Abstract
Pathological activation of digestive zymogens within the pancreatic acinar cell initiates acute pancreatitis. Cytosolic events regulate this activation within intracellular compartments of unclear identity. In an in vivo model of acute pancreatitis, zymogen activation was detected in both zymogen granule-enriched and microsomal cellular fractions. To examine the mechanism of this activation in vitro, a reconstituted system was developed using pancreatic cytosol, a zymogen granule-enriched fraction, and a microsomal fraction. Addition of cytosol to either particulate fraction resulted in a prominent increase in both trypsin and chymotrypsin activities. The percentage of the pool of trypsinogen and chymotrypsinogen activated was about twofold and sixfold greater, respectively, in the microsomal than in the zymogen granule-enriched fraction. Activation of chymotrypsinogen but not trypsinogen was significantly enhanced by ATP (5 mM) but not by the inactive ATP analog AMP-PNP. The processing of procarboxypeptidase B to its mature form also demonstrated a requirement for ATP and cytosol. E64d, an inhibitor of cathepsin B, a thiol protease that can activate trypsin, completely inhibited trypsin activity but did not affect chymotrypsin activity or carboxypeptidase B generation. These studies demonstrate that both zymogen granule-enriched and microsomal fractions from the pancreas can support cytosol-dependent zymogen activation. A component of the activation of some zymogens, such as chymotrypsinogen and procarboxypeptidase, may depend on ATP but not on trypsin or cathepsin B.
Keywords: trypsin(ogen), chymotrypsin(ogen), procarboxypeptidase B, cathepsin B, zymogen granule-enriched fraction, microsomal fraction
Exocrine pancreatic digestive zymogens are proproteins that are secreted by pancreatic acinar cells and proteolytically processed to their lower-molecular-weight active forms in the small intestine. Although pancreatic amylase and lipase are secreted as active enzymes, all pancreatic digestive proteases and several other hydrolases are zymogens. In the small intestine, pancreatic zymogen activation begins with the conversion of trypsinogen to trypsin by the intestinal brush-border protease enterokinase. Trypsin then converts the remaining pancreatic zymogens, including the chymotrypsinogens and procarboxypeptidases, to the active forms required for nutrient digestion. Under pathological conditions that cause pancreatitis, digestive zymogens undergo premature activation within the pancreatic acinar cell (2, 6, 14, 15, 27). High levels of activation overcome protective mechanisms, causing cell injury and acute pancreatitis. The cellular compartment and molecular mechanisms linked to this pathological proteolytic processing remain unclear.
Studies have described pancreatic acinar cell zymogen activation taking place in zymogen granules and other vesicular compartments. Two studies have suggested that initial zymogen activation might occur in zymogen granules (18, 22). Recent work from our laboratory and others concluded that initial zymogen activation takes place in a vesicular, nonzy-mogen granule compartment (13, 20). The latter studies used antibodies that selectively label the trypsinogen activation peptide, a surrogate for trypsinogen activation, in confocal and immunoelectron microscopy to detect the site of trypsinogen activation in the caerulein hyperstimulation model of acute pancreatitis. Trypsinogen activation peptide was initially localized to small vesicles in a supranuclear region of the acinar cell that contained markers of lysosomes and recycling endosomes. Whether the dissimilar findings relating to the site of zymogen activation are due to differences in detection methods or experimental systems remains unclear.
Similar to the in vivo caerulein hyperstimulation model of acute pancreatitis, zymogen activation can be initiated by supraphysiological stimulation of G protein-coupled receptors on the pancreatic acinar cell. Thus supraphysiological concentrations of agonists such as cholecystokinin and acetylcholine analogs cause pathological zymogen activation within acinar cells (13, 15, 20). In this study, we report that, in the in vivo caerulein hyperstimulation model, this activation occurs in zymogen granule-enriched and a microsomal fraction.
Although reconstituted cell systems have been used to define many molecular mechanisms, technical limitations have precluded their use to study zymogen activation in the pancreatic acinar cell. To further explore the mechanisms responsible for the activation and define relevant cellular compartments, we have reconstituted zymogen activation in vitro. Using this system, we showed that zymogen activation 1) requires both cytosol and cellular organelles, 2) can take place in both zymogen-granule enriched and microsomal fractions, and 3) exhibits ATP-dependent and independent mechanisms.
Materials and Methods
In Vivo Studies
All experiments and procedures using animals were approved by the Veterans Affairs International Animal Care and Use Committee (West Haven, CT; approved protocol FG0006). Experimental pancreatitis was initiated in 150- to 200-g Sprague-Dawley rats (anesthetized intraperitoneally with ketamine-xylazine) via a tail vein injection of 40 μg/kg caerulein (hyperstimulatory dose), a cholecystokinin analog used to cause pancreatitis, and pathologic pancreatic zymogen activation (4, 26). Controls received saline. After 15 min, rats were euthanized in a CO2 chamber, and the pancreas was harvested. Particulate fractions and cytosolic fractions were assayed for enzyme content as described below.
Pancreatic Fractionation: Isolation of Zymogen Granule-Enriched, Microsomal and Cytosolic Fractions
Pancreatic fractionation followed the protocol of Tartakoff and Jamieson (28) with several modifications. The pancreas was divided in two; one-half was homogenized on ice in 10 vol (∼5 ml) of 0.3 M sucrose and 1 mM DTT in the absence of protease inhibitors and used for isolation of the zymogen granule-enriched and microsomes (preparation A). The other one-half was homogenized in 10 vol (∼5 ml) of 0.3 M sucrose, 1 mM DTT, 25 mM Tris (pH 7.2), 5 mM aprotonin, soybean trypsin inhibitor (0.1 mg/ml), 5 mM benzamidine, 0.1 mM 4-(2-aminoethyl)benzenesulfonyl fluoride, and complete EDTA-free protease inhibitor cocktail (20 μl/ml of 25× stock solution). All reagents were purchased from Sigma, except for the Roche protease inhibitor cocktail, used for isolation of cytosol (cytosolic buffer; denoted preparation B). The high-molecular-weight protease inhibitors were added to inhibit enzyme activity in the cytosol and to ensure that enzyme activity observed in the assay was within the organelles. Homogenates were centrifuged at 500 g for 5 min. All preparations and storage were at 4°C unless otherwise stated.
Preparation of Membrane Fractions
The pellet from the 500-g centrifugation was discarded, and the resulting postnuclear supernatant was centrifuged at 3,000 g for 10 min over a 2 M sucrose cushion. The sucrose cushion was necessary to prevent organelle rupture during centrifugation. After the 3,000-g centrifugation, the layer at the interface of preparation A was retained, diluted with 2 vol of 0.3 M sucrose, and washed at 3,000 g over a 2 M sucrose cushion (further described below in KCl and Sucrose Wash). The layer at the interface was retained; this was the zymogen granule-enriched fraction, which was subsequently diluted with distilled water to give a concentration of 0.3 M sucrose (using a refractometer; Leica, Buffalo, NY). The supernatant from the initial 3,000-g centrifugation (preparation A) was then centrifuged at 15,000 g for 15 min over a 2 M sucrose cushion. The layer at the interface (mitochondrial fraction) was retained. The remaining supernatant was centrifuged at 180,000 g for 60 min over a 2 M sucrose cushion. The layer at the interface was removed, diluted with 2 vol of 0.3 M sucrose, and washed at 288,000 g over a 2 M sucrose cushion. The layer at the interface was retained; this was the microsomal fraction, which was subsequently diluted with water to give a concentration of 0.3 M sucrose.
KCl and Sucrose Wash
To eliminate associated proteins from the organelle surface, zymogen granule-enriched and microsomal fractions were diluted fivefold in 0.3 M sucrose (control) or 0.5 M KCl (21). They were centrifuged at 3,000 g and 180,000 g, respectively, over a 2 M sucrose cushion. The layer at each interface was collected and diluted with water to 0.3 M sucrose. These were denoted as “washed” fractions.
Preparation of the Cytosolic Fraction
After the 500-g centrifugation, the supernatant (preparation B) was retained and centrifuged at 180,000 g for 60 min on a 2 M sucrose cushion. The supernatant was removed, and the layer at the interface was discarded. Endogenous ATP and low-molecular-weight protease inhibitors were removed from the cytosol by dialysis (molecular weight cut-off 3,500) in 0.3 M sucrose, 25 mM Tris (pH 7.2), and 100 mM KCl for 60 min with a dialysis cassette (Slide-A-Lyzer dialysis cassette, 3,500 molecular weight cut off, 0.5- to 3-ml capacity; Pierce Biotech). After dialysis, the fraction was centrifuged at 288,000 g for 15 min in tubes precoated with cytosolic buffer to remove small vesicles.
The zymogen granule-enriched fraction and the microsomal fraction from preparation A (lacking protease inhibitors) and the cytosolic fraction (containing high-molecular-weight protease inhibitors) from preparation B were used in activation assays described elsewhere.
Zymogen Activation Assays
In this study, the conversion of zymogens to mature enzymes is taken as either an increase in enzyme activity, as determined with fluorogenic assays (16, 17), or the appearance of mature enzyme forms, as determined with immunoblot analysis (15).
Enzymatic Assays
Assay buffer (350 μl) [50 mM Tris (pH 7.6) and 150 mM KCl] was added to each well of a 24-well plate (Greiner Bio-one Cellstar TC-plate; Greiner Bio-One, Monroe, NC) at room temperature (25°C). Zymogen granule-enriched or microsomal fractions (50 μl), previously stored on ice, were then added to the assay buffer in each well at room temperature. Next, 50 μl of 400 μM enzyme substrate (trypsin or chymotrypsin substrate; trypsin was MAA3135-v and chymotrypsin was MAA3114-v, from Peptides International, Louisville, KY) diluted in assay buffer at room temperature (40 μM final) were added to each well. The plate was lightly agitated from side to side to mix the contents of each well and then left at room temperature under normal lighting conditions. In preliminary studies, incubation times of 0–60 min were used to allow uptake of fluorogenic substrate by the organelles; 15 min was found to be the optimal value. For in vivo experiments, stimulated zymogen granule-enriched and microsomal fractions were assayed alone, in the absence of any cytosol or ATP, after immediate addition of substrate.
For in vitro experiments, after the 15-min incubation of substrates with organelles, 50 μl of buffer (control) or cytosol (50 μl) were added to each well and incubated for a further 15 min. In preliminary studies, these conditions resulted in a linear increase in fluorometric emission. Preliminary studies also demonstrated that the cytosol-dependent generation of protease activity was linear when 50 μl was added to the assay; this amount was used unless otherwise stated. After a total incubation of 30 min, ATP (5 mM) or AMP-PNP (5 mM) was added, and fluorescence emissions were recorded at an excitation wavelength of 380 nm and an emission of 440 nm for 11 min (20 reads) with an HTS 7000 fluorimeter (Perkin-Elmer Analytical Instruments, Shelton, CT). The fluorometric emission slope (relative fluorescence units/s) represented enzymatic activity. Organelles were lysed with Triton X-100, and amylase content was determined (Phaebadas kit; Pharmacia, Rochester, NY). The enzyme activity was normalized to amylase content (expressed as fold activation vs. the zymogen granule/microsomal fraction plus cytosol plus ATP condition). Because preliminary studies demonstrated that normalization to amylase or protein content were equivalent, data are normalized to amylase content.
Pancreatic Zymogens
For determination of total pancreatic trypsinogen and chymotrypsinogen in the zymogen granule-enriched and microsomal fractions, samples were incubated with excess enterokinase (5 μg/100 μl) in the presence of 10 mM CaCl2 at 37°C for 60 min, allowing for complete activation of trypsinogen and chymotrypsinogen in the sample (18). The samples were assayed with fluorogenic substrate for either trypsin or chymotrypsin (values for total activity determined as relative fluorescence units per second as a function of total protein content, with protein determined by the Bradford assay). To determine the percentage activation induced by cytosol and ATP in the zymogen granule-enriched and microsomal fractions, the rfu/sec values obtained in these samples were expressed as a function of total protein content and the values divided by the total activity in that fraction.
Organelle Lysis
To determine whether active enzymes from lysed organelles contributed to measured enzyme activities, the cytosolic preparation was modified. High-molecular-weight protease inhibitors (0.1 mg/ml soybean trypsin inhibitor, 5 mM aprotinin) were replaced with lower-molecular-weight inhibitors (5 mM benzamidine, 5 mM leupeptin). Before dialysis, the soybean trypsin inhibitor (0.1 mg/ml) and 5 mM aprotonin were added to one-half of the preparation. Both cytosolic preparations were dialyzed to remove low-molecular-weight inhibitors, centrifuged at 288,000 g, and used in trypsin and chymotrypsin activity assays. Hence, the cytosolic preparation with soybean trypsin inhibitor has the capacity to inhibit free protease and the other does not. Furthermore, cytosol treated with membrane-permeable complete EDTA-free protease inhibitor cocktail (20 μl/ml of 25 × stock solution; Roche) was used in additional assays to demonstrate that zymogen activation was occurring within membrane-bound compartments.
Proteasomal Inhibitors
Proteasomal inhibitors lactacystin (20 μM) and MG-132 (0.75 μM) (Calbiochem) were added at the same time as substrate in enzyme assays to examine the possible involvement of the proteasome in chymotrypsin activation.
Immunoblot Assays for Zymogen Activation
Zymogen granule-enriched fraction or microsomal fraction was incubated alone, in the presence of cytosol, or in the presence of cytosol and ATP. After 1 min, 10 μl of complete EDTA-free protease inhibitor cocktail and 100 μl of 6× Laemmli sample buffer were added to each well. Because carboxypeptidase degraded rapidly in this system, procarboxypeptidase processing was assayed after 1-min incubations. Samples were heated to 95°C for 5 min. Equal protein was loaded as determined by the Bradford assay, and proteins were separated by SDS-PAGE (12.5% polyacrylamide) and processed for immunoblot analysis on polyvinylidene difluoride membranes as described (15). To detect proteolytic zymogen activation, membranes were probed with an antibody that detected the zymogen and active forms of procarboxypeptidase B (gift from G. Scheele).
Protein Kinase Inhibitors
The general protein kinase inhibitors K252a (100 nM) and H-89 (100, 30, and 1 μM) (Calbiochem) and the phosphatidylinositol 3-kinase inhibitors wortmannin (100 nM) and LY-294002 (25 μM) (Sigma) were added at the same time as substrate in enzyme assays to determine the effects of protein kinases in trypsinogen and chymotrypsinogen activation. Controls were incubated with the same concentration of drug vehicle (DMSO).
Role of Cathepsin B in Zymogen Activation
To examine the effects of cathepsin B inhibition on zymogen activation, trypsin and chymotrypsin were assayed in the presence of the cathepsin B inhibitor E64d (Sigma). E64d (1 mM) was added at time 0 (24). Control experiments used an equal volume of the vehicle (1:1 methanol-water). Effects of E64d on 100 ng of pure trypsin and chymotrypsin (Sigma) were evaluated using the same assay technique as described above.
Statistical Analysis
Statistical significance was determined using the Wilcoxon rank sum test. Significance was assumed for P < 0.05. Deviation bars in all figures represent 25th and 75th quartiles.
Results
Detection of Zymogen Activation in Both Zymogen Granule-Enriched and Microsomal Fractions In Vivo
Zymogen activation in particulate fractions
Zymogen granule-enriched and microsomal fractions were harvested from rat pancreas stimulated in vivo with supraphysiological concentrations of caerulein for 15 min. This treatment caused approximately threefold and fivefold increases in trypsin activity in both the zymogen granule-enriched and microsomal fractions, respectively, when compared with unstimulated pancreas. (Fig. 1 A). Similarly, chymotrypsin activity was ∼10- and 6-fold higher in the zymogen granule-enriched and microsomal fractions, respectively, when compared with controls from unstimulated pancreas (normalized to amylase content) (Fig. 1B). On the basis of evidence that zymogen activation might occur in distinct cellular organelles, we developed a cell-free assay system to determine whether activation could be reconstituted using isolated pancreatic fractions.
Fig. 1.
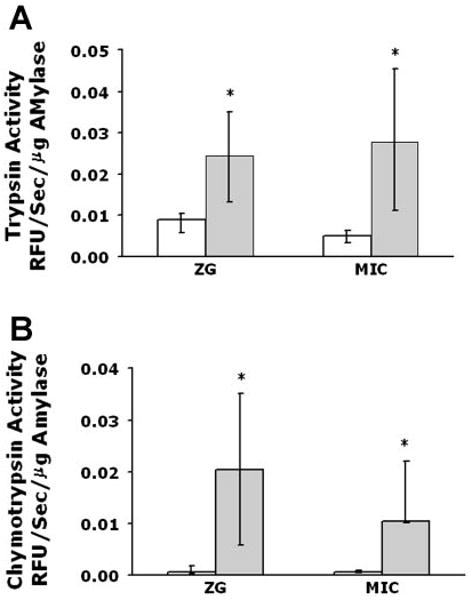
Zymogen activation is observed in both a zymogen granule-enriched (ZG) and microsomal (Mic) fraction following supraphysiological caerulein treatment in vivo. Rats were treated for 15 min with caerulein (40 μg/kg body wt) via tail vein injection. Enhanced trypsin (A) and chymotrypsin (B) activities are seen in both ZG and Mic fractions at 15 min. *P < 0.05 vs. ZG or Mic from unstimulated pancreas (control).
Reconstitution of Zymogen Activation in Particulate Fractions
Total zymogen and amylase content of particulate fractions
The relative trypsinogen, chymotrypsinogen, and amylase contents were similar between the zymogen granule-enriched and microsomal fractions (Fig. 2). Therefore, results were normalized to amylase content unless noted otherwise.
Fig. 2.
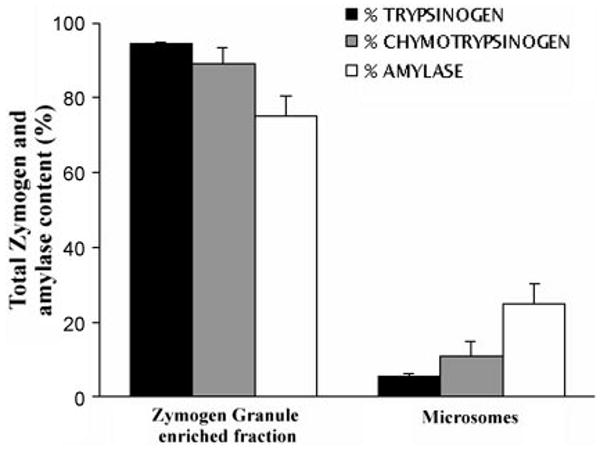
Zymogen and amylase distribution in ZG and Mic pancreatic fractions. Trypsinogen, chymotrypsinogen, and amylase content were assayed as described in MATERIALS AND METHODS. Values are expressed as percentages of the total of these 2 fractions.
Trypsinogen activation
Zymogen granule-enriched, mitochondrial, and microsomal fractions were isolated from unstimulated pancreas and assayed for trypsin activity. The mitochondrial fraction and cytosol showed negligible activities. A small amount of basal activity was observed in zymogen granule-enriched and microsomal fractions alone (Fig. 3). However, the activities in these fractions were significantly enhanced by the presence of cytosol (Fig. 3). Addition of 5 mM ATP did not affect trypsin activity in either the zymogen granule-enriched fraction or microsomes in the presence or absence of cytosol (Fig. 3), indicating that trypsinogen processing to trypsin is ATP independent.
Fig. 3.
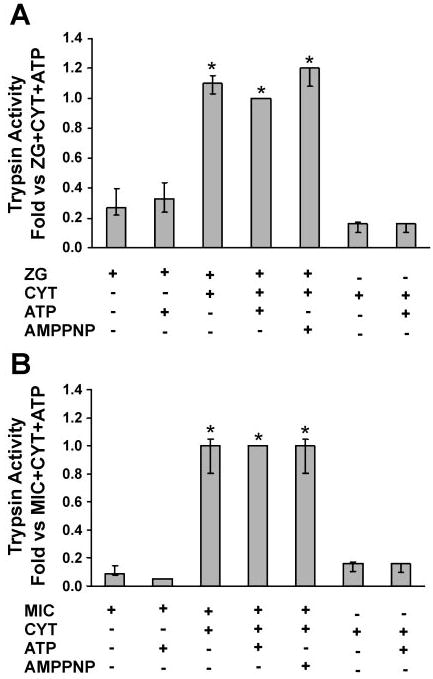
Trypsinogen activation reconstituted in ZG (A) and Mic (B) pancreatic fractions is dependent on cytosol (Cyt). Activity is enhanced by the addition of Cyt but not by 5 mM ATP (see MATERIALS AND METHODS). Data are normalized to amylase content and expressed as fold vs. ZG/Mic + Cyt + ATP; n = 3. AMP-PNP, nonhydrolyzable ATP analog. *P < 0.05 vs. no ATP condition.
When the trypsin activity was normalized, the relative activity was about sixfold greater in microsomes than in the zymogen granule-enriched fraction, indicating more efficient processing in the microsomes. To validate this finding, trypsinogen content was estimated by its conversion to trypsin by enterokinase (Fig. 2). Values for the cytosol-dependent activation were compared with the total trypsinogen activation for each fraction. Cytosol stimulated activation of 2% and 5% of total trypsinogen in the zymogen granule-enriched fraction and microsomes, respectively, further suggesting that processing may be more efficient in the microsomes (Table 1). However, because the total trypsinogen content of the zymogen granule-enriched fraction was much greater, the 2% of the pool activated yielded higher overall activity than the 5% of the smaller microsomal trypsinogen pool. Thus, when trypsinogen activation was normalized to its distribution between the two compartments, the activities contributed by the zymogen-granule enriched fraction were about eightfold higher than those seen in microsomal compartments (not shown).
Table 1. Percent zymogen activation in reconstituted system.
| ZG | ZG/Cyt | ZG/Cyt/ATP | Mic | Mic/Cyt | Mic/Cyt/ATP |
|---|---|---|---|---|---|
| Trypsin activity | |||||
| 0 | 2.2±0.8 | 2.4±0.9 | 1.5±<0 | 5.0±1.0 | 4.2±0.8 |
| Chymotrypsin activity | |||||
| 0 | 1.0±0.2 | 7.0±1.1 | 0.5±<0 | 6.0±3.1 | 10.0±3.8 |
Values are means ± SE. Activities are calculated as rfu/s per total protein content from each assay condition and expressed as a percentage of the total activated zymogen as determined in Fig. 2. ZG, zymogen granule-enriched fraction; Mic, microsomal fraction; Cyt, cytosol fraction.
Chymotrypsinogen activation
Isolated zymogen granule-enriched, mitochondrial, and microsomal fractions from unstimulated pancreas were assayed for chymotrypsin activity. Similar to trypsin, no chymotrypsin activity was observed in the mitochondrial fraction. In the cytosolic fraction alone, activity was negligible. Zymogen granule-enriched and microsomal fractions alone showed little activity (Fig. 4). Furthermore, addition of ATP to each fraction did not affect enzyme activity. However, there was a prominent increase in chymotrypsin activity when cytosol was combined with either fraction (Fig. 4). Chymotrypsin activities increased further with the addition of ATP. Thus, when both ATP and cytosol were added, chymotrypsin activity increased 5-fold and 2.5-fold in zymogen granules and microsomes, respectively. In preliminary studies, the effects of 0.25, 0.5, 1, and 5 mM ATP were examined. Little enhancement of activation was seen until the concentration of 5 mM was reached; this value was used in all studies. Addition of the nonhydrolyzable ATP analog AMP-PNP did not enhance zymogen activation, confirming that ATP hydrolysis is required. Similar to trypsin, chymotrypsin activity (normalized to amylase content) was up to sixfold higher in the microsomal fraction than in the zymogen granule-enriched fraction (not shown). When normalized to the zymogen content, cytosol-dependent activation was 1 and 6% of total chymotrypsinogen content for zymogen granule-enriched and microsomal fractions, respectively. In the presence of cytosol and ATP, these values increased to 7 and 10%, respectively (Table 1). Similar to trypsinogen, more efficient processing of chymotrypsinogen was seen in microsomes than in the zymogen granule-enriched fraction. However, when chymotrypsinogen activation was normalized to the distribution of zymogens between the two fractions, the activities contributed by the zymogen granule-enriched fraction were about sixfold higher than those seen in microsomal compartments (not shown). In contrast to trypsinogen, these studies demonstrate that cytosol-dependent chymotrypsinogen processing in isolated organelles exhibits both ATP-independent and ATP-dependent components.
Fig. 4.
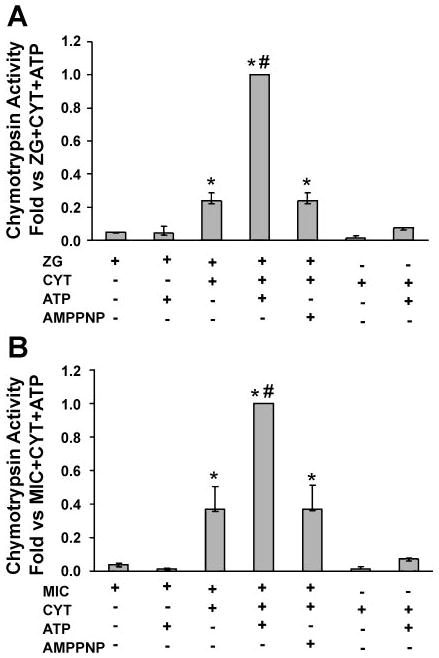
Chymotrypsinogen activation reconstituted in ZG (A) and Mic (B) pancreatic fractions is dependent on Cyt and ATP. It is enhanced by the addition of Cyt and ATP and requires ATP hydrolysis (experimental conditions described in MATERIALS AND METHODS). Data are normalized to amylase content and expressed as fold vs. ZG/Mic + Cyt + ATP; n = 3. *P < 0.05 vs. no ATP condition; #P < 0.05 vs. ZG plus Cyt condition.
Contribution of free enzyme from lysed organelles
To test the possibility that organelles were being lysed during the assay, an event that might lead to an underestimation of zymogen activation, additional studies were performed. With the use of two different cytosolic preparations, one with high-molecular-weight protease inhibitors and the other without, the contribution of enzyme from lysed organelles was determined. If significant lysis occurred, then there would be more activity in preparations lacking protease inhibitors in the cytosol than in those with the inhibitors. Because the effects of the two cytosol preparations were similar, it is unlikely that organelles are lysed during the assay (Table 2). As a control, membrane-permeable protease inhibitors did inhibit enzyme activity (Table 2).
Table 2. Effects of membrane-impermeable and -permeable protease inhibitors on zymogen activation.
| Inhibitor in Cyt | ZG | ZG/Cyt | ZG/Cyt/ATP | Mic | Mic/Cyt | Mic/Cyt/ATP |
|---|---|---|---|---|---|---|
| Trypsin activity | ||||||
| −STI/PIC | 0 | 101±12 | 100 | 0 | 110±10 | 100 |
| +STI | 0 | 114±18 | 111±4 | 0 | 107±4 | 91±2 |
| +PIC | 0 | 44±14* | 57±10* | 0 | 54±3* | 58±8* |
| Chymotrypsin activity | ||||||
| −STI/PIC | 0 | 39±8 | 100 | 0 | 67±10 | 100 |
| +STI | 0 | 42±5 | 119±10 | 0 | 69±13 | 97±4 |
| +PIC | 0 | 13±5* | 41 ±12* | 0 | 22±13* | 44±6* |
Values are means of %activity ± SE. For ZG data, maximal activity (100%) is set as ZG/Cyt/ATP using −STI/PIC Cyt. For Mic data, maximal activity (100%) is set as Mic/Cyt/ATP using −STI/PIC Cyt. In the absence or presence of membrane-impermeable protease inhibitor in the Cyt, no major difference in activity is seen, indicating that free enzyme is not contributing to activity. When PIC (containing membrane-permeant inhibitors) is present in Cyt, there is a significant reduction in activity (*P < 0.05). STI, soybean trypsin inhibitor; PIC, protease inhibitor cocktail.
Effects of membrane-associated proteins and the proteasome
The proteasome is an organelle with trypsin and chymotryptic activities that can be associated with membrane bound compartments. Although its major role is ATP-dependent protein degradation, it can sometimes catalyze limited proteolysis (1, 21). To exclude a role for the proteasome in zymogen activation or a direct contribution to our enzymatic assays, two studies were performed. First, to remove the proteasome and other peripherally associated proteins from membranes, the zymogen granule-enriched and microsomal fractions were washed in 0.5 M KCl or 0.3 M sucrose (control) (3). Neither wash condition affected enzyme activities (Table 3). Next, chymotrypsin activity was assayed in the presence or absence of the proteasomal inhibitors lactacystin (20 μM) and MG-132 (0.75 μM); neither reduced chymotrypsinogen activation (Table 4). Interestingly, lactacystin causes a slight but significant increase in activity in both zymogen granule-enriched and microsomal fractions when cytosol and ATP were present. This is not a direct effect on the enzyme because lactacystin had no effect on purified chymotrypsin (not shown). These studies demonstrate that membrane-associated proteins, including the proteasome, do not contribute to the ATP-dependent activation of chymotrypsinogen.
Table 3. Effects of sucrose and KCl wash of particulate fractions on zymogen activation.
| Wash Condition | ZG | ZG/Cyt | ZG/Cyt/ATP | Mic | Mic/Cyt | Mic/Cyt/ATP |
|---|---|---|---|---|---|---|
| Trypsin activity | ||||||
| Sucrose | 35±12 | 94±12 | 100 | 4 | 94±4 | 100 |
| KCI | 42±9 | 103±10 | 106 ± 9 | 3 | 98±3 | 104 |
| Chymotrypsin activity | ||||||
| Sucrose | 0 | 40±14 | 100 | 0 | 53±12 | 100 |
| KCI | 0 | 28±17 | 111 ± 2 | 0 | 43±14 | 82 ± 17 |
Values are means of %activity ± SE. For ZG data, maximal activity (100%) is set as ZG/Cyt/ATP using sucrose-washed ZG fractions. For Mic data, maximal activity (100%) is set as Mic/Cyt/ATP using sucrose-washed microsomal fractions. Comparison of like conditions show no significant differences between sucrose or KCl.
Table 4. Effects of proteasome inhibitors on chymotrypsinogen activation.
| Condition | ZG | ZG/Cyt | ZG/Cyt/ATP | Mic | Mic/Cyt | Mic/Cyt/ATP |
|---|---|---|---|---|---|---|
| Chymotrypsin activity | ||||||
| Control | 0 | 37±10 | 100 | 0 | 44±5 | 100 |
| Lactacystin | 0 | 48±4 | 130 ± 7* | 0 | 43±6 | 123 ± 2* |
| Chymotrypsin activity | ||||||
| Control | 0 | 28±2 | 100 | 0 | 44±5 | 100 |
| MG-132 | 0 | 38±7 | 121 ± 4 | 0 | 46±4 | 118 ± 9 |
Values are means of %activity ± SE. For ZG data, maximal activity (100%) is set as ZG/Cyt/ATP control. For Mic data, maximal activity (100%) is set as Mic/Cyt/ATP control. Comparison of like conditions show no significant differences except where indicated. *P < 0.05. It is interesting to note that these significant differences do not refer to an inhibitory event.
Procarboxypeptidase B activation
To confirm the results of assays that used fluorogenic protease substrates, the generation of carboxypeptidase B was assayed by immunoblot (Fig. 5). No carboxypeptidase B was seen in either the zymogen granule-enriched or microsomal fraction alone or in the presence of 5 mM ATP. Carboxypeptidase B was detected in cytosol alone; when we compared bands with densitometry, cytosol alone accounted for 6–9% of maximal activity (not shown). When cytosol was added to either particulate fraction, an immunore-active band corresponding to carboxypeptidase B was observed. A more intense band was seen in the presence of cytosol and ATP (this condition denotes maximal activity; 100%). This result indicates that, similar to chymotrypsinogen, procarboxypeptidase B processing is dependent on cytosol and is further enhanced by ATP.
Fig. 5.
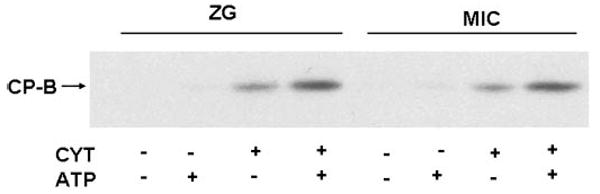
Procarboxypeptidase B activation reconstituted in a ZG and Mic fractions is dependent on cytosol and ATP. Immunoblot assay demonstrating that carboxypeptidase B (CP-B), the active form of procarboxypeptidase B, is generated in both ZG and Mic fractions with added Cyt and further enhanced by ATP. Results are representative of 3 separate experiments.
Potential role of kinases in chymotrypsinogen activation
The requirement of ATP for conversion of chymotrypsinogen and procarboxypeptidase to their active forms suggests a potential role for protein kinases. To investigate this further, the effects of protein kinase inhibitors (K252a, H-89, wortmannin, and LY-294002) were examined. Although the first two inhibitors can affect a broad range of protein kinases (11), the latter two were used at concentrations known to specifically inhibit phosphatidylinositol 3-kinase, an enzyme involved in acinar cell zymogen activation (8, 25).
Neither wortmannin nor LY-294002 inhibited cytosol-dependent or cytosol/ATP-dependent activation of chymotrypsin. Trypsinogen activation, although ATP insensitive, was studied, and no inhibition of activity was seen. Studies with K252a at 100 nM, a concentration that inhibits many kinases, showed no inhibition of activity. However, the general protein kinase inhibitor H-89 caused a concentration-dependent inhibition of cytosol/ATP-dependent chymotrypsin activity in both the zymogen granule-enriched and microsomal fractions (Fig. 6). These data indicate that ATP-dependent mechanisms in zymogen granule-enriched and microsomal fractions may be mediated by protein kinases.
Fig. 6.
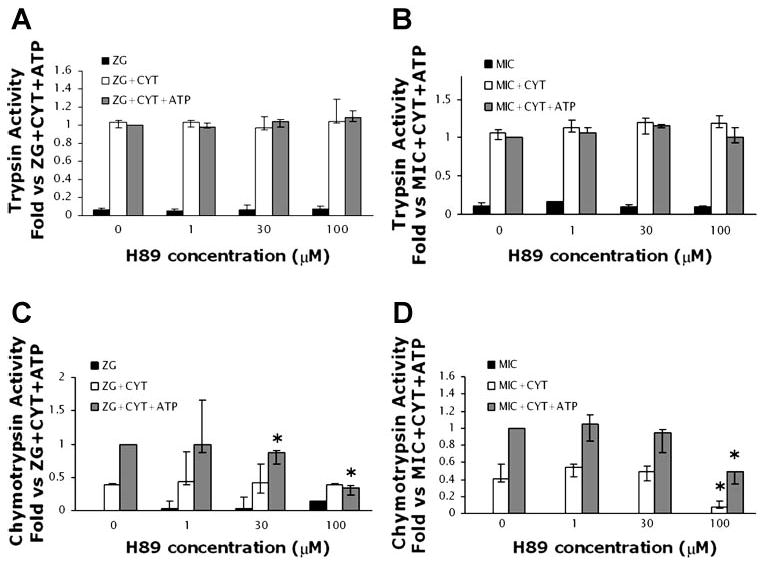
The protein kinase inhibitor H-89 does not affect trypsinogen activation (A and B) but decreases chymotrypsinogen activation (C and D) in ZG and Mic fractions. Assays were repeated as for Figs. 3 and 4 with the exception that some samples were preincubated with H-89 (1, 30, and 100 μM) for 30 min. Control experiments (i.e., in the absence of H-89) were preincubated with 1% DMSO, the vehicle for H-89. Data are normalized to amylase content and expressed as fold vs. ZG/Mic + Cyt + ATP; n = 3. Activation in Cyt alone was negligible (data not shown). *P < 0.05 vs. ZG/Mic + Cyt + ATP.
Selective effects of E64d on zymogen activation
The thiol protease, cathepsin B, has been proposed as the mediator of pathologic trypsinogen activation, which initiates the proteolytic cascade activating other zymogens in acute pancreatitis (5, 7, 23, 24). Fluorogenic protease activity assays were repeated in the presence of the cathepsin B inhibitor, E64d (9, 10, 12). The inhibitor blocked cytosol-dependent generation of trypsin activity in both the zymogen granule-enriched and microsomal fractions (Fig. 7). In control studies, E64d did not affect the activity of purified trypsin (not shown). These findings suggest that, similar to other models, cathepsin B converts trypsinogen to trypsin in our reconstitution system. However, cytosol and ATP-dependent chymotrypsin activity and carboxypeptidase B generation in the particulate fractions were not inhibited by E64d (Figs. 8 and 9). The latter findings suggest that chymotrypsinogen and procarboxypeptidase activation might be independent from cathepsin B and trypsin.
Fig. 7.
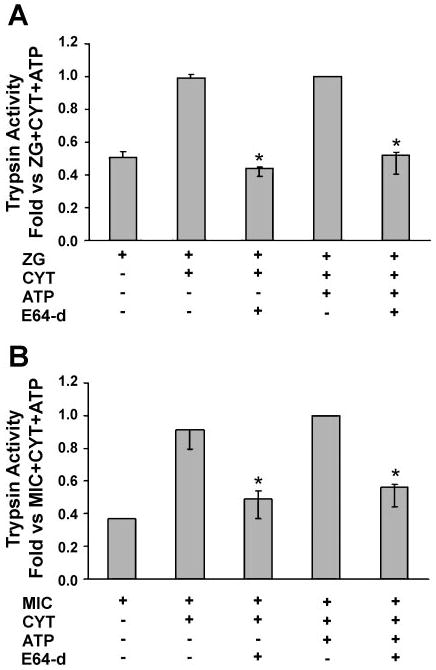
The cathepsin B inhibitor, E64d, prevents Cyt-dependent trypsinogen activation in ZG (A) and Mic (B) fractions. Assays were performed as for Fig. 3, but some samples were preincubated with E64d (1 mM) for 30 min. Control experiments (i.e., in the absence of E64d) were preincubated with 1:1 methanol-water, the vehicle for E64d. Data are normalized to amylase content and expressed as fold vs. ZG/Mic + Cyt + ATP; n = 3. Activation in Cyt alone was negligible (data not shown). *P < 0.05 vs. Cyt or Cyt/ATP condition.
Fig. 8.
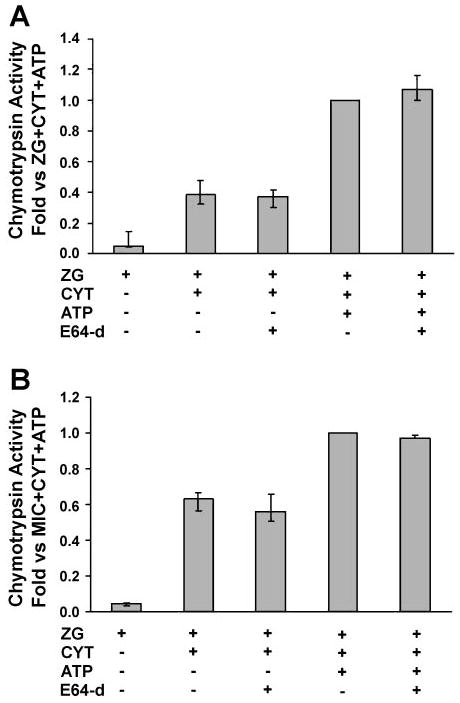
The cathepsin B inhibitor E64d does not prevent chymotrypsinogen activation in ZG (A) and Mic (B) fractions. Assays were performed as for Fig. 4, but some samples were preincubated with E64d (1 mM) for 30 min. Control experiments (i.e., in the absence of E64d) were preincubated with 1:1 methanol-water, the vehicle for E64d. Data are normalized to amylase content and expressed as fold vs. ZG/Mic + Cyt + ATP; n = 3. Activation in Cyt alone was negligible (data not shown).
Fig. 9.
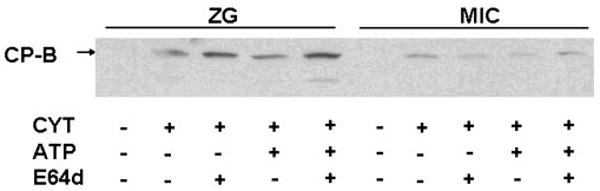
CP-B generation in ZG and Mic fractions is not inhibited by preincubation with E64d. Immunoblot assay demonstrating that Cyt- and ATP-dependent generation of CP-B is not inhibited by preincubation with E64d (1 mM). Results are representative of 3 separate experiments.
Discussion
The pathological activation of digestive zymogens, particularly proteases, appears to have a central role in the initiation of acute pancreatitis. Our initial studies suggested that zymogen activation could occur in distinct cellular organelles. After the induction of caerulein pancreatitis, we observed enhanced protease activities in both zymogen granule-enriched and microsomal fractions, suggesting that zymogen activation could proceed in several pancreatic organelles.
To further investigate the mechanism of this activation, we developed a reconstitution system using pancreatic cytosol and isolated organelles. In this system, we used fractions from unstimulated tissue and introduced factors to determine whether they could induce activation within organelles. Although it could be argued that activating zymogens in subcellular fractions from unstimulated tissue may not fully reflect pathological enzyme activation seen in pancreatitis, the approach of using inactive proteins and adding back stimulatory factors has been used in many reconstitution systems (29–31).
With the use of this system, we have made several key observations. First, zymogen activation occurs within intact organelles and is cytosol dependent. This is supported by the finding that little activation is observed in either isolated organelles or cytosol. When isolated organelles are incubated with cytosol, a significant increase in protease activity is seen (Figs. 3 and 4). The presence of high-molecular-weight membrane-impermeable serine protease inhibitors in the cytosol ensures that the protease activities detected by the fluorogenic assays are not being generated in the cytosol but within organelles (Table 2). The low level of activity detected in cytosol is possibly due to contamination from light vesicles, as this activity was markedly diminished by very high-speed centrifugation. Furthermore, there is a lack of an effect on activity by removal of adherent proteins from membrane fractions with potassium chloride or treatment with proteasomal inhibitors. Because cytosol-dependent zymogen activation is observed with isolated organelles from unstimulated cells, it might represent basal cellular activity (as seen in whole cell studies; Refs. 16, 17), or it could result from activation of the machinery, such as protein kinases, required for zymogen activation during preparation of cytosol.
Second, cytosol-stimulated zymogen activation occurs in both zymogen granule-enriched and microsomal compartments, suggesting that more than one compartment might support zymogen activation. Although the levels of activation seen in these compartments represent a small percentage of the total zymogen content (Table 1), the values are consistent with those from studies that have used intact cells (9, 15, 24). Our data suggest a more efficient processing of zymogens in microsomes than with zymogen granule-enriched fraction and a more efficient processing of chymotrypsinogen than of trypsinogen (Table 1). The proportions of total zymogen in zymogen granule-enriched fraction are greater than those seen in the microsomal fraction (Fig. 2) and comparable to levels described in another study (18). Thus, when normalized to the distribution of active enzymes between the two compartments, the greater contribution to enzyme activity comes from the zymogen granule-enriched fraction. Whether a similar distribution is observed in intact cells remains unclear.
Studies from our laboratory and another group have both reported that initial zymogen processing is localized to a vesicular compartment containing markers of lysosomes and recycling endosomes (13, 20). In our present study, immunoblot analysis demonstrates that the lysosomal and endosomal marker GRAMP-92 is present predominantly in the zymogen granule-enriched fraction and to a lesser extent in the microsomal fraction (data not shown). Because these two have substantially different densities, it is unlikely that GRAMP-92 is associated with the same organelle in the zymogen granules and microsomes. Additional fractionation studies will be required to identify the compartments ultimately responsible for zymogen processing.
Third, on the basis of the effects of ATP and E64d in our system, the requirements for trypsinogen activation may be distinct from those necessary for activation of chymotrypsinogen and procarboxypeptidase. The ATP requirement for conversion of chymotrypsinogen and procarboxypeptidase and kinase inhibitor studies indicates a potential role for one or more protein kinases. Our data with protein kinase inhibitors indicates a lack of involvement for phosphatidylinositol 3-kinase in the cell-free system. The results with the inhibitor H-89 suggest that a protein kinase inhibited in the range from 30 to 100 μM may regulate processing. Protein kinase A and protein kinase G are inhibited at submicromolar concentrations of H-89 and are therefore unlikely candidates (11). However, other protein kinases, such as casein kinases I and II, calmodulin-dependent kinases, and protein kinase C, require inhibitory H-89 concentrations in the ranges observed in this study (11). Notably, preliminary studies suggest that H-89 also selectively inhibits chymotrypsin but not trypsin activity in acini simulated by supraphysiological concentrations of caerulein (unpublished data). Studies with selective protein kinase inhibitors and specific agonists may identify the protein kinases that mediate this activation.
The inhibitory effects of E64d on trypsinogen activation are consistent with the published effects of cathepsin B on trypsinogen activation, but its lack of effectiveness in inhibiting chymotrypsinogen or procarboxypeptidase B processing is surprising. Together with the selective effects of ATP on chymotrypsinogen or procarboxypeptidase B activation, these findings strongly suggest that the activation of some zymogens is trypsin independent. Indeed, one study even suggests that trypsin has a more relevant role in autodegradation rather than in activation of itself and other zymogens (10). However, we cannot exclude the possibility that undetectable increases in trypsin activity might be responsible for the activation of the other zymogens. Most other studies with pancreatic cathepsin B and trypsinogen activation have not examined activation of other zymogens (10, 12, 24). In this context, in cathepsin B knockout mice, despite a significant reduction in trypsinogen activation, cell injury and other measures of pancreatitis are still prominent (9). Furthermore, in transgenic mice with increased levels of pancreatic secretory trypsin inhibitor 1, active trypsin is inhibited and the severity of caerulein-induced pancreatitis is reduced but not eliminated (19). One possible explanation for the persistent injury seen in both the cathepsin B knockout and the transgenic pancreatic secretory trypsin inhibitor 1 animals is that activation of proteases other than trypsinogen may not be reduced.
In summary, this report used an in vivo model of acute pancreatitis and a reconstituted system to examine the mechanisms of zymogen activation in the pancreatic acinar cell. From these studies, we conclude that pathological zymogen activation can occur in more than one cellular organelle and requires cytosolic factors. Furthermore, these studies demonstrate that zymogen activation is complex and that distinct pathways may lead to activation of specific subsets of proteases. In this context, activation within the acinar cell may not follow the pattern that has been established for intestinal activation, with trypsinogen activation leading to proteolytic activation of other zymogens. Elucidation of the cytosolic factors and pathways that mediate zymogen activation may lead to the rational design of preventive therapies.
Acknowledgments
We thank Christine Shugrue, Shaconna Smalls, and Sergey Babakov for expert advice and helpful discussions.
Grants: This work was supported by National Institute of Diabetes and Digestive and Kidney Diseases Grants R21 DK-69702 (to E. C. Thrower) and RO1 DK-54021 (to F. S. Gorelick) and a Veterans Affairs Merit and Senior Career Development Award (to F. S. Gorelick)
References
- 1.Adams J. The proteasome: a suitable antineoplastic target. Nat Rev Cancer. 2004;4:349–360. doi: 10.1038/nrc1361. [DOI] [PubMed] [Google Scholar]
- 2.Cavallini G, Tittobello A, Frulloni L, Masci E, Mariani A, DiFrancesco V. Gabexate for the prevention of pancreatic damage related to endoscopic retrograde cholangiopancreatography. Gabexate in digestive endoscopy—Italian Group. N Engl J Med. 1996;335:919–923. doi: 10.1056/NEJM199609263351302. [DOI] [PubMed] [Google Scholar]
- 3.Elkabetz Y, Shapira I, Rabinovich E, Bar-Nun S. Distinct steps in dislocation of luminal endoplasmic reticulum-associated degradation substrates: roles of endoplasmic reticulum-bound p97/Cdc48p and proteasome. J Biol Chem. 2004;279:3980–3989. doi: 10.1074/jbc.M309938200. [DOI] [PubMed] [Google Scholar]
- 4.Fallon M, Gorelick F, Anderson J, Mennone A, Saluja A, Steer M. Effect of cerulein hyperstimulation on the paracellular barrier of rat exocrine pancreas. Gastroenterology. 1995;108:1863–1872. doi: 10.1016/0016-5085(95)90151-5. [DOI] [PubMed] [Google Scholar]
- 5.Figarella C, Miszczuk-Jamska B, Barrett A. Possible lysozomal activation of pancreatic zymogens. Biol Chem. 1988;369:293–298. [PubMed] [Google Scholar]
- 6.Grady T, Mah'moud M, Otani T, Rhee S, Lerch MM, Gorelick FS. Zymogen proteolysis within the pancreatic acinar cell is associated with cellular injury. Am J Physiol Gastrointest Liver Physiol. 1998;275:G1010–G1017. doi: 10.1152/ajpgi.1998.275.5.G1010. [DOI] [PubMed] [Google Scholar]
- 7.Greenbaum LM, Hirshkowitz A, Shoichet I. The activation of trypsinogen by cathepsin B. J Biol Chem. 1959;234:2885–2890. [PubMed] [Google Scholar]
- 8.Gukovsky I, Cheng JH, Nam KJ, Lee OT, Lugea A, Fischer L, Penninger JM, Pandol SJ, Gukovskaya AS. Phosphatidylinositide 3-kinase γ regulates key pathologic responses to cholecystokinin in pancreatic acinar cells. Gastroenterology. 2004;126:554–566. doi: 10.1053/j.gastro.2003.11.017. [DOI] [PubMed] [Google Scholar]
- 9.Halangk W, Lerch MM, Brandt-Nedelev B, Roth W, Ruthenbuerger M, Reinheckel T, Domschke W, Lippert H, Peters C, Deussing J. Role of cathepsin B in intracellular trypsinogen activation and the onset of acute pancreatitis. J Clin Invest. 2000;106:773–781. doi: 10.1172/JCI9411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Halangk W, Kruger B, Ruthenberger M, Sturzebecher J, Albrecht E, Lippert H, Lerch MM. Trypsin activity is not involved in premature, intrapancreatic trypsinogen activation. Am J Physiol Gastrointest Liver Physiol. 2002;282:G367–G374. doi: 10.1152/ajpgi.00315.2001. [DOI] [PubMed] [Google Scholar]
- 11.Hidaka H, Kobayashi R. Pharmacology of protein kinase inhibitors. Annu Rev Pharmacol Toxicol. 1992;32:377–397. doi: 10.1146/annurev.pa.32.040192.002113. [DOI] [PubMed] [Google Scholar]
- 12.Hietaranta AJ, Saluja AK, Bhagat L, Singh VP, Song AM, Steer M. Relationship between NF-κB and trypsinogen activation in rat pancreas after supramaximal caerulein stimulation. Biochem Biophys Res Commun. 2001;280:388–395. doi: 10.1006/bbrc.2000.4120. [DOI] [PubMed] [Google Scholar]
- 13.Hofbauer B, Saluja AK, Lerch MM, Bhagat L, Bhatia M, Lee HS, Frossard JL, Adler G, Steer ML. Intra-acinar cell activation of trypsinogen during caerulein-induced pancreatitis in rats. Am J Physiol Gastrointest Liver Physiol. 1998;275:G352–G362. doi: 10.1152/ajpgi.1998.275.2.G352. [DOI] [PubMed] [Google Scholar]
- 14.Lasson A, Ohlsson K. Protease inhibitors in acute pancreatitis: correlation between biochemical changes and clinical course. Scand J Gastroenterol. 1984;19:779–786. [PubMed] [Google Scholar]
- 15.Leach SD, Modlin IM, Scheele GA, Gorelick FS. Intracellular activation of digestive zymogens in rat pancreatic acini. Stimulation by high doses of cholecystokinin. J Clin Invest. 1991;87:362–366. doi: 10.1172/JCI114995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lu Z, Karne S, Kolodecik T, Gorelick FS. Alcohols enhance caerulein-induced zymogen activation in pancreatic acinar cells. Am J Physiol Gastrointest Liver Physiol. 2002;282:G501–G507. doi: 10.1152/ajpgi.00388.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lu Z, Kolodecik TR, Karne S, Nyce M, Gorelick F. Effect of ligands that increase cAMP on caerulein-induced zymogen activation in pancreatic acini. Am J Physiol Gastrointest Liver Physiol. 2003;285:G822–G828. doi: 10.1152/ajpgi.00213.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mithofer K, Fernandex-del Castillo C, Rattner D, Warshaw AL. Subcellular kinetics of early trypsinogen activation in acute rodent pancreatitis. Am J Physiol Gastrointest Liver Physiol. 1998;274:G71–G79. doi: 10.1152/ajpgi.1998.274.1.G71. [DOI] [PubMed] [Google Scholar]
- 19.Nathan JD, Romac J, Peng RY, Peyton M, MacDonald RJ, Liddle RA. Transgenic expression of pancreatic secretory trypsin inhibitor-I ameliorates secretagogue-induced pancreatitis in mice. Gastroenterology. 2005;128:717–727. doi: 10.1053/j.gastro.2004.11.052. [DOI] [PubMed] [Google Scholar]
- 20.Otani T. Codistribution of TAP and the granule membrane protein GRAMP-92 in rat caerulein-induced pancreatitis. Am J Physiol Gastrointest Liver Physiol. 1998;275:G999–G1009. doi: 10.1152/ajpgi.1998.275.5.G999. [DOI] [PubMed] [Google Scholar]
- 21.Piotrowski Z, Mys´ liwiec P, Gryko M, Ostrowska H, Baltaziak M. Chymotrypsin-like activity in rat tissues in experimental acute pancreatitis. Rocz Akad Med Bialymst. 2003;48:61–65. [PubMed] [Google Scholar]
- 22.Raraty M, Ward J, Erdemli G, Vaillant C, Neoptolemos JP, Sutton R, Petersen OH. Calcium-dependent enzyme activation and vacuole formation in the apical granular region of pancreatic acinar cells. Proc Natl Acad Sci USA. 2000;97:13126–13131. doi: 10.1073/pnas.97.24.13126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Saluja A, Hashimoto S, Saluja M, Powers RE, Meldolesi J, Steer ML. Subcellular redistribution of lysosomal enzymes during caerulein-induced pancreatitis. Am J Physiol Gastrointest Liver Physiol. 1987;253:G702–G710. doi: 10.1152/ajpgi.1987.253.4.G508. [DOI] [PubMed] [Google Scholar]
- 24.Saluja AK, Donovan EA, Yamanaka K, Yamaguchi Y, Hofbauer B, Steer ML. Caerulein-induced in vitro activation of trypsinogen in rat pancreatic acini is mediated by cathepsin B. Gastroenterology. 1997;113:304–310. doi: 10.1016/s0016-5085(97)70108-2. [DOI] [PubMed] [Google Scholar]
- 25.Singh VP, Saluja AK, Bhagat L, van Acker GJ, Song AM, Soltoff SP, Cantley LC, Steer ML. Phosphatidylinositol 3-kinase-dependent activation of trypsinogen modulates the severity of acute pancreatitis. J Clin Invest. 2001;108:1387–1395. doi: 10.1172/JCI12874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Steer ML, Meldolesi J. The cell biology of experimental pancreatitis. N Engl J Med. 1987;316:144–150. doi: 10.1056/NEJM198701153160306. [DOI] [PubMed] [Google Scholar]
- 27.Suzuki M, Isaji S, Stanten R, Frey CF, Ruebner B. Effect of protease inhibitor FUT-175 on acute hemorrhagic pancreatitis in mice. Int J Pancreatol. 1992;11:59–65. [PubMed] [Google Scholar]
- 28.Tartakoff AM, Jamieson JD. Subcellular fractionation of the pancreas. Methods Enzymol. 1974;31:41–59. doi: 10.1016/0076-6879(74)31006-3. [DOI] [PubMed] [Google Scholar]
- 29.Thrower EC. Measurement of Ca2+ flux through Ins(1,4,5)P3 receptor-Ca2+ channels in lipid bilayers (“dip-tip” and “Schindler” methodology) Methods Mol Biol. 1999;114:221–233. doi: 10.1385/1-59259-250-3:221. [DOI] [PubMed] [Google Scholar]
- 30.Thrower EC, Park HY, Yoo SH, Ehrlich BE. Effects of chromogranin A on IP3 receptor activity. J Biol Chem. 2002;277:15801–15806. doi: 10.1074/jbc.M110139200. [DOI] [PubMed] [Google Scholar]
- 31.Thrower EC, Choe CU, So SH, Jeon SH, Ehrlich BE, Yoo SH. A functional interaction between chromogranin B and the inositol 1,4,5-trisphosphate receptor/Ca2+ channel. J Biol Chem. 2003;278:49699–49706. doi: 10.1074/jbc.M309307200. [DOI] [PubMed] [Google Scholar]


