Abstract
During eukaryotic transcription, RNA-polymerase activity generates torsional stress in DNA, having a negative impact on the elongation process. Using our previous studies of chromatin fiber structure and conformational transitions, we suggest that this torsional stress can be alleviated, thanks to a tradeoff between the fiber twist and nucleosome conformational transitions into an activated state named “reversome”. Our model enlightens the origin of polymerase pauses, and leads to the counterintuitive conclusion that chromatin-organized compaction might facilitate polymerase progression. Indeed, in a compact and well-structured chromatin loop, steric hindrance between nucleosomes enforces sequential transitions, thus ensuring that the polymerase always meets a permissive nucleosomal state.
Introduction
Transcription is a fundamental biological process during which a dedicated protein, the RNA-polymerase (RNAP), achieves the synthesis of a RNA stretch from a genomic DNA template. It can be divided into three phases: initiation, elongation, and termination. Initiation provides RNAP with an access to the promoter sequence. In eukaryotic cells, this requires the assembly of transcription factors together with RNAP into the transcription initiation complex. We here do not address problems related to the initiation phase, which are by far the most complex ones, inasmuch as they are involved at the heart of the transcriptional regulation. We focus on the elongation phase, which starts once the elongation complex has been completed, and progresses until a termination sequence is encountered. The elongation complex consists of a denaturation bubble of length ∼10 nucleotides, enclosed within RNAP (1). During elongation, RNAP tracks along the genomic sequence, swallowing the DNA double helix.
However, in eukaryotic species, genomic DNA is wrapped around octamers of histone proteins, forming nucleosomes in turn organized at a higher level into a chromatin fiber. This complex architecture is bound to hinder both the initiation and the elongation phases (2). In the standard paradigm, transcription elongation requires a decondensed state of chromatin to take place. This is questionable for at least two reasons:
-
1.
In vivo, chromatin decondensation remains elusive, all the more because chromosome structure is not yet elucidated. Whereas it is generally assumed that the fiber itself is decondensed in regions that have to be transcribed, the fiber structure has never been resolved, neither in condensed nor in decondensed chromatin—and we do not even know whether there is any difference between both structures.
-
2.
In vitro, even in decondensed fibers, nucleosomes constitute nearly absolute obstacles to RNAP progression (3).
We wish to examine here whether elongation could take place within a condensed chromatin fiber, and if so, according to which scenario.
Biological Setting
Our approach is based on a modeling study of the interplay between conformational dynamics of the chromatin fiber and RNAP processing along the fiber (4). We recall here the main biological features of eukaryotic transcription, focusing on recent biophysical results.
RNAP or DNA: which is moving?
There are three types of RNAP according to the type of RNA they synthesize. RNAP I is dedicated to ribosomal RNA synthesis and occurs in a particular environment—the nucleolus—probably devoid of nucleosomes because of its very high transcription rate. RNAP II transcribes RNA encoding proteins. The corresponding transcripts are much longer than the transcripts delivered by RNAP III, i.e., tRNAs and other small RNAs. Entanglement problems are therefore much more stringent for RNAP II than for RNAP III. As a matter of fact, RNAP progression along the genomic sequence requires a relative rotation of the RNAP together with its transcript around the DNA. Then there are two possibilities: either the DNA is kept fixed and the RNAP turns around it, thus following the DNA helical groove and producing a RNA strand coiled around the DNA double helix; or the RNAP is kept fixed and the DNA double helix has to screw inside it. In the first case, long RNA transcripts would have difficulty getting untangled and their further migration would thus be impeded. That is why we favor the second case, where the RNAP is jammed into some nuclear structure (e.g., transcription factory (5)).
The twin-supercoiled-domain (TSD) model
The above assumption implies in turn a topological problem because eukaryotic transcription occurs within chromatin loops, i.e., genomic segments ∼50–200 kilobases long, that partition chromatin into functionally independent domains (6); the loop ends are clamped by insulator elements (7), not necessarily tightly tethered to a matrix but enough constrained to make each loop a topologically insulated domain that traps DNA supercoiling and ensures the conservation of the linking number in the loop. We recall that the linking number is roughly the number of times one DNA strand is coiled around the other one (8). This topological quantity is conserved in the absence of topoisomerase activity, or before the topoisomerases act efficiently (see below). As the elongation complex progresses along the genomic sequence, the DNA double helix in front of it becomes overwound (positively supercoiled) whereas the DNA behind it becomes underwound (negatively supercoiled). This is the so-called twin-supercoiled-domain (TSD) model, first introduced by Liu and Wang (9) and extensively acknowledged since (for a review, see (10)).
Nucleosome conformations in a transcribing loop
The TSD model has been shown to be potentially relevant for eukaryotes as well (11,12). More recently Matsumoto and Hirose directly visualized (by fluorescence imaging) transcription-coupled negative supercoiling in chromatin even in the presence of active topoisomerases (13), thus strongly supporting the model. However, what kind of structural rearrangement of the chromatin loop should occur jointly with the absorption of positive (respectively, negative) supercoiling downstream (respectively, upstream)? We recently revisited the TSD model in the chromatin context by means of a single chromatin fiber nanomanipulation by magnetic tweezers and we proposed that nucleosomes may act as a topological buffer. This feature relies on the existence of three stable nucleosome states evidenced by the nanomanipulation, namely: N (negatively crossed), O (open), and P (positively crossed), according to the relative position and orientation of the linkers, one with respect to the other (14). In higher eukaryotes, linker histones H1/H5 presumably play a role both in stabilizing the states N and P against O, and channeling the transition in between them by acting as a pivot (15,16).
The reversome hypothesis
A convergent set of experimental observations (17–19) tends to indicate that RNAP II can transcribe through a nucleosome only if the nucleosome is in an activated conformation.
Using the same setup as in Bancaud et al. (14), we found that a fiber submitted to a large positive torsional stress can trap positive turns at a rate of two turns per nucleosome (20). This trapping has been shown to reflect a nucleosome chiral transition to a metastable state, called “reversome” (alternatively by the name of R-octasome (21)). This new state has been claimed to be a good candidate for the required activated conformation. Indeed, the transition to reversome is accompanied by the undocking of both H2A-H2B dimers from the (H3-H4)2 tetramer (22) that relieves the hindrance to RNAP progression. The free-energy landscape of a nucleosome under physiological ionic conditions is schematically represented in Fig. 1. It presents three minima N, P, and R, corresponding respectively to the negative, positive, and reversome states, with FR > FP ≈ FN, and a maximum B corresponding to the top of the barrier encountered during the transition between states P and R, with a corresponding free energy FB.
Figure 1.
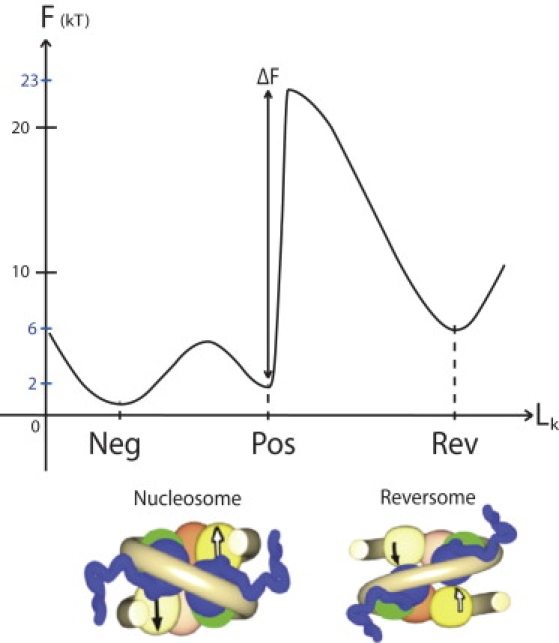
(Color online) Free-energy landscape for the nucleosome conformation. The reaction coordinate (abscissa Lk) is the linking number of nucleosomal DNA; this choice appears relevant to investigate the landscape changes when a torque Γ is applied to the DNA (see subsection Linking Number Conservation: Accounting for Mechanical Constraints). The two main states are sketched here: the current nucleosome, with two substates N and P according to the relative positions of the linkers (negative or positive crossing); and an activated state, the reversome, in which the histone core partially unfolds and the nucleosomal DNA adopts a right-handed path around the histone core. (Courtesy of Hua Wong.)
The less stable structure of the reversome arguably facilitates the RNAP progression through the reversome particles during transcription. Moreover, this auxiliary transcriptional mechanism avoids the need for a complete disassembling of the nucleosome into single histones, hence epigenetic marks can be preserved.
The fiber structure in a transcribing loop
After more than 30 years of effort, the structure of the chromatin fiber is still a matter of debate, both in vitro (where the path of the linker DNA remains elusive) and in vivo (where it is expected to vary considerably according to the cell cycle period and functional status of the fiber)—with possibly several different structures coexisting along the chromosome (23). The fiber structure is no better assessed in a transcribing loop.
Because we focus here on the transcription elongation within a condensed fiber, we favor regular fiber structures. These are indeed energetically favored by stacking interactions between nucleosomes and possibly functionally too, hence selected during (spontaneous) self-organization or (active) remodeling of the fiber. It has been shown in vitro that a small amount of nucleosome positioning is enough to get a regular structure (24). Accordingly, we shall consider as the generic setting the regular model structure of chromatin fiber established in a previous work (25), presenting a strong nucleosome stacking, hence strong steric hindrance (see Fig. 2).
Figure 2.
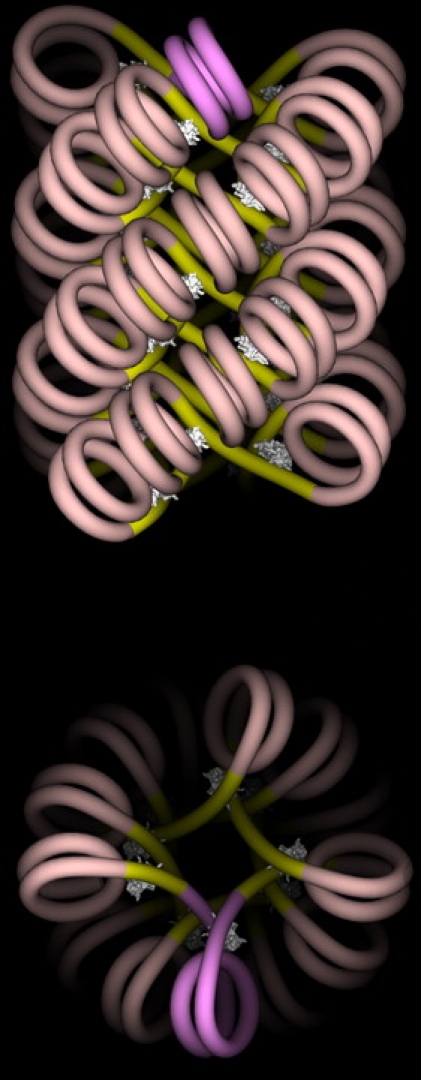
(Color online) The n-start fiber structure (with n = 4, corresponding to a repeat length of nrepeat = 87 bps (25)). Note the close and regular nucleosome stacking along each start, preventing the transition to reversome of a single nucleosome, and instead enforcing a concerted sequential transition. (Courtesy of Julien Mozziconacci.)
Our Modeling Framework
Let us sum up the biophysical bases of our model of transcription elongation in a chromatin loop:
-
1.
RNAP is jammed into some nuclear structure and exerts a torque inducing the rotation of DNA on itself that can be estimated from experimental data to occur at a constant rate of ω0 ≈ 4π rad/s (two turns per second) (26), which provides a first boundary condition in our model.
-
2.
The DNA is turning inside RNAP, inducing positive (respectively, negative) supercoiling in the downstream (respectively, upstream) part of the loop.
-
3.
The chromatin fiber within the loop is assumed to be condensed enough to ensure nucleosome stacking.
-
4.
Given that the average linking number of chromosomes in vivo has been evaluated to ∼–1 (16), we assume that the starting nucleosome state in the fiber is an appropriate mix A of positive and negative nucleosome states, of linking number LkA = –1.
-
5.
The positive torque exerted by RNAP on the loop downstream may induce the transition of nucleosomes into reversomes.
We shall adopt a continuous medium modeling of the fiber as a homogeneous elastic rod (27,28) and evaluate the role of chromatin fiber rigidity, the transmission of the torque exerted by RNAP along the fiber and the dissipation in the surrounding viscous medium. This continuous description is supported by the high and regular nucleosome density Λ along the fiber, varying between 0.5 and 1 nm−1. To be valid, this framework mainly requires the description of the fiber behavior at the level of a few nucleosomes, with an elementary length dX along the fiber axis such that 1/Λ ≪ dX ≪ N/Λ, with N the number of nucleosomes per chromatin loop. This amounts to smoothing out single-nucleosome inhomogeneities and describing the average fiber behavior at a suprananometer scale. In this setting, the local state of the fiber is described by means of one or more continuous deterministic fields as, for instance, the local fraction of reversomes at point x and time t, denoted below ξ(x, t) (29). We will switch to a discrete description in the section Transition Kinetics and Critical Torque to take into account the reversome transition kinetics and to evaluate the torque exerted by RNAP on the fiber.
Typical values of the relevant parameters of the model are summed up in Table 1.
Table 1.
Biological setting
| Entity | Parameter | Typical value | Definition |
|---|---|---|---|
| RNAP | ω0 | 4π rad/s | Angular velocity induced by RNAP to DNA (or equivalently to the fiber). |
| V | 20 bps/s | RNAP velocity, i.e., number of transcribed bps per second. | |
| Loop | N | 250 | Number of nucleosomes per chromatin loop. |
| L | 250–500 nm | Loop length. | |
| l0 | 100–500 nm | Length of the loop region downstream of the initiation site. | |
| Fiber | Λ | 0.5–1 nm−1 | Linear density of nucleosomes in a fiber. |
| Lp | 30–300 nm | Persistence length of the fiber. | |
| R | 15 nm | Fiber radius. | |
| DNA | lpitch | 3.4 nm | Pitch of the DNA-double helix in B-form. |
| npitch | 10.5 bps | Number of base pairs corresponding to the pitch. | |
| Nucleosome | nrepeat | 200 bps | Nucleosome repeat length, i.e., number of bps per nucleosome. |
| lrepeat | 70 nm | Length of DNA per nucleosome lrepeat = nrepeat.lpitch. | |
| LkN | −1.4 | Linking number (per nucleosome) of the negative N state. | |
| LkP | −0.4 | Linking number (per nucleosome) of the positive P state. | |
| LkA | −1.0 | Linking number (per nucleosome) of the average A state in condensed fibers. | |
| LkB | −0.25 to 0 | Linking number at the barrier B position.∗ | |
| LkR | 1.0 | Linking number (per nucleosome) of the reversome R state. | |
| Energy and kinetics | FN | 0.7 kT | Free energy of the P state. |
| FP | 2 kT | Free energy of the P state. | |
| FB | 23 kT | Free energy of the barrier between P and R. | |
| FR | 6 kT | Free energy of the R state. | |
| k0 | 3 106 s−1 | Preexponential factor for spontaneous fluctuation between P and R states. |
Summary of the notations and typical values of the parameters. The main parameters for the different states of the nucleosome have been obtained in Bancaud et al. (20).
The value 0 is that used in Bancaud et al. (20). The value −0.25 is obtained by fitting the experimental hysteresis curves of Bancaud et al. (20) with a kinetic model similar to the one described in Appendix S1 in the Supporting Material (H. Wong, J. Mozziconacci, M. Barbi, J. M. Victor, unpublished results).
Preliminary Investigations: Several Time- and Space Scales
Kinematic notations
We shall denote X the arc-length (curvilinear abscissa) measured along the chromatin fiber denoting the position of the RNAP with respect to the transcription initiation site (TIS). If the relative DNA-RNAP angular velocity is ω0 ≈ 4π rad/s, then the distance X(t) traveled by the RNAP measured along the fiber is
| (1) |
where lpitch is the pitch of the DNA double helix, Λ the number of nucleosomes per nm along the fiber, and lrepeat the repeat length, i.e., the DNA length per nucleosome. A length ΔX along the chromatin fiber corresponds to a length ΔX Λ lrepeat along the embedded DNA. The length of the chromatin loop downstream of the RNAP is l(t) = l0 – X(t), with l0 the length of the loop region downstream of the TIS. In the following, we will also introduce the variable x, defined as the arc-length downstream of the RNAP, again measured along the chromatin fiber (see Fig. 3).
Figure 3.
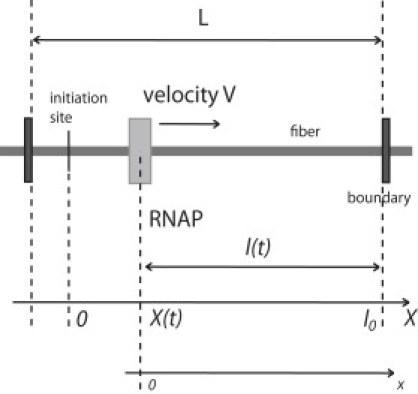
Relative positions along the chromatin fiber. The polymerase moves to the right, X(t) being its position at time t. The value l0 is the length of the loop region downstream of the initiation site X(0) = 0, and l(t) = l0 – X(t) is the length remaining at time t between the polymerase and the downstream boundary.
Propagation of torsional stress
A preliminary issue is to investigate the propagation of the torsional stress generated by the polymerase through a fiber with a given local nucleosome state. Let us assume here that the fiber is exclusively composed of nucleosomes, with no allowed transition into reversomes (i.e., ξ(x, t) = 0 over the whole fiber at any time). At the chromatin scale, inertial effects can be ignored, hence it is relevant to restrict ourselves to the overdamped regime, in which external forces and torques are fully balanced by viscous dissipation. We introduce the torsional shear strain τ(x, t) and the integrated torsion
such that Θ(x, t) is the angle by which a fixed point on the chromatin fiber surface at abscissa x has turned around the fiber axis at time t. By equating the elastic torque (torsional shear stress) and the viscous (Stokes) torque, we get
| (2) |
where Γ(x, t) is the elastic torque exerted at x on the part of the loop downstream of x; (∂Γ/∂x)(x, t) is the net elastic torque experienced by an element dx of the elastic rod of radius R modeling the chromatin fiber; and η is the dynamic viscosity of the surrounding solvent (water or crowded chromatin, but in any case, η does not exceed 10-times the viscosity of pure water η ≈ 10−3 N.s.m−2). The elastic torque can be determined within linear response theory and is proportional to the torsional shear strain,
| (3) |
where k is the Boltzmann constant, T the temperature, and Lp the twist persistence length of the fiber. The value of Lp varies from ∼30 nm in a loosely condensed fiber (Λ = 0.5 nm−1) up to ∼300 nm in a tightly condensed fiber (Λ = 1 nm−1) because of steric hindrance (30). Jointly, these two equations lead to a plain diffusion equation for Θ(x, t),
| (4) |
where D = kT Lp /ηR2 (yielding D = 1.8 10−10 m2/s for Lp ≈ 30 nm).
The relevant boundary conditions in our context are those of a finite chromatin fiber of length l(t) with one end fixed (on the downstream boundary) and one end rotating (on the RNAP side) at constant angular velocity ω0. In this case we have Θ(0, t) ≡ ω0t and, on the downstream boundary, Θ(l(t), t) ≡ 0. Anticipating that the torsional shear stress propagates much faster than the RNAP progresses, we start by keeping the RNAP fixed at x = 0 (quasistationary approximation), hence fixing l(t) = l0. We can then look for a stationary solution in the form Θ(x, t) = f(x)ω0t: substituting into Eq. 4, we obtain
| (5) |
The scaling form of this expression means that the torsional shear strain spreads along the fiber in a diffusive way roughly as . It thus takes no more than t0 ∼ l02/D ≈ 10−2 s for the torsional strain to invade the whole loop, whereas the polymerase progresses by no more than 1.6 10−2 turn, i.e., 0.16 bp, during this time. This validates the quasistationary approximation made in investigating the stress propagation, while still considering that the RNAP stays fixed at x = 0.
For t ≫ t0, the function f(x) reduces to the simple linear equivalent expression f(x) ∼1 – x/l0, leading to
| (6) |
| (7) |
| (8) |
so that the torsional strain τ(x, t) and the torque Γ(x, t) become very quickly homogeneous all along the fiber and then increase linearly with time. We conclude that the torque Γ(0, t) that RNAP should exert on the downstream part of the loop, to progress at a constant angular velocity, would rapidly exceed its maximum value. This has been estimated on Escherichia coli RNAP to be <40 pN·nm (31). Considering the typical values given in Table 1, the maximum torque would be reached after RNAP has progressed by less than half a turn, i.e., 5 bp. This feature shows that RNAP cannot progress simply this way through a topologically constrained fiber, thus requiring either topoisomerase activity, if available, or a more sophisticated scenario involving conformational changes within the fiber, strain exchange, and ensuing stress relaxation. In the next section, we examine such a scenario and check its validity.
Elongation within a Condensed Fiber
Mechanical control of the nucleosome-reversome transition
Let us now consider the RNAP activity specifically within a condensed fiber. The most relevant feature of the fiber structure (25) is the regular and close nucleosome stacking into helical piles. (Helical piles are also known as “starts”; the helical axis of each “start” being, by definition, transverse to the dyad axis of the stacked nucleosomes, there is only one way of decomposing the 30-nm fiber into a bunch of a variable number n of nucleosomal piles: one thus speaks of n-start fiber structure (25)). See Fig. 2. The closeness of stacked nucleosome faces along the start axis generates geometrical (hence mechanical) constraints on the conformational changes of single nucleosomes. The conversion of a single nucleosome into a reversome within a stacked pile is prevented due to steric hindrance. With the progression of the RNAP, the supercoiling constraint increases. The torsional constraint is then essentially applied to the last nucleosome in the pile, although the rest of the fiber remains rigidly packed. Steric hindrance thus favors a domino effect where, under the effect of the applied torque, the nucleosomes pass to their altered reversome R state one by one, forming a progressive wavefront. The fiber response to the torsional constraint imposed by the RNAP activity is now controlled by the direct interaction between the border layer of the reversome wavefront and the adjacent layer of the stacked nucleosomes, and essentially by what happens in the linker relating the most downstream reversome and its neighboring nucleosome: here is the basic step in the propagation of the mechanical constraints that triggers the transition of the said nucleosome into a reversome and later stabilizes it in an irreversible way. In this model, steric constraints prevent the relaxation to chemical equilibrium and actually maintain the fiber in a far-from-equilibrium state.
Linking number conservation: a naive model
At which speed does the reversome wavefront progress? A naive model of the process can be introduced that immediately leads to an approximate but quite accurate estimation. The previous qualitative analysis leads us to assume, as a closure relation, a steplike profile for the local fraction of reversomes ξ(x, t) (see Fig. 4),
| (9) |
with x∗(t) the position of the reversome wavefront with respect to the RNAP location.
Figure 4.
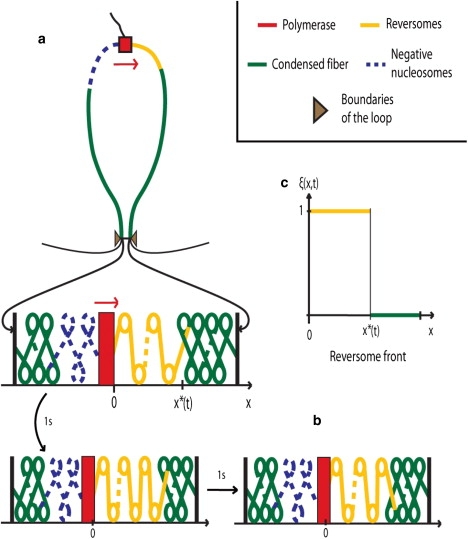
(Color online) RNA-polymerase processing within condensed chromatin fiber. (a) The supercoiling generated by the polymerase activity is trapped within the loop delineated by topological boundaries (the thin black regions are outside the loop). The ensuing torsional constraints trigger the sequential transition of nucleosomes (in green) into reversomes (in yellow). (b) Illustration of the domino effect: after 1 s, the fifth nucleosome downstream of the polymerase (green in panel a) has turned into a reversome (yellow in panel b); after one more second, the sixth nucleosome has turned into a reversome. (c) Reversome density profile: in the bold yellow region [0, x∗], the reversome density ξ(x, t) equals 1. The wavefront is located at x∗ and propagates downstream ∼10 times faster than the polymerase progression. In the polymerase wake, the nucleosomes turn to the negative state (dashed blue in panel a) to ensure the conservation of the total linking number of the loop.
As RNAP moves forward, the linking number variation in the fiber, ω0t/2π, i.e., the additional number of turns of one fiber end imposed by RNAP at time t, is mainly absorbed into the A→P→R transitions that have occurred in the fiber region x ≤ x∗(t). Explicitly, the linking number conservation condition writes
| (10) |
where we have introduced the linking number difference between the R and A states ΔLkRA = LkR – LkA. This leads to an approximate estimation of the reversome wavefront motion,
| (11) |
which therefore progresses at constant speed
| (12) |
By using the values of Table 1, the last two expressions give 1 nucl./s and 200 bps/s, respectively. Note that RNAP progresses in a much slower way, because V is ∼10 times slower than vest.
Of course, the previous derivation is oversimplified, insofar as it neglects the torsional shear strain induced by the applied torque. Nevertheless, as it will be confirmed through a more precise model, the obtained estimate for the reversome wavefront speed is rather good for a large choice of realistic fiber parameters.
Linking number conservation: accounting for mechanical constraints
Fiber torsion contribution
In naked DNA, the linking number conservation expresses itself in a balanced interchange between DNA twist and plectoneme formation (DNA writhe). In a chromatin loop, a different tradeoff will take place between chromatin fiber torsion and nucleosome conformational transitions. As discussed in the subsection Propagation of Torsional Stress, the applied torque spreads extremely rapidly through the whole fiber extent and becomes therefore homogeneous in a very short lapse of time. However, at this point, the torsional strain τ(x, t) now has a discontinuity at x = x∗(t) because of the change in the persistence length of the fiber; indeed, the part of the fiber upstream of x∗(t) is exclusively composed of reversomes whereas the part downstream is composed of nucleosomes. Inasmuch as reversomes have a more open structure than nucleosomes (see Fig. 1), reversome fibers are expected to be loosely condensed, with a persistence length close to 30 nm, ∼10 times smaller than in a tightly condensed fiber. Hence, the torsional strain τ(x, t) downstream of x∗(t) is negligible with respect to the one upstream and will be put to zero. We get, therefore,
| (13) |
This torsional strain should be accounted for in the fiber linking number balance.
The linking number of the fiber Lkfiber can be decomposed into fiber writhe and twist contributions (8):
| (14) |
In practice, the fiber persistence length and the fiber diameter are such that the fiber axis can only bend smoothly, so that its writhe is practically negligible (8), at Wrfiber ∼0. Moreover, for a relaxed and homogeneous fiber, the twist can be written as the sum of the single nucleosome linking numbers (8): for a fiber of length x∗(t) with N nucleosomes in the state X, Twfiber = N LkX = ΛLkX x∗(t). If such a fiber is now subjected to a torque Γ(t), a torsional contribution τ(t) x∗(t)/2π should be added to the relaxed fiber twist, and we finally get
| (15) |
The conservation of the linking number is therefore more correctly expressed by
| (16) |
On the right-hand side of Eq. 16, we recognize the fiber twist (coming from the contribution of all the transitions into the reversome state) that has occurred in the loop at time t, which has added to the fiber torsion Θ[0, t]/2π.
Equation 16 leads to a correction to our initial naive estimation of Eq. 10. To solve this equation for x∗(t), we now need to calculate the torque Γ(t) during RNAP progression. This requires a detailed description of the A→R transition kinetics.
Transition kinetics and critical torque
Whereas the A→P transition is rapid and occurs with almost no energetic cost (16), the P→R transition implies the crossing of a large free energy barrier. It is therefore a kinetic process, described by the rate equation
| (17) |
with PR (respectively, PP) the probability of being in the R (respectively, P) state. The forward and backward rate constants are given by k = k0 exp(– (GB – GP)/kT) and k′ = k0 exp(– (GB – GR)/kT), respectively, with GX = FX – 2πLkXΓ the Gibbs potential for the X state (20). In practice, however, the reverse transition R→P is highly improbable for typical RNAP velocities (with k0 given in Table 1 and the torque Γ = Γc obtained in Appendix S1 in the Supporting Material, we get k ∼6 s−1 and k′ ∼10−51 s−1) so that the term –k′PR in Eq. 17 can be neglected, thus leading to the simplified kinetic equation
| (18) |
Each P→R transition should occur within a typical transition time matching the RNAP velocity. Due to the kinetic character of the transition, the torque should therefore reach a critical threshold Γc (and the torsional strain a corresponding critical value τc to allow the transition into reversome to occur within this typical time). The critical torque Γc can be calculated following Evans (32), as done in Appendix S1 in the Supporting Material. It results to be constant with very good approximation, and writes
| (19) |
where we have introduced the constants ΔF = FB – FP and B = 2π(LkB − LkP), with LkB the linking number at the barrier position. Numerically, the value of the critical torque strongly depends on LkB. Using the parameters listed in Table 1, we obtain Γc in the interval 3–9 kT, or, equivalently, 15–35 pN·nm.
The reversome wavefront velocity is slightly reduced with respect to the estimation vest of Eq. 12, and writes
| (20) |
Equation 20 indicates that the additional fiber torsion introduced by the RNAP progression is not fully absorbed by nucleosome state transition, but partially used in twisting the fiber rod itself. As a consequence, the RNAP should apply greater than two turns for each A→P→R transition, which leads to the observed decrease in the wavefront velocity. In any case, with Γc in the interval 15–35 pN·nm, the estimated vest always matches the exact velocity within 6% (and down to 0.3% in the best case).
A complete picture of the stepping progression of the reversome wavefront, that takes into account its discrete nature, is given in Appendix S2 in the Supporting Material.
Biological Interpretation and Predictions
The torque exerted by RNAP
In the scenario that emerges from previous considerations, the transcription of every 20 bp induces two positive coils downstream that can be absorbed by the formation of one reversome. A reversome wavefront progresses downstream of an elongating RNAP II at a rate ∼200 bp/s. Moreover, this reversome wavefront is expected to stop at boundary elements, because they act as topological insulators. To ensure the relevance of this model, however, RNAP should be able to exert a positive torque sufficient to trigger the transition. We have found in the section Transition Kinetics and Critical Torque that the maximum value of the torque predicted by the model amounts to Γc = 15–35 pN·nm.
The torque necessary to trigger the chiral transition of a nucleosome into a reversome has been recently measured (33). The authors reported a value close to 10 pN·nm in very low salt conditions (10 mM phosphate buffer). On the other hand, the torque exerted by E. coli RNAP has been estimated to be at least 6 pN.nm and always lower than 40 pN·nm (31). The interval obtained in our model is therefore included in the one proposed by Harada et al. (31). Moreover, recent experiments (34) gave the first in vivo evidence for torque generation by elongating RNAP II in eukaryotes, indicating that mechanical stresses, constrained by architectural features of DNA and chromatin, may broadly contribute to gene regulation. Transcription-generated dynamic DNA supercoiling may be propagated over thousands of basepairs through chromatin and contribute to the control of a variety of DNA transactions (34).
These data demonstrate that RNAP is a powerful molecular motor, likely to exert sufficiently high torque for inducing the A→R transition and generating a reversome wavefront, as described in this article. Of course, any measure of the torque exerted by RNAP in physiological conditions would be highly valuable and would, moreover, provide a critical test of our model.
Transcription in a compact fiber
An important feature of the presented model is that the progressing RNAP encounters only nucleosomes in an activated state (here identified with the reversome state). This process achieves twist relaxation and at the same time ensures that the RNAP progresses in a locally open and transcriptionally permissive configuration, encountering only transparent reversomes. Steric constraints prevent the chemical equilibrium from being reached (a kind of frustration phenomenon) and enforce the sequential transition of nucleosomes into reversomes.
We are thus led to the following quite counterintuitive prediction: RNAP progression is facilitated in a compact chromatin fiber, because steric constraints between nucleosomes enforce a steplike reversome profile, ensuring that the RNAP will always face reversomes during its progression. In other words, RNAP activity within a compact fiber modifies its surroundings in such a way as to ensure that each nucleosome encountered by the RNAP as it moves along the fiber will be in the reversome conformation. The spreading of the reversome phase appears as a precursor extending farther and farther ahead of the processing RNAP, and moving ∼10 times faster than the RNAP.
Interestingly, there is evidence that transcription of siRNAs occurs in highly condensed chromatin (35).
The loop decondensation indirectly observed in vivo in yeast (36), where a chromatin locus moves toward a nuclear pore upon transcription, is a consequence of the conversion of fiber twist into fiber writhe (37). Arguably, only the decondensation associated with the conformational change of nucleosomes into reversomes is required for polymerase processing. It is nevertheless important to emphasize here that we consider only the elongation phase; a local decondensation of the 30-nm fiber is required for the transcription initiation.
Comparison with recent experiments
The reversome wavefront proposed by our model progresses downstream of an elongating RNAP II at a rate ∼200 bp/s. Strikingly, recent experiments by Petesch and Lis (38) give evidence for a rapid wavefront of nucleosome disruption, progressing at a comparable rate and stopping at the loop boundary. This wavefront arises immediately after heat shock induction and before productive elongation.
We propose that heat-shock transcription factor binding triggers a rapid productive elongation phase, during which RNAP II translocates over some genomic distance. Its progression in a topologically constrained environment creates positive torque in the downstream portion of the template, high enough to convert, at a distance, a fraction of nucleosomes into reversomes, through a domino effect. Arguably, this first productive elongation phase is too fast for topoisomerases to come into play. Reversomes are expected to be much less stable and to easily lose H2A/H2B dimers to form hexasomes or tetrasomes. Some reversomes may be lost altogether because H3/H4 tetramers prefer to bind negatively supercoiled DNA (21). Thus, the positive torque in front of the advancing RNAP will produce a complex (random) mixture of integral or partially disrupted reversomes, or will disrupt the nucleosome particles altogether. Our model thus explains straightforwardly why, in the Petesch and Lis experiments, nucleosome disruption observed downstream of the RNAP is much faster than the rate of elongation, and why it occurs over the entire downstream region and is limited to it.
Transcription initiation and RNAP pauses
Another interesting feature that has emerged from our analysis is the need of a DNA stretch free of nucleosomes at the beginning of the transcribed region (see Appendix S2). This free DNA length should ensure that at least two turns of supercoiling have been accumulated downstream of the RNAP before it arrives in front of the first nucleosome, so that the A→R transition can be achieved without inducing any negative torque. Relevant to our modeling, an initial region free of nucleosomes, immediately downstream of the TIS, is often observed (39,40). It is also interesting to note that, in the Petesch and Lis (38) experiment, and even under non-heat-shock conditions, the gene harbors a paused molecule of RNAP II, at position (+20)–(+40). RNAP stalling at this position occurs even after the gene is induced, even if its residence time dramatically decreases upon gene activation (41–43).
As soon as all the nucleosomes in the loop have turned into reversomes, the additional supercoiling due to further elongation fully accumulates in the form of fiber torsion; then the strain rapidly becomes too large, hence the resisting torque too strong, for the RNAP to progress any further, and a pause in the transcriptional activity is observed. Our model thus predicts that RNAP pausing will occur soon after the reversome wavefront has reached the loop boundary, i.e., when t = t∗, while the RNAP has traveled ∼l0/10. For typical values of l0 as given in Table 1, this leads to the rough estimate that pauses will occur after RNAP has transcribed 1–5 kb corresponding to a duration between 50 s and 250 s of nonstop elongation. This is in striking agreement with the elongation residence time recently evaluated by Darzacq et al. (44): these authors reported the first complete set of kinetic parameters of RNAP II transcription in physiological conditions (see Table 1 in their article); they found in particular an elongation residence time of ∼30 s with pausing occurring ∼1 kb downstream from the promoter. It remains to be seen whether there is a boundary 10 kb downstream from the TIS. More generally we suggest measuring nonstop elongation times for different loci together with the length of the corresponding genomic region downstream of the TIS.
Conclusion
Based on the facts that polymerase transcribes only through an activated nucleosome state and that its progression modifies the DNA linking number, we have proposed a scenario elucidating how transcription elongation can proceed within condensed chromatin. At odds with current views, this scenario does not require a decondensation of the 30-nm fiber. Our modeling study of the interplay between the RNAP activity and the chromatin fiber conformational dynamics evidences that, on the contrary, the presence of steric, mechanical, and topological constraints enforce an ordered preactivation of the fiber downstream of the RNAP. More precisely, within a condensed fiber loop with closely stacked nucleosomes, the very RNAP activity and the torsional constraints it generates at a distance along the chromatin fiber trigger the propagation of a conformational transition of the nucleosomes into a transcription-prone structure, more permissive to RNAP processing and transcriptional activity; we identify this nucleosomal structure with a recently proposed reversome conformation. Importantly, such an allosteric mechanism is relevant only in a condensed chromatin fiber. Obviously, alternative scenarios are to be searched for in other contexts likely to involve decondensed chromatin, e.g., for elongating RNAP I or III, or even RNAP II in highly transcribed genes.
Of note, we stress that all the relevant parameters—which are listed in Table 1—are taken from the literature, hence there are no fitted parameters in our model.
Finally, let us underline that it is a general fact that topological constraints induce long-range couplings along the fiber that coordinate fiber transactions and processes at the scale of a chromatin loop (typically embedding exons and introns associated to one gene); topological invariants play a channeling role in strongly constraining the possible deformations of the fiber. Conversely, functional constraints strongly condition the structure and dynamics of the fiber. Presumably, chromatin structure and function have coevolved so as to reach a good, if not optimal, consistency and efficiency.
Supporting Material
Two appendices and one figure are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(09)01734-2.
Supporting Material
Acknowledgments
We thank the reviewers for their recommendations, which improved the manuscript significantly.
C.B. holds a Doctoral Fellowship from the Agence Nationale de Recherche sur le Syndrome d'Immuno Déficience Acquise et les Hépatites Virales. Work at the Institut des Hautes Études Scientifiques has also been funded by a grant from the Génopole Evry, France. M.B. and J.-M.V. are supported in part by Agence Nationale de Recherche grant No. 05-NANO-062-03.
References
- 1.Shilatifard A., Conaway R.C., Conaway J.W. The RNA polymerase II elongation complex. Annu. Rev. Biochem. 2003;72:693–715. doi: 10.1146/annurev.biochem.72.121801.161551. [DOI] [PubMed] [Google Scholar]
- 2.Orphanides G., Reinberg D. RNA polymerase II elongation through chromatin. Nature. 2000;407:471–475. doi: 10.1038/35035000. [DOI] [PubMed] [Google Scholar]
- 3.Chang C.H., Luse D.S. The H3/H4 tetramer blocks transcript elongation by RNA polymerase II in vitro. J. Biol. Chem. 1997;272:23427–23434. doi: 10.1074/jbc.272.37.23427. [DOI] [PubMed] [Google Scholar]
- 4.Wolffe A.P. Academic Press; New York: 1998. Chromatin: Structure and Function. [Google Scholar]
- 5.Cook P.R. The organization of replication and transcription. Science. 1999;284:1790–1795. doi: 10.1126/science.284.5421.1790. [DOI] [PubMed] [Google Scholar]
- 6.Byrd K., Corces V.G. Visualization of chromatin domains created by the gypsy insulator of Drosophila. J. Cell Biol. 2003;162:565–574. doi: 10.1083/jcb.200305013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Labrador M., Corces V.G. Setting the boundaries of chromatin domains and nuclear organization. Cell. 2002;111:151–154. doi: 10.1016/s0092-8674(02)01004-8. [DOI] [PubMed] [Google Scholar]
- 8.Barbi M., Mozziconacci J., Victor J.M. How the chromatin fiber deals with topological constraints. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 2005;71:031910. doi: 10.1103/PhysRevE.71.031910. [DOI] [PubMed] [Google Scholar]
- 9.Liu L.F., Wang J.C. Supercoiling of the DNA template during transcription. Proc. Natl. Acad. Sci. USA. 1987;84:7024–7027. doi: 10.1073/pnas.84.20.7024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lavelle C. Transcription elongation through a chromatin template. Biochimie. 2007;89:519–527. doi: 10.1016/j.biochi.2006.09.019. [DOI] [PubMed] [Google Scholar]
- 11.Giaever G.N., Wang J.C. Supercoiling of intracellular DNA can occur in eukaryotic cells. Cell. 1988;55:849–856. doi: 10.1016/0092-8674(88)90140-7. [DOI] [PubMed] [Google Scholar]
- 12.Ljungman M., Hanawalt P.C. Localized torsional tension in the DNA of human cells. Proc. Natl. Acad. Sci. USA. 1992;89:6055–6059. doi: 10.1073/pnas.89.13.6055. (Erratum in Proc. Natl. Acad. Sci. USA, 89:E9364) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Matsumoto K., Hirose S. Visualization of unconstrained negative supercoils of DNA on polytene chromosomes of Drosophila. J. Cell Sci. 2004;117:3797–3805. doi: 10.1242/jcs.01225. [DOI] [PubMed] [Google Scholar]
- 14.Bancaud A., Conde e Silva N., Viovy J.L. Structural plasticity of single chromatin fibers revealed by torsional manipulation. Nat. Struct. Mol. Biol. 2006;13:444–450. doi: 10.1038/nsmb1087. [DOI] [PubMed] [Google Scholar]
- 15.Sivolob A., Prunell A. Linker histone-dependent organization and dynamics of nucleosome entry/exit DNAs. J. Mol. Biol. 2003;331:1025–1040. doi: 10.1016/s0022-2836(03)00831-3. [DOI] [PubMed] [Google Scholar]
- 16.Prunell A., Sivolob A. Paradox lost: nucleosome structure and dynamics by the DNA minicircle approach. In: Zlatanova J., Leuba S., editors. Chromatin Structure and Dynamics: State of the Art, New Comprehensive Biochemistry. Vol. 39. Elsevier; Amsterdam, The Netherlands: 2004. [Google Scholar]
- 17.Kireeva M.L., Walter W., Studitsky V.M. Nucleosome remodeling induced by RNA polymerase II: loss of the H2A/H2B dimer during transcription. Mol. Cell. 2002;9:541–552. doi: 10.1016/s1097-2765(02)00472-0. [DOI] [PubMed] [Google Scholar]
- 18.Lee M.S., Garrard W.T. Positive DNA supercoiling generates a chromatin conformation characteristic of highly active genes. Proc. Natl. Acad. Sci. USA. 1991;88:9675–9679. doi: 10.1073/pnas.88.21.9675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bondarenko V.A., Steele L.M., Studitsky V.M. Nucleosomes can form a polar barrier to transcript elongation by RNA polymerase II. Mol. Cell. 2006;24:469–479. doi: 10.1016/j.molcel.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 20.Bancaud A., Wagner G., Prunell A. Nucleosome chiral transition under positive torsional stress in single chromatin fibers. Mol. Cell. 2007;27:135–147. doi: 10.1016/j.molcel.2007.05.037. [DOI] [PubMed] [Google Scholar]
- 21.Zlatanova J., Bishop T.C., van Holde K. The nucleosome family: dynamic and growing. Structure. 2009;17:160–171. doi: 10.1016/j.str.2008.12.016. [DOI] [PubMed] [Google Scholar]
- 22.Mozziconacci J., Victor J.M. Nucleosome gaping supports a functional structure for the 30nm chromatin fiber. J. Struct. Biol. 2003;143:72–76. doi: 10.1016/s1047-8477(03)00102-3. [DOI] [PubMed] [Google Scholar]
- 23.van Holde K., Zlatanova J. Chromatin fiber structure: where is the problem now? Semin. Cell Dev. Biol. 2007;18:651–658. doi: 10.1016/j.semcdb.2007.08.005. [DOI] [PubMed] [Google Scholar]
- 24.Weidemann T., Wachsmuth M., Langowski J. Counting nucleosomes in living cells with a combination of fluorescence correlation spectroscopy and confocal imaging. J. Mol. Biol. 2003;334:229–240. doi: 10.1016/j.jmb.2003.08.063. [DOI] [PubMed] [Google Scholar]
- 25.Wong H., Victor J.M., Mozziconacci J. An all-atom model of the chromatin fiber containing linker histones reveals a versatile structure tuned by the nucleosomal repeat length. PLoS One. 2007;2:e877. doi: 10.1371/journal.pone.0000877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Uptain S.M., Kane C.M., Chamberlin M.J. Basic mechanisms of transcript elongation and its regulation. Annu. Rev. Biochem. 1997;66:117–172. doi: 10.1146/annurev.biochem.66.1.117. [DOI] [PubMed] [Google Scholar]
- 27.Ben-Haïm E., Lesne A., Victor J.M. Chromatin: a tunable spring at work inside chromosomes. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 2001;64:051921. doi: 10.1103/PhysRevE.64.051921. [DOI] [PubMed] [Google Scholar]
- 28.Love A.E.H. Dover; Mineola, NY: 1944. Treatise on the Mathematical Theory of Elasticity Theory. [Google Scholar]
- 29.Lesne A., Victor J.M. Chromatin fiber functional organization: some plausible models. Eur Phys J E Soft Matter. 2006;19:279–290. doi: 10.1140/epje/i2005-10050-6. [DOI] [PubMed] [Google Scholar]
- 30.Mergell B., Everaers R., Schiessel H. Nucleosome interactions in chromatin: fiber stiffening and hairpin formation. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 2004;70:011915. doi: 10.1103/PhysRevE.70.011915. [DOI] [PubMed] [Google Scholar]
- 31.Harada Y., Ohara O., Kinosita K. Direct observation of DNA rotation during transcription by Escherichia coli RNA polymerase. Nature. 2001;409:113–115. doi: 10.1038/35051126. [DOI] [PubMed] [Google Scholar]
- 32.Evans E. Probing the relation between force—lifetime—and chemistry in single molecular bonds. Annu. Rev. Biophys. Biomol. Struct. 2001;30:105–128. doi: 10.1146/annurev.biophys.30.1.105. [DOI] [PubMed] [Google Scholar]
- 33.Celedon A., Nodelman I.M., Sun S.X. Magnetic tweezers measurement of single molecule torque. Nano Lett. 2009;9:1720–1725. doi: 10.1021/nl900631w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kouzine F., Sanford S., Levens D. The functional response of upstream DNA to dynamic supercoiling in vivo. Nat. Struct. Mol. Biol. 2008;15:146–154. doi: 10.1038/nsmb.1372. [DOI] [PubMed] [Google Scholar]
- 35.Grewal S.I., Elgin S.C. Transcription and RNA interference in the formation of heterochromatin. Nature. 2007;447:399–406. doi: 10.1038/nature05914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cabal G.G., Rodriguez-Navarro S., Nehrbass U. Molecular analysis of SAGA mediated nuclear pore gene gating activation in yeast. Nature. 2006;441:770–773. [Google Scholar]
- 37.Mozziconacci J., Lavelle C., Victor J.M. A physical model for the condensation and decondensation of eukaryotic chromosomes. FEBS Lett. 2006;580:368–372. doi: 10.1016/j.febslet.2005.12.053. [DOI] [PubMed] [Google Scholar]
- 38.Petesch S.J., Lis J.T. Rapid, transcription-independent loss of nucleosomes over a large chromatin domain at Hsp70 loci. Cell. 2008;134:74–84. doi: 10.1016/j.cell.2008.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vaillant C., Audit B., Arneodo A. Experiments confirm the influence of genome long-range correlations on nucleosome positioning. Phys. Rev. Lett. 2007;99:2181035. doi: 10.1103/PhysRevLett.99.218103. [DOI] [PubMed] [Google Scholar]
- 40.Miele V., Vaillant C., Grange T. DNA physical properties determine nucleosome occupancy from yeast to fly. Nucleic Acids Res. 2008;36:3746–3756. doi: 10.1093/nar/gkn262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Saunders A., Core L.J., Lis J.T. Breaking barriers to transcription elongation. Nat. Rev. Mol. Cell Biol. 2006;7:557–567. doi: 10.1038/nrm1981. [DOI] [PubMed] [Google Scholar]
- 42.Lis J.T. Imaging Drosophila gene activation and polymerase pausing in vivo. Nature. 2007;450:198–202. doi: 10.1038/nature06324. [DOI] [PubMed] [Google Scholar]
- 43.Nechaev S., Adelman K. Promoter-proximal Pol II: when stalling speeds things up. Cell Cycle. 2008;7:1539–1544. doi: 10.4161/cc.7.11.6006. [DOI] [PubMed] [Google Scholar]
- 44.Darzacq X., Shav-Tal Y., Singer R.H. In vivo dynamics of RNA polymerase II transcription. Nat. Struct. Mol. Biol. 2007;14:796–806. doi: 10.1038/nsmb1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


