Abstract
The RNA polymerase II core promoter is a structurally and functionally diverse transcriptional regulatory element. There are two main strategies for transcription initiation – focused and dispersed initiation. In focused initiation, transcription starts from a single nucleotide or within a cluster of several nucleotides, whereas in dispersed initiation, there are several weak transcription start sites over a broad region of about 50 to 100 nucleotides. Focused initiation is the predominant means of transcription in simpler organisms, whereas dispersed initiation is observed in approximately two thirds of vertebrate genes. Regulated genes tend to have focused promoters, and constitutive genes typically have dispersed promoters. Hence, in vertebrates, focused promoters are used in a small but biologically important fraction of genes. The properties of focused core promoters are dependent upon the presence or absence of sequence motifs such as the TATA box and DPE. For example, Caudal, a key regulator of the homeotic gene network, preferentially activates transcription from DPE- versus TATA-dependent promoters. The basal transcription factors, which act in conjunction with the core promoter, are another important component in the regulation of gene expression. For instance, upon differentiation of myoblasts to myotubes, the cells undergo a switch from a TFIID-based transcription system to a TRF3-TAF3-based system. These findings suggest that the core promoter and basal transcription factors are important yet mostly unexplored components in the regulation of gene expression.
Introduction
The core promoter lies at the center of the transcription process, yet it is often an overlooked component in the regulation of gene expression (for reviews, see: Smale and Kadonaga, 2003; Thomas and Chiang, 2006; Heintzman and Ren, 2007; Juven-Gershon et al., 2008b). The core promoter is generally defined to be the DNA region that directs the accurate initiation of transcription by RNA polymerase II. In the past, the core promoter has often been presumed to be a generic entity that functions by a single universal mechanism, but it is now clearly apparent that there is widespread diversity in core promoter structure and function. In this review, we will discuss some key features of the RNA polymerase II core promoter, and provide some examples of the role of the core promoter and the basal transcription machinery in the regulation of gene expression.
Focused versus Dispersed Transcription Initiation
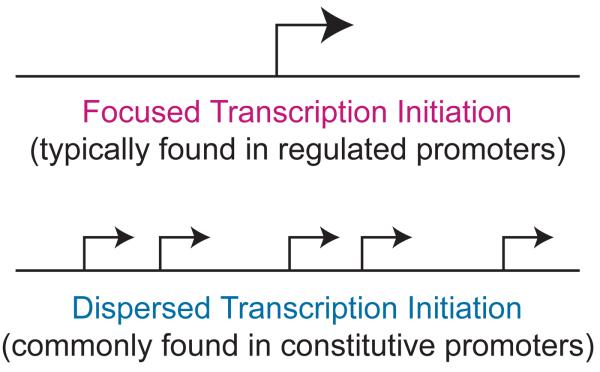
Examination of the patterns of transcription initiation reveals two different modes of transcription initiation – focused and dispersed (Fig. 1) (see, for example: Smale and Kadonaga, 2003; Carninci et al., 2006; Juven-Gershon et al., 2006a, 2008b). In focused initiation, transcription starts at a single nucleotide or within a narrow region of several nucleotides, whereas in dispersed initiation, there are multiple weak start sites over a broad region of about 50 to 100 nucleotides. Some promoters exhibit the combined qualities of both focused and dispersed promoters – for instance, a promoter might have multiple dispersed start sites with one particularly strong start site. In this essay, however, we will limit the discussion to focused and dispersed promoters rather than the mixed mode promoters.
Fig. 1.
Focused versus dispersed transcription initiation. In focused transcription, there is either a single major transcription start site or several start sites within a narrow region of several nucleotides. Focused transcription is the predominant mode of transcription in simpler organisms. In dispersed transcription, there are several weak transcription start sites over a broad region of about 50 to 100 nucleotides. Dispersed transcription is the most common mode of transcription in vertebrates. For instance, dispersed transcription is observed in about two-thirds of human genes. In vertebrates, focused transcription tends to be associated with regulated promoters, whereas dispersed transcription is typically observed in constitutive promoters in CpG islands.
Focused transcription initiation occurs in all organisms, and appears to be the predominant or exclusive mode of transcription in simpler organisms. In vertebrates, however, about 70% of genes have dispersed promoters, which are typically found in CpG islands. It generally appears that focused promoters are associated with regulated genes, whereas dispersed promoters are used in constitutive genes. From a teleological standpoint, this arrangement is consistent with the notion that it would be easier to regulate the transcription of a gene with a single transcription start site than one with multiple start sites. Conversely, variations in the expression of a constitutive gene would be minimized by the use of multiple start sites.
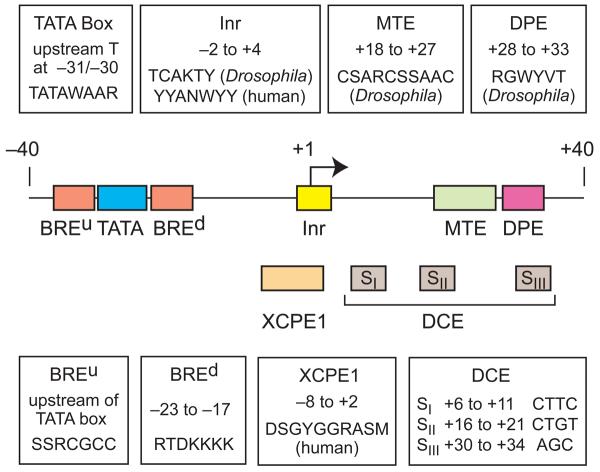
Most mechanistic studies of RNA polymerase II transcription have been carried out with focused promoters. Although focused promoters comprise a minority of all promoters in vertebrates, there is an inordinate amount of effort devoted to focused promoters relative to dispersed promoters because of the biological significance of the regulated genes with which the focused promoters are associated. The analysis of focused core promoters has led to the discovery of sequence motifs such as the TATA box, BREu (upstream TFIIB Recognition Element), Inr (Initiator), MTE (Motif Ten Element), DPE (Downstream Promoter Element), DCE (Downstream Core element), and XCPE1 (X Core Promoter Element 1) (Fig. 2). In contrast, dispersed promoters generally lack BRE, TATA, DPE, and MTE motifs (Sandelin et al., 2007; Carninci et al., 2006). It is likely that there are fundamental differences in the mechanisms of transcription from focused versus dispersed promoters. For the remainder of this review, we will mainly describe studies of focused core promoters.
Fig. 2.
Some core promoter motifs for transcription by RNA polymerase II. This diagram is roughly to scale. These motifs are typically found in focused core promoters. There are no universal core promoter elements. It is likely that additional core promoter motifs remain to be discovered. The properties of any particular core promoter are dictated by the presence or absence of specific core promoter elements. For instance, as discussed in the text, a TATA-dependent core promoter with TATA + Inr motifs has different properties than a DPE-dependent core promoter with Inr + DPE motifs.
Basal Transcription Factors
The focused core promoter, which typically encompasses −40 to +40 relative to the +1 transcription start site, is the location at which the RNA polymerase II machinery initiates transcription. Purified RNA polymerase II can synthesize RNA from a DNA template, but is not able to recognize the core promoter. This process requires additional factors that are commonly known as the “general” or “basal” transcription factors, which include TFIIA (Transcription Factor for RNA polymerase II A), TFIIB, TFIID, TFIIE, TFIIF, and TFIIH. These factors do not act in a “general” manner at all core promoters, and hence, we will refer to them as the “basal” transcription factors.
With TATA-driven core promoters, transcription can be achieved in vitro with purified RNA polymerase II, TFIIB, TFIID, TFIIE, TFIIF, and TFIIH. However, these same factors are not able to mediate transcription from a DPE-driven promoter (Lewis et al., 2005). In addition, NC2 (Negative Cofactor 2; also known as Dr1-Drap1), which was identified as repressor of TATA-dependent transcription, was found to be an activator of DPE-dependent transcription (Willy et al., 2000; Hsu et al., 2008). TATA- versus DPE-dependent transcription appears to be controlled, at least in part, by a simple circuit in which TBP (TATA box-Binding Protein) activates TATA transcription and represses DPE transcription, whereas NC2 and Mot1 (an ATPase that removes TBP from DNA) block TBP function (Hsu et al., 2008; van Werven et al., 2008) and thus promote DPE transcription and repress TATA transcription (Hsu et al., 2008).
TFIID is a key basal transcription factor that is involved in the recognition of focused core promoters (for review, see Thomas and Chiang, 2006). TFIID is a multisubunit complex that comprises TBP and about a dozen TAFs (TBP-Associated Factors). There are multiple potential points of interaction of TFIID with the core promoter. The TBP subunit binds to the TATA box, the TAF1 and TAF2 subunits recognize the Inr, the TAF1 subunit is in close proximity to the DCE, and the TAF6 and TAF9 subunits appear to interact with the DPE.
There are also mechanisms of core promoter recognition that do not involve the canonical TFIID complex. For instance, as discussed below, there are TRFs (TBP-Related Factors) with functions that are distinct from those of TBP.
The other known basal transcription factors participate in the early steps in transcription as follows. TFIIB interacts with TBP and assists in the recruitment of polymerase to the core promoter. TFIIB can bind to core promoter sequences at the BREu and BREd motifs in a manner that is dependent upon the binding of TBP to the TATA box (for review, see Deng and Roberts, 2007). TFIIA appears to promote the binding of TBP to the TATA box. TFIIE, TFIIF, and TFIIH act subsequent to the binding of TFIID and TFIIB to the core promoter, and mediate the unwinding of DNA and the early steps in the transcription process.
Core Promoter Motifs
The focused core promoter is diverse in terms of its structure and function. There are several known sequence motifs that can contribute to core promoter activity (Fig. 2), and it is likely that many other core promoter elements remain to be discovered. There are no universal core promoter elements. A brief overview of several core promoter motifs is as follows.
The Initiator (Inr)
The Inr encompasses the transcription start site, and is probably the most commonly occurring core promoter motif (Ohler et al., 2002; FitzGerald et al., 2006; Gershenzon et al., 2006). The function of the Inr as a distinct core promoter element was articulated by Smale and Baltimore (1989). Although several factors have been found to interact with the Inr, the binding of TFIID correlates best with Inr activity (discussed in Smale and Kadonaga, 2003). Functional analyses have determined that the Inr consensus is YYANWYY (IUPAC nucleotide code) in humans and TCAKTY in Drosophila. Inriceand Arabidopsis, a YR Inr motif (with R+1) has been identified (Yamamoto et al., 2007b). Inr-like sequences have also been described in Saccharomyces cerevisiae (Yang et al., 2007).
Computational analyses of Drosophila promoters have suggested an Inr consensus of TCAGTY (Ohler et al., 2002; FitzGerald et al., 2006), which is nearly identical to the Drosophila Inr consensus of TCAKTY determined via functional studies, such as the binding of TFIID (Purnell et al., 1994; Chalkley and Verrijzer, 1999). In contrast, computational analyses of mammalian promoters (both focused and dispersed) have led to a broader mammalian Inr consensus of YR (where R is +1; Carninci et al., 2006; Frith et al., 2008), which differs from the functional mammalian Inr consensus (YYANWYY). This difference is probably due to the high frequency (perhaps around 70%; Carninci et al., 2006; Kim et al., 2005; Cooper et al., 2006) of dispersed promoters in mammals and the inclusion of both dispersed and focused promoters in the computational analyses. The Inr consensus for focused mammalian promoters may more closely resemble the mammalian functional Inr consensus (YYANWYY) or even perhaps the more restrictive Drosophila Inr consensus (TCAKTY).
Focused transcription typically initiates within the Inr, and the A nucleotide in the Inr consensus is usually designed as the “+1” position, whether or not transcription actually initiates at that particular nucleotide. This convention is useful because other core promoter motifs, such as the MTE and DPE, function with the Inr in a manner that exhibits a strict spacing dependence with the Inr consensus sequence (and hence, the A+1 nucleotide) rather than the actual transcription start site (Burke and Kadonaga, 1997; Kutach and Kadonaga, 2000; Lim et al., 2004).
The TATA Box and BRE Motifs
The TATA box is the first core promoter motif that was discovered (Goldberg, 1979) as well as the best known core promoter element. The metazoan TATA box consensus is TATAWAAR, where the upstream T is usually located at −31 or −30 relative to the A+1 (or G+1) position in the Inr (Carninci et al., 2006; Ponjavic et al., 2006). As noted above, the TATA box is recognized and bound by the TBP subunit of the TFIID complex. Both the TATA box and TBP are conserved from archaebacteria to humans (Reeve, 2003). The TATA box is also present in plants (Molina and Grotewold, 2005; Yamamoto et al., 2007a, 2007b). Although the TATA box is a well known core promoter motif, it is present in only about 10-15% of mammalian core promoters (Carninci et al., 2006; Kim et al., 2005; Cooper et al., 2006).
The BRE (TFIIB Recognition Element) was initially identified as a TFIIB binding sequence that is immediately upstream of a subset (∼10-30%) of TATA box elements (Lagrange et al., 1998). In addition, a second TFIIB recognition site, the BREd (downstream TFIIB recognition element), was found immediately downstream of the TATA box (Deng and Roberts, 2005). The discovery of the BREd led to the renaming of the original BRE as BREu, for upstream BRE (reviewed in Deng and Roberts, 2007). Both the BREu and BREd function in conjunction with a TATA box, and have been found to increase as well as to decrease the levels of basal transcription (Lagrange et al., 1998; Evans et al., 2001; Deng and Roberts, 2005). More recent studies suggest a distinct role for the BREu in transcriptional regulation (Juven-Gershon et al., 2008a; discussed below).
DPE and MTE Motifs
The DPE (downstream core promoter element) was identified as a TFIID recognition site that is downstream of the Inr (Burke and Kadonaga, 1996). The DPE is located precisely from +28 to +33 relative to the A+1 and is conserved from Drosophila to humans (Burke and Kadonaga, 1997). The DPE does not appear to be present in Saccharomyces cerevisiae. The DPE is a recognition site for TFIID, which binds cooperatively to the Inr and DPE motifs. The spacing between the Inr and DPE is critical for transcriptional activity of DPE-dependent promoters (Kutach and Kadonaga, 2000). DPE-dependent promoters typically contain only DPE and Inr motifs. In some cases, however, TATA, Inr, and DPE motifs can be found in the same core promoter.
The MTE (motif ten element) was found to be a functionally active core promoter element that corresponds to an overrepresented sequence (termed motif 10) that was identified in Drosophila core promoter regions (Ohler et al., 2002; Lim et al., 2004). The MTE is located immediately upstream of the DPE at precisely +18 to +27 relative to the A+1 in the Inr, and is conserved from Drosophila to humans. DNase I footprinting analyses suggest that the MTE, like the DPE, is a recognition site for TFIID. The MTE functions cooperatively with the Inr, but can act independently of the DPE as well as the TATA box. There is, however, synergy between the MTE and DPE as well as between the MTE and TATA box.
These studies led to the design of a super core promoter (SCP) that contains a TATA, Inr, MTE, and DPE in a single promoter (Juven-Gershon et al., 2006b). The SCP is the strongest core promoter observed in vitro and in cultured cells, and yields high levels of transcription in conjunction with transcriptional enhancers. These findings indicate that gene expression levels can be modulated via the core promoter.
Role of the Core Promoter in the Regulation of Gene Expression
Transcriptional regulation is achieved not only by diversity in enhancers, but also by diversity in core promoter structure (see, for example: Smale, 2001; Butler and Kadonaga, 2002). This effect is seen, in particular, in the area of enhancer-promoter communication. For instance, the Drosophila AE1 and IAB5 enhancers preferentially activate the TATA-containing even-skipped core promoter relative to the TATA-less and DPE-containing white core promoter (Ohtsuki et al., 1998). In addition, DPE- as well as TATA-specific enhancers were identified in an enhancer-trapping screen in Drosophila (Butler and Kadonaga, 2001). Thus, some activators prefer TATA-dependent promoters, whereas others prefer DPE-dependent promoters.
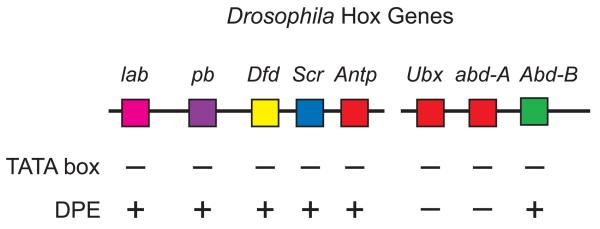
More recently, the analysis of the Drosophila homeotic (Hox) genes has revealed new insights into the role of the core promoter in a regulatory network (Juven-Gershon et al., 2008a). In this study, it was found that nearly all of the Hox genes, which were previously known to have TATA-less promoters, contain DPE-dependent core promoters (Fig. 3). This observation suggested that at least some of the transcription factors that regulate the Hox gene network might be DPE-specific activators. Following this hypothesis, it was found that Caudal, a sequence-specific DNA-binding transcription factor and key regulator of the Hox genes, is a DPE-specific activator. In addition, Caudal-mediated activation of the Antennapedia P2 enhancer-promoter region as well as the Sex combs reduced enhancer-promoter region was observed to be dependent upon the DPE motifs in their respective core promoters. These findings collectively indicate an important role of the DPE in the regulation of the Hox genes.
Fig. 3.
Nearly all of the Drosophila Hox genes contain TATA-less, DPE-dependent core promoters. The two most evolutionarily recent Hox genes, Ubx and abd-A, lack both TATA and DPE motifs. Both the upstream (P1) and downstream (P2) promoters of the Antp gene contain TATA-less, DPE-dependent core promoters. In the indicated DPE-dependent core promoters, the Inr and DPE sequences as well as the Inr-to-DPE spacing are conserved from Drosophila melanogaster to Drosophila virilis, which are separated by an evolutionary period of about 40 to 60 million years. lab, labial; pb, proboscipedia; Dfd, Deformed; Scr, Sex combs reduced; Antp, Antennapedia; Ubx, Ultrabithorax; abd-A, abdominal-A; Abd-B, Abdominal-B. Results taken from Juven-Gershon et al. (2008a).
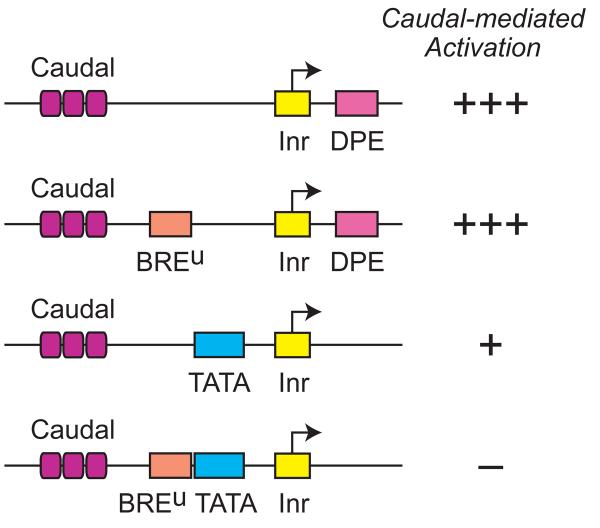
Further investigation showed that the function of Caudal is more complex than a simple matter of specificity for the DPE relative to the TATA box. Specifically, the BREu motif suppresses the ability of Caudal to activate transcription via the TATA box but not the DPE. Hence, as depicted in Fig. 4, there are three levels of Caudal activation – strong activation via the DPE (in the presence or absence of a BREu), weaker activation via the TATA box in the absence of a BREu, and little or no activation via the TATA box with a BREu.
Fig. 4.
Caudal is a DPE-specific activator. Caudal preferentially activates transcription from DPE-dependent core promoters relative to TATA-dependent core promoters. In addition, the presence of a BREu motif upstream of the TATA box further suppresses the ability of Caudal to activate transcription. The BREu motif does not affect the ability of Caudal to activate a DPE-dependent core promoter. The TATA box also does not alter the ability of Caudal to activate transcription via the DPE motif (not shown). Results taken from Juven-Gershon et al. (2008a).
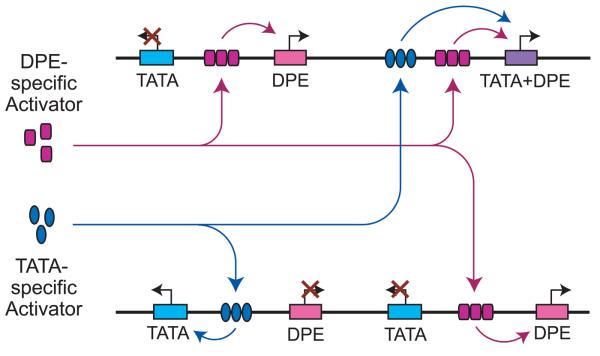
Hence, these studies of Caudal and the Hox genes reveal how specific core promoter motifs can play a central role in an important biological network. Yet, it is important to consider why Caudal might act as a DPE-specific activator. In a simple sense, it could be imagined that DPE specificity would be useful in the construction of regulatory networks. As in the wiring of a printed circuit board, there could be connections between transcriptional enhancers and their cognate core promoters. The use of DPE- and TATA-specific activators would enable the construction of more sophisticated and effective connections between enhancers and promoters (Fig. 5).
Fig. 5.
A simplified, hypothetical diagram of activation by DPE- and TATA-specific factors. Transcription factors bind to enhancers, but only activate transcription from promoters with the appropriate core promoter elements. The core promoter containing both TATA and DPE motifs can be activated by either DPE- or TATA-specific activators. Transcription levels can be further regulated by the presence of the BREu as well as other core promoter motifs.
TBP-related Factors (TRFs) and Transcriptional Regulation
There is diversity not only in core promoter elements, but also in the basal transcription machinery. This concept is nicely exemplified in studies of the TBP-related factors (TRFs) (for reviews, see: Jones, 2007; Müller et al., 2007; Reina and Hernandez, 2007; Torres-Padilla and Tora, 2007). There are three TRFs, which are generally termed TRF1, TRF2, and TRF3.
TRF1 does not exist in yeast and humans, but is present in Drosophila. In many eukaryotes, including yeast and humans, TBP participates in transcription by RNA polymerases I, II, and III. However, in Drosophila, TRF1 is used instead of TBP for RNA polymerase III transcription (Takada et al., 2000).
TRF2 (also known as TLF, TLP, TRF, and TRP) is present in most eukaryotes, and is involved in transcription by RNA polymerase II. TRF2 does not bind to TATA box sequences, and cannot replace TBP in vitro. It appears that many genes are regulated by TRF2 instead of TBP – one such example is the Drosophila histone H1 gene (Isogai et al., 2007). The TATA-less H1 linker histone gene is in a cluster of genes that also includes the four TATA-containing core histone genes, which are transcribed with TBP. These findings suggest the use of different transcriptional mechanisms within a cluster of genes.
TRF3 (also known as TBP2 and TBPL2) appears to be present only in vertebrates, and is the TRF that is most closely related to TBP. TRF3 can bind to TATA boxes and support TATA-dependent transcription (Bártfai et al., 2004; Jallow et al., 2004). TRF3 was found to be important for embryonic development (Bártfai et al., 2004; Jallow et al., 2004). In addition, zebrafish embryos that are depleted of TRF3 exhibit multiple developmental defects and fail to undergo hematopoiesis (Hart et al., 2007).
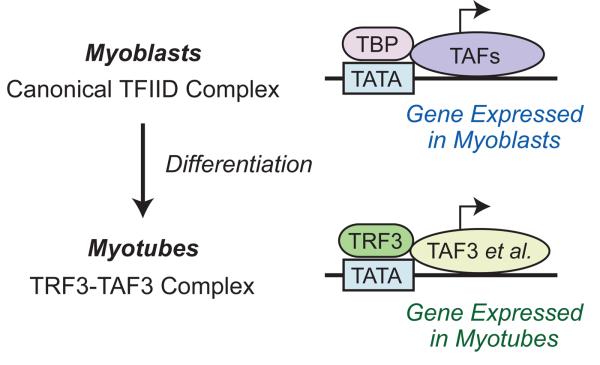
A particularly striking function of TRF3 was discovered during the analysis of the differentiation of myoblasts to myotubes (Deato and Tjian, 2007; Deato et al., 2008). Myoblasts were found to contain the canonical TBP-containing TFIID complex; however, upon terminal differentiation into myotubes, the TFIID complex was replaced by a TRF3-TAF3-containing complex (Fig. 6). These findings suggest that terminally differentiated cells may employ specialized transcription systems that are dedicated to the particular functions of the cells. It will be interesting to see if an analogous effect is observed in the differentiation of other cell types.
Fig. 6.
Replacement of the canonical TFIID complex by a TRF3-TAF3-containing complex upon terminal differentiation of myoblasts into myotubes. Both complexes bind to TATA box motifs via the TBP or TRF3 subunits. These findings exemplify the establishment of a new basal transcription system upon cell differentiation, and suggest that analogous processes may occur in other cell types. Results taken from Deato and Tjian (2007).
Conclusions and Perspectives
The core promoter and the basal transcriptional machinery are two important yet relatively unexplored dimensions in the regulation of gene expression. It is now apparent that diversity in the structure and function of core promoters and basal transcription factors contributes to developmental processes that lead to organismal complexity (Levine and Tjian, 2003). Thus, in the future, it will be essential to consider and to incorporate these factors in the analysis of gene regulation. For instance, transcriptional enhancers would ideally be studied in conjunction with their cognate core promoters. Alternatively, a new generation of reporter vectors could be designed with core promoters that function with both TATA- and DPE-specific enhancers (see, for example, Pfeiffer et al., 2008). The increased appreciation and understanding of core promoter motifs and basal transcription factors will lead to new and exciting discoveries, and ultimately, provide a more complete and accurate view of biological regulation.
Acknowledgments
We are grateful to Joshua Theisen, Timur Yusufzai, Jer-Yuan Hsu, Barbara Rattner, Moriah Eustice, Sharon Torigoe, and Trevor Parry for critical reading of this manuscript. Our research on the RNA polymerase II core promoter is supported by a grant from the National Institutes of Health (GM041249).
References
- Bártfai R, Balduf C, Hilton T, Rathmann Y, Hadzhiev Y, Tora L, Orbán L, Müller F. TBP2, a vertebrate-specific member of the TBP family, is required in embryonic development of zebrafish. Curr. Biol. 2004;14:593–598. doi: 10.1016/j.cub.2004.03.034. [DOI] [PubMed] [Google Scholar]
- Burke TW, Kadonaga JT. Drosophila TFIID binds to a conserved downstream basal promoter element that is present in many TATA-box-deficient promoters. Genes Dev. 1996;10:711–724. doi: 10.1101/gad.10.6.711. [DOI] [PubMed] [Google Scholar]
- Burke TW, Kadonaga JT. The downstream core promoter element, DPE, is conserved from Drosophila to humans and is recognized by TAFII60 of Drosophila. Genes Dev. 1997;11:3020–3031. doi: 10.1101/gad.11.22.3020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler JEF, Kadonaga JT. Enhancer-promoter specificity mediated by DPE or TATA core promoter motifs. Genes Dev. 2001;15:2515–2519. doi: 10.1101/gad.924301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler JEF, Kadonaga JT. The RNA polymerase II core promoter: a key component in the regulation of gene expression. Genes Dev. 2002;16:2583–2592. doi: 10.1101/gad.1026202. [DOI] [PubMed] [Google Scholar]
- Carninci P, Sandelin A, Lenhard B, Katayama S, Shimokawa K, Ponjavic J, Semple CA, Taylor MS, Engström PG, Frith MC, Forrest AR, Alkema WB, Tan SL, Plessy C, Kodzius R, Ravasi T, Kasukawa T, Fukuda S, Kanamori-Katayama M, Kitazume Y, Kawaji H, Kai C, Nakamura M, Konno H, Nakano K, Mottagui-Tabar S, Arner P, Chesi A, Gustincich S, Persichetti F, Suzuki H, Grimmond SM, Wells CA, Orlando V, Wahlestedt C, Liu ET, Harbers M, Kawai J, Bajic VB, Hume DA, Hayashizaki Y. Genome-wide analysis of mammalian promoter architecture and evolution. Nat. Genet. 2006;38:626–635. doi: 10.1038/ng1789. [DOI] [PubMed] [Google Scholar]
- Chalkley GE, Verrijzer CP. DNA binding site selection by RNA polymerase II TAFs: a TAF(II)250-TAF(II)150 complex recognizes the initiator. EMBO J. 1999;18:4835–4845. doi: 10.1093/emboj/18.17.4835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper SJ, Trinklein ND, Anton ED, Nguyen L, Myers RM. Comprehensive analysis of transcriptional promoter structure and function in 1% of the human genome. Genome Res. 2006;16:1–10. doi: 10.1101/gr.4222606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deato MDE, Tjian R. Switching of the core transcription machinery during myogenesis. Genes Dev. 2007;21:2137–2149. doi: 10.1101/gad.1583407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deato MDE, Marr MT, 2nd, Sottero T, Inouye C, Hu P, Tjian R. MyoD targets TAF3/TRF3 to activate myogenin transcription. Mol. Cell. 2008;32:96–105. doi: 10.1016/j.molcel.2008.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng W, Roberts SG. A core promoter element downstream of the TATA box that is recognized by TFIIB. Genes Dev. 2005;19:2418–2423. doi: 10.1101/gad.342405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng W, Roberts SG. TFIIB and the regulation of transcription by RNA polymerase II. Chromosoma. 2007;116:417–429. doi: 10.1007/s00412-007-0113-9. [DOI] [PubMed] [Google Scholar]
- Evans R, Fairley JA, Roberts SG. Activator-mediated disruption of sequence-specific DNA contacts by the general transcription factor TFIIB. Genes Dev. 2001;15:2945–2949. doi: 10.1101/gad.206901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FitzGerald PC, Sturgill D, Shyakhtenko A, Oliver B, Vinson C. Comparative genomics of Drosophila and human core promoters. Genome Biol. 2006;7:R53. doi: 10.1186/gb-2006-7-7-r53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frith MC, Valen E, Krogh A, Hayashizaki Y, Carninci P, Sandelin A. A code for transcription initiation in mammalian genomes. Genome Res. 2008;18:1–12. doi: 10.1101/gr.6831208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gershenzon NI, Trifonov EN, Ioshikhes IP. The features of Drosophila core promoters revealed by statistical analysis. BMC Genomics. 2006;7:161. doi: 10.1186/1471-2164-7-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg ML. Stanford University; Stanford, CA, U.S.A.: 1979. Ph.D. Thesis. [Google Scholar]
- Hart DO, Raha T, Lawson ND, Green MR. Initiation of zebrafish haematopoiesis by the TATA-box-binding protein-related factor Trf3. Nature. 2007;450:1082–1085. doi: 10.1038/nature06349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintzman ND, Ren B. The gateway to transcription: identifying, characterizing and understanding promoters in the eukaryotic genome. Cell. Mol. Life Sci. 2007;64:386–400. doi: 10.1007/s00018-006-6295-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu JY, Juven-Gershon T, Marr MT, 2nd, Wright KJ, Tjian R, Kadonaga JT. TBP, Mot1, and NC2 establish a regulatory circuit that controls DPE-dependent versus TATA-dependent transcription. Genes Dev. 2008;22:2353–2358. doi: 10.1101/gad.1681808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isogai Y, Keles S, Prestel M, Hochheimer A, Tjian R. Transcription of histone gene cluster by differential core-promoter factors. Genes Dev. 2007;21:2936–2949. doi: 10.1101/gad.1608807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jallow Z, Jacobi UG, Weeks DL, Dawid IB, Veenstra GJ. Specialized and redundant roles of TBP and a vertebrate-specific TBP paralog in embryonic gene regulation in Xenopus. Proc. Natl. Acad. Sci. U.S.A. 2004;101:13525–13530. doi: 10.1073/pnas.0405536101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones KA. Transcription strategies in terminally differentiated cells: shaken to the core. Genes Dev. 2007;21:2113–2117. doi: 10.1101/gad.1598007. [DOI] [PubMed] [Google Scholar]
- Juven-Gershon T, Hsu J-Y, Kadonaga JT. Perspectives on the RNA polymerase II core promoter. Biochem. Soc. Trans. 2006a;34:1047–1050. doi: 10.1042/BST0341047. [DOI] [PubMed] [Google Scholar]
- Juven-Gershon T, Cheng S, Kadonaga JT. Rational design of a super core promoter that enhances gene expression. Nat. Methods. 2006b;3:917–922. doi: 10.1038/nmeth937. [DOI] [PubMed] [Google Scholar]
- Juven-Gershon T, Hsu J-Y, Kadonaga JT. Caudal, a key developmental regulator, is a DPE-specific transcriptional factor. Genes Dev. 2008a;22:2823–2830. doi: 10.1101/gad.1698108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juven-Gershon T, Hsu J-Y, Theisen JW, Kadonaga JT. The RNA polymerase II core promoter - the gateway to transcription. Curr. Opin. Cell Biol. 2008b;20:253–259. doi: 10.1016/j.ceb.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TH, Barrera LO, Zheng M, Qu C, Singer MA, Richmond TA, Wu Y, Green RD, Ren B. A high-resolution map of active promoters in the human genome. Nature. 2005;436:876–880. doi: 10.1038/nature03877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutach AK, Kadonaga JT. The downstream promoter element DPE appears to be as widely used as the TATA box in Drosophila core promoters. Mol. Cell Biol. 2000;20:4754–4764. doi: 10.1128/mcb.20.13.4754-4764.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagrange T, Kapanidis AN, Tang H, Reinberg D, Ebright RH. New core promoter element in RNA polymerase II-dependent transcription: sequence-specific DNA binding by transcription factor IIB. Genes Dev. 1998;12:34–44. doi: 10.1101/gad.12.1.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine M, Tjian R. Transcription regulation and animal diversity. Nature. 2003;424:147–151. doi: 10.1038/nature01763. [DOI] [PubMed] [Google Scholar]
- Lewis BA, Sims RJ, 3rd, Lane WS, Reinberg D. Functional characterization of core promoter elements: DPE-specific transcription requires the protein kinase CK2 and the PC4 coactivator. Mol. Cell. 2005;18:471–481. doi: 10.1016/j.molcel.2005.04.005. [DOI] [PubMed] [Google Scholar]
- Lim CY, Santoso B, Boulay T, Dong E, Ohler U, Kadonaga JT. The MTE, a new core promoter element for transcription by RNA polymerase II. Genes Dev. 2004;18:1606–1617. doi: 10.1101/gad.1193404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molina C, Grotewold E. Genome wide analysis of Arabidopsis core promoters. BMC Genomics. 2005;6:25. doi: 10.1186/1471-2164-6-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller F, Demény MA, Tora L. New problems in RNA polymerase II transcription initiation: matching the diversity of core promoters with a variety of promoter recognition factors. J. Biol. Chem. 2007;282:14685–14689. doi: 10.1074/jbc.R700012200. [DOI] [PubMed] [Google Scholar]
- Ohler U, Liao GC, Niemann H, Rubin GM. Computational analysis of core promoters in the Drosophila genome. Genome Biol. 2002;3:RESEARCH0087. doi: 10.1186/gb-2002-3-12-research0087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtsuki S, Levine M, Cai HN. Different core promoters possess distinct regulatory activities in the Drosophila embryo. Genes Dev. 1998;12:547–556. doi: 10.1101/gad.12.4.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer BD, Jenett A, Hammonds AS, Ngo TT, Misra S, Murphy C, Scully A, Carlson JW, Wan KH, Laverty TR, Mungall C, Svirskas R, Kadonaga JT, Doe CQ, Eisen MB, Celniker SE, Rubin GM. Tools for neuroanatomy and neurogenetics in Drosophila. Proc. Natl. Acad. Sci. U.S.A. 2008;105:9715–9720. doi: 10.1073/pnas.0803697105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponjavic J, Lenhard B, Kai C, Kawai J, Carninci P, Hayashizaki Y, Sandelin A. Transcriptional and structural impact of TATA-initiation site spacing in mammalian core promoters. Genome Biol. 2006;7:R78. doi: 10.1186/gb-2006-7-8-r78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purnell BA, Emanuel PA, Gilmour DS. TFIID sequence recognition of the initiator and sequences farther downstream in Drosophila class II genes. Genes Dev. 1994;8:830–842. doi: 10.1101/gad.8.7.830. [DOI] [PubMed] [Google Scholar]
- Reeve JN. Archaeal chromatin and transcription. Mol. Microbiol. 2003;48:587–598. doi: 10.1046/j.1365-2958.2003.03439.x. [DOI] [PubMed] [Google Scholar]
- Reina JH, Hernandez N. On a roll for new TRF targets. Genes Dev. 2007;21:2855–2860. doi: 10.1101/gad.1623207. [DOI] [PubMed] [Google Scholar]
- Sandelin A, Carninci P, Lenhard B, Ponjavic J, Hayashizaki Y, Hume DA. Mammalian RNA polymerase II core promoters: insights from genome-wide studies. Nat. Rev. Genet. 2007;8:424–436. doi: 10.1038/nrg2026. [DOI] [PubMed] [Google Scholar]
- Smale ST, Baltimore D. The “initiator” as a transcription control element. Cell. 1989;57:103–113. doi: 10.1016/0092-8674(89)90176-1. [DOI] [PubMed] [Google Scholar]
- Smale ST. Core promoters: active contributors to combinatorial gene regulation. Genes Dev. 2001;15:2503–2508. doi: 10.1101/gad.937701. [DOI] [PubMed] [Google Scholar]
- Smale ST, Kadonaga JT. The RNA polymerase II core promoter. Annu. Rev. Biochem. 2003;72:449–479. doi: 10.1146/annurev.biochem.72.121801.161520. [DOI] [PubMed] [Google Scholar]
- Takada S, Lis JT, Zhou S, Tjian R. A TRF1:BRF complex directs Drosophila RNA polymerase III transcription. Cell. 2000;101:459–469. doi: 10.1016/s0092-8674(00)80857-0. [DOI] [PubMed] [Google Scholar]
- Thomas MC, Chiang CM. The general transcription machinery and general cofactors. Crit. Rev. Biochem. Mol. Biol. 2006;41:105–178. doi: 10.1080/10409230600648736. [DOI] [PubMed] [Google Scholar]
- Torres-Padilla ME, Tora L. TBP homologues in embryo transcription: who does what? EMBO Rep. 2007;8:1016–1018. doi: 10.1038/sj.embor.7401093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Werven FJ, van Bakel H, van Teeffelen HA, Altelaar AF, Koerkamp MG, Heck AJ, Holstege FC, Timmers HT. Cooperative action of NC2 and Mot1p to regulate TATA-binding protein function across the genome. Genes Dev. 2008;22:2359–2369. doi: 10.1101/gad.1682308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willy PJ, Kobayashi R, Kadonaga JT. A basal transcription factor that activates or represses transcription. Science. 2000;290:982–985. doi: 10.1126/science.290.5493.982. [DOI] [PubMed] [Google Scholar]
- Yamamoto YY, Ichida H, Abe T, Suzuki Y, Sugano S, Obokata J. Differentiation of core promoter architecture between plants and mammals revealed by LDSS analysis. Nucleic Acids Res. 2007a;35:6219–6226. doi: 10.1093/nar/gkm685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto YY, Ichida H, Matsui M, Obokata J, Sakurai T, Satou M, Seki M, Shinozaki K, Abe T. Identification of plant promoter constituents by analysis of local distribution of short sequences. BMC Genomics. 2007b;8:67. doi: 10.1186/1471-2164-8-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C, Bolotin E, Jiang T, Sladek FM, Martinez E. Prevalence of the initiator over the TATA box in human and yeast genes and identification of DNA motifs enriched in human TATA-less core promoters. Gene. 2007;389:52–65. doi: 10.1016/j.gene.2006.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]