Abstract
Molecular imaging of separate but still incompletely resolved spots on high-performance thin-layer chromatography (HPTLC) plates is used for the direct analysis of porcine brain lipids by desorption electrospray ionization mass spectrometry (DESI-MS). Seven class-specific spots were imaged in the negative ion mode and shown to contain more than fifty lipids. A low lateral resolution of 400 × 400 μm allowed simple, rapid and incomplete separation to be combined with DESI imaging for the identification of many components of these extremely complex mixtures. In this work, tandem mass spectrometry (MS/MS) was also employed to confirm the identity of particular lipids directly on HPTLC plates.
Keywords: Ambient ionization, 2D chromatography, lipids, derivatization, imaging mass spectrometry, in-situ analysis, screening
Introduction
Lipids and their metabolites play a vital role in a variety of biological processes including cell–cell adhesion, storage of energy, cell signaling, localization of membrane proteins and enzymatic modulation as cofactors. Various classes of lipids have characteristic distributions in different tissues, cell membranes and organelles. Lipids in biological specimens can sometimes be used as biomarkers for disease1. The brain, in which the lipids account for almost half of the organ dry weight, contains a large variety of lipids of widely differing polarity. The polar lipids are predominantly represented by phospholipids (50–60 mol% of lipid mass) and by sphingolipids and glycolipids which represent approximately 5–10 mol% of total lipid mass.2
Several methods are available for lipid analysis, among the most common are those based on chromatography coupled to mass spectrometry (MS).3, 4 Electron ionization (EI), electrospray ionization (ESI), and matrix-assisted laser desorption ionization (MALDI) are well established ionization techniques that allow interrogation of a wide range of lipids. The very large number of related lipids in biological materials means that a separation step is usually required before MS identification although categorization studies, including tissue imaging to determine disease state can be done without accompanying separations.5, 6
Thin-layer chromatography (TLC) is a robust, rapid, inexpensive and reasonably powerful technique for lipid separation. It is routinely used in synthetic labs and it can be used to perform the separation of many samples in parallel.7 Normally, after TLC separation the compounds are scraped off the TLC plate, then extracted using a solvent and ionized and analyzed by MS. This procedure is time-consuming, and the reproducibility and recovery of lipids is sometimes poor, due mainly to spot overlap and/or loss of sample.
Direct coupling of TLC with MS analysis goes back to work in the early 1980’s.8 Alkaloids from mushroom extracts (I. napipes) were separated on cellulose thin-layer plates and directly ionized using secondary ion mass spectrometry (SIMS) followed by quadrupole mass analysis. The thin-layer plates were introduced in the vacuum chamber and the spots subjected to keV energy argon ion bombardment. Since the chromatographic support is an insulator, the surface was flooded with low-energy electrons in order to neutralize surface positive charge build up. In a related experiment, Francel’s group analyzed antiprotozoal agents separated on TLC by Fast Atom Bombardment (FAB-MS).9 In their experiments, the tip of a FAB probe, coated with a piece of double-faced masking tape, was pressed against the TLC spot of interest, transferring it to the coated adsorbent. A suitable solvent (dichloromethane or methanol) and a FAB matrix liquid (glycerine or thioglycerine) were added to the TLC adsorbent adhering to the probe tip. The probe tip was then introduced into the mass spectrometer to acquire FAB spectra in the usual manner. The coupling of TLC to MALDI was introduced by Hercules’ group10 and imaging capabilities were also demonstrated.10, 11
Contemporary interest in these direc-analysis methods continues, following improvements in both TLC and MS technologies.12–18 The introduction of the ambient ionization techniques19–21 has made possible the direct analysis of TLC plates outside of the MS vacuum without the requirement for matrix addition. For instance, Van Berkel et al. introduced a combined surface sampling probe/ESI emitter for the direct analysis of TLC plates. In this device, a solvent is pumped through a probe onto the TLC surface where it creates a liquid film. The chemicals present on the TLC plate dissolve at this microjunction and they are aspirated back into a sampling capillary. At the end of the sampling capillary, the sample is sprayed toward the mass spectrometer.22, 23
Desorption electrospray ionization (DESI)16, 24–27 and the closely related ambient sonic spray ionization (EASI)18, 28 method have also been successfully used to characterize compounds on TLC plates. In DESI, a solvent is electrosprayed onto the surface where it forms a thin film; secondary scattered droplets are generated as subsequently arriving primary droplets splash into the film. As the analyte-containing droplets are carried through the atmosphere into the vacuum interface the solvent evaporates generating ionized molecules that can be mass analyzed29, 30 (Figure 1).
Figure 1.
DESI source schematics. Adapted from Ref 50
DESI is carried out in the native environment at ambient pressure without addition of matrices, which can cause interference in the low molecular range. Ionization by DESI is soft, occurring without or with minimum fragmentation, especially when compared to SIMS. One noteworthy feature of DESI is that reagents can be added to the solvent. These reagents can be used to selectively tag analytes of interest making them easier to detect, e.g. by generating preformed ions which yield gas phase ions with high efficiency. This experiment is usually called reactive-DESI.31–34
Recently, the possibility of analyzing phospholipids by coupling high-performance (HP) TLC with MALDI-MS35–37 has been investigated. The critical step in this experiment is the addition of the matrix to the TLC plate. In other recent work, MALDI-ToF-MS was used to characterize cyanobacterial toxins (cyclopeptides) after HPTLC separation.38
Here, we present the analysis of complex lipid mixtures based on separation by 2D-HPTLC followed by direct interrogation of the components on the TLC plates using DESI-MS. The polar lipids are identified by the position of the TLC spot which identifies the lipid class and then by the main features of the mass spectrum with confirmation of the identification of individual lipids coming from the MS/MS product ion spectra of putative molecular ions. The possibility of mapping the distribution of non-polar lipids is briefly explored by reactive DESI using betaine aldehyde to derivatize cholesterol so generating the quaternary which can be observed directly from the plate.34 The work is motivated by the desire to simplify mapping for the study of inborn errors of metabolism involving lipids that can require multiple sample preparation and or detection steps such as scrapping TLC plates, radioactivity measurements and gas-chromatography.39,40
Material and Methods
Material
Chloroform, methanol, triethylamine, and acetic acid were purchased from Mallinckrodt (Phillipsburg, NJ). Water was purified (18MΩ cm) using a PureLab ultra system by Elga LabWater (High Wycombe, UK) before use. Betaine aldheyde and primuline were from Sigma-Aldrich (St. Louis, MO). Total porcine brain extract was provided by Avanti Polar Lipids (Alabaster, AL). The extract was provided and diluted in chloroform (see supporting material for more information).
Thin Layer Chromatography
Separation by HPTLC was performed on 5×5cm Nano-Silica XHL HPTLC plates, glass backed, 200 μm stationary phase, (Sorbent Technologies, Inc., Atlanta). Aliquots containing 10 μg of total lipids brain porcine extract were applied at the lower-left corner of the HPTLC plates. The plates were developed in the first dimension in solvent system I [chloroform-triethylamine-methanol-water 35:35:35:7 (v/v/v/v)]. After drying the plates in air, the second dimension was developed in solvent system II [chloroform-methanol-acetic acid 65:35:8 (v/v/v)]. The plates were air dried again and some of them sprayed with the dye primuline (0.05% in water:acetone 8:2 (v/v)), to allow visualization of the spots under a UV lamp. Retention factors for each spot, for the first and the second dimension (Rf1 and Rf2) were calculated as the distance traveled by the compound divided by the distance traveled by the solvent. All plates were then subjected to MS analysis.
Desorption Electrospray Ionization Mass Spectrometry
DESI-MS and reactive DESI-MS were performed in both positive and negative ion modes, using a Thermo Finnigan LTQ (San Jose, CA) linear ion trap mass spectrometer equipped with the custom-built, automated DESI ion source, which is described in detail elsewhere.41, 42 The experiments were conducted in the full scan mode; methanol was the spray solvent for conventional (non-reactive) DESI while acetonitrile containing betaine aldheyde (20 ppm) was employed for reactive-DESI-MS. In both cases the flow rate of solvent was set at 20 μL/min. The DESI source was operated under conditions such that the incoming droplets were sprayed from the emitter placed 2 mm from the surface. The incident angle was set at 55° to the surface plane and the collection angle was 20°.
Imaging of TLC plates
Imaging of the TLC plates was performed by continuously scanning the surface in the x-direction at a surface velocity of 300 μm/s while acquiring mass spectra every 1.33s in full scan mode over the range m/z 200–1500. At the end of each x-direction scan line, a 400μ step was made in the y-direction and the cycle restarted until the entire area (area 4cm×4cm) had been covered. This procedure resulted in the collection of 10,000 mass spectra from an array of 100×100 pixels. More details about image acquisition can be found in the literature. 25, 41
Results and Discussion
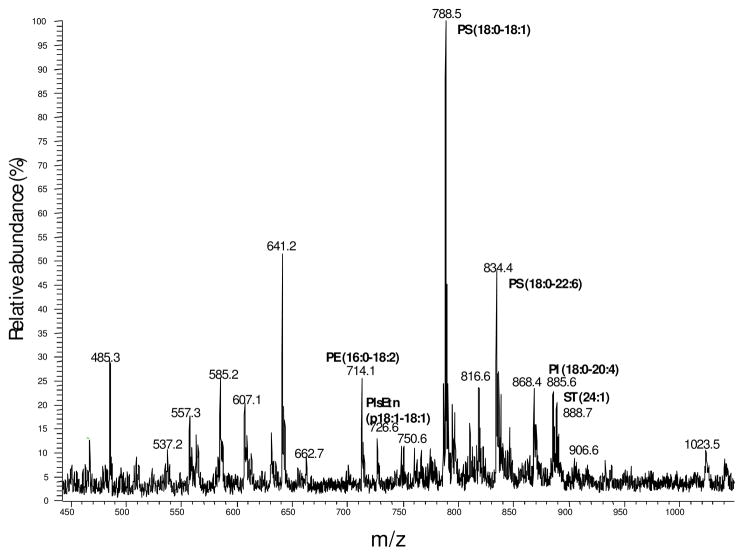
The DESI-MS ion source used in this study is depicted in Figure 1. The analyte, a porcine brain extract, was initially interrogated by DESI-MS without prior HPTLC separation. The presence of phosphatidylserine, PS (18:0–18:1) and PS (18:0–22:6); phosphatidylethanolamine, PE(16:0–18:2); phosphatidylinositol, PI(18:0–20:4); and sulfatide, ST(24:1) in the extract can be seen in the spectrum shown in Figure 2. However, the identification of the other lipids is very difficult, due to peak overlap of isobaric compounds, 13C isotopic contributions, and the fact that signal intensities for each lipid depend on ionization efficiencies as well as differences in concentration.
Figure 2.
Negative ion mode DESI-MS spectrum of total lipid brain porcine extract without HPTLC separation.
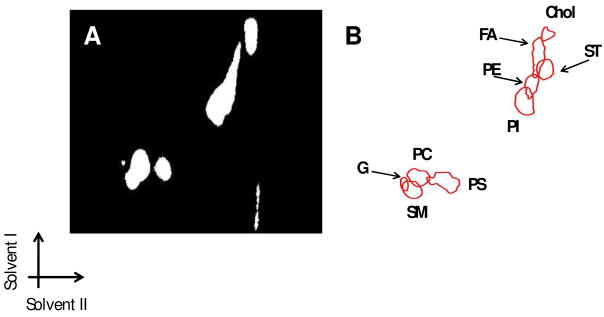
HPTLC images were acquired both by DESI-MS and using optical/UV methods, as shown in Fig. 3. The advantage of DESI-MS imaging is that it provides chemical information, allowing exact recognition of lipids by their mass to charge ratios (m/z) and confirmation by their collision-induced dissociation MS/MS data. Although the spots corresponding to different lipid classes are not completely resolved, the specificity and selectivity achieved by the MS and MS/MS detection allows the resolution of overlapping spots in dozens of cases, as discussed below. The derivatization of the lipids on the HPTLC plate with primuline allows visualization of the spot using UV light (Fig. 3a). In this way the DESI experiment can be performed in the spot sampling mode, by directly positioning the sprayer on a spot which one wishes to investigate. Neither products of reaction nor clusters involving primuline and lipids were observed in the mass spectra. The background was not subtracted.
Figure 3.
(A) UV/optical image of the HPTLC plate and (B) DESI-MS overlayed drawing of the HPTLC plate (see Fig. 4 for specific representative ions). The bottom left arrows show the direction of the solvent system applied to the plate. Chol, cholesterol; FA, fatty acids; ST, sulfatides; PE, phosphatidylethanolamine; PI, phosphatidylinositol; PS, phosphatidylserine; PlsEtn, Plamenylethanolamine; PC*, Phosphatidylcholine; SM*, Sphingomyelin; G, Ganglioside (*acetate adducts).
The HPTLC plate was spotted with analyte and developed in two dimensions, placed on the sampling stage and interrogated using the imaging DESI-MS system. During the HPTLC process, lipids were reasonably well separated according to polarity; the less polar species exhibit the highest Rf value. The more polar spots contain lipids such as gangliosides (G), phosphatidylcholine (PC) and sphingomyelin (SM), which are more hydrophilic than the lipids which have higher Rf values in both directions. Especially for the polar lipids the flow rate of the solvent sprayed was a critical parameter to obtain efficient desorption/ionization. Low flow rates, from 1–8 μl/min, as usually used during DESI-MS experiments, do not allow desorption of Gs and provide only a weak signal for the PCs. Flow rates in the range of 15–20 μl/min (spot size ~1 mm), are necessary to produce adequate signal intensity, especially for the more polar compounds. High flow rates (higher than 20 μl/min) produce too large spot sizes which are not suitable for the imaging experiments.
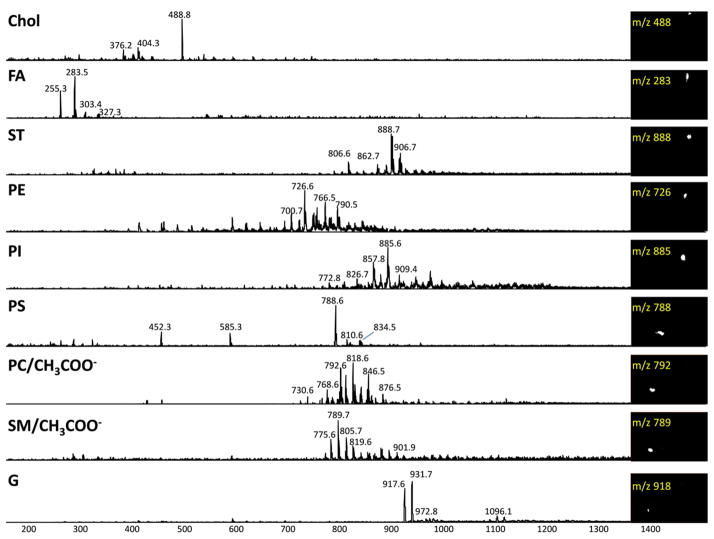
In Figure 4 we present images of each spot and the corresponding MS spectra in the negative ion mode; eight different spots were obtained corresponding to G (Rf1=0.18; Rf2=0.15), SM (Rf1=0.15; Rf2=0.19), PC (Rf1=0.20; Rf2=0.23), PS (Rf1=0.19; Rf2=0.35), PI (Rf1=0.58; Rf2=0.73), PE (Rf1=0.65; Rf2=0.78), ST (Rf1=0.73; Rf2=0.83) and fatty acids (FA) (Rf1=0.79; Rf2=0.80).
Figure 4.
Mass spectra of TLC spots. Except for Chol, mapped in positive ion mode, all spectra were acquired in negative ion mode. Each class of lipids was mapped by selecting one specific representative ion (showed in the inset). Chol, cholesterol; FA, fatty acids; ST, sulfatides; PE, phosphatidylethanolamine; PI, phosphatidylinositol; PS, phosphatidylserine; PlsEtn, Plamenylethanolamine; PC*, Phosphatidylcholine; SM*, Sphingomyelin; G, Ganglioside (*acetate adducts).
The spot containing cholesterol, Chol (Rf1=0.90; Rf2= 0.88) was also mapped by reactive-DESI in the positive ion mode. Due to its low proton affinity, this compound cannot be detected when ordinary spray solvents are used. However, when betaine aldehyde is added to the spray, the cholesterol derivative, generated in situ, can be monitored easily.34
Although the possibility of exploiting the HPTLC/DESI-MS methodology for quantitative studies was not investigated, it was possible to obtain the percent composition of the molecular species in each spot using the peak intensity normalized to the sum of the intensity of all peaks. These data were obtained by interrogating, in the spot sampling mode, three different sample areas from each spot, resulting in three different spectra from each lipid class. Similarities in ionization efficiency between members of the same class probably make this a better measurement than it would otherwise be if interclass comparisons were involved. More than fifty different lipid species were identified. Table 1 reports the phospholipids and sphingolipids detected in the negative mode and their percent composition. Note that the relative standard deviation (RSD%) just reports the precision of the method, since the real (theoretical) values for the lipid concentration (or their ratios) are not known; the relative error or bias (measurement of accuracy) could not be estimated.
Table 1.
Phospholipids and Sphingolipids Identified in the Negative Ion Mode
| PC | m/z | Molecular species | % ± SD (n=3) |
|---|---|---|---|
| 790.4 | 16:0-16:1 | 3.6 ± 1.7 | |
| 792.5 | 16:0-16:0 | 18.8 ± 3.9 | |
| 818.5 | 16:0-18:1 | 36.2 ± 2.6 | |
| 820.5 | 16:0-18:0 | 11.4 ± 0.7 | |
| 846.5 | 18:1-18:2 | 13.8 ± 1.4 | |
| 848.4 | 18:1-18:1 | 12.3 ± 3.5 | |
| 864.3 | 16:0-22:6 | 6.0 ± 1.1 | |
| SM | m/z | Molecular species | % ± SD |
| 747.5 | d33:1 | 3.2 ± 1.6 | |
| 759.7 | d34:2 | 2.5 ± 0.6 | |
| 761.7 | d34:1 | 3.7 ± 2.3 | |
| 775.7 | d35:1 | 14.2 ± 2.4 | |
| 777.7 | d35:0 | 5.2 ± 1.6 | |
| 789.5 | d36:1 | 42.1 ± 4.2 | |
| 791.5 | d36:0 | 7.5 ± 1.6 | |
| 815.7 | d38:2 | 2.0 ± 0.6 | |
| 817.7 | d38:1 | 9.9 ± 2.6 | |
| 819.7 | d38:0 | 9.6 ± 2.0 | |
| PE | m/z | Molecular Species | % ± SD |
| 714.5 | D16:0-18:2 | 0.7 ± 0.2 | |
| 716.5 | D16:0-18:1 | 2.9 ± 1.7 | |
| 742.6 | D18:1-18:1 | 5.1 ± 0.7 | |
| 744.6 | D18:0-18:1 | 4.3 ± 2.8 | |
| 764.5 | D18:1-20:4 | 2.6 ± 0.4 | |
| 766.5 | D18:0-20:4 | 6.6 ± 3.9 | |
| 790.5 | D18:0-22:6 | 9.5 ± 1.1 | |
| 792.5 | D18:0-22:5 | 5.1 ± 1.1 | |
| 794.6 | D20:0-20:4 | 5.3 ± 1.0 | |
| PS | m/z | Molecular species | % ± SD |
| 760.6 | 16:0-18:1 | 4.0 ± 1.0 | |
| 786.5 | 18:0-18:2 | 7.6 ± 0.2 | |
| 788.5 | 18:0-18:1 | 55.6 ± 1.2 | |
| 790.5 | 18:0-18:0 | 7.0 ± 0.1 | |
| 810.6 | 18:0-20:4 | 7.1 ± 2.4 | |
| 834.4 | 18:0-22:6 | 8.2 ± 0.7 | |
| 836.6 | 18:0-22:5 | 5.5 ± 1.3 | |
| 838.5 | 18:0-22:4 | 5.0 ± 0.5 | |
| PI | m/z | Molecular Species | % ± SD |
| 857.8 | 16:0-20:4 | 16.3 ± 1.3 | |
| 859.7 | 18:0-18:3 | 7.7 ± 0.7 | |
| 861.6 | 18:0-18:2 | 8.3 ± 0.4 | |
| 883.6 | 18:1-20:4 | 19.1 ± 1.4 | |
| 885.6 | 18:0-20:4 | 28.2 ± 2.2 | |
| 887.6 | 18:0-20:3 | 9.8 ± 0.2 | |
| 909.4 | 18:0-22:6 | 6.4 ± 0.6 | |
| 911.5 | 18:0-22:5 | 6.3 ± 0.8 | |
| PlsEtn | m/z | Molecular Species | % ± SD |
| 700.7 | P16:0-18:1 | 5.3 ± 1.8 | |
| 722.6 | P16:0-20:4 | 2.8 ± 0.8 | |
| 726.6 | P18:1-18:1 | 13.5 ± 0.1 | |
| 728.6 | P18:0-18:1 | 5.6 ± 0.2 | |
| 748.6 | P18:1-20:4 | 6.6 ± 2.9 | |
| 750.6 | P18:0-20:4 | 9.6 ± 3.3 | |
| 774.6 | P18:0-22:6 | 4.9 ± 1.0 | |
| 776.5 | P18:0-22:5 | 4.5 ± 0.6 | |
| 778.7 | P20:0-20:4 | 5.2 ± 1.2 | |
Table S2 reports the FA percent composition in each phospholipids class (specifically, the % of each FA moiety was calculated as the sum of % of phospholipids containing the respective FA moiety, followed by normalization to the sum of all FA % moieties. The FA % corresponding to symmetric phospholipids was multiplied by a factor of 2 to account for two identical FA moieties). The predominant PC acyl chains were found to be those represented by C16:0 and C18:1, while high contents of C18:0 and C18:1 were detected for PS and C20:4 for PE and PI. Saturated FA’s were found in higher percentages for PC and PS, while PE and PI contain greater amounts of polyunsaturated fatty acids (PUFA) (Table S2b).
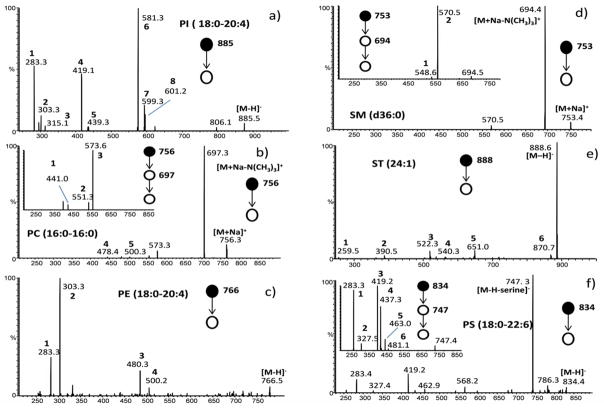
Tandem mass spectrometry (MSn) was employed in the spot sampling mode to confirm and characterize the lipid composition of each spot. The assignment was performed by comparison between the spectra of unknown lipids and those acquired from standards or, when available, those described in the literature. For systematic studies on collision induced dissociation of phospholipids ionized by ESI and DESI see references 42 and 43. Briefly, in the negative ion mode DESI-MS/MS for the phospholipids undergoes the loss of neutral carboxylic acid (RCOOH) and the loss of the corresponding ketene (R′CH=C=O). The products of these two neutral fragment losses and the fatty acid anions, which were also observed, allow acyl chain assignment and the structural characterization for each phospholipids species. Some further information can be obtained by the loss of the head group such as the loss of serine from PS (Fig. 5).
Figure 5.
Product ion MS/MS spectra for (a) PI (18:0–20:4) (1) [(C18:0) R1COO]− ion, (2) [(C20:4) R2COO]− ion, (3) [glyceroinositolphosphate-H2O-H]− ion, (4) [M - H - (R′2CH=C=O) - inositol] ion, (5) [M - H - (R′1CH=C=O) - inositol]− ion, (6) [M - H - (R2COOH)]− ion, (7) [M - H - (R′1CH=C=O)]− ion, (8) [M - H - (R1COOH)]− ion. (b) MS/MS and MS3 of PC (16:0–16:0), (1) [M + Na – N(CH3)3 – (C16 RCOOH)]+, (2) [M – N(CH3)3 - (C2H5 PO4)]+, (3) [M + Na – N(CH3)3 - (C2H5 PO4)]+, (4) [M – (C16 RCOOH)]+, (4) [M + Na – (C16 RCOOH)]+. (c) MS/MS of PE (18:0–20:4) (1) [(C18:0) R1COO]− ion, (2) [(C20:4) R2COO]− ion, (3) [M – H - (R′2CH=C=O)]− ion, (3) [M – H - (R′1CH=C=O)]−. (d) MS/MS and MS3 of SM (d36:0) (1) [M – N(CH3)3 - (C2H5 PO4)]+, (2) [M + Na – N(CH3)3 - (C2H5 PO4)]+. (e) MS/MS of ST (24:1) (1) [galactose sulfate –H]−, (2) [M-H - galactose sulfate - C16H30O]−, (3) [M-H - H2O - C22H43CH=C=O]−, (4) [M - H - C22H43CH=C=O]−, (5) [M - H - C16H30O]−, (6) [M -H-H2O]−. (f) MS/MS and MS3 of PS (18:0–22:), (1) [(C18:0) R1COO]− ion, (2) [(C22:6) R2COO]− ion, (3) [M – H – (C3H5NO2) – R2COOH]−, (4) [M – H – (C3H5NO2) – R′2C=C=O]−, (5) [M – H – (C3H5NO2) – R1COOH]−, (6) [M – H – (C3H5NO2) – R′1C=C=O]−.
PI consists of a phosphatidic acid backbone, linked via the phosphate group to inositol (hexahydroxycyclohexane). It is acidicity allows it to be successfully ionized in the negative mode. Figure 5 (panel a) shows the product ion spectra obtained by the selection of the anions m/z 885. The most abundant ion recorded at m/z 581 corresponds to the loss of ketene of C20:4. While the ion of m/z 599 corresponds to the loss of C18:0 ketene. The peaks at m/z 419 were generated by the loss of the inositol portion and ketene from C20:4. Other peaks at m/z 283 and 303 are related to the fatty acid anions.
The choline-containing species generally ionize better in the positive ion mode than in the negative ion mode, due to the permanent positive charge located on their head group. Nevertheless adduct formation can be used to detect these species in negative mode DESI (and ESI) experiments, by adding acetate to the sprayer (or sample) solution.42, 44 During the HPTLC-DESI-MS experiment, PC and SM were recorded both in positive and in negative modes. Acetate adduct ions were obtained for PC and SM in the negative ion mode (Fig. 4), and mass-selection and collisional activation of the acetate adducts yielded one major product ion that corresponds to the loss of methyl acetate (data not shown). No acetate was added to the spray solvent but acetic acid was used as component of solvent mixture during the HPTLC separation. In the positive ion mode PC and SM were recorded principally as sodiated adducts; the most abundant fragment ions generated from the sodiated adducts are [M + Na - N(CH3)3]+ corresponding to the loss of trimethylamine; another characteristic fragment also observed is the ion [M + Na - N(CH3)3 - (C2H5 PO4)]+ corresponding to the loss of phosphocholine (Fig. 5, panels b and d).
Phosphatidylethanolamines were better ionized in the negative mode, although the ion positive mode also provides PE profiles. Structural elucidation was achieved by collision induced dissociation (Fig. 5 panel c), which revealed fragments of the fatty acid anions at m/z 283 (C18:0) and m/z 303 (C20:4) and the loss of their related ketenes (m/z 500 and m/z 480).
Sulfatides are glycosphingolipids which are formed from 3-sulfate esters of galactosylcerebrosides. These compounds are easily ionized in the negative ion mode and different molecular species were identified during the HPTLC-DESI-MS experiments (Table 1). STs give structural information upon collision-induced dissociation since the neutral loss of ketene from the fatty acid chain and the generation of the galactose sulfate anion, provide key structural information. (Fig. 5, panel e) negative ion product spectra of each of the PSs generated a characteristic fragment ion [M-H-serine]−, which is diagnostic for this class, as depicted in Fig. 5 (panel f) where the ion m/z 747 is the peak corresponding to the loss of serine. PS acyl chains were identified by the assignment of fatty acids anions and by the loss of neutral carboxylic acid selecting the ion [M-H-serine]− in the MS3 study.
Gangliosides, which contain sugar chains with different numbers of sialic acid residues, are often denoted by the “Svennerholm” nomenclature that is based on the number of sialic acid residues and a number reflecting the relative position of the gangliosides upon thin layer chromatography.45, 46 Fig. S1 (panel b) shows full scan MS spectra on spot G in the negative ion mode. Two main peaks were detected at m/z 917 and m/z 931 and assigned as doubly charged disialotetrahexosylgangliosides (GD1), GD1(d36:1) and GD1(d38:1), respectively. These two gangliosides were also identified as their sodium adducts at m/z 1857 and m/z 1885 while the peaks at m/z 1836 and 1864 represent the same two gangliosides with single charges. Monosialotetrahexosylgangliosides (GM1) containing d36:1 and d38:1 chains were also identified as singly charged ions at m/z 1544 and 1572, respectively. To identify these molecules, MS/MS experiments were performed and they showed the typical fragmentation pathway of these gangliosides. In Figure S1 (panel a) the product ion spectra of m/z 917 is shown. The typical fragment ion at m/z 290, designated C1β - H2O, reflects the fragment of sialic acid less water while cleavage at the glycosidic bond of sialic acid in α, provides the fragment Y2β at m/z 1544. The cleavage of both the sialic acid in β and in α, generates the fragment at m/z 1253 (Y4α/Y2β). The product ion spectrum obtained in Figure 4 is consistent with that presented by Merrill et al.47
Conclusion
Total brain porcine extract is a complex mixture containing a very large variety of lipids. As such it provides a most stringent test of analytical performance even at the qualitative identification level. Direct DESI-MS analysis of such an extract is used to identify the more abundant lipids. However, due to spectral congestion, ion-suppression and/or peak overlapping (present in unit-resolution mass spectrometers) only a partial characterization is achieved. Two-dimensional HPTLC provides modest but sufficient clean up and allows separation of class-specific lipids. This allows identification of a much larger number of lipids. For the lipid mixture evaluated here, eight different spots corresponding to class-specific lipids were obtained in the negative ion mode during the imaging of the plates. The identification of the particular lipids present in each class of lipids was achieved by tandem mass spectrometry using product ion spectra. The lateral resolution used in these studies was only 400 × 400 μm in order to allow more rapid analysis than a higher spatial resolution and it seems to be an appropriate choice for TLC-MS imaging applications. The resolution can be changed as needed in order to customize applications of the technique.25, 41
Extensive adduct formation can have deleterious consequences in more complex samples. Nevertheless, during this work this phenomenon has been exploited for the identification/ionization of such phospholipids as choline containing species, which, in this way, could be detected in the negative mode as their acetate adducts and in the positive mode as sodium adducts. For most other lipids, the formation of adducts did not negatively influence the analysis.
Cholesterol was mapped in the positive ion mode by reactive-DESI adding betaine aldehyde to the spray, and monitoring the in situ generated cholesterol derivative.34 Quantitation or determination of lower limits of detection was not evaluated in this work although the use of internal or external standards could make this task feasible as demonstrated earlier in the case of DESI48 or MALDI-TLC49.
Besides phospholipids and sphingolipids, the detection of some gangliosides was achieved and they were characterized by MS/MS. HPTLC-DESI-MS is a useful tool to characterize complex mixture of lipids, as shown by the total porcine brain extracts studied here polar lipids are successfully ionized by DESI. This study illustrates the application of DESI directly on TLC plates for a better characterization of complex matrices.
Supplementary Material
Figure S1. (a) Product ion spectra of GD1 (m/z 917) obtained in spot sampling mode. (b) Full scan mass spectra obtained by DESI analysis in spot sampling mode of gangliosides.
Table S2. Relative fatty acid percentage composition of phopspholipids detected during HPTLC-DESI-MS analysis
Acknowledgments
This work was supported by U.S. National Institutes of Health (NIH grant 1R21EB009459-01) and the Office of Naval Research (N00014-05-1-0405).
References
- 1.Vence DE, Vence J. Biochemistry of Lipids, Lipoproteins and Membranes. Elsevier; Amsterdam: 1996. [Google Scholar]
- 2.Han XL, Holtzman DM, McKeel DW, Kelley J, Morris JC. J Neurochem. 2002;82:809–818. doi: 10.1046/j.1471-4159.2002.00997.x. [DOI] [PubMed] [Google Scholar]
- 3.Watson AD. J Lipid Res. 2006;47:2101–2111. doi: 10.1194/jlr.R600022-JLR200. [DOI] [PubMed] [Google Scholar]
- 4.Han XL, Gross RW. Mass Spectrom Rev. 2005;24:367–412. doi: 10.1002/mas.20023. [DOI] [PubMed] [Google Scholar]
- 5.Wiseman JM, Puolitaival SM, Takats Z, Cooks RG, Caprioli RM. Angew Chem-Int Edit. 2005;44:7094–7097. doi: 10.1002/anie.200502362. [DOI] [PubMed] [Google Scholar]
- 6.Dill AL, Ifa DR, Manicke NE, Costa AB, Ramos-Vara JA, Knapp DW, Cooks RG. Anal Chem. 2009 doi: 10.1021/ac901028b. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Touchstone JC. J Chrom B. 1995;671:169–195. doi: 10.1016/0378-4347(95)00232-8. [DOI] [PubMed] [Google Scholar]
- 8.Unger SE, Vincze A, Cooks RG, Chrisman R, Rothman LD. Anal Chem. 1981;53:976–981. [Google Scholar]
- 9.Chang TT, Lay JO, Francel RJ. Anal Chem. 1984;56:109–111. [Google Scholar]
- 10.Gusev AI, Proctor A, Rabinovich YI, Hercules DM. Anal Chem. 1995;67:1805–1814. [Google Scholar]
- 11.Gusev AI, Vasseur OJ, Proctor A, Sharkey AG, Hercules DM. Anal Chem. 1995;67:4565–4570. [Google Scholar]
- 12.Mehl JT, Hercules DM. Anal Chem. 2000;72:68–73. doi: 10.1021/ac990003y. [DOI] [PubMed] [Google Scholar]
- 13.Chai WG, Leteux C, Lawson AM, Stoll MS. Anal Chem. 2003;75:118–125. doi: 10.1021/ac025833h. [DOI] [PubMed] [Google Scholar]
- 14.Santos LS, Haddad R, Hoehr NF, Pilli RA, Eberlin MN. Anal Chem. 2004;76:2144–2147. doi: 10.1021/ac035387d. [DOI] [PubMed] [Google Scholar]
- 15.Dreisewerd K, Muthing J, Rohlfing A, Meisen I, Vukelic Z, Peter-Katalinic J, Hillenkamp F, Berkenkamp S. Anal Chem. 2005;77:4098–4107. doi: 10.1021/ac048373w. [DOI] [PubMed] [Google Scholar]
- 16.Van Berkel GJ, Ford MJ, Deibel MA. Anal Chem. 2005;77:1207–1215. doi: 10.1021/ac048217p. [DOI] [PubMed] [Google Scholar]
- 17.Dreisewerd K, Lemaire R, Pohlentz G, Salzet M, Wisztorski M, Berkenkamp S, Fournier I. Anal Chem. 2007;79:2463–2471. doi: 10.1021/ac061768q. [DOI] [PubMed] [Google Scholar]
- 18.Haddad R, Milagre HMS, Catharino RR, Eberlin MN. Anal Chem. 2008;80:2744–2750. doi: 10.1021/ac702216q. [DOI] [PubMed] [Google Scholar]
- 19.Venter A, Nefliu M, Cooks RG. TrACs. 2008;27:284–290. [Google Scholar]
- 20.Van Berkel GJ, Pasilis SP, Ovchinnikova O. J Mass Spectrom. 2008;43:1161–1180. doi: 10.1002/jms.1440. [DOI] [PubMed] [Google Scholar]
- 21.Harris GA, Nyadong L, Fernandez FM. Analyst. 2008;133:1297–1301. doi: 10.1039/b806810k. [DOI] [PubMed] [Google Scholar]
- 22.Ford MJ, Van Berkel GJ. Rapid Commun Mass Spectrom. 2004;18:1303–1309. doi: 10.1002/rcm.1486. [DOI] [PubMed] [Google Scholar]
- 23.Van Berkel GJ, Sanchez AD, Quirke JME. Anal Chem. 2002;74:6216–6223. doi: 10.1021/ac020540+. [DOI] [PubMed] [Google Scholar]
- 24.Pasilis SP, Kertesz V, Van Berkel GJ, Schulz M, Schorcht S. Anal Bioanal Chem. 2008;391:317–324. doi: 10.1007/s00216-008-1874-6. [DOI] [PubMed] [Google Scholar]
- 25.Van Berkel GJ, Kertesz V. Anal Chem. 2006;78:4938–4944. doi: 10.1021/ac060690a. [DOI] [PubMed] [Google Scholar]
- 26.Van Berkel GJ, Tomkins BA, Kertesz V. Anal Chem. 2007;79:2778–2789. doi: 10.1021/ac0622330. [DOI] [PubMed] [Google Scholar]
- 27.Pasilis SP, Kertesz V, Van Berkel GJ, Schulz M, Schorcht S. J Mass Spectrom. 2008;43:1627–1635. doi: 10.1002/jms.1431. [DOI] [PubMed] [Google Scholar]
- 28.Eberlin LS, Abdelnur PV, Passero A, de Sa GF, Daroda RJ, de Souza V, Eberlin MN. Analyst. 2009;134:1652–1657. doi: 10.1039/b817847j. [DOI] [PubMed] [Google Scholar]
- 29.Costa AB, Cooks RG. Chem Phys Let. 2008;464:1–8. [Google Scholar]
- 30.Costa AB, Cooks RG. Chem Comm. 2007:3915–3917. doi: 10.1039/b710511h. [DOI] [PubMed] [Google Scholar]
- 31.Chen H, Cotte-Rodriguez I, Cooks RG. Chem Comm. 2006:597–599. doi: 10.1039/b516448f. [DOI] [PubMed] [Google Scholar]
- 32.Nyadong L, Green MD, De Jesus VR, Newton PN, Fernandez FM. Anal Chem. 2007;79:2150–2157. doi: 10.1021/ac062205h. [DOI] [PubMed] [Google Scholar]
- 33.Huang G, Chen H, Zhang X, Cooks RG, Ouyang Z. Anal Chem. 2007;79:8327–8332. doi: 10.1021/ac0711079. [DOI] [PubMed] [Google Scholar]
- 34.Wu C, Ifa DR, Manicke NE, Cooks RG. Anal Chem. 2009;81:7618–7624. doi: 10.1021/ac901003u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rohlfing A, Muthing J, Pohlentz G, Distler U, Peter-Katalinic J, Berkenkamp S, Dreisewerd K. Anal Chem. 2007;79:5793–5808. doi: 10.1021/ac070633x. [DOI] [PubMed] [Google Scholar]
- 36.Fuchs B, Schiller J, Suss R, Schurenberg M, Suckau D. Anal Bioanal Chem. 2007;389:827–834. doi: 10.1007/s00216-007-1488-4. [DOI] [PubMed] [Google Scholar]
- 37.Fuchs B, Schiller J, Suess R, Zscharnack M, Bader A, Mueller P, Schuerenberg M, Becker M, Suckau D. Anal Bioanal Chem. 2008;392:849–860. doi: 10.1007/s00216-008-2301-8. [DOI] [PubMed] [Google Scholar]
- 38.Meisen I, Distler U, MuÌt̂hing J, Berkenkamp S, Dreisewerd K, Mathys W, Karch H, Mormann M. Anal Chem. 2009;81:3858–3866. doi: 10.1021/ac900217q. [DOI] [PubMed] [Google Scholar]
- 39.Brunetti-Pierri N, Corso G, Rossi M, Ferrari P, Balli F, Rivasi F, Annunziata I, Ballabio A, Dello Russo A, Andria G, Parenti G. Am J Hum Genet. 2002;71:952–958. doi: 10.1086/342668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pianese P, Salvia G, Campanozzi A, D’Apolito O, Dello Russo A, Pettoello-Mantovani M, Corso G. J Pediatr Gastroenterol Nutr. 2008;47:645–651. doi: 10.1097/MPG.0b013e318170956a. [DOI] [PubMed] [Google Scholar]
- 41.Ifa DR, Wiseman JM, Song QY, Cooks RG. Int J Mass Spectrom. 2007;259:8–15. [Google Scholar]
- 42.Manicke NE, Wiseman JM, Ifa DR, Cooks RG. J Am Soc Mass Spectrom. 2008;19:531–543. doi: 10.1016/j.jasms.2007.12.003. [DOI] [PubMed] [Google Scholar]
- 43.Merrill AH, Stokes TH, Momin A, Park H, Portz BJ, Kelly S, Wang E, Sullards MC, Wang MD. J Lipid Res. 2009;50:S97–S102. doi: 10.1194/jlr.R800073-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kerwin JL, Tuininga AR, Ericsson LH. J Lipid Res. 1994;35:1102–1114. [PubMed] [Google Scholar]
- 45.Sonnino S, Mauri L, Chigorno V, Prinetti A. Glycobiology. 2007;17:1030–1030. doi: 10.1093/glycob/cwl052. [DOI] [PubMed] [Google Scholar]
- 46.IUPAC-IUB Joint Commision on Biochemical Nomenclature (JBCN) Adv. Carbohydr Chem Biochem. 2000;55:311–326. Available at: http://www.chem.qmul.ac.uk/iupac/misc/glylp.html.
- 47.Merrill AH, Sullards MC, Allegood JC, Kelly S, Wang E. Methods. 2005;36:207–224. doi: 10.1016/j.ymeth.2005.01.009. [DOI] [PubMed] [Google Scholar]
- 48.Ifa DR, Manicke NE, Rusine AL, Cooks RG. Rapid Commun Mass Spectrom. 2008;22:503–510. doi: 10.1002/rcm.3377. [DOI] [PubMed] [Google Scholar]
- 49.Nicola AJ, Gusev AI, Hercules DM. Appl Spectrosc. 1996;50:1479–1482. [Google Scholar]
- 50.Manicke NE, Kistler T, Ifa DR, Cooks RG, Ouyang Z. J Am Soc Mass Spectrom. 2009;20:321–325. doi: 10.1016/j.jasms.2008.10.011. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. (a) Product ion spectra of GD1 (m/z 917) obtained in spot sampling mode. (b) Full scan mass spectra obtained by DESI analysis in spot sampling mode of gangliosides.
Table S2. Relative fatty acid percentage composition of phopspholipids detected during HPTLC-DESI-MS analysis