly seen in GISTs regardless of tumor risk, while other changes (e.g., losses of 9p and 13q and gains of 8q, 17q, and 20q) are observed more frequently in malignant and/or metastatic GISTs (El-Rifai, et al. 2000; Gunawan, et al. 2007; Meza-Zepeda, et al. 2006). These large-scale chromosomal copy-number (CN) imbalances are infrequent in GISTs from pediatric patients (Janeway, et al. 2007). Pediatric GISTs comprise ~1–2% of total GIST cases, and only ~15% of these tumors possess mutations in KIT and even more rarely in PDGFRA. Most pediatric GISTs occur in females with a predilection towards multi-focal gastric location, and exhibit differential patterns of gene expression (Agaram, et al. 2008a; Miettinen, et al. 2005b; Prakash, et al. 2005). The genetic or epigenetic event(s) driving oncogenesis in KIT/PDGFRA mutation-negative GISTs, whether adult or pediatric, is unknown; however, several recent studies have implicated components of the insulin-like growth factor signaling pathway in the pathogenesis of GIST (Braconi, et al. 2008; Prakash, et al. 2005; Tarn, et al. 2008; Trent, et al. 2006), and recent work by us and others has suggested that adult and pediatric GISTs that lack mutations in KIT or PDGFRA may share aberrations in this pathway (Agaram, et al. 2008a; Belinsky, et al. 2008; Tarn, et al. 2008). Based on these similarities between KIT/PDGFRA mutation-negative adult and pediatric GISTs, we examined both mutant and non-mutant GISTs for chromosomal copy number imbalances using the Affymetrix GeneChip® Human Mapping 50K Xba Array as well as the higher-density Affymetrix Genome-Wide Human SNP Array 6.0. We found that adult mutation-negative GISTs, like pediatric GISTs, generally do not manifest the large-scale chromosomal aberrations that are seen in mutant GISTs.
Materials and Methods
Tumor Samples
Tumor specimens obtained from surgically treated GIST patients were snap-frozen in liquid nitrogen and stored at −80°C until use. When possible, tumors were evaluated for potential risk of recurrence based on tumor size, mitotic count, and anatomical site of occurrence (Miettinen and Lasota 2006). All samples were obtained following institutional review board guidelines.
Immunohistochemical Analysis
Immunohistochemistry for CD117 (KIT) was performed on 5 μM sections using the Benchmark® XT System (Ventana Medical Systems, Tucson, AZ). Paraffin removal and antigen retrieval was performed using Ventana's CC1 buffer. Blocking, secondary and ABC steps were carried out using Ventana's IVIEW DAB Kit. The primary Anti-Human CD117, KIT polyclonal rabbit antibody A4502 (Dako, Carpinteria, CA) was used at a 1:1000 dilution.
Genomic DNA Isolation for Genotyping and SNP Analysis
Frozen tumor samples ~2 mm in diameter were homogenized in lysis buffer (50 mM Tris-HCl/150 mM NaCl/5 mM EDTA/0.5% NP40), and genomic DNA isolated with the use of the Easy-DNA kit (Invitrogen, Carlsbad, California). Histopathological evaluation of hematoxylin/eosin (H&E)- and/or KIT-stained paraffin-embedded sections indicated that these specimens were composed of 70–95 % tumor tissue. For the isolation of DNA from optimal cutting temperature (OCT) medium-embedded tissue, tumor tissue was flash-frozen in OCT and cut into sections. H&E- and KIT-stained sections were used as guides for macrodissection of tumor tissue. Prior to DNA extraction as above, slides were washed several times in ice-cold ethanol followed by water to remove OCT medium. DNA concentration was determined using the NanoDrop 1000 (ThermoFischer Scientific, Waltham MA), and agarose-gel electrophoresis was used to check the integrity of DNA preparations.
Mutational Analyses of KIT, PDGFRA, BRAF, KRAS, and HRAS
PCR amplification of genomic DNA and mutational analysis of KIT (exons 9, 11, 13, and 17) and PDGFRA (exons 12, 14, and 18) was performed as previously described (Tarn, et al. 2005). For a number of samples in which exon-based screening showed no mutations, KIT sequences were further confirmed by complete sequencing of the KIT cDNA. RNA was isolated using Trizol reagent (Invitrogen, Carlsbad, CA), and 2 μg of RNA was reverse transcribed with SuperScript II (Invitrogen, Carlsbad, CA) according to the manufacturer's specifications. Primers located within the KIT coding sequence were used to amplify and sequence overlapping fragments of KIT cDNA (Supplementary Table 1). Primer sequences for amplification of genomic DNA and sequencing of B-RAF exon 15, KRAS exon 5, HRAS exon 1, and HRAS exon 2 are also detailed in Supplementary Table 1. All PCR products were column-purified, and sequencing was carried out on an ABI PRISM 3100 Genetic Analyzer (Applied Biosystems, Foster City, CA).
Single Nucleotide Polymorphism Arrays
Samples 1–17 were analyzed using Affymetrix GeneChip® Human Mapping 50K Xba Arrays. Samples 18–31 were analyzed using the Affymetrix Genome-Wide Human SNP Array 6.0. Probe datasheets are found at http://www.affymetrix.com/products/arrays/index.affx. Briefly, genomic DNA (250 μg) from each tumor was digested with the appropriate restriction enzyme, adaptor-ligated, and PCR-amplified using a generic primer that recognizes the adaptor sequence. PCR conditions were optimized for each array, and the purified PCR products were fragmented with DNase I, biotin-labeled, and hybridized to arrays according to the manufacturer's recommendations. Intensities of probe hybridization were analyzed using Affymetrix GeneChip Operating Software version 1.4. Genotyping for the 50K arrays was performed using Affymetrix GeneChip® Genotyping Analysis Software 4.1 with Dynamic Model Mapping Analysis, using default settings for both homozygote and heterozygote calls, and unpaired CN and LOH analysis was carried out using the Affymetrix Chromosome Copy Number Analysis Tool (CNAT) 4.0, with default genomic smoothing window settings. For the 6.0 arrays, the Affymetrix Genotyping Console 2.1 with Birdseed genotype calling algorithm was used for genotyping, and CN analysis was carried out with CNAT 5.0 using default configurations. The Affymetrix segment-reporting tool was used to define the magnitude, position, and size of regions of CN gain or loss. For samples analyzed with the SNP 6.0 array, focal copy-number (CN) aberrations were defined as changes involving <10% of a chromosomal arm. In tabulating losses and gains, all discrete large-scale CN aberrations along a chromosome were counted.
Results
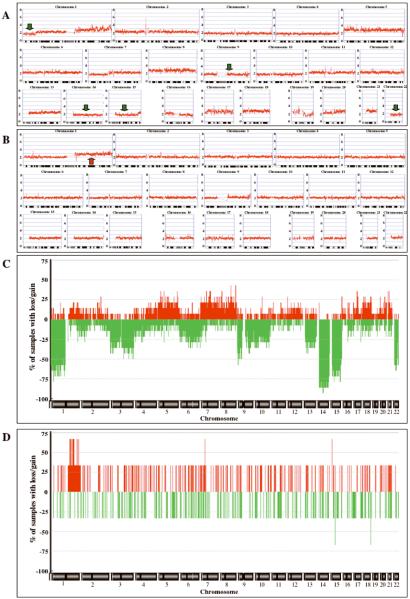
Evaluation of GIST specimens from the Fox Chase Cancer Center Biosample Repository identified 14 samples with KIT or PDGFRA mutations and 3 samples that lacked detectable mutations (Table 1). Most of these archival samples (including all 3 mutation-negative tumors) were IM-naïve. Nine tumors harbored mutations in KIT exon 11, but the sample set also included tumors with mutations in KIT exons 9, 13, or 17, and in PDGFRA exons 14 and 18. The wild-type status for KIT and PDGFRA in samples #15–17 was confirmed by exon-based screening for hot-spot mutations (KIT exons 9, 11, 13, and 17; PDGRFA exons 12 and 14) as well as whole KIT cDNA sequencing. Immunohistochemical analysis confirmed KIT expression in the KIT/PDGFRA mutation-negative GIST samples (Figure 1). Copy number (CN) and allele-specific genotyping analysis was performed on these 17 samples using the Affymetrix GeneChip® Human Mapping 50 K Xba Array. Genome-scale plots of chromosomal CN for a selected mutant GIST (case #8) and a mutation-negative GIST (case #15) are shown in Figure 2. Case #8, which harbors an exon 11 KIT deletion, exhibits large-scale CN loss on multiple chromosomes typically implicated in GISTs (Assamaki, et al. 2007; Meza-Zepeda, et al. 2006; Wozniak, et al. 2007) (e.g., chromosomes 1p, 9, 14q, 15q, and 22q, indicated by green arrows in Figure 2A), along with additional losses and gains on other chromosomes (Figure 2A, Table 1). A frequency plot of chromosomal changes across the mutant sample set indicates frequent CN loss in chromosomes 1p, 3, 9, 13q, 14q, 15q, and 22q (Figure 2C). All KIT/PDGFRA mutant samples demonstrated regions of chromosomal CN losses (mean of 5.4 losses, range 2–13, Table 1), and 50% of the mutant samples also demonstrated regions of gain (mean of 1.6 gains, range 0–8). This limited sample set exhibited trends in the patterns of losses versus anatomic site of tumor occurrence that have been previously described {Wozniak, 2007 #578}. For example, 14q losses were more frequent in gastric versus non-gastric GISTs (86% versus 71%) while 1p and 15q losses were more predominant in non-gastric GISTs (100%/86%) than in gastric GISTs (14%/43%). In contrast, the 3 mutation-negative cases displayed very little cytogenetic progression. Case #15 shows a single region of CN gain (chromosome 1q, red arrow in Figure 2B, Table 1), while no large-scale chromosome losses were seen in the 3 KIT/PDGFRA mutation-negative samples (Figure 2D, Table 1). The single region of copy-number (CN) gain in sample #15 has been infrequently documented in GISTs (Gunawan, et al. 2007, Assamaki, et al. 2007, Wozniak, et al. 2007). We also identified two regions of CN-neutral loss-ofheterozygosity (LOH) using the 50K array. KIT/PDGFRA mutation-negative case #15 contained a region of LOH extending from 5p13.2-pter, while an additional region of LOH encompassing chromosome 19q was identified in mutant sample #3 (data not shown).
Table 1.
Clinical and molecular findings and large-scale genomic copy number changes detected by Affymetrix GeneChip 50K Xba array.
| No. | Age/Sex | Site | Risk | Typeb | Genotypec | Copy number losses | Copy number gains |
|---|---|---|---|---|---|---|---|
| 1 | 66/F | sm. int. | low | M | KIT exon 11: p.M552_Y553del | 1p; 6; 14q; 15cen-q21.1; 20q13.12-q13.2 | |
| 2 | 70/F | stomach | high | Sp | KIT exon 11:p.V559G | 3q; 14q | |
| 3 | 77/M | stomach | n/aa | n/aa | KIT exon 11:p.V560D | 6q; 9p; 10p; 13q; 14q; 15q; 21q; 22q | 5; 12; 16p; 19pd; 20 |
| 4 | 63/M | rectum | inter. | Sp | KIT exon 11:p.W557F, p.K558_V559del | 1p; 2p; 4q22.1-pter; 9p21.1-pter, 9cen-q31.2; 13q; 14q; 22q | 4q22.1-qter; 9cen -p21.1; 9q31.2-q31.3; 12; 16p; 18 |
| 5 | 37/M | omentum | high | Ep | KIT exon 11: p.K550_W557delinsIL | 1p; 3p21.1-24.2; 9; 10; 14q; 15q; 17p11.2-pter; 22q | 20q |
| 6 | 56/F | stomach | low | Sp | KIT exon 11:p.V560D | 3;14q | |
| 7 | 71M | colon | high | Sp | KIT exon 11: p.N567_L576delinsKE | 1p; 5p; 9p; 15q; 22q | |
| 8 | 60/F | stomach | high | Sp | KIT exon 11: p.Q556_W557del, p.K558N | 1p34.3-pter; 3; 4; 7p; 9; 10; 14q; 15cen-q26.1; 19ql3.12-ql3.41;22q | 1q; 5; 8; 17p31.1-qter; 18; 19p; 20q; 21q |
| 9 | 36/F | stomach | high | M | KIT exon 11:p.D579del | 1p34.2-p34.3, 1p33-p34.1, 1p32.1-p32.3, 1p22.1-p31.1, 1q32.2-q32.3, 1q41-qter; 3; 9cen-q31.3; 10; 14cen-q12, 14q21.1-qter; 15q; 22q | |
| 10 | 43/M | abd. wall | high | Sp | KIT exon 9: p. N512_N513insAY | 1p; 13q; 15q; 18; 22q | |
| 11 | 47/M | sm. int. | low | Sp | KIT exon 13:p.K642E | 1p; 2p25.1-pter, 2p24.3-p25.1, 2p23.2-p22.3, 2q32.3-q36.1, 2q36.1-qter; 14q; 15q | 7 |
| 12 | 63/M | sm. int. | low | Sp | KIT exon 13:p.K642E | 1p; 6; 14q; 15q | 7 |
| 13 | 81/F | stomach | high | M | KIT exon 17:p.Y823D | 3; 9p22.2-pter, 9p13.2-p13.3, 9q21.1-q34.13; 14q; 15q; 22q | |
| 14 | 71/M | stomach | high | Ep | PDGFRA exon 14: p.N659Y; exon 18: p.Y849C | 12p; 13q; 14q | 8q |
| 15 | 33/F | stomach | inter. | M | none identified | 1q | |
| 16 | 53/M | stomach | low | Ep | none identified | ||
| 17 | 48/M | rectum | high | Sp | none identified |
not available.
Sp, spindle; Ep, epithelial; M, mixed.
Nomenclature conforms to guidelines from the Human Genetic Variation Society (http://www.hgvs.org/mutnomen).
CN gain > 3
Figure 1.
Immunohistochemical staining showing KIT expression in KIT/PDGFRA mutation-negative samples #15–17 (A–C).
Figure 2.
Genome-wide plots of SNP CN copy data (A, B) and frequency plots of CN alterations (C, D) for samples analyzed with the Affymetrix GeneChip® Human Mapping 50K Xba Array. (A) KIT-mutant GIST #8. (B) KIT/PDGFRA-mutation-negative GIST #15. Copy-number values (0–8) are indicated to the left of each panel, chromosome labels and ideograms are shown at the top and bottom of each panel, respectively. Green and red arrows indicate selected regions of CN loss and gain, respectively, that are discussed in the text. (C) Frequency plots of CN alterations in 14 mutant GISTs. (D) Frequency plots of CN alterations in 3 mutation-negative GISTs. Chromosome labels are shown at the bottom of each panel, frequency values to the left of the panel. Determination of CN gains (red) and losses (green) was done using the ACE algorithm (Analysis of Copy Error, CGH-Explorer, University of Oslo, Norway), which was used with a False Discovery Rate (FDR) of 0.0001.
To expand upon these observations, a series of 32 fresh-frozen GIST samples from the multi-institutional RTOG-0132 trial of neoadjuvant/adjuvant IM for advanced primary/metastatic GIST (Eisenberg, et al. 2009) were evaluated for mutations in KIT and PDGFRA. A total of 28 KIT/PDGFRA mutant and four mutation-negative tumors were identified. From these, seven KIT mutant samples (#18–24) and the four KIT/PDGFRA mutation-negative GISTs (#28–31) were selected for SNP analysis (Table 2). Three additional independent samples (#25–27) lacking mutations in KIT and PDGFRA, including one pediatric GIST patient (case # 27) that was referred to our institute, were also included in this analysis. These 14 samples were analyzed using the high-density Affymetrix Genome-Wide Human SNP Array 6.0. As many of the specimens included in this analysis (noted in Table 2) were obtained following 8–12 weeks of neo-adjuvant IM therapy, the frozen specimens were first evaluated for tumor content and cellularity. The samples were embedded in optimal-cutting temperature (OCT) medium, sectioned, and H&E- and KIT-stained. For inclusion, the tissues were required to consist of at least 40% tumor cellularity. Adjacent normal tissue and highly acellular areas were then macrodissected away prior to DNA isolation. Figure 3 shows a genome-scale heat map (log2 ratios of SNP CN) for GIST cases #18–31 analyzed by the Affymetrix Genome-Wide Human SNP Array 6.0. All KIT-mutant cases (#18–24) exhibited multiple regions of large-scale CN loss (shown in red), with frequent (>50%) losses in chromosomes 1p, 9, 14q, 15q, and 22q. Multiple regions of CN gain (in blue) were also evident in cases 20, 21, and 23. The 7 KIT-mutant samples exhibited a mean of 7.0 losses (range of 5–10) and 3.0 gains (range 2–11) per tumor (Table 2). In confirmation of our previous results seen with the lower-density arrays, chromosomal CN changes were minimal in 5 of 7 GISTs for which mutations in KIT and PDGFRA were not identified (#27–31), including the pediatric case #27 (Figure 3, Table 2). The single region of CN loss across chromosome 16q in case #31 was not seen in any of the mutant samples analyzed in this report. Regions of CN-neutral LOH were infrequent in KIT-mutated GISTs; case #21 exhibited CN-neutral LOH across chromosome 4 (data not shown). Large-scale regions of LOH without CN change were not seen in the mutation-negative GIST samples analyzed by the SNP 6.0 array
Table 2.
Clinical and molecular findings and large-scale genomic copy number changes detected by Affymetrix Genome-Wide Human SNP Array 6.0.
| Case | Sex | Site | Disease status | Genotypec | Copy number losses | Copy number gains |
|---|---|---|---|---|---|---|
| 18a, f | M | stomach | primary | KIT exon 11: p.P551_Q556delinsT | 1p36.12-34.3; 4pter-p16.1; 9; 14q; 15q; 22q12.1-qter | |
| 19a, f | F | other | recurrent | KIT exon 11: p.W567_V559delinsF | 1p; 4q12-q25; 9; 10, 12pter-p11.22; 15q; 19q12-qter; 21q22.2-qter; 22q | |
| 20a, f | F | stomach | primary | KIT exon 11: p.V560_I571del | 2q32.1-qter; 6q13-q21, 6q21-q25.3; 9; 14q; 15q; 21q11.2-q22.3; 22q | 2q24.3-q31.2; 5; 6pter-q13, 6q25.3-qter; 7; 8; 13q; 16p; 18; 19; 20 |
| 21a | M | stomach | primary | KIT exon 11: p.E554_K558del | 1pter-p34.3; 3pter-p14.1; 6p21.31-p21.1, 6p12.3-p12.1, 6q; 9; 10q; 14q; 21q; 22q | 1q; 3p14.1-qter; 5; 6p22.2-p21.31; 8; 10pter-p11.21; 12; 13q; 16p; 17; 19 |
| 22a, f | F | other | recurrent | KIT exon 11: p.K550_W557del | 2q12.1-q21.1; 5pter-p13.3; 9; 14q; 15q; 22q | |
| 23a, f | M | sm. int | primary | KIT exon 11: p.M552_Y553del | 1p; 6; 13q31.1-qter; 15q; 19pter-p13.12 | 8qe; 21q |
| 24a, f | M | other | recurrent | KIT exon 11:p.V560E | 1p; 2p; 11; 13q; 14q; 15q; 22q | |
| 25 | M | sm. int | recurrent | BRAF exon 15: p.V600E | 1p; 6q; 9p; 11q; 13q13.2-qter; 14q; 15q13.2-qter; 17q12-q21.32; 18; 19q13.12-qter; 22q11.21-q12.3, 22q13.1-qter | 1q; 4; 5; 6p; 7; 11p; 13q12.2-q13.3; 15q11.2-q13.1; 16; 17pe, 17q21.33-q24.3e, 17q25.1-qtere; 19pter-q13.12e; 21q |
| 26 | F | sm. int | recurrent | none identified | 1pter-p36.21, 1p36.13-p22.2, 1p21.3-pter; 4q; 6q; 9pter-p21.3, 9p21.3-p21.1d, 9p21.1-pcen; 10; 11q12.1-qter; 13q; 14q; 15q; 17pter-q12; 18; 19q12-q13.42; 22q | 1q; 2; 3; 4p; 5pter-p13.2e, 5p13.1-qter; 6p; 7pe, 7q; 8; 9qe; 11pter-p14.2e, 11p14.2-q12.1; 12; 16; 17q21.33-qter; 19pter-q12, 19q13.42-qter; 20p13e, 20p13-qter; 21q |
| 27b | F | stomach | recurrent | none identified | ||
| 28a | M | other | recurrent | none identified | ||
| 29a | F | stomach | recurrent | none identified | ||
| 30a, f | M | other | recurrent | none identified | ||
| 31a, f | F | stomach | primary | none identified | 16q |
Case from RTOG S0132/ACRIN 6665 clinical trial.
Pediatric case.
Nomenclature conforms to guidelines from the Human Genetic Variation Society (http://www.hgvs.org/mutnomen).
Homozygous deletion.
Portions with CN gain > 3.
Tumor specimens were obtained following 8–12 weeks neo-adjuvant IM therapy.
Figure 3.
Heat map (log2 ratios of SNP CN) for GIST cases #18–31 analyzed by Affymetrix Genome-Wide Human SNP Array 6.0. Heat maps were created in Affymetrix Genotyping Console 3.0 and exported as portable network graphics files. For ease of visualization, image brightness and contrast were adjusted simultaneously for heat maps and scale. The color scale ranges from bright red (CN loss) to white to bright blue (CN gain). Case numbers and mutation status are indicated at the top. Chromosome numbers are shown to the left of the figure, p and q arms are separated by hash marks. The red arrow indicates the position of the chromosome 9p homozygous deletion identified in mutation-negative sample #26 and mutant samples #18, 20, and 22.
Unlike the other wild-type samples in this study, 2 of the 7 KIT/PDGFRA mutation-negative cases (#25 and #26) exhibited an extremely high level of cytogenetic progression, with numerous (>10) regions of CN loss as well as gain (Figure 3, Table 2). Case #25 was a surgically resected tumor from a patient whose disease had recurred following treatment with IM. Based on a recent report (Agaram, et al. 2008b), we examined all GIST samples (KIT/PDGFRA mutant and mutation-negative) for mutations in HRAS, KRAS, and BRAF. No mutations in HRAS or KRAS were found. However, case #25, while wild type for KIT and PDGFRA, was found to harbor a kinase-domain mutation in BRAF (exon 15, V600E). We did not identify a BRAF mutation in any other of ~80 GISTs examined (data not shown). The second KIT/PDGFRA mutation-negative tumor with significant CN aberrations, case #26, was a metastatic GIST that recurred after treatment with IM, sunitinib, and several other biologic therapeutics. Although no BRAF mutation was found, this sample contained a homozygous deletion of chromosome 9p21.2–21.3 (position indicated by arrow in Figure 3) and several regions with higher-level amplifications (CN > 3) (Table 2). Small homozygous deletions on chromosome 9p were also found in KIT-mutant samples #18, 20, and 22 (Figure 3, Supplementary Table 2). The minimal region of overlap between the 4 samples was ~0.28 Mbp and encompassed the cell-cycle kinase inhibitors CDKN2A (encoding p16INK4/p14ARF) and CDKN2B (encoding p15INK4B).
Finally, we searched for focal regions of CN aberration within the KIT/PDGFRA/BRAF mutation-negative samples that otherwise lacked large-scale changes (e.g. samples 27–31). The SNP array 6.0 consists of ~1.8 million markers genome wide with a median inter-marker distance of ~700 bp, which should be expected to increase confidence in reporting for small regions of CN gain or loss. Excluding regions that fully overlapped known regions of copy number variation (CNV) or fell within intragenic regions in the genome, we found few focal changes (defined as <10% of a chromosome arm) within these samples. Case #30 exhibited an ~0.6 Mbp region of low-level CN gain on chromosome 7p14.1 (encompassing the proteosome subunit gene PSMA2 and the mitochondrial ribosomal subunit MRPL32) as well as an ~0.2 Mbp region of CN loss on 1q25.2 encompassing a portion of the PAPPA2 gene (Supplementary Table 2). No other focal changes were identified within cases 27–31. In contrast, we were able to detect multiple focal regions of CN gain and/or loss, ranging from 0.11–4.7 Mbp, in the 7 KIT/PDGFRA-mutated samples (#18–24), the single BRAF-mutated sample (#25), as well as in the cytogenetically progressed case #26 (Supplementary Table 2).
Discussion
All 21 KIT/PDGFRA-mutant GISTs as well as a BRAF-mutated GIST analyzed in this study exhibit CN loss in multiple chromosomes, including at least one chromosome typically deleted in GISTs (i.e., chromosome 1p, 9, 13q, 14q, 15q, or 22q). In contrast, of the 8 adult KIT/PDGFRA/BRAF mutation-negative GISTs we analyzed using low- and high-density SNP arrays, 7 demonstrated very few genomic aberrations. We also found no evidence of cytogenetic progression in the single pediatric case we examined. The isolated regions of CN aberration we did see in these mutation-negative samples (i.e., case #15, 1q gain; case #31, 16q loss) are relatively rare in GISTs, occurring in about 5–20% of mutant GISTs (Assamaki, et al. 2007; Gunawan, et al. 2007; Wozniak, et al. 2007). We found only a single instance of CN-neutral LOH in the mutation-negative samples (case #15, chromosome 5p13.2-pter), as well as several instances in the samples harboring mutations. Using the SNP 6.0 array we found multiple regions of focal CN aberrations in the KIT/PDGFRA/BRAF-mutated samples, as well as in one sample for which no mutation was identified, but among the 5 mutation-negative samples lacking large-scale changes, only one case displayed focal CN changes overlapping known genes. These results suggest that adult mutation-negative GISTs demonstrate minimal cytogenetic progression. Several studies using CGH approaches have described a few GIST tumors with few or no large-scale losses, but these studies either did not provide KIT/PDGFRA mutational status {El-Rifai, 2000 #440}{Gunawan, 2007 #604} or did not include mutation-negative GISTs {Wozniak, 2007 #578}. A recent array-CGH study {Assamaki, 2007 #576}, however, did describe four KIT/PDGFRA mutation-negative GISTs with deletions in chromosomes 1, 14 and/or 22 {Assamaki, 2007 #576}. It is unclear whether the discrepancy between our results for adult mutation-negative GISTs and those of Assamaki and colleagues are due to some unappreciated sampling bias or due to technical differences in the studies. Notably, many of the tumor samples analyzed in the CGH study were from paraffin-embedded tissue, which has been suggested to decrease the detection rate of mutations (Corless, et al. 2004; Lasota, et al. 2007).
Our findings of minimal cytogenetic changes in adult mutation-negative GISTs are similar to what has been described for pediatric GISTs (Janeway, et al. 2007). Although the oncogenic driving force in KIT/PDGFRA mutation-negative GISTs has not been identified, there is some evidence that aberrations in the insulin-like growth factor receptor (IGF1R) pathway may play a role. We have previously demonstrated through a combination of immunoblot, immunohistochemical, and qPCR approaches that IGF1R is over-expressed in adult KIT/PDGFRA mutation-negative v. mutant GISTs (Belinsky, et al. 2008; Tarn, et al. 2008). We did not, however, identify mutations in IGF1R in any of our GIST cases. Overexpression of IGF1R has also been described in pediatric GISTs (Agaram, et al. 2008a; Tarn, et al. 2008), which suggested to us that both pediatric and adult mutation-negative GISTs may be driven at least in part by signaling through the IGF pathway in the absence of activating RTK mutations. The studies described here indicate that a lack of cytogenetic progression is another shared characteristic of these tumor subsets. However expression-profiling analyses have suggested that GISTs from children and young adults have transcriptional profiles that are distinct from adult GISTs (Prakash, et al. 2005), even when compared to adult mutation-negative GISTs (Agaram, et al. 2008a). GISTs in children also have rather distinct clinopathological features, occurring almost exclusively in females with a gastric origin (Miettinen, et al. 2005a; Prakash, et al. 2005; Price, et al. 2005). In contrast, of our 8 KIT/PDGFRA/BRAF mutation-negative adult GISTs, 4 were diagnosed in males, and 4 cases presented in the small intestine or other non-gastric location.
Two KIT/PDGFRA mutation-negative cases that we examined demonstrated an exceptionally high level of cytogenetic changes for GISTs. Case #25, which contains >10 regions of CN gain and loss, was found to harbor a point mutation, V600E, in exon 15 of the BRAF gene. BRAF, a RAF-family serine/threonine protein kinase, is a known effector of the MAP-kinase signaling pathway. Mutations in BRAF are found in ~8% of all human cancers, and the V600E kinase-domain mutation accounts for perhaps 80% of these (Pratilas and Solit 2007). This mutation was recently reported for 3 cases of a set of 41 adult KIT/PDGFRA mutation-negative GISTs (Agaram, et al. 2008b). These cases exhibited a specific spectrum of clinicopathological characteristics, namely high-risk tumors of small bowel origin, occurring in women between the ages of 49–55. The case reported herein is a small bowel tumor that recurred in a 57-year old male patient following IM therapy. As we found this BRAF mutation in only one of ~80 GISTs we examined, the connection between this activating kinase mutation and significant cytogenetic progression is tentative and requires further confirmation. The other KIT/PDGFRA mutation-negative tumor that exhibited a high degree of cytogenetic progression (case #26) is a metastatic GIST that recurred after treatment with IM, sunitinib, and several other biological therapies. Although no BRAF mutation was found in this tumor, several regions of higher-level (CN > 3) aberration were detected, as well a homozygous deletion at chromosome 9p21.2–p21.3. Assamaki and colleagues identified 9p loss mainly in GISTs with high biological potential, and established a minimal overlapping region of 9p loss of ~16 Mbp including the CDKN2A/2B gene locus (Assamaki, et al. 2007). The current study refines the minimal overlapping region of deletion to ~0.28 Mbp in 9p21.3, a region that contains only the CDKN2A/2B genes. Whether inactivation of these genes due to homozygous deletion contributed to the cytogenetic progression in the current case is unclear. The CDNK2A/ARF locus encodes both the p16INK4A protein, which functions as an inhibitor of the CDK4 kinase, and the p14ARF product, which functions as a stabilizer of the p53 tumor suppressor gene. CDKN2B encodes the p15INK4B protein, which also functions as an inhibitor of cyclin-dependent kinases. The connection between p16/p15 expression and GIST progression has been a topic of considerable interest in recent years (Huang, et al. 2006; Sabah, et al. 2006; Schmieder, et al. 2008; Schneider-Stock, et al. 2005; Steigen, et al. 2008).
In summary, this study demonstrates that mutation-negative GISTs from adult patients exhibit a markedly lower degree of cytogenetic progression than GISTs with activating mutations in KIT or PDGFRA. Moreover, for the first time a GIST with an alternate activating mutation in the serine/threonine kinase BRAF has been demonstrated to exhibit multiple secondary cytogenetic changes, some of which have also been seen in RTK-mutated GISTs. Identifying molecular targets for the successful treatment of adult and pediatric mutation-negative GISTs remains a challenge for the field.
Supplementary Material
Acknowledgments
Grant support: This work was supported in part by NIH grants (CA106588 and a supplement to U10 CA21661) to A.K.G., an award by the Fox Chase Cancer Center (FCCC) Translational Research Committee to M.v.M. and A.K.G as part of the FCCC core grant (P30 CA006927), a Translational Research Program seed grant by the Radiation Therapy Oncology Group funded by the National Cancer Institute to A.K.G (U10CA21661), an NIH Training Grant (Institutional NRSA) Appointment (CA009035-31) to L.R, and an award by Tania Stutman and the GIST Cancer Research Fund.
Abbreviations
- GIST
gastrointestinal stromal tumor
- RTK
receptor tyrosine kinase
- PDGFR
platelet-derived growth factor receptor
- IM
imatinib mesylate
- SNP
single-nucleotide polymorphism
- ICC
interstitial cells of Cajal
- CGH
comparative genomic hybridization
- LOH
loss-of-heterozygosity
- CN
copy-number
- IGF
insulin-like growth factor
Footnotes
Conflict of interest statement: The authors declare no conflicts of interest.
References
- Agaram NP, Laquaglia MP, Ustun B, Guo T, Wong GC, Socci ND, Maki RG, DeMatteo RP, Besmer P, Antonescu CR. Molecular characterization of pediatric gastrointestinal stromal tumors. Clin Cancer Res. 2008a;14(10):3204–15. doi: 10.1158/1078-0432.CCR-07-1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agaram NP, Wong GC, Guo T, Maki RG, Singer S, Dematteo RP, Besmer P, Antonescu CR. Novel V600E BRAF mutations in imatinib-naive and imatinib-resistant gastrointestinal stromal tumors. Genes Chromosomes Cancer. 2008b;47(10):853–9. doi: 10.1002/gcc.20589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assamaki R, Sarlomo-Rikala M, Lopez-Guerrero JA, Lasota J, Andersson LC, Llombart-Bosch A, Miettinen M, Knuutila S. Array comparative genomic hybridization analysis of chromosomal imbalances and their target genes in gastrointestinal stromal tumors. Genes, Chromosomes & Cancer. 2007;46(6):564–76. doi: 10.1002/gcc.20439. [DOI] [PubMed] [Google Scholar]
- Belinsky MG, Rink L, Cai KQ, Ochs MF, Eisenberg B, Huang M, von Mehren M, Godwin AK. The insulin-like growth factor system as a potential therapeutic target in gastrointestinal stromal tumors. Cell Cycle. 2008;7(19):2949–55. doi: 10.4161/cc.7.19.6760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohringer S, Godde R, Bohringer D, Schulte T, Epplen J. A software package for drawing ideograms automatically. Online Jornal of Bionformatics. 2002;1:51–61. [Google Scholar]
- Braconi C, Bracci R, Bearzi I, Bianchi F, Sabato S, Mandolesi A, Belvederesi L, Cascinu S, Valeri N, Cellerino R. Insulin-like growth factor (IGF) 1 and 2 help to predict disease outcome in GIST patients. Ann Oncol. 2008;19(7):1293–8. doi: 10.1093/annonc/mdn040. [DOI] [PubMed] [Google Scholar]
- Corless CL, Fletcher JA, Heinrich MC. Biology of gastrointestinal stromal tumors. J Clin Oncol. 2004;22(18):3813–25. doi: 10.1200/JCO.2004.05.140. [DOI] [PubMed] [Google Scholar]
- Corless CL, Heinrich MC. Molecular Pathobiology of Gastrointestinal Stromal Sarcomas. Annu Rev Pathol. 2007 doi: 10.1146/annurev.pathmechdis.3.121806.151538. [DOI] [PubMed] [Google Scholar]
- Corless CL, McGreevey L, Haley A, Town A, Heinrich MC. KIT mutations are common in incidental gastrointestinal stromal tumors one centimeter or less in size. Am J Pathol. 2002;160(5):1567–72. doi: 10.1016/S0002-9440(10)61103-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debiec-Rychter M, Dumez H, Judson I, Wasag B, Verweij J, Brown M, Dimitrijevic S, Sciot R, Stul M, Vranck H. Use of c-KIT/PDGFRA mutational analysis to predict the clinical response to imatinib in patients with advanced gastrointestinal stromal tumours entered on phase I and II studies of the EORTC Soft Tissue and Bone Sarcoma Group. Eur J Cancer. 2004;40(5):689–95. doi: 10.1016/j.ejca.2003.11.025. and others. [DOI] [PubMed] [Google Scholar]
- Demetri GD, von Mehren M, Blanke CD, Van den Abbeele AD, Eisenberg B, Roberts PJ, Heinrich MC, Tuveson DA, Singer S, Janicek M. Efficacy and safety of imatinib mesylate in advanced gastrointestinal stromal tumors. N Engl J Med. 2002;347(7):472–80. doi: 10.1056/NEJMoa020461. and others. [DOI] [PubMed] [Google Scholar]
- Eisenberg BL, Harris J, Blanke CD, Demetri GD, Heinrich MC, Watson JC, Hoffman JP, Okuno S, Kane JM, von Mehren M. Phase II trial of neoadjuvant/adjuvant imatinib mesylate (IM) for advanced primary and metastatic/recurrent operable gastrointestinal stromal tumor (GIST): early results of RTOG 0132/ACRIN 6665. J Surg Oncol. 2009;99(1):42–7. doi: 10.1002/jso.21160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Rifai W, Sarlomo-Rikala M, Andersson LC, Knuutila S, Miettinen M. DNA sequence copy number changes in gastrointestinal stromal tumors: tumor progression and prognostic significance. Cancer Res. 2000;60(14):3899–903. [PubMed] [Google Scholar]
- Fletcher CD, Berman JJ, Corless C, Gorstein F, Lasota J, Longley BJ, Miettinen M, O'Leary TJ, Remotti H, Rubin BP. Diagnosis of gastrointestinal stromal tumors: a consensus approach. Int J Surg Pathol. 2002;10(2):81–9. doi: 10.1177/106689690201000201. and others. [DOI] [PubMed] [Google Scholar]
- Gunawan B, von Heydebreck A, Sander B, Schulten HJ, Haller F, Langer C, Armbrust T, Bollmann M, Gasparov S, Kovac D. An oncogenetic tree model in gastrointestinal stromal tumours (GISTs) identifies different pathways of cytogenetic evolution with prognostic implications. Journal of Pathology. 2007;211(4):463–70. doi: 10.1002/path.2128. and others. [DOI] [PubMed] [Google Scholar]
- Heinrich MC, Corless CL, Demetri GD, Blanke CD, von Mehren M, Joensuu H, McGreevey LS, Chen CJ, Van den Abbeele AD, Druker BJ. Kinase mutations and imatinib response in patients with metastatic gastrointestinal stromal tumor. J Clin Oncol. 2003a;21(23):4342–9. doi: 10.1200/JCO.2003.04.190. and others. [DOI] [PubMed] [Google Scholar]
- Heinrich MC, Corless CL, Duensing A, McGreevey L, Chen CJ, Joseph N, Singer S, Griffith DJ, Haley A, Town A. PDGFRA activating mutations in gastrointestinal stromal tumors. Science. 2003b;299(5607):708–10. doi: 10.1126/science.1079666. and others. [DOI] [PubMed] [Google Scholar]
- Hirota S, Isozaki K, Moriyama Y, Hashimoto K, Nishida T, Ishiguro S, Kawano K, Hanada M, Kurata A, Takeda M. Gain-of-function mutations of c-kit in human gastrointestinal stromal tumors. Science. 1998;279(5350):577–80. doi: 10.1126/science.279.5350.577. and others. [DOI] [PubMed] [Google Scholar]
- Huang HY, Huang WW, Lin CN, Eng HL, Li SH, Li CF, Lu D, Yu SC, Hsiung CY. Immunohistochemical expression of p16INK4A, Ki-67, and Mcm2 proteins in gastrointestinal stromal tumors: prognostic implications and correlations with risk stratification of NIH consensus criteria. Ann Surg Oncol. 2006;13(12):1633–44. doi: 10.1245/s10434-006-9188-4. [DOI] [PubMed] [Google Scholar]
- Janeway KA, Liegl B, Harlow A, Le C, Perez-Atayde A, Kozakewich H, Corless CL, Heinrich MC, Fletcher JA. Pediatric KIT wild-type and platelet-derived growth factor receptor alpha-wild-type gastrointestinal stromal tumors share KIT activation but not mechanisms of genetic progression with adult gastrointestinal stromal tumors. Cancer Res. 2007;67(19):9084–8. doi: 10.1158/0008-5472.CAN-07-1938. [DOI] [PubMed] [Google Scholar]
- Kindblom LG, Remotti HE, Aldenborg F, Meis-Kindblom JM. Gastrointestinal pacemaker cell tumor (GIPACT): gastrointestinal stromal tumors show phenotypic characteristics of the interstitial cells of Cajal. Am J Pathol. 1998;152(5):1259–69. [PMC free article] [PubMed] [Google Scholar]
- Meza-Zepeda LA, Kresse SH, Barragan-Polania AH, Bjerkehagen B, Ohnstad HO, Namlos HM, Wang J, Kristiansen BE, Myklebost O. Array comparative genomic hybridization reveals distinct DNA copy number differences between gastrointestinal stromal tumors and leiomyosarcomas. Cancer Research. 2006;66(18):8984–93. doi: 10.1158/0008-5472.CAN-06-1972. [DOI] [PubMed] [Google Scholar]
- Miettinen M, Lasota J. Gastrointestinal stromal tumors: review on morphology, molecular pathology, prognosis, and differential diagnosis. Arch Pathol Lab Med. 2006;130(10):1466–78. doi: 10.5858/2006-130-1466-GSTROM. [DOI] [PubMed] [Google Scholar]
- Miettinen M, Lasota J, Sobin LH. Gastrointestinal stromal tumors of the stomach in children and young adults: a clinicopathologic, immunohistochemical, and molecular genetic study of 44 cases with long-term follow-up and review of the literature. Am J Surg Pathol. 2005a;29(10):1373–81. doi: 10.1097/01.pas.0000172190.79552.8b. [DOI] [PubMed] [Google Scholar]
- Miettinen M, Sobin LH, Lasota J. Gastrointestinal stromal tumors of the stomach: a clinicopathologic, immunohistochemical, and molecular genetic study of 1765 cases with long-term follow-up. Am J Surg Pathol. 2005b;29(1):52–68. doi: 10.1097/01.pas.0000146010.92933.de. [DOI] [PubMed] [Google Scholar]
- Prakash S, Sarran L, Socci N, DeMatteo RP, Eisenstat J, Greco AM, Maki RG, Wexler LH, LaQuaglia MP, Besmer P. Gastrointestinal stromal tumors in children and young adults: a clinicopathologic, molecular, and genomic study of 15 cases and review of the literature. J Pediatr Hematol Oncol. 2005;27(4):179–87. doi: 10.1097/01.mph.0000157790.81329.47. and others. [DOI] [PubMed] [Google Scholar]
- Pratilas CA, Solit DB. Therapeutic strategies for targeting BRAF in human cancer. Rev Recent Clin Trials. 2007;2(2):121–34. doi: 10.2174/157488707780599393. [DOI] [PubMed] [Google Scholar]
- Price VE, Zielenska M, Chilton-MacNeill S, Smith CR, Pappo AS. Clinical and molecular characteristics of pediatric gastrointestinal stromal tumors (GISTs) Pediatr Blood Cancer. 2005;45(1):20–4. doi: 10.1002/pbc.20377. [DOI] [PubMed] [Google Scholar]
- Rubin BP, Singer S, Tsao C, Duensing A, Lux ML, Ruiz R, Hibbard MK, Chen CJ, Xiao S, Tuveson DA. KIT activation is a ubiquitous feature of gastrointestinal stromal tumors. Cancer Res. 2001;61(22):8118–21. and others. [PubMed] [Google Scholar]
- Sabah M, Cummins R, Leader M, Kay E. Altered expression of cell cycle regulatory proteins in gastrointestinal stromal tumors: markers with potential prognostic implications. Hum Pathol. 2006;37(6):648–55. doi: 10.1016/j.humpath.2006.01.023. [DOI] [PubMed] [Google Scholar]
- Schmieder M, Wolf S, Danner B, Stoehr S, Juchems MS, Wuerl P, Henne-Bruns D, Knippschild U, Hasel C, Kramer K. p16 expression differentiates high-risk gastrointestinal stromal tumor and predicts poor outcome. Neoplasia. 2008;10(10):1154–62. doi: 10.1593/neo.08646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider-Stock R, Boltze C, Lasota J, Peters B, Corless CL, Ruemmele P, Terracciano L, Pross M, Insabato L, Di Vizio D. Loss of p16 protein defines high-risk patients with gastrointestinal stromal tumors: a tissue microarray study. Clin Cancer Res. 2005;11(2 Pt 1):638–45. and others. [PubMed] [Google Scholar]
- Sircar K, Hewlett BR, Huizinga JD, Chorneyko K, Berezin I, Riddell RH. Interstitial cells of Cajal as precursors of gastrointestinal stromal tumors. Am J Surg Pathol. 1999;23(4):377–89. doi: 10.1097/00000478-199904000-00002. [DOI] [PubMed] [Google Scholar]
- Steigen SE, Bjerkehagen B, Haugland HK, Nordrum IS, Loberg EM, Isaksen V, Eide TJ, Nielsen TO. Diagnostic and prognostic markers for gastrointestinal stromal tumors in Norway. Mod Pathol. 2008;21(1):46–53. doi: 10.1038/modpathol.3800976. [DOI] [PubMed] [Google Scholar]
- Tarn C, Merkel E, Canutescu AA, Shen W, Skorobogatko Y, Heslin MJ, Eisenberg B, Birbe R, Patchefsky A, Dunbrack R. Analysis of KIT mutations in sporadic and familial gastrointestinal stromal tumors: therapeutic implications through protein modeling. Clin Cancer Res. 2005;11(10):3668–77. doi: 10.1158/1078-0432.CCR-04-2515. and others. [DOI] [PubMed] [Google Scholar]
- Tarn C, Rink L, Merkel E, Flieder D, Pathak H, Koumbi D, Testa JR, Eisenberg B, von Mehren M, Godwin AK. Insulin-like growth factor 1 receptor is a potential therapeutic target for gastrointestinal stromal tumors. Proc Natl Acad Sci U S A. 2008;105(24):8387–92. doi: 10.1073/pnas.0803383105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trent JC, Ramdas L, Dupart J, Hunt K, Macapinlac H, Taylor E, Hu L, Salvado A, Abbruzzese JL, Pollock R. Early effects of imatinib mesylate on the expression of insulin-like growth factor binding protein-3 and positron emission tomography in patients with gastrointestinal stromal tumor. Cancer. 2006;107(8):1898–908. doi: 10.1002/cncr.22214. and others. [DOI] [PubMed] [Google Scholar]
- van Oosterom AT, Judson I, Verweij J, Stroobants S, Donato di Paola E, Dimitrijevic S, Martens M, Webb A, Sciot R, Van Glabbeke M. Safety and efficacy of imatinib (STI571) in metastatic gastrointestinal stromal tumours: a phase I study. Lancet. 2001;358(9291):1421–3. doi: 10.1016/s0140-6736(01)06535-7. and others. [DOI] [PubMed] [Google Scholar]
- Wozniak A, Sciot R, Guillou L, Pauwels P, Wasag B, Stul M, Vermeesch JR, Vandenberghe P, Limon J, Debiec-Rychter M. Array CGH analysis in primary gastrointestinal stromal tumors: cytogenetic profile correlates with anatomic site and tumor aggressiveness, irrespective of mutational status. Genes, Chromosomes & Cancer. 2007;46(3):261–76. doi: 10.1002/gcc.20408. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.