Abstract
Mutations in miRNA genes have been implicated in hearing loss in human families and mice. It is also possible that mutations in miRNA binding sites of inner ear targets alter gene expression levels and lead to hearing loss. To investigate these possibilities we screened predicted target genes of the miR-183 miRNA family known to be expressed in the inner ear sensory epithelium. In one Iranian family segregating autosomal recessive non-syndromic hearing loss (ARNSHL), we identified a homozygous variant in a predicted miR-96/182 binding site in the 3′UTR of the RDX (DFNB24) gene. However, in vitro functional studies showed that this site is not a functional target for miR-96/182. We extended our study to include the miR-183 genes themselves and 24 additional predicted target genes of the miRNA-183 family. Screening these miRNAs and target sequences in numerous families segregating either autosomal dominant non-syndromic deafness (ADNSHL) or ARNSHL did not identify any potential deafness-causing mutations. These results suggest that mutations disrupting gene regulation by the miR-183 family are not a common cause of human hearing loss.
Keywords: radixin, ERM protein family, miRNA, ADNSHL, ARNSHL
INTRODUCTION
Deafness is the most common human sensory deficit, and its social, economic and quality-of-life consequences are severe [Morton, 1991]. It is estimated that globally 4 of every 10,000 children are born with profound sensorineural hearing loss (SNHL) [Smith et al., 2005]. Non-syndromic forms of SNHL (NSHL) account for ~70% of hereditary hearing loss.
Most cases of inherited deafness are monogenic. That global dysregulation of numerous genes can also result in human NSHL has only recently been discovered. Mencia and colleagues screened NSHL families with dominant, recessive and unknown patterns of inheritance and identified separate mutations in two ADNSHL DFNA50 families in miR-96 [Mencia et al., 2009], a member of the miR-183 miRNA family that is expressed in the inner ear sensory epithelium [Soukup et al., 2009; Weston et al., 2006; Wienholds et al., 2005]. Interestingly, expression of the miR-183 family is not limited to the ear, but is highly expressed in other sensory organs as well [Xu et al., 2007].
We therefore hypothesized that dysregulation of inner ear genes known to be regulated by the miR-183 family could lead to ARNSHL. We focused on RDX, which encodes the radixin protein and causes DFNB24 ARNSHL [Khan et al., 2007]. Radixin and its related ezrin-radixin-moesin (ERM) protein family member ezrin are present in hair cell stereocilia of the mouse inner ear [Kitajiri et al., 2004; Pataky et al., 2004]. Although no deafness-causing mutations in ezrin have been identified to date, targeting of ezrin by miR-183 has been reported for some lung cancers [Wang et al., 2008], raising the possibility of miR-183 family involvement in the regulation of radixin.
We identified a variant in a predicted miR-96/182 binding site in the 3′UTR of RDX in an Iranian family segregating ARNSHL. While analysis with three algorithms predicted that this region is a miR-96/182 binding site, we performed in vitro functional studies that excluded this possibility. We then extended our study to include 24 additional predicted target genes. Screening the miR-183 family and their predicted targets in American ADNSHL and Iranian ARNSHL families, respectively, did not identify any potential deafness-causing variants. It appears that mutations disrupting gene regulation by the miR-183 family are not a common cause of human hearing loss.
MATERIALS AND METHODS
Clinical Evaluation of Families
One hundred fifty American ADNSHL and 576 Iranian ARNSHL families were studied. To document the degree of hearing loss audiologic testing was completed on consenting family members. A detailed family history was taken including any reported balance or visual problems. In some cases caloric testing and funduscopy were completed. Ten milliliters of whole blood was obtained from family members by venipuncture and genomic DNA was extracted as described previously [Grimberg et al., 1989]. Human research institutional review boards at the Welfare Science and Rehabilitation University and Iran University of Medical Sciences, Tehran, Iran, the National Centre of Excellence in Molecular Biology, Lahore, Pakistan, the Quaid-I-Azam University, Islamabad, Pakistan, the Combined Neuroscience Institutional Review Board (IRB) at the National Institutes of Health, Bethesda, Maryland, USA, and the University of Iowa, Iowa City, Iowa, USA approved all procedures.
Target Gene Prediction
Target genes of the miRNA-183 miRNA family were chosen as all three family members are expressed in the inner ear sensory epithelium [Friedman et al., 2009; Weston et al., 2006; Wienholds et al., 2005]. Three algorithms – miRanda (http://www.microrna.org/microrna/); PicTar (http://pictar.mdc-berlin.de/); and TargetScan (http://www.targetscan.org/) – that base predictions on thermodynamics, evolutionary conservation and target site-seed complementarity were used to select mRNA targets of miR-183/96/182 regulation. These algorithms identified hundreds of mRNA targets for each miRNA, although only some were common to all three algorithms. Inner ear expression and function were additional criteria used to select candidate target genes for screening (Table I).
Table 1.
Candidate miR-183/96/182 target genes
| Gene Symbol | Gene Name | OMIM Number‡ | Predicted miRNA Target Sites† | Inner Ear Expression * | Known Function | Number of Families Screened (ARNSHL) |
|---|---|---|---|---|---|---|
| AQP5 | Aquaporin 5 | 600442 | miR-96 | Yes | Transmembrane water transport | 192 |
| ATP2B4 | Plasma membrane calcium pump | 108732 | miR-183 | Yes | Calcium ion transport | 192 |
| BTG1 | B-cell translocation gene 1 | 109580 | miR-183 | Yes | Negative regulator of cell proliferation | 192 |
| CaBP1 | Calcium binding protein 1 | 605563 | miR-96 | Yes | Competitively inhibits calmodulin binding to calcium | 192 |
| Cav1.2 | Voltage-dependent calcium channel 1.2. | 114205 | miR-96 miR-182 |
Yes | Calcium ion transport | 192 |
| Cav1 | Caveolin 1 | 601047 | miR-96 | ? | Integral membrane protein that interacts with G-proteins in cell signaling | 192 |
| CELSR1 | Cadherin EGF receptor 1 | 604523 | miR-96 | ? | Receptor involved in contact-mediated communication | 192 |
| CELSR2 | Cadherin EGF receptor 2 | 604265 | miR-96 | Yes | Receptor involved in contact-mediated communication | 192 |
| EZRIN | Cytovillin | 123900 | miR-183 | Yes | Component of microvilli and substrate for protein-tyrosine kinases | 576 |
| GNG5 | Guanine nucleotide-binding protein 5 | 600874 | miR-183 | Yes | G-protein cell signaling | 192 |
| GRID1 | Glutamate receptor delta 1 | 610659 | miR-96 | ? | Synaptic transmission | 192 |
| KCC2 | Solute carrier 12A5 | 606726 | miR-96 | Yes | Potassium/chloride transport | 192 |
| KCNJ14 | Potassium inwardly-rectifying channel (Kir2.4) | 603953 | miR-183 | Yes | Potassium transport | 192 |
| MITF | Microphthalmia-associated transcription factor | 156845 | miR-96 miR-182 |
Yes | Basic helix-loop-helix transcription factor involved in regulation of development | 192 |
| MYRIP | Myosin VIIa and Rab-interacting protein | 611790 | miR-96 miR-182 (2) |
Yes | Actin transport and cytoskeleton | 192 |
| NCS1 | Neuronal calcium sensor 1 | 603315 | miR-183 | Yes | Calcium-ion sensor that modulates synaptic activity | 192 |
| NEFL | Neurofilament protein | 162280 | miR-183 | Yes | Neurofilament assemby and axonal transport | 192 |
| PEX19 | Peroxisome biogenesis factor 19 | 600279 | miR-183 | Yes | Cellular protein sorting | 192 |
| PPP2CA | Protein phosphatase, catalytic subunit, alpha | 176915 | miR-183 | Yes | Regulation of protein phosphorylation | 192 |
| RDX | Radixin | 179410 | miR-96 miR-182 |
Yes | Cytoskeletal protein that may link actin to plasma membrane | 576 |
| RYK | Receptor-like tyrosine kinase | 600524 | miR-96 | ? | Growth factor receptor tyrosine kinase | 192 |
| SLC6A6 | Solute carrier family 6 | 186854 | miR-183 | Yes | Taurine (neurotransmitter) transporter | 192 |
| TFCP2L3 | Transcription factor CP2-like 3 | 608576 | miR-96 (3) | Yes | Transcriptional regulation | 192 |
| TREK-1 | Potassium channel KCNK2 | 603219 | miR-183 | Yes | Central nervous system (CNS) potassium transport | 192 |
| ZCCHC3 | Zinc finger CCHC containing 3 | - | miR-183 | Yes | Transcriptional regulation | 192 |
Online Mammalian Inheritance in Man identification number
Predictions from miRanda, PicTar and TargetScan algorithms. Number of sites if multiple sites (brackets).
Reported in database or publication
- Not available
PCR, DHPLC and Sequencing
miRNA-183/96/182 genes and the 3′UTR of predicted target genes were amplified using gene-specific primers (Table II). Amplification reactions were cycled using a standard protocol on a GeneMate Genius thermocycler (ISC BioExpress, UT, USA). For denaturing high performance liquid chromatography (DHPLC), all amplicons were pooled post-PCR and heteroduplexes were formed by denaturing at 95°C for 5 min in a thermal cycler and cooling at a rate of 1°C/min to room temperature as described previously [Prasad et al., 2004]. DHPLC analysis of each amplicon was performed at three different temperatures. The analysis was conducted using Navigator™ Software (Transgenomic™, Omaha, NE) to estimate optimal temperature, run time and acetonitrile gradient. The best predicted temperature was bracketed by ±2°C to optimize sensitivity. Sequencing was completed with a BigDye™ v3.1 Terminator Cycle Sequencing kit (Applied Biosystems, Foster City, CA), according to the manufacturer’s instructions. Sequencing products were read using an ABI 3730s sequencer (Perkin Elmer, Waltham, MA). All sequencing chromatograms were compared to published cDNA sequence; nucleotide changes were detected using Sequencher v4.5 (Gene Code Corporation, Ann Arbor, MI).
Table 2.
Oligonucleotides used for amplification of human miRNAs and target gene binding sites
| Gene# | Forward Primer (5′-3′) | Reverse Primer (5′-3′) |
|---|---|---|
| miR-96 | TACCGAAGGGCCATAAACAG | AGGCAGTGTAAGGCGATCTG |
| miR-182 | AGCAGGAAGGGGGACTGT | GCAGGGAAACACAGAGTGTCA |
| miR-183 | AAGGTCATCTTGGGCTGATG | GGCTCTCTGGGGACACACT |
| AQP5 | TGGCTGCACAGTTAGAGAGG | CGGCATTCAATGAACCAGTC |
| ATP2B4 | TCCTTGGTAGTCACTTGTCATTTT | TCACACTACTGGCGGATTTT |
| BTG1 | TCTTGGGGATGGATTATGGA | TCTGGGAGAAACTGAAACCA |
| CaBP1 | CAGGATGTACTGGCGGATG | GACATGCTGTGTGGGGTCT |
| Cav1.2 (96) | CAAGAAGGCATTTTGCTTCA | AAAGCTTGTCACACTCCAAATACA |
| Cav1.2 (182) | TTGACAGCATGTTGCAGTTTC | TTGGGCATACACAATGGTTG |
| Cav1 | TGCATCAGCCGTGTCTATTC | TCAGACTGCCAAAAATAGATGAA |
| CELSR1 | CTGCCCTTGAAGTGGAGTG | CCCTCTCAGTTCTGGCTTTG |
| CELSR2 | CCTGCTCCTGTCTTGTGCTT | GGGAGTCAATTTCCAGCGTA |
| EZRIN | GAAACTTCATGCTGGCCTGT | CTGTGTGTGCGAGAGTGCTT |
| GNG5 | TTCCCAAACCACTCCTTATGA | ATTGTATGCTGCTGCCAGTG |
| GRID1 | TTCCCAATTTTCGAAGTCAG | GGTTCCTCGTCTTCCCTTCT |
| KCC2 | GGTTGCAAACCAAATCAAGAG | ATTTTGTGCAGACGGGAGTC |
| KCNJ14 | GTAGAGCACCCAGCCAAGAG | CCTTTTGGCATTACAGAACCA |
| MITF (96) | TTGGACTAGCACTGACTGAACTG | AGCATCACCAATGTTTCCAAG |
| MITF (182) | ATTTCTGCAGGTGGCAGGT | TTCCTTTGTGCTTTTAACTTCCTA |
| MYRIP (96) | TTGACAAAAATGTGTACTGTGTAAGC | GATCAAAATCACTTGATGACAAAA |
| MYRIP (182) | AAGTGCCTGCTCTGAAGGAG | GGAAATGCACATAGCAGCAA |
| MYRIP (182) | CAAGTGATTTTGATCTTTAGTGTCA | ATCTTGGCCCCTCCAGTTAC |
| NCS1 | TTGCCATCTATCGACCTTCC | CAGGACAGGGGAGAGGAGAG |
| NEFL | CAGATGCAAGCTATGTGCAA | GTTAAAAGGGGCACTGACCA |
| PEX19 | CAGCTATGGGGAACATCTGG | GGCAGAAACCACAATGGAGT |
| PPP2CA | CCTAATGGAAATGGGAAGAGC | TCCAATGATTGTTTGCTGCT |
| RDX | AGCTGAACCACCAACAGAGAA | TGGAAAAGAGGCAATGGAAC |
| RYK | TTGGACTAGGGGTACATTCTTACA | CAAGGCAGACCAGGTATCTTTT |
| SLC6A6 | GATCAAGGGCCTTATGTGGA | TGTGAAAATTCTGCGGTCTG |
| TFCP2L3 (96) | GCCATGTGAGAGCTGTGAAC | GCATGTAGCAGGAGACCACA |
| TFCP2L3 (96) | GCCCTAAGGCAGAAGATGAA | TGTTTCGCTCAGGAAATTTTG |
| TFCP2L3 (96) | GCCCAGAAACTTAGGAAGCA | GATTCCCCCTCTCCCATTTA |
| TREK-1 | GCTTGTTGAACGGTCCACTT | TGGTCCATATCTAGGCTCAGTT |
| ZCCHC3 | GTCCCAAAGCAGTGCACAAT | TTTTAAAAGGGAGGGGCAAC |
Round brackets: miRNA binding site
Luciferase Assays
We utilized RT-PCR to amplify and subclone a fragment (SpeI/HindIII) of the 3′UTR of RDX (nucleotides 2041–4247 of GeneBank™ accession number NM_002906, containing a potential binding site for miR-96 and miR-182) into the luciferase reporter vector, pMIR-REPORT (Ambion, Austin, TX), 3′ to the firefly luciferase cassette. To introduce the c.*95C>A variation we used the QuickChange II Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA) following the manufacturer’s guidelines. We plated HEK293 cells at a density of 5 × 105 cells/well in 24-well plates coated with poly-D-lysine (Sigma, St. Louis, MI) and transfected them with 150 ng of pMIR reporter construct (pMIR-REPORT-3′UTR/Rdx *95C or pMIR-REPORT-3′UTR/Rdx *95A), 15 ng of hpRL-SV40 (Promega, Madison, WI), and 5–50 pmol of the specified miRNA mimics or control oligonucleotide with a scrambled sequence (Dharmacom, Lafayette, CO) using Liopfectamine 2000 (Invitrogen, Carlsbad, CA). Using the dual luciferase assay kit (Promega, Madison, WI), we measured firefly luciferase 48 hrs post-transfection and normalized to Renilla activity. We performed three independent experiments for each assay.
Western Blots
We plated HeLa cells at a density of 8 × 104 cells/well in 12-well plates and transfected them with 20–200 pmol of the specified miRNA mimics or control oligonucleotide with a scrambled sequence (Dharmacom, Lafayette, CO) using Lipofectamine 2000 (Invitrogen, Carlsbad, CA). We lysed cells 72 hrs post-transfection in RIPA buffer, resolved extracts by SDS-Page, and transferred them to Hybond-P membranes (GE Healthcare, Piscataway, NJ). We used commercially available primary antibodies raised against Radixin (ab52495; 1:10,000; Abcam, Cambridge, MA) and acetylated alpha-tubulin (sc-23950; 1:1000; Santa Cruz Biotechnology, Santa Cruz, CA). Secondary antibodies conjugated to HRP (Santa Cruz Biotechnology, Santa Cruz, CA) and ECL reagents (GE Healthcare, Piscataway, NJ) were used for detection as specified by the manufacturer. Three independent experiments were completed for each assay.
RESULTS
Radixin – A Potential Target of miR-96/182 Regulation
We screened the predicted miR-96/182 binding site in the RDX 3′UTR in probands from 192 unmapped Iranian ARNSHL families (Table I). In one family, L-1007, we identified a homozygous c.*95C>A variation in the predicted binding site in affected individual II:1 (Figs 1A and 1B). The c.*95C nucleotide is highly conserved between species (Figs 1C and 1D). This variant was not present in 64 (128 chromosomes) Iranian controls or 191 (382 chromosomes) CEPH (Centre d’Etude du Polymorphisme Humain) controls.
Figure 1. Variation in the predicted miR-96/182 binding site of the RDX 3′UTR.
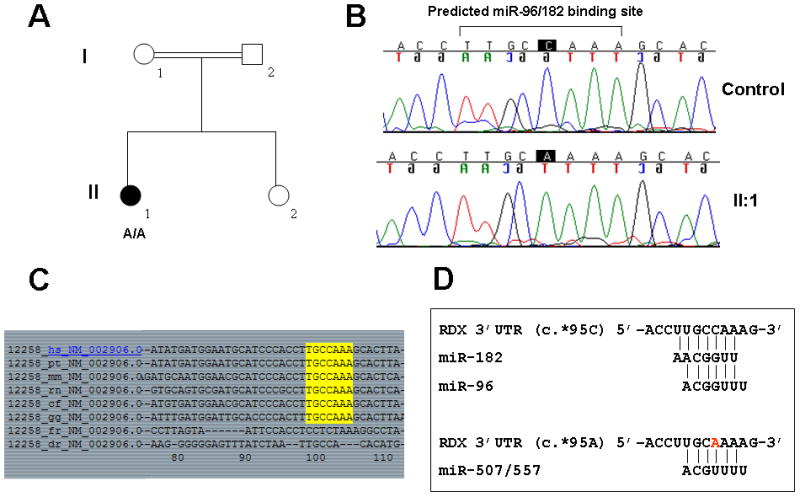
A Pedigree of Iranian family L-1007 with ARNSHL. Open symbols = unaffected; filled symbols = ARNSHL. The c.*95C>A genotype for affected proband II:1 is shown. B Direct sequencing revealed a homozygous alteration (c.*95C>A) in the miR-96/182 seed sequence in proband II:1. C Multisequence alignment generated using PicTar (http://pictar.mdc-berlin.de/) showing that the predicted miR-96/182 binding site in the 3′UTR of the RDX gene is highly conserved across species. hs, human; pt, primate; mm, mouse; rn, rat; cf, dog; gg, chicken; fr, pufferfish; dr, drosophila. D Consequential pairing of the reference RDX target region (c.*95C) with miR-182/96 seeds (top), and the mutant RDX target region (c.*95A) with miR-507/557 seeds (bottom), predicted using TargetScan (http://www.targetscan.org/).
Besides altering the predicted binding site for miR-96/182, the c.*95C>A variant also creates a novel binding site for miR-507/557. Thus, we hypothesized that the c.*95C>A mutation in RDX could result in hearing loss by (i) disrupting the binding of miR-96 and miR-182, and/or (ii) producing a new binding site for miR-507 and miR-557. The former has been demonstrated for a number of diseases including irritable bowel syndrome [Kapeller et al., 2008], while the latter has recently been described in Tourette syndrome, muscularity in sheep and in Parkinson disease [Chou et al., 2007; Clop et al., 2006; Wang et al., 2008].
Activity of the Predicted RDX miR-96/182 Binding Site In Vitro
To determine whether the predicted miR-86/182 binding site in the RDX 3′UTR is a biologically relevant target of regulation we performed luciferase assays (Fig 2A). Neither miR-96 nor miR-182 affect luciferase activity of the chimeric luciferase reporter/RDX constructs as compared to scrambled control oligonucleotides. We also failed to observe any decrease in activity for the mutant construct when co-transfected with miR-507 or miR-557 mimics.
Figure 2. Radixin is not targeted by miR-96 and miR-182 in vitro.
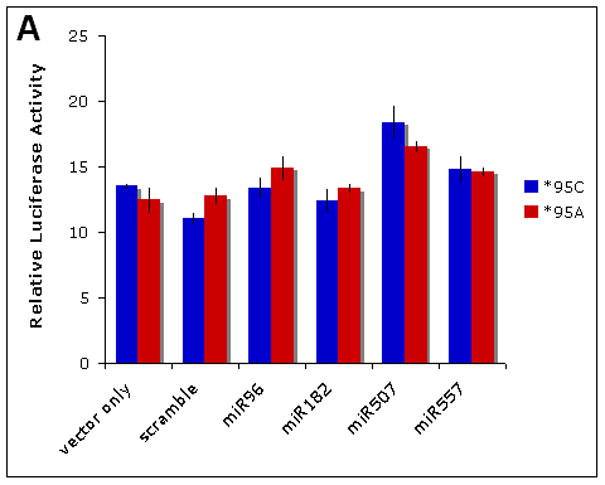
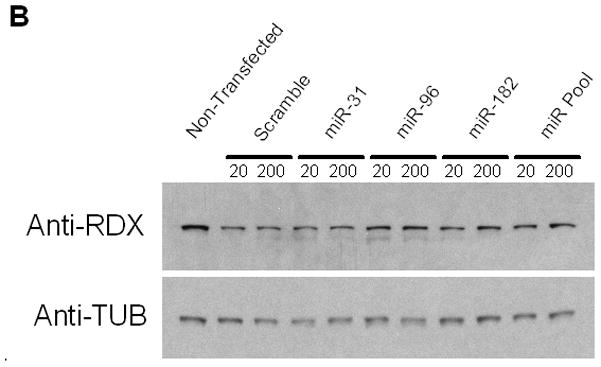
A Luciferase reporter activity of reference (*95C) and mutant (*95A) RDX 3′UTR constructs co-transfected with miRNA mimics in HEK293 cells. Blue bars = reference sequence; red bars = mutant sequence. Data are averages of six replicates. Error bars denote standard deviation (SD). B Western blot analysis of endogenous radixin protein levels in HeLa cells treated with miRNA mimics (20, 200 pmol). miR-31 was included based on similarity to miR-96 and -182. miR Pool denotes cocktail of all three miR mimics. Cell lysates were collected 72 hrs after transfection. Blots were stripped and reprobed with alpha-tubulin antibody as a loading control.
We repeated transfections using a 10-fold increase in mimic but failed to observe any evidence for miRNA targeting (data not shown). In addition, we tested whether miRNA mimics could affect the endogenous expression of RDX in HeLa cells (Fig 2B). As shown by western blot, no significant reduction in RDX protein levels is detected in the presence of miRNA mimics compared to control. We also screened a family of Pakistani origin generously provided by Dr. Thomas Friedman and Dr. Zubair Ahmed (Laboratory of Molecular Genetics, National Institute on Deafness and Other Communication Disorders) that mapped to the DFNB24 locus. However, no 3′UTR variants were identified.
Screening of the miR-183 miRNA Family and Additional Predicted Targets
In addition to the RDX gene, we identified a large number of other potential targets of miR-96/182 regulation (Table I). Predicted miR-183 binding sites in a total of 24 candidate target genes were screened in 192 ARNSHL families, however we identified no potential pathogenic variants in any of these genes. Since mutations in the miR-96 seed sequence have been linked to ADNSHL in humans and mice [Lewis et al., 2009; Mencia et al., 2009], we also screened 150 American ADNSHL families for mutations in the miR-96, miR-182 and miR-183 genes (Table I). However, we found no potential pathogenic variants.
DISCUSSION
Radixin was selected as a potential target of miR-183 miRNA family regulation based on its association with DFNB24 ARNSHL, its interesting temporospatial expression pattern in the inner ear [Khan et al., 2007; Kitajiri et al., 2004], and the presence of a predicted miR-96/182 target site in its 3′UTR. Despite this, our in vitro assays results do not support a direct role for the miR-183 miRNA family in the regulation of radixin.
Menica and colleagues did identify two mutations in adjacent nucleotides of the miR-96 seed sequence in two families segregating progressive ADNSHL [Mencia et al., 2009]. Supporting the disease-causing nature of these sequence variations was the simultaneous discovery by Lewis and colleagues of a single-base change in the miR-96 seed of the diminuendo (Dmdo) mouse model that also results in progressive hearing loss [Lewis et al., 2009]. However, investigation of families with hereditary deafness by Mencia and colleagues [2009], and now also in our laboratory has failed to find causative mutations in either miR-182 or miR-183. A possible explanation is that expression of other members of this cluster compensates for the loss of either miR-182 or miR-183 but not for the loss of miR-96. The identification of miR-96 mutations in only 2/568 genetically undiagnosed Spanish families with hereditary hearing loss in the Mencia et al study and in 0/150 American ADNSHL families in our study suggests that mutations in this miRNA gene are a relatively rare cause of NSHL.
Detailed investigation of the downstream effects on gene regulation of the human and mouse miR-96 mutations revealed five genes containing predicted miR-96 binding sites that were upregulated in the presence of mutant miR-96 siRNA mimics [Lewis et al., 2009; Mencia et al., 2009]. The task of identifying true targets is difficult. Software algorithms such as miRanda, TargetScan and Pictar can be used to identify candidate genes by calculating the statistical weighting of matches with the seed region of miRNAs [John et al., 2004], but these tools must be coupled with in vitro reporter assays. For example, Lewis et al identified 132 potential target genes of miR-96 using miRanda and chose 13 for further characterization based on known inner ear expression and gene function. Of these 13 genes, only 5 - Aqp5, Celsr2, Odf2, Myrip and Ryk - were upregulated in the presence of siRNA mimicking the Dmdo miR-96 mutation, indicating loss of repression. Despite this data, analysis of human homologues of these genes in 192 hearing loss families in this study did not reveal any mutations.
The implications of miRNA involvement in the human auditory system are profound. Xu and colleagues discovered a cluster of miRNA genes whose expression is limited to sensory tissue including the inner ear [Xu et al., 2007]. Mencia et al. and Lewis et al. showed that mutations in one member of this cluster, miR-96, lead to inherited deafness. These results identify a novel miRNA-mediated regulatory system essential to mammalian hearing. By studying animal models of miRNA-induced deafness, we hope to build on this foundation by understanding more about miRNA regulation and how its dysregulation leads to disease. The challenge remains to decipher whether over-expression of miR-96 target genes represents a dominant-negative effect or subtle, widespread dysregulation of gene expression in the pathogenesis of hearing loss.
Acknowledgments
Drs Thomas Friedman and Zubair Ahmed (Laboratory of Molecular Genetics, National Institute on Deafness and Other Communication Disorders) are thanked for generously providing DNA from the Pakistani family. We also thank Tychele Turner (McKusick-Nathans Institute of Genetic Medicine, Johns Hopkins University School of Medicine) for technical assistance. RJH Smith is the Sterba Hearing Research Professor, University of Iowa College of Medicine, who supported this project in part with National Institutes of Health (NIH)-NIDCD RO1 DC003544 and RO1 DC002842. M. Hildebrand is supported by an Australian National Health and Medical Research Council (NHMRC) Postdoctoral Training Fellowship. No authors involved in this study report a conflict of interest.
Funding: This project was supported by NIH-NIDCD grants RO1 DC03544 and RO1 DC02842 (RJHS)
Abbreviations
- ADNSHL
autosomal dominant non-syndromic hearing loss
- ARNSHL
autosomal recessive non-syndromic hearing loss
- SNHL
sensorineural hearing loss
References
- Chou IC, Wan L, Liu SC, Tsai CH, Tsai FJ. Association of the Slit and Trk-like 1 gene in Taiwanese patients with Tourette syndrome. Pediatr Neurol. 2007;37:404–6. doi: 10.1016/j.pediatrneurol.2007.06.017. [DOI] [PubMed] [Google Scholar]
- Clop A, Marcq F, Takeda H, Pirottin D, Tordoir X, Bibe B, Bouix J, Caiment F, Elsen JM, Eychenne F, Larzul C, Laville E, Meish F, Milenkovic D, Tobin J, Charlier C, Georges M. A mutation creating a potential illegitimate microRNA target site in the myostatin gene affects muscularity in sheep. Nat Genet. 2006;38:813–8. doi: 10.1038/ng1810. Epub 2006 Jun 4. [DOI] [PubMed] [Google Scholar]
- Friedman LM, Dror AA, Mor E, Tenne T, Toren G, Satoh T, Biesemeier DJ, Shomron N, Fekete DM, Hornstein E, Avraham KB. MicroRNAs are essential for development and function of inner ear hair cells in vertebrates. Proc Natl Acad Sci U S A. 2009;106:7915–20. doi: 10.1073/pnas.0812446106. Epub 2009 Apr 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimberg J, Nawoschik S, Belluscio L, McKee R, Turck A, Eisenberg A. A simple and efficient non-organic procedure for the isolation of genomic DNA from blood. Nucleic Acids Res. 1989;17:8390. doi: 10.1093/nar/17.20.8390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- John B, Enright AJ, Aravin A, Tuschl T, Sander C, Marks DS. Human MicroRNA targets. PLoS Biol. 2004;2:e363. doi: 10.1371/journal.pbio.0020363. Epub 2004 Oct 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapeller J, Houghton LA, Monnikes H, Walstab J, Moller D, Bonisch H, Burwinkel B, Autschbach F, Funke B, Lasitschka F, Gassler N, Fischer C, Whorwell PJ, Atkinson W, Fell C, Buchner KJ, Schmidtmann M, van der Voort I, Wisser AS, Berg T, Rappold G, Niesler B. First evidence for an association of a functional variant in the microRNA-510 target site of the serotonin receptor-type 3E gene with diarrhea predominant irritable bowel syndrome. Hum Mol Genet. 2008;17:2967–77. doi: 10.1093/hmg/ddn195. Epub 2008 Jul 9. [DOI] [PubMed] [Google Scholar]
- Khan SY, Ahmed ZM, Shabbir MI, Kitajiri S, Kalsoom S, Tasneem S, Shayiq S, Ramesh A, Srisailpathy S, Khan SN, Smith RJ, Riazuddin S, Friedman TB. Mutations of the RDX gene cause nonsyndromic hearing loss at the DFNB24 locus. Hum Mutat. 2007;28:417–23. doi: 10.1002/humu.20469. [DOI] [PubMed] [Google Scholar]
- Kitajiri S, Fukumoto K, Hata M, Sasaki H, Katsuno T, Nakagawa T, Ito J, Tsukita S. Radixin deficiency causes deafness associated with progressive degeneration of cochlear stereocilia. J Cell Biol. 2004;166:559–70. doi: 10.1083/jcb.200402007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis MA Quint E, Glazier AM, Fuchs H, De Angelis MH, Langford C, van Dongen S, Abreu-Goodger C, Piipari M, Redshaw N, Dalmay T, Moreno-Pelayo MA, Enright AJ, Steel KP. An ENU-induced mutation of miR-96 associated with progressive hearing loss in mice. Nat Genet. 2009;41:614–8. doi: 10.1038/ng.369. Epub 2009 Apr 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mencia A, Modamio-Hoybjor S, Redshaw N, Morin M, Mayo-Merino F, Olavarrieta L, Aguirre LA, del Castillo I, Steel KP, Dalmay T, Moreno F, Moreno-Pelayo MA. Mutations in the seed region of human miR-96 are responsible for nonsyndromic progressive hearing loss. Nat Genet. 2009;41:609–13. doi: 10.1038/ng.355. Epub 2009 Apr 12. [DOI] [PubMed] [Google Scholar]
- Morton NE. Genetic epidemiology of hearing impairment. Ann N Y Acad Sci. 1991;630:16–31. doi: 10.1111/j.1749-6632.1991.tb19572.x. [DOI] [PubMed] [Google Scholar]
- Pataky F, Pironkova R, Hudspeth AJ. Radixin is a constituent of stereocilia in hair cells. Proc Natl Acad Sci U S A. 2004;101:2601–6. doi: 10.1073/pnas.0308620100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad S, Kolln KA, Cucci RA, Trembath RC, Van Camp G, Smith RJ. Pendred syndrome and DFNB4-mutation screening of SLC26A4 by denaturing high-performance liquid chromatography and the identification of eleven novel mutations. Am J Med Genet A. 2004;124A:1–9. doi: 10.1002/ajmg.a.20272. [DOI] [PubMed] [Google Scholar]
- Smith RJ, Bale JF, Jr, White KR. Sensorineural hearing loss in children. Lancet. 2005;365:879–890. doi: 10.1016/S0140-6736(05)71047-3. [DOI] [PubMed] [Google Scholar]
- Soukup GA, Fritzsch B, Pierce ML, Weston MD, Jahan I, McManus MT, Harfe BD. Residual microRNA expression dictates the extent of inner ear development in conditional Dicer knockout mice. Dev Biol. 2009;328:328–41. doi: 10.1016/j.ydbio.2009.01.037. Epub 2009 Feb 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, van der Walt JM, Mayhew G, Li YJ, Zuchner S, Scott WK, Martin ER, Vance JM. Variation in the miRNA-433 binding site of FGF20 confers risk for Parkinson disease by overexpression of alpha-synuclein. Am J Hum Genet. 2008;82:283–9. doi: 10.1016/j.ajhg.2007.09.021. Epub 2008 Jan 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weston MD, Pierce ML, Rocha-Sanchez S, Beisel KW, Soukup GA. MicroRNA gene expression in the mouse inner ear. Brain Res. 2006;1111:95–104. doi: 10.1016/j.brainres.2006.07.006. Epub 2006 Aug 10. [DOI] [PubMed] [Google Scholar]
- Wienholds E, Kloosterman WP, Miska E, Alvarez-Saavedra E, Berezikov E, de Bruijn E, Horvitz HR, Kauppinen S, Plasterk RH. MicroRNA expression in zebrafish embryonic development. Science. 2005;309:310–1. doi: 10.1126/science.1114519. Epub 2005 May 26. [DOI] [PubMed] [Google Scholar]
- Xu S, Witmer PD, Lumayag S, Kovacs B, Valle D. MicroRNA (miRNA) transcriptome of mouse retina and identification of a sensory organ-specific miRNA cluster. J Biol Chem. 2007;282:25053–66. doi: 10.1074/jbc.M700501200. Epub 2007 Jun 27. [DOI] [PubMed] [Google Scholar]


