Abstract
TGFβ induces epithelial-mesenchymal transdifferentiation (EMT) accompanied by cellular differentiation and migration. Despite extensive transcriptomic profiling, identification of TGFβ-inducible, EMT-specific genes has met with limited success. Here, we identify a post-transcriptional pathway by which TGFβ modulates expression of EMT-specific proteins, and EMT itself. We show that heterogeneous nuclear ribonucleoprotein E1 (hnRNP E1) binds a structural, 33 nucleotides (nt) TGF beta-activated translation (BAT) element in the 3’-UTR of disabled-2 (Dab2) and interleukin-like EMT inducer (ILEI) transcripts, and repress their translation. TGFβ activation leads to phosphorylation at Ser43 of hnRNP E1 by protein kinase Bβ/Akt2, inducing its release from the BAT element and translational activation of Dab2 and ILEI mRNAs. Modulation of hnRNP E1 expression or its post-translational modification alters TGFβ-mediated reversal of translational silencing of the target transcripts and EMT. These results suggest the existence of a TGFβ-inducible post-transcriptional regulon that controls EMT during development and metastatic progression of tumors.
Epithelial-mesenchymal transition (EMT), in which cells undergo a switch from a polarized, epithelial phenotype to a highly motile fibroblastic or mesenchymal phenotype is fundamental during embryonic development and can be reactivated in a variety of diseases including fibrosis and cancer (ref. 1–5). TGFβ is one of the growth factors implicated in EMT (ref. 1–5). Using normal murine mammary gland epithelial (NMuMG) cells6,7 and mouse mammary epithelial cells, EpH4, transformed with oncogenic Ras (EpRas)8 as in vitro models for TGFβ-induced EMT two candidate EMT genes were defined, Disabled-2 (Dab2)9 and FAM3C or interleukin like EMT inducer (ILEI)10. Dab2 is a putative tumor suppressor gene, but modulates late stages of tumor progression by promoting EMT-dependent metastasis9. ILEI was initially identified as a candidate gene for autosomal recessive nonsyndromic hearing loss locus 17 (DFNB17)11 and was subsequently shown to belong to the FAM3A-D gene family12. ILEI was shown to be translationally upregulated during EMT in EpRas cells10. Short hairpin RNA (shRNA)-mediated silencing of Dab2 in NMuMG cells inhibits TGFβ-mediated EMT and re-expression of human Dab2 in Dab2 knock-down cells restores TGFβ-mediated EMT9. Stable knockdown of ILEI inhibits TGFβ-mediated EMT in EpRas cells, whereas ILEI expression induces epithelial plasticity changes and tumor formation in non-tumorigenic NMuMG cells and 3T3 fibroblasts10. Cumulatively, these data suggest that both Dab2 and ILEI are required, but not sufficient (i.e., in a TGFβ-independent fashion) to induce EMT. However, the molecular mechanism by which expression of Dab2 and ILEI is regulated by TGFβ remains elusive.
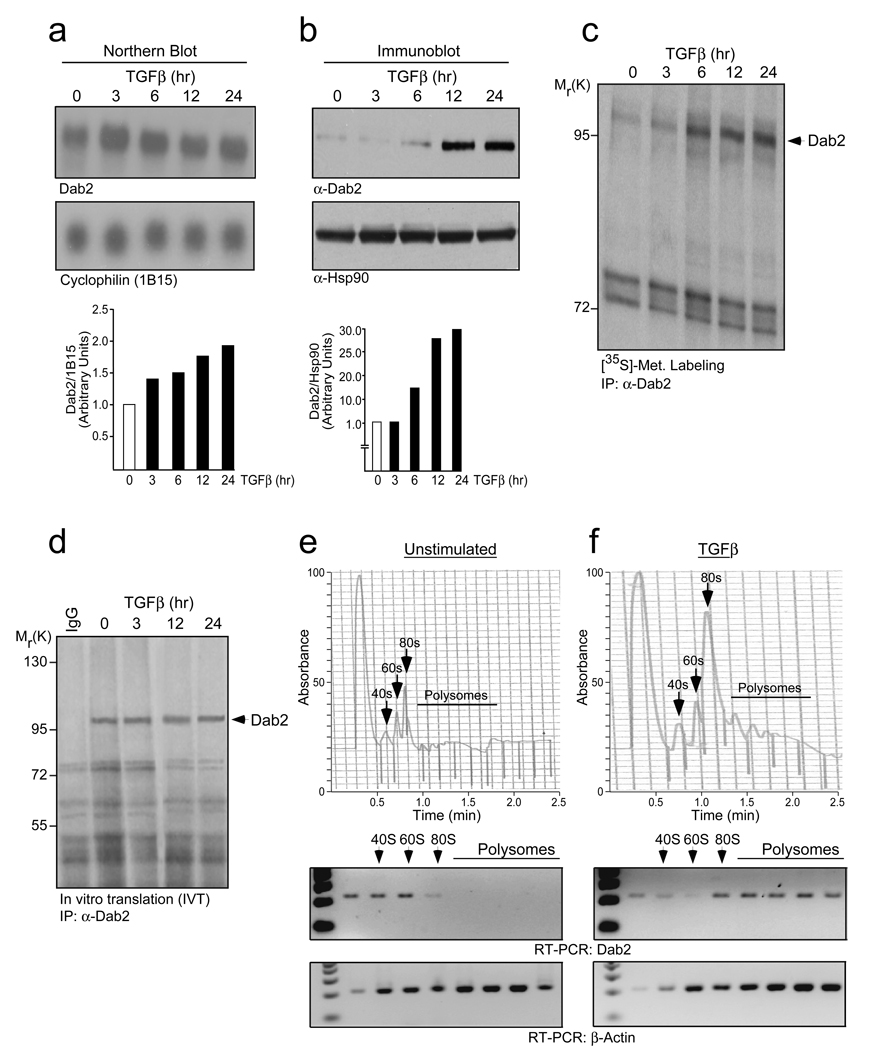
Despite intensive transcriptional array analysis of human tumors, the identity and validation of ‘EMT signature genes’ remains elusive (ref. 13)14. Our recent experiments suggest that post-transcriptional regulation of gene expression plays an important role in TGFβ-mediated EMT. We initially observed that TGFβ treatment of NMuMG and EpRas cells led to increased expression of Dab2 protein without a concomitant increase in its mRNA (Fig. 1a, b; SI 1a, b). Unstimulated cells, despite having abundant Dab2 mRNA, had low levels of Dab2 protein (Fig. 1b; SI 1b). De novo Dab2 synthesis increased significantly only after 3–6 hr of TGFβ stimulation and peaked at ~12 hr (Fig. 1c). In vitro translation efficiencies of total RNA isolated from TGFβ-treated cells showed that lack of Dab2 protein expression was not due to decreased mRNA stability (Fig. 1d). We next monitored the translocation of Dab2 mRNA from the non-translating, non-polysomal pool to the actively translating, polysomal pool in unstimulated and TGFβ-treated cells. In unstimulated cells, Dab2 mRNA was absent from the polysomal fractions (Fig. 1e), but was abundant in actively translating polysomes after 24 hr of TGFβ treatment (Fig. 1f). Translation of β-actin was unaffected indicating transcript selective translation of Dab2 (Fig. 1e, f). Further, polysome release experiments confirmed that Dab2 is translationally regulated in a TGFβ-dependent fashion (SI 1c–e).
Figure 1.
TGFβ translationally up-regulates Dab2 expression. (a) Northern blot analysis examining Dab2 expression levels in NMuMG cells treated with TGFβ for the times indicated. Lower panel represents the quantification of band intensities analyzed by NIH Image J software. Dab2 band intensity was normalized to cyclophilin (1B15), then normalized to the t=0 unstimulated. (b) Immunoblot (IB) analysis examining Dab2 protein levels in NMuMG cells treated with TGFβ for the indicated times. Lower panel represents the quantification of band intensities analyzed by NIH Image J software. Dab2 band intensity was normalized to Hsp90, then normalized to the t=0 unstimulated. (c) Metabolic labeling with [35S]-methionine analyzing the de novo rate of Dab2 synthesis post-TGFβ stimulation. (d) Dab2 mRNA stability analysis by in vitro translation (IVT) of total RNA isolated from NMuMG cells treated with TGFβ for the times indicated followed by immunoprecipitation (IP) with α-Dab2 antibody and mouse IgG. (e) & (f) Translocation of Dab2 mRNA from the non-polysomal to polysomal pool was analyzed by semi-quantitative RT-PCR of RNA isolated from each fraction following polysome profiling. Full scans of (a), (b), (c) and (d) are shown in SI Fig. 7.
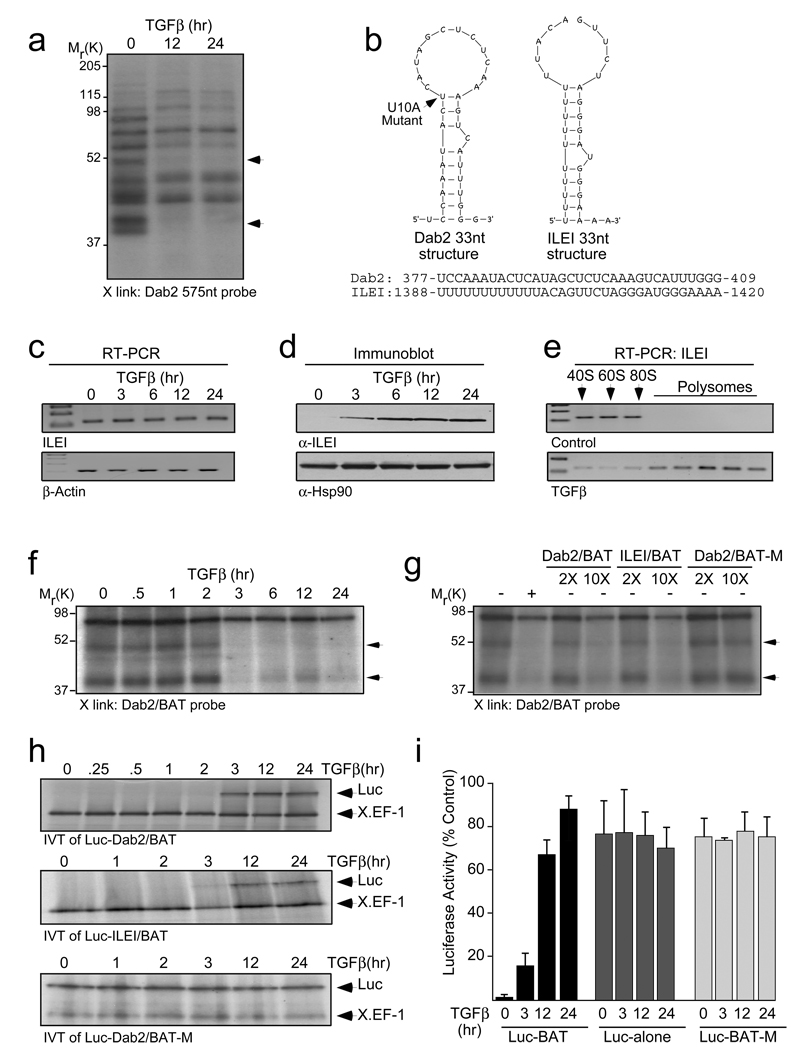
We hypothesized that the conserved first 575 nt of Dab2 3’-UTR harbors a cis regulatory element which regulates its expression. UV-crosslinking analysis using this region as a probe revealed two proteins, which showed TGFβ-dependent loss of binding (Fig. 2a). Fine mapping subsequently defined a 33-nt region as the cis element (SI 2a). We named this region ‘BAT’ for TGFβeta activated translational element and its secondary structure reveals a stem-loop with an asymmetric bulge. A U10A mutant was predicted to destroy this secondary structure using ‘Mfold’ analysis15 (Fig. 2b). A ‘PatSearch’ algorithm16 driven search of a non-redundant 3’-UTR database for similar structures reconfirmed the Dab2 3’-UTR to harbor the BAT element (UTRdb ID: 3MMU027375), and additionally identified the 3’-UTR of ILEI (UTRdb ID: 3MMU039724) (Fig. 2b). Examination of the temporal relationship between ILEI mRNA and protein expression levels showed a pattern similar to Dab2 (Fig. 2c, d; SI 1a, b) and polysome profiling reaffirmed that TGFβ translationally upregulates ILEI (Fig. 2e). UV-crosslinking analysis and decoy experiments using Dab2/BAT, its U10A mutant and ILEI/BAT showed that the binding of the 50 and 40 kDa proteins were TGFβ-dependent (Fig. 2f) and confirmed the specificity of the element (Fig. 2g).
Figure 2.
The 3’-UTR of Dab2 mRNA contains a cis regulatory (BAT) element, which is also present in ILEI mRNA. (a) UV crosslinking (X-link) analysis to characterize regulatory element(s) in the 3’-UTR of Dab2 mRNA using [α-32P]-labeled Dab2 3’-UTR 575-nt probe (10 fmol) and S100 cytosolic extract from NMuMG cells treated with TGFβ for the times indicated. (b) Secondary structure of the mouse Dab2/BAT (dG = −5.0 Kcal/mol) and ILEI/BAT (dG = −2.5 Kcal/mol) elements as predicted by the Mfold algorithm. Substituted nucleotide (U10A), indicated in boldface, represents a mutant form. ILEI/BAT element was folded under (F 5 0 2)/ (F 9 0 2)/ (P 11 0 2) constraints. (c) Semi-quantitative RT-PCR and (d) IB analyses examining mRNA and protein expression levels of ILEI in NMuMG cells treated with TGFβ. (e) Translocation of ILEI mRNA from the non-polysomal to polysomal pool was analyzed by semi-quantitative RT-PCR of RNA isolated from each fraction following polysome profiling. (f) X-link analysis was performed with [α-32P]-labeled Dab2/BAT probe (10 fmol) and S100 cytosolic extract from NMuMG cells treated with TGFβ. The arrows indicate the positions of two proteins that fail to bind the probe following TGFβ treatment. (g) Specificity of the BAT element was examined by decoy X-link using [α-32P]-labeled Dab2/BAT probe and a 2- or 10-fold molar excess (2X or 10X) of unlabeled Dab2/BAT, ILEI/BAT, and mutant (U10A) Dab2/BAT-M cRNA. (h) IVT analyses with chimeric Luc-Dab2/BAT, Luc-ILEI/BAT and Luc-Dab2/BAT-M shows that TGFβ treatment relieves translational silencing conferred by the WT and not the mutant BAT element following 3 hr of TGFβ treatment. (i) Dual-luciferase assay examining the in vivo translational silencing activity conferred by the BAT element by co-transfecting with wild-type, mutant (Luc-BAT, Luc-BAT-M) or luciferase alone (Luc-alone) and CMV-driven renilla luciferase constructs. The firefly luciferase values were normalized to renilla luciferase values (which were checked for uniformity to monitor equal transfection efficiency). Results are shown as means ± s.d. for three independent sets of experiments (n=3), each experiment done in triplicates. Full scans of (a), (d), (f), (g) and (h) are shown in SI Fig. 7.
Unstimulated cytosolic extracts inhibited the translation, in a dose-dependent fashion, of a chimeric luciferase construct carrying wild-type BAT (Luc-Dab2/BAT) (SI 2b) but not that of the construct carrying the U10A mutant (Luc-Dab2/BAT-M), suggesting that proteins in these extracts bind the BAT element and functionally silence translation (SI 2c). In vitro, translational silencing of Luc-Dab2/BAT and Luc-ILEI/BAT was reversed after 3 hr of TGFβ stimulation (Fig. 2h). Decoy experiments further confirmed the BAT-specific translation silencing of the chimeric luciferase cRNA by unstimulated cytosolic extracts (SI 2d). Similarly, in vivo translation was found to be silenced in unstimulated cells using the WT BAT chimera (Luc-BAT), but not the mutant chimera (Luc-BAT-M), or the luciferase construct with no 3’-UTR (Luc-alone) (Fig. 2i). TGFβ relieved translation silencing as early as 3 hr and by greater than 80% at 24 hr. These results established BAT as a novel, structural element sufficient to mediate translational silencing in vitro and in vivo.
Size-exclusion chromatography of unstimulated extracts was used to isolate the BAT binding mRNP complex responsible for translational silencing. Fractions #36–38 showed maximum translation silencing activity (Fig. 3a; SI 3a). These fractions were pooled, affinity purified using the BAT element, and visualized by silver staining (Fig. 3b). The lower band (Fig. 3b, arrowhead), present in both the active chromatographic fractions and unstimulated cytosolic extracts, was identified, through mass spectrometric analysis, as heterogeneous nuclear ribonucleoprotein E1 (hnRNP E1). Immunoblot analysis confirmed the presence of hnRNP E1 in the fractions with maximal translation silencing activity (Fig. 3c). TGFβ induced the loss of binding of hnRNP E1 to both Dab2 and ILEI BAT elements after 3 hr of treatment (Fig. 3d), and the kinetics of hnRNP E1 release from the BAT element correlated with the kinetics of reversal of translational silencing in vitro (Fig. 2h) and Dab2 and ILEI protein induction by TGFβ (Fig. 1b, 2d).
Figure 3.
hnRNP E1 is an integral functional component of the mRNP complex. (a) IVT assay for translation inhibitory activity of chimeric Dab2/BAT-Luc cRNA using size exclusion chromatographic fractions. (b) Chromatographic fractions (# 36–38) harboring translational silencing activity were subjected to pull-down with Dab2/BAT cRNA bound to cyanogens bromide (CNBr)-activated sepharose beads after pre-clearing with U10A Dab2/BAT-M cRNA. Precipitated mRNP complex was visualized by silver staining (left panel) and the band (arrowhead) which migrated similarly to the band that does not bind the BAT element after TGFβ treatment (shown by arrowhead in right panel) was analyzed by LC-MS. (c) IB analysis of chromatographic fractions with α-hnRNP E1 antibody exclusively detected hnRNP E1 in fractions harboring translational silencing activity. (d) RNA affinity pull-down and IB analyses using S100 cytosolic extracts for the times indicated to define the temporal association of hnRNP E1 with the Dab2 and ILEI BAT element. (e) RNA affinity pull-down IB analyses using BAT, BAT-M and DICE cRNAs of unstimulated and TGFβ-treated S100 cytosolic extracts examining the selective binding of hnRNP E1, and not hnRNP K to the BAT element. (f), (g) & (h) hnRNP E1 interacts with the BAT element in vivo. Immunoprecipitation with α-hnRNP E1 (f) or mouse IgG (g) of cytosolic extracts from NMuMG cells treated with TGFβ for the times indicated followed by semi-quantitative RT-PCR (using Dab2, ILEI, and β-actin specific primers) analyses of RNA isolated from the immunoprecipitates to examine in vivo association of hnRNP E1 with the BAT element. RNA isolated from input extracts were also analyzed by semi-quantitative RT-PCR (h). Full scans of (a), (b), (c), (d) and (e) are shown in SI Fig. S7.
hnRNP E1, together with heterogeneous nuclear ribonucleoprotein K (hnRNP K), bind to poly r(C) regions, called differentiation control elements (DICE) in 3’-UTR of 15-lipoxygenase and L2 mRNAs and mediate their translational regulation17. However, in pull down experiments, despite both hnRNP E1 and hnRNP K being present in input extracts, only hnRNP E1 from unstimulated extracts bound the WT BAT cRNA, whereas hnRNP E1 in extracts from TGFβ-treated cells did not bind. The BAT-M did not pull down either hnRNP E1 or hnRNP K, whereas a DICE cRNA pulled down both proteins in a TGFβ-independent fashion (Fig. 3e). Immunodepletion of hnRNP E1 from unstimulated cytosolic extracts caused loss of translational silencing activity as assayed by in vitro translation of Luc-Dab2/BAT (SI 3b,c). In vitro binding assays showed that GST-hnRNP E1 could be precipitated in a dose-dependent manner by both Dab2 and ILEI BAT elements, but not by the mutant (SI 3d). In vivo interaction studies revealed that although Dab2 and ILEI mRNAs were steadily expressed, hnRNP E1 interacted with them only in unstimulated cells (Fig. 3f–h). Hence, hnRNP E1 is a functional component of the mRNP complex, binding to the BAT element in a TGFβ-dependent manner, which correlates with the kinetics of reversal of translational silencing of ILEI and Dab2 mRNAs.
We observed that pre-treatment with calf intestinal alkaline phosphatase (CIP) renders translational silencing activity to TGFβ-treated extracts, suggesting a necessity of TGFβ-dependent phosphorylation for the release of translational inhibition (SI 4a). We next evaluated TGFβ-mediated phosphorylation of hnRNP E1 as a possible mechanism for loss of translational silencing following TGFβ treatment. TGFβ induced phosphorylation of hnRNP E1 at serine residue(s), with phospho-hnRNP E1 detected as early as 30 min after TGFβ treatment and maximal effects observed at 3 and 6 hr (Fig. 4a). Sequence analysis revealed that mouse hnRNP E1 contains an Akt consensus phosphorylation site at Ser43 (SI 4b). We therefore postulated that hnRNP E1 might be a substrate of Akt. As shown by others18,19.20, TGFβ was found to activate Akt (Fig. 4b); furthermore, using a substrate-directed phospho-specific antibody, Akt-mediated phosphorylation of hnRNP E1 was demonstrated to be TGFβ-dependent (Fig. 4c). Use of PI3K inhibitor, LY294002, showed robust inhibition of TGFβ-induced phospho-hnRNP E1 (Fig. 4d) and attenuated release of hnRNP E1 from the Dab2/BAT element following TGFβ treatment (Fig. 4e). Selective inhibition of either TGFβ signaling, with the type I receptor inhibitor SB-431542, and of Akt kinase, with Akt IV, inhibited TGFβ-dependent hnRNP E1 phosphorylation and Akt activation, without affecting total Akt levels (SI 4c), confirming the direct correlation between TGFβ signaling and hnRNP E1 phosphorylation. Importantly, inhibiting either TGFβ signaling or Akt also inhibited the reversal of translational silencing (SI 4c, bottom panel), hence suggesting a direct relationship between hnRNP E1 phosphorylation and reversal of translational silencing post-TGFβ stimulation.
Figure 4.
Phosphorylation of hnRNP E1 at serine-43 by TGFβ-mediated activation of Akt2 disrupts its binding to the BAT element and activates translation of Dab2 and ILEI. (a) IB analysis of immunoprecipitates derived from NMuMG WCLs with α-phospho-serine (p-ser) antibody (top panel) and α-hnRNP E1 antibody (bottom panel) to examine TGFβ-dependent hnRNP E1 phosphorylation. (b) IB analysis of WCLs to examine TGFβ-mediated Akt activation, revealed by phospho-Akt (pS473). (c) IB analysis of α-hnRNP E1 immunoprecipitates derived from NMuMG WCLs were probed with the phospho-Akt substrate antibody that recognizes the RXRXXpS/pT motif. (d) IB analysis of α-hnRNP E1 immunoprecipitates from LY294002 treated and untreated WCLs with α-phospho-serine (p-ser) antibody (top panel) and α-hnRNP E1 antibody (bottom panel) to confirm Akt as the kinase. (e) RNA affinity pull-down and IB analysis of cytosolic extracts from unstimulated and LY294002-treated cells to examine temporal association of hnRNP E1 and the BAT element. (f) Phosphorylated hnRNP E1 does not bind the BAT element. Increasing amounts of phosphorylated-GST-hnRNP E1 protein was subjected to pull-down with Dab2/BAT cRNA. The precipitates and the supernatants post pull-down were analyzed by IB. (j) Akt phosphorylates hnRNP E1 at Ser43. Activated kinases were recovered by anti-p-Akt (pSer473) or PAK1 immunoprecipitation and incubated with 5 µg of GST-hnRNP E1 or serine-43-alanine (S43A) mutant GST-hnRNP E1 in the presence of [γ-32P]-ATP. The kinase reaction products were detected by autoradiography. (g) IB analysis of WCLs derived from NMuMG cells post insulin and TGFβ stimulation to examine insulin and TGFβ-mediated Akt activation (top panel). IB analysis of immunoprecipitates derived from NMuMG WCLs with α-phospho-serine (p-ser) antibody (bottom panel) to examine insulin and TGFβ-dependent hnRNP E1 phosphorylation. (h) IB analysis of Akt1 and Akt2 immunoprecipitates derived from NMuMG WCLs with α-phospho-Akt (pS473) antibody to examine insulin and TGFβ-dependent isoform specific Akt activation. (i) TGFβ activated Akt2 specifically phosphorylates hnRNP E1. Activated Akt1 or Akt2 was recovered by anti-Akt1 or anti-Akt2 immunoprecipitation following TGFβ stimulation and incubated with 5 µg of GST-hnRNP E1 in the presence of [γ-32P]-ATP. Scans of (a), (b), (c), (d), (e), (f), (g), (h) and (i) are shown in SI Fig. S7.
Additionally, recombinant Akt phosphorylated GST-hnRNP E1, but not GST in an in vitro kinase assay (SI 4d) and in vitro Akt-phosphorylated GST-hnRNP E1 no longer bound the Dab2/BAT element (SI 4e). In vivo phosphorylation of hnRNP E1 by Akt was investigated by using immunoprecipitated Akt (pan Akt antibody) as the kinase source to phosphorylate hnRNP E1. TGFβ-activated Akt was capable of phosphorylating a WT hnRNP E1 fusion protein but not a S43A mutant, confirming Ser43 as the Akt phosphorylation site (Fig. 4f). Since p21-activated kinase 1 (PAK1) can phosphorylate hnRNP E1 on Thr60 and Thr12721, we examined the phosphorylating effects of PAK1 following TGFβ stimulation. PAK1 immunoprecipitates phosphorylated both WT and the S43A mutant of hnRNP E1 indicating that phosphorylation at Ser43 is specific to TGFβ signaling (Fig. 4f).
We investigated if phosphorylation of hnRNP E1 by activated Akt was specific to TGFβ stimulation. Both insulin, previously shown to activate Akt (refs. 22 & 23), and TGFβ induced Akt activation in NMuMG cells, albeit with different kinetics (Fig. 4g, top and middle panels); however, insulin-mediated Akt activation did not result in hnRNP E1 phosphorylation (Fig. 4g, bottom panel). Insulin stimulation also failed to induce either Dab2 or ILEI protein expression (SI 4f) or reversal of in vitro translation silencing activity (SI 4g). To determine whether different Akt isoforms24 were activated by insulin and TGFβ, we immunoprecipitated lysates with the three Akt isoforms, Akt1, Akt2 and Akt3 and probed them with α-p-Akt (pSer473) antibody (Fig. 4h). Insulin selectively activated Akt1 (Fig. 4h, top panel), whereas TGFβ activated Akt2 (Fig. 4h, third panel). Neither insulin nor TGFβ activated Akt3 (data not shown). Similar isoform specific Akt2 activation was observed in EpRas cells (SI 4h). Substrate specificity of Akt2 for hnRNP E1 was further demonstrated by using immunoprecipitated Akt1 or Akt2 from TGFβ-treated cells as the kinase source to phosphorylate GST-hnRNP E1 in vitro. Only TGFβ-activated Akt2, and not Akt1, was capable of phosphorylating the GST-hnRNPE1 protein in vitro (Fig. 4i). The fact that immunoprecipitated Akt1 fails to phosphorylate hnRNP E1 suggests that it is not being activated by TGFβ since both Akt1 and Akt2 share the same phosphorylation target sequence and purified Akt1 and Akt2 can phosphorylate GST-hnRNP E1 in vitro (data not shown). Hence, phosphorylation of hnRNP E1 on Ser43 by TGFβ-activated Akt2 disrupts its binding to the BAT element and causes reversal of translation silencing.
We examined whether modulating hnRNP E1 levels altered TGFβ-mediated EMT. We stably overexpressed (E23) or silenced (E2KD) hnRNP E1 in NMuMG cells (SI 5a), and compared effects on EMT. NMuMG cells underwent EMT after TGFβ treatment (24 hr), while the process was blocked or constitutively active in E23 and E2KD cells, respectively (Fig. 5a). Expression of Dab2, ILEI and mesenchymal cell markers N-cadherin and vimentin were constitutively active in E2KD cells and completely blocked in E23 cells (Fig. 5b). Parental NMuMG cells showed classical mesenchymal cells features following 24 hr TGFβ treatment, including loss of E-cadherin expression, actin reorganization at cell junctions and re-localization of ZO-1 from tight junctions (ref. 5), whereas such changes were absent in E23 cells and visible in E2KD unstimulated cells, demonstrating that hnRNP E1 is an important component of the TGFβ-mediated translational regulation of Dab2 and ILEI, and EMT.
Figure 5.
Modulation of hnRNP E1 expression or its posttranslational modification alters translation of Dab2 and ILEI and sensitivity of NMuMG cells to TGFβ-induced EMT. (a) Phase contrast images of unstimulated and TGFβ-treated (24 hr) WT, E23 and E2KD cells examining morphological changes post TGFβ-stimulation. Images were taken at 10× magnification. (b) IB analysis monitoring Dab2, ILEI, N-cadherin, vimentin and b-actin protein levels in WT, E23 and E2KD cells treated with TGFβ for the times indicated. (c) In vivo validation of Ser43 as the hnRNP E1 phosphorylation site. WCLs derived from NMuMG, KIM2 and KIWT6 cells were immunoprecipitated with α-hnRNP E1 antibody and analyzed by IB with α-phospho serine antibody (top panel) and α-hnRNP E1 antibody (second panel). TGFβ-dependent Akt activation analyzed by IB analysis of WCLs derived from NMuMG, KIWT6 and KIM2 cells treated with TGFβ for the times indicated (third and bottom panel). (d) IB analysis examining Dab2, ILEI, N-cadherin, vimentin and β-actin protein levels in cells treated with TGFβ for the times indicated. (e) & (f) IVT and RNA pull-down assays with cytosolic extracts from SH14, KIWT6 and KIM2 cells treated with TGFβ for the times indicated to examine translational silencing of chimeric Luc-Dab2/BAT cRNA (d) and temporal association of the modified hnRNP E1 with the Dab2/BAT cRNA (e). (g) Role of hnRNP E1 on EMT is mediated by Dab2 and ILEI. IB analysis of WCLs derived from SH14 cells, un-transfected or transiently transfected with ILEI, Dab2 or control-A siRNA to confirm knockdown of Dab2 and ILEI, respectively (first and second panel, respectively). IB analysis examining N-cadherin, vimentin and Hsp90 protein levels in these cells (third, fourth and bottom panel, respectively). Full scans of (b), (c), (d), (e), (f) and (g) are shown in SI Fig. S7.
To confirm that hnRNP E1, and specifically the Ser43 phosphorylation of hnRNP E1, was regulating TGFβ-mediated EMT, we knocked in either WT (KIWT6 cells) or a phospho-mutant (KIM2 cells) version of hnRNP E1 into stable hnRNP E1 knockdown cells, SH14 (shRNA directed against the 3’-UTR of hnRNP E1) (SI 6a). Stable knockdown of hnRNP E1 rendered mesenchymal phenotype to cells even in the absence of TGFβ, whereas knock-in of either the WT or S43A mutant hnRNP E1 rescued the epithelial phenotype (SI 6b). TGFβ stimulation induced EMT in KIWT6, but failed to do so in KIM2 cells (SI 6b). Correspondingly, TGFβ-induced hnRNP E1 phosphorylation in KIWT6 cells, but not in KIM2 cells (Fig. 5c, top panel) further confirming Ser43 as the phosphorylation site. In both KIWT6 and KIM2 cells, TGFβ activated Akt similarly to that observed in parental NMuMG cells (Fig. 5c, third panel). Vimentin, N-cadherin, Dab2 and ILEI expression corroborated the morphological analysis (Fig. 5d). Cytosolic extracts from these cells confirmed that Ser43 phosphorylation of hnRNP E1 also regulates translational silencing activity (Fig. 5e). In addition, RNA pull-down showed that hnRNP E1 is not released from the BAT element following TGFβ treatment in KIM2 cells as in NMuMG and KIWT6 cells (Fig. 5f). These results confirm that TGFβ-activated Akt2 phosphorylates hnRNP E1 at Ser43, a prerequisite for its release from the BAT element and concurrent translational activation of Dab2 and ILEI mRNAs.
Since Dab2 and ILEI are required but not sufficient to induce EMT it is difficult to precisely define their function downstream of hnRNP E1. Overexpression of Dab2 or ILEI alone does not induce any morphological changes associated with EMT (SI 6c & data not shown) or up-regulate N-cadherin expression (SI 6d) independent of TGFβ stimulation. We hypothesized that if Dab2 and ILEI are required for EMT, then silencing the expression level of either one will rescue epithelial cell properties in the SH14 cells. Silencing of either Dab2 or ILEI through siRNA attenuated induction of EMT as evident by loss of expression of mesenchymal cell markers, N-cadherin and vimentin (Fig. 5g) and loss of morphological features associated with mesenchymal cells (data not shown) Cumulatively, these results clearly support our hypothesis that the role of hnRNP E1 in EMT is mediated through induction of Dab2 and ILEI and that they are critical mediators of EMT.
We have identified a transcript-selective translational regulation pathway by which TGFβ modulates expression of mRNAs required for EMT. hnRNP E1 binds to a structural, 33-nucleotide TGFbeta-activated translation (BAT) element in the 3’-UTR of Dab2 and ILEI, thereby repressing their translation. TGFβ activates a kinase cascade terminating in phosphorylation of Ser43 of hnRNP E1, by isoform-specific stimulation of protein kinase Bβ/Akt2, inducing its release from the BAT element and loss of translational silencing of Dab2 and ILEI mRNAs. Modulation of hnRNP E1 expression, or of its Ser43 site, alters TGFβ-mediated loss of translational silencing and EMT. Although TGFβ causes global translational upregulation by activation of the mTOR pathway25, this is the first mechanistic demonstration of transcript-specific translational activation of transcripts regulating EMT by TGFβ. This is also the first evidence of isoform specific Akt activation by TGFβ and Akt2-mediated phosphorylation of hnRNP E1, and confirms recent findings that Akt2 is involved in promoting EMT, invasiveness and metastasis26.
The BAT element in the 3’-UTR of Dab2 and ILEI provides further insights into the importance of regulatory elements in maintenance of homeostasis. Coordinated translational regulation of Dab2 and ILEI may constitute a post-transcriptional regulon inhibiting the expression of related EMT genes27. The fact that ectopic overexpression of Dab2 or ILEI does not induce TGFβ-independent EMT suggests that there might be other candidates regulated by this pathway. Post-transcriptional regulons may have evolved as mechanisms to rapidly and coordinately suppress multiple EMT genes and downregulate metastatic progression27. The autocrine response of cells to TGFβ-induced Akt2 activation, and subsequent translational activation of transcripts involved in EMT, may represent a mechanism by which increased TGFβ expression in tumor cells contributes to cancer progression and provides avenues for novel anti-cancer therapeutic strategies.
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by Grants CA55536 and CA80095 from the National Cancer Institute to P.H.H. A.C. is supported by an American Heart Association (Ohio Valley Affiliate) Pre-doctoral Fellowship 075080B. We are grateful to Drs. Donna M Driscoll, Barsanjit Mazumder and members of our lab for helpful discussions and critical insights. We appreciate the assistance of Drs. Michael T Kinter and Belinda Willard for LC-MS; Drs. Judith A Drazba and John Peterson for imaging; and Dr. Michael Budiman for FPLC. We acknowledge Dr. Rakesh Kumar for the generous gift of GST-hnRNP E1 construct and Dr. Takashi Kobayashi for the generous gifts of mouse pCMV14-hnRNP E1-FLAG and psiRNA-hH1neo-mouse hnRNP E1. We are grateful to Dr. Harold Moses for giving us the EpRas cell line.
Footnotes
AUTHOR CONTRIBUTIONS
P.H.H. directed the project. G.J. made the initial observation of uncoupled Dab2 mRNA and protein expression levels. G.S.H. performed the experiments in EpRas cell line. P.S.R. contributed in the polysome profiling and PatSearch analyses. P.L.F. provided critical insights and expertise throughout. G.J., G.S.H. and A.C. made all the reagents. A.C. performed most of the experiments. P.H.H. and A.C. analyzed the data and wrote the paper. All authors reviewed the manuscript.
REFERENCES
- 1.Massague J. TGFβeta in Cancer. Cell. 2008;134:215–230. doi: 10.1016/j.cell.2008.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bierie B, Moses HL. TGF-beta and cancer. Cytokine Growth Factor Rev. 2006;17:29–40. doi: 10.1016/j.cytogfr.2005.09.006. [DOI] [PubMed] [Google Scholar]
- 3.Derynck R, Akhurst RJ, Balmain A. TGF-beta signaling in tumor suppression and cancer progression. Nature Genet. 2001;29:117–129. doi: 10.1038/ng1001-117. [DOI] [PubMed] [Google Scholar]
- 4.Zavadil J, Bottinger EP. TGF-beta and epithelial-to-mesenchymal transitions. Oncogene. 2005;24:5764–5774. doi: 10.1038/sj.onc.1208927. [DOI] [PubMed] [Google Scholar]
- 5.Thiery JP, Sleeman JP. Complex networks orchestrate epithelial-mesenchymal transitions. Nature Rev. Mol. Cell. Biol. 2006;7:131–142. doi: 10.1038/nrm1835. [DOI] [PubMed] [Google Scholar]
- 6.Miettinen PJ, Ebner R, Lopez AR, Derynck R. TGF-beta induced transdifferentiation of mammary epithelial cells to mesenchymal cells: involvement of type I receptors. J. Cell Biol. 1994;127:2021–2036. doi: 10.1083/jcb.127.6.2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thuault S, et al. Transforming growth factor-beta employs HMGA2 to elicit epithelial-mesenchymal transition. J. Cell Biol. 2006;174:175–183. doi: 10.1083/jcb.200512110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Oft M, et al. TGF-beta1 and Ha-Ras collaborate in modulating the phenotypic plasticity and invasiveness of epithelial tumor cells. Genes Dev. 1996;10:2462–2477. doi: 10.1101/gad.10.19.2462. [DOI] [PubMed] [Google Scholar]
- 9.Prunier C, Howe PH. Disabled-2 (Dab2) is required for transforming growth factor beta-induced epithelial to mesenchymal transition (EMT) J. Biol. Chem. 2005;280:17540–17548. doi: 10.1074/jbc.M500974200. [DOI] [PubMed] [Google Scholar]
- 10.Waerner T, et al. ILEI: a cytokine essential for EMT, tumor formation, and late events in metastasis in epithelial cells. Cancer Cell. 2006;10:227–239. doi: 10.1016/j.ccr.2006.07.020. [DOI] [PubMed] [Google Scholar]
- 11.Greinwald JH, Jr, et al. Localization of a novel gene for nonsyndromic hearing loss (DFNB17) to chromosome region 7q31. Am. J. Med. Genet. 1998;78:107–113. [PubMed] [Google Scholar]
- 12.Zhu Y, et al. Cloning, expression, and initial characterization of a novel cytokine-like gene family. Genomics. 2002;80:144–150. doi: 10.1006/geno.2002.6816. [DOI] [PubMed] [Google Scholar]
- 13.Pradet-Balade B, Boulme F, Beug H, Mullner EW, Garcia-Sanz JA. Translation control: bridging the gap between genomics and proteomics? Trends Biochem Sci. 2001;26:225–229. doi: 10.1016/s0968-0004(00)01776-x. [DOI] [PubMed] [Google Scholar]
- 14.Kang Y, Massague J. Epithelial-mesenchymal transitions: twist in development and metastasis. Cell. 2004;118:277–279. doi: 10.1016/j.cell.2004.07.011. [DOI] [PubMed] [Google Scholar]
- 15.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grillo G, Licciulli F, Liuni S, Sbisa E, Pesole G. PatSearch: A program for the detection of patterns and structural motifs in nucleotide sequences. Nucleic Acids Res. 2003;31:3608–3612. doi: 10.1093/nar/gkg548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ostareck DH, et al. mRNA silencing in erythroid differentiation: hnRNP K and hnRNP E1 regulate 15-lipoxygenase translation from the 3' end. Cell. 1997;89:597–606. doi: 10.1016/s0092-8674(00)80241-x. [DOI] [PubMed] [Google Scholar]
- 18.Bakin AV, Tomlinson AK, Bhowmick NA, Moses HL, Arteaga CL. Phosphatidylinositol 3-kinase function is required for transforming growth factor beta-mediated epithelial to mesenchymal transition and cell migration. J. Biol. Chem. 2000;275:36803–36810. doi: 10.1074/jbc.M005912200. [DOI] [PubMed] [Google Scholar]
- 19.Kattla JJ, Carew RM, Heljic M, Godson C, Brazil DP. Protein kinase B/Akt activity is involved in renal TGF-beta1-driven epithelial-mesenchymal transition in vitro and in vivo. Am. J. Physiol. Renal Physiol. 2008;295:F215–F225. doi: 10.1152/ajprenal.00548.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kato M, et al. TGF-β activates Akt kinase through a microRNA-dependent amplifying circuit targeting PTEN. Nature Cell Biol. 2009;11:881–889. doi: 10.1038/ncb1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Meng Q, et al. Signaling-dependent and coordinated regulation of transcription, splicing, and translation resides in a single coregulator, PCBP1. Proc. Natl. Acad. Sci. U. S. A. 2007;104:5866–5871. doi: 10.1073/pnas.0701065104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Datta SR, Brunet A, Greenberg ME. Cellular survival: a play in three Akts. Genes Dev. 1999;13:2905–2927. doi: 10.1101/gad.13.22.2905. [DOI] [PubMed] [Google Scholar]
- 23.Brazil DP, Hemmings BA. Ten years of protein kinase B signaling: a hard Akt to follow. TRENDS Biochem. Sci. 2001;26:657–664. doi: 10.1016/s0968-0004(01)01958-2. [DOI] [PubMed] [Google Scholar]
- 24.Kato S, Ding J, Du K. Differential activation of CREB by Akt1 and Akt2. Biochem. Biophys. Res. Commun. 2007;354:1061–1066. doi: 10.1016/j.bbrc.2007.01.094. [DOI] [PubMed] [Google Scholar]
- 25.Lamouille S, Derynck R. Cell size and invasion in TGF-beta-induced epithelial to mesenchymal transition is regulated by activation of the mTOR pathway. J. Cell Biol. 2007;178:437–451. doi: 10.1083/jcb.200611146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Irie HY, et al. Distinct roles of akt1 and Akt2 in regulating cell migration and epithelial-mesenchymal transition. J. Cell Biol. 2005;171:1023–1034. doi: 10.1083/jcb.200505087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Keene JD, Tenenbaum SA. Eukaryotic mRNPs may represent posttranscriptional operons. Mol. Cell. 2002;9:1161–1167. doi: 10.1016/s1097-2765(02)00559-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.