Abstract
The vaccinia virus E6R gene encodes a late protein that is packaged into virion cores. A temperature sensitive mutant was used to study the role of this protein in viral replicative cycle. Cts52 has a P226L missense mutation in the E6R gene, shows a two-log reduction in plaque formation, but displays normal patterns of gene expression, late protein processing and DNA replication during infection. Mutant virions produced at 40°C were similar in their morphology to wt virions grown at 40°C. The particle to infectivity ratio was 50 times higher in purified Cts52 grown at 40°C when compared to the mutant grown at permissive temperature. In vitro characterization of Cts-52 particles grown at 40 °C revealed no differences in protein composition or in DNA content and the mutant virions could bind and enter cells. However, core particles prepared from Cts52 grown at 40 °C failed to transcribe in vitro. Our results show that E6 in the virion has either a direct or an indirect role in viral transcription.
Keywords: Vaccinia virus, Poxvirus, Temperature-sensitive mutant, Virus genetics, Virus assembly, Virus Transcription
Introduction
Vaccinia virus (VACV), the prototypic member of the family Poxviridae, has a 192-kb genome, encoding more than 200 polypeptides that function in viral transcription, genome replication, virion assembly and host defense (for review see (Moss, 2007)). Over one third of the virus coded genes are incorporated into the virion particle, including enzymes involved in early viral RNA synthesis. The vaccinia virion is a membraned, brick-shaped particle with a unique and complex internal structure featuring a biconcave core and flanking lateral bodies (for review see (Condit et al., 2006)). The replicative cycle of VACV occurs exclusively in the cytoplasm and the virions utilize their own encapsidated transcription apparatus to initiate the process. Throughout viral replicative cycle virus genes are expressed in a controlled cascade where each stage of the cycle depends on the completion of the previous phase (Moss, 1994; Condit and Niles, 2002).
Vaccinia virion assembly is a complex and poorly understood process that is of significant interest (Condit et al., 2006). The first evidence of virus morphogenesis is the formation of virus “factories”, which appear soon after virus penetration, the expression of early virus genes, and the first rounds of viral DNA replication. Electron microscopy of infected cells reveals factories as specific sites in the cytoplasm depleted of cellular organelles and sometimes delimited by ER-derived cisternae; these factories represent the location of viral DNA replication (Tolonen et al., 2001). The appearance of crescents, membrane-like structures, marks the initial steps of viral morphogenesis, followed by formation of enclosed membrane vesicles known as immature virions (IV) (Dales and Mosbach, 1968; Risco et al., 2002). During the final stages of IV formation, viral DNA is encapsidated to form dense substructures within the IV, yielding immature virions with nucleoids, or IVN (Dales, 1963; Ericsson et al., 1995). The final step of morphogenesis requires proteolysis of some viral proteins and comprises reorganization of the internal and external particle structure, resulting in the formation of an infectious mature virion (MV) (Dales and Siminovith, 1961; Byrd and Hruby, 2006). The majority of MV remain in the cytoplasm, however a small fraction are transported through the trans-Golgi apparatus and thus acquire a two new membrane layers forming wrapped virions (WV) (Sodeik et al., 1993; Smith and Vanderplasschen, 1998). Finally WV move to the periphery of the cell, lose one membrane layer by fusion with the plasma membrane and are released as an alternate, double membraned form of infectious virus, the extracellular virion (EV) (Smith and Law, 2004). Specific functions and localizations within the particle have been determined for less than half of the over 70 proteins in the virion.
Recently our group has completed the mapping and preliminary characterization of temperature sensitive alleles in the Condit/Dales collection of VACV mutants (Kato et al., 2008). Among the genes affected by the mutants in this collection, E6R is the least well characterized, described only as a core protein and with no other function ascribed to the gene. Three ts mutants, Cts52, Dts40 and Dts81, have been placed in the same complementation group and later mapped in the E6R ORF. In this report we present a complete characterization of Cts52. Our results show that under non-permissive conditions the mutant virus displayed normal patterns of gene expression, late protein processing and DNA replication. Virions produced at the non-permissive temperature were similar in morphology to wt virions produced at 40°C, but the particle to infectivity ratio was 50 times higher in purified Cts52 grown at 40°C when compared to the mutant grown at permissive temperature. Purified Cts52 particles grown at 40°C did not reveal any differences in protein composition or in the quantity of DNA encapsidated and were able to enter the cell. However, cores prepared from Cts52 grown at 40°C failed to transcribe in vitro.
Results
Characteristics of the E6R gene
The VACV E6R gene is 1704 nucleotides in length and encodes a 567 amino acid protein of MW 66,670 that is present in purified virions. It is the 13th most abundant protein in the virion and it is conserved among all the Chordopoxvirinae (Chung et al., 2006). A distant homolog with 20.4% identity and 37.9% similarity has been described in Melanoplus sanguinipes entomopoxvirus (Afonso et al., 1999). A phylogenetic analysis of the sequenced E6 genes groups all the Orthopoxviruses in the same sub-clade (data not shown). It is also noteworthy that viruses grouped using E6R gene homology clustered into sub-clades which correlate precisely with Chordopoxvirinae genera. DNA sequence analysis predicts that E6R is a late gene: the translation initiation ATG codon is preceded by a TAAAA late gene promoter signature and the body of the gene contains three early gene transcription termination sequences. Our own results confirm that E6 is expressed late during infection since in the absence of DNA replication we were not able to detect its presence by western blot (data not shown). The three E6R ts mutants described here are in the same complementation group and have the following mutations: Cts52: P226L; Dts 41: P150L; Dts 80: L260F (Kato et al., 2008).
One step growth, DNA replication and protein synthesis in E6R ts mutant infected cells
To analyze the growth characteristics of Cts52 we performed one-step growth experiments.
BSC40 cells were infected with an MOI of 10 with either wild type (wt) or mutant virus and incubated at permissive (31°C) and non-permissive temperatures (40°C). At different times post-infection, infected cells were harvested and the amount of virus in each sample determined by plaque assay. As shown in Fig. 1A wt VACV (WR strain) was able to grow at both temperatures, producing nearly 800 pfu/cell at 31°C and almost 400 pfu/cell at 40°C. On the other hand, cells infected with Cts52 at 31°C produced 100 pfu/cell but at 40°C only 2 pfu/cell were formed. This result represents a reduction of almost two logs in the production of infectious virus at the non-permissive temperature.
Fig. 1. One-step growth and DNA replication in cells infected with wt and mutants in the E6R gene.
A) BSC40 cells were infected at moi = 10 pfu/cell with wt (VACV-WR) or Cts52 incubated at 31 or 40°C. At indicated times post-infection samples were removed and virus yields were determined by plaque titration at 31°C. B) BSC40 cells were infected wt (VACV-IHD-W), or Dts41or Dts80 as described above. Virus yield was determined by plaque assay at 31°C after 24 and 48 hpi. C) BSC-40 cells were infected wt (VACV-WR) or Cts52, incubated at 31 or 40 °C as described above. At indicated times post-infection, cell extracts were prepared and applied in triplicate on nylon membranes, and hybridization to a 32P labeled vaccinia virus DNA probe was performed as described under Methods. The numbers (arbitrary units) express the average value of three different samples. wt-31°C (◆), wt-40 °C (∎), Cts52-31 °C (▴), Cts52-40 °C (●).
A similar effect was observed when BSC40 cells were infected with the two other ts mutants in this same complementation group, Dts41 and Dts80. Since these mutants were in the VACV IHD-W background this strain was used as the wt virus control. As shown in Fig. 1B, wt virus grown at the permissive temperature produced 55 and 114 pfu/cell after 24 and 48 hour of infection respectively, and at 40°C, the wt virus produced 42 and 16 pfu/cell after 24 and 48 hour of infection. The two Dts mutants presented a similar pattern of one-step growth when analyzed under permissive and non-permissive conditions. At 31°C, Dts41 generated 88 and 100 pfu/cell after 24 and 48 hours of infection respectively and Dts80 produced 140 and 180 pfu/cell during the same period. However, under non-permissive conditions the total amount of infectious particles in Dts41 infections was reduced to 0.6 and 0.7 pfu/cell and Dts80 was reduced to 1.4 and 0.4 pfu/cell after the same period of incubation. These results correspond to a two-log reduction in the titer of infectious particles produced at 40°C compared to 31°C, similar to what was described for Cts52. Since Cts52, Dts41 and Dts80 comprise a single complementation group and display similar growth characteristics, most of the remaining phenotypic characterization was focused on Cts52.
Since mutants in the E6R gene were not able to produce infectious progeny, we next determined whether Cts52 was altered in synthesis of macromolecules. DNA accumulation was measured by infecting BSC40 cells with wt or Cts52 viruses and incubating at 31°C and 40°C. At different times after infection, the cells were harvested and the cell lysates were applied to a slot-blot membrane, hybridized to a 32P-labeled vaccinia DNA probe, and quantified using a phosphor-imager. The data presented in Fig. 1C show that both wt and Cts52 viruses were able to accumulate DNA to the same level regardless the temperature of incubation. Thus, the temperature sensitivity of Cts52 does not result from a defect in DNA replication.
Next, we investigated gene expression in Cts52 virus infections under non-permissive conditions by analyzing the global synthesis of virus protein, the processing of one of the major core proteins, and the accumulation of the E6 protein. To evaluate the global profile of protein synthesis BSC40 cells were infected with wt and Cts52 virus at permissive and non-permissive temperatures. At different times after infection, total protein synthesis was analyzed by metabolic labeling with 35S-methionine in a 30-minute pulse. The 35S-labeled proteins were separated by SDS-PAGE and analyzed by autoradiography. The patterns of protein synthesis in wt and mutant infections incubated at both permissive and non-permissive conditions were indistinguishable from one another (Fig 2A). At 3 hours post infection early viral protein synthesis is apparent concomitant with a decrease in the synthesis of host proteins. By 6 hours post infection cellular protein synthesis had been reduced to a minimal level and late viral protein synthesis is observed and persists throughout the course of the infection. Thus, Cts52 displays no apparent defect in protein synthesis at the non-permissive temperature which could account for its temperature sensitivity.
Fig. 2. Virus protein synthesis, processing of p4a, and accumulation of E6.
BSC40 cells were infected at moi = 10 pfu/cell, with wt or Cts52 and incubated at 31 or 40 °C. A) At different times post-infection, the cells were pulse-labeled with 35S methionine, cells lysates were prepared, the proteins separated on SDS-PAGE, and gels were dried and autoradiographed. The virus used in the infection and the hpi of the pulse label is indicated at the top of each column. The incubation temperature is indicated to the left of the figure and the approximate molecular weights, in kDa, are indicated to the right of each autoradiogram. B-C) At indicated times after infection, cells lysates were prepared, the proteins separated by SDS-PAGE, transferred to a nitrocellulose membrane and probe against viral proteins: p4a /A10 (B), or E6 (C). The virus, the temperature of incubation, and the hpi are indicated on the top of the figure, the estimated molecular weights in kDa are indicated on the left, and the arrow on the right represents the relevant proteins.
The cleavage of specific structural proteins is an important process in VACV morphogenesis and a mandatory step in the formation of a MV (Byrd and Hruby, 2006). One of the proteins that is processed during morphogenesis is the product of the A10L gene, p4a. This gene is expressed as a 102-kDa precursor that it is cleaved into three new proteins of 62, 9 and 23 kDa. To analyze if A10 was processed during Cts52 infection BSC40 cells were infected with wt and mutant viruses at 31 or 40°C and after 12 and 24 hours of infection the cells were harvested, proteins were separated by SDS-PAGE, transferred to a nitrocellulose membrane, and the western blot probed with antibody to the A10 protein. In Fig. 2B, we show that wt and Cts52 viruses grown at permissive and non-permissive temperatures accumulate the A10 protein with no visible difference. Furthermore, the processing of the 102-kDa precursor p4a protein into the 62-kDa 4a mature form was not affected. This result demonstrates that the protein processing in Cts52 grown at 40°C was not impaired.
To investigate the accumulation of E6 protein during Cts52 infection under non-permissive conditions, western blots of infected cell extracts prepared as described above were probed with antibody to the E6 protein. The accumulation of E6 was very similar in both wt and mutant infections and dependent on the temperature of incubation (Fig. 2C). At 31°C, both wt and Cts52 accumulated less E6 when compared to the amount present in infected cells when the viruses were grown 40°C. Nevertheless equivalent amounts of E6 protein accumulated in wt and Cts52 infections incubated at 40°C. This result shows that the temperature sensitivity of Cts52 is not associated with a reduction in the amount of E6 protein.
Electron microscopic analysis of mutants in the E6 gene grown at the non-permissive temperature
VACV morphogenesis begins after viral DNA replication, extends throughout infection, and is sufficiently asynchronous such that the full spectrum of assembly intermediates and mature virus forms can be detected at late times of infection using transmission electron microscopy (Condit et al., 2006). To determine whether virus formation was affected during infection, BSC40 cells were infected with VACV wt or Cts52 mutant viruses, incubated at 31 °C and 40 °C and processed for electron microscopy after 24 and 48 hours of infection (Fig. 3). Viral membrane crescents, IV, IVN and normal looking MV were observed in wt and Cts52 virus infected cells incubated at permissive temperature (data not shown). As shown in Fig. 3A, wt virus grown at 40 °C for 24 hours produced normal progeny as has been well documented (Hooda-Dhingra et al., 1989; Kato et al., 2004). Cts52 grown at the non-permissive temperature produced MV-like particles that were indistinguishable from wt virions (Fig. 3B-D). Specifically, we were able to identify crescents, IV, IVN and MV-like particles. Some of the MV-like particles observed in Cts 52 infections grown at 40° C appeared round rather than the expected ovoid shape and possessed lucent centers rather than an electron dense core, suggesting that Cts52 particles formed at the non-permissive temperature may not be completely normal in structure. However these same features are apparent among MV in wt infections incubated at 40°C (Fig.3A), thus we suspect that many if not all of these abnormalities may result generally from the high temperature incubation and are not specifically associated with the temperature sensitive mutation in Cts52. In short, we were not able reliably to distinguish the particles formed in Cts52 infections from those formed in wt infections. Likewise, BSC40 cells infected Dts41 and Dts80 grown at 40°C were able to form particles with a morphology that was similar wt virus grown at 40°C (Fig. 3 E-F), consistent with previous observations (Dales et al., 1978). These results show that the ts E6R mutants were able to morph into particles that resemble MV but are non-infectious.
Fig. 3. Electron micrographs of wt and mutants-infected cells.
BSC40 cells were infected with wt or mutant virus at a moi = 10 pfu/cell, incubated at 40 °C for 24 (A-C) and 48 hs (D -F) and processed for electron microscopy as described in Methods. (A) wt; (B - D) Cts52; (E) Dts41; (F) Dts80. IV= immature virions; MV= mature virions; C= crescents; N= nucleus; F= DNA factories.
Characteristics of the Cts52 purified particles
To obtain better insight into the mutant particles, BSC40 cells were infected with wt and Cts52 viruses and incubated at 40°C for 24 hours or at 31 °C for 48 hours. Infected cells were then disrupted and the virus was purified using velocity sedimentation on sucrose gradients. As shown in Table 1 no major difference was observed in the yield for each virus grown at the same temperature of incubation. At 31 °C more than 1000 particles were produced in each infection while at 40 °C approximately 700 particles were formed per cell. However, a difference in the specific infectivity (particles/pfu) was evident when we compared the same virus grown under different conditions. In this comparison, the specific infectivity for wt was1.9 times lower for particles formed at 40 °C while for Cts52 the specific infectivity was reduced 50 times for viruses grown under non-permissive condition. Cts52 grown at the permissive temperature was nearly 11 times less infectious than the wt grown under the same conditions and the mutant virus grown at 40°C was more than 350 times less infectious than the wt grown at the same temperature. Thus Cts52 particles grown at 40°C, while normal in appearance, are severely reduced in infectivity.
Table 1.
Characteristics of purified wt and mutant Vaccinia virus particles
| Virus-Temperature a | Yield (Particles/cell)b |
Infectivity (particles/pfu) |
|---|---|---|
| Wt- 31°C | 1308 | 25 |
| Wt- 40°C | 732 | 47.5 |
| Cts52-31 °C | 1464 | 293 |
| Cts52-40 °C | 696 | 14,745 |
Wt and Cts52 were isolated from high moi infections. Viruses grown at 40 °C were incubated for 24 hours; viruses grown at 31 °C were incubated for 72 hours.
Calculated from an A260nm reading of purified virus, assuming a 1 A260nm =1.2 × 1010 particles/ml.
To characterize the macromolecular content of the virions, we compared the DNA and protein composition of purified virions grown under different conditions. To measure the amount of DNA, different quantities of purified viruses were blotted onto a nylon membrane, hybridized with 32P-labeled VACV DNA Hind III E fragment, and quantified using a phosphorimager. The phosphorimage presented in Fig. 4A and quantified in Fig. 4B demonstrates that the amount of DNA present in the virions was comparable among all samples, independent of the growth condition. The protein profile of virions was examined using two different particle concentrations analyzed by SDS-PAGE and stained with Coomassie blue. The result presented in Fig. 4C shows that the protein profiles among the viruses grown at different temperatures were indistinguishable.
Fig. 4. Analysis of the DNA content and protein composition of purified wt and mutant particles.
A) Samples of purified wt and Cts52 were diluted and the amount of virions determined by A260nm and indicated on the top of each lane were applied onto a nylon membrane, processed and hybridized to 32P VACV DNA fragment. B) The blot was analyzed with a phosphor-imager apparatus, the data were normalized and graphed as described in Methods. C) The polypeptide composition of purified virions was analyzed by SDS-PAGE and Coomassie blue staining. The amount of virions determined by A260nm is indicated on the top of each lane and the approximate molecular weight marker is indicated on the left of the gel. D) Partition of purified virions into core and membrane fraction and Western blot analysis. 0.06 A260 nm units (~0.7 μg) of each virion sample was resuspended in core buffer and processed as described under Methods. The gene product corresponding to each antiserum used with the predicted molecular weight is indicated to the left of each row, the virus sample probed is indicated on the top of the figure. C= core fraction, M = membrane fraction. Wt31, wt grown at 31°C; Wt-40, wt grown at 40 °C; Cts- 31, Cts52 grown at 31°C; Cts40, Cts52 grown at 40 °C.
After treatment of purified VACV with a non-ionic detergent and a reducing agent, the disrupted virions can be separated by centrifugation in soluble and particulate fractions (Ichihashi et al., 1984). The soluble fraction is composed of proteins that constitute the virus membrane while the particulate fraction contains proteins and DNA from the virus core. The E6 protein has been designated as a core protein. To analyze the presence of E6 in virions and the integrity of the mutant particles, purified viruses were disrupted with NP40 and DTT, core and membrane fractions were separated by centrifugation, and the presence of specific virion proteins was analyzed by western blot. The results presented in Fig. 4D show no alteration in the amount E6 protein contained in virions grown under different conditions. We also did not observe any major difference in the content of other virion proteins analyzed. The core proteins H4, a subunit of the early RNA polymerase, D1 and D12, large and small subunits capping enzyme, L4, an RNA and ssDNA binding protein, E8, an essential virion core protein, and the membrane protein A27 were present in similar amounts and partitioned in the predicted virion fraction. Thus the data presented so far suggest that the defect in Cts52 particles produced at 40 °C was not directly related to the DNA or protein content of the virions or to the integrity of the particles.
Cts52 can bind and enter BSC40 cells
To investigate whether purified Cts52 grown at the non-permissive temperature was competent to initiate an infectious cycle we examined the ability of the virus to bind and penetrate cells and induce a cytopathic effect. The binding process was examined by infecting BSC40 cells for 1 hour at 4 °C. After this period, the monolayer was washed to remove the unadsorbed virus and the monolayer processed for immunofluorescence. The presence of virus associated with the cell membrane was detected by labeling the cells with anti-A27 antibody, using a fluorescence confocal microscope. As can be seen in Fig. 5 A-D, both wt and mutant viruses grown at permissive and non-permissive temperatures were able to bind to cells. The entry process was analyzed by infecting BSC40 as described above but after removing the unadsorbed viruses, media containing cyclohexamide was added to the monolayer and the cells were incubated for 2 hours at 31 °C. Infected cells were then processed as described and the presence of viral cores inside the cells was determined by labeling the monolayer with anti-A4 antibody, a core-specific antibody that detects uncoated virions. The result presented in Fig. 5 E-G showed that both wt and Cts52 grown at permissive and non-permissive temperature were able to enter the cells with no major differences. The identification of the A27 membrane protein attached to the cell and the A4 core protein in the cytoplasm of the cell shows that the Cts52 mutation did not interfere with binding, uptake, or uncoating of virions.
Fig. 5. Analysis of the binding and entry process of wt and mutant virions.
BSC40 cells were infected with moi= 1000 particles of wt (A, C, E, G) or Cts52 (B, D, F, H) virus grown at 31 °C (A, B, E, F) or 40 °C (C, D, G, H). The cells were processed and stained for viral membrane protein A27 (green) (A-D) or viral core protein A4 (red) (E-H), and DNA was stained with Dapi (blue). The cells were analyzed by confocal microscopy as described in Methods.
During the first hours of infection, cells go through a profound transformation in their morphology in a phenomenon described as cytopathic effect (CPE). This effect is characterized by the induction of cell rounding, inhibition of host RNA and protein synthesis, and death of the infected cells (Bablanian et al., 1978; Tsung et al., 1996). To analyze whether Cts52 grown at 40°C was able to induce CPE, BSC40 cells were infected for 1 hour at 4 °C. After this period, the unabsorbed virus was removed, fresh media was added, cells were incubated for 6 hours at 31 °C, and the induction of CPE was observed by differential interference contrast microscopy and by confocal microscopy labeling the cells with DAPI and phalloidin. The results showed that at this time wt viruses grown at both temperatures and Cts52 grown at 31°C were able to induce intense CPE in infected cell (data not shown). In all three situations, the cells were rounded, there was a rearrangement of the actin cytoskeleton, and DNA factories were observed in the cytoplasm. On the other hand, in cells infected with Cts52 grown at 40 °C the cells appeared slightly detached from each other but few cells showed cytoplasmic retraction, and in most cells, the actin cytoskeleton did not appear to be rearranged, and no DNA factories were observed (data not shown). These results suggest that although Cts52 virus grown at the non-permissive temperature was able to enter BSC40 cells, the virus was unable to initiate replication.
Cts52 virions are defective in core transcription
All the enzymes necessary for RNA synthesis, addition of a 5′ mRNA cap and a 3′ poly-A tail, and extrusion of the viral transcript from the core are packaged in the virion (Moss, 1994). Virions need to be permeabilezed with NP40 and DTT to transcribe the DNA and extrude the RNA from the core. To test the ability of permeabilezed virions to synthesize RNA, purified virions were permeabilezed with NP40 and DTT and incubated with a mixture of the four ribonucleoside-triphosphates as described in Methods. The result presented in Fig. 6A shows that wt virions grown at permissive or non-permissive temperatures were able to synthesize RNA at the same rate. Cts52 viruses grown at 31 °C showed linear synthesis of RNA up to 10 minutes and after this time, the rate of transcription declined and ceased after 30 minutes. Cts52 virions grown at 40 °C were unable to synthesize RNA.
Fig 6. RNA synthesis directed by purified wt and mutants particles.
A) Total core RNA synthesis. Purified virions grown at different temperature were permeabilized with NP40 and DTT treatment, incubated in a reaction mixture that contains [α32P] CTP, unlabelled GTP, ATP and UTP, MgCl2 as described in Methods. At indicated times, samples were removed, and the acid precipitable radioactivity was determined in a liquid scintillation counter. wt-31°C, (◆); wt-40°C, (∎); Cts52-31 °C, (●); Cts52-40 °C, (▴). B) RNA extrusion from cores. Purified virions grown at 31 °C were activated for RNA as described above. At indicated times samples were removed and the core and the soluble fraction were separated by centrifugation as described in Methods. wt-core, (∎); wt-supernatant, (◆); Cts52-core (▴); Cts-52-supernantant, (●).
One possible explanation for the observed cessation of transcription in Cts52 virions grown at 31 °C is inhibition of RNA extrusion from the virions. The release of the newly synthesized RNA from cores is an essential step for maintenance of continuous transcription in permeabilezed virions. Under certain assay conditions, if RNA accumulates in the core RNA synthesis will cease (Shuman and Moss, 1989). To analyze whether RNA extrusion was affected in Cts52 virions grown at 31 °C, permeabilized cores were incubated in the presence of ribonucleoside triphosphates. At indicated times an aliquot of the reaction was removed, the core and soluble fractions were separated by centrifugation, and the amount of RNA present in both fractions was determined. As shown in Fig 6B the RNA synthesized by wt viruses was distributed preferentially in the soluble fraction and a small proportion stayed in the insoluble fraction. Similar to the wt, the RNA synthesized by Cts52 virions was released into the soluble fraction and only a small amount remained in the cores. Nevertheless, the rate of RNA extrusion from the cores was reduced as the incubation proceeded.
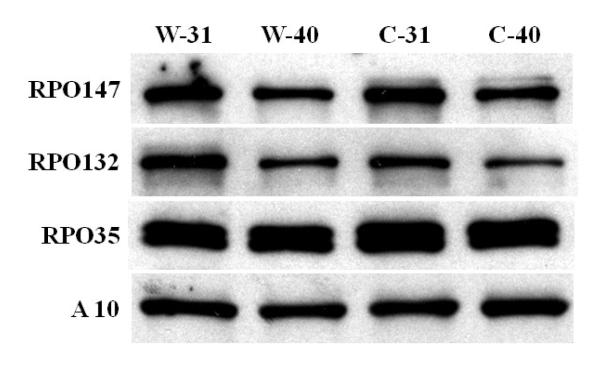
The inability of Cts 52 to synthesize RNA could also be related to a reduction or absence of RNA polymerase associated with the core (Zhang et al., 1994). To analyze the presence of the RNA polymerase subunits, purified virions were solubilized in SDS-sample buffer, the proteins separated by SDS-PAGE, and the presence of RPO 147(J6), RPO 132(A24) and RPO 35 (A29) were determined by western blot. As shown in Fig. 7 both wt and Cts52 viruses grown under permissive and non-permissive conditions contain equivalent amount of These RNA polymerase subunits associated with the particles. Therefore, the inability of Cts 52 to synthesize RNA is not directly related to the amount of RNA polymerase associated with the viral particle.
Fig 7. Western blot analysis of the RNA polymerases subunits in wt and mutants particles.
0 .06 A260 nm units (~0.7 μg) of each virion sample was resuspended in SDS-sample buffer and processed as described under Methods. The gene product corresponding to each antiserum used is indicated to the left of each row, the virus sample probed is indicated on the top of the figure. The core protein A10 was used as a gel loading control.
Discussion
We have conducted a phenotypic analysis of temperature sensitive mutants which map to the vaccinia virus E6R gene, encoding a virion core protein. Mutants in three different ts alleles in two different virus strains all present similar phenotypes under non-permissive conditions, namely production of mature virions (MV) that are morphologically similar to wt virions grown at 40 °C, but which are nevertheless non infectious. Detailed analysis of one allele, Cts52, reveals that viral gene expression, DNA replication and processing of virion structural protein precursors is unimpaired in mutant infections, that the non-infectious virions produced can bind and enter cells, but that they are defective in an in vitro core directed transcription reaction. Thus, the most likely explanation for the reduced infectivity of the virions produced under non-permissive conditions is that they are unable to direct early viral gene expression after entering cells. The E6R gene has no homologs outside of the family Poxviridae and no known motifs suggestive of a specific function. Mutants in three other poxvirus core protein genes, L3L, L4R and E8R, present a similar phenotypic profile, namely production under non permissive conditions of typical pox virions that are defective in virion transcription (Wilcock and Smith, 1994; Wilcock and Smith, 1996; Resch and Moss, 2005; Kato et al., 2007). In these cases as well, bioinformatic analysis provides no insights into the function of the gene products. The next most fundamental question concerning the function of these genes is whether they play a direct role in the virion core transcription reaction, or whether they play a subtle role in virion structure the impacts indirectly on the virion transcription reaction.
Studies with inducible knockout mutants in the E6R gene suggest a possible role for E6 in virion assembly or structure. Specifically, in the absence of E6 protein, virus assembly is defective in association of viroplasm with membrane crescents, such that normal IV, IVN and MV are absent and membrane crescents and unencapsidated viroplasm accumulate (Resch et al., 2009; Boyd et al., 2010). While such a phenotype might on the surface suggest a role for E6 in structure or assembly, it is noteworthy that defects in enzymes that play a known role in transcription can also result in virion assembly phenotypes. Specifically, temperature sensitive alleles in either subunit of the mRNA capping enzyme and inducible knockout alleles in either subunit of VETF, the viral early transcription initiation factor, are defective in MV formation and accumulate IV and IVN (Carpenter and DeLange, 1991; Hu et al., 1996; Hassett et al., 1997; Hu et al., 1998). These studies suggest that virus assembly is coupled to proper assembly of the virion transcription complex. Thus, the observation of a morphogenesis phenotype for any given mutants does not exclude the possibility that the gene in question has a direct role in virion transcription.
Mutants in several genes with known or likely roles in virion transcription and/or RNA metabolism present phenotypes similar or identical to the E6R ts mutants described here. These include mutants affecting H4 (rap94), the early gene transcription specificity subunit of the viral RNA polymerase, H1, a tyrosine/serine protein phosphatase, H6, a DNA topoisomerase and I8, an RNA helicase (Kane and Shuman, 1992; Zhang et al., 1994; Liu et al., 1995; Gross and Shuman, 1996; Da Fonseca and Moss, 2003). In each case in mutant infections under non-permissive conditions, non-infectious virions are produced which are morphologically indistinguishable from wt virions by electron microscopy. Thus, the phenotype of the E6R mutants is not inconsistent with a direct role of E6 in the virion transcription reaction.
Previous characterization of mutants in the A3L gene demonstrates clearly that the virion transcription reaction may be compromised by defects in virion structure. The A3L gene encodes a major virion structural protein that is thought to be a component of the virion core wall. Temperature sensitive mutants in A3L produce morphologically aberrant MV that contain a wt complement of virion proteins but are defective in transcription, despite encapsidation of active transcription enzymes (Kato et al., 2004). While A3 mutants MV are obviously structurally aberrant, other mutants with significant known defects in virion protein composition appear deceptively normal using conventional electron microscopic techniques. Specifically, the H4L (rap94) mutant virions described above lack all of the major known or suspected enzymes involved in early viral transcription, including the viral RNA polymerase, the capping enzyme/termination factor, the poly(A) polymerase large subunit, DNA-dependent ATPase I, RNA helicase, and topoisomerase, and yet these virions are indistinguishable from wt virions using conventional electron microscopy (Zhang et al., 1994). Application of higher resolution electron microscopy techniques or approaches to controlled degradation of virions may yet reveal structural defects in virions produced in E6R infections or infections with phenotypically similar mutants in genes such as E8R, L3L and L4R (Ichihashi et al., 1984; Szajner et al., 2005; Heuser, 2005). Based on the precedent set by A3L mutant infections, such structural defects could impact indirectly on virion core directed transcription.
Lastly, we must consider the intriguing possibility that E6 and other vaccinia virion core proteins might play a dual role in structure and transcription. Precedent for this concept is firmly established through studies of the reovirus virion, showing that several RNA metabolism enzymes, notably the viral RNA polymerase and mRNA capping enzymes, are integral components of the virus capsid (Schiff et al., 2007). It seems not unlikely that elements of the vaccinia virion substructure, for example the core wall, may play dual roles in RNA metabolism and virion core structure. This seems particularly likely in the case of a gene such as E6R, in which inducible knockout mutants result in a morphogenesis phenotype while temperature sensitive mutants result in production of transcription defective virions.
In summary, the phenotypic characterization of E6R mutants presented here is consistent with a role for E6 in either virion structure, transcription, or both. Precise definition of E6 function awaits further studies that include higher resolution examination of the substructure of the vaccinia virion.
Materials and Methods
Virus, cell culture, plaque assay, one-step growth
Wild-type vaccinia virus strains WR and IHDW were used as controls for the studies of the temperature mutants Cts52, Dts41 and Dts80 (Kato et al., 2008). Throughout this work, viruses were grown in BSC40 cells, an African green monkey cell line. The methodology for cell culture, virus infection, purification, plaque assay and one-step growth have been described before (Condit and Motyczka, 1981; Condit et al., 1983; Kato et al., 2004). The non-permissive temperature for analysis of ts mutants was 39.7°C but in the text is designated as 40°C for simplicity.
Analyses of VV DNA by slot-blot hybridization
BSC-40 cells were infected with VACV and at different times post-infection cells were harvested and extracts were prepared and processed as described previously (Damaso et al., 2002). For quantitation, the blots were analyzed with a phosphor-imager apparatus (Storm 860, GE-Healthcare) and the average of the read of the triplicates was calculated and plotted. For the analysis of the DNA content present in purified virions, the data obtained from the phosphor-imager were normalized for the same amount of virus and an average number was calculated and plotted.
Metabolic labeling of viral proteins
For metabolic labeling of proteins, infected cells were incubated with100 μCi/ ml of 35S-labeled amino acids mixture (ProMix – GE Healthcare) in a methionine free media for 30 minutes. After this period, a cell extract was prepared, proteins separated in a sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE), and analyzed by autoradiography as described before (Kato et al., 2004).
Vaccinia virus purification and analysis of virus particles
Ten 150-mm dishes containing confluent monolayers of BSC40 cells were infected with either VACV-WR or Cts52 viruses at a moi of 10 at the permissive or the non-permissive temperature. Infected cells incubated were for 24 h at 40°C or for 72 h at 31 °C and harvested for virus purification. Cell-associated virus was purified by differential centrifugation through a 36% sucrose cushion followed by banding on a 24–40% sucrose gradient as previously described (Ausubel et al., 1994; Kato et al., 2004).
Western blot analysis
Virion core and envelope extracts were prepared and analyzed by incubating 0.06 A260nm units (0.7 μg) of wt or mutant particles with core buffer (100 mM DTT and 1 % NP40 in 50 mM Tris–HCl pH 8.0) for 30 minutes at 37 °C. Core and membrane fractions were partitioned by a 15 minutes centrifugation at 14,000 × g. The fractions were resuspended in SDS-sample buffer and the proteins were separated by a 11 % SDS-PAGE and transferred to a nitrocellulose membrane as described before (Kato et al., 2007). The membrane was blocked with a solution of TBS-T (50 mM Tris-HCl pH 7.5; 150 mM NaCl; 0.01% Tween 20) and 5% non-fat dry milk (TBS-TM) for 1 hour incubated with an appropriate dilution of the primary antibody in TBS-TM overnight at 8°C. The membrane was washed 5 times for 5 minutes with TBS-T and incubated with a horseradish conjugated secondary antibodies (GE Healthcare) diluted in TBS-TM for 2 hours at 8°C. The membrane was washed 5 times for 5 minutes and the protein complex detected by chemiluminescence (GE Healthcare). A His-tag E6 recombinant protein (amino acids 67 to 430) was constructed, purified and used as source for raising a rabbit polyclonal antibody that was prepared by Alpha Diagnostic International, and used at a dilution of 1:1000. For the analysis of the RNA polymerase subunits unfractionated virions were resuspended in SDS-sample buffer and the proteins were separated by a 10% SDS-PAGE and processed as described above. The dilution and source of the other antibodies were the following: anti-L4 (1:40,000) rabbit serum was supplied by Dr. Dennis Hruby (Oregon State University); anti-H4 (1:5,000) and anti- A29 (1:2,000) rabbit serum was supplied by Dr. Bernard Moss (NIAID); anti-A10 (1:12,000) and anti-A4 (1:1,000) rabbit sera were supplied by Dr Mariano Esteban (Universidad Autónoma, Madrid), anti-A27 (1:30,000) monoclonal antibody supplied by Dr. David Ulaeto (Dstl Porton Down, Salisbury); anti-D1 (1:1,000) and anti-D12 (1:6,000) rabbit sera were supplied by Dr Edward Niles (SUNY at Buffalo); anti-J6 (1:2,000), anti –A24 (1:2,000) rabbit serum supplied by Dr Stewart Shuman (Sloan-Kettering Institute).
Electron microscopy
BSC40 cells were infected with either wt or mutant viruses at a moi of 10 at permissive or non-permissive temperature. Samples from 24 to 48 post-infection were prepared for transmission electron microscopy as described (Kato et al., 2004). The samples were analyzed with the assistance of the University of Florida ICBR Electron Microscopy Core Laboratory.
Attachment and entry assay
BSC40 cell monolayers in 8 chamber slides (Lab-Tek) were infected with 1,000 particles per cell for 1 hour at 4°C as described before (Kato et al., 2007). After the incubation the unabsorbed virus was removed, the cells were washed once with PBS and processed for virus binding assay or and incubated at 31°C in DMEM containing100 μg/ml of cycloheximide for 2 hours for entry assay. All subsequent steps were performed at room temperature. Cells were fixed with 4% paraformaldehyde in PBS, washed once with PBS, quenched for 5 min with PBS containing 20 mM glycine, and washed once again with PBS. Cells were then permeabilized for 2 min with 0.1% triton X-100, washed twice with PBS, and blocked for 10 min with PBS containing 10% fetal bovine serum. Antibodies were diluted in PBS containing 10% fetal bovine serum. Cells were then incubated for 1 hour with anti-A27 antibody for the binding assay or anti-A4 antibody for the entry assay. Cells were washed three times with PBS and then incubated for 30 min with secondary antibodies. Alexafluor 594 labeled goat-anti-rabbit antibody (Invitrogen) was used as secondary antibody for visualization of A4; Alexafluor 488 labeled goat-anti-mouse antibody (Invitrogen) was used as secondary antibody for A27 visualization. Cells were washed twice with PBS and stained with 0.5 μg/ml DAPI for 15 min. Finally, cells were washed three times with PBS and observed directly on a Leica TCS SP2 Confocal Microscope. Images shown are Z-stacks.
Transcription by permeabilized virions
0.4 A260nm units of virus were incubated in a reaction mixture containing 60 mM Tris-HCl pH 8.0, 0.05% Nonidet P40 (NP40), 10 mM dithiothreitol (DTT), 10 mM MgCl2, 5 mM ATP, 1 mM UTP, 1 mM GTP, 0.2 mM α-32P-CTP (100 μCi/μmol), at 31°C as described previously (Kato et al., 2004). At various times 50-μl aliquots were removed, precipitated directly with 5% TCA, filtered onto a glass microfiber filters (934-AH, Whatman), and the radioactivity was determined by liquid scintillation counting. To analyze the RNA extruded or core associated, 50-μl aliquots were removed at different time points and centrifuged at 14,000 × g for 3 minutes and the supernatant
Acknowledgments
We thank Bernard Moss, Stewart Shuman, Mariano Esteban, Dennis Hruby, David Ulaeto and Edward Niles for antisera; Karen Kelley and the University of Florida ICBR Electron Microscopy Core Laboratory for excellent advice and technical assistance; Douglas E. Smith and the University of Florida Cell and Tissue Analysis Core for technical assistance with the confocal microscope; and Nicole Kay, Ricky Ortiz, Sandya Shivashankar for technical support. This work was supported by NIH grant R01 AI055560 to RCC.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference List
- Afonso CL, Tulman ER, Lu Z, Oma E, Kutish GF, Rock DL. The genome of Melanoplus sanguinipes entomopoxvirus. J.Virol. 1999;73:533–552. doi: 10.1128/jvi.73.1.533-552.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. Current protocols in molecular biology. John Wiley & Sons; New York: 1994. [Google Scholar]
- Bablanian R, Baxt B, Sonnabend JA, Esteban M. Studies on the mechanisms of vaccinia virus cytopathic effects. II. Early cell rounding is associated with virus polypeptide synthesis. J.Gen.Virol. 1978;39:403–413. doi: 10.1099/0022-1317-39-3-403. [DOI] [PubMed] [Google Scholar]
- Boyd O, Turner P, Moyer RW, Condit RC, Moussatche N. Involvement of E6 protein in vaccinia virus morphogenesis. Virology. 2010 doi: 10.1016/j.virol.2010.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrd CM, Hruby DE. Vaccinia virus proteolysis--a review. Rev.Med.Virol. 2006;16:187–202. doi: 10.1002/rmv.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter MS, DeLange AM. A temperature-sensitive lesion in the small subunit of the vaccinia virus-encoded mRNA capping enzyme causes a defect in viral telomere resolution. J.Virol. 1991;65:4042–4050. doi: 10.1128/jvi.65.8.4042-4050.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung CS, Chen CH, Ho MY, Huang CY, Liao CL, Chang W. Vaccinia virus proteome: identification of proteins in vaccinia virus intracellular mature virion particles. J.Virol. 2006;80:2127–2140. doi: 10.1128/JVI.80.5.2127-2140.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Condit RC, Motyczka A. Isolation and preliminary characterization of temperature-sensitive mutants of vaccinia virus. Virology. 1981;113:224–241. doi: 10.1016/0042-6822(81)90150-1. [DOI] [PubMed] [Google Scholar]
- Condit RC, Motyczka A, Spizz G. Isolation, characterization, and physical mapping of temperature-sensitive mutants of vaccinia virus. Virology. 1983;128:429–443. doi: 10.1016/0042-6822(83)90268-4. [DOI] [PubMed] [Google Scholar]
- Condit RC, Moussatche N, Traktman P. In a nutshell: structure and assembly of the vaccinia virion. Adv.Virus Res. 2006;66:31–124. doi: 10.1016/S0065-3527(06)66002-8. [DOI] [PubMed] [Google Scholar]
- Condit RC, Niles EG. Regulation of viral transcription elongation and termination during vaccinia virus infection. Biochim.Biophys.Acta. 2002;1577:325–336. doi: 10.1016/s0167-4781(02)00461-x. [DOI] [PubMed] [Google Scholar]
- Da Fonseca F, Moss B. Poxvirus DNA topoisomerase knockout mutant exhibits decreased infectivity associated with reduced early transcription. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:11291–11296. doi: 10.1073/pnas.1534874100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dales S. The uptake and development of vaccinia virus in strain L cells followed with labeled viral deoxyribonucleic acid. J.Cell Biol. 1963;18:51–72. doi: 10.1083/jcb.18.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dales S, Milovanovitch V, Pogo BG, Weintraub SB, Huima T, Wilton S, McFadden G. Biogenesis of vaccinia: isolation of conditional lethal mutants and electron microscopic characterization of their phenotypically expressed defects. Virology. 1978;84:403–428. doi: 10.1016/0042-6822(78)90258-1. [DOI] [PubMed] [Google Scholar]
- Dales S, Mosbach EH. Vaccinia as a model for membrane biogenesis. Virology. 1968;35:564–583. doi: 10.1016/0042-6822(68)90286-9. [DOI] [PubMed] [Google Scholar]
- Dales S, Siminovith L. The development of vaccinia virus in Earle’s L strain cells as examined by electron microscopy. J.Biophys.Biochem.Cytol. 1961;10:475–503. doi: 10.1083/jcb.10.4.475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damaso CR, Oliveira MF, Massarani SM, Moussatche N. Azathioprine inhibits vaccinia virus replication in both BSC-40 and RAG cell lines acting on different stages of virus cycle. Virology. 2002;300:79–91. doi: 10.1006/viro.2002.1534. [DOI] [PubMed] [Google Scholar]
- Ericsson M, Cudmore S, Shuman S, Condit RC, Griffiths G, Locker JK. Characterization of ts 16, a temperature-sensitive mutant of vaccinia virus. J.Virol. 1995;69:7072–7086. doi: 10.1128/jvi.69.11.7072-7086.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross CH, Shuman S. Vaccinia virions lacking the RNA helicase nucleoside triphosphate phosphohydrolase II are defective in early transcription. J.Virol. 1996;70:8549–8557. doi: 10.1128/jvi.70.12.8549-8557.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassett DE, Lewis JI, Xing X, DeLange L, Condit RC. Analysis of a temperature-sensitive vaccinia virus mutant in the viral mRNA capping enzyme isolated by clustered charge-to-alanine mutagenesis and transient dominant selection. Virology. 1997;238:391–409. doi: 10.1006/viro.1997.8820. [DOI] [PubMed] [Google Scholar]
- Heuser J. Deep-etch EM reveals that the early poxvirus envelope is a single membrane bilayer stabilized by a geodetic “honeycomb” surface coat. J.Cell Biol. 2005;169:269–283. doi: 10.1083/jcb.200412169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooda-Dhingra U, Thompson CL, Condit RC. Detailed phenotypic characterization of five temperature-sensitive mutants in the 22- and 147-kilodalton subunits of vaccinia virus DNA-dependent RNA polymerase. J.Virol. 1989;63:714–729. doi: 10.1128/jvi.63.2.714-729.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu X, Carroll LJ, Wolffe EJ, Moss B. De novo synthesis of the early transcription factor 70-kilodalton subunit is required for morphogenesis of vaccinia virions. J.Virol. 1996;70:7669–7677. doi: 10.1128/jvi.70.11.7669-7677.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu X, Wolffe EJ, Weisberg AS, Carroll LJ, Moss B. Repression of the A8L gene, encoding the early transcription factor 82-kilodalton subunit, inhibits morphogenesis of vaccinia virions. J.Virol. 1998;72:104–112. doi: 10.1128/jvi.72.1.104-112.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichihashi Y, Oie M, Tsuruhara T. Location of DNA-binding proteins and disulfidelinked proteins in vaccinia virus structural elements. J.Virol. 1984;50:929–938. doi: 10.1128/jvi.50.3.929-938.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kane EM, Shuman S. Temperature-sensitive mutations in the vaccinia virus H4 gene encoding a component of the virion RNA polymerase. J.Virol. 1992;66:5752–5762. doi: 10.1128/jvi.66.10.5752-5762.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato SE, Condit RC, Moussatche N. The vaccinia virus E8R gene product is required for formation of transcriptionally active virions. Virology. 2007;367:398–412. doi: 10.1016/j.virol.2007.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato SE, Moussatche N, D’Costa SM, Bainbridge TW, Prins C, Strahl AL, Shatzer AN, Brinker AJ, Kay NE, Condit RC. Marker rescue mapping of the combined Condit/Dales collection of temperature-sensitive vaccinia virus mutants. Virology. 2008;375:213–222. doi: 10.1016/j.virol.2008.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato SE, Strahl AL, Moussatche N, Condit RC. Temperature-sensitive mutants in the vaccinia virus 4b virion structural protein assemble malformed, transcriptionally inactive intracellular mature virions. Virology. 2004;330:127–146. doi: 10.1016/j.virol.2004.08.038. [DOI] [PubMed] [Google Scholar]
- Liu K, Lemon B, Traktman P. The dual-specificity phosphatase encoded by vaccinia virus, VH1, is essential for viral transcription in vivo and in vitro. J.Virol. 1995;69:7823–7834. doi: 10.1128/jvi.69.12.7823-7834.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss B. Vaccinia virus transcription. In: Conaway RC, Conaway JW, editors. Transcription: Mechanisms and Regulation. Raven Press; Ltda, New York: 1994. pp. 185–205. [Google Scholar]
- Moss B. Poxviridae: the viruses and their replication. In: Knipe DM, Howley PM, editors. Fields Virology. Wolters Kluwer-Lippincott Williams and Wilkins; Philadelphia: 2007. pp. 2906–2945. [Google Scholar]
- Resch W, Moss B. The conserved poxvirus L3 virion protein is required for transcription of vaccinia virus early genes. Journal of Virology. 2005;79:14719–14729. doi: 10.1128/JVI.79.23.14719-14729.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resch W, Weisberg AS, Moss B. Expression of the highly conserved vaccinia virus E6 protein is required for virion morphogenesis. Virology. 2009;386:478–485. doi: 10.1016/j.virol.2009.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risco C, Rodriguez JR, Lopez-Iglesias C, Carrascosa JL, Esteban M, Rodriguez D. Endoplasmic reticulum-Golgi intermediate compartment membranes and vimentin filaments participate in vaccinia virus assembly. J.Virol. 2002;76:1839–1855. doi: 10.1128/JVI.76.4.1839-1855.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiff LA, Nibert ML, Tyler KL. Orthoreoviruses and their replication. In: Knipe DM, Howley PM, editors. Fields Virology. Wolters Kluwer-Lippincott Williams and Wilkins; Philadelphia: 2007. pp. 1853–1915. [Google Scholar]
- Shuman S, Moss B. Bromouridine Triphosphate Inhibits Transcription Termination and Messenger-RNA Release by Vaccinia Virions. Journal of Biological Chemistry. 1989;264:21356–21360. [PubMed] [Google Scholar]
- Smith GL, Law M. The exit of vaccinia virus from infected cells. Virus Res. 2004;106:189–197. doi: 10.1016/j.virusres.2004.08.015. [DOI] [PubMed] [Google Scholar]
- Smith GL, Vanderplasschen A. Extracellular enveloped vaccinia virus. Entry, egress, and evasion. Adv.Exp.Med.Biol. 1998;440:395–414. [PubMed] [Google Scholar]
- Sodeik B, Doms RW, Ericsson M, Hiller G, Machamer CE, van ’t HW, van MG, Moss B, Griffiths G. Assembly of vaccinia virus: role of the intermediate compartment between the endoplasmic reticulum and the Golgi stacks. J.Cell Biol. 1993;121:521–541. doi: 10.1083/jcb.121.3.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szajner P, Weisberg AS, Lebowitz J, Heuser J, Moss B. External scaffold of spherical immature poxvirus particles is made of protein trimers, forming a honeycomb lattice. J.Cell Biol. 2005;170:971–981. doi: 10.1083/jcb.200504026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolonen N, Doglio L, Schleich S, Krijnse LJ. Vaccinia virus DNA replication occurs in endoplasmic reticulum-enclosed cytoplasmic mini-nuclei. Mol.Biol.Cell. 2001;12:2031–2046. doi: 10.1091/mbc.12.7.2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsung K, Yim JH, Marti W, Buller RM, Norton JA. Gene expression and cytopathic effect of vaccinia virus inactivated by psoralen and long-wave UV light. J.Virol. 1996;70:165–171. doi: 10.1128/jvi.70.1.165-171.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilcock D, Smith GL. Vaccinia virus core protein VP8 is required for virus infectivity, but not for core protein processing or for INV and EEV formation. Virology. 1994;202:294–304. doi: 10.1006/viro.1994.1346. [DOI] [PubMed] [Google Scholar]
- Wilcock D, Smith GL. Vaccinia virions lacking core protein VP8 are deficient in early transcription. Journal of Virology. 1996;70:934–943. doi: 10.1128/jvi.70.2.934-943.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Ahn BY, Moss B. Targeting of a multicomponent transcription apparatus into assembling vaccinia virus particles requires RAP94, an RNA polymerase-associated protein. J.Virol. 1994;68:1360–1370. doi: 10.1128/jvi.68.3.1360-1370.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]