Abstract
CD4 T cells selected by MHC class-II expressing thymocytes (T-CD4 T cells) have distinct effector functions compared to that of epithelial cell-selected CD4 T cells (E-CD4 T cells). T-CD4 T cells produce both Th1 and Th2 effector cytokines immediately after stimulation and also express IL-4 in addition to IFN-γ under the Th1 differentiation condition. In the present study, we investigated the capability of T-CD4 T cells to become IL-17-producing cells. We found that T-CD4 T cells express reduced IL-17 under Th17 inducing conditions. T-CD4 T cells express very low levels of receptor for TGF-β and IL-21 that are essential to induce IL-17 expression. In addition, the induction of RORγt, a key transcription factor for IL-17 gene expression, was compromised in T-CD4 T cells under Th17 skewing conditions and ectopic expression of RORγt restored IL-17 expression. The defect of IL-17 and RORγt expression in T-CD4 T cells is cell intrinsic and not due to effects of a secreted factor. Thus, the developmental pathway of CD4 T cells in the thymus plays a critical role in controlling an immune response by suppressing the generation of the Th17 lineage.
Introduction
Effector CD4 T cells including T helper cell (Th) 1, Th2 and Th17 are essential to mount efficient adaptive immune responses by producing a unique set of cytokines (Glimcher and Murphy, 2000; Harrington et al., 2005; Mosmann and Coffman, 1989; Murphy et al., 2000; Park et al., 2005; Weaver et al., 2007). Th1 cells mainly produce IFN-γ and mediate cellular immune responses to intracellular pathogens, whereas Th2 cells typically produce IL-4 and regulate humoral immune responses to extra cellular pathogens. Th17 cells are responsibe for host defense by producing pro-inflammatory cytokines most notably IL-17 (Bauquet et al., 2009; Langrish et al., 2005; Manel et al., 2008; Murphy et al., 2003; Murphy et al., 2000). Accumulating data suggest that Th17-mediated immune responses also promote chronic inflammation and autoimmunity (Jin et al., 2008). Cerebral IL-17-producing T cells played an important role in the delayed phase of ischemic brain injury (Shichita et al., 2009). In addition, a study suggested that Th17 cells were preferentially induced in response to fungal infections and may play an important role in orchestrating host defense against certain fungi (Acosta-Rodriguez et al., 2007). Consistent with these observations, mice deficient in IL-17 production showed lower neutrophil infiltration than wild-type mice in H. pylori infection (Sakamoto, 2008). In addition, IL-17 receptor-deficient mice are susceptible to bacterial pneumonias and are unable to form abdominal abscesses (Ye et al., 2001). Thus, Th17 cells are essential effector cells in host defense against certain pathogens including fungi and extracellular bacteria. However, the broad distribution of IL-17R seems to be the basis for Th17 cell-mediated tissue inflammation and autoimmunity.
Th17 cell differentiation requires TGF-β1 and IL-6 (Bettelli et al., 2006; Mangan et al., 2006; Veldhoen et al., 2006). IL-1 also plays a vital and selective role in promoting IL-17 cells as evidenced by a failure to induce EAE in IL-1RI−/− mice (Matsuki et al., 2006). In conjunction with T cell receptor stimulation, these cytokines induce the expression of the orphan nuclear receptor RORγt, which is critical for IL-17 expression (Ivanov et al., 2006; Manel et al., 2008). IL-21 is also involved in Th17 differentiation and mice deficient in IL-21 are defective in generating Th17 cells (Wei et al., 2007). In addition to IL-21, IL-21 receptor-deficient mice are also defective in generating Th17 cells (Wei et al., 2007). Therefore, Th17 cells secrete IL-21, which in turn amplifies the Th17 response in an autocrine fashion (Wei et al., 2007).
Recently, we and others have identified an alternate pathway for CD4 T cell development, a pathway mediated by MHC class II-expressing thymocytes (Choi et al., 2005; Li et al., 2005). Unlike epithelial cell-selected CD4 (E-CD4) T cells, thymocyte-selected CD4 (T-CD4) T cells produce IL-4 even under Th1 differentiating condition in a Stat6-independent manner. Moreover, mice that have T-CD4 T cells produced IL-4 shortly after in vivo stimulation in the absence of iNKT cells that are known to be the source of initial IL-4 production in vivo (Li et al., 2007b). T-CD4 cells also appear to protect mice from the development of EAE (Park et al., 2004). We also have demonstrated that mice with T-CD4 cells exhibited reduced airway inflammation in an allergen-induced allergic model (Li et al., 2007b). Although the underlying mechanisms for the protection are not well characterized, IL-17 expression was greatly reduced in CD4 T cells isolated from the lungs of allergen challenged mice when the mice had T-CD4 T cells (Li et al., 2007b). These observations prompted us to investigate whether IL-17 expression is controlled by the thymic selection pathway in the thymus.
In the present study, we showed that T-CD4 T cells expressed reduced IL-17 and that the decrease in IL-17 expression is not due to the elevated level of IL-4 production by T-CD4 T cells. However, TGF-β responsiveness of T-CD4 T cells was reduced and correlated with decreased expression of all three subunits of TGF-β receptor. Moreover, T-CD4 T cells produced more IL-21 than E-CD4 T cells but IL-21R expression was not induced upon Th17 differentiation. Th17 cells differentiated from T-CD4 T cells also failed to induce the level of RORγt that is the key transcription factor for IL-17 gene expression and ectopic expression of RORγt was able to restore IL-17 production in T-CD4 T cells. These data suggest that the CD4 T cell developmental process in the thymus plays an important role in shaping CD4 T cell fate and function.
Materials and Methods
Mice
Mice carrying the human type III CIITA transgene (CIITATg), CIITATg mice on the CIITA-deficient background (CIITATg/CIITA−/−), Stat6−/− and IL-4−/− mice were previously described (Chang and Flavell, 1995; Gourley et al., 1999; Kaplan et al., 1996; Li et al., 2005; Li et al., 2007b; Patel et al., 2005). C57BL/6 (B6) mice, B6SJL (CD45.1) and the MHC class II deficient Aβ−/− (CD45.1) mice were purchased from the Jackson Laboratory (Bar Harbor, Maine) and Taconic (Albany, New York), respectively. All mice were housed under specific pathogen free conditions at the University of Michigan Medical School in accordance with institutional guidelines. Mice were used at 8–12 weeks of age and all animal experiments were performed under protocols approved by the Animal Care and Use Committee of the University of Michigan.
Bone marrow chimeras
Recipient B6 SJL (CD45.1) or Aβ−/− (CD45.1) mice were lethally irradiated (950 rad) and rested for 24 h before receiving BM cells. Total BM cells were prepared from the femurs and tibias of donor mice and depleted of mature T cells, B cells, and MHC class II-positive lymphocytes by using a cocktail of antibodies containing anti-CD4 (RL172) and anti-CD8 (TIB105, TIB210), anti-CD19 (1D3), and anti-MHC class II (M5/114), and followed by complement-mediated lysis. These cells were subsequently referred to as T-depleted BM cells. Each recipient mouse received 3 × 106 T-depleted BM cells in 500 μl of 1x PBS via tail vein injection. Reconstituted mice were analyzed 2–3 months later. E- and T-CD4 T cells were generated by transferring BM from WT and Tg mice into lethally irradiated WT or Aβ−/− mice, respectively. CD4 T cells from chimeras were enriched for subsequent experiments.
Flow cytometry
All antibodies for flow cytometry were purchased from BD Biosciences (Franklin Lakes, NJ) and eBioscience (San Diego, CA), and cells were pre-incubated with the anti-FcγR mAb 2.4G2 to block nonspecific antibody binding. The following FITC-, PE-, PerCP-, cychrome-, or biotin-conjugated antibodies were used: CD4 (L3T4), CD45.1 (A20), CD45.2 (104), IL-17, IFN-γ (XMG1.2), IL-4 (11B11) and Foxp3 (FJK-16s). Events were acquired on a flow cytometer (FACS-Caliber and FACS canto; Beckman Dickinson), and the data were analyzed with the CELL Quest Pro or FlowJo software.
Cell preparation and stimulation of CD4 T cells
Total peripheral CD4 T cells were enriched from single-cell suspensions from spleens and lymph nodes (auxiliary, brachial, inguinal, and mesenteric) with anti-CD4 microbeads (Miltenyi Biotec, Auburn, CA). To obtain naïve CD4 T cells, enriched CD4 T cells were stained with anti-CD4, CD45RB, and CD44 and electronically sorted for CD4+CD45RBhiCD44lo cells. In some experiments, enriched CD4 T cells from splenocytes were used. To induce Th cell differentiation under Th17 inducing conditions, total or naïve CD4 T cells at 106/ml were stimulated with 5 μg/ml of plate-bound anti-CD3ε (145-2C11), 1 μg/ml anti-CD28 (37.51), and 50 U IL-2 (Roche), 5ng/ml TGF-β, 10ng/ml IL-23 (R&D), 10ng/ml IL-6 (eBioscience), 10 μg/ml anti-IL-4 (11B11) and 10 μg/ml anti-IFN-γ (R4-6A2) (BD Bioscience) for 5-7 days.
Cytokine assays
For the ELISA assays, differentiated cells were restimulated overnight with 5 μg/ml of the plate-bound anti-CD3ε antibody at a cell density of 1 × 106/ml. IL-17 and IL-21 in the supernatants was quantified by paired cytokine-specific antibodies (BD Biosciences) and (R&D system) respectively. Recombinant cytokines were used as standards. For intracellular cytokine staining, cells were restimulated with 50 ng/ml phorbol myristyl acetate and 1.5 μM ionomycin (Calbiochem) for 5 h. Monensin (Sigma-Aldrich) at 3 μM was added during the last 3 h of stimulation. Cells were fixed in 2–4% paraformaldehyde and permeabilized with 0.2% saponin (Sigma-Aldrich), and followed by intracellular staining with anti-IL-17 and anti-IFN-γ. Events were acquired on a FACSCalibur or FACSCanto (Becton Dickinson) flow cytometer, and the data were analyzed with the CELLQuest Pro or FlowJo software.
RNA analysis and quantitative real time PCR
Total RNA from CD4 T cells was extracted using TRIZOL-mediated lysis (Invitrogen) according to the manufacturer’s recommendations and reverse transcribed using the SuperScript First-Strand cDNA Synthesis System (Invitrogen). Quantitative real-time PCR was performed with SYBR Green PCR Master Mix (Applied Biosystems). All PCR reactions were done in triplicate, and the data were analyzed by the comparative threshold cycle (ΔCT) method and normalized to GAPDH. The primer pairs used for GAPDH were 5′-CCAGGTTGTCTCCTGCGACT-3′ and 5′ATACCAGGAAATGAGCTTGACAAAGT3′, for Gata3 were 5′AGAACCGGCCCCTTATCAA3′ and 5′AGTTCGCGCAGGATGTCC3′, for IL-17A were 5′-GGGTCTTCATTGCGGTGG-3′ and 5′-CTCCAGAAGGCCCTCAGACTAC-3′ for RORγt were 5′-CCGCTGAGAGGGCTTCAC-3′ and 5′-TGCAGGAGTAGGCCACATTACA-3′, for IL-21 5′-AAGATTCCTGAGGATCCGAGAAG-3′ and 5′-GCATTCGTGAGCGTCTATAGTGTC-3′, for IL-21R 5′-CTCCCCCCTTGAACGTGACT-3′ and TTGCCCCTCAGCACGTAGTT-3′, for TGFβR-I 5′-CATTCACCACCGTGTGCCAAATGA-3′ and 5′-ACCTGATCCAGACCCTGATGTTGT-3′, for TGFβR-II 5′-TCCCAAGTCGGATGTGGAAATGGA-3′ and TCGCTGGCCATGACATCACTGTTA-3′, for TGFβR-III 5′-CCTCCTCCACAGATTTTCCA-3′ and 5′-CCCAGATCAAGCCTTCTGAG-3′
ChIP analysis
ChIP analysis was performed according to the ChIP assay protocol. In brief, 2–3 × 106 CD4 T cells from WT and Tg mice were fixed in 1% formaldehyde for 10 min at room temperature, washed, lysed, and sonicated with three pulses to generate chromatin fragments of 200–500-bp in length. Anti-acetylated histone H3 antibody (Upstate Biotechnology) was added (3 μl per immuno-precipitation) to the diluted lysates and incubated overnight. No antibody group was used as a negative control. Protein A-sepharose CL-4B beads (GE Healthcare) were added for 1 h. After washes, the immunocomplexes were eluted, the cross-links were reversed, and DNA was purified by phenol/chloroform extraction and resuspended in 50 μl TE buffer. The primers used for IL-17 internal enhancer were 5′-5′CAATGAAGACCCTGATAGATATCCCTC 3′ and 5′CAGCCCCCTGCCTCCACTTTTAATAAG 3′and for CD3ε were 5′-CATTTCCAAGTGACGTGG-3′ and 5′-AACACACTGGCTGCATGC-3′ (Avni et al., 2002).
Retroviral Transduction
The Phoenix-Eco packaging cells were transfected with either RV-RORγt/GFP or RV- as described (Sisk et al., 2000). The MIG/GFP (gift from Dr. Dan Littman) using CaPO4 transfected Phoenix-Eco cells were cultured at 37°C for 24 h, then 32°C for an additional 24 h to allow efficient viral production. The supernatants containing either RV-RORγt/GFP or RV-MIG/GFP viruses were filtered through a 0.45-μM filter and used immediately to infect primary CD4 T cells. The protocol for retroviral transduction was described previously (Gourley et al., 2002). Briefly, CD4 T cells were stimulated 20–28 h under Th17-inducing conditions. Cells were spin infected with the retroviral supernatant and cultured additional 4 days in the presence of Th17-inducing cytokines. On day 5, cells were also restimulated with 50 ng/ml PMA and 1.5μM ionomycin (Calbiochem) for 5 h. Monensin (Sigma-Aldrich) at 3 μM was added during the last 3 h of stimulation. Events were acquired on a FACSCalibur (Becton Dickinson) flow cytometer, and the data was analyzed with the CELL Quest Pro software.
Results
Reduced IL-17 expression in CD4 T cells isolated from CIITATg mice
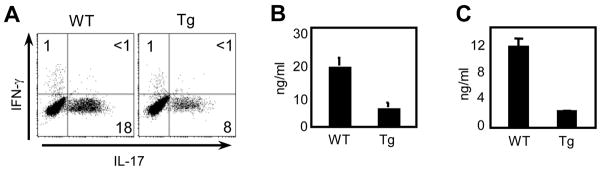
To investigate IL-17 expression in T-CD4 T cells, we first compared IL-17 expression in CD4 T cells prepared from the wild type littermate control (hereon WT) and CIITATg (hereon Tg) mice that have E-CD4 T cells only and E- and T-CD4 T cells together, respectively. We differentiated CD4 T cells in vitro under the Th17 inducing condition as described in the Materials and Methods. On day 5, cells were restimulated with PMA and ionomycin for 5h and then analyzed for IL-17 and IFN-γ production by intracellular staining (ICS). As shown in Figure 1A, the percentage of IL-17 producing cells were lower in the culture of Tg CD4 T cells compared to that of WT cells. Consistent with this, the amount of IL-17 protein secreted to the culture supernatants measured by ELISA was also reduced in Tg cells (Fig. 1B). It is known that Tg CD4 T cells have a greater number of effector/memory-like cells than control mice (Li et al., 2007b). Therefore, we considered the possibility that a large number of Tg CD4 T cells are already committed effector cells and thus they become resistant to Th17 cell inducing signaling. If this were the case, we anticipated that naïve CD4 T cells would differentiate to Th17 cells normally. To test this, we prepared naïve CD4 T cells by sorting cells that were CD45RBhiCD44lo, differentiated under Th17 inducing conditions, and compared IL-17 amounts in the culture supernatants by ELISA. As shown in Figure 1C, a considerable decrease of IL-17 in the Tg culture was observed, suggesting that this property was associated with being T-CD4 T development.
Figure 1. Thymocyte-selected CD4 T cells express less IL-17.
(A, B) Enriched splenic CD4 T cells from WT and Tg mice were differentiated under the Th17 inducing condition for five days as described in the Materials and Methods. Cells were then re-stimulated for 5 h with PMA and ionomycin and stained for IFN-γ and IL-17 (A) or restimulated with plate bound anti-CD3 overnight and the culture supernatant was collected and used to measure IL-17 by ELISA (B). (C) Sorted naïve (CD45RBhi and CD44lo) CD4 T cells from WT and Tg mice were cultured and analyzed as in (B). Data shown are representative of at least three independent experiments.
IL-4 or Stat6 deficiency does not enhance IL-17 expression in Tg CD4 T cells
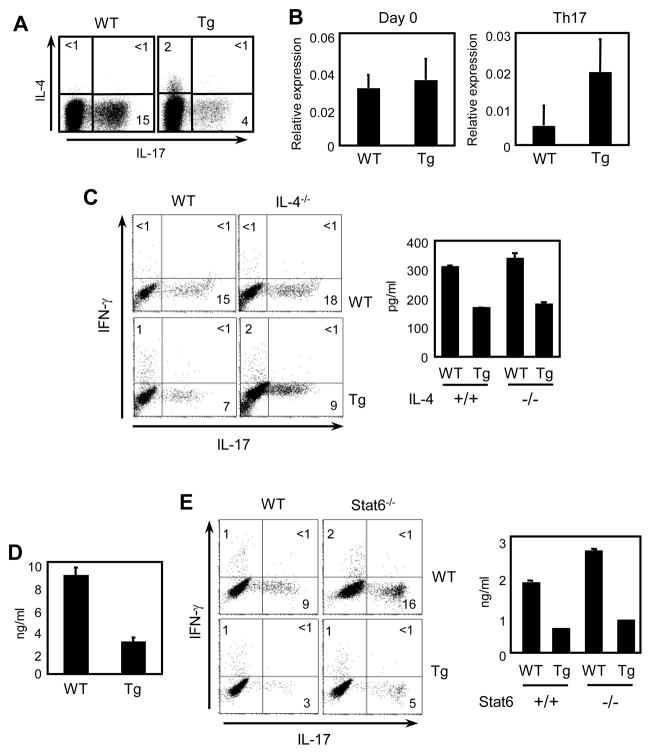
Previous reports have shown that differentiation of Th17 cells is reduced in the presence of IFN-γ or IL-4 (McGeachy et al., 2007). Because T-CD4 T cells produce IL-4 even under the Th1 condition (Li et al., 2007b), we asked if they produce IL-4 under Th17 inducing conditions. A small number of Th17 differentiated Tg CD4 T cells expressed IL-4 (Fig 2A). In addtion, the mRNA level of GATA3 was higher in Tg than WT CD4 T cells although the amounts of GATA3 were decreased upon Th17 cell differentation (Fig 2B). Having observed the expession of IL-4 together with GATA3 in Tg cells, we asked if reduced expression of IL-17 is due to IL-4 produced by T-CD4 T cells. To address this issue, CD4 T cells from WT, Tg, IL-4−/−, and IL-4−/− Tg mice were prepared, differentiated to Th17 cells and examined. The absence of IL-4 did not change IL-17 expression in either WT or Tg CD4 T cells (Fig 2B). Naïve CD4 T cells also did not show a difference in their IL-17 production (Fig 2C). To test the possibility that other Th2 cytokines may contribute to the different phenotype of Tg CD4 T cells, we utilized Stat6−/− mice in which the production of all Th2 cytokines is compromised (Kaplan et al., 1996). As shown in Figure 2D, decreased expression of IL-17 measured by both ICS and ELISA was observed in the Tg culture regardless of the Stat6 expression status. Together, these data suggest that the reduced expression of IL-17 is not due to the difference in Th2 cytokine production by Tg CD4 T cells.
Figure 2. Deficiency of IL-4 or Stat6 does not change IL-17 expression.
(A) Enriched splenic CD4 T cells from WT and Tg mice were differentiated under the Th17 inducing condition for five days as described in the Materials and Methods. Cells were then re-stimulated for 5 h with PMA and ionomycin and stained for IL-4 and IL-17. (B) RNA was extracted from freshly isolated (Day 0) and Th17 cells of WT and Tg CD4 T cells and gata3 expression was quantified by qRT-PCR and results were expressed as ratios relative to the housekeeping gene GAPDH. (C and D) Freshly isolated splenic CD4 T cells (C) or sorted naïve (CD45RBhi and CD44lo) CD4 T cells (D) from WT and Tg mice that were sufficient (control) or deficient (IL-4−/−) in the IL-4 gene were differentiated to Th17 cells and analyzed IL-17 and IFN-γ by ICS and ELISA. (E) Freshly isolated splenic CD4 T cells from WT and Tg mice with or without Stat6 expression were differentiated and analyzed for IL-17 and IFN-γ by ICS or ELISA. Data shown are representative of at least three independent experiments.
IL-17 expression is regulated by the CD4 T cell selection pathway in the thymus
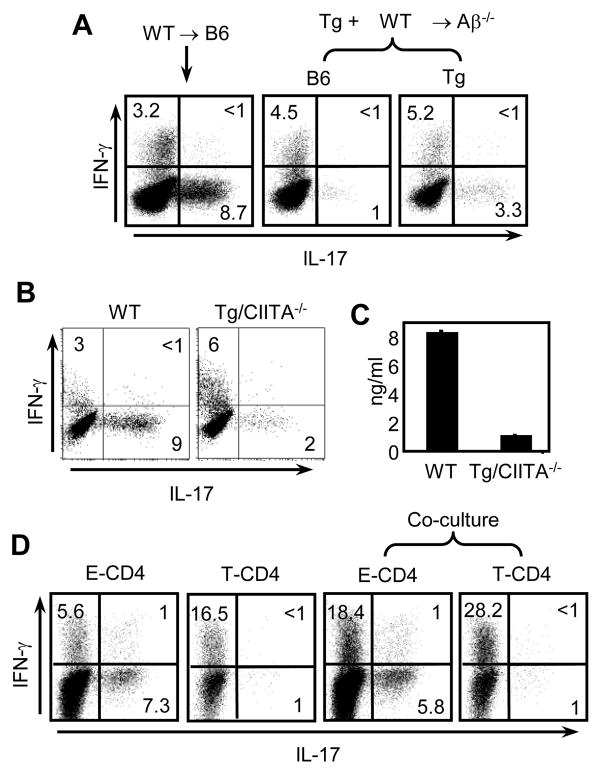
The observations so far suggest that the impairment of IL-17 expression in Tg CD4 T cells is not likely caused by cytokines or environmental factors during the culture in vitro. The results therefore prompted us to visit the issue of CIITA transgene expression in CD4 T cells. There are at least two possible ways that CIITA can affect IL-17 expression. First, CIITA is a transcription factor and thus CIITA directly suppresses the expression of the IL-17 gene. Alternatively, CIITA transgene expression in thymocytes induces MHC class II expression, which then allows thymocytes to mediate development of CD4 T cells. Thus, thymocyte-selected CD4 T cells have an intrinsic defect in IL-17 expression. To distinguish these possibilities, we performed mixed bone marrow chimera experiments. We constructed two groups of chimeras; WT+Tg → Aβ−/− and WT→B6. In WT+Tg →Aβ−/− mice WT cells cannot be selected by thymic epithelial cell (TEC) due to the deficiency of Aβ expression in host mice. Therefore, WT cells are forced to undergo positive selection mediated by CIITA-expressing and thus, MHC class II+ thymocytes. We have demonstrated that this selection pathway is efficiently operated in this type of chimera (Li et al., 2005). The same WT BM cells were transferred to WT host in which WT cells are selected by host TEC. After 8–12 weeks of reconstitution, we isolated CD4 T cells from the chimeras and stimulated them under the Th17 differentiation condition. The two sources of donor cells and host cells were identified with CD45 alleles. The results showed that thymocyte-selected WT cells that do not express CIITA produced less IL-17 expressing cells than the WT cells selected by TEC (Fig 3A). IFN-γ expression was comparable among three groups of mice. The data therefore indicate that the thymic selection pathway, but not CIITA per se, plays a role in determining IL-17 expression of the resulting CD4 T cells. We next tested mice that express the CIITA transgene but not the endogenous CIITA gene. In these mice, CIITA and thus MHC class II is expressed in the T cell compartment but not in APC or thymic epithelial cells due to the deficiency of endogensous CIITA (Li et al., 2005). Therefore, in these mice, CD4 T cells are generated by MHC class II expressed on thymocytes in the thymus (Li et al., 2005). When CD4 T cells from the WT or Tg CIITA −/− mice were compared for their IL-17 expression, Tg CIITA−/− CD4 T cells produced decreased IL-17 (Fig 3A and 3B).
Figure 3. T cell selection pathway determines the potential to express IL-17.
(A) Mixed bone marrow chimeras were constructed as indicated at the top. BM from B6 were transferred to B6 recipients or co-transferred with those from Tg mice into Aβ−/− recipients. 8–10 weeks after BM transplantation, splenic CD4 T cells were enriched and differentiated under the same condition as in Figure 1. Before fixation and permeabilization, CD4 T cells were stained with anti-CD45.1 and anti-CD45.2 antibodies in order to distinguish CD4 T cells from two different sources of donors and the recipient. Cytokine production profiles were subsequently assayed by ICS. (B and C) Freshly isolated splenic CD4 T cells from WT or Tg mice lacking the endogenous CIITA gene (Tg/CIITA−/−) were differentiated under the same condition as in Figure 1 and analyzed by ICS (B) or by ELISA (C). (D) E-and T-CD4 T cells were generated by using bone marrow chimeras as described in the Material and Methods. An equal number of E- and T-CD4 T cells were cultured alone or cultured together under the Th17 inducing condition. Cells were then re-stimulated for 5 hours with PMA and ionomycin followed by ICS. Data shown are representative of at least three independent experiments.
Although our data strongly suggest that thymic selection determines the potential to produce IL-17 it is still possible that CIITA transgene-expressing CD4 T cells influence WT cells in trans during the culture in vitro. To test this, we generated E- and T-CD4 T cells using BM chimeras (details in the Material and Methods) and mixed the two in equal proportions to generate Th17 cells. As shown in Figure 3D, culturing E- or T-CD4 T cells alone showed the expected IL-17 expression. When E-CD4 T cells were co-cultured with T-CD4 T cells, E-CD4 T cells maintained IL-17 expression, albeit at a slightly lower than singly cultured E-CD4 T cells. In contrast, T-CD4 T cells in the same culture showed reduced IL-17. Therefore, our data demonstrate that the potential to express IL-17 expression is regulated primarily by the thymic selection pathway and that reduced IL-17 production seems to be an intrinsic property of thymocyte-selected CD4 T cells.
Reduced expression of receptors for TGF-β and IL-21 in T-CD4 T cells
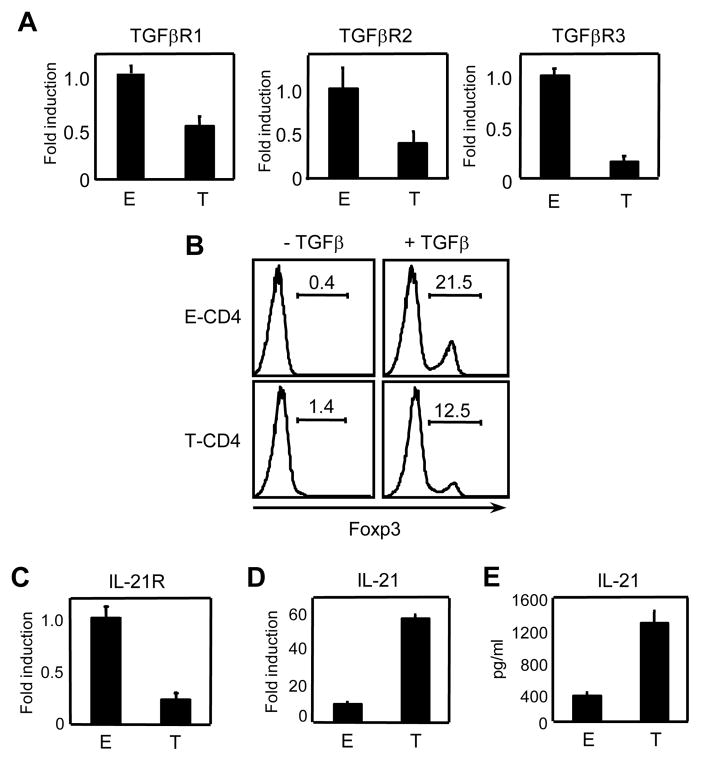
Next we looked into possible mechanisms by which IL-17 expression is diminished in T-CD4 T cells. Studies have shown that TGF-β and IL-6 are necessary to promote Th17 differentiation (Bettelli et al., 2006; Mangan et al., 2006; Veldhoen et al., 2006). We therefore tested the expression level of receptors for IL-6 and TGF-β. IL-6 receptor levels were comparable between WT and Tg CD4 T cells (data not shown). We then examined TGF-β receptors by assessing the mRNA levels of three subunits of TGF-β receptor by qRT-PCR. A considerable decrease of all three subunits TGF-β receptor was observed as shown in Figure 4A. The decrease indicated a possible defect in TGF-β responsiveness of T-CD4 T cells. To test this, we measured the level of Foxp3 expression, which requires TGF-β signaling (Zheng et al., 2008). E- and T-CD4 T cells were stimulated with anti-CD3 in the presence or absence of TGF-β for 3 days and Foxp3 expression was analyzed. As shown in figure 4B, the induction of foxp3 levels by TGF-β was much greater on E-CD4 T cells than T-CD4 T cells suggesting that TGF-β signaling is compromised in T-CD4 T cells.
Figure 4. Expression of receptors for TGFβ and IL-21 is reduced in T-CD4 T cells.
(A) RNA was extracted from freshly isolated E- and T-CD4 T cells and TGFβR1, TGFβR2 and TGFβR3 were quantified by qRT-PCR. Fold induction was performed by using the comparative threshold cycle (ΔCT) method and normalized to GAPDH. The shown is the relative expression level in T-CD4 T cells compared that in E-CD4 T cells. (B) Enriched E- and T-CD4 T cells were stimulated with anti-CD3 and CD28 in presence or absence of 5μg/ml of TGFβ for 3 days and analyzed for Foxp3 induction. (C and D) RNA was extracted from freshly isolated E- and T-CD4 T cells and analyzed for IL-21R (C) IL-21 (D) by qRT-PCR. Fold induction was calculated by using the comparative threshold cycle (ΔCT) method and normalized to GAPDH. The shown is the relative expression level in T-CD4 T cells compared that in E-CD4 T cells. (E) Freshly isolated E- and T-CD4 T cells were differentiated under the Th17 inducing condition for 5 days followed by re-stimulation with plate bound anti-CD3. The culture supernatant was analyzed for IL-21 by ELISA. Data shown are representative of at least three independent experiments.
In addition to TGF-β, IL-21 has shown to induce the expression of IL-17 and mice deficient in both IL-21 and IL-21R are defective in generating Th17 cells (Nurieva et al., 2007; Wei et al., 2007; Zhou et al., 2007). Thus, we asked if expression of IL-21R and IL-21 is impaired in T-CD4 T cells. We found that IL-21R expression was greatly diminished in T-CD4 T cells but IL-21 was expressed at higher levels in T-CD4 T cells than E-CD4 T cells (Fig 4C& 4D). IL-21 protein levels measured by ELISA supported the difference in mRNA levels (Fig 4E). Therefore, T-CD4 T cells express reduced levels of key receptors that are necessary to differentiate to Th17 cells.
T-CD4 T cells express little RORγt
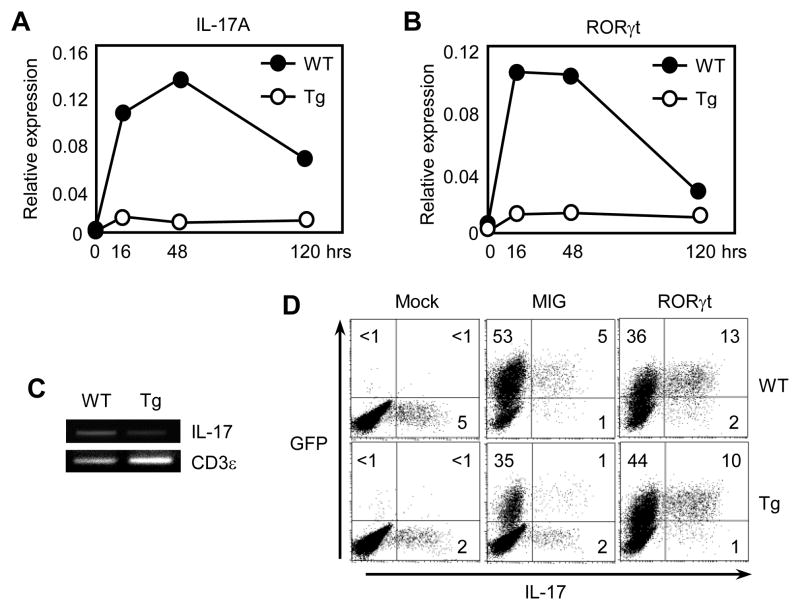
To study IL-17 gene regulation in T-CD4 T cells in detail, we examined the kinetics of IL-17 gene expression over time under the Th17 skewing condition. CD4 T cells from WT mice showed the induction of IL-17 mRNA by peaking at 48 hours (Fig. 5A). In contrast, the level of IL-17 transcripts in Tg CD4 T cells was barely increased. We also studied the expression of RORγt that is a key transcription factor for IL-17 expression during the same time period (Ivanov et al., 2006; Manel et al., 2008). RORγt expression was induced at a faster rate than IL-17 and peaked at 16 hours after stimulation in WT CD4 T cells. However, Tg CD4 T cells showed a very small increase of RORγt mRNA (Fig. 5B).
Figure 5. Ectopic expression of RORγt enhances IL-17 expression in T-CD4 T cells.
Freshly isolated CD4 T cells from both WT and Tg mice were cultured under Th17 inducing conditions for different time points and RNA was extracted and analyzed for IL-17 (A) and RORγt (B) by qRT-PCR. (C) CD4 T cells from WT and Tg mice were used for the ChIP assay with an anti-acetylated histone H3 antibody. PCR primers specific for the IL-17 enhancer were used to amplify the precipitated DNA Primers specific for CD3ε were used as an internal loading control. (D) WT and Tg CD4 T cells were transduced with viruses encoding IRES-GFP (MIG) or RORγt-IRES-GFP (RORγt) under Th17 inducing condition. Cells were analyzed after 5 days in culture. Data shown are representative of at least three independent experiments.
To understand a possible mechanism by which IL-17 gene expression is not induced in Tg CD4 T cells, we examined the accessibility of the IL-17 locus using a chromatin immunoprecipitation assay. Freshly isolated CD4 T cells from WT and Tg mice were lysed and immunoprecipated with an anti-acetylated histone antibody bound to the internal enhancer region of the IL-17 locus. As shown in Figure 5C, the amount of histone acetylation in the enhancer region was greatly reduced in Tg CD4 T cells compared to that of WT CD4 T cells, indicating that the IL-17 locus in Tg CD4 T cells is not as accessible as in WT CD4 T cells. If the inefficient expression of RORγt causes reduced IL-17, ectopic expression of RORγt in Tg CD4 T cells could restore IL-17 production. Indeed, expressing RORγt via the retrovirus-mediated introduction resulted in the induction of IL-17 in a transduced (GFP+) population of WT as well as Tg cells (Fig. 5D), indicating that RORγt is a limiting factor.
Discussion
CD4 T cells educated on MHC class II expressing thymocytes show different properties compared to that by epithelial cell-selected CD4 T cells. Unlike E-CD4 T cells, T-CD4 T cells express Th1 and Th2 effector cytokines immediately after stimulation (Li et al., 2007b). The most striking feature is Stat6-independent IL-4 expression even under Th1 differentiation conditions (23). In addition to these differences, our current study demonstrated that IL-17 expression in T-CD4 T cells is much lower than E-CD4 T cells. Thus, thymic selection pathway controls the effector cell fate of CD4 T cells.
Currently, it is not understood how thymic selection process regulates Th cell fate. Nevertheless, we suspect that signaling given to developing thymocytes plays a key role in shaping the programming of gene expression. CD4 T cell development requires cell to cell interaction and therefore cell surface molecules on thymic epithelial cells (TEC) and thymocytes likely contribute to the decision making process. Among many different receptors expressed differently between TEC and thymocytes, one notable receptor is the SLAM receptor family that is expressed on thymocytes but not TEC (Wang et al., 2004). We have shown that SLAM signaling is essential to generate T-CD4 T cells, whereas E-CD4 T cells develop normally in the absence of the same signaling (Li et al., 2007a). Therefore, it is tempting to speculate that SLAM receptor-mediated signaling changes the intracellular environment, which in turn results in chromatin remodeling. The signal can enhance the accessibility of transcriptional machinery to certain cytokine gene loci including the IL-4 locus allowing IL-4 expression soon after positive selection of T-CD4 T cells. The same signal prevents the RORγt gene from being expressed. Lack of RORγt induction was also accompanied by the reduced expression of cytokine receptors including TGF-β and IL-21. IL-6 receptor expression was comparable between E- and T-CD4 T cells (data not shown). Moreover, we have shown that T-CD4 T cells produce IL-6 and respond to IL-6 normally, if not better (Sofi et al., 2009). In addition, phosphorylation of Stat3 in Tg CD4 T cells was comparable to that of WT cells upon IL-6 stimulation further supports comparable IL-6 signaling between the two (data not shown). Thus, T-CD4 T cells appear to be specifically impaired in TGF-β and IL-21 signals necessary to become Th17 cells. Although T-CD4 T cells produce more IL-4 than E-CD4 T cells, IL-4 itself does not change IL-17 gene expression because T-CD4 T cells deficient in the IL-4 gene showed the similar reduction in IL-17 expression.
A recent study revealed that IRF4-deficient T cells are completely impaired in Th17 cell polarization (Huber et al., 2008). IRF4-deficient mice are resistant to induction of EAE that is primarily mediated by IL-17 producing cells (Brustle et al., 2007). A factor(s) regulating IRF4 induction during Th17 polarization has not been identified. However, IRF4 expression was not altered in T-CD4 T cells suggesting the defect in the IL-17 expression is not due to inadequate IRF4 expression (data not shown). IL-1 is also known to play a unique, non-redundant role during murine Th17 cell polarization (Chung et al., 2009). We found that adding IL-1 to the culture could not improve IL-17 expression in T-CD4 T cells (data not shown). We also ruled out that low expression of IL-17 is not caused by an extrinsic factor. E-CD4 T cells produced a comparable amount of IL-17 when they were cultured in the absence or presence of T-CD4 T cells. These observations indicate that the effector cell fate of T-CD4 T cells is imprinted during the selection process. A similar imprinting scenario could be responsible for naturally occurring regulatory T cells (Treg) which fate is determined by the expression of Foxp3 in the thymus (Lio and Hsieh, 2008).
T-CD4 T cells share similar development requirements and functional properties with invariant NKT (iNKT) cells that are also selected by thymocytes (Bendelac, 1995). However, the capability to produce IL-17 seems to be dissimilar between these two cell types. iNKT cells can make IL-17 upon stimulation in the absence of Th17 inducing cytokines TGF-β and IL-6 (Rachitskaya et al., 2008). Moreover, a study has shown that iNKT cells regulate Th17 cell development of E-CD4 T cells (Mars et al., 2009). CD4 T cells isolated from iNKT-cell deficient mice produced more IL-17 suggesting a negative effect of iNKT cells on Th17 lineage development in vivo (Mars et al., 2009). Although it is not clear how iNKT cells regulate Th17 cell generation, iNKT cells seem to have a property of inhibiting IL-17 expression of E-CD4 T cells in trans. In contrast, T-CD4 T cells did not show any effect on E-CD4 T cells when they cultured together (Fig 3D). Instead, T-CD4 T cells made reduced IL-17 under the Th17 inducing condition. Therefore, T-CD4 T cells are distinct from iNKT cells in IL-17 expression although the two share the same developmental pathway.
We have shown that mice that can generate T-CD4 T cells were less susceptible to airway inflammation induced by an experimental allergen (Li et al., 2007b). Mice protected from airway inflammation expressed reduced IL-17 in the inflamed lungs. In addition, it was also reported that Tg mice were protected from EAE (Park et al., 2004). Although the study reporting EAE did not measure the IL-17 it is possible that IL-17 level may be low as well because IL-17 is known to contribute to severity of EAE (Park et al., 2004). Together, the data presented here suggest a regulatory role of T-CD4 T cells during an immune response. Further investigation is warranted to elucidate the role of T-CD4 T cells in the modulation of immune related diseases.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Acosta-Rodriguez EV, Rivino L, Geginat J, Jarrossay D, Gattorno M, Lanzavecchia A, Sallusto F, Napolitani G. Surface phenotype and antigenic specificity of human interleukin 17-producing T helper memory cells. Nat Immunol. 2007;8:639–46. doi: 10.1038/ni1467. [DOI] [PubMed] [Google Scholar]
- Avni O, Lee D, Macian F, Szabo SJ, Glimcher LH, Rao A. T(H) cell differentiation is accompanied by dynamic changes in histone acetylation of cytokine genes. Nat Immunol. 2002;3:643–51. doi: 10.1038/ni808. [DOI] [PubMed] [Google Scholar]
- Bauquet AT, Jin H, Paterson AM, Mitsdoerffer M, Ho IC, Sharpe AH, Kuchroo VK. The costimulatory molecule ICOS regulates the expression of c-Maf and IL-21 in the development of follicular T helper cells and TH-17 cells. Nat Immunol. 2009;10:167–75. doi: 10.1038/ni.1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendelac A. Positive selection of mouse NK1+ T cells by CD1-expressing cortical thymocytes. J Exp Med. 1995;182:2091–6. doi: 10.1084/jem.182.6.2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bettelli E, Carrier Y, Gao W, Korn T, Strom TB, Oukka M, Weiner HL, Kuchroo VK. Reciprocal developmental pathways for the generation of pathogenic effector TH17 and regulatory T cells. Nature. 2006;441:235–8. doi: 10.1038/nature04753. [DOI] [PubMed] [Google Scholar]
- Brustle A, Heink S, Huber M, Rosenplanter C, Stadelmann C, Yu P, Arpaia E, Mak TW, Kamradt T, Lohoff M. The development of inflammatory T(H)-17 cells requires interferon-regulatory factor 4. Nat Immunol. 2007;8:958–66. doi: 10.1038/ni1500. [DOI] [PubMed] [Google Scholar]
- Chang CH, Flavell RA. Class II transactivator regulates the expression of multiple genes involved in antigen presentation. J Exp Med. 1995;181:765–7. doi: 10.1084/jem.181.2.765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi EY, Jung KC, Park HJ, Chung DH, Song JS, Yang SD, Simpson E, Park SH. Thymocyte-thymocyte interaction for efficient positive selection and maturation of CD4 T cells. Immunity. 2005;23:387–96. doi: 10.1016/j.immuni.2005.09.005. [DOI] [PubMed] [Google Scholar]
- Chung Y, Chang SH, Martinez GJ, Yang XO, Nurieva R, Kang HS, Ma L, Watowich SS, Jetten AM, Tian Q, Dong C. Critical regulation of early Th17 cell differentiation by interleukin-1 signaling. Immunity. 2009;30:576–87. doi: 10.1016/j.immuni.2009.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glimcher LH, Murphy KM. Lineage commitment in the immune system: the T helper lymphocyte grows up. Genes Dev. 2000;14:1693–711. [PubMed] [Google Scholar]
- Gourley T, Roys S, Lukacs NW, Kunkel SL, Flavell RA, Chang CH. A novel role for the major histocompatibility complex class II transactivator CIITA in the repression of IL-4 production. Immunity. 1999;10:377–86. doi: 10.1016/s1074-7613(00)80037-0. [DOI] [PubMed] [Google Scholar]
- Gourley TS, Patel DR, Nickerson K, Hong SC, Chang CH. Aberrant expression of Fas ligand in mice deficient for the MHC class II transactivator. J Immunol. 2002;168:4414–9. doi: 10.4049/jimmunol.168.9.4414. [DOI] [PubMed] [Google Scholar]
- Harrington LE, Hatton RD, Mangan PR, Turner H, Murphy TL, Murphy KM, Weaver CT. Interleukin 17-producing CD4+ effector T cells develop via a lineage distinct from the T helper type 1 and 2 lineages. Nat Immunol. 2005;6:1123–32. doi: 10.1038/ni1254. [DOI] [PubMed] [Google Scholar]
- Huber M, Brustle A, Reinhard K, Guralnik A, Walter G, Mahiny A, von Low E, Lohoff M. IRF4 is essential for IL-21-mediated induction, amplification, and stabilization of the Th17 phenotype. Proc Natl Acad Sci U S A. 2008;105:20846–51. doi: 10.1073/pnas.0809077106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov, McKenzie BS, Zhou L, Tadokoro CE, Lepelley A, Lafaille JJ, Cua DJ, Littman DR. The orphan nuclear receptor RORgammat directs the differentiation program of proinflammatory IL-17+ T helper cells. Cell. 2006;126:1121–33. doi: 10.1016/j.cell.2006.07.035. [DOI] [PubMed] [Google Scholar]
- Jin D, Zhang L, Zheng J, Zhao Y. The inflammatory Th 17 subset in immunity against self and non-self antigens. Autoimmunity. 2008;41:154–62. doi: 10.1080/08916930701776605. [DOI] [PubMed] [Google Scholar]
- Kaplan MH, Schindler U, Smiley ST, Grusby MJ. Stat6 is required for mediating responses to IL-4 and for development of Th2 cells. Immunity. 1996;4:313–9. doi: 10.1016/s1074-7613(00)80439-2. [DOI] [PubMed] [Google Scholar]
- Langrish CL, Chen Y, Blumenschein WM, Mattson J, Basham B, Sedgwick JD, McClanahan T, Kastelein RA, Cua DJ. IL-23 drives a pathogenic T cell population that induces autoimmune inflammation. J Exp Med. 2005;201:233–40. doi: 10.1084/jem.20041257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Kim MG, Gourley TS, McCarthy BP, Sant’Angelo DB, Chang CH. An alternate pathway for CD4 T cell development: thymocyte-expressed MHC class II selects a distinct T cell population. Immunity. 2005;23:375–86. doi: 10.1016/j.immuni.2005.09.002. [DOI] [PubMed] [Google Scholar]
- Li W, Sofi MH, Rietdijk S, Wang N, Terhorst C, Chang CH. The SLAM-associated protein signaling pathway is required for development of CD4+ T cells selected by homotypic thymocyte interaction. Immunity. 2007a;27:763–74. doi: 10.1016/j.immuni.2007.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Sofi MH, Yeh N, Sehra S, McCarthy BP, Patel DR, Brutkiewicz RR, Kaplan MH, Chang CH. Thymic selection pathway regulates the effector function of CD4 T cells. J Exp Med. 2007b;204:2145–57. doi: 10.1084/jem.20070321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lio CW, Hsieh CS. A two-step process for thymic regulatory T cell development. Immunity. 2008;28:100–11. doi: 10.1016/j.immuni.2007.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manel N, Unutmaz D, Littman DR. The differentiation of human T(H)-17 cells requires transforming growth factor-beta and induction of the nuclear receptor RORgammat. Nat Immunol. 2008 doi: 10.1038/ni.1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangan PR, Harrington LE, O’Quinn DB, Helms WS, Bullard DC, Elson CO, Hatton RD, Wahl SM, Schoeb TR, Weaver CT. Transforming growth factor-beta induces development of the T(H)17 lineage. Nature. 2006;441:231–4. doi: 10.1038/nature04754. [DOI] [PubMed] [Google Scholar]
- Mars LT, Araujo L, Kerschen P, Diem S, Bourgeois E, Van LP, Carrie N, Dy M, Liblau RS, Herbelin A. Invariant NKT cells inhibit development of the Th17 lineage. Proc Natl Acad Sci U S A. 2009;106:6238–43. doi: 10.1073/pnas.0809317106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuki T, Nakae S, Sudo K, Horai R, Iwakura Y. Abnormal T cell activation caused by the imbalance of the IL-1/IL-1R antagonist system is responsible for the development of experimental autoimmune encephalomyelitis. Int Immunol. 2006;18:399–407. doi: 10.1093/intimm/dxh379. [DOI] [PubMed] [Google Scholar]
- McGeachy MJ, Bak-Jensen KS, Chen Y, Tato CM, Blumenschein W, McClanahan T, Cua DJ. TGF-beta and IL-6 drive the production of IL-17 and IL-10 by T cells and restrain T(H)-17 cell-mediated pathology. Nat Immunol. 2007;8:1390–7. doi: 10.1038/ni1539. [DOI] [PubMed] [Google Scholar]
- Mosmann TR, Coffman RL. TH1 and TH2 cells: different patterns of lymphokine secretion lead to different functional properties. Annu Rev Immunol. 1989;7:145–73. doi: 10.1146/annurev.iy.07.040189.001045. [DOI] [PubMed] [Google Scholar]
- Murphy CA, Langrish CL, Chen Y, Blumenschein W, McClanahan T, Kastelein RA, Sedgwick JD, Cua DJ. Divergent pro- and antiinflammatory roles for IL-23 and IL-12 in joint autoimmune inflammation. J Exp Med. 2003;198:1951–7. doi: 10.1084/jem.20030896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy KM, Ouyang W, Farrar JD, Yang J, Ranganath S, Asnagli H, Afkarian M, Murphy TL. Signaling and transcription in T helper development. Annu Rev Immunol. 2000;18:451–94. doi: 10.1146/annurev.immunol.18.1.451. [DOI] [PubMed] [Google Scholar]
- Nurieva R, Yang XO, Martinez G, Zhang Y, Panopoulos AD, Ma L, Schluns K, Tian Q, Watowich SS, Jetten AM, Dong C. Essential autocrine regulation by IL-21 in the generation of inflammatory T cells. Nature. 2007;448:480–3. doi: 10.1038/nature05969. [DOI] [PubMed] [Google Scholar]
- Park H, Li Z, Yang XO, Chang SH, Nurieva R, Wang YH, Wang Y, Hood L, Zhu Z, Tian Q, Dong C. A distinct lineage of CD4 T cells regulates tissue inflammation by producing interleukin 17. Nat Immunol. 2005;6:1133–41. doi: 10.1038/ni1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park WS, Bae Y, Chung DH, Choi YL, Kim BK, Sung YC, Choi EY, Park SH, Jung KC. T cell expression of CIITA represses Th1 immunity. Int Immunol. 2004;16:1355–64. doi: 10.1093/intimm/dxh132. [DOI] [PubMed] [Google Scholar]
- Patel DR, Li W, Park JS, Sofi MH, Gourley TS, Hangoc G, Kaplan MH, Chang CH. Constitutive expression of CIITA directs CD4 T cells to produce Th2 cytokines in the thymus. Cell Immunol. 2005;233:30–40. doi: 10.1016/j.cellimm.2005.03.006. [DOI] [PubMed] [Google Scholar]
- Rachitskaya AV, Hansen AM, Horai R, Li Z, Villasmil R, Luger D, Nussenblatt RB, Caspi RR. Cutting edge: NKT cells constitutively express IL-23 receptor and RORgammat and rapidly produce IL-17 upon receptor ligation in an IL-6-independent fashion. J Immunol. 2008;180:5167–71. doi: 10.4049/jimmunol.180.8.5167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto KTA, Imamura S, Kita M. IL-17 is involved in Helicobacter pylori-induced gastric inflammatory responses in a mouse model. Helicobacter. 2008:13. doi: 10.1111/j.1523-5378.2008.00629.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shichita T, Sugiyama Y, Ooboshi H, Sugimori H, Nakagawa R, Takada I, Iwaki T, Okada Y, Iida M, Cua DJ, Iwakura Y, Yoshimura A. Pivotal role of cerebral interleukin-17-producing gammadeltaT cells in the delayed phase of ischemic brain injury. Nat Med. 2009;15:946–50. doi: 10.1038/nm.1999. [DOI] [PubMed] [Google Scholar]
- Sisk TJ, Gourley T, Roys S, Chang CH. MHC class II transactivator inhibits IL-4 gene transcription by competing with NF-AT to bind the coactivator CREB binding protein (CBP)/p300. J Immunol. 2000;165:2511–7. doi: 10.4049/jimmunol.165.5.2511. [DOI] [PubMed] [Google Scholar]
- Sofi MH, Li W, Kaplan MH, Chang CH. Elevated IL-6 expression in CD4 T cells via PKCtheta and NF-kappaB induces Th2 cytokine production. Mol Immunol. 2009;46:1443–50. doi: 10.1016/j.molimm.2008.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Hamburg JP, de Bruijn MJ, Ribeiro de Almeida C, van Zwam M, van Meurs M, de Haas E, Boon L, Samsom JN, Hendriks RW. Enforced expression of GATA3 allows differentiation of IL-17-producing cells, but constrains Th17-mediated pathology. Eur J Immunol. 2008;38:2573–86. doi: 10.1002/eji.200737840. [DOI] [PubMed] [Google Scholar]
- Veldhoen M, Hocking RJ, Atkins CJ, Locksley RM, Stockinger B. TGFbeta in the context of an inflammatory cytokine milieu supports de novo differentiation of IL-17-producing T cells. Immunity. 2006;24:179–89. doi: 10.1016/j.immuni.2006.01.001. [DOI] [PubMed] [Google Scholar]
- Wang N, Satoskar A, Faubion W, Howie D, Okamoto S, Feske S, Gullo C, Clarke K, Sosa MR, Sharpe AH, Terhorst C. The cell surface receptor SLAM controls T cell and macrophage functions. J Exp Med. 2004;199:1255–64. doi: 10.1084/jem.20031835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver CT, Hatton RD, Mangan PR, Harrington LE. IL-17 family cytokines and the expanding diversity of effector T cell lineages. Annu Rev Immunol. 2007;25:821–52. doi: 10.1146/annurev.immunol.25.022106.141557. [DOI] [PubMed] [Google Scholar]
- Wei L, Laurence A, Elias KM, O’Shea JJ. IL-21 is produced by Th17 cells and drives IL-17 production in a STAT3-dependent manner. J Biol Chem. 2007;282:34605–10. doi: 10.1074/jbc.M705100200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye P, Rodriguez FH, Kanaly S, Stocking KL, Schurr J, Schwarzenberger P, Oliver P, Huang W, Zhang P, Zhang J, Shellito JE, Bagby GJ, Nelson S, Charrier K, Peschon JJ, Kolls JK. Requirement of interleukin 17 receptor signaling for lung CXC chemokine and granulocyte colony-stimulating factor expression, neutrophil recruitment, and host defense. J Exp Med. 2001;194:519–27. doi: 10.1084/jem.194.4.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng SG, Wang J, Horwitz DA. Cutting edge: Foxp3+CD4+CD25+ regulatory T cells induced by IL-2 and TGF-beta are resistant to Th17 conversion by IL-6. J Immunol. 2008;180:7112–6. doi: 10.4049/jimmunol.180.11.7112. [DOI] [PubMed] [Google Scholar]
- Zhou L, Ivanov, Spolski R, Min R, Shenderov K, Egawa T, Levy DE, Leonard WJ, Littman DR. IL-6 programs T(H)-17 cell differentiation by promoting sequential engagement of the IL-21 and IL-23 pathways. Nat Immunol. 2007;8:967–74. doi: 10.1038/ni1488. [DOI] [PubMed] [Google Scholar]