Abstract
Fluorescence resonance energy transfer (FRET) provides a powerful means to study protein conformational changes. However, the incorporation of an exogenous FRET pair into a protein could lead to undesirable structural perturbations of the native fold. One of the viable strategies to minimizing such perturbations is to use non-natural amino acid based FRET pairs. Previously we have shown that p-cyanophenylalanine (PheCN) and tryptophan (Trp) constitute such a FRET pair, useful for monitoring protein folding-unfolding transitions. Herein, we further show that 7-azatryptophan (7AW) and 5-hydroxytryptophan (5HW) can also serve as a FRET acceptor to PheCN, and the resultant FRET pairs offer certain advantages over PheCN-Trp. For example, the fluorescence spectrum of 7AW is sufficiently separated from that of PheCN, making it straightforward to decompose the FRET spectrum into donor and acceptor contributions. Moreover, we show that PheCN, Trp and 7AW can be used together to form a multi-FRET system, allowing more structural information to be extracted from a single FRET experiment. The applicability of these FRET systems is demonstrated in a series of studies wherein they are employed to monitor the urea-induced unfolding transitions of the villin headpiece subdomain (HP35), a designed ββα motif (BBA5), and the human Pin1 WW domain.
Keywords: Protein folding, FRET, p-cyanophenylalanine, 7-azatryptophan, 5-hydroxytryptophan
Introduction
Fluorescence resonance energy transfer (FRET) is a powerful and widely used technique in chemistry and biology due to its sensitivity to the separation distance between the donor and acceptor [1–3]. However, the application of FRET to protein conformational studies typically requires labeling of the protein in question with exogenous dye fluorophores, which is both inconvenient and potentially disruptive to the parent molecule. In addition, this labeling requirement makes the simultaneous use of more than one donor-acceptor pair difficult, limiting the amount of structural information attainable via FRET spectroscopy. Recently, we have shown that the non-natural amino acid p-cyanophenylalanine (PheCN) and tryptophan (Trp) constitute a useful FRET pair and have applied it to the study of the conformational distribution of an unstructured peptide in aqueous solution [4], membrane-mediated helix folding [5], and urea-induced unfolding transitions of the villin headpiece subdomain (HP-35) and the LysM domain [6]. Herein, we further show that PheCN can be used as a FRET donor to two Trp analogues, 7-azatryptophan (7AW) and 5-hydroxytryptophan (5HW), and that PheCN-Trp-7AW forms a multi-FRET system [7]. The latter offers the ability to simultaneously monitor relative distance changes among three points in a protein, and thus is advantageous for protein folding studies.
PheCN is a versatile spectroscopic probe, as it exhibits unique infrared [8–12] and fluorescence [13] properties. In particular, the peak position (~300 nm) of the PheCN fluorescence spectrum is insensitive to environment, making it a good candidate to serve as a FRET donor to Trp analogues, which often show a high molar absorptivity near that wavelength. In addition, the molar absorptivity of PheCN at 240 nm is about 6.5 times larger that of Trp, making selective excitation of the donor fluorescence feasible. In this regard, PheCN has certain advantages over other amino acid FRET donors to Trp, such as tyrosine (Tyr) [14, 15]. Furthermore, as a non-natural amino acid, PheCN can be incorporated into a peptide sequence via standard solid-phase peptide synthesis protocol or into a protein through in vivo methods [16]. Perhaps more importantly, PheCN is expected to only minimally perturb the native structure of the protein in question, especially when it is used to substitute a phenylalanine (Phe) or tyrosine (Tyr) residue. For example, Raleigh and coworkers [17] have recently shown that when the Phe5 residue in the hydrophobic core of protein NTL9 was replaced with PheCN, the mutant protein exhibited almost identical folding thermodynamic properties as the parent.
7AW was first incorporated into proteins 40 years ago [18], and has since been widely used as a fluorescent reporter of protein structure, stability and dynamics [19–30]. Herein, we further explore its utility as a FRET acceptor to PheCN. The absorption and fluorescence spectra of 7AW are red-shifted compared to those of Trp [31]. As a result, the fluorescence spectrum of PheCN overlaps almost completely with the red portion of the absorption spectrum of 7AW, while the fluorescence spectrum of 7AW, centered at around 390 nm in water, is almost completely separated from that of PheCN. The former manifests as an increase in the Förster distance, and the latter makes it easier to decompose the FRET spectrum into the constituent donor and acceptor contributions. Therefore, in practice 7AW could be a better choice over Trp to serve as a FRET acceptor to PheCN.
5HW is a naturally occurring amino acid that is not found in proteins but is a precursor of the neurotransmitter serotonin [32]. Similar to 7AW, 5HW has also been extensively used as a fluorescent probe in protein conformational studies [21, 26, 27, 33–36], due to its higher fluorescence quantum yield compared to that of Trp [28]. Furthermore, and perhaps more important for our purposes, the fluorescence intensity of 5HW has been shown to be less sensitive to solvent polarity than Trp [32]. Thus, the PheCN-5HW FRET pair offers certain advantages over PheCN-Trp when used to investigate denaturant-induced protein folding-unfolding transitions.
To demonstrate the utility of these non-natural amino acid FRET pairs in protein conformational studies, we use them to monitor the urea-induced unfolding transitions of two model systems, HP35 and BBA5. As shown (Figure 1a), HP35 is a three-helix bundle [37, 38] and its equilibrium and kinetic folding properties have been extensively studied [39–44]. Incorporation of the PheCN-7AW FRET pair into the protein is accomplished by mutating the C-terminal Phe residue to PheCN and the single Trp residue to 7AW, respectively. Since the Trp residue is conveniently located at the N-terminal end of the C-terminal helix, this arrangement of the donor-acceptor pair will allow us to directly and specifically monitor the unfolding transition of the C-terminal helix upon HP35 denaturation. BBA5 is a small designed protein [45], which folds into a ββα motif (Figure 1b). Here, we use this peptide to examine the utility of the PheCN-5HW FRET pair by mutating the N-terminal Gly residue to 5HW and the C-terminal Tyr residue to PheCN. Finally, we use two mutants of the hPin1 WW domain to test the applicability of PheCN-Trp-7AW as a multi-FRET system. The hPin1 WW domain consists of residues 6–39, or the N-terminal domain, of human Pin1 (hPin1) [46] and folds into a three-stranded β-sheet structure (Figure 1c) in aqueous solution [47]. In the first mutant, the single Phe residue (Phe25), which is located in the center β-strand, is substituted with PheCN, and one of the two Trp residues, Trp34, which is located on the third β-strand, is replaced with 7AW. In the second mutant, besides the aforementioned Trp34 → 7AW mutation, all Tyr residues are eliminated via Tyr23 → PheCN and Tyr24 → Phe mutations. Because in both cases the donor and acceptors are located on different β-strands of the WW domain, we believe that these mutants are ideally suited to validate the usefulness of PheCN-Trp-7AW as a multi-FRET system.
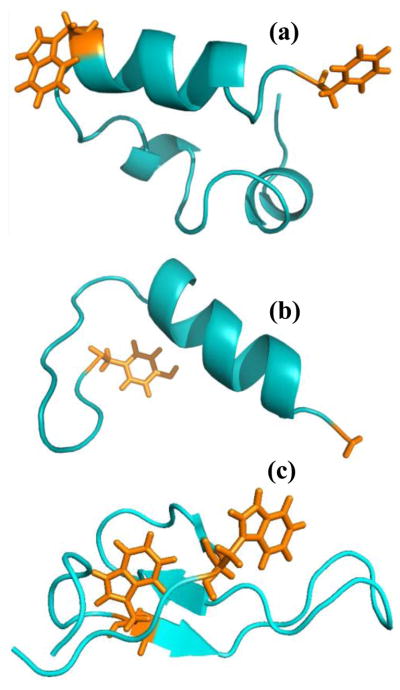
Figure 1.
(a) NMR structure of HP35 (PDB code 1VII), (b) NMR structure of BBA5 (PDB code 1T8J), and (c) NMR structure of the hPin1 WW domain (PDB code 2KCF). The sidechains involved in the FRET mutations in HP35 and BBA5 are shown, whereas for WW domain, only the Trp sidechains are shown.
Materials and Methods
Materials
p-cyanophenylalanine (Bachem), D, L-7-azatryptophan (Sigma Aldrich), and 5-hydroxy-L-tryptophan (Anaspec) were used as received. All peptides were synthesized on a peptide synthesizer (Protein Technologies, AZ) using Rink resin and then purified to homogeneity by reverse-phase HPLC. The identity of each peptide was further verified by either matrix-assisted laser desorption ionization mass spectrometry or electrospray mass spectrometry. For protein samples containing D- and L-7-azatryptophans (i.e., mutants of HP35 and the hPin1 WW domain), the corresponding diastereomers were further separated by reverse-phase HPLC following previous studies [20, 48] and the more stable diastereomer was used. All peptide and protein samples were prepared in 50 mM phosphate buffer (pH 7) and their final concentrations were determined optically using ε278 = 7840 cm−1M−1 for BBA5-PH, ε280 = 6820 cm−1M−1 for HP35-AP, and ε280 = 15490 cm−1M−1 for the WW mutants.
Absorption Measurement
All UV-Vis spectra were measured on a Lambda 25 UV-Vis spectrometer (Perkin Elmer, MA).
CD Measurements
The far-UV circular dichroism (CD) spectra were measured on a 62A DS spectropolarometer (Aviv Associates, NJ) with a 1 mm sample holder. The peptide or protein concentration was in the range of 30 – 90 μM.
Fluorescence Measurements
The fluorescence spectra at 2 nm resolution were collected on a Fluorolog 3.10 spectrofluorometer (Jobin Yvon Horiba, NJ) using a 1 cm quartz sample holder whose temperature was maintained at 20 °C. To minimize self-quenching, the optical density (OD) of each sample at the excitation wavelength was adjusted to be in the range of 0.1 – 0.2. The peptide or protein concentration was about 10 μM.
Förster Distance Calculation
We assumed that the mechanism of the observed PheCN fluorescence quenching by 7AW and 5HW can be described within the framework of Förster theory. Accordingly, the Förster distance (R0) for all FRET pairs was determined using the following equation [1]:
| (1) |
where κ2 is the orientation factor, assumed to be 2/3, QD is the fluorescence quantum yield of the donor in the absence of the acceptor, N is Avogadro’s number, η is the refractive index of the medium, assumed to be 1.33, and J(λ) is the overlap integral, which was determined according to the following equation:
| (2) |
where FD(λ) is the area-normalized emission spectrum of the donor and εA(λ) is the wavelength-dependent molar absorption coefficient of the acceptor. All of the reported Förster distances were calculated using the fluorescence and absorption spectra of the respective free amino acids.
Results and Discussion
PheCN-7AW FRET pair
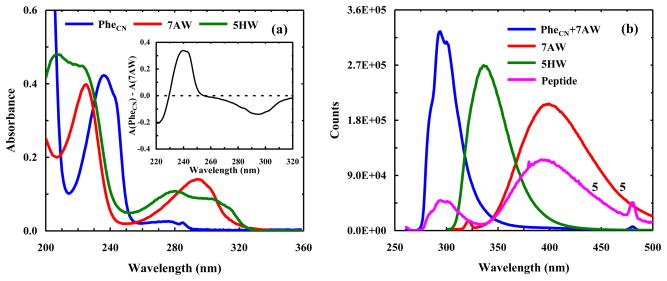
The absorption spectrum of 7AW is red shifted by ~10 nm in comparison to that of Trp (Figure 2a), resulting in a stronger overlap with the fluorescence spectrum of PheCN. As a result, the Förster distance (R0) of this FRET pair is increased to 18.5 ± 0.5 Å from the 16 Å of the PheCN-Trp FRET pair [4]. In addition, the molar absorptivity of 7AW at 240 nm is approximately 7 times smaller than that of PheCN (Figure 2a), making selective excitation of the donor possible by using an excitation wavelength near 240 nm (Inset of Figure 2a). Indeed, the fluorescence spectrum of a solution containing equal molar amounts of free PheCN and 7AW indicates that the 7AW emission is too weak to be detected when the mixture is excited at 240 nm (Figure 2b). On the other hand, the fluorescence spectrum of a peptide containing both PheCN and 7AW (sequence: G-7AW-K-PheCN-T-V), also obtained with an excitation wavelength of 240 nm, shows characteristics of a FRET spectrum wherein the donor/acceptor fluorescence is significantly quenched/enhanced (Figure 2b). While the fluorescence intensity of 7AW (the acceptor) is relatively low due to its low fluorescence quantum yield in water [31], it is clear that its fluorescence spectrum (Figure 2b) is well separated from that of PheCN. Thus, compared with Trp, 7AW is a more convenient FRET acceptor to PheCN because there is no need to perform spectral decomposition [6] in order to determine the fluorescence intensities of the donor and acceptor. This feature may prove to be particularly useful in time-resolved FRET measurements.
Figure 2.
(a) Absorption spectra of PheCN, 7AW, and 5HW in water, as indicated. These spectra were collected at room temperature and the concentration of the amino acid was 20 μM. Also shown in the inset is the difference between these spectra, which indicates that PheCN and 7AW can be selectively excited, e.g., at 240 and 290 nm, respectively. (b) Fluorescence spectra of 7AW, 5HW, a 1:1 mixture of PheCN and 7AW, and a control peptide (sequence: G-7AW-K-PheCN-T-V) in water at 20 °C. For 7AW and 5HW, the excitation wavelength was 290 nm, whereas for the peptide and the mixture of PheCN and 7AW, the excitation wavelength was 240 nm. The concentration of the amino acids only was 20 μM, and that of the peptide and each amino acid in the PheCN + 7AW mixture was 16 μM. For easy comparison, the peptide and 7AW spectra have been multiplied by a factor of five.
Using PheCN-7AW to probe urea-induced unfolding of HP35
We test the utility of the PheCN-7AW FRET pair by applying it to monitor the urea-induced unfolding transition of an individual α-helix within a well studied helical protein, HP35 (sequence: L42SDEDFKAVF-GMTRSAFANL-PLWKQQNLKK-EKGLF76). As shown (Figure 1a), HP35 folds into a three-helix bundle, which is stabilized by a hydrophobic core composed of three Phe residues (i.e., Phe47, Phe51, and Phe58), Leu69, and Val50 [37, 38]. In addition, the single Trp residue (Trp64) and the C-terminal Phe residue (Phe76) are conveniently located at the two ends of the C-terminal helix, and are approximately 18.5 Å apart from each other (Cα to Cα). Thus, we incorporate the PheCN-7AW FRET pair into HP35 by mutating Trp64 to 7AW and Phe76 to PheCN (the resultant mutant is hereafter referred to as HP35-AP) to allow for the direct assessment of the denaturant-induced unfolding transition of the C-terminal helix through FRET measurements. The far-UV CD spectrum of HP35-AP is practically identical to that of HP35 (Figure S1, Supplementary data), suggesting that these mutations do not disrupt the native helical fold of the protein.
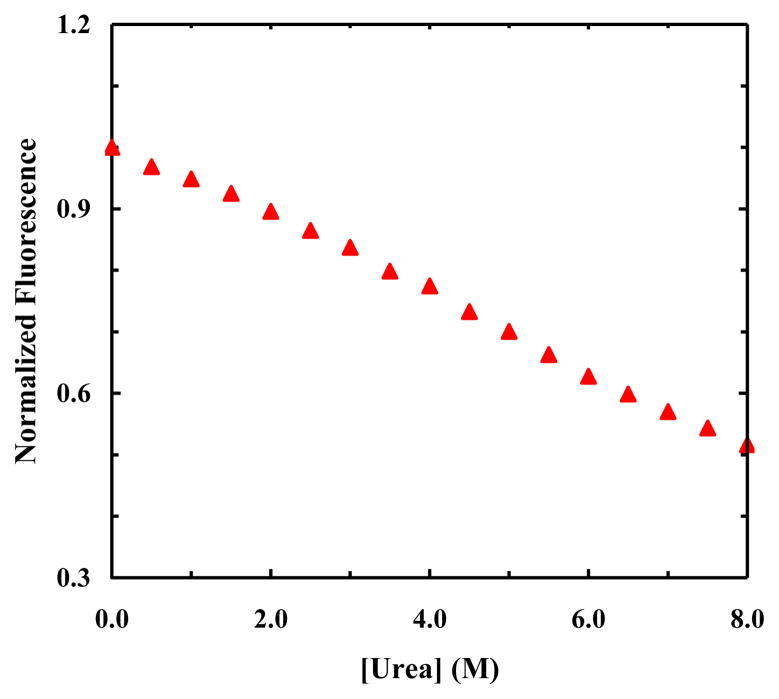
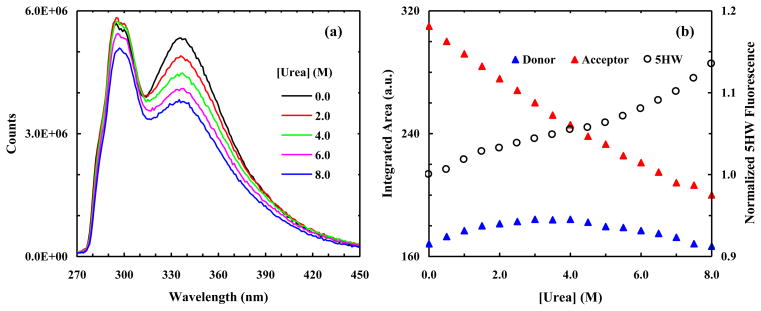
As shown, the fluorescence spectra of HP35-AP measured at various denaturant concentrations show that FRET indeed occurs between PheCN and 7AW (Figure 3a), and that the fluorescence intensity of the donor/acceptor increases/decreases with increasing urea concentration (Figure 3b). While these FRET results indicate a trend that is expected for an unfolding process in which the separation distance between the donor and acceptor is increased, a quantitative assessment of these data requires the consideration of other factors. For example, if the fluorescence quantum yield of the acceptor or donor is sensitive to environment, the trends presented in Figure 3b cannot be taken as a quantitative measure of how the FRET efficiency changes with increasing urea concentration. Fortunately, in the current case the intrinsic fluorescence intensity of the acceptor can be independently measured by directly exciting 7AW at 290 nm, where the absorbance of PheCN is negligible (Figure 2a). As shown (Figure 3b), the fluorescence intensity of 7AW thus obtained shows a rather strong dependence on urea concentration, indicating that the fluorescence quantum yield of 7AW depends on denaturant concentration. Therefore, to eliminate this dependence, we divided the fluorescence intensity of 7AW obtained via FRET excitation at 240 nm by that obtained via direct excitation at 290 nm. As indicated (Figure 4), the resultant ratio shows a sigmoid, albeit broad, unfolding transition as a function of urea concentration.
Figure 3.
(a) Representative fluorescence spectra of HP35-AP obtained with λex = 240 nm and at different urea concentrations, as indicated. (b) Normalized fluorescence intensities of the donor (PheCN) and acceptor (7AW) versus urea concentration, as indicated. These data correspond to the maximum values of the respective PheCN and 7AW fluorescence emissions in the FRET spectra, e.g., those in (a). Also shown (open circle) is the normalized fluorescence intensity of 7AW obtained with λex = 310 nm using the same sample as that used in the corresponding FRET measurement.
Figure 4.
Normalized fluorescence intensity of 7AW in HP35-AP versus urea concentration. These data correspond to the ratio between the fluorescence intensity of 7AW obtained with λex = 240 nm and that obtained with λex = 310 nm.
The fluorescence quantum yield of PheCN shows a modest dependence on urea concentration, as indicated by the slight decrease in its fluorescence intensity in the concentration range of 6–8 M (Figure 3b). However, if we were to assume that the quantum yield of PheCN and hence the Förster distance of the PheCN-7AW FRET pair are independent of urea concentration, we would be able to estimate how the ensemble-averaged separation distance between the donor and acceptor changes upon protein unfolding using the FRET efficiency determined from the PheCN fluorescence of HP35-AP and that of a reference peptide (G-K-PheCN-T-V). For example, the separation distance between PheCN and Trp is calculated using this method to be about 18 Å in 0 M urea, which is close to that (18.5 Å, Cα to Cα) estimated based on the NMR structure [38]. On the other hand, the FRET efficiency in 8 M urea, determined using the fluorescence intensities of the protein and the reference peptide collected under the same conditions, suggests that the ensemble-averaged separation distance (because an ensemble of conformations exists in the denatured state) between the donor and acceptor shows only a modest increase, to about 22 Å. Since in this case the donor and acceptor are located on the same helix (i.e., the C-terminal helix), these results suggest that this helix does not entirely unfold even in 8 M urea, a picture that is consistent with previous studies indicating that native and nonnative residual structures exist in the denatured state of HP35 [41, 42].
PheCN-5HW FRET pair
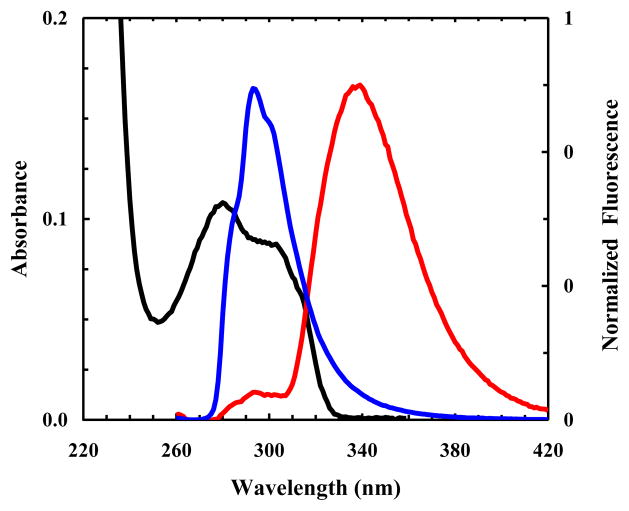
The fluorescence quantum yield of 5HW is higher than that of Trp and is also much less sensitive to solvent polarity, thus making it a useful fluorescence probe of proteins [31]. Herein, we explore the feasibility of using PheCN-5HW as a FRET pair to monitor protein conformational changes. As shown (Figures 2 and 5), the fluorescence emission spectrum of PheCN almost completely overlaps with the red portion of the absorption spectrum of 5HW, indicating that they form an ideal FRET pair. Indeed, the fluorescence spectrum of a short peptide that contains both PheCN and 5HW (i.e., G-5HW-K-PheCN-T-V), excited at 240 nm, shows that the fluorescence of PheCN is significantly quenched (Figure 5), as expected for a scenario wherein strong FRET occurs. Similarly, the Förster distance of this FRET pair is determined to be 18.5 ± 0.5 Å using the absorption and fluorescence spectra of the free amino acids (Figure 2).
Figure 5.
Absorption spectrum of 5HW (black). Also shown are the fluorescence spectra of free PheCN (blue) and a control peptide (G-5HW-K-PheCN-T-V) (red) obtained with λex = 240 nm. For easy comparison, the fluorescence data have been normalized.
Using PheCN-5HW to probe urea-induced unfolding of BBA5
We use a designed mini-protein, BBA5 [45], to test the applicability of this FRET pair in protein folding studies. As shown (Figure 1b), BBA5 (sequence: GAHQRLLKALEDSRSFDYSpVRY, where ‘p’ represents D-Pro) folds into a ββα conformation that is stabilized by a hydrophobic core. Because of its small size, BBA5 has been a favorite folding model for both experimental and computational studies [49]. To use FRET to monitor the denaturant-induced unfolding transition of BBA5, we mutated the N-terminal Gly to 5HW and the C-terminal Tyr to PheCN (the resultant mutant, 5HW-AHQRLLKALEDSRSFDYSpVR-PheCN, is referred to as BBA5-PH). Both CD and IR measurements (Figure S2, Supplementary data) indicate that BBA5-PH is folded and maintains the ββα conformation.
As shown (Figure 6a), the fluorescence spectra of BBA5-PH obtained at different urea concentrations indicate that FRET occurs between PheCN and 5HW. Moreover, the fluorescence intensity (integrated area) of the acceptor, which was obtained by decomposing the FRET spectrum into the respective donor and acceptor components, decreases monotonically with increasing urea concentration (Figure 6b), as expected for an unfolding process. The fluorescence intensity of the donor, on the other hand, does not show a monotonic increase with increasing urea concentration. Instead, the integrated fluorescence intensity of PheCN shows a concave downward dependence on urea concentration (Figure 6b). There are two possible explanations for this trend. One possibility is that at higher urea concentrations, where the unfolded state of BBA5-PH dominates, the immediate environment of PheCN becomes more hydrophobic as it participates in the formation of a nonnative hydrophobic cluster. As a result, the fluorescence quantum yield of PheCN decreases [13, 17] upon protein unfolding. The other possibility is that this concave downward trend arises entirely from the dependence of the fluorescence quantum yield of the donor on urea concentration. Finally, the fluorescence intensity of 5HW obtained via selective excitation at 290 nm indicates that its fluorescence quantum yield is relatively insensitive to the presence of urea (Figure 6b). Thus, compared to Trp, whose fluorescence shows a stronger dependence on urea concentration [6], 5HW is more convenient to use in FRET applications.
Figure 6.
(a) Representative fluorescence spectra of BBA5-PH obtained with λex = 240 nm and at different urea concentrations, as indicated. (b) Fluorescence intensities (integrated area) of the donor (PheCN) and acceptor (5HW) of BBA5-PH versus urea concentration. Also shown (open cycle) is the maximum fluorescence intensity of 5HW obtained with λex = 290 nm using the same sample as that used in the corresponding FRET measurement.
Using PheCN-Trp-7AW as a novel parallel FRET system to probe urea-induced unfolding transition of the hPin1 WW domain
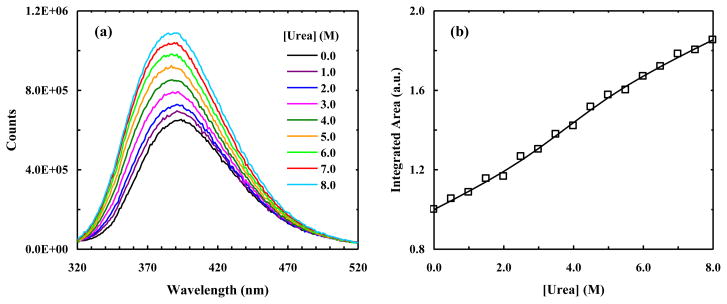
The absorption and fluorescence spectra of 7AW are sufficiently separated from those of Trp, making it possible to selectively excite and probe 7AW fluorescence when Trp is present [31]. These separations also make the simultaneous use of PheCN-Trp and PheCN-7AW for multi-FRET applications feasible. Compared with conventional FRET experiments, wherein only one FRET pair is used, the concurrent use of two FRET pairs in either a parallel or step-wise fashion is more advantageous because it is capable of yielding more structural information from a single FRET measurement [7]. To test the applicability of PheCN-Trp-7AW as a parallel FRET system in studying protein folding-unfolding transitions, we use it to probe the relative motion among the three β-strands of a well studied protein [50–52], the WW domain (Figure 1c), upon urea denaturation. The hPin1 WW domain (sequence: K6LPPG WEKRM SRSSG RVYYF NHITN ASQWE RPSG39) contains a single Phe residue (i.e., Phe25) and two Trp residues (i.e., Trp11 and Trp34), which are conveniently located on the center, first and third strands, respectively. In addition, the distances between Phe25 and Trp11 (~5 Å) and between Phe25 and Trp34 (~8 Å) are well within the Förster distances of the PheCN-Trp and PheCN-7AW FRET pairs. Thus, we introduce the PheCN-Trp-7AW FRET system into the hPin1 WW domain by mutating Phe25 to PheCN and Trp34 to 7AW (the resultant mutant is hereafter referred to as W7AW-P). The thermal melting temperature (Tm) of W7AW-P is determined by CD spectroscopy to be 60 °C (Figure S3, Supplementary data), which is almost identical to that (59 °C) of the wild-type hPin1 WW domain [50], thus indicating that the mutations do not disrupt the native fold.
It has been shown that the 7AW fluorescence of a system containing both Trp and 7AW can be selectively excited using an excitation wavelength longer than 305 nm [19]. Indeed, the fluorescence spectrum of W7AW-P thus obtained is dominated by 7AW emission (Figure 7a), which is peaked at about 390 nm. In addition, the fluorescence intensity (integrated area) of 7AW, when excited at 310 nm, increases with increasing urea concentration and shows a sigmoidal, albeit broad, unfolding transition (Figure 7b). Fitting this transition to an apparent two-state model, which assumes that the fluorescence quantum yields of the fluorophore in the folded and unfolded states both depend linearly on urea concentration, yields the following thermodynamic parameters for unfolding: ΔG(0) = 2.4 ± 0.3 kcal/mol, m = 0.62 ± 0.06 kcal/mol, and Cm = 3.8 M ± 0.4 at 293 K, where ΔG(0) is the free energy of unfolding in water, m measures the dependence of ΔG on denaturant concentration, and Cm is the urea concentration at the midpoint of the unfolding transition. These values compare well with those (i.e., ΔG(0) = 2.24 kcal/mol, m = 0.58 kcal/mol, and Cm = 3.86 M) obtained for the WW domain of human Yes-associated protein by Kelley and coworkers [50].
Figure 7.
(a) Representative fluorescence spectra of W7AW-P obtained with λex = 310 nm and at different urea concentrations, as indicated. (b) Integrated area of the fluorescence spectrum of W7AW-P obtained with λex = 310 nm versus urea concentration. The solid line represents the best fit of these data to a two-state model.
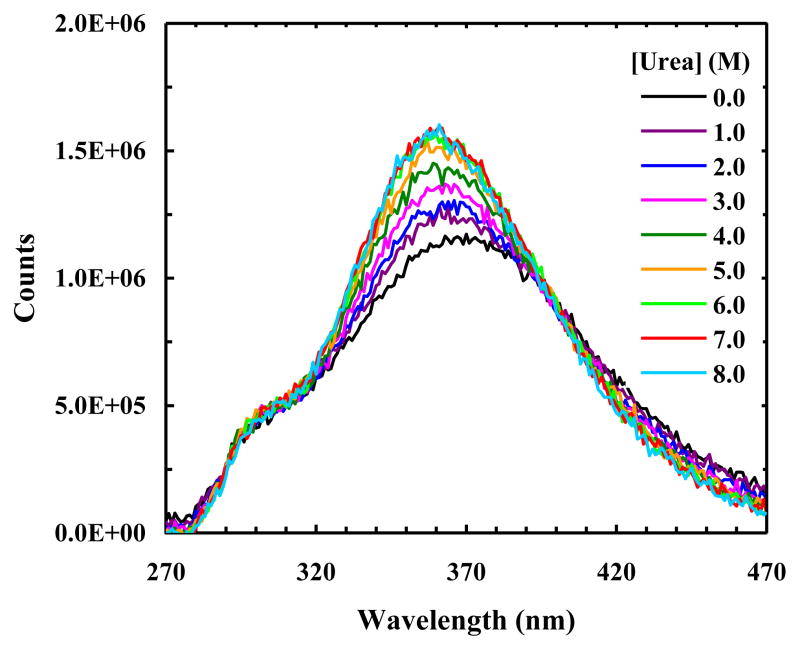
As shown (Figure 8), the fluorescence spectra of W7AW-P obtained with an excitation wavelength of 240 nm indicate that FRET occurs between the donor and acceptors when the protein is folded, as expected. What is interesting is that the PheCN fluorescence remains significantly quenched even at high concentrations of urea, indicating that Trp and 7AW are not the sole quenchers of the PheCN fluorescence. Based on their structural similarity and close proximity, we speculate that the additional fluorescence quenching arises from the nearby Tyr residues. This speculation is confirmed by a recent study of Raleigh and coworkers [53] which showed that an adjacent Tyr residue reduces the PheCN fluorescence by approximately 80–85%.
Figure 8.
Fluorescence spectra of W7AW-P obtained with λex = 240 nm and at different urea concentrations, as indicated.
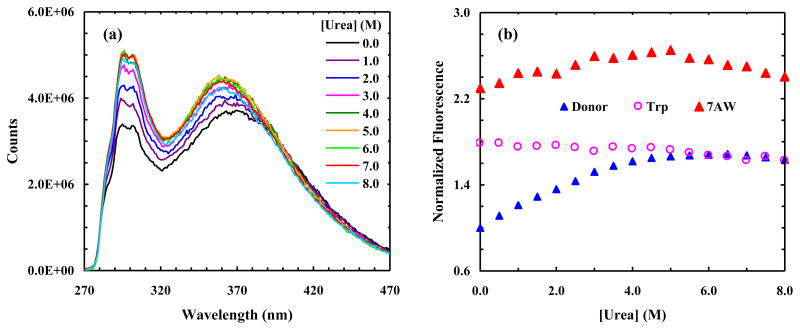
To provide further evidence supporting the usefulness of PheCN-Trp-7AW as a multi-FRET system, we studied a second mutant of the hPin1 WW domain wherein all Tyr residues are removed via site-specific mutations. Specifically, this mutant, which is hereafter referred to as W7AW-P2, contains the following three mutations: Trp34 → 7AW, Tyr23 → PheCN and Tyr24 → Phe. CD studies show that this mutant is folded at low temperature (Figure S4, Supplementary data). As indicated (Figure 9a), the PheCN fluorescence intensity of W7AW-P2 is significantly higher than that of W7AW-P, thus confirming the role of Tyr in quenching the PheCN fluorescence. In addition, it is evident that the PheCN fluorescence intensity increases upon addition of urea, as expected if unfolding increases the assemble-averaged distance between the donor and acceptor(s). However, in order to confirm that FRET indeed occurs between PheCN and Trp and also between PheCN and 7AW, one needs to show that the corresponding FRET spectrum contains contributions from all three fluorophores. To estimate such contributions, we fit each FRET spectrum, F(λ), to the following equation:
| (3) |
Figure 9.
(a) Representative fluorescence spectra of W7AW-P2 obtained at 4 °C with λex = 240 nm and different urea concentrations, as indicated. (b) Normalized fluorescence intensities (integrated area) of the donor (PheCN) and acceptors (7AW and Trp) of W7AW-P2 versus urea concentration, as indicated. All fluorescence intensities are normalized to that of the PheCN obtained in 0 M urea solution.
In the above equation, FD (λ) represents the contribution of PheCN, which is modeled by αFPheCN (λ), where α is a constant and FPheCN (λ) is the fluorescence spectrum of free PheCN measured under the same experimental conditions. Similarly, F7AW (λ) represents the contribution of 7AW, which is approximated by βF310 (λ), where β is a constant and F310 (λ) is the fluorescence spectrum of W7AW-P2 obtained with an excitation wavelength of 310 nm. Finally, FTrp (λ) represents the contribution of Trp and is modeled here by a Gaussian function. We found eq. 3 fits all FRET spectra well (Figure S5, Supplementary data) and, as shown (Figure 9b), the results indicate that they contain contributions from both acceptors. In addition, the 7AW fluorescence intensity (i.e., the integrated area of the corresponding spectrum) is larger than that of Trp in 0 M urea solution, which is consistent with the fact that in the native hPin1 WW domain Tyr23 (mutated to PheCN in W7AW-P2) is closer to Trp34 (mutated to 7AW in W7AW-P2) than it is to Trp11. What is more, the fluorescence intensities of 7AW and Trp show different dependences on urea concentration (Figure 9b). While several factors, which include the ensemble-averaged distances between the donor and the two acceptors and the urea-dependence of the fluorescence quantum yield of the acceptors, determine how the fluorescence intensity of 7AW or Trp changes as a function of urea concentration, this difference nevertheless substantiates the utility of this multi-FRET system in protein folding studies. Furthermore, these FRET data also provide interesting insight into the urea-denatured state of the WW domain; they suggest that significant FRET still occurs even under conditions wherein the protein is unfolded. This finding appears to be consistent with the study of Kelley and coworkers [50], who have shown that the WW domain retains a hydrophobic cluster in the chemically denatured state that likely involves aromatic residues and is unable to bind 1-anilinonaphaline-8-sulfonic acid.
In summary, we demonstrate the utility of the PheCN-Trp-7AW FRET system by applying it to monitor the urea-induced unfolding transition of the hPin1 WW domain. While such multi-FRET systems are useful in equilibrium protein folding/unfolding studies, we believe that they are even more effective in probing the timing and sequence of chain motions in proteins, such as those occurring during a protein folding event. In addition, the FRET system demonstrated here can be easily modified to include other non-natural amino acid based FRET pairs [54, 55].
Conclusions
Previous studies have established the utility of PheCN and Trp as a FRET pair in protein conformational studies. Herein, we show that 7AW and 5HW can substitute Trp to form new FRET pairs with PheCN. These FRET pairs have a Förster distance of 18.5 ± 0.5 Å and offer certain advantages over the PheCN-Trp FRET pair. For example, the fluorescence spectra of PheCN and 7AW are almost completely separated from each other, making it straightforward to assess the donor and acceptor contributions to a given FRET spectrum. More importantly, we demonstrate that PheCN, Trp, and 7AW can be used together to form a multi-FRET system, allowing more structural information to be extracted from a single FRET experiment. We believe that such multi-FRET systems will become particularly useful in studies concerning relative motions of peptide chains, such as those taking place in protein folding.
Supplementary Material
Acknowledgments
We gratefully acknowledge financial support from the NIH (GM-065978). JMGR is a trainee of the NIH Structural Biology Training Program at Penn (T32-GM008275).
Abbreviation used
- CD
Circular dichroism
- FRET
Fluorescence resonance energy transfer
- HP35
villin headpiece subdomain
- hPin1
human Pin1
- PheCN
p-cyanophenylalanine
- 7AW
7-azatryptophan
- 5HW
5-hydroxytryptophan
Appendix A. Supplementary data
Supplementary data associated with this article can be found, in the online version, at
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Förster T. Annalen der Physik. 1948;2:55–75.; English translation in Mielczarek EV, Greenbaum E, Knox RS, editors. Biological Physics. American Institute of Physics; New York: 1993. pp. 148–160.
- 2.Kang JS, Piszczek G, Lakowicz JR. Enhanced emission induced by FRET from a long-lifetime, low quantum yield donor to a long-wavelength, high quantum yield acceptor. J Fluoresc. 2002;12:97–103. doi: 10.1023/A:1015375622992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hass E. The study of protein folding and dynamics by determination of intermolecular distance distributions and their fluctuations using ensemble and single-molecule FRET measurements. ChemPhysChem. 2005;6:858–870. doi: 10.1002/cphc.200400617. [DOI] [PubMed] [Google Scholar]
- 4.Tucker MJ, Oyola R, Gai F. Conformational distribution of a 14-residue peptide in solution: A fluorescence resonance energy transfer study. J Phys Chem B. 2005;109:4788–4795. doi: 10.1021/jp044347q. [DOI] [PubMed] [Google Scholar]
- 5.Tucker MJ, Tang J, Gai F. Probing the kinetics of membrane-mediated helix folding. J Phys Chem B. 2006;110:8105–8109. doi: 10.1021/jp060900n. [DOI] [PubMed] [Google Scholar]
- 6.Glasscock JM, Zhu Y, Chowdhury P, Tang J, Gai F. Using an amino acid fluorescence resonance energy transfer pair to probe protein unfolding: Application to the villin headpiece subdomain and the LysM domain. Biochemistry. 2008;47:11070–11076. doi: 10.1021/bi8012406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Watrob HM, Pan CP, Barkley MD. Two-step FRET as a structural tool. J Am Chem Soc. 2003;125:7336–7343. doi: 10.1021/ja034564p. [DOI] [PubMed] [Google Scholar]
- 8.Getahun Z, Huang CY, Wang T, De Leon B, DeGrado WF, Gai F. Using nitrile-derivatized amino acids as infrared probes of local environment. J Am Chem Soc. 2003;125:405–411. doi: 10.1021/ja0285262. [DOI] [PubMed] [Google Scholar]
- 9.Huang CY, Wang T, Gai F. Temperature dependence of the CN stretching vibration of a nitrile-derivatized phenylalanine in water. Chem Phys Lett. 2003;371:731–738. [Google Scholar]
- 10.Ghosh A, Remorino A, Tucker MJ, Hochstrasser RM. 2D IR photon echo spectroscopy reveals hydrogen bond dynamics of aromatic nitriles. Chem Phys Lett. 2009;469:325–330. doi: 10.1016/j.cplett.2008.12.094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aschaffenburg DJ, Moog RS. Probing hydrogen bonding environments: Solvatochromic effects on the CN vibration of benzonitrile. J Phys Chem B. 2009;113:12736–12743. doi: 10.1021/jp905802a. [DOI] [PubMed] [Google Scholar]
- 12.Liu L, Strzalka J, Tronin A, Johansson JS, Blasie JK. Mechanism of interaction between the general anesthetic halothane and a model ion channel protein, II: Fluorescence and vibrational spectroscopy using a cyanophenylalanine probe. Biophys J. 2009;96:4176–4187. doi: 10.1016/j.bpj.2009.01.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tucker MJ, Oyola R, Gai F. A novel fluorescent probe for protein binding and folding studies: p-cyano-phenylalanine. Biopolymers. 2006;83:571–576. doi: 10.1002/bip.20587. [DOI] [PubMed] [Google Scholar]
- 14.Schiller PW, Natarajan S, Bodanszky M. Determination of the intramolecular tyrosine-tryptophan distance in a 7-peptide related to the C-terminal sequence of cholecystokinin. Int J Peptide Protein Res. 1978;12:139–142. doi: 10.1111/j.1399-3011.1978.tb02877.x. [DOI] [PubMed] [Google Scholar]
- 15.Edelhoch H, Brand L, Wilchek M. Fluorescence studies with tryptophanyl peptides. Biochemistry. 1969;6:547–559. doi: 10.1021/bi00854a024. [DOI] [PubMed] [Google Scholar]
- 16.Schultz KC, Supekova L, Ryu Y, Xie J, Perera R, Schultz PG. A genetically engineered infrared probe. J Am Chem Soc. 2006;128:13984–13985. doi: 10.1021/ja0636690. [DOI] [PubMed] [Google Scholar]
- 17.Aprilakis KN, Taskent H, Raleigh DP. Use of the novel fluorescent amino acid p-cyanophenylalanine offers a direct probe of hydrophobic core formation during the folding of the N-terminal domain of the ribosomal protein L9 and provides evidence for two-state folding. Biochemistry. 2007;46:12308–12313. doi: 10.1021/bi7010674. [DOI] [PubMed] [Google Scholar]
- 18.Schlesinger S. The effect of amino acid analogues on alkaline phosphatase formation in Escherichia coli K-12. J Biol Chem. 1968;243:3877–3883. [PubMed] [Google Scholar]
- 19.Negrerie M, Bellefeuille SM, Whitham S, Petrich JW, Thornburg RW. Novel noninvasive in situ probe of protein structure and dynamics. J Am Chem Soc. 1990;112:7419–7421. [Google Scholar]
- 20.Rich RL, Negrerie M, Elliott S, Thornburg RW, Petrich JW. The photophysical probe, 7-azatryptophan, in synthetic peptides. Photochem Photobiol. 1993;58:28–30. [Google Scholar]
- 21.Hogue CWV, Szabo AG. Character of aminoacyl-adenylates in B. subtilis tryptophanyl-tRNA synthase, by the fluorescence of tryptophan analogues 5-hydroxytryptophan and 7-azatryptophan. Biophys Chem. 1993;48:159–169. doi: 10.1016/0301-4622(93)85007-5. [DOI] [PubMed] [Google Scholar]
- 22.Cornish VW, Benson DR, Altenbach CA, Hideg K, Hubbell WL, Schultz PG. Site-specific incorporation of biophysical probes into proteins. Proc Nat Acad Sci USA. 1994;91:2910–2914. doi: 10.1073/pnas.91.8.2910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rich RL, Gai F, Lane JW, Petrich JW, Schwabacher AW. Using 7-azatryptophan to probe small molecule-protein interactions on the picosecond timescale: The complex of avidin and biotinylated 7-azatryptophan. J Am Chem Soc. 1995;117:733–739. [Google Scholar]
- 24.Soumillon P, Jespers L, Vervoot J, Fastrez J. Biosynthetic incorporation of 7-azatryptophan into the phage lambda lysozyme: Estimation of tryptophan accessibility, effect on enzymatic activity and protein stability. Protein Eng. 1995;8:451–456. doi: 10.1093/protein/8.5.451. [DOI] [PubMed] [Google Scholar]
- 25.Shen F, Triezenberg SJ, Hensley P, Porter D, Knutson JR. Transcriptional activation domain of the herpiesvirus protein VP16 becomes conformationally constrained upon interaction with basal transcription factors. J Biol Chem. 1996;271:4827–4837. doi: 10.1074/jbc.271.9.4827. [DOI] [PubMed] [Google Scholar]
- 26.Wong CY, Efink MR. Incorporation of tryptophan analogues into staphylococcal nuclease: Stability toward thermal and guanidine-HCl induced unfolding. Biochemistry. 1998;37:8947–8953. doi: 10.1021/bi971863g. [DOI] [PubMed] [Google Scholar]
- 27.Das K, Ashby KD, Smirnov AV, Reinach FC, Petrich JW, Farah CS. Fluorescence properties of recombinant tropomyosin containing trytophan, 5-hydroxytryptophan and 7-azatryptophan. Photochem Photobiol. 1999;70:719–730. [PubMed] [Google Scholar]
- 28.Scott DJ, Leejeerajumnean S, Brannigan JA, Lewis RJ, Wilkinson AJ, Hoggett JG. Quaternary re-arrangement analyzed by spectral enhancement: The interaction of sporulation repressor with its antagonist. J Mol Biol. 1999;293:997–1004. doi: 10.1006/jmbi.1999.3221. [DOI] [PubMed] [Google Scholar]
- 29.Blouse GE, Perron MJ, Thompson JH, Day DE, Link CA, Shore JD. A concerted structural transition in the plasminogen activator inhibitor-1 mechanism of inhibition. Biochemistry. 2002;41:11997–12009. doi: 10.1021/bi025967p. [DOI] [PubMed] [Google Scholar]
- 30.Filippis VD, Boni SD, Dea ED, Dalzoppo D, Grandi C, Fontana A. Incorporation of the fluorescent amino acid 7-azatryptophan into the core domain 1–47 of hirudin as a probe of hirudin folding and thrombin recognition. Protein Sci. 2004;13:1489–1502. doi: 10.1110/ps.03542104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Twine SM, Szabo AG. Fluorescent amino acid analogs. Methods Enzymol. 2003;360:104–127. doi: 10.1016/s0076-6879(03)60108-4. [DOI] [PubMed] [Google Scholar]
- 32.Schröder P, Abele C, Gohr P, Stuhlfauth-Roisch U, Grosse W. Latest on enzymology of seratonin biosynthesis in walnut seeds. Adv Exp Med Biol. 1999;467:637–644. doi: 10.1007/978-1-4615-4709-9_81. [DOI] [PubMed] [Google Scholar]
- 33.Hogue CWV, Rasquinha I, Szabo AG, MacManus JP. A new intrinsic fluorescent probe for proteins: biosynthetic incorporation of 5-hydroxytryptophan into oncomodulin. FEBS Lett. 1992;310:269–272. doi: 10.1016/0014-5793(92)81346-n. [DOI] [PubMed] [Google Scholar]
- 34.Ross JBA, Senear DF, Waxman E, Kombo BB, Rusinova E, Huang YT, Laws WR, Hasselbacher CA. Spectral enhancement of proteins: Biological incorporation and fluorescence characterization of 5-hydroxytryptophan in bacteriophage λ cI repressor. Proc Natl Acad Sci USA. 1992;89:12023–12027. doi: 10.1073/pnas.89.24.12023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Farah CS, Reinach FC. Regulatory properties of recombinant tropomyosons containing 5-hydroxytryptophan: Ca2+-binding to troponin results in a conformational change in a region of tropomyosin outside the troponin binding site. Biochemistry. 1999;38:10543–10551. doi: 10.1021/bi982813u. [DOI] [PubMed] [Google Scholar]
- 36.Paulucci AA, Hicks L, Machado A, Miranda MTM, Kay CM, Farah CS. Specific sequences determine the stability and cooperativity of folding of the C-terminal half of tropomyosin. J Biol Chem. 2002;277:39574–39584. doi: 10.1074/jbc.M204749200. [DOI] [PubMed] [Google Scholar]
- 37.McKnight CJ, Doering DS, Matsudaira PT, Kim PS. A thermostable 35-residue subdomain within villin headpiece. J Mol Biol. 1996;260:126–134. doi: 10.1006/jmbi.1996.0387. [DOI] [PubMed] [Google Scholar]
- 38.McKnight CJ, Matsudiara PT, Kim PS. NMR structure of the 35-residue villin headpiece subdomain. Nat Struct Biol. 1997;4:180–184. doi: 10.1038/nsb0397-180. [DOI] [PubMed] [Google Scholar]
- 39.Wang M, Tang Y, Sato S, Vugmeyster L, McKnight CJ, Raleigh DP. Dynamic NMR line-shape analysis demonstrates that the villin headpiece subdomain folds in the microsecond time scale. J Am Chem Soc. 2003;125:6032–6033. doi: 10.1021/ja028752b. [DOI] [PubMed] [Google Scholar]
- 40.Kubelka J, Eaton WA, Hofrichter J. Experimental tests of villin subdomain folding simulations. J Mol Biol. 2003;329:625–630. doi: 10.1016/s0022-2836(03)00519-9. [DOI] [PubMed] [Google Scholar]
- 41.Tang Y, Rigotti DJ, Fairman R, Raleigh DP. Peptide models provide evidence for significant structure in the denatured state of a rapidly folding protein: the villin headpiece subdomain. Biochemistry. 2004;43:3264–3272. doi: 10.1021/bi035652p. [DOI] [PubMed] [Google Scholar]
- 42.Brewer SH, Vu DM, Tang Y, Li Y, Franzen S, Raleigh DP, Dyer RB. Effect of modulating unfolded state structure on the folding kinetics of the villin headpiece subdomain. Proc Natl Acad Sci USA. 2005;102:16662–16667. doi: 10.1073/pnas.0505432102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brewer SH, Song B, Raleigh DP, Dyer RB. Residue specific resolution of protein folding dynamics using isotope-edited infrared temperature jump spectroscopy. Biochemistry. 2007;46:3279–3285. doi: 10.1021/bi602372y. [DOI] [PubMed] [Google Scholar]
- 44.Gao JM, Kelly JW. Toward quantification of protein backbone-backbone hydrogen bonding energies: An energetic analysis of an amide-to-ester mutation in an alpha-helix within a protein. Protein Sci. 2008;17:1096–1101. doi: 10.1110/ps.083439708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Struthers M, Ottensen JJ, Imperiali B. Design and NMR analysis of compact, independently folded BBA motifs. Fold Des. 1998;3:95–103. doi: 10.1016/S1359-0278(98)00015-7. [DOI] [PubMed] [Google Scholar]
- 46.Ranganathan R, Lu KP, Hunter T, Noel JP. Structural and functional analysis of the mitotic rotamase Pin1 suggests substrate recognition is phosphorylation dependent. Cell. 1997;89:875–886. doi: 10.1016/s0092-8674(00)80273-1. [DOI] [PubMed] [Google Scholar]
- 47.Kowalski JA, Lui K, Kelly JW. NMR solution structure of the isolated apo Pin1 WW Domain: Comparison to the X-ray crystal structures of Pin 1. Biopolymers. 2002;63:111–121. doi: 10.1002/bip.10020. [DOI] [PubMed] [Google Scholar]
- 48.Brennen JD, Clark ID, Hogue CWV, Ito AS, Juliano L, Paiva ACM, Rajendran B, Szabo AG. Interaction of enantiomers of lysl-7-azatryptophyl-lysine with acidic phospholipids vesicles: A fluorescence study. Appl Spectrosc. 1995;49:51–59. [Google Scholar]
- 49.Snow CD, Nguyen H, Pande VS, Gruebele M. Absolute comparison of simulated and experimental protein-folding dynamics. Nature. 2002;420:102–106. doi: 10.1038/nature01160. [DOI] [PubMed] [Google Scholar]
- 50.Koepf EK, Petrassi M, Sudol M, Kelly JW. WW: an isolated three-stranded antiparallel β-sheet domain that unfolds and refolds reversibly; evidence for a structured hydrophobic cluster in urea and GdnHCl and a disordered thermal unfolded state. Protein Sci. 1999;8:841–853. doi: 10.1110/ps.8.4.841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ferguson N, Johnson CM, Macias M, Oschkinat H, Fersht A. Ultrafast folding of WW domains without structured aromatic clusters in the denatured state. Proc Natl Acad Sci USA. 2001;98:13002–13007. doi: 10.1073/pnas.221467198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sharpe T, Jonsson AL, Rutherford TJ, Daggett V, Fersht AR. The role of the turn in beta-hairpin formation during WW domain folding. Protein Sci. 2007;16:2233–2239. doi: 10.1110/ps.073004907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Taskent-Sezgin H, Chung J, Patsalo V, Miyake-Stoner SJ, Miller AM, Brewer SH, Mehl RA, Green DF, Raleigh DP, Carrico I. Interpretation of p-cyanophenylalanine fluorescence in proteins in terms of solvent exposure and contribution of side-chain quenchers: A combined fluorescence. IR and molecular dynamics study. Biochemistry. 2009;48:9040–9046. doi: 10.1021/bi900938z. [DOI] [PubMed] [Google Scholar]
- 54.Miyake-Stoner SJ, Miller AM, Hammill JT, Peeler JC, Hess KR, Mehl RA, Brewer SH. Probing protein folding using site-specifically encoded unnatural amino acids as FRET donors with Tryptophan. Biochemistry. 2009;48:5953–5962. doi: 10.1021/bi900426d. [DOI] [PubMed] [Google Scholar]
- 55.Waegele MM, Tucker MJ, Gai F. 5-Cyanotryptophan as an infrared probe of local hydration status of proteins. Chem Phys Lett. 2009;478:249–253. doi: 10.1016/j.cplett.2009.07.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.