Abstract
An IPTG-inducible mutant in the E6R gene of vaccinia virus was used to study the role of the E6 virion core protein in viral replication. In the absence of the inducer, the mutant exhibited a normal pattern DNA replication, concatemer resolution and late gene expression, but it showed an inhibition of virion structural protein processing it failed to produce infectious particles. Electron microscopic analysis showed that in the absence of IPTG viral morphogenesis was arrested before IV formation: crescents, aberrant or empty IV-like structures, and large aggregated virosomes were observed throughout the cytoplasm. The addition of IPTG to release a twelve-hour block showed that virus infectious particles could be formed in the absence of de novo DNA synthesis. Our observations show that in the absence of E6 the association of viroplasm with viral membrane crescents is impaired.
Introduction
Poxviridae comprise a family of viruses that can infect both vertebrates and invertebrates. These viruses are characterized by their large genomic DNA, cytoplasmic site of replication, the encapsidation of an active transcription apparatus, and their complex morphology (Moss, 2007). One of the great challenges in the study of the biology of poxviruses is to understand the morphogenesis steps that culminate with the production of the infectious virus particle (Condit et al., 2006). Electron microscopy studies using vaccinia virus (VACV) as a model have contributed to an understanding of the temporal sequence of events that culminates with the formation of the infectious virion. During the second hour of infection, a cytoplasmic region appears that is depleted of cellular organelles and can be delimited by an ER derived membrane (Dales and Siminovith, 1961; Dales, 1963; Tolonen et al., 2001). This domain, also known as a factory, viral factory or DNA factory, is thought to be the site of viral DNA replication. The first indication of virion formation is the appearance of crescent-shaped membrane structures within the factories (Dales and Mosbach, 1968; Sodeik et al., 1993; Risco et al., 2002). Apparently, the crescent structures grow in length, and shortly afterwards they become closed circles (spheres in three dimensions) named immature virions (IV). During this transformation step, IVs become filled with material, called viroplasm, that is uniform in density but discernibly more electron dense than the surrounding factory. Some IV contain an electron dense, DNA containing “nucleoid” substructure and thus are called immature virions with nucleoid (IVN) (Dales, 1963; Ericsson et al., 1995). Mature virions (MV) appear after a profound transformation of the internal IVN structure to form a biconcave core flanked by lateral bodies, a process that is dependent on proteolysis. During this process MV migrate away from the factories (Katz and Moss, 1970; Ansarah-Sobrinho and Moss, 2004; Byrd and Hruby, 2006).
Although the VACV genome was sequenced almost two decades ago, functional analysis of the poxvirus genome is not complete (Goebel et al., 1990; Condit et al., 2006; Moss, 2007). Temperature sensitive (ts) mutants or inducible knockout viruses have been used to identify genes implicated in the formation of crescents, assembly of IV and the maturation of IV to MV. Inducible knockout and ts viruses often differ mechanistically. In inducible knockout viruses the target gene product is absent under non-permissive whereas in ts mutants the target gene product may be stably synthesized but nevertheless inactive. One might therefore predict that ts and inducible knockout viruses could present two distinct phenotypes when grown under non-permissive conditions. However, for most of the genes studied to date ts and inducible alleles have resulted in a similar phenotypes or phenotypes with only subtle differences (Kane and Shuman, 1993; Szajner et al., 2001a; Byrd et al., 2002; Szajner et al., 2003; Unger and Traktman, 2004; Szajner et al., 2004a; Mercer and Traktman, 2005). In at least one case however, specifically phenotypic analysis of the G7L gene, the phenotype of an inducible knockout mutant was found to be significantly and provocatively different compared to a ts mutant in the same gene (Mercer and Traktman, 2005). Thus, the comparison of viruses bearing a ts mutation or an inducible allele may contribute to a fuller understanding of gene function.
The complete mapping of the Condit/Dales collection of VACV temperature sensitive mutants to single gene resolution mapped three ts mutants to the E6R gene, a virion protein of unknown function (Kato et al., 2008). E6 is conserved in all chordopoxviruses but it has no distinguishing functional motifs and no homologs in non-poxvirus organisms. In the preceding article (Boyd et al., 2010), we showed that a ts mutant in the E6R gene (Cts52) grown under non permissive conditions exhibited normal patterns of gene expression, late protein processing and DNA replication, and produced virions that were indistinguishable in their morphology and protein composition from wt virus. However, purified Cts52 virions grown under non-permissive conditions were non-infectious and failed to transcribe in vitro. Concurrently with the characterization of Cts52, we constructed a mutant virus bearing an inducible E6R gene (vE6i) by substituting the natural E6R promoter with a bacteriophage T7 promoter regulated by the lac operator. We show here that in the presence of the inducer, IPTG, vE6i produced a normal yield of infectious particles but formed smaller than wt plaques. In the absence of IPTG, vE6i displayed a normal pattern of DNA replication and late gene expression, but the processing of the virion proteins was impaired. Resolution of concatemeric viral DNA replication intermediates in the inducible mutant was not affected in the absence of the inducer. Analyses by transmission electron microscopy revealed that under non-permissive conditions morphogenesis was arrested before IV formation, and crescents, aberrant or empty IV-like structures, and dense unencapsidated viroplasm were observed in the cytoplasm. When the inducer block was released at a late time post infection, numerous filled IVs and a few normal MVs could be formed even when the DNA replication inhibitor cytosine arabinoside was added simultaneously with IPTG. Our results suggest that in the absence of E6 the association of viroplasm with viral membrane crescents is defective. In conjunction with studies using ts mutants presented in the accompanying paper, these results suggest that the E6 protein functions at multiple steps during vaccinia morphogenesis.
Results
Construction and characterization of vE6i
To better understand the role of E6R in VACV replication, we engineered a recombinant VACV in which E6R expression was strictly controlled by a bacteriophage T7 promoter and the Escherichia coli lac operator and repressor. The parent virus for the construction was vT7LacOI, a recombinant VACV which constitutively expresses the lac repressor and which also expresses the bacteriophage T7 RNA polymerase under control of a VACV promoter and the lac operator (Ward et al., 1995). The construction was made by substituting the promoter region of the endogenous E6R gene with a T7 promoter-lac operator-EMC IRES cassette coupled to a gpt-cassette regulated by the VACV early-late 7.5-promoter (Fig 1A). Recombinant virus was selected for growth in the presence of mycophenolic acid and IPTG. The presence of the insertion in the recombinant virus was confirmed by sequencing of the E5R-E6R region, and inducer independent virus could be recovered when vE6i was rescued with a PCR product that spanned the E5-E6 region (data not shown).
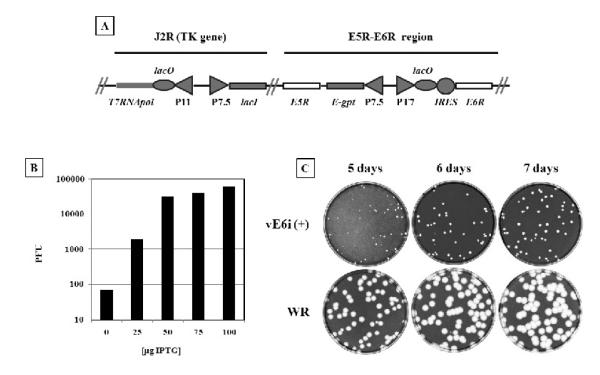
Fig. 1.
Construction of vE6i inducible mutant.
A diagram of the modified regions in vE6i is shown in (A): the upper lines represent the J2R and E5R - E6R regions that were modified. The white rectangles represent the unmodified E5R and E6R ORF. The shaded rectangles represent the DNA insertions encoding for the bacteriophage T7 RNA polymerase (T7RNApol) regulated by the late P11 promoter (P11) and lac operator (lacO); E. coli lac repressor (lacI) regulated by the VACV p7.5 early/late promoter (P7.5); E. coli xanthine-guanine phosphoribosyltransferase (E-gpt) regulated by p7.5. The E6R natural promoter was substituted by a T7 promoter (PT7), lacO, and encephalomyocarditis internal ribosomal entry site (IRES) regulatory sequence. The IPTG requirement for VACV replication and the optimal concentration of the inducer was determined by titering vE6i in the presence of increasing concentration of IPTG. After 72 hours, the cells were stain with crystal violet and the viral plaques counted and graphed (B). To verify is the small plaque phenotype was constant with time the plaque assay was incubated up 7 days before the cells were stained with crystal violet (C).
To determine the IPTG requirement for plaque formation, plaque assays were conducted in the presence of varying concentrations of inducer and the quantification of the assays is shown in Fig. 1B. Virus plaqued in the absence of IPTG contains a low concentration of inducer independent virus, which we interpret as revertants, likely formed by mutational inactivation of the lac repressor in vE6i. Addition of increasing amounts of IPTG to plaque assays reveals inducer-dependent plaque formation with a maximum titer and plaque size achieved at IPTG concentrations of 50 μM and higher. The apparent virus titer increased more than 10 fold by addition of presence of 25μM of IPTG and nearly 1000 fold with higher concentration of the inducer. At concentrations of 50μM of IPTG and above the apparent titers of vE6i are equivalent but the virus formed small plaques regardless of IPTG concentration. All subsequent experiments were carried in the presence of 50μM of IPTG.
To determine whether the small plaque phenotype of vE6i was an effect of the length of the incubation period, BSC40 cells were infected with 100 pfu of either VACV-WR or vE6i virus under permissive conditions and incubated from 5 to 7 days. As shown in Fig. 1C, although plaques produced by both viruses increased in size with time, vE6i plaques were consistently smaller than wt regardless of incubation time. The reasons for the small plaque phenotype of vE6i in the presence of inducer are not clear. It may be a direct consequence of altered E6R expression resulting from manipulation E6R transcriptional regualtion, or it could result from collateral damage acquired during construction of the mutant. Importantly, Resch et al (2009) have also constructed an E6R inducible virus which displays a normal plaque size in the presence of inducer, however that virus was constructed differently from ours. As described above, in our construction the endogenous E6R promoter was substituted with a T7-LacOi promoter region while in the Resch et al virus the endogenous E6R gene was inactivated with a GFP insertion and a new copy of E6R under the control of T7-LacOi promoter was inserted in the A56R gene region. It is possible that the 5′ end modifications made to the E6R gene in our construction affect E6 expression in time or amount in a fashion that results in small plaque size during vE6i infection under permissive conditions. It is also possible that vE6i has sustained some collateral damage resulting (for example) from the expression of the GPT gene that in our construction was placed in opposite direction to the E5R gene, and could therefore theoretically influence E5R expression. Finally, is also possible that in the Resch et al (2009) construction the plaque size would also be small but that the inactivation A56 (HA) permits cell fusion and the appearance of large plaques (Turner and Moyer, 2008), thus masking a small plaque phenotype. Regardless, the fundamental features of our vE6i mutant are similar to the Resch et al mutant, and in our experiments the vE6i + IPTG controls behave as wt viruses in all respects measured aside from plaque size and slightly delayed protein processing.
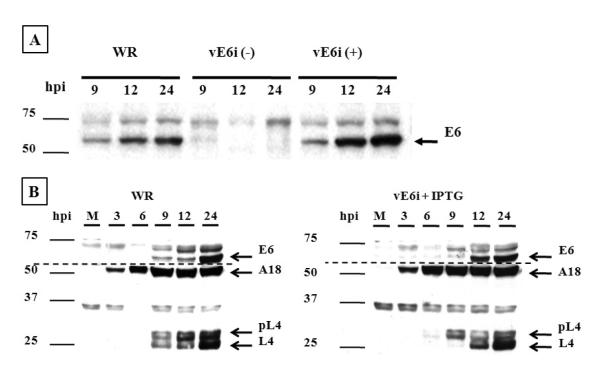
To confirm the lack of E6 synthesis in vE6i infection in the absence of IPTG, BSC40 cells were infected at a MOI of 10 and at varying times the cells were harvested, the proteins separated by SDS-PAGE, transferred to a nitrocellulose membrane and E6 was identified by western blot. In the presence of the inducer no difference in the expression of E6 was observed between WR and vE6i viruses. E6 is first detected at 9 h post infection and the protein continues to accumulate during the infection. However, in the absence of IPTG there was no expression of E6 in the vE6i infection (Fig 2A).
Fig. 2.
Expression and accumulation of E6.
To analyze the accumulation of E6 protein and the expression of VACV early and late genes, BSC40 cells were infected with moi =10 with VACV-WR or vE6i virus in the absence or in the presence of IPTG. At indicated times, cell extracts were prepared and the proteins were separated on 11% SDS-PAGE and transferred to a nitrocellulose membrane. (A) The blot was probed with E6 rabbit polyclonal antibody and developed by chemiluminescence. (B) The blots were probed with E6 and L4 rabbit polyclonal antibodies and A18 mouse monoclonal antibody. The dashed line indicates the separation of two different membranes. The lane (M) represents the mock-infected cells.
The E6R gene is expressed late during the VACV replicative cycle and its synthesis is inhibited by cytosine arabinoside (Assarsson et al., 2008; Boyd et al., 2010). To verify that the temporal expression of VACV genes was not altered in infections by the vE6i recombinant, the synthesis of two other viral proteins were analyzed: A18, which is expressed both early and late during infection (Simpson and Condit, 1994; Assarsson et al., 2008), and L4, which is expressed late during infection as a 28 kDa precursor and processed to a 25 kDa mature form (Yang et al., 1988). As shown in Fig. 2B, during both WR and vE6i infection A18 expression started at 3 h post- infection and the amount of protein increased as the infection proceeded, consistent with the previously described constitutive expression pattern. L4 protein synthesis was also normal in vE6i infections done in the presence of IPTG but processing was slightly delayed: a band corresponding to the precursor L4 protein was initially detected at 9 h post infection but the mature form appears later and the protein accumulated throughout the infection.
One step growth, DNA replication and protein synthesis on vE6i infected cells
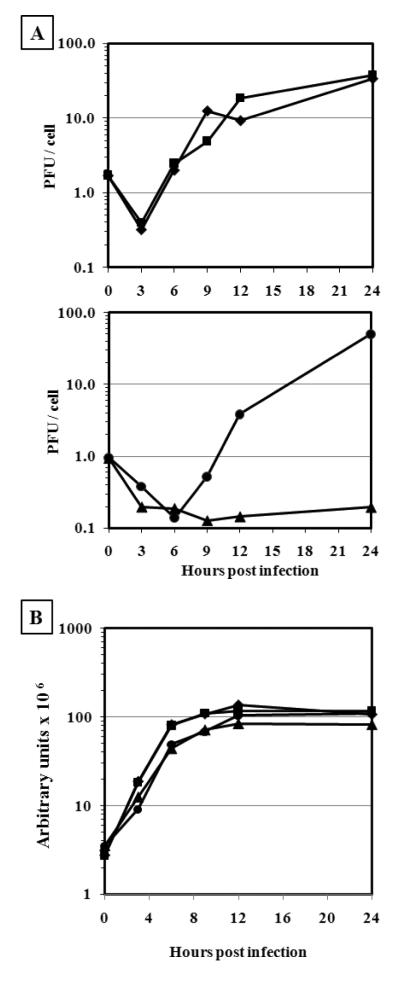
To investigate the different aspects of vE6i replication, we compared with WR infections the production of infectious virus, synthesis of DNA, and protein synthesis in vE6i-infected cells in the absence or presence of inducer. For one-step growth analysis, BSC40 cells were infected at a MOI of 10 with either WR or vE6i in the absence or presence of IPTG, and at different times after infection cells were harvested and the amount of virus in each sample was measured by plaque assay (Fig. 3A). No difference was observed in the total amount of infectious virus formed in WR infection in the absence or presence of IPTG since 34 and 38 pfu/cell respectively were produced after 24 h of infection. Similar results were observed in cells infected with vE6i in the presence of IPTG since 50 pfu/cell was generated in the same period. In the absence of the inducer, the amount of infectious virus formed in vE6i infection declined and after 24h of infection and only 0.2 pfu/cell was recovered. This result confirms that E6 is essential for VACV replication and that the vE6i mutation is suitably tight for further phenotypic characterization.
Fig. 3.
One-step growth and DNA replication.
BSC40 cells were infected at moi = 10 pfu/ml with VACV-WR or vE6i virus in the absence or in the presence of IPTG. At indicated hours post infection samples were removed and the virus yield was determined by plaque assay (A) and the amount of DNA measured by slot blot hybridization (B). WR (−) IPTG (◆); WR (+) IPTG (■); vE6i (−) IPTG (▲);vE6i (+) IPTG (●).
We next investigated whether synthesis of viral DNA and proteins was altered in vE6i infections done under non-permissive conditions. For measuring the accumulation of DNA, BSC40 cells were infected in the absence or presence of the inducer and at varying times the cells were harvested, the cell lysates were applied to a slot-blot membrane, hybridized to a 32P-labeled vaccinia DNA probe, and quantified using a phosphor-imager. As shown in Fig. 3B, both WR and vE6i viruses were able to synthesize DNA to the same extent and with identical kinetics, regardless of the presence or absence of IPTG. This result shows that the inability of vE6i to form infectious virus is not related to the synthesis and accumulation of DNA.
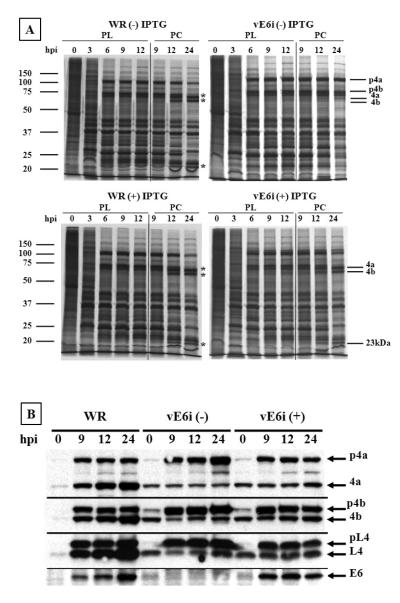
Next, we investigated whether in the absence of IPTG the pattern of viral gene expression and protein processing was altered in vE6i-infected cells. Protein processing is an important step in morphogenesis; inhibition of proteolysis arrests virus morphogenesis at a premature stage and conversely, treatments that interrupt virus morphogenesis at the IV/IVN stage also inhibit proteolysis (Condit et al., 2006). To analyze the synthesis and processing of viral proteins BSC40 cells were infected in the absence or presence of IPTG and at varying times total protein synthesis was evaluated by pulse labeling with 35S-methionine for 30 minutes. After the metabolic labeling, the cells were harvested, the proteins were separated by SDS-PAGE and the radioactively labeled proteins were visualized by autoradiography (Fig. 4A). In the pulse-labeling experiment, a normal pattern of viral protein synthesis was observed in WR infected cells in the absence or presence of IPTG. Soon after the infection, shut-off of host protein synthesis began and early viral proteins were observed at 3 h post-infection. After 6 h of infection a new protein profile composed of late protein synthesis emerged and continued throughout the experiment. In the pulse labeling experiment, vE6i-infected cells in the absence or presence the inducer presented a pattern of protein synthesis that was indistinguishable from WR. To examine the processing of viral proteins, infected cells were labeled with 35S-methionine at 9 h post-infection for 30 minutes and the radioactive proteins were chased in the presence of unlabeled methionine before analysis by SDS-PAGE and detection by autoradiography (Fig. 4A). In the pulse-chase experiment, cells infected with WR virus presented a typical profile of protein processing at 12 and 24 h post infection. The precursor proteins p4a and p4b, with molecular weights of 102 kDa and 73 kDa respectively, present at 9 h post infection were cleaved to a smaller size with molecular weights of 62 kDa and 60 kDa respectively. A 23-kDa peptide that is formed from the processing of p4a can also be observed. A similar profile was observed in vE6i infected-cells in the presence of IPTG where the precursor proteins were cleaved to the same sizes observed in the wt infection. A different picture emerges when vE6i infection is done in the absence of the inducer since in this experimental condition p4a and p4 were not processed to the smaller size. This observation was confirmed by western blot. As shown in Fig. 4B, in cells infected with WR virus the precursor proteins p4a, p4b and pL4 were processed into 4a, 4b and L4 proteins that accumulate during the infection. Protein processing was observed in vE6i-infected cells the presence of IPTG but appeared somewhat reduced in efficiency compared to the wt infection. In the absence of the inducer in vE6i infections, precursor proteins accumulate during infection and the low molecular weight products were not formed. A small amount of the processed proteins was observed in vE6i-infected cells in the absence of IPTG. This likely results from input virions, since it was observed at zero hpi and the quantity was constant during the course of infection. Finally, in the absence of IPTG vE6i-infected cells were unable to synthesize E6, which accumulates in WR infection or in vE6i-infected cells in the presence of the inducer. These results suggest that the absence of E6 interferes directly or indirectly the processing of the major core proteins.
Fig. 4.
Protein synthesis and proteolytic processing of viral proteins.
BSC40 cells were infected at moi = 10 pfu/cell and incubated in the presence or absence of IPTG. (A) Cells were pulse-labeled (PL) with 35S methionine at various times post-infection, indicated in hours at the top of each autoradiogram. The pulse-chase (PC) was performed by incubating the cells with an excess of non-radioactive methionine, for the times indicated at the top of each lane. Proteins were separated on an 11% SDS-PAGE, and gels were dried and exposed to film. The virus and the incubation condition is indicated at the top of each gel. Approximate molecular weights, in kDa, are indicated to the left of each autoradiograms. The major viral core proteins p4a/4a, p4b/4b and 23 kDa are indicated on the right of the autoradiograms. (B) Western blot analysis was performed on infected cell extracts prepared as described above. The cells were harvested, a cell extract was prepared, and the proteins were separated on an 11% SDS-PAGE, transferred to a nitrocellulose membrane, and blotted against specific vaccinia virus proteins, as indicated on the right side of the figure.
No mature virions are formed in the absence of E6
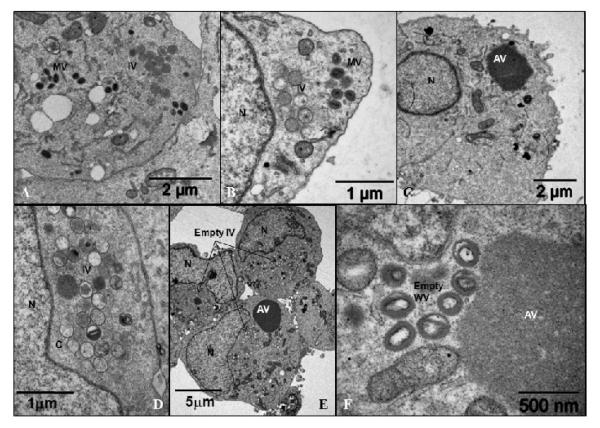
As shown above, in the absence of E6R expression the proteolytic processing of VACV proteins was inhibited. Defects in proteolysis of this sort are often diagnostic of a defect in virion morphogenesis (Szajner et al., 2003; Unger and Traktman, 2004; Szajner et al., 2004a; Mercer and Traktman, 2005; Byrd and Hruby, 2005; Condit et al., 2006). To determine whether virus morphogenesis was compromised BSC40 cells were infected with vE6i in the absence or presence of the inducer. At 24 or 48 h post infection, cells were harvested and prepared for electron microscopic analysis (Fig. 5). In the presence of IPTG, normal looking IV and MV were formed in the cell cytoplasm at 24 (A) and 48 (B) h post infection. A different picture emerged when vE6i was grown in the absence of IPTG. After 24 (C-D) or 48 (E-F) h post infection large aggregated virosomes (AV) were formed, and crescents and empty IV were found in the cytoplasm. A few IV and IVN were also present. Structures that resemble a MV could be found but they were devoid of the core and lateral bodies (Fig. 5F). These results suggest that in the absence of E6 virions were not able to efficiently encapsidate viroplasm and DNA.
Fig. 5.
Electron micrograph of the vE6i infected cells.
BSC40 cells were infected with vE6i in the presence of IPTG (A, B) or in the absence of the inducer (C – F). After 24 hpi (A – D) or 48 hpi (E – F) the cells were processed for microscopy as described in Methods. IV= immature virions; MV= mature virions; WV= wrapped virions; C= crescents; AV = aggregated virosome; N=nucleus.
Viral DNA processing is not inhibited in the absence of E6
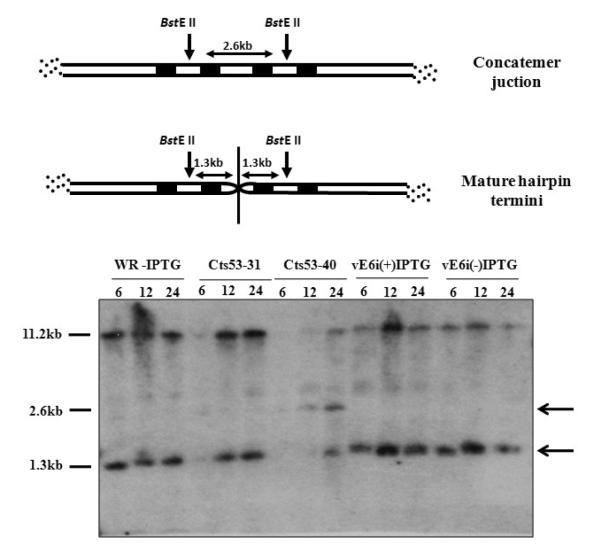
During VACV genome replication, DNA concatemers are formed and resolved into monomers before being encapsidated into mature virions (Moyer and Graves, 1981). This phenomenon requires expression of late genes and complete morphogenesis does not occur if the viral resolvase gene (A22R) is not active (Merchlinsky and Moss, 1989; Garcia and Moss, 2001). In the absence of A22, both concatemer resolution and maturation beyond IVN formation are blocked. The ability of vE6i to resolve DNA concatemers was analyzed by Southern blot as described (Merchlinsky and Moss, 1989). DNA isolated from virus-infected cells was digested with BstEII, the fragments separated in a 0.8% agarose gel, transferred to a nylon membrane and hybridized to a VACV DNA probe containing the genomic terminal sequences. The digestion of mature monomeric DNA with BstEII results in a 1.3-kb terminal fragment, whereas DNA in the concatemeric form yields a 2.6-kb junction fragment (Fig. 6 scheme on top). Our results show that in wt-infected cells (WR-IPTG) viral concatemer junctions were completely resolved since only the 1.3-kb fragment was observed (Fig .6). Cts53, a mutant in rpo147 (J6R) which is defective in late protein synthesis and telomere resolution at non-permissive temperature, was used as a control and as a marker for the 2.6-kb concatemer junction fragment (Hooda-Dhingra et al., 1989; Merchlinsky and Moss, 1989). At the permissive temperature in a Cts53 infection (Cts53-31), the 1.3-kb resolved fragment is formed while at non-permissive temperature (Cts53-40) the 2.6-kb concatemer junction fragment accumulates, as expected. In vE6i-infected cells, in the absence or presence of IPTG, only the 1.3-kb fragment is formed. This result shows that E6 is not involved in the formation of a monomeric mature viral DNA molecule.
Fig. 6.
Telomere resolution.
A diagram showing relevant features of the viral genome concatemer junction and mature DNA hairpin ends is shown in the top of the figure. Shaded areas represent direct repeats within the viral termini. The arrows represent the site of cleavage of the BstE II restriction enzyme and the double arrows represent the size of the fragment that is generated in both resolved and non-resolved condition. The autoradiogram of the Southern blot is shown in the bottom part of the figure. BSC40 cells were infected with WR or mutant virus at moi = 10 pfu/cell and incubated in absence or presence of IPTG. Cts53 virus, used a positive control, was grown at 31 and 40°C. DNA was extracted at various times post-infection, indicated in h at the top of each lane, cleaved with BstE II, the fragments separated in an 0.8% agarose gel, blotted, and probed with a 32P-labeled DNA containing concatemer junction sequences. The 2.6- and 1.3-kb bands, indicated to the right of the autoradiogram, represent unresolved and resolved end fragments, respectively. The slower migrating bands represent internal fragments hybridized to the probe due to the presence of tandem repeat sequences.
Induction of E6 during a non-permissive infection restores the ability of vE6i to produce infectious particles
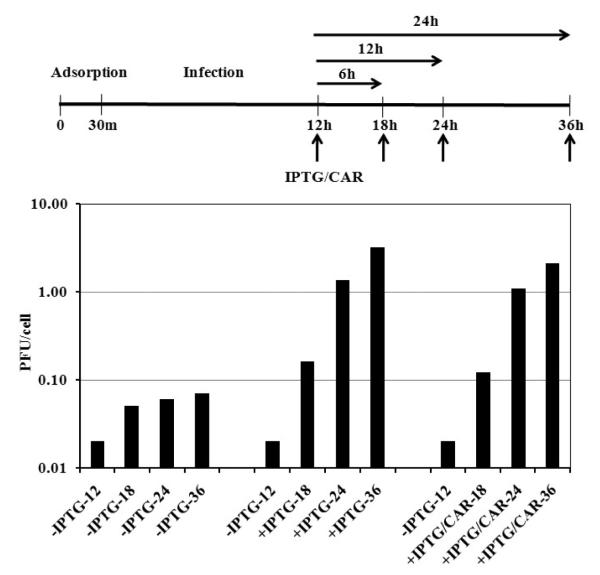
As described above, in the absence of the inducer, vE6i was not able to process the virion core proteins and virus morphogenesis was arrested at an early stage of virion assembly characterized by formation of empty IVs and aggregated virosomes. We were interested to know whether the accumulated structures were viable for continued morphogenesis or were dead-end products of an aborted assembly process. Thus to determine whether mature virions could be formed late in a non-permissive vE6i infection following IPTG induction of E6 expression, BSC40 cells were infected with vE6i in the absence of IPTG. Twelve hours later, IPTG was added to the infected cells in the absence or presence 40 μg/ml of cytosine arabinoside (CAR), an inhibitor of DNA synthesis. At 6, 12 and 24 h after the addition of the drugs (18, 24 and 36 hpi) samples were removed and the viral titer was determined. In the absence of the inducer, vE6i was not able to form infectious particles and the virus titer slightly increased from 0.02 (12 h) to 0.07 (36 h) pfu/cell (Fig. 7). By contrast, as early as six hours after the addition of IPTG, the virus titer increased from 0.02 to 0.16 pfu/cell and after 24 and 36 h, the virus titer increased to 1.37 and 3.2pfu/cell respectively. A similar result was observed when CAR and IPTG were added to the infected cells simultaneously. Within 6 h after addition of the drugs, the virus titer increased from 0.02 to 0.12 pfu/cell and after 24 and 36 h of incubation the virus titer increased to 1.11 and 2.13pfu/cell.
Fig. 7.
Release of the IPTG blockage.
A diagram on the top shows the scheme for the procedure. The vertical arrows represent the hpi where samples were collected for plaque assay. The horizontal arrows represent the hours of the blockage release. The result of the plaque assay is presented in the bottom part of the figure. BSC40 cells were infected with vE6i at moi = 10 pfu/cell and incubated in absence IPTG. At 12 hpi, IPTG or IPTG + CAR in fresh media were added to the cells. Cells that did not receive the drugs were also supplemented with fresh media. At the indicated hpi, samples were removed and the virus yield was measured by plaque titration.
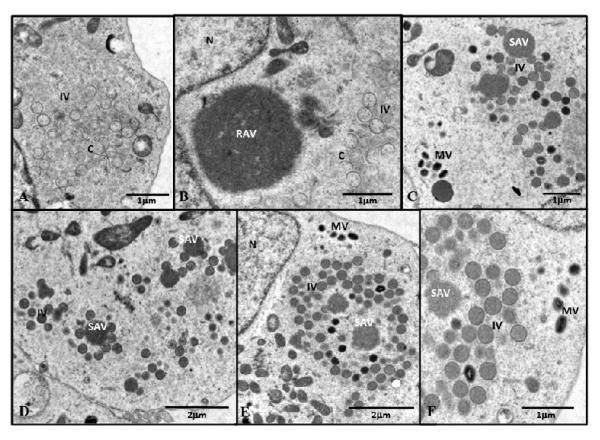
The formation of virus was also followed by electron microscopy (Fig. 8 A-F). After 12 h of vE6i infection in the absence of IPTG crescents, empty IV and aggregated virosomes were found in the cytoplasm of infected cells (data not shown). When the infection in the absence of IPTG was extended to 36 h, a similar picture was observed with morphogenesis was still arrested so that crescents and empty IV accumulated in the cytoplasm (A-B). Furthermore, “rough aggregated virosomes” (RAV) were observed that were not associated with any viral membrane structures, either rigid crescents or flaccid membranes, as have been observed after rifampicin treatment (Grimley et al., 1970). When the E6 expression block was released by the addition of IPTG and the infection was extended for an additional 24 h (36 hpi), all the steps in virus assembly could be identified and normal looking IV, dense IV and a few MV were present throughout the cytoplasm (C-D). The rough aggregated virosomes apparently became smoother (SAV) and were surrounded by crescents that were filled with viroplasm. A similar picture was observed with vE6i-infected cells when the E6 block was released by addition of IPTG in the presence of CAR and the infection extended for 24 h. As in the absence of CAR, crescents filled with viroplasm surrounding smooth aggregated virosomes, and normal looking IV, dense IV and a few MV were present throughout the cytoplasm. We note that while induction of E6 synthesis late during infection results in appearance of numerous normal looking IV, the numbers of MV as measured by electron microscopy or plaque titration, while significantly increased, are relatively few compared to a normal wt infection. Our results suggest that upon the release of block of E6R gene and the synthesis of E6 protein the virus was able to resume morphogenesis and efficiently package viroplasm into membranes, and that a few of these new IV may morph further to form new infectious virions, even in the absence of de novo DNA synthesis.
Fig. 8.
Electron micrograph after the IPTG blockage release.
BSC40 cells were infected with vE6i at moi = 10 pfu/cell and incubated in absence IPTG. At 12 hpi, fresh media with no drugs (A, B) or containing IPTG (C, D) or IPTG + CAR (E, F) were added to the cells. After 24h of the blockage release (36 hpi), the cells were processed for microscopy as described in Methods. EIV = empty immature virions; IV= immature virions; MV= mature virions; WV= wrapped virions; C= crescents; RAV = rough aggregated virosome; SAV = smooth aggregated virosome; N=nucleus.
E6 protein is unstable in the absence of any one of the 7-protein complex proteins
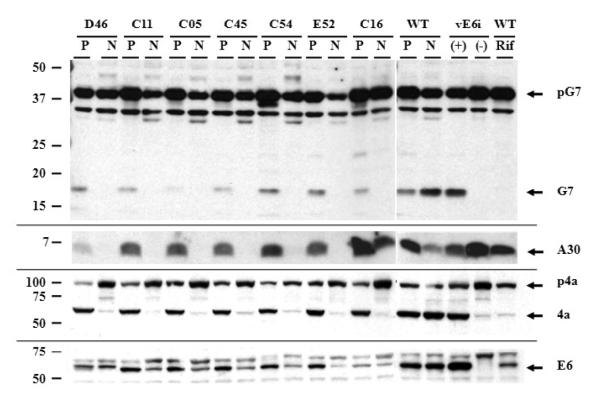
VACV proteins F10, A30, G7, J1, D2, and D3 form a 7-protein complex (7PC) that is responsible for the association of membrane crescent structures with viroplasm (Szajner et al., 2004a). In the absence of any one of these proteins, morphogenesis is arrested at an early stage, empty crescents and aggregated virosomes are formed, and the other six proteins associated with the complex become unstable (Dyster and Niles, 1991; Szajner et al., 2001a; Szajner et al., 2001b; Szajner et al., 2003; Szajner et al., 2004a; Szajner et al., 2004b; Szajner et al., 2004c; Mercer and Traktman, 2005). Since a similar picture of virus morphogenesis was observed with vE6i in the absence of the inducer, we decided to compare the fate of two proteins from the 7PC during vE6i infection. BSC40 cells were infected with vE6i and the following ts mutants from the 7PC: Dts46 (A30), Cts11 (G7), Cts5 (D3), Cts45 (J1), Cts54 (F10) and Ets52 (D2). Cts16, a mutant from the viral protease I7, was included as a negative control. Infected cells were incubated for 24 h under permissive conditions (31°C or + IPTG) or non-permissive conditions (40°C, or −IPTG or + rifampicin). Infected cells were then harvested, the proteins were separated by SDS-PAGE, transferred to a nitrocellulose membrane, and the western blots were probed with antibodies to the G7, A30, A10 and E6 proteins (Fig. 9). In all samples from cells infected with the mutants under permissive conditions (P), the 42 kDa precursor protein G7 (pG7) was processed into a 15 kDa protein (G7), similar to the wt infection. Under non-permissive conditions (N), the accumulation of pG7 was reduced in all the 7PC mutants and no G7 was produced by proteolysis. A different result was obtained in cells infected with the mutant viruses Cts16, vE6i or with wt virus in the presence of rifampicin. Under these conditions, there was a slight increase on the accumulation of pG7 and the mature G7 protein was not formed. A similar result was obtained when the expression of A30 and A10 (p4a) were analyzed. Under permissive conditions, the 7PC mutants were able to express the proteins and p4a was processed into 4a. However, under non-permissive conditions, the accumulation of A30 and the processing of A10 were severely repressed. In cells infected with Cts16, vE6i or wt under non-permissive conditions the expression of A30 was not repressed but there was a significant inhibition of the processing of the p4a protein. The fate of E6 protein was also analyzed in cells infected under permissive and non-permissive conditions. In cells infected with the 7PC-mutants at 40°C, we observed a decrease in the accumulation of E6 when compared with virus grown at 31°C, suggesting that E6 was unstable. In Cts16 infected cells at 40°C or in wt infection in the presence of rifampicin, the accumulation of E6 was reduced. No E6 expression was observed in vE6i-infected cells in the absence of IPTG. (Note that E6 made in the Dts46 infection has a slower mobility compared to E6 made in other infections. We believe this results from the fact that Dts 46 is derived from the IHD-W strain of VACV while the other viruses were derived from the WR VACV strain. DNA sequencing (not shown) demonstrates that IHD-W contains two E6R polymorphisms compared to the WR strain, T102A and K124E, which could theoretically retard the migration of the IHD-W protein.) Taken together, these results show that the behavior E6 is different from the proteins from the7PC, suggesting that E6 has a role in virus morphogenesis that is distinct from the 7PC.
Fig. 9.
Stability of E6 in cells infected with mutants of the seven-protein complex.
BSC40 cells were infected with VACV mutants of the seven-protein complex (7PC) at moi = 10 pfu/cell and incubated 31°C (P) or 40°C (N) for 24 hours. After this period cell extracts were prepared, proteins separated in a 11% SDS-PAGE, transferred to a nitrocellulose membrane and blotted using rabbit antiserum against the VACV proteins G7, A30, A10 (p4a/4a) and E6. A 15% SDS-PAGE was used for the analysis of the A30 protein. The mutant viruses indicated in the lanes correspond to following genes: D46 (A30), C11 (G7), C05 (D3), C45 (J1), C54 (F10), E52 (D2), and C16 (I7). vE6i was incubated in the absence (−) or in the presence (+) of IPTG. The arrows in the right of the figure indicate the VACV precursor (p) or the protein. The numbers in the left of the figure represents the molecular weight markers in kDa.
Discussion
Genetic analysis of poxvirus morphogenesis reveals four distinct stages of virion assembly: 1) membrane crescent formation, 2) association of viroplasm with membrane crescents to form IV, 3) DNA packaging to form IVN and 4) morphogenesis of IVN to MV (Condit et al., 2006). Phenotypically, mutants in any stage of virion morphogenesis show no apparent defects in late virus protein synthesis, so that theoretically the full complement of structural proteins and assembly enzymes (save the mutant protein) are present. Transition from stage 3, IVN, to stage 4, MV, is accompanied by and in fact requires cleavage of major structural precursor proteins, notably the major core proteins p4a and p4b, and several others. The results presented here demonstrate that when the virion core protein E6 is absent from infection, vaccinia morphogenesis is arrested at stage 2, IV formation via association of viroplasm with membrane crescents. Specifically, in the absence of E6R gene expression, late viral proteins are synthesized and viral DNA is replicated and processed normally, but precursor structural proteins are not processed, normal IV, IVN and MV are rare or absent, and instead infected cells accumulate two significant aberrant structures, empty IV and aggregated virosomes. Empty IV consist of membrane crescents and spheres that are devoid of the normal complement of intermediate density viroplasm, and aggregated virosomes appear as very large accretions of material resembling the intermediate density viroplasm that is normally packaged into IV. A similar result has been observed recently by Moss and co-workers with an independently constructed inducible mutant in the E6R gene (Resch et al., 2009). The simplest interpretation of these results is that the E6 protein is required for the normal association of viroplasm with viral membranes.
Mutants in several other virion proteins yield a phenotype virtually identical to the vE6i phenotype observed here. These include mutants in genes A15L, A30L, D2L, D3R, G7L and J1R (Szajner et al., 2001a; Szajner et al., 2001b; Chiu and Chang, 2002; Szajner et al., 2003; Szajner et al., 2004a; Szajner et al., 2004b; Mercer and Traktman, 2005; Chiu et al., 2005). Interestingly, these six proteins form a complex, dubbed the “seven protein complex” (7PC), along with the product of the F10R gene, a viral serine/threonine protein kinase (Szajner et al., 2004a; Resch et al., 2005). In contrast to other proteins in the complex, mutation of the F10R gene yields a very early morphogenesis phenotype characterized by the complete absence of crescents or other virion precursors, suggesting a pivotal early regulatory role for F10 in virion morphogenesis (Traktman et al., 1995; Wang and Shuman, 1995; Szajner et al., 2004b). Importantly, repression of synthesis of any of the proteins in the 7PC results in destabilization of the remaining proteins in the complex (Szajner et al., 2004a). We therefore considered the hypothesis that the vE6i phenotype was in fact a pleiotropic effect of destabilization of the 7PC rather than a true reflection of E6 action. However, we found that two members of the 7PC, A30 and pG7, remained stable during vE6i infections. Since repression of any one of the 7PC complex proteins results in destabilization of the others, this result implies that the entire 7PC complex remains stable during a vE6i infection. Interestingly however, we also found that the E6 protein was destabilized during infection with 7PC mutants. One provocative interpretation of this result is that in fact the E6 protein is the pivotal player in the 7PC/E6R mutant phenotype and that the common phenotype of 7PC complex mutants reflects the destabilization of the E6 protein. In any case, our results show that E6R acts on the same pathway as the 7PC, promoting the association of viroplasm with viral membrane crescents.
Structures resembling aggregated virosomes are frequently observed in mutant infections in which early stages of assembly, specifically any step preceding formation of IVN, is disrupted. For example mutants in the virion membrane proteins A14 or A17 or in gene A11R disrupt normal membrane crescent formation and at the same time cause abnormal accumulation of aggregated virosomes strikingly similar to the structures formed in a vE6i infection (Rodriguez et al., 1995; Wolffe et al., 1996; Rodriguez et al., 1998; Resch et al., 2005). Likewise, as detailed above, mutants in any of the 7PC complex proteins, like the vE6i mutants, result in accumulation of aggregated virosomes. Lastly, mutants in several genes thought to interrupt DNA packaging accumulate not only aggregated virosomes but also sometimes “crystalloid” structures thought to represent deposits of unpackaged viral DNA (DeMasi et al., 2001; Garcia and Moss, 2001; Grubisha and Traktman, 2003; Unger and Traktman, 2004). Traktman et al. (2000) have demonstrated that the aggregated virosomes that accumulate when membrane crescent formation is prevented by repression of synthesis of the A14 viral membrane protein contain concentrations of two DNA binding proteins, L4 and F17, that are normally found in both IV and MV. These studies together suggest that the initial steps in vaccinia assembly can be segregated into three subassembly processes: membrane crescent formation, assembly of viroplasm, and DNA packaging. Under normal conditions these processes are coordinated, however interruption of any one process may cause abnormal accumulation of other sub-assemblies. Thus disruption of association of viroplasm with viral membrane crescents, as observed in vE6i mutant infections, results in accumulation of both membrane crescent and viroplasm sub-assemblies.
The intermediates that accumulate in vE6i infections could represent normal intermediates in virus assembly or they could be dead-end products of an abortive assembly process. To discriminate between these two hypotheses we conducted experiments to reverse the block in E6 expression in the presence or absence of a DNA replication inhibitor. We found that even in the absence of DNA replication, induction of E6 resulted in the resumption of morphogenesis, efficient packaging of viroplasm into membranes to form IV, and the assembly of some mature infectious particles. Following induction we observed by electron microscopy viral membrane crescents associated with the large aggregated virosomes, apparently “biting off” portions of viroplasm for packaging into virions. This implies that the viroplasm in aggregated virosomes represents an accretion of normal, functional viroplasm competent for packaging and subsequent morphogenesis. Likewise, it seems likely that the crescents and empty IV that accumulate under non-permissive conditions during a vE6i infection are recycled for use in assembly rather than disassembled and reassembled de novo. Preliminary confocal microscopy studies (not shown) suggest that viral DNA is not concentrated in aggregated virosomes but remains distributed normally throughout factories in a vE6i infection under non-permissive conditions. Thus, repression of E6R gene function results in accumulation of functionally competent intermediates in virus assembly, which may be used to resume assembly upon induction of E6 synthesis. The resumption of assembly results in efficient IV formation. MV formation is observed but is relatively inefficient. It remains to be determined whether the apparent inefficiency of MV formation represents a downstream block to morphogenesis or is a consequence of overall stress to the infection imposed by the experimental protocol.
In the accompanying manuscript we described three temperature sensitive (ts) alleles of the E6 gene which had identical phenotypes and which differed dramatically from vE6i (Boyd et al., 2010). Under non-permissive conditions, association of viroplasm with membrane crescents occurred normally during infection with all of the ts mutants, and MV were formed which were indistinguishable from wt virus in appearance but which were nevertheless non-infectious. Characterization of one of these ts alleles, Cts52, demonstrated that the non-infectious particles formed under non-permissive conditions could bind and enter cells, but that the virions were defective in virus core particle directed transcription in vitro. The dramatically different phenotypes of the ts and inducible E6R mutants most likely relates to the mechanism of conditional lethality in each case. Specifically, during infection with vE6i under non-permissive conditions E6 protein is absent whereas in ts mutant infections E6 protein is present but not functional. This suggests that the E6 protein functions in at least two steps during virus morphogenesis, first for association of viroplasm with crescents and secondly for some subtle aspect of MV formation that impacts either directly or indirectly on the virion transcription reaction. In the absence of E6 protein as in vE6i infections, association of membrane crescents with viroplasm cannot occur, however if the E6 polypeptide is present but functionally compromised as in ts mutant infections, the membrane association with viroplasm and appears normal, but some downstream event required for function of morphologically normal MV is defective. Numerous other examples exist of vaccinia genes in which both ts and inducible alleles have been studied and in the majority of these cases the phenotypes of the two types of mutation are similar or identical (Condit et al., 2006). However in the case of gene G7L, which encodes one of the 7PC proteins, a ts allele caused a defect in membrane crescent formation while repression of G7 synthesis resulted in a defect in association of morphologically normal membrane crescents with viroplasm (Mercer and Traktman, 2005). The authors in this case also interpreted this result to mean that G7 functioned in multiple steps in virion morphogenesis. Thus comparison of ts and inducible alleles of a given gene can reveal subtleties of gene function not gleaned from study of either mutant type alone. The precise role of E6 in association of membrane crescents with viroplasm and the formation of transcriptionally competent MV, and the potential relationship between the two processes, remains to be determined.
Materials and Methods
Cell culture, virus, plaque assay, one-step growth
Viruses were grown in BSC-40 cells, an African green monkey cell line. The procedures for cell culture, virus infection, purification, plaque assay and one-step growth have been described previously (Condit and Motyczka, 1981; Condit et al., 1983; Kato et al., 2004). Wildtype vaccinia virus strain WR was used as a control for the studies. For the construction of vE6i the E6 ORF was PCR amplified and inserted into pVOTE.1 (Ward et al., 1995) to create pE6 with E6 downstream from the T7 promoter and lac operator, and adjacent to a P7.5-gpt cassette. The E5 flank was synthesized by PCR, and joined to a restriction fragment from pE6 consisting of the P7.5-gpt cassette and the pT7-lacO-E6. The resulting linear DNA consisted of the pT7-lacO promoter/operator and the P7.5-gpt cassette flanked by the E5 and E6 flanks, and was recombined into vT7lacOI (Alexander et al., 1992) by selection for mycophenolic acid resistant plaques in the presence of IPTG. The resulting construct was verified by sequencing to confirm that unintended mutations had not been introduced into the E5 or E6 genes. Temperature sensitive viruses Cts53 (J6R), Dts46 (A30R), Cts11 (G7L), Cts5 (D3R), Ct 45 (J1R), Cts 54 (F10L), Ets52 (D2L), Cts16 (I7L) were from our ts mutant collection and have been described elsewhere (Hooda-Dhingra et al., 1989; Dyster and Niles, 1991; Traktman et al., 1995; Szajner et al., 2001a; Byrd et al., 2002; Mercer and Traktman, 2005; Chiu et al., 2005).
Analyses of VV DNA slot-blot hybridization
BSC-40 cells were infected with VACV at moi of 10 pfu/cell and at different times post-infection cell extracts were prepared and processed as described previously (Damaso et al., 2002). The blots were analyzed on a Storm 860 phosphorimager (GE-Healthcare) using Image Quant 5.1 software (Molecular Dynamics).
Metabolic labeling of viral proteins
Infected BSC-40 cells were incubated with 100 μCi/ ml of 35S-labeled amino acids mixture (ProMix – GE Healthcare) in a methionine free media for 30 minutes. After this period, a cell extract was prepared, proteins separated in a sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE), and analyzed by autoradiography as described before (Kato et al., 2004).
Western blot analysis
BSC-40 cells were infected with wt VACV or vE6i in the absence or presence of 50μM of IPTG. At varying times post-infection the cells were harvested and western blots were prepared and analyzed as described previously (Kato et al., 2007). The E6 antibody has been described previously and was used at a dilution of 1:1,000 (Boyd et al., 2010). The dilution and source of the other antibodies were: anti-A18 (1:20,000) monoclonal (Black et al., 1998); anti-L4 (1:100,000) rabbit serum was supplied by Dr. Dennis Hruby (Oregon State University); anti-H4 (1:5,000) and anti-A3 (1:30,000) rabbit serum was supplied by Dr. Bernard Moss (NIAID); anti-A10 (1:30,000) rabbit serum were supplied by Dr. Mariano Esteban (CNB-CSIC, Spain), anti-G7 (1: 10,000) and anti-A30 (1:5,000) rabbit sera were supplied by Dr. Paula Traktman (Wisconsin Medical School)
Electron microscopy
BSC40 cells were infected with either wt VACV or vE6i at a moi of 10 in the absence or presence 50 μM of IPTG. Samples from 24 to 48 post-infection were fixed with 2% glutaraldehyde in 0.1 M cacodylate buffer and post-fixed with 1% osmium tetroxide. After dehydration, the samples were embedded in Embed 812 resin and visualized in a Hitachi H-7000 electron microscope with assistance of the University of Florida ICBR Electron Microscopy & Bioimaging Core Laboratory.
Telomere Resolution
The telomere resolution assay was done as described previously (Sekiguchi and Shuman, 1997). Briefly, monolayers of BSC40 cells infected with wt, Cts 53 or vE6i in the absence or presence of 50 μM IPTG, at a moi of 10. The cells were harvested after 6, 12 and 24 hours post infection, DNA was extracted, cleaved with BstE II (New England BioLabs), and fractionated in a 1% agarose gel. Southern blots were prepared and probed with 32P-labeled pBD6 (Merchlinsky et al., 1988) as described (Sekiguchi and Shuman, 1997).
Acknowledgments
We thank Bernard Moss, Paula Traktman, Mariano Esteban and Dennis Hruby for antisera; Karen Kelley and the University of Florida ICBR Electron Microscopy Core Laboratory for excellent advice and technical assistance, and Nicole Kay for technical support. This work was supported by NIH grant R01 AI055560 to RCC.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference List
- Alexander WA, Moss B, Fuerst TR. Regulated expression of foreign genes in vaccinia virus under the control of bacteriophage T7 RNA polymerase and the Escherichia coli lac repressor. J.Virol. 1992;66:2934–2942. doi: 10.1128/jvi.66.5.2934-2942.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ansarah-Sobrinho C, Moss B. Role of the I7 protein in proteolytic processing of vaccinia virus membrane and core components. J.Virol. 2004;78:6335–6343. doi: 10.1128/JVI.78.12.6335-6343.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assarsson E, Greenbaum JA, Sundstrom M, Schaffer L, Hammond JA, Pasquetto V, Oseroff C, Hendrickson RC, Lefkowitz EJ, Tscharke DC, Sidney J, Grey HM, Head SR, Peters B, Sette A. Kinetic analysis of a complete poxvirus transcriptome reveals an immediate-early class of genes. Proc.Natl.Acad.Sci.U.S.A. 2008;105:2140–2145. doi: 10.1073/pnas.0711573105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black EP, Moussatche N, Condit RC. Characterization of the interactions among vaccinia virus transcription factors G2R, A18R, and H5R. Virology. 1998;245:313–322. doi: 10.1006/viro.1998.9166. [DOI] [PubMed] [Google Scholar]
- Boyd O, Strahl AL, Rodeffer C, Condit RC, Moussatche N. Temperature-sensitive mutant in the vaccinia virus E6 protein produce virions that are transcriptionally inactive. Virology. 2010 doi: 10.1016/j.virol.2010.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrd CM, Bolken TC, Hruby DE. The vaccinia virus I7L gene product is the core protein proteinase. J.Virol. 2002;76:8973–8976. doi: 10.1128/JVI.76.17.8973-8976.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrd CM, Hruby DE. A conditional-lethal vaccinia virus mutant demonstrates that the I7L gene product is required for virion morphogenesis. Virol.J. 2005;2:4. doi: 10.1186/1743-422X-2-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrd CM, Hruby DE. Vaccinia virus proteolysis--a review. Rev.Med.Virol. 2006;16:187–202. doi: 10.1002/rmv.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu WL, Chang W. Vaccinia virus J1R protein: a viral membrane protein that is essential for virion morphogenesis. J.Virol. 2002;76:9575–9587. doi: 10.1128/JVI.76.19.9575-9587.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu WL, Szajner P, Moss B, Chang W. Effects of a temperature sensitivity mutation in the J1R protein component of a complex required for vaccinia virus assembly. J.Virol. 2005;79:8046–8056. doi: 10.1128/JVI.79.13.8046-8056.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Condit RC, Motyczka A. Isolation and preliminary characterization of temperature-sensitive mutants of vaccinia virus. Virology. 1981;113:224–241. doi: 10.1016/0042-6822(81)90150-1. [DOI] [PubMed] [Google Scholar]
- Condit RC, Motyczka A, Spizz G. Isolation, characterization, and physical mapping of temperature-sensitive mutants of vaccinia virus. Virology. 1983;128:429–443. doi: 10.1016/0042-6822(83)90268-4. [DOI] [PubMed] [Google Scholar]
- Condit RC, Moussatche N, Traktman P. In a nutshell: structure and assembly of the vaccinia virion. Adv.Virus Res. 2006;66:31–124. doi: 10.1016/S0065-3527(06)66002-8. [DOI] [PubMed] [Google Scholar]
- Dales S. The uptake and development of vaccinia virus in strain L cells followed with labeled viral deoxyribonucleic acid. J.Cell Biol. 1963;18:51–72. doi: 10.1083/jcb.18.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dales S, Mosbach EH. Vaccinia as a model for membrane biogenesis. Virology. 1968;35:564–583. doi: 10.1016/0042-6822(68)90286-9. [DOI] [PubMed] [Google Scholar]
- Dales S, Siminovith L. The development of vaccinia virus in Earle’s L strain cells as examined by electron microscopy. J.Biophys.Biochem.Cytol. 1961;10:475–503. doi: 10.1083/jcb.10.4.475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damaso CR, Oliveira MF, Massarani SM, Moussatche N. Azathioprine inhibits vaccinia virus replication in both BSC-40 and RAG cell lines acting on different stages of virus cycle. Virology. 2002;300:79–91. doi: 10.1006/viro.2002.1534. [DOI] [PubMed] [Google Scholar]
- DeMasi J, Du S, Lennon D, Traktman P. Vaccinia virus telomeres: interaction with the viral I1, I6, and K4 proteins. J.Virol. 2001;75:10090–10105. doi: 10.1128/JVI.75.21.10090-10105.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyster LM, Niles EG. Genetic and biochemical characterization of vaccinia virus genes D2L and D3R which encode virion structural proteins. Virology. 1991;182:455–467. doi: 10.1016/0042-6822(91)90586-z. [DOI] [PubMed] [Google Scholar]
- Ericsson M, Cudmore S, Shuman S, Condit RC, Griffiths G, Locker JK. Characterization of ts 16, a temperature-sensitive mutant of vaccinia virus. J.Virol. 1995;69:7072–7086. doi: 10.1128/jvi.69.11.7072-7086.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia AD, Moss B. Repression of vaccinia virus Holliday junction resolvase inhibits processing of viral DNA into unit-length genomes. J.Virol. 2001;75:6460–6471. doi: 10.1128/JVI.75.14.6460-6471.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goebel SJ, Johnson GP, Perkus ME, Davis SW, Winslow JP, Paoletti E. The complete DNA sequence of vaccinia virus. Virology. 1990;179:247–263. doi: 10.1016/0042-6822(90)90294-2. [DOI] [PubMed] [Google Scholar]
- Grimley PM, Rosenblum EN, Mims SJ, Moss B. Interruption by Rifampin of an early stage in vaccinia virus morphogenesis: accumulation of membranes which are precursors of virus envelopes. J.Virol. 1970;6:519–533. doi: 10.1128/jvi.6.4.519-533.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grubisha O, Traktman P. Genetic analysis of the vaccinia virus I6 telomere-binding protein uncovers a key role in genome encapsidation. J.Virol. 2003;77:10929–10942. doi: 10.1128/JVI.77.20.10929-10942.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooda-Dhingra U, Thompson CL, Condit RC. Detailed phenotypic characterization of five temperature-sensitive mutants in the 22- and 147-kilodalton subunits of vaccinia virus DNA-dependent RNA polymerase. J.Virol. 1989;63:714–729. doi: 10.1128/jvi.63.2.714-729.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kane EM, Shuman S. Vaccinia virus morphogenesis is blocked by a temperature-sensitive mutation in the I7 gene that encodes a virion component. J.Virol. 1993;67:2689–2698. doi: 10.1128/jvi.67.5.2689-2698.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato SE, Condit RC, Moussatche N. The vaccinia virus E8R gene product is required for formation of transcriptionally active virions. Virology. 2007;367:398–412. doi: 10.1016/j.virol.2007.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato SE, Moussatche N, D’Costa SM, Bainbridge TW, Prins C, Strahl AL, Shatzer AN, Brinker AJ, Kay NE, Condit RC. Marker rescue mapping of the combined Condit/Dales collection of temperature-sensitive vaccinia virus mutants. Virology. 2008;375:213–222. doi: 10.1016/j.virol.2008.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato SE, Strahl AL, Moussatche N, Condit RC. Temperature-sensitive mutants in the vaccinia virus 4b virion structural protein assemble malformed, transcriptionally inactive intracellular mature virions. Virology. 2004;330:127–146. doi: 10.1016/j.virol.2004.08.038. [DOI] [PubMed] [Google Scholar]
- Katz E, Moss B. Vaccinia virus structural polypeptide derived from a high-molecular-weight precursor: formation and integration into virus particles. J.Virol. 1970;6:717–726. doi: 10.1128/jvi.6.6.717-726.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer J, Traktman P. Genetic and cell biological characterization of the vaccinia virus A30 and G7 phosphoproteins. J.Virol. 2005;79:7146–7161. doi: 10.1128/JVI.79.11.7146-7161.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merchlinsky M, Garon CF, Moss B. Molecular cloning and sequence of the concatemer junction from vaccinia virus replicative DNA. Viral nuclease cleavage sites in cruciform structures. J.Mol.Biol. 1988;199:399–413. doi: 10.1016/0022-2836(88)90613-4. [DOI] [PubMed] [Google Scholar]
- Merchlinsky M, Moss B. Resolution of vaccinia virus DNA concatemer junctions requires late-gene expression. J.Virol. 1989;63:1595–1603. doi: 10.1128/jvi.63.4.1595-1603.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss B. Poxviridae: the viruses and their replication. In: Knipe DM, Howley PM, editors. Fields Virology. Wolters Kluwer-Lippincott Williams and Wilkins; Philadelphia: 2007. pp. 2906–2945. [Google Scholar]
- Moyer RW, Graves RL. The mechanism of cytoplasmic orthopoxvirus DNA replication. Cell. 1981;27:391–401. doi: 10.1016/0092-8674(81)90422-0. [DOI] [PubMed] [Google Scholar]
- Resch W, Weisberg AS, Moss B. Vaccinia virus nonstructural protein encoded by the A11R gene is required for formation of the virion membrane. J.Virol. 2005;79:6598–6609. doi: 10.1128/JVI.79.11.6598-6609.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resch W, Weisberg AS, Moss B. Expression of the highly conserved vaccinia virus E6 protein is required for virion morphogenesis. Virology. 2009;386:478–485. doi: 10.1016/j.virol.2009.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risco C, Rodriguez JR, Lopez-Iglesias C, Carrascosa JL, Esteban M, Rodriguez D. Endoplasmic reticulum-Golgi intermediate compartment membranes and vimentin filaments participate in vaccinia virus assembly. J.Virol. 2002;76:1839–1855. doi: 10.1128/JVI.76.4.1839-1855.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez D, Esteban M, Rodriguez JR. Vaccinia virus A17L gene product is essential for an early step in virion morphogenesis. J.Virol. 1995;69:4640–4648. doi: 10.1128/jvi.69.8.4640-4648.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez JR, Risco C, Carrascosa JL, Esteban M, Rodriguez D. Vaccinia virus 15-kilodalton (A14L) protein is essential for assembly and attachment of viral crescents to virosomes. J.Virol. 1998;72:1287–1296. doi: 10.1128/jvi.72.2.1287-1296.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekiguchi J, Shuman S. Novobiocin inhibits vaccinia virus replication by blocking virus assembly. Virology. 1997;235:129–137. doi: 10.1006/viro.1997.8684. [DOI] [PubMed] [Google Scholar]
- Simpson DA, Condit RC. The vaccinia virus A18R protein plays a role in viral transcription during both the early and the late phases of infection. J.Virol. 1994;68:3642–3649. doi: 10.1128/jvi.68.6.3642-3649.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sodeik B, Doms RW, Ericsson M, Hiller G, Machamer CE, van ’t HW, van MG, Moss B, Griffiths G. Assembly of vaccinia virus: role of the intermediate compartment between the endoplasmic reticulum and the Golgi stacks. J.Cell Biol. 1993;121:521–541. doi: 10.1083/jcb.121.3.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szajner P, Jaffe H, Weisberg AS, Moss B. Vaccinia virus G7L protein Interacts with the A30L protein and is required for association of viral membranes with dense viroplasm to form immature virions. J.Virol. 2003;77:3418–3429. doi: 10.1128/JVI.77.6.3418-3429.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szajner P, Jaffe H, Weisberg AS, Moss B. A complex of seven vaccinia virus proteins conserved in all chordopoxviruses is required for the association of membranes and viroplasm to form immature virions. Virology. 2004a;330:447–459. doi: 10.1016/j.virol.2004.10.008. [DOI] [PubMed] [Google Scholar]
- Szajner P, Weisberg AS, Moss B. Unique temperature-sensitive defect in vaccinia virus morphogenesis maps to a single nucleotide substitution in the A30L gene. J.Virol. 2001a;75:11222–11226. doi: 10.1128/JVI.75.22.11222-11226.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szajner P, Weisberg AS, Moss B. Evidence for an essential catalytic role of the F10 protein kinase in vaccinia virus morphogenesis. J.Virol. 2004b;78:257–265. doi: 10.1128/JVI.78.1.257-265.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szajner P, Weisberg AS, Moss B. Physical and functional interactions between vaccinia virus F10 protein kinase and virion assembly proteins A30 and G7. J.Virol. 2004c;78:266–274. doi: 10.1128/JVI.78.1.266-274.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szajner P, Weisberg AS, Wolffe EJ, Moss B. Vaccinia virus A30L protein is required for association of viral membranes with dense viroplasm to form immature virions. J.Virol. 2001b;75:5752–5761. doi: 10.1128/JVI.75.13.5752-5761.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolonen N, Doglio L, Schleich S, Krijnse LJ. Vaccinia virus DNA replication occurs in endoplasmic reticulum-enclosed cytoplasmic mini-nuclei. Mol.Biol.Cell. 2001;12:2031–2046. doi: 10.1091/mbc.12.7.2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traktman P, Caligiuri A, Jesty SA, Liu K, Sankar U. Temperature-sensitive mutants with lesions in the vaccinia virus F10 kinase undergo arrest at the earliest stage of virion morphogenesis. J.Virol. 1995;69:6581–6587. doi: 10.1128/jvi.69.10.6581-6587.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traktman P, Liu K, DeMasi J, Rollins R, Jesty S, Unger B. Elucidating the essential role of the A14 phosphoprotein in vaccinia virus morphogenesis: construction and characterization of a tetracycline-inducible recombinant. J.Virol. 2000;74:3682–3695. doi: 10.1128/jvi.74.8.3682-3695.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner PC, Moyer RW. The vaccinia virus fusion inhibitor proteins SPI-3 (K2) and HA (A56) expressed by infected cells reduce the entry of superinfecting virus. Virology. 2008;380:226–233. doi: 10.1016/j.virol.2008.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unger B, Traktman P. Vaccinia virus morphogenesis: A13 phosphoprotein is required for assembly of mature virions. J.Virol. 2004;78:8885–8901. doi: 10.1128/JVI.78.16.8885-8901.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Shuman S. Vaccinia virus morphogenesis is blocked by temperature-sensitive mutations in the F10 gene, which encodes protein kinase 2. J.Virol. 1995;69:6376–6388. doi: 10.1128/jvi.69.10.6376-6388.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward GA, Stover CK, Moss B, Fuerst TR. Stringent chemical and thermal regulation of recombinant gene expression by vaccinia virus vectors in mammalian cells. Proc.Natl.Acad.Sci.U.S.A. 1995;92:6773–6777. doi: 10.1073/pnas.92.15.6773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolffe EJ, Moore DM, Peters PJ, Moss B. Vaccinia virus A17L open reading frame encodes an essential component of nascent viral membranes that is required to initiate morphogenesis. J.Virol. 1996;70:2797–2808. doi: 10.1128/jvi.70.5.2797-2808.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang WP, Kao SY, Bauer WR. Biosynthesis and post-translational cleavage of vaccinia virus structural protein VP8. Virology. 1988;167:585–590. [PubMed] [Google Scholar]