Abstract
Flaviviruses have a positive-sense, single-stranded RNA genome of ∼11 kb, encoding a large polyprotein that is cleaved to produce ∼10 mature proteins. Cell fusing agent virus, Kamiti River virus, Culex flavivirus and several recently discovered flaviviruses have no known vertebrate host and apparently infect only insects. We present compelling bioinformatic evidence for a 253–295 codon overlapping gene (designated fifo) conserved throughout these insect-specific flaviviruses and immunofluorescent detection of its product. Fifo overlaps the NS2A/NS2B coding sequence in the − 1/+ 2 reading frame and is most likely expressed as a trans-frame fusion protein via ribosomal frameshifting at a conserved GGAUUUY slippery heptanucleotide with 3′-adjacent RNA secondary structure (which stimulates efficient frameshifting in vitro). The discovery bears striking parallels to the recently discovered ribosomal frameshifting site in the NS2A coding sequence of the Japanese encephalitis serogroup of flaviviruses and suggests that programmed ribosomal frameshifting may be more widespread in flaviviruses than currently realized.
Keywords: Flavivirus, Sequence analysis, Ribosomal frameshifting, Translation, Insect-specific flavivirus, Overlapping gene, NS2A
Introduction
The genus Flavivirus (reviewed in Lindenbach et al., 2007) includes species such as dengue virus (DENV), Japanese encephalitis virus (JEV), West Nile virus, yellow fever virus and tick-borne encephalitis virus. The positive-sense, single-stranded genomic RNA is ∼11 kb in length and contains a long open reading frame, which is translated as a polyprotein and cleaved by virus-encoded and host proteases to produce ∼10 mature proteins. While the majority of flaviviruses are transmitted from one vertebrate host to another by hematophagous arthropod vectors, a number of flaviviruses are apparently insect-specific. Such flaviviruses include cell fusing agent virus (CFAV; Cammisa-Parks et al., 1992, Cook et al., 2006, Cook et al., 2009, Stollar and Thomas, 1975), Kamiti River virus (KRV; Crabtree et al., 2003, Sang et al., 2003), Culex flavivirus (CxFV; Blitvich et al., 2009, Cook et al., 2009, Farfan-Ale et al., 2009, Hoshino et al., 2007, Kim et al., 2009, Morales-Betoulle et al., 2008), Quang Binh virus (QBV; Crabtree et al., 2009), Aedes flavivirus (AEFV; Hoshino et al., 2009) and Nakiwogo virus (NAKV; Cook et al., 2009). In addition, flavivirus-like sequences have been found integrated into mosquito genomes (e.g., cell silent agent or CSA; Crochu et al., 2004).
Many viruses harbour sequences that induce a portion of ribosomes to shift − 1 nt and continue translating in the new reading frame (Brierley and Pennell, 2001). The eukaryotic − 1 frameshift site typically consists of a ‘slippery’ heptanucleotide fitting the consensus motif N NNW WWH, where NNN represents any three identical nucleotides, WWW represents AAA or UUU, H represents A, C or U, and spaces separate zero-frame codons. This is followed by a ‘spacer’ region of 5–9 nt and then a stable RNA secondary structure such as a pseudoknot or hairpin. Recently, we identified a ribosomal frameshift site in the JEV serogroup of flaviviruses (Balmori Melian et al., 2009, Firth and Atkins, 2009) that gives rise to the NS1′ protein, whose origin had previously been an unsolved enigma. In the JEV serogroup, frameshifting takes place just 8 codons into the NS2A coding sequence at a highly conserved Y CCU UUU slippery site (where Y represents U or C; G:U anticodon:codon re-pairing occurs at position 1 of the heptanucleotide when Y is U), while a very stable 3′-adjacent RNA pseudoknot provides an additional, and in this case essential, stimulatory element. The frameshifting efficiency in virus-infected cells is estimated to be ∼20–50%. Ribosomes that frameshift translate a 45-codon ORF (termed foo, for Flavivirus Overlapping ORF) in the − 1/+ 2 frame relative to the polyprotein coding sequence before terminating. This produces a 52 amino acid NS2AN-term-FOO trans-frame fusion peptide that, in contrast to zero-frame NS2A, fails to mediate cleavage at the NS1|NS2A boundary. Thus, the mature protein product is NS1-NS2AN-term-FOO (i.e., a 52 amino acid extension of NS1) which equates to the previously identified NS1′ protein (Balmori Melian et al., 2009, Blitvich et al., 1999).
The identification of programmed frameshifting in the JEV serogroup of flaviviruses prompted us to investigate other flavivirus clades. Analysis of the polyprotein coding sequences of the insect-specific flaviviruses revealed the presence of a much longer − 1/+ 2 frame ORF (which we termed fifo; ‘Fairly Interesting Flavivirus ORF’) overlapping the NS2A and NS2B coding sequences. Fifo contains nearly 300 codons and detailed bioinformatic analysis provides overwhelming evidence that it is indeed a coding sequence, most likely translated as a trans-frame fusion via programmed ribosomal frameshifting. Here, we describe the bioinformatic analysis of the fifo ORF, immunofluorescent detection of its product, and experimental characterization of the proposed frameshift site.
Results and discussion
Identification and bioinformatic analysis of fifo in Culex flavivirus and Quang Binh virus
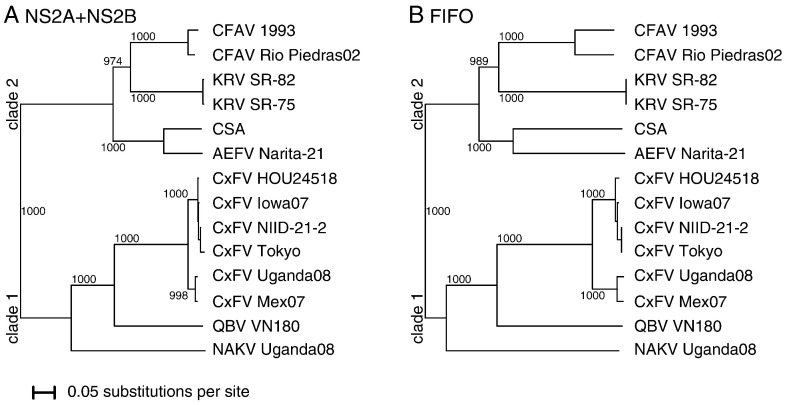
The available insect-specific flavivirus NS2A/NS2B sequences divide into two major phylogenetic clades (Fig. 1A). The first clade (hereafter Clade 1) comprises CxFV, QBV and NAKV, while the second clade (hereafter Clade 2) encompasses CFAV, KRV and AEFV. It should be noted, however, that in phylogenetic studies performed using envelope gene sequences, CFAV clusters with CxFV instead of KRV (Hoshino et al., 2007). CSA also belongs to Clade 2.
Fig. 1.
Simple phylogenetic trees based on (A) NS2A+NS2B and (B) FIFO. The amino acid neighbor-joining tree was produced with CLUSTALX (Larkin et al., 2007). Columns with alignment gaps were excluded. Numbers indicate bootstrap support (out of 1000), while the scale bar represents the number of substitutions per site.
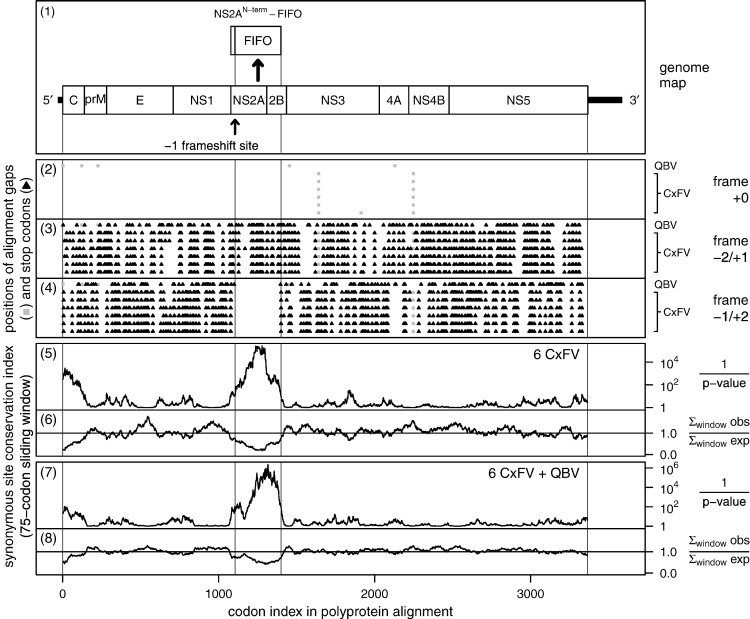
We first identified fifo as an unusually long ORF conserved throughout all available CxFV sequences with coverage of the NS2A/NS2B region of the genome (six sequences; Fig. 2 , panel 4). In the CxFV Tokyo strain (GenBank ID: AB262759), the polyprotein coding sequence encompasses nucleotides 92–10183 and the maximal (i.e., stop codon to stop codon) coordinates for the fifo ORF are 3328–4290, giving rise to a 321-codon ORF in the + 2 frame relative to the polyprotein coding sequence. The entire polyprotein coding sequence of AB262759 has 162 stop codons in the + 2 frame out of a total of 3363 codons so, on average, a 321-codon region in the + 2 frame would be expected to contain 15.5 stop codons. Thus, the probability of obtaining an uninterrupted 321-codon + 2 frame ORF simply by chance is extremely small. Indeed, if + 2 frame stop codons within the polyprotein coding sequence are assumed to be randomly distributed, then the probability is of order p ∼ 2 × 10–5. To guard against the possibility that the absence of stop codons in the + 2 frame could be due to an unusual amino acid composition in the NS2A/NS2B region of the polyprotein (e.g., these proteins are particularly hydrophobic), we used a simplified (i.e., single-sequence) version of the CCRT statistic of Chung et al. (2007), as follows. A codon usage table for AB262759 was calculated using the entire polyprotein coding sequence. Then zero-frame codons in the fifo region were replaced with codons drawn randomly according to the frequencies in the codon usage table, while maintaining the zero-frame amino acid sequence. This randomization procedure was repeated 10,000 times, and the number of + 2 frame stop codons (in the fifo region) was counted for each randomization. Only 0.02% of the randomized sequences had an uninterrupted + 2 frame ORF, and the mean number of + 2 frame stop codons was 8.3. Thus, although the amino acid composition of the NS2A/NS2B proteins does favour a reduction in the number of + 2 frame stop codons (8.3 expected, compared with 15.5 expected if whole-polyprotein frequencies are assumed), the reduction is not enough to explain the presence of such a long + 2 frame ORF as a chance event.
Fig. 2.
Conservation at synonymous sites in CxFV and QBV. Conservation was calculated for an input alignment comprising the polyprotein coding sequences from the CxFV sequences AB262759, AB377213, FJ663034, FJ502995, GQ165808 and EU879060, and the QBV sequence FJ644291. Panel 1 shows a map of the CxFV genome, along with the proposed overlapping gene fifo and the proposed frameshift site. Panels 2–4 show the positions of stop codons in the three forward reading frames. The + 0 frame is the polyprotein frame and is therefore devoid of stop codons. Note the conserved absence of stop codons in the − 1/+ 2 frame within the fifo ORF. Panels 5–8 depict the conservation at polyprotein-frame synonymous sites. Panels 5 and 7 depict the probability that the degree of conservation within a given window could be obtained under a null model of neutral evolution at synonymous sites, while panels 6 and 8 depict the absolute amount of conservation as represented by the ratio of the observed number of substitutions within a given window to the number expected under the null model (see Firth and Atkins, 2009, for details). Note that the large sliding window size (75 codons) is responsible for the broad smoothing of the conservation scores at either end of the fifo ORF. The range of features detected by this type of analysis depends partly on the sliding window size − 75 codons is optimal for features with a width of 75 codons. If features cover fewer codons (e.g., many non-coding functional elements), then the corresponding conservation peaks may be diluted to such an extent that they are not distinguishable from background noise.
Further inspection showed that a long ORF was also present at this genomic location in the related QBV (GenBank ID: FJ644291; Fig. 2, panel 4). Here, the maximal fifo ORF comprises 296 codons. Alignment with the CxFV sequences showed that the maximal QBV fifo ORF is 22 codons shorter at the 5′ end and 3 codons shorter at the 3′ end than the maximal CxFV fifo ORF. In fact (see below) the proposed ribosomal frameshift site is 5 codons into the QBV ORF and 27 codons into the CxFV ORF, so that the frameshift sites align and frameshifting would give access to a 295-codon ORF in CxFV and a 292-codon ORF in QBV. Within this ORF there are 286 point nucleotide differences (33%) between QBV FJ644291 and CxFV AB262759, and yet the open reading frame is preserved in both viruses.
Next, the polyprotein coding sequences were analyzed for conservation at synonymous sites, as described in Firth and Atkins (2009). The polyprotein coding sequences from QBV and six CxFV isolates were extracted, translated, aligned with CLUSTALW (Larkin et al., 2007), and back-translated to a nucleotide sequence alignment. Beginning with pairwise sequence comparisons, conservation at synonymous sites (only) was evaluated by comparing the observed number of base substitutions with the number expected under a neutral evolution model. The procedure takes into account whether synonymous site codons are 1-, 2-, 3-, 4- or 6-fold degenerate and the differing probabilities of transitions and transversions. Statistics were then summed over a phylogenetic tree as described in Firth and Brown (2006), and averaged over a sliding window. The analysis revealed a striking, and highly statistically significant (p ∼ 4 × 10–16 for the total conservation within the whole fifo ORF), peak in polyprotein-frame synonymous site conservation in a region corresponding precisely to the conserved open reading frame, fifo (Fig. 2, panels 5–8). Peaks in synonymous site conservation are generally indicative of functionally important overlapping elements. Although, in general, such elements may be either coding or non-coding, the extent (∼300 codons) and degree (Fig. 2, panel 6) of the conservation here would be unusual for a non-coding element.
One other explanation for extended conservation could be recombination (though apparently relatively rare in the flaviviruses; Simmonds, 2006, Taucher et al., 2010). However, recombination is expected to suppress the observed frequency of non-synonymous substitutions at least as much as the observed frequency of synonymous substitutions whereas the opposite effect is observed in this case. For example, in a comparison between QBV FJ644291 and CxFV AB262759, the relative frequencies of identical, distinct but synonymous, and non-synonymous polyprotein-frame aligned codon pairs were 36 ± 3%, 27 ± 3% and 37 ± 4% within the fifo region, and 28 ± 1%, 40 ± 1% and 32 ± 1% outside of the fifo region. (The observed frequencies are, of course, exact numbers. The ‘error’ ranges – based on simple Poisson statistics – are simply for assessing the statistical significance of the observed differences.) Thus, although the frequency of polyprotein-frame synonymous substitutions is greatly reduced within the fifo region (27 ± 3% compared with 40 ± 1% outside of fifo), the frequency of non-synonymous substitutions is in fact slightly increased (37 ± 4% compared with 32 ± 1% outside of fifo). Similarly, application of a variety of recombination–detection programs from the RDP2 package (Martin et al., 2005) failed to identify potential recombination in this region of the genome. Finally, recombination does not provide an explanation for the other evidence presented herein (e.g., a conserved long open reading frame, a conserved and well-defined translation mechanism, and presence in all other sequenced insect-specific flaviviruses).
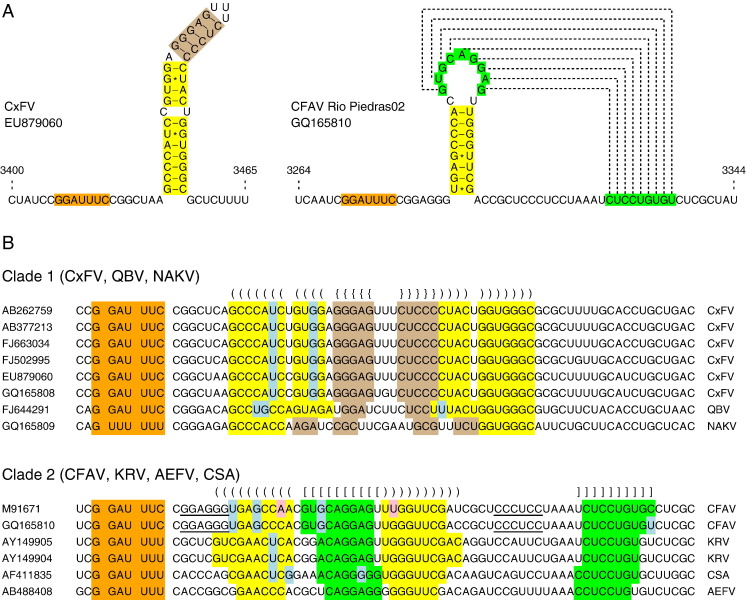
We next inspected the sequences for a potential translation mechanism. It is unlikely that independent initiation (e.g., via an internal ribosome entry site or shunting mechanism) occurs since there is no positionally conserved AUG codon in the CxFV/QBV alignment, and some sequences have no AUG codon until 119 codons into the ORF. However, near the 5′ end of the fifo ORFs of both CxFV and QBV we found a conserved slippery heptanucleotide motif G GAU UUC (spaces separate polyprotein or zero-frame codons). The slippery site was followed by a 7-nt spacer and then a large RNA hairpin structure containing one unpaired residue and, in CxFV, a 1-bp symmetric bulge (Fig. 3 ). The hairpin structure was essentially conserved in CxFV and QBV — albeit with some minor differences near the apex, and some G:C to G:U or A:U to G:U substitutions in the stem that preserved the base pairings. Allowing for G:U codon:anticodon re-pairing at position 2 of the heptanucleotide, the combination of the G GAU UUC heptanucleotide and the 3′-adjacent RNA hairpin structure fit the consensus motif for − 1 ribosomal frameshifting. In fact G GAU UUH is a recognized shifty site. Frameshifting in Red clover necrotic mosaic virus and other dianthoviruses, besides certain umbraviruses, occurs at G GAU UUU (Bekaert and Rousset, 2005, Gibbs et al., 1996, Kim and Lommel, 1994). Brierley et al. (1992) measured an in vitro frameshifting efficiency of 15.5% for G GAU UUA in the context of a 'minimal' infectious bronchitis coronavirus (IBV) based 3′-adjacent pseudoknot, compared with 16.1% for G GGU UUA in the same context. Thus, all other things being equal, G GAU UUH may be expected to stimulate frameshifting as efficiently as G GGU UUH. Brierley et al. (1992) measured frameshifting efficiencies of 38.3% and 15.8%, respectively, for G GGU UUU and G GGU UUC. Although there is no reason to expect that these values (measured in vitro in rabbit reticulocyte lysate, and in the context of the minimal IBV-based 3′-adjacent pseudoknot) are representative of values in insect cells infected with insect-specific flaviviruses, it nonetheless seems reasonable to suppose that frameshifting at this site has the potential to be highly efficient. Moreover, the functional importance of the G GAU UUC motif in CxFV and QBV is supported by the conservation of ‘G’ at position one, despite the corresponding polyprotein-frame codon encoding different amino acids in the two viruses (ccG/proline in CxFV; caG/glutamine in QBV). Frameshifting at this location would result in the translation of a 295-codon (CxFV) or 292-codon (QBV) − 1/+ 2 frame ORF (see Fig. 4 for an amino acid alignment), fused to the N-terminal region of NS2A.
Fig. 3.
Potential stimulatory RNA secondary structures for − 1 ribosomal frameshifting in insect-specific flavivirus sequences. (A) Structure predictions for representative CxFV and CFAV sequences. (B) Sequence alignments for the 14 available insect-specific flavivirus sequences, showing the conserved presence of a slippery heptanucleotide (G GAU UUY, or G UUU UUU in NAKV; orange) and potential to form a 3′-adjacent RNA hairpin (Clade 1) or pseudoknot (Clade 2). Base pairings in stem 1 are indicated with ‘()’s or ‘{}’s and yellow or tan background, while base pairings in stem 2 of the pseudoknot are indicated with ‘[]’s and green background. Paired base substitutions that preserve base pairings are highlighted in pink, while single base substitutions that replace canonical Watson-Crick base pairings with G:U base pairings are highlighted in pale blue. A potential extension to stem 1 in CFAV is indicated by underscores, although these base pairings are expected to be disrupted once the ribosome is positioned on the slippery site.
Fig. 4.
Amino acid alignment for CxFV, QBV and NAKV FIFO. Note that amino acids ‘DF’ at positions 1–2 come from the zero-frame at the shift site. FIFO is predicted to be expressed as part of a trans-frame fusion protein with the N-terminal region of NS2A (not shown). Predicted transmembrane regions are underlined.
Identification and bioinformatic analysis of fifo in Nakiwogo virus
In NAKV – the most divergent member of Clade 1 (Fig. 1) – the G GAU UUY motif is absent, but the presence of an ∼300 codon ORF in the − 1/+ 2 reading frame strongly suggests that frameshifting also occurs in NAKV. The most likely slippery site appears to be the nucleotides G UUU UUU, which align with the slippery heptanucleotide sequence in the CxFV and QBV genomes (Fig. 3B). Frameshifting at this position would result in the translation of a 282-codon fifo ORF. Within this region there are 447 nucleotide differences (53%) between NAKV GQ165809 and CxFV AB262759 — again illustrating the immense improbability of such a long ORF being preserved unless it is indeed coding. Although G UUU UUU does not allow canonical codon:anticodon re-pairing at position 1, it is consistent with one of a handful of known non-canonical shift sites, viz. the shift site G UUA AAC utilized by Equine arteritis virus (family Arteriviridae; Den Boon et al., 1991). In fact, Brierley et al. (1992) measured a frameshifting efficiency of 13.7% for G UUU UUC (in the context of the minimal IBV-based 3′-adjacent pseudoknot) and comparison of the frameshifting efficiencies measured for G GGU UUU and G GGU UUC or U UUU UUU and U UUU UUC suggest that the frameshifting efficiency of G UUU UUU should be of the order of 20–33% in the minimal IBV-based context, i.e., as efficient as G GAU UUY.
Identification and bioinformatic analysis of fifo in cell fusing agent virus, Kamiti River virus, Aedes flavivirus and cell silent agent
Analysis of the CFAV, KRV, AEFV and CSA sequences also revealed the presence of a long − 1/+ 2 frame fifo ORF overlapping the NS2A/NS2B region in addition to a potential ribosomal frameshift site near the 5′ end of the fifo ORF. Similar to CxFV and QBV, the slippery heptanucleotide was G GAU UUY (Y represents U or C) but, instead of a 3′-adjacent potential stem-loop structure, there was a 3′-adjacent potential pseudoknot (Fig. 3). Support for the functionality of the G GAU UUY motif comes from the presence of both ucG (serine; 6-fold degenerate) and gcG (alanine; 4-fold degenerate) codons that preserve the ‘G’ at position one of the heptanucleotide, even though the polyprotein-frame amino acid is not conserved, while support for the pseudoknot comes from a number of A:U to G:U and G:C to G:U substitutions, besides one C:G to A:U paired substitution (between the two strains of CFAV) that preserve the predicted base pairings. Frameshifting at the G GAU UUY motif would result in translation of a 259-codon fifo ORF in KRV, AEFV and CFAV-RioPiedras02 (Fig. 5 ).
Fig. 5.
Amino acid alignment for CFAV, KRV, AEFV and CSA FIFO. Note that amino acids ‘DF’ at positions 1–2 come from the zero-frame at the shift site. FIFO is predicted to be expressed as part of a trans-frame fusion protein with the N-terminal region of NS2A (not shown). Predicted transmembrane regions are underlined. The three premature termination codons in the fifo-defective CFAV-1993 sequence are indicated with ‘⁎’s on black rectangles.
There are currently two distinct CFAV sequences in GenBank with NS2A coverage. One isolate (CFAV-Rio Piedras02) was obtained from wild-caught mosquitoes (Cook et al., 2009), while the other isolate (herein referred to as CFAV-1993) was obtained from a laboratory mosquito cell line (Cammisa-Parks et al., 1992). CFAV-RioPiedras02 contains the proposed frameshift site, including the 3′-adjacent pseudoknot, and the 259-codon fifo ORF (Figs. 3, 5). In CFAV-1993, however, the fifo ORF is disrupted by three premature termination codons (Fig. 5). We propose that propagation in cell culture has resulted in the loss of FIFO in CFAV-1993. Nonetheless, the historic relic of a 259-codon fifo ORF is clearly visible in the CFAV-1993 sequence: the frameshift slippery site and 3′-adjacent pseudoknot are still present, and the region has very few + 2 frame stop codons (3 out of 259 codons, compared with 190 out of 3341 for the full polyprotein coding sequence which equates to a mean of 14.7 for a 259-codon region; Figs. 3, 5). Frameshifting likely still occurs but would produce a truncated trans-frame protein lacking nearly 90% of FIFO. Although the occurrence of frameshifting may, in itself, be advantageous for the virus irrespective of whether or not the full FIFO protein is produced (see The evolution of FIFO and implications for frameshifting in other flaviviruses), the apparent viability of CFAV-1993 indicates that the full-length FIFO protein is non-essential for replication in cell culture. It is interesting to note that JEV-serogroup NS1′ is also non-essential for replication – at least for West Nile virus (subtype Kunjin virus) – although abolishing NS1′ attenuates the virus (Balmori Melian et al., 2009). Nonetheless, the fact that the fifo ORF is so widely conserved in wild-type insect-specific flaviviruses suggests that it plays an important role in nature.
CSA is an Aedes albopictus chromosome-integrated sequence of insect-specific flavivirus origin (Crochu et al., 2004). The consequences, if any, of chromosome-integrated flavivirus sequences for the host and/or for subsequent viral infection are not known. However, it cannot be assumed that selection on CSA will operate to preserve the coding potential of the fifo ORF. On the other hand, if chromosome integration was a relatively recent event, then the coding signature of the fifo ORF prior to integration may remain relatively intact since substitutions in chromosomal sequences occur at a much slower rate than in RNA virus genomes. Indeed, estimates of the time to the most recent common ancestor of CSA and CFAV and the divergence rate of Aedes species (Crochu et al., 2004) suggest that the expected total number of nucleotide substitutions within the fifo ORF since CSA integration is likely to be zero. Moreover, analysis of the CSA sequence, GenBank ID: AF411835, revealed that the fifo ORF is present and contains an apparently intact ribosomal frameshift site at its 5′ end, allowing access to a 253-codon fifo ORF (Figs. 3 and 5).
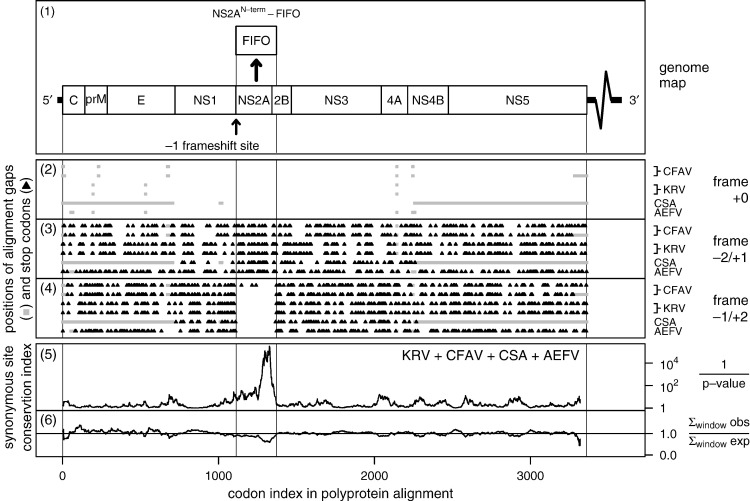
The conservation at polyprotein-frame synonymous sites was also determined for an alignment of the CFAV, KRV, AEFV and CSA sequences (Fig. 6 ). This time, the conservation plot revealed highly significant polyprotein-frame synonymous site conservation in just the 3′ half of the fifo ORF, while the 5′ half was relatively unconstrained. This indicates that the C-terminal half of FIFO is subject to stronger functional constraints than the N-terminal half. One possible reason why the conservation score in the 5′ half of fifo is much less significant in this alignment (Fig. 6) than the CxFV/QBV alignment (Fig. 2) may simply be that the CxFV/QBV alignment includes a number of closely related sequences (the six CxFV strains) while the CFAV/KRV/AEFV/CSA alignment includes only highly divergent sequences.
Fig. 6.
Conservation at synonymous sites in CFAV, KRV, AEFV and CSA. See Fig. 2 for details. Here, the alignment comprises the polyprotein coding sequences from AY149905 (KRV), AY149904 (KRV), AB488408 (AEFV), AF411835 (CSA; chromosome-integrated sequence), M91671 (presumed fifo-defective CFAV-1993) and GQ165810 (CFAV-RioPiedras02). AY149905 and AY149904 are nearly identical. Only AY149905, AB488408, AF411835 and GQ165810 were used to calculate the synonymous site conservation statistics (panels 5–6). There is considerable variation in the 3′ UTR lengths between the different sequences (e.g., 559, 1208 and 945 nt, respectively, in CFAV M91671, KRV AY149905 and AEFV AB488408; see Gritsun and Gould, 2006, for a discussion) — hence the wiggly line in the genome diagram. The CFAV-1993 sequence (isolated from a laboratory mosquito cell line) has lost the fifo ORF, indicating that the product is non-essential in cell culture.
Insect-specific flaviviruses were recently isolated from phlebotomine sandflies (Moureau et al., 2009). However, the currently available sequence data for these viruses do not cover the NS2A/NS2B region and thus it remains to be seen whether the genomes of these viruses also harbour the fifo ORF.
Sequence analysis of the predicted FIFO protein
NS2A and NS2B are among the least conserved proteins in an amino acid comparison between CxFV and CFAV (23% and 19% amino acid identity, respectively; Hoshino et al., 2007), and the FIFO amino acid sequences are even more divergent (Fig. 1). Within the Flavivirus genus, FIFO has a restricted phylogenetic distribution and presumably evolved more recently than NS2A/NS2B, via the process of ‘overprinting’ of the preexisting NS2A/NS2B coding sequence (Keese and Gibbs, 1992). Such ‘de novo’ proteins often have ‘non-essential’ or ‘secondary’ functions. Thus, it is not surprising that FIFO is subject to weaker functional constraints (as reflected by lower amino acid conservation) than NS2A/NS2B. In this case, it seems that the particular nature of the FIFO protein is compatible with a substantial degree of amino acid variation. In fact, de novo proteins encoded by overlapping genes often assume particular characteristics – perhaps as a result of their evolutionary origin enforcing an ‘unusual’ amino acid composition and/or evolutionary competition from the overlapping ancestral gene – that are compatible with a high degree of evolutionary flexibility in their amino acid sequences (Rancurel et al., 2009). For example structurally disordered domains are a common feature of many overlapping genes (Rancurel et al., 2009).
Application of blastp (Altschul et al., 1990) to FIFO revealed no similar sequences in GenBank (15 Aug 2009). This result was not surprising because genes created de novo via overprinting usually have no sequence similarity to other genes (Keese and Gibbs, 1992). Application of InterProScan (Zdobnov and Apweiler, 2001) resulted in the identification of two potential transmembrane domains in each sequence in Clade 1 and one potential transmembrane domain in each sequence in Clade 2. In all cases, the transmembrane domains occupied a similar position — near the N-terminal end of FIFO (typically commencing at or close to amino acid 58 of FIFO; Figs. 4, 5). Thus, despite the considerable divergence between the two insect-specific flavivirus clades (in fact, taken over all 14 sequences, there are more than 1000 phylogenetically independent base substitutions within the fifo ORF), both maintain a similarly long fifo ORF and a similarly located predicted membrane-spanning region. Analysis with MLOGD (Firth and Brown, 2006), besides the synonymous site conservation graphs (Figs. 2 and 6), indicated that the C-terminal region of FIFO was subject to the strongest amino acid constraints.
The flavivirus NS1|NS2A cleavage site is non-standard, being signalase-like with respect to the ‘-1, -3’ rule, but lacking the upstream hydrophobic domain (Lindenbach et al., 2007). Previous work has demonstrated that, at least in DENV, efficient NS1|NS2A cleavage requires translation of substantial parts of NS2A (Falgout et al., 1989, Falgout and Markoff, 1995, Leblois and Young, 1995). Although the exact mechanism has not been defined, it has been proposed that the presence of NS2A results in a conformation that presents the NS1|NS2A cleavage site to an endoplasmic reticulum-resident host protease (Falgout and Markoff, 1995). When frameshifting occurs in the JEV-serogroup flaviviruses, it appears that the 52 amino acid NS2AN-term-FOO fusion is insufficient to mediate NS1|NS2A cleavage, resulting in the NS1-NS2AN-term-FOO product known as NS1′ (Balmori Melian et al., 2009, Firth and Atkins, 2009). This mechanism appears to function in both insect and vertebrate cells as NS1′ is produced in both (Balmori Melian et al., 2009). At this stage, it is unclear whether or not NS1|NS2A cleavage in the insect-specific flaviviruses requires the synthesis of NS2A. If NS1|NS2A cleavage does require NS2A synthesis, then it is unknown whether or not NS2AN-term-FIFO synthesis will substitute. Thus, FIFO may be produced as a NS1-NS2AN-term-FIFO fusion or as a NS2AN-term-FIFO fusion or perhaps in both forms. Moreover, the two predicted transmembrane domains in Clade 1 may allow a suitable topology for further signalase cleavage within FIFO itself (potential cleavage sites were identified with SignalP 3.0 [Bendtsen et al., 2004], though it remains to be seen whether they are actually utilized).
So far as we are aware, the exact location of the NS1|NS2A cleavage site has not been experimentally verified in any of the insect-specific flaviviruses. If the published predictions (summarized in Hoshino et al., 2009) are correct, then it is interesting to note that the location of the frameshift site relative to the cleavage site differs substantially between the two insect-specific flavivirus clades (NS2A codons 28 and 29 in CxFV, but NS2A codons 3 and 4 in CFAV, KRV and AEFV). However, it is possible that NS1|NS2A cleavage in CxFV occurs at an alternative site 24 codons downstream, so that the frameshift site would be at NS2A codons 4 and 5 in CxFV. In favour of this new site, the − 3 and − 1 amino acids are V and G in CxFV and A and A in QBV, while the old site has − 3 and − 1 amino acids I and A in CxFV and I and N in QBV; thus, the new site appears to be more consistent with the ‘-1, -3’ rule for signalase cleavage.
Immunodetection of FIFO
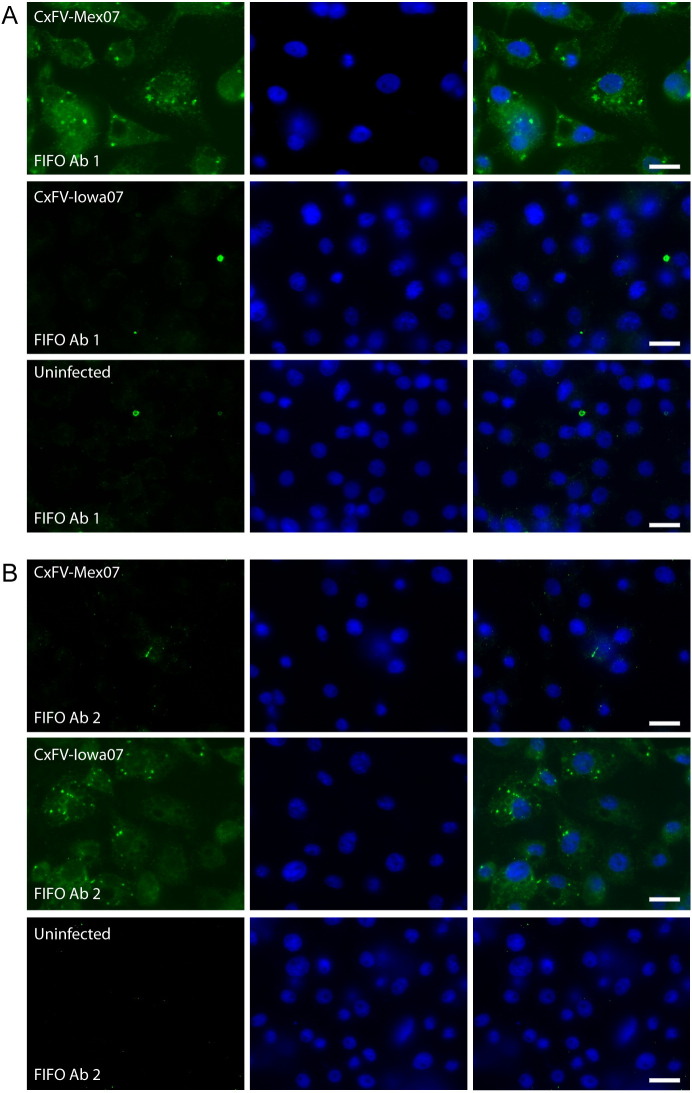
Separate Abs were raised against two predicted 14-aa CxFV FIFO antigens — one within the C-terminal region of CxFV-Mex07 FIFO (FIFO Ab 1) and the other within the N-terminal region of CxFV-Iowa07 FIFO (FIFO Ab 2). The peptide used to generate FIFO Ab 1 differs in one amino acid position from the homologous region of CxFV-Iowa07, and the peptide used to generate FIFO Ab 2 differs in two amino acid positions from the homologous region of CxFV-Mex07 (see Methods). To determine whether proteins containing FIFO are produced during CxFV infection, C6/36 cells were infected with CxFV or mock infected, incubated for 4 days, fixed with methanol, and then analyzed by immunofluorescence assay (IFA) using the Abs described above.
FIFO Ab 1 detected proteins in the CxFV-Mex07-infected cells, but not the mock-infected or CxFV-Iowa07-infected cells (Fig. 7A). FIFO Ab 2 detected proteins in the CxFV-Iowa07-infected cells, but not the mock-infected or CxFV-Mex07-infected cells (Fig. 7B). Thus the Abs are apparently strain-specific and it is highly unlikely that the Abs are simply recognizing a host protein that just happens to be up-regulated by virus-infection. Similar findings were observed in IFAs performed using paraformaldehyde-fixed cells (data not shown). Western blotting with FIFO Ab 1 resulted in the faint detection of a product migrating at ∼38 kDa when lysates from CxFV-Mex07-infected cells were used (data not shown). This is similar to the predicted size of NS2AN-term-FIFO (i.e., ∼34 kDa in the absence of post-translational modification). However, this product was not consistently observed with these lysates and nor was it detected in Western blots performed with FIFO Ab 2 and lysates from CxFV-Iowa07-infected cells. Thus, it is not clear whether the ∼38 kDa band is a non-specific artifact or if FIFO is not readily detected by western blot due to the relatively low frameshifting efficiency estimated for CxFV (∼6%; see Analysis of the proposed site of frameshifting).
Fig. 7.
Detection of FIFO by immunofluorescence in CxFV-infected mosquito cells. C6/36 cells were infected with CxFV-Mex07 (first row) or CxFV-Iowa07 (second row) or mock infected (third row). At 4 days p.i., cells were fixed with methanol and immunostained with rabbit polyclonal antibodies against FIFO (FIFO Ab 1 and FIFO Ab 2; panels A and B, respectively), followed by Alexa 594-conjugated donkey anti-rabbit IgG. Scale bars = 10 μm.
Analysis of the proposed site of frameshifting
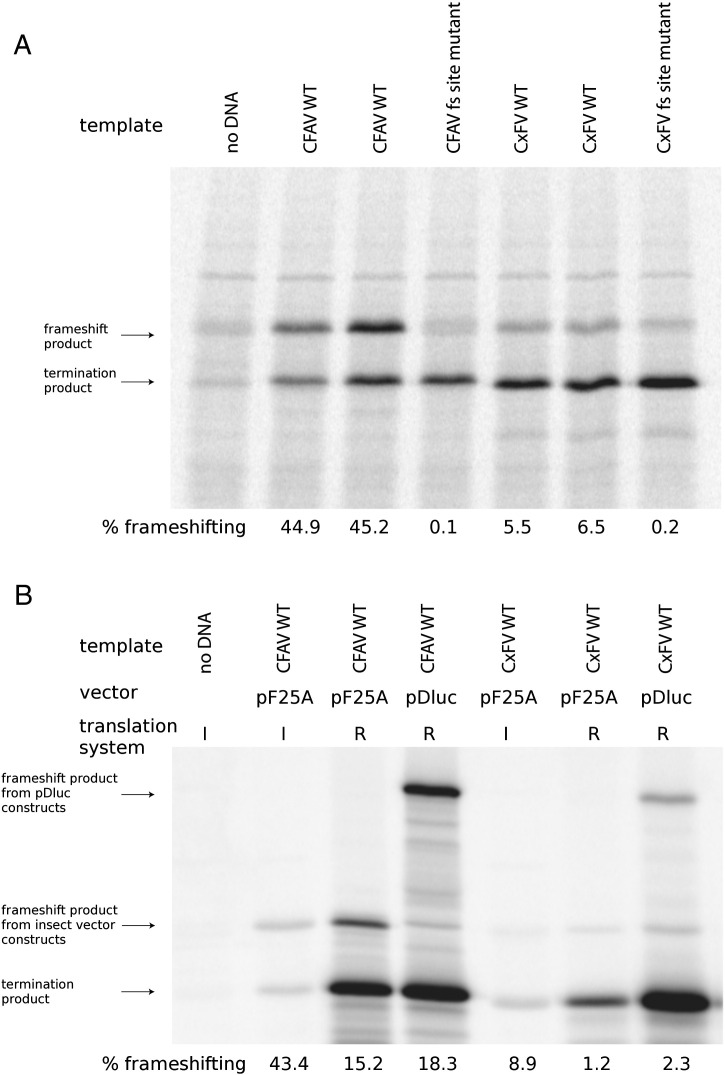
In order to test the proposed site of frameshifting, the slippery heptanucleotide and 3′-adjacent local sequence from CxFV and CFAV were cloned between two ORFs in vector pF25A ICE such that − 1 frameshifting produces a fusion product of the two ORFs. Frameshifting efficiencies were determined by comparison of the termination and frameshift products generated in an insect cell-free translation system (the exact inserts used correspond to the sequences shown in Fig. 3A). Frameshifting efficiencies were ∼6% and ∼45% for WT CxFV and CFAV sequences, respectively (Fig. 8A). In contrast, when the G GAU UUC slippery heptanucleotide was mutated to A GAC UUC (synonymous zero-frame codons but disrupted potential for codon:anticodon re-pairing in the − 1 frame) the frameshifting efficiencies dropped to 0.1–0.2%. Thus, it is clear that the G GAU UUY motif and 3′-adjacent sequence are capable of stimulating significant levels of frameshifting in both viruses.
Fig. 8.
SDS PAGEs for frameshifting assays. (A) Translation of products from pF25A constructs in the insect cell-free system. The frameshifting cassette was fused between upstream and downstream ORFs such that translation of the downstream ORF – as a fusion with the upstream ORF – occurs only if frameshifting occurs. The predicted sizes of the termination product (upstream ORF, no frameshifting) and frameshift product (fusion of both ORFs) are marked. Frameshifting efficiencies were calculated from the intensities of labelled bands after normalization for the number of methionine residues in each product. WT sequence (especially CFAV) stimulates high levels of frameshifting, while frameshifting is reduced to background levels in the frameshift (fs) site knockout mutants. Unfortunately background bands (first lane) at the same positions as both the termination and frameshift products makes precise measurement difficult for the shift site mutants, although it is clear that frameshifting is greatly diminished in the mutants. (B) Comparison of frameshifting efficiencies between the insect system (I) and rabbit reticulocyte lysate (R). Both vectors pF25A and pDluc were tested in rabbit reticulocyte lysate but only the insect-optimized pF25A vector was usable in the insect system.
The considerable difference in the efficiencies measured for CxFV and CFAV is of interest, and may be related to the 3′-adjacent stimulatory structure — a simple hairpin in CxFV but a pseudoknot in CFAV. However, these measurements are based on just 66 nt (CxFV) or 81 nt (CFAV) of local sequence fused into the reporter vector, so it is not certain that these measurements are representative of the frameshifting efficiencies in virus-infected cells (for example, longer-range interfering RNA structures or nascent peptide effects may result in different frameshifting efficiencies in the context of the full virus genome). The possibility that the 5′-terminal regions of the downstream vector sequence might be interfering with the predicted frameshift-stimulatory hairpin structure in CxFV by forming competing RNA structures was investigated by folding WT CxFV sequence, and CxFV sequence in the vector context, with pknotsRG (Reeder et al., 2007). However no evidence was found for interference with the predicted hairpin structure. Given the substantial variation in FIFO between the two insect-specific flavivirus clades, it is quite possible that the FIFO protein is simply required in much lower quantities in CxFV than in CFAV.
Although there is no evidence to suggest that vertebrates are hosts of the insect-specific flaviviruses, the frameshift assays were repeated using a mammalian expression vector, pDluc, to determine whether the striking difference in frameshifting efficiency between CxFV and CFAV would be preserved. With the same sequences cloned into pDluc and translated in rabbit reticulocyte lysate, the overall frameshifting efficiencies were 3-fold lower (∼2% for CxFV and ∼18% for CFAV; Fig. 8B) though the relative difference between CxFV and CFAV was similar. The overall 3-fold reduction was apparently not due to the different vector since translation of the inserts in the insect vector in rabbit reticulocyte lysate produced similar values (Fig. 8B). It is also not a temperature effect since all experiments were performed at the same temperature (30 °C). Possible reasons for the 3-fold difference between the insect cell-free system and rabbit reticulocyte lysate include potential differences in free monovalent and divalent cation concentrations, tRNA abundances, or perhaps other trans-acting factors.
With the exception of NAKV, the frameshift site (G GAU UUY) is essentially identical between the two insect-specific flavivirus clades and it seems probable that it was present ancestrally. Although the 3′-adjacent stimulatory structure differs between the two clades (a hairpin in Clade 1 and a pseudoknot in Clade 2), this degree of variation is quite common in other cases of frameshifting in RNA viruses. For example, frameshifting in HIV-1 utilizes a 3′-adjacent hairpin in Group M but a pseudoknot in Group O (Baril et al., 2003a, Baril et al., 2003b). Similarly, frameshifting in the alphaviruses utilizes a wide variety of 3′-adjacent hairpin and pseudoknot structures in the different species (Firth et al., 2008; B.Y. Chung et al, manuscript in preparation). In contrast, the slippery site itself tends to be more conserved. For example, nearly all alphaviruses and all HIV-1 isolates utilize a U UUU UUA slippery site.
Potential for programmed frameshifting in other flaviviruses
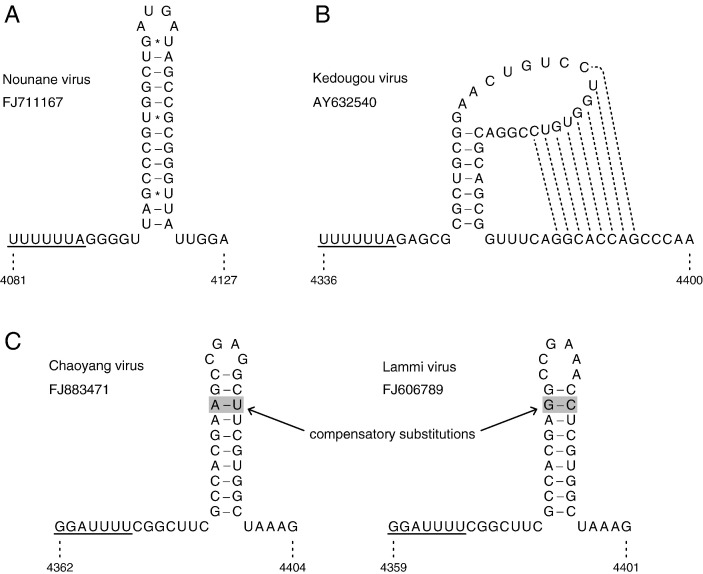
A cursory search revealed a number of potential ribosomal frameshift sites in certain other flaviviruses, most of which give access only to very short out-of-frame ORFs. The most promising candidates were found in the genomes of four poorly characterized flaviviruses: Nounane virus (NOUV), Lammi virus (LAMV), Kedougou virus (KEDV) and Chaoyang virus. The genome of NOUV (GenBank ID: FJ711167; Junglen et al., 2009) contains a U UUU UUA slippery site followed by a 5 nt spacer and a potential 13 bp hairpin structure (Fig. 9A) 24–25 codons upstream of the NS2A/NS2B predicted cleavage site. The motif is curiously similar to the experimentally confirmed frameshift cassette in sleeping disease alphavirus (Firth et al., 2008; B.Y. Chung et al, manuscript in preparation) which also has a U UUU UUA slippery site and a 3′-adjacent 13 bp hairpin structure. Moreover, the five nucleotides immediately following the U UUU UUA motif (viz. GGGGU) are identical between the two viruses. In NOUV, the out-of-frame ORF has just 2 codons and frameshifting at this location would result in a truncated form of NS2A. The KEDV genome (GenBank ID: AY632540; Kuno and Chang, 2005) contains a U UUU UUA slippery site followed by a 5 nt spacer and a stable potential pseudoknot (though, perhaps unusually for frameshift-stimulating structures, the second loop region of the pseudoknot has > 1 nt; Fig. 9B; Brierley and Pennell, 2001). The slippery site is located 52–53 codons downstream of the NS2A/NS2B predicted cleavage site, and the out-of-frame ORF has 21 codons. The Chaoyang virus genome (GenBank ID: FJ883471) contains a G GAU UUU slippery site followed by a 7 nt spacer and a potential 10 bp hairpin structure (Fig. 9C) 83–84 codons downstream of the NS2A/NS2B predicted cleavage site. Here, the out-of-frame ORF has 107 codons and frameshifting at this location would result in an elongated version of NS2B. An homologous site is also present in the LAMV genome (GenBank ID: FJ606789; Huhtamo et al., 2009). Here, the shift site and out-of-frame ORF lengths are identical to those in Chaoyang virus, and the hairpin – although 1 bp shorter – is supported by an A:U to G:C substitution that preserves the base pairing (Fig. 9C). Although the sequence data is very limited, a comparison of FJ883471 with FJ606789 indicates that the potential shift site and 3′-adjacent hairpin, though not the downstream out-of-frame ORF, correspond to a peak in polyprotein-frame synonymous site conservation (data not shown).
Fig. 9.
Potential slippery heptanucleotides and stimulatory RNA secondary structures for − 1 ribosomal frameshifting in Nounane virus, Kedougou virus, Chaoyang virus and Lammi virus. In contrast to the insect-specific flavivirus fifo frameshift site, these cases are merely candidate frameshift sites and currently have either no (Nounane, Kedougou) or only very modest (Chaoyang, Lammi) phylogenetic support. Structures were predicted with pknotsRG (Reeder et al., 2007).
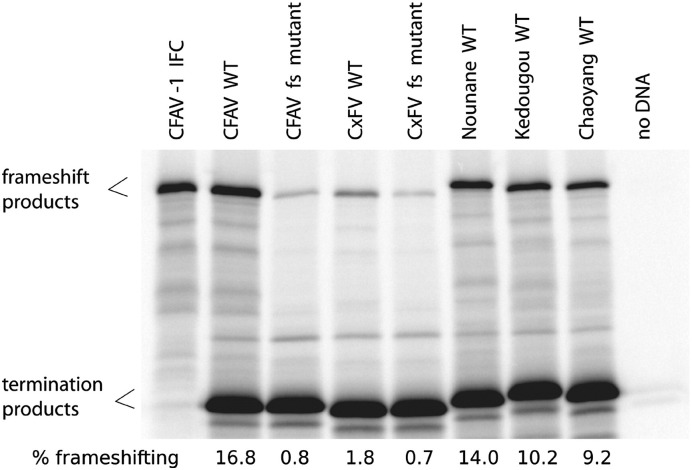
The putative shift sites and 3′-adjacent sequence including the predicted RNA structures from NOUV, KEDV and Chaoyang virus were tested in the pDluc vector in rabbit reticulocyte lysate. In the case of NOUV, the G:U base pairing near the base of the predicted stem (Fig. 9A) was changed to a U:G pair (which is not predicted to alter the structure) in order to remove the − 1 frame termination codon and allow translation of the downstream reporter gene. Similarly, a − 1 frame UGA termination codon at nucleotides + 10 to + 12 with respect to the 3′ end of the predicted pseudoknot in KEDV (Fig. 9B) was changed to UUA which, again, is not predicted to alter the structure. All three inserts stimulated high levels of frameshifting: ∼14% for NOUV, ∼10% for KEDV, and ∼9% for Chaoyang virus (Fig. 10 ).
Fig. 10.
SDS PAGE of translation products from pDluc constructs in rabbit reticulocyte lysate. See Fig. 8 for further details. Lane 1 shows translation products for an in-frame control (IFC) – in which an extra nucleotide has been inserted next to the slippery heptanucleotide to put both ORFs into the same reading frame – to show the expected size of the frameshift fusion product. Note that the faint band corresponding to the frameshift product for the CFAV and CxFV frameshift site (fs) mutants may represent a background of low-level slippage occurring anywhere within the overlap region between the upstream and downstream ORFs and does not necessarily represent slippage just at the mutated shift site.
It is important to note that the presence of a slippery heptanucleotide and 3′-adjacent RNA structure does not automatically imply that frameshifting will occur, as even minor mutations to known frameshift-stimulating structures can greatly diminish frameshifting efficiency (Chen et al., 1995, Chen et al., 1996). Even if frameshifting does occur (as indicated by the reporter assays), it may be accidental rather than functionally important and subject to purifying selection. Thus, the key to identifying biologically relevant frameshift sites from sequence analysis alone is the phylogenetic conservation of potential frameshift-stimulatory elements over a significantly divergent alignment. This is particularly true if the out-of-frame ORF is too short to obtain an independent measure of its ‘coding potential’. However, without more sequence data for these and/or closely related species for comparative sequence analysis, and in the absence of experimental evidence for function in virus-infected cells, it is not clear at present whether or not the NOUV, LAMV, KEDV and Chaoyang virus candidate frameshift sites have any biological significance. Future sequencing data should help to clarify their status.
The evolution of FIFO and implications for frameshifting in other flaviviruses
The discovery of fifo raises a number of interesting evolutionary questions. Does FIFO have similar or different functions in the two insect-specific flavivirus clades? Although the frameshift site and at least some of the fifo ORF was presumably present in the ancestral virus, was the full fifo ORF also present in the ancestral virus or did it evolve through two independent elongation events? Was frameshifting at the 5′ end of the NS2A coding sequence present in the last common ancestor of both the JEV-serogroup of flaviviruses and the insect-specific flaviviruses? If so, then it was surely also present in the ancestor of DENV, even though it has now apparently been lost in DENV and many other flaviviruses (transfer of the frameshift cassette via recombination, after the divergence of the JEV-serogroup from DENV, appears highly unlikely since in this genomic region, as elsewhere, JEV is much more similar to DENV than to CxFV or CFAV). If not then, given the general rarity of programmed ribosomal frameshifting, what selective forces drove the evolution on two separate occasions of frameshifting in this particular region of the flavivirus genome? Some possible such selective forces may be (i) to produce an alternative version of NS1 with a distinct C-terminal extension; (ii) to produce an NS1-like protein, viz. NS1′, that is not C-terminally linked to the downstream polyprotein (perhaps as a mechanism to modulate the post-translational processing pathway for a portion of NS1); (iii) to act as a ‘ribosome sink’, i.e., to reduce the quantity of ribosomes translating the downstream polyprotein products; and (iv) as part of a regulatory mechanism (especially if the frameshifting efficiency is temporally modulated, e.g., by the build-up of a viral protein or by changing cellular conditions; cf. Goff, 2004, Baranov et al., 2002). In scenarios (ii)–(iv), the initial selective force would simply be to have a frameshift site, with the actual amino acid sequence encoded by the out-of-frame ORF being essentially irrelevant. This could then have been co-opted as a suitable site to begin the evolution of an overlapping gene via incremental elongation by substitutions of intervening − 1/+ 2 frame stop codons with sense codons and selection on the encoded amino acid sequence (cf. Belshaw et al., 2007), resulting in a short 45-codon ORF in the JEV-serogroup and the much longer fifo ORF in the insect-specific flaviviruses.
While scenarios (i) and (ii) would explain independent evolution of both frameshifting and the conserved location of frameshifting near the 5′ end of the NS2A coding sequence, scenario (iii) appears to explain just the evolution of frameshifting rather than the precise location of frameshifting. Since the flavivirus structural proteins are encoded by the 5′ end of the polyprotein coding sequence, and since only a fraction (∼5% or fewer) of the translated non-structural proteins that comprise the components of the viral replication complex and, in particular, the viral polymerase or RdRp (NS5 in the flaviviruses) appear to be actually utilized for replication (Ahlquist, 2006, Quinkert et al., 2005, Uchil and Satchidanandam, 2003), there may be relatively little selective pressure against the accidental evolution of slippery sites throughout the middle regions of the polyprotein coding sequence. Indeed reducing the level of nonstructural protein synthesis, and simultaneously freeing up ribosomes, may even be beneficial for the virus. However, the common location of the JEV-serogroup and insect-specific flavivirus frameshift sites could also result from scenario (iii) if the NS2A/NS2B amino acid composition just happens to be particularly amenable to harbouring overlapping genes (e.g., it is unusually hydrophobic: codons for hydrophobic amino acids tend to have a ‘U’ or ‘C’ at position 2 which is incompatible with a − 1/+ 2 frame stop codon, thus hydrophobic regions are compatible with a reduced frequency of stop codons in the − 1/+ 2 frame). In this case, the close proximity of both frameshift sites to the 5′ end of the NS2A coding sequence may have evolved simply to avoid splitting protein domains.
It is interesting to note that Quinkert et al. (2005) observed a factor of three- to six-fold fewer NS4B and NS5B proteins than core proteins in cells infected with Hepatitis C virus (family Flaviviridae) which uses a similar polyprotein expression strategy and genome organization to the flaviviruses, implying that 66–83% of translating ribosomes may terminate early — albeit 3′ of the core protein coding sequence. What remains uncertain is whether early termination is ‘programmed’ (i.e., occurring at specific sites, under purifying selection for this function) or whether it occurs randomly throughout the polyprotein coding sequence. While most polyprotein cleavage products of well-studied members of the Flaviviridae family have been well-characterized (including N- and C-terminal sequencing), there are a number of ‘alternative’ cleavage products in various flaviviruses that have been less well characterized and the potential for C-termini produced as a result of ribosomal frameshifting, as opposed to enzymatic cleavage, perhaps merits investigation. Furthermore, selection may favour frameshift sites that are located near to polyprotein cleavage sites so as to avoid splitting protein domains. (In, for example, those alphaviruses that utilize stop codon read-through to express the RdRp [alphavirus NSP4] as a fusion with NSP123, the C-terminal of NSP3 is defined in two ways — either by termination at the read-through stop codon or by cleavage from NSP4. However the NSP3|NSP4 cleavage site is just seven codons downstream of the read-through stop codon so that the two forms of NSP3 differ by only seven amino acids [Strauss and Strauss, 1994].) Such trans-frame products – particularly if the out-of-frame ORF is very short – may have escaped notice as they may be difficult to distinguish from the analogous cleavage products.
Conclusions
We have presented compelling evidence for a 253- to 295-codon overlapping gene, fifo, in the genomes of all known insect-specific flaviviruses. Evidence includes (i) the conserved presence of a long − 1/+ 2 frame ORF overlapping the NS2A/NS2B region of the polyprotein coding sequence despite > 1000 phylogenetically independent base substitutions within the region, (ii) a statistically highly significant enhancement in the conservation at polyprotein-frame synonymous sites in both a CxFV-QBV alignment and a CFAV-KRV-AEFV-CSA alignment; (iii) a well-defined and conserved translation mechanism via programmed ribosomal frameshifting at the 5′ end of the ORF; (iv) reporter assays confirm the viability of the proposed translation mechanism; and (v) immunofluorescence assays with two separate Abs raised against different regions of FIFO reveal the specific presence of proteins containing FIFO antigens in CxFV-infected cells.
This discovery adds to a small number of known cases of overlapping genes that are internal to large polyprotein coding sequences in RNA viruses and accessed via well-defined programmed ribosomal frameshifting sites (as opposed to low level accidental translation of alternative reading frames; cf. Yewdell and Hickman, 2007). Other examples include potyvirus PIPO (Chung et al., 2008), Alphavirus TF (Firth et al., 2008) and JEV-serogroup FOO/NS1′ (Balmori Melian et al., 2009, Firth and Atkins, 2009). The new ORF fifo, however, is unusual in the length of the overlap region. In fact, only a handful of known overlapping genes in virus genomes are longer (e.g., ones accessed via leaky scanning or alternative transcripts; Rancurel et al., 2009).
Overlapping genes are difficult to identify and are often overlooked. However, it is important to be aware of such genes as early as possible. Undetected overlapping genes can cause considerable and persistent confusion since their functions may be wrongly ascribed to the genes they overlap. Furthermore, only once it has been identified, can the functions of an overlapping gene be investigated in their own right. Although characterization of the insect-specific flavivirus polyprotein products has barely commenced, to a certain extent their functions may be inferred from the more well-studied mosquito-borne flaviviruses such as DENV, JEV and yellow fever virus. In contrast, the FIFO product (or products) represents a completely novel protein of as yet unknown function. A full characterization of the FIFO protein is, however, beyond the scope of this report of its discovery and will, instead, be addressed in future work.
Methods
Bioinformatics
Mosquito flavivirus sequences with NS2A/NS2B coverage were identified by applying tblastn (Altschul et al., 1990) to the NS2A amino acid sequences derived from CxFV AB262759 and KRV AY149905. This revealed the following sequences in GenBank (as of 10 Sep. 2009): AB262759.2 (CxFV), AB377213.1 (CxFV), FJ663034.1 (CxFV), FJ502995.1 (CxFV), GQ165808.1 (CxFV), EU879060.1 (CxFV), FJ644291.1 (QBV) and GQ165809.1 (NAKV) were detected when tblastn was applied to the NS2A sequence from AB262759, while AY149905.1 (KRV), AY149904.1 (KRV), GQ165810.1 (CFAV), M91671.1 (CFAV), AF411835.1 (chromosome-integrated sequence, CSA), AB488408.1 (AEFV) and DQ181510.1 (CFAV, partial sequence) were detected when tblastn was applied to the NS2A sequence from AY149905. The partial sequence DQ181510.1 is locally identical to GQ165810.1, and was therefore discarded. Polyprotein-encoding sequences were extracted, translated, aligned with CLUSTALW (Larkin et al., 2007) and back-translated to produce nucleotide alignments.
Antibodies
Polyclonal antibodies to two 14-aa predicted antigens within FIFO were prepared by GenScript Inc., Piscataway, NJ. FIFO Ab 1 was raised against peptide sequence CRNLRSGWSGIHELD (‘C’ + CxFV-Mex07 FIFO amino acids 279 to 292; CxFV-Iowa07 has a ‘T’ instead of the italicized ‘S’). FIFO Ab 2 was raised against peptide sequence CPTGGRAFAPADHSN (‘C’ + CxFV-Iowa07 FIFO amino acids 14 to 27; CxFV-Mex07 has an ‘S’ and an ‘S’ instead of the italicized ‘A’ and ‘P’). Rabbits were injected with one of the two peptides, and antibodies were affinity-purified from immune sera.
Immunofluorescence assays
Aedes albopictus (C6/36) cells were seeded on six-well (9.6-cm2) dishes containing 18 mm diameter coverslips at a density of 1 × 105 cells/well and incubated at 28 °C until they reached confluency. Cells were infected with CxFV or mock infected, then incubated for 4 days. Cells were fixed either at − 20 °C for 3 min with 100% methanol or at room temperature for 10 min with 2% paraformaldehyde in PBS then washed three times with PBS. Fixed cells were permeabilized by incubation with 0.2% Triton X-100 in PBS for 5 min and then washed three times with PBS. Samples were blocked for 10 min with 2% bovine serum albumin in PBS. Primary and secondary antibodies were diluted in 2% bovine serum albumin in PBS. After being blocked, cells were incubated for 1 h with primary antibody, washed three times with PBS, and then incubated for an additional hour with secondary antibody. Immunostained cells were washed a final three times with PBS and mounted on slides with ProLong reagent with DAPI (4′,6-diamidino-2-phenylindole dihydrochloride) (Invitrogen). Immunostained samples were examined with a Zeiss Axiovert 200 inverted microscope equipped with fluorescence optics. Images were prepared using Photoshop and Illustrator software (Adobe Systems).
Frameshift assays
The sequence encompassing the predicted frameshift site and 3′ stimulatory structure (frameshift cassette) for each virus was generated using overlapping synthetic oligonucleotides and cloned into vector pDluc (kindly supplied by Dr. M. Howard, University of Utah), a modified version of the dual luciferase vector described by Grentzmann et al. (1998). The firefly luciferase gene is in the − 1 frame relative to the renilla luciferase gene such that − 1 frameshifting within the inserted sequence results in a renilla-firefly luciferase fusion product. To introduce the CxFV and CFAV frameshift cassettes into the insect vector, pF25A ICE T7 Flexi (Promega), primers specific to the 5′ end of renilla luciferase (TATAAAGCGATCGCCATGGCTTCCAAGGTGTACGACCCC) and an internal region of firefly luciferase (AATTATGTTTAAACTTACCCATAGCGCTTCATAGCTTCTGCC) were used for PCR amplification. These fragments were cloned into the Sgf I and Pme I sites (italicized). All constructs were verified by DNA sequencing.
Plasmid DNAs were used as templates in the reticulocyte lysate TNT® T7 Quick Coupled Transcription/Translation System (Promega) for pDluc constructs or the TNT® T7 Insect Cell Extract for pF25A ICE constructs (Promega). 35S-methionine (GE Healthcare) was added to the reactions and protein products were separated by SDS PAGE. Dried gels were analyzed using a Typhoon PhosphorImager (GE Healthcare) and the amount of radioactivity in products was determined using the ImageQuant 5.2 program (Molecular Dynamics). After normalization for the number of methionines in the termination and frameshift products, the frameshifting efficiencies were calculated as [frameshift / (frameshift + termination)].
Acknowledgments
This work was supported by National Institutes of Health Grant R01 GM079523 and an award from Science Foundation Ireland to JFA and National Institutes of Health Grant 5R21AI067281-02 to BJB.
Contributor Information
Andrew E. Firth, Email: a.firth@ucc.ie.
Bradley J. Blitvich, Email: blitvich@iastate.edu.
Norma M. Wills, Email: nwills@genetics.utah.edu.
Cathy L. Miller, Email: clm@iastate.edu.
John F. Atkins, Email: j.atkins@ucc.ie.
References
- Ahlquist P. Parallels among positive-strand RNA viruses, reverse-transcribing viruses and double-stranded RNA viruses. Nat. Rev. Microbiol. 2006;4:371–382. doi: 10.1038/nrmicro1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Balmori Melian, E., Hinzman, E., Nagasaki, T., Firth, A.E., Wills, N.M., Nouwens, A.S., Blitvich, B.J., Leung, J., Funk, A., Atkins, J.F., Hall, R., Khromykh, A.A., 2009. NS1′ of flaviviruses in the Japanese encephalitis serogroup is a product of ribosomal frameshifting and plays a role in viral neuro-invasiveness, J. Virol. doi:10.1128/JVI.01979-09. [DOI] [PMC free article] [PubMed]
- Baranov P.V., Gesteland R.F., Atkins J.F. Recoding: translational bifurcations in gene expression. Gene. 2002;286:187–201. doi: 10.1016/s0378-1119(02)00423-7. [DOI] [PubMed] [Google Scholar]
- Baril M., Dulude D., Steinberg S.V., Brakier-Gingras L. The frameshift stimulatory signal of human immunodeficiency virus type 1 group O is a pseudoknot. J. Mol. Biol. 2003;331:571–583. doi: 10.1016/S0022-2836(03)00784-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baril M., Dulude D., Gendron K., Lemay G., Brakier-Gingras L. Efficiency of a programmed -1 ribosomal frameshift in the different subtypes of the human immunodeficiency virus type 1 group M. RNA. 2003;9:1246–1253. doi: 10.1261/rna.5113603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bekaert M., Rousset J.P. An extended signal involved in eukaryotic -1 frameshifting operates through modification of the E site tRNA. Mol. Cell. 2005;17:61–68. doi: 10.1016/j.molcel.2004.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belshaw R., Pybus O.G., Rambaut A. The evolution of genome compression and genomic novelty in RNA viruses. Genome Res. 2007;17:1496–1504. doi: 10.1101/gr.6305707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendtsen J.D., Nielsen H., Von Heijne G., Brunak S. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 2004;340:783–795. doi: 10.1016/j.jmb.2004.05.028. [DOI] [PubMed] [Google Scholar]
- Blitvich B.J., Scanlon D., Shiell B.J., Mackenzie J.S., Hall R.A. Identification and analysis of truncated and elongated species of the flavivirus NS1 protein. Virus Res. 1999;60:67–79. doi: 10.1016/s0168-1702(99)00003-9. [DOI] [PubMed] [Google Scholar]
- Blitvich B.J., Lin M., Dorman K.S., Soto V., Hovav E., Tucker B.J., Staley M., Platt K.B., Bartholomay L.C. Genomic sequence and phylogenetic analysis of Culex flavivirus, an insect-specific flavivirus, isolated from Culex pipiens (Diptera: Culicidae) in Iowa. J. Med. Entomol. 2009;46:934–941. doi: 10.1603/033.046.0428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brierley I., Pennell S. Structure and function of the stimulatory RNAs involved in programmed eukaryotic -1 ribosomal frameshifting. Cold Spring Harbor Symp. Quant. Biol. 2001;66:233–248. doi: 10.1101/sqb.2001.66.233. [DOI] [PubMed] [Google Scholar]
- Brierley I., Jenner A.J., Inglis S.C. Mutational analysis of the “slippery-sequence” component of a coronavirus ribosomal frameshifting signal. J. Mol. Biol. 1992;227:463–479. doi: 10.1016/0022-2836(92)90901-U. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cammisa-Parks H., Cisar L.A., Kane A., Stollar V. The complete nucleotide sequence of cell fusing agent (CFA): homology between the nonstructural proteins encoded by CFA and the nonstructural proteins encoded by arthropod-borne flaviviruses. Virology. 1992;189:511–524. doi: 10.1016/0042-6822(92)90575-a. [DOI] [PubMed] [Google Scholar]
- Chen X., Chamorro M., Lee S.I., Shen L.X., Hines J.V., Tinoco I., Jr., Varmus H.E. Structural and functional studies of retroviral RNA pseudoknots involved in ribosomal frameshifting: nucleotides at the junction of the two stems are important for efficient ribosomal frameshifting. EMBO J. 1995;14:842–852. doi: 10.1002/j.1460-2075.1995.tb07062.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Kang H., Shen L.X., Chamorro M., Varmus H.E., Tinoco I., Jr. A characteristic bent conformation of RNA pseudoknots promotes -1 frameshifting during translation of retroviral RNA. J. Mol. Biol. 1996;260:479–483. doi: 10.1006/jmbi.1996.0415. [DOI] [PubMed] [Google Scholar]
- Chung W.Y., Wadhawan S., Szklarczyk R., Pond S.K., Nekrutenko A. A first look at ARFome: dual-coding genes in mammalian genomes. PLoS Comput. Biol. 2007;3:e91. doi: 10.1371/journal.pcbi.0030091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung B.Y., Miller W.A., Atkins J.F., Firth A.E. An overlapping essential gene in the Potyviridae. Proc. Natl. Acad. Sci. U. S. A. 2008;105:5897–5902. doi: 10.1073/pnas.0800468105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook S., Bennett S.N., Holmes E.C., De Chesse R., Moureau G., De Lamballerie X. Isolation of a new strain of the flavivirus cell fusing agent virus in a natural mosquito population from Puerto Rico. J. Gen. Virol. 2006;87:735–748. doi: 10.1099/vir.0.81475-0. [DOI] [PubMed] [Google Scholar]
- Cook S., Moureau G., Harbach R.E., Mukwaya L., Goodger K., Ssenfuka F., Gould E., Holmes E.C., De Lamballerie X. Isolation of a novel species of flavivirus and a new strain of Culex flavivirus (Flaviviridae) from a natural mosquito population in Uganda. J. Gen. Virol. 2009;90:2669–2678. doi: 10.1099/vir.0.014183-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crabtree M.B., Sang R.C., Stollar V., Dunster L.M., Miller B.R. Genetic and phenotypic characterization of the newly described insect flavivirus, Kamiti River virus. Arch. Virol. 2003;148:1095–1118. doi: 10.1007/s00705-003-0019-7. [DOI] [PubMed] [Google Scholar]
- Crabtree M.B., Nga P.T., Miller B.R. Isolation and characterization of a new mosquito flavivirus, Quang Binh virus, from Vietnam. Arch. Virol. 2009;154:857–860. doi: 10.1007/s00705-009-0373-1. [DOI] [PubMed] [Google Scholar]
- Crochu S., Cook S., Attoui H., Charrel R.N., De Chesse R., Belhouchet M., Lemasson J.J., De Micco P., De Lamballerie X. Sequences of flavivirus-related RNA viruses persist in DNA form integrated in the genome of Aedes spp. mosquitoes. J. Gen. Virol. 2004;85:1971–1980. doi: 10.1099/vir.0.79850-0. [DOI] [PubMed] [Google Scholar]
- Den Boon J.A., Snijder E.J., Chirnside E.D., De Vries A.A., Horzinek M.C., Spaan W.J. Equine arteritis virus is not a togavirus but belongs to the coronavirus-like superfamily. J. Virol. 1991;65:2910–2920. doi: 10.1128/jvi.65.6.2910-2920.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falgout B., Markoff L. Evidence that flavivirus NS1-NS2A cleavage is mediated by a membrane-bound host protease in the endoplasmic reticulum. J. Virol. 1995;69:7232–7243. doi: 10.1128/jvi.69.11.7232-7243.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falgout B., Chanock R., Lai C.J. Proper processing of dengue virus nonstructural glycoprotein NS1 requires the N-terminal hydrophobic signal sequence and the downstream nonstructural protein NS2a. J. Virol. 1989;63:1852–1860. doi: 10.1128/jvi.63.5.1852-1860.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farfan-Ale J.A., Loroño-Pino M.A., Garcia-Rejon J.E., Hovav E., Powers A.M., Lin M., Dorman K.S., Platt K.B., Bartholomay L.C., Soto V., Beaty B.J., Lanciotti R.S., Blitvich B.J. Detection of RNA from a novel West Nile-like virus and high prevalence of an insect-specific flavivirus in mosquitoes in the Yucatan Peninsula of Mexico. Am. J. Trop. Med. Hyg. 2009;80:85–95. [PMC free article] [PubMed] [Google Scholar]
- Firth A.E., Brown C.M. Detecting overlapping coding sequences in virus genomes. BMC Bioinformatics. 2006;7:75. doi: 10.1186/1471-2105-7-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firth A.E., Atkins J.F. A conserved predicted pseudoknot in the NS2A-encoding sequence of West Nile and Japanese encephalitis flaviviruses suggests NS1′ may derive from ribosomal frameshifting. Virol. J. 2009;6:14. doi: 10.1186/1743-422X-6-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firth A.E., Chung B.Y., Fleeton M.N., Atkins J.F. Discovery of frameshifting in Alphavirus 6K resolves a 20-year enigma. Virol. J. 2008;5:108. doi: 10.1186/1743-422X-5-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs M.J., Cooper J.I., Waterhouse P.M. The genome organization and affinities of an Australian isolate of carrot mottle umbravirus. Virology. 1996;224:310–313. doi: 10.1006/viro.1996.0533. [DOI] [PubMed] [Google Scholar]
- Goff S.P. Genetic reprogramming by retroviruses: enhanced suppression of translational termination. Cell Cycle. 2004;3:123–125. [PubMed] [Google Scholar]
- Grentzmann G., Ingram J.A., Kelly P.J., Gesteland R.F., Atkins J.F. A dual-luciferase reporter system for studying recoding signals. RNA. 1998;4:479–486. [PMC free article] [PubMed] [Google Scholar]
- Gritsun T.S., Gould E.A. The 3′ untranslated regions of Kamiti River virus and Cell fusing agent virus originated by self-duplication. J. Gen. Virol. 2006;87:2615–2619. doi: 10.1099/vir.0.81950-0. [DOI] [PubMed] [Google Scholar]
- Hoshino K., Isawa H., Tsuda Y., Yano K., Sasaki T., Yuda M., Takasaki T., Kobayashi M., Sawabe K. Genetic characterization of a new insect flavivirus isolated from Culex pipiens mosquito in Japan. Virology. 2007;359:405–414. doi: 10.1016/j.virol.2006.09.039. [DOI] [PubMed] [Google Scholar]
- Hoshino K., Isawa H., Tsuda Y., Sawabe K., Kobayashi M. Isolation and characterization of a new insect flavivirus from Aedes albopictus and Aedes flavopictus mosquitoes in Japan. Virology. 2009;391:119–129. doi: 10.1016/j.virol.2009.06.025. [DOI] [PubMed] [Google Scholar]
- Huhtamo E., Putkuri N., Kurkela S., Manni T., Vaheri A., Vapalahti O., Uzcátegui N.Y. Characterization of a novel flavivirus from mosquitoes in northern europe that is related to mosquito-borne flaviviruses of the tropics. J. Virol. 2009;83:9532–9540. doi: 10.1128/JVI.00529-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junglen S., Kopp A., Kurth A., Pauli G., Ellerbrok H., Leendertz F.H. A new flavivirus and a new vector: characterization of a novel flavivirus isolated from uranotaenia mosquitoes from a tropical rain forest. J. Virol. 2009;83:4462–4468. doi: 10.1128/JVI.00014-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keese P.K., Gibbs A. Origins of genes: “big bang” or continuous creation? Proc. Natl. Acad. Sci. U. S. A. 1992;89:9489–9493. doi: 10.1073/pnas.89.20.9489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K.H., Lommel S.A. Identification and analysis of the site of -1 ribosomal frameshifting in red clover necrotic mosaic virus. Virology. 1994;200:574–582. doi: 10.1006/viro.1994.1220. [DOI] [PubMed] [Google Scholar]
- Kim D.Y., Guzman H., Bueno R., Jr., Dennett J.A., Auguste A.J., Carrington C.V., Popov V.L., Weaver S.C., Beasley D.W., Tesh R.B. Characterization of Culex Flavivirus (Flaviviridae) strains isolated from mosquitoes in the United States and Trinidad. Virology. 2009;386:154–159. doi: 10.1016/j.virol.2008.12.034. [DOI] [PubMed] [Google Scholar]
- Kuno G., Chang G.J. Biological transmission of arboviruses: reexamination of and new insights into components, mechanisms, and unique traits as well as their evolutionary trends. Clin. Microbiol. Rev. 2005;18:608–637. doi: 10.1128/CMR.18.4.608-637.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin M.A., Blackshields G., Brown N.P., Chenna R., McGettigan P.A., McWilliam H., Valentin F., Wallace I.M., Wilm A., Lopez R., Thompson J.D., Gibson T.J., Higgins D.G. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Leblois H., Young P.R. Maturation of the dengue-2 virus NS1 protein in insect cells: effects of downstream NS2A sequences on baculovirus-expressed gene constructs. J. Gen. Virol. 1995;76:979–984. doi: 10.1099/0022-1317-76-4-979. [DOI] [PubMed] [Google Scholar]
- Lindenbach B.D., Thiel H.J., Rice C.M. Flaviviridae: the viruses and their replication. In: Knipe D.M., Howley P.M., editors. Fields Virology. 5th ed. Lippincott-Raven Publishers; Philadelphia: 2007. pp. 1101–1152. [Google Scholar]
- Martin D.P., Williamson C., Posada D. RDP2: recombination detection and analysis from sequence alignments. Bioinformatics. 2005;21:260–262. doi: 10.1093/bioinformatics/bth490. [DOI] [PubMed] [Google Scholar]
- Morales-Betoulle M.E., Monzón Pineda M.L., Sosa S.M., Panella N., López M.R., Cordón-Rosales C., Komar N., Powers A., Johnson B.W. Culex flavivirus isolates from mosquitoes in Guatemala. J. Med. Entomol. 2008;45:1187–1190. doi: 10.1603/0022-2585(2008)45[1187:cfifmi]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Moureau, G., Ninove, L., Izri, A., Cook, S., De Lamballerie, X., Charrel, R.N., 2009. Flavivirus RNA in Phlebotomine Sandflies. Vector Borne Zoonotic. Dis. doi:10.1089/vbz.2008.0216. [DOI] [PMC free article] [PubMed]
- Quinkert D., Bartenschlager R., Lohmann V. Quantitative analysis of the hepatitis C virus replication complex. J. Virol. 2005;79:13594–13605. doi: 10.1128/JVI.79.21.13594-13605.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rancurel C., Khosravi M., Dunker K.A., Romero P.R., Karlin D. Overlapping genes produce proteins with unusual sequence properties and offer insight into de novo protein creation. J. Virol. 2009;83:10719–10736. doi: 10.1128/JVI.00595-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeder J., Steffen P., Giegerich R. pknotsRG: RNA pseudoknot folding including near-optimal structures and sliding windows. Nucleic Acids Res. 2007;35:W320–W324. doi: 10.1093/nar/gkm258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sang R.C., Gichogo A., Gachoya J., Dunster M.D., Ofula V., Hunt A.R., Crabtree M.B., Miller B.R., Dunster L.M. Isolation of a new flavivirus related to cell fusing agent virus (CFAV) from field-collected flood-water Aedes mosquitoes sampled from a dambo in central Kenya. Arch. Virol. 2003;148:1085–1093. doi: 10.1007/s00705-003-0018-8. [DOI] [PubMed] [Google Scholar]
- Simmonds P. Recombination and selection in the evolution of picornaviruses and other mammalian positive-stranded RNA viruses. J. Virol. 2006;80:11124–11140. doi: 10.1128/JVI.01076-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stollar V., Thomas V.L. An agent in the Aedes aegypti cell line (Peleg) which causes fusion of Aedes albopictus cells. Virology. 1975;64:367–377. doi: 10.1016/0042-6822(75)90113-0. [DOI] [PubMed] [Google Scholar]
- Strauss J.H., Strauss E.G. The alphaviruses: gene expression, replication, and evolution. Microbiol. Rev. 1994;58:491–562. doi: 10.1128/mr.58.3.491-562.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taucher C., Berger A., Mandl C.W. A trans-complementing recombination trap demonstrates a low propensity of flaviviruses for intermolecular recombination. J. Virol. 2010;84:599–611. doi: 10.1128/JVI.01063-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchil P.D., Satchidanandam V. Architecture of the flaviviral replication complex. Protease, nuclease, and detergents reveal encasement within double-layered membrane compartments. J. Biol. Chem. 2003;278:24388–24398. doi: 10.1074/jbc.M301717200. [DOI] [PubMed] [Google Scholar]
- Yewdell J.W., Hickman H.D. New lane in the information highway: alternative reading frame peptides elicit T cells with potent antiretrovirus activity. J. Exp. Med. 2007;204:2501–2504. doi: 10.1084/jem.20071986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zdobnov E.M., Apweiler R. InterProScan — an integration platform for the signature-recognition methods in InterPro. Bioinformatics. 2001;17:847–848. doi: 10.1093/bioinformatics/17.9.847. [DOI] [PubMed] [Google Scholar]