Abstract
Nucleoside analogs that project substituents into the minor groove when incorporated into duplex RNA perturb the binding of proteins and can affect base pairing specificity. The synthesis of 2-aminopurine ribonucleoside analogs and their phosphoramidites, their incorporation into duplex RNA, their post-synthetic modification via Cu-catalyzed azide-alkyne cycloaddition (CuAAC) and their effect on duplex stability and base pairing specificity is described.
Nucleoside analogs incorporated into RNA have a variety of uses such as investigating reaction mechanisms involving RNA1,2, imparting favorable properties on siRNAs3,4, probing RNA structure and function5,6 and exploring interaction between RNAs and proteins or small molecules7. In our previous studies, we showed that site-specific incorporation of the nucleoside analogs N2-benzylguanosine or N2-benzyl-2’-deoxyguanosine into duplex RNA blocked the binding of dsRBMs (double-stranded RNA binding motifs, 65–70 amino acid sequence motifs found in many cellular proteins that bind duplex RNA8–11). Since dsRBMs bind RNA through minor groove contacts12,13, and the 2-amino group of guanosine is directed into the minor groove, the introduction of the N2-benzyl analogs at specific sites controlled the binding of the dsRBM-containing proteins. This allowed us to characterize the binding of the RNA-dependent protein kinase (PKR) to siRNA9 and the binding of the RNA editing enzyme ADAR2 to model substrates10. However, since these early studies only employed N2-benzylguanosine as the modified base, the approach was limited to the modification of G•C pairs and to a single N2 substituent. Therefore, as an extension of this work it was desirable to develop new analogs that would allow for the modification of the RNA minor groove at A•U pairs and with a variety substituents. In addition, nucleoside analogs that block dsRBM binding may improve siRNA performance, since dsRBM proteins such as PKR and the ADARs can impede siRNAs from causing the desired specific RNAi effect14–17. New ways of directing functionality into the minor groove of duplex RNA will therefore also be of interest in siRNA design.
To create new nucleoside analogs that could be used to modify A•U pairs in RNA with minor groove substituents, we chose to synthesize derivatives of 2-aminopurine ribonucleoside bearing different substituents at N2. Like guanosine, 2-aminopurine in duplex RNA projects the 2-amino group into the minor groove. Furthermore, 2-aminopurine is an effective replacement for adenosine as it is anticipated to form a stable, two hydrogen bond pair with uridine. However, there are no reports on the effect of N2-modified 2-aminopurine ribonucleoside substitution for adenosine in duplex RNA. Here we describe the synthesis of 2-aminopurine ribonucleoside analogs and their phosphoramidites, their incorporation into RNA, their post-synthetic modification via Cu-catalyzed azide-alkyne cycloaddition (CuAAC) and their effect on duplex stability and base pairing specificity.
The preparation of the analog N2,N2-dimethyl-2-aminopurine ribonucleoside has previously been reported via SNAr reaction between a 2-chloropurine ribonucleoside precursor and dimethylamine.19 Since SNAr reactions typically proceed faster with fluoride leaving groups than other halide leaving groups20, we prepared a 2-fluoropurine ribonucleoside precursor to evaluate it as a substrate for substitution reactions with various amines. For the purposes of comparison, we also investigated a 2-bromopurine ribonucleoside intermediate as an alternative precursor to 2-aminopurine derivatives.
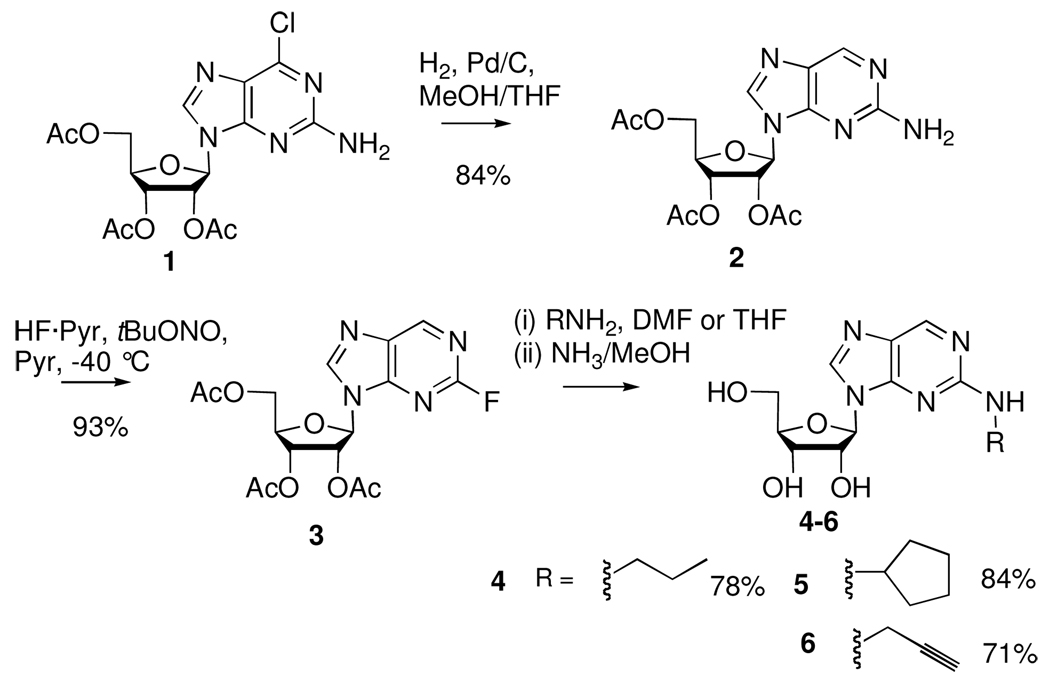
We chose to protect the ribose hydroxyl groups of the 2-fluoropurine ribonucleoside intermediate in acetate esters to allow for convenient deprotection at the amine substitution step. Thus, 2’,3’,5’-tri-O-acetyl-2-fluoropurine ribonucleoside (3) was prepared by de-halo hydrogenolysis and diazotization/fluorination of 2’,3’,5’-tri-O-acetyl-6-chloro-2-aminopurine (1). The 6-chloro hydrogenolysis substrate was readily prepared from guanosine via the method of Robins and Uznanski21. Hydrogenolysis of 1 has previously been reported in methanol22, however we found the reaction was more efficient upon addition of 10% THF to the solvent, delivering 2’,3’,5’-tri-O-acetyl-2-aminopurine (2) in 84% yield. Diazotization/fluorination of 2 proceeded in excellent yield to afford the acetyl protected 2-fluoropurine intermediate. We evaluated the 2-fluoropurine intermediate as a substrate for SNAr reactions by treating it with amines in DMF or THF. Cyclopentyl and propyl groups were chosen for incorporation at N2 in order to investigate the effect of varying the size of the minor groove obstruction in an RNA duplex on duplex stability and protein binding. In order to prepare RNA amenable to post-synthetic modification via CuAAC reaction, we also introduced a propargyl group. For the cyclopentylamine and propylamine substitutions, the substitution reactions occurred rapidly at ambient temperature with an excess of alkylamine in DMF, consuming the fluoropurine starting material in less than 30 min. After completion of the substitution reaction it was necessary to add NH3/methanol to the reaction mixture to achieve complete acetyl deprotection. Chromatographic purification yielded free nucleoside derivatives 4 and 5 in good yield. The rate of substitution with propargylamine was slightly slower. After 3 h in THF at 65 °C the fluoropurine was completely consumed. Subsequent NH3/methanol treatment and purification afforded the desired N2-modified 2-aminopurine ribonucleoside derivative 6 in good yield.
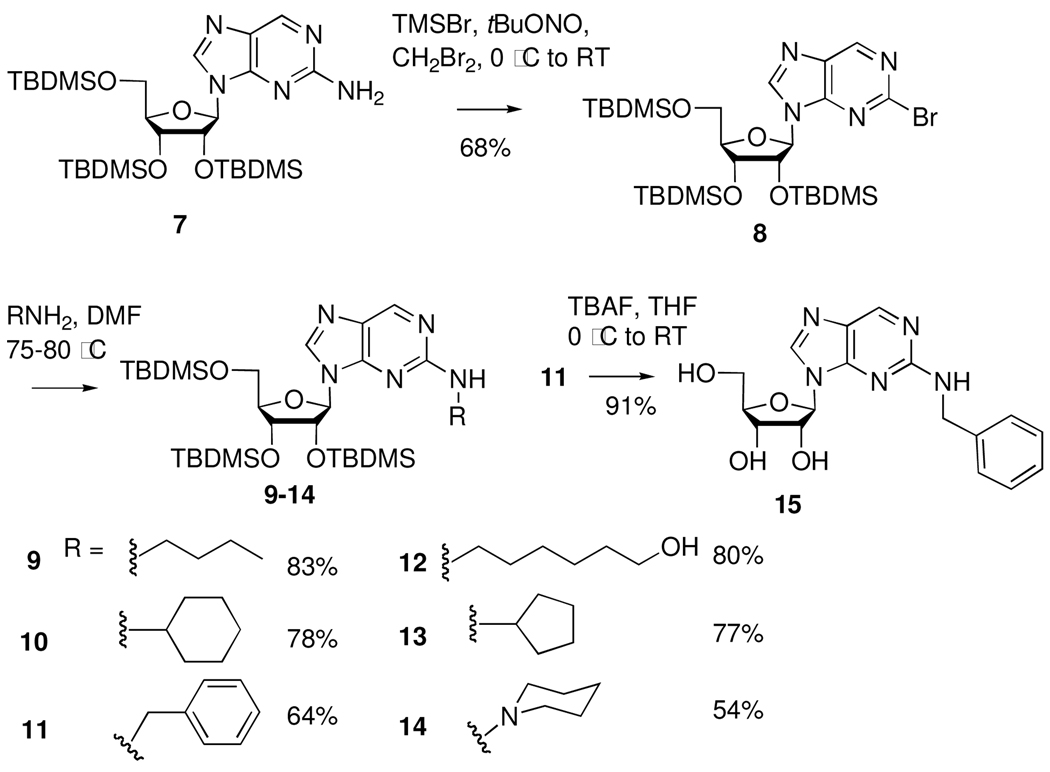
The SNAr reaction with amines was also investigated using 2’,3’,5’-tri-O-tert-butyldimethylsilyl-2-bromopurine ribonucleoside (8) as a substrate. The bromopurine intermediate was prepared by diazotization/bromination23 of tri-O-TBDMS protected 2-aminopurine ribonucleoside, which was generated in one step from commercially available 6-mercapto-2-aminopurine ribonucleoside24. SNAr reactions with excess amine occurred in 1–5 hours at 75–80 °C in DMF in good yields to afford the tri-O-TBDMS N2-modified ribonucleosides 9–14. Deprotection to the free nucleoside was demonstrated on the TBDMS-protected N2-benzyl derivative 11 by treatment with TBAF, isolating the free nucleoside 15. Both the tri-O-acetyl fluoropurine intermediate and the tri-O-TBDMS bromopurine intermediate gave acceptable yields in convenient reaction times. However, retention of ribose protection by the tri-O-TBDMS substrate allows for further elaboration of the nucleoside structure.
Nucleosides 4–6 were converted to 5’-O-DMTr, 2’-O-TBDMS, β-cyanoethyl phosphoramidites 22–24 following conventional procedures, and incorporated into a 12-mer RNA by solid-phase synthesis. The N2-propargyl amidite 24 was also incorporated at two positions in a 21-mer RNA which forms the sense strand of an siRNA targeting the human caspase 2 message.
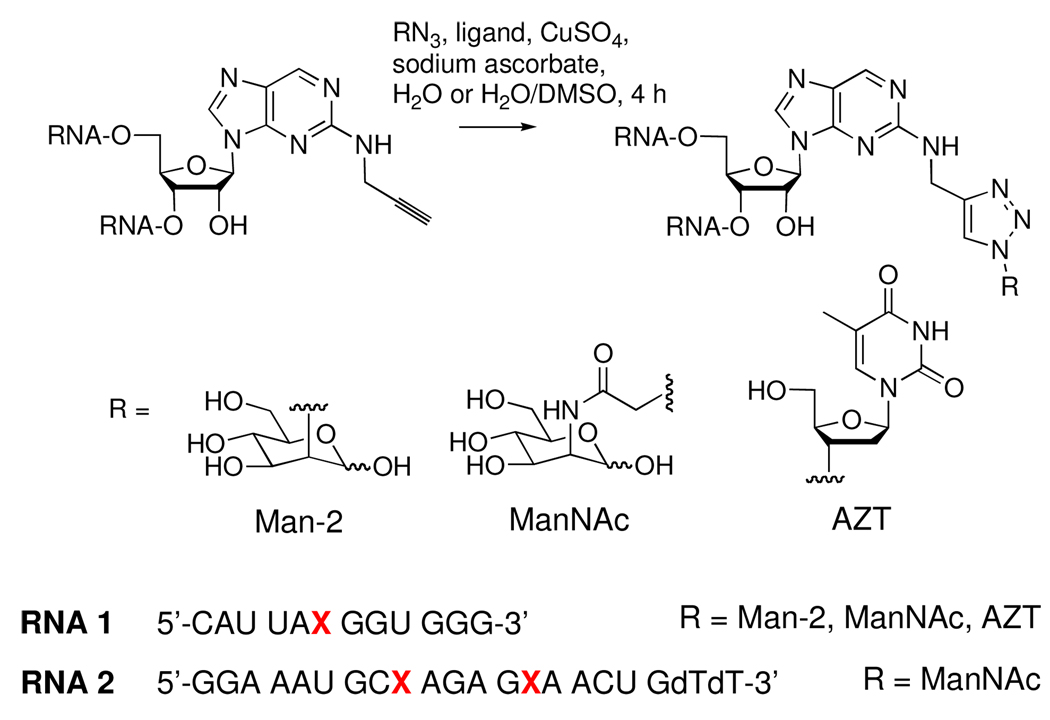
The RNAs incorporating N2-propargyl-2-aminopurine were further modified via CuAAC reaction25–27 with three azides of interest. The azides were chosen based on their potential to deliver steric bulk to the minor groove and to impart favorable delivery properties on siRNAs. We chose to employ the water-soluble tris-(hydroxypropyltriazolylmethyl)amine CuI ligand, which has previously been reported in the CuAAC reaction with oligonucleotide substrates28,29. Its solubility offers the advantage of completely aqueous reaction media, which will be beneficial in future applications where native RNA structure must be maintained during the reaction. We prepared the ligand from tripropargylamine and 3-azido-propanol via CuBr catalysis, as detailed in the supporting information. Click products of the 12-mer RNA were generated by incubating an aqueous solution of the single-stranded RNA with tris-(hydroxypropyltriazolylmethyl)amine, CuSO4, sodium ascorbate and azide (5 equiv) for 4 h at ambient temperature. The click products were purified from the reaction mixture by denaturing polyacrylamide gel electrophoresis (PAGE) and confirmed by ESI-MS. CuAAc reaction with N-azidoacetyl-D-mannosamine30 (ManNAc-N3) went to completion, as determined by a quantitative shift in mobility of the RNA band on PAGE. However, the reaction of the same RNA with 2-azido-2-deoxy-D-mannose30 (Man-2-N3) and AZT, both secondary azides, went to approximately 50% completion. In each case, the click product was purified by extraction of the slower gel band. The lower conversion of alkynyl RNA in the presence of these two azides might be due to the relatively short length of the alkynyl linker. Incomplete conversion of alkyne-containing oligonucleotides via CuAAC reactions has been previously been observed with short alkynyl linkers connected to nucleobases in single-stranded DNA31. The ManNAc click product was formed from the 21-mer via the same protocol. The reaction consumed all alkynyl RNA and yielded approximately equal amounts of mono-and di-click products, which were readily separated by PAGE.
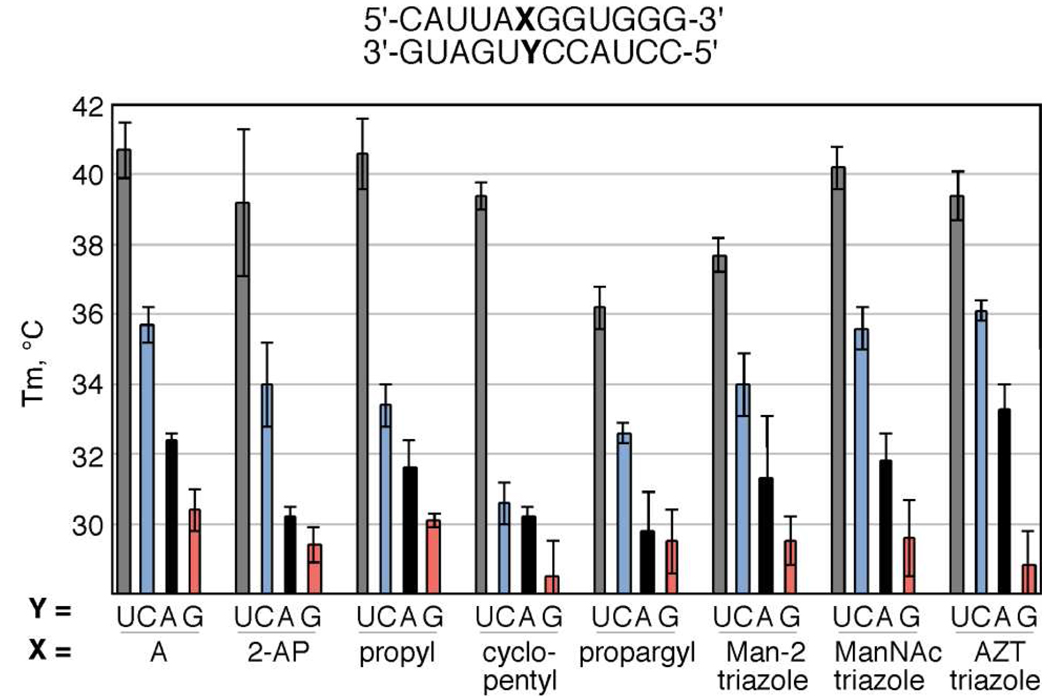
The effect of the N2-amino substitutions on duplex stability was investigated via thermal denaturation (Tm) studies of a 12 base pair RNA duplex (Figure 2). The nucleoside analogs were incorporated into the duplex opposite each of the four common bases (adenine (A), guanine (G), cytosine (C), and uracil (U)). 2-Aminopurine was also incorporated opposite each of the common bases for comparison. Firstly, the Tm of each of the modifications opposite U was examined to determine the ability of the modifications to replace A in an A•U base pair (grey bars). The duplex containing an A•U base pair had a measured Tm of 40.7 ± 0.8 °C. Replacement of A with the N2-cyclopentyl and N2-propyl 2-aminopurine analogs resulted in a small destabilization, within 1.3 ± 0.8 °C of the natural base. The Tm analysis also indicated that two of the bulky triazole-containing click products, ManNAc and AZT, were well-accommodated in the minor groove, both within 1.3 ± 0.8 °C of the natural A•U pair. Therefore, these four analogs are excellent replacements of A in duplex RNA. Furthermore, they project substituents with a range of size and structure in to the minor groove and can therefore be used to study minor groove contacts at A•U sites and to disrupt complexes with dsRBMs. The greatest destabilization opposite U was presented by the N2-propargyl modification, at 4.5 ± 0.7 °C, for reasons yet to be defined.
Figure 2.
Thermal denaturation (Tm) analysis of 12-mer RNA duplexes containing adenosine, 2-aminopurine, and N2-modified 2-aminopurine bases opposite the four common nucleobases.
The change in Tm of each of the modifications in mismatches against C, A and G was next examined to determine the effect of the modifications on pairing specificity (Figure 2, blue, black and red bars, respectively). A substantial effect on specificity was presented by the N2-cyclopentyl modification. This modification causes a 5.1 ± 0.6 °C destabilization when placed opposite C relative to the natural A•C mismatch, but its stability opposite U was within 1.3 ± 0.8 °C of the A•U base pair. Therefore, this new modified nucleoside is more specific for U than natural A in the context of the 12-mer RNA studied. This increased specificity profile of the N2-cyclopentyl modification is important since its incorporation into the antisense strand of an siRNA could make the siRNA more specific for its intended target, generating RNAi reagents with fewer off-target effects. Experiments to explore this effect are currently underway in our laboratory. The three click products showed similar specificity profiles to A. Given the similar base pairing stability and specificity of the ManNAc click product compared to A and the substantial steric bulk this analog introduces into the RNA minor groove, it was chosen for incorporation into siRNA as detailed above for future studies on protein binding and RNAi activity.
In summary, N2-modified 2-aminopurine analogs can be generated in good yield via SNAr reaction between amines and 2-fluoro and 2-bromo precursors, readily incorporated into duplex RNA and modified post-synthesis via CuAAC reaction with azides. Four of six modifications were well-tolerated as adenosine replacements, including two exceptionally bulky triazole-containing click products. The base pairing specificity profile was also affected by the minor groove projections, with the N2-cyclopentyl derivative showing increased specificity for uracil relative to adenine. Results of protein binding and RNAi studies with RNAs incorporating these new nucleoside analogs will be reported in due course.
Supplementary Material
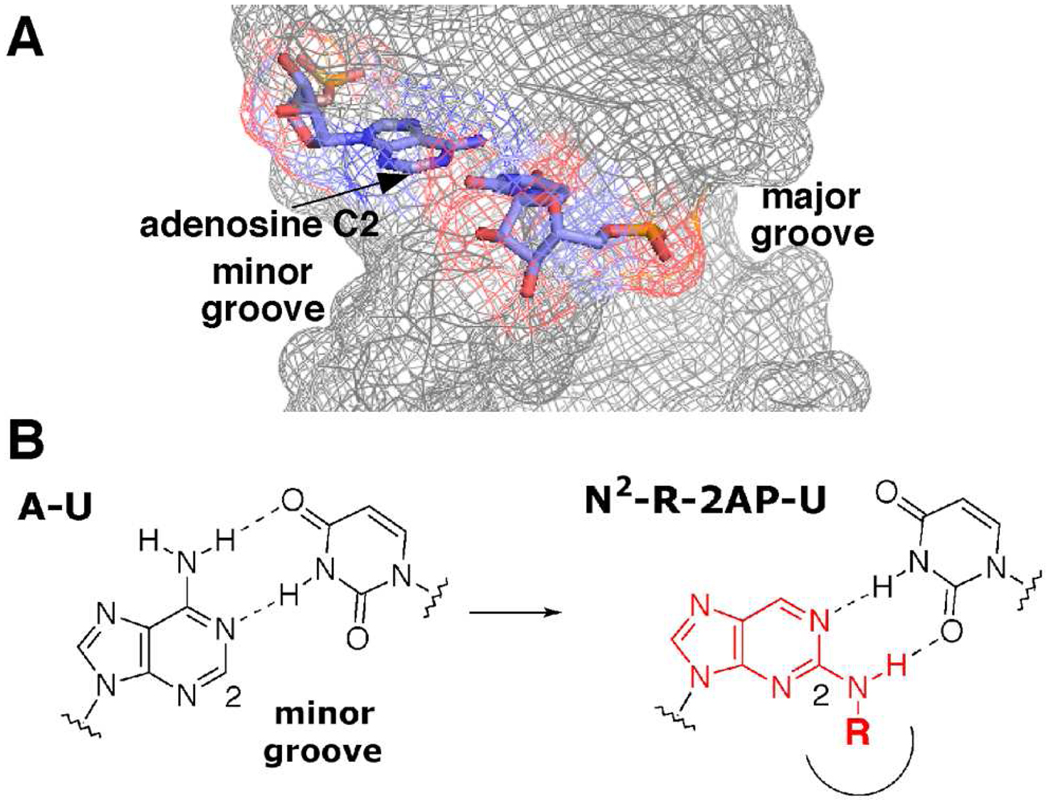
Figure 1.
A Structure of an A–U base pair in duplex RNA, showing location of adenosine C2.18 B N2-Modified 2-aminopurines place steric bulk (R) in the duplex RNA minor groove.
Scheme 1.
Preparation of N2-modified 2-aminopurine ribonucleosides via 2-fluoropurine intermediate.
Scheme 2.
Preparation of N2-modified 2-aminopurine ribonucleosides via 2-bromopurine intermediate.
Scheme 3.
Preparation of phosphoramidites.
Scheme 4.
Post-synthetic RNA modification via CuAAC.
Acknowledgment
The authors thank Professor Xi Chen, Dr. Hongzhi Cao and Saddam Muthana (Department of Chemistry, University of California, Davis) for gifts of 2-azido-2-deoxy-D-mannose and Nazidoacetyl-D-mannosamine. P. A. B acknowledges support from the National Institutes of Health (R01 GM080784).
Footnotes
Supporting Information Available: Synthetic procedures and characterization data for 2–6 and 8–24 and Cu ligand used in CuAAC; Procedures concerning RNA preparation, CuAAC reaction, duplex formation and Tm analysis; Table of Tm data.
References
- 1.Maydanovych O, Easterwood LM, Cui T, Veliz EA, Pokharel S, Beal PA. Methods Enzymol. 2007;424:369–386. doi: 10.1016/S0076-6879(07)24017-0. [DOI] [PubMed] [Google Scholar]
- 2.Suydam IT, Strobel SA. J. Am. Chem. Soc. 2008;130:13639–13648. doi: 10.1021/ja803336y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Watts J, Deleavey G, Damha M. Drug Discov. Today. 2008;13:842–855. doi: 10.1016/j.drudis.2008.05.007. [DOI] [PubMed] [Google Scholar]
- 4.Terrazas M, Kool ET. Nucleic Acids Res. 2009;37:346–353. doi: 10.1093/nar/gkn958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rist MJ, Marino JP. Curr. Org. Chem. 2002;6:775–793. [Google Scholar]
- 6.Hougland JL, Piccirilli JA. Methods Enzymol. 2009;468:107–125. doi: 10.1016/S0076-6879(09)68006-X. [DOI] [PubMed] [Google Scholar]
- 7.Tor Y. Pure Appl. Chem. 2009;81:263–272. [Google Scholar]
- 8.Doyle M, Jantsch MF. J. Struct. Biol. 2002;140:147–153. doi: 10.1016/s1047-8477(02)00544-0. [DOI] [PubMed] [Google Scholar]
- 9.Puthenveetil S, Whitby L, Ren J, Kelnar K, Krebs JF, Beal PA. Nucleic Acids Res. 2006;34:4900–4911. doi: 10.1093/nar/gkl464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stephens OM, Haudenschild BL, Beal PA. Chem. Biol. 2004;11:1239–1250. doi: 10.1016/j.chembiol.2004.06.009. [DOI] [PubMed] [Google Scholar]
- 11.Puthenveetil S, Véliz EA, Beal PA. ChemBioChem. 2004;5:383–386. doi: 10.1002/cbic.200300816. [DOI] [PubMed] [Google Scholar]
- 12.Ryter JM, Schultz SC. EMBO J. 1998;17:7505–7513. doi: 10.1093/emboj/17.24.7505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu HH, Henras A, Chanfreau G, Feigon J. P Natl Acad Sci USA. 2004;101:8307–8312. doi: 10.1073/pnas.0402627101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Armstrong ME, Gantier M, Li LL, Chung WY, McCann A, Baugh JA, Donnelly SC. J. Immunol. 2008;180:7125–7133. doi: 10.4049/jimmunol.180.11.7125. [DOI] [PubMed] [Google Scholar]
- 15.Nishikura K. Nat Rev Mol Cell Biol. 2006;7:919–931. doi: 10.1038/nrm2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang ZR, Weinschenk T, Gu KT, Schluesener HJ. J. Cell. Biochem. 2006;97:1217–1229. doi: 10.1002/jcb.20716. [DOI] [PubMed] [Google Scholar]
- 17.Richardt-Pargmann D, Vollmer J. In: Oligonucleotide Therapeutics. Rossi JJ, Gait MJ, Eckstein F, editors. Vol. 1175. Oxford: Blackwell Publishing; 2009. pp. 40–54. [Google Scholar]
- 18.Dock-Bregeon AC, Chevrier B, Podjarny A, Johnson J, de Bear JS, Gough GR, Gilham PT, Moras D. J. Mol. Biol. 1989;209:459–474. doi: 10.1016/0022-2836(89)90010-7. [DOI] [PubMed] [Google Scholar]
- 19.Schaeffer HJ, Thomas HJ. J. Am. Chem. Soc. 1958;80:4896–4899. [Google Scholar]
- 20.Liu J, Robins MJ. J. Am. Chem. Soc. 2007;129:5962–5968. doi: 10.1021/ja070021u. [DOI] [PubMed] [Google Scholar]
- 21.Robins MJ, Uznanski B. Can. J. Chem. 1981;59:2601–2607. [Google Scholar]
- 22.Zagorowska I, Adamiak RW. Biochimie. 1996;78:123–130. doi: 10.1016/0300-9084(96)82644-7. [DOI] [PubMed] [Google Scholar]
- 23.Francom P, Robins M. J Org Chem. 2003;68:666–669. doi: 10.1021/jo020625a. [DOI] [PubMed] [Google Scholar]
- 24.Fox JJ, Wempen I, Hampton A, Doerr IL. J. Am. Chem. Soc. 1956;80:1669–1675. [Google Scholar]
- 25.Rostovtsev V, Green L, Fokin V, Sharpless K. Angew Chem Int Edit. 2002;41:2596. doi: 10.1002/1521-3773(20020715)41:14<2596::AID-ANIE2596>3.0.CO;2-4. −+ [DOI] [PubMed] [Google Scholar]
- 26.Meldal M, Tornøe CW. Chem. Rev. 2008;108:2952–3015. doi: 10.1021/cr0783479. [DOI] [PubMed] [Google Scholar]
- 27.Huisgen R. In: 1,3-Dipolar Cycloaddition Chemistry. Padwa A, editor. New York: Wiley; 1984. pp. 1–176. [Google Scholar]
- 28.Jacobsen MF, Ravnsbaek JB, Gothelf KV. Org. Biomol. Chem. 2010;8:50–52. doi: 10.1039/b919387a. [DOI] [PubMed] [Google Scholar]
- 29.Kocalka P, El-Sagheer AH, Brown T. ChemBioChem. 2008;9:1280–1285. doi: 10.1002/cbic.200800006. [DOI] [PubMed] [Google Scholar]
- 30.Chokhawala HA, Huang SS, Lau K, Yu H, Cheng JS, Thon V, Hurtado-Ziola N, Guerrero JA, Varki A, Chen X. ACS Chem. Biol. 2008;3:567–576. doi: 10.1021/cb800127n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gierlich J, Burley GA, Gramlich PME, Hammond DM, Carell T. Org. Lett. 2006;8:3639–3642. doi: 10.1021/ol0610946. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.