Abstract
Background
Animal models are increasingly being used for the assessment of fetal cell microchimerism in maternal tissue. We wished to determine the optimal transgenic mouse strain and analytic technique to facilitate the detection of rare transgenic microchimeric fetal cells amongst large numbers of maternal wild-type cells.
Methods
We evaluated two strains of mice transgenic for the enhanced green fluorescent protein (EGFP): a commercially available, commonly used strain (C57BL/6-Tg(ACTB-EGFP)10sb/J) (CAG) and a newly created strain (ROSA26-EGFP) using three different techniques: in vivo and ex vivo fluorescent imaging (for whole body and dissected organs, respectively), PCR amplification of gfp, and FCM.
Results
By fluorescent imaging, organs from CAG mice were 10-fold brighter than organs from ROSA26-EGFP mice (p<0.0001). By PCR, more transgene from CAG mice was detected compared to ROSA26-EGFP mice (p=0.04). By FCM, ROSA26-EGFP cell fluorescence was more uniform than CAG cells. A greater proportion of cells from ROSA26-EGFP organs were positive for EGFP than cells from CAG organs, but CAG mice had a greater proportion of cells with the brightest fluorescent intensity.
Conclusions
Each transgenic strain possesses characteristics that make it useful under specific experimental circumstances. The CAG mouse model is preferable when experiments require brighter cells, whereas ROSA26-EGFP is more appropriate when uniform or ubiquitous expression is more important than brightness. Investigators must carefully select the transgenic strain most suited to the experimental design to obtain the most consistent and reproducible data. In vivo imaging allows for phenotypic evaluation of whole animals and intact organs; however, we did not evaluate its utility for the detection of rare, fetal microchimeric cells in the maternal organs. Finally, while PCR amplification of a paternally inherited transgene does allow for the quantitative determination of rare microchimeric cells, FCM allows for both quantitative and qualitative evaluations of fetal cells at very high sensitivity in a plethora of maternal organs.
Keywords: transgenic, EGFP, methodology, microchimerism
INTRODUCTION
During pregnancy, fetal cells enter the maternal circulation (1) and can persist in the maternal blood and tissues for decades (2). This phenomenon, defined as a stable state of apparent engraftment of a small number of allogenic fetal cells in the maternal body, is known as fetal cell microchimerism (3,4). While many studies of microchimerism in humans have been undertaken to investigate the role of retained fetal cells on maternal health, they are often limited by the number of subjects and the availability of healthy and diseased tissues for analysis. Therefore, to better study the biological implications of fetal cell microchimerism, animal models have been developed.
The presence of fetal cell microchimerism during and after mouse pregnancy is well known (4). Our laboratory has reported on the use of transgenic male mice mated to wild-type female mice for studies of fetal cell microchimerism, in which the transgene represents a unique, paternally inherited sequence that can be used to identify fetal cells in maternal tissues (5,6). Once expressed, the protein product of the transgene can also be used as a marker of fetal cells. There are a number of transgenic animal models that may be suitable for use in studies of microchimerism. In addition, a variety of techniques can be used to locate, quantify and characterize microchimeric fetal cells. The optimal strain and technique will facilitate the detection of rare transgenic microchimeric fetal cells amongst large numbers of maternal wild-type cells.
Here, we evaluated two strains of mice transgenic for the green fluorescent protein: a commercially available, commonly used strain (7) and a newly created strain (8). We assessed each of these two strains using three different techniques: PCR, in vivo imaging (9,10) as well as the imaging of explanted organs using the same technique (ex vivo imaging), and flow cytometry. We compared transgenic strains to each other and to their respective wild-type background. Our aim was to determine the relative advantages and disadvantages of each strain and fetal DNA/cell detection technique in order to identify the best methods to distinguish between transgenic and wild-type cells.
MATERIALS AND METHODS
Mice
The Institutional Animal Care and Use Committee (IACUC) of the Tufts University School of Medicine Division of Laboratory Animal Medicine (DLAM) approved the protocol described here. All institutional guidelines regarding the ethical use and care of experimental animals were followed. The enhanced green fluorescent protein (EGFP) transgenic C57BL/6-Tg(ACTB-EGFP)10sb/J (CAG)(stock no. 003291) and wild-type C57BL/6J mice (stock no. 000664) were purchased from Jackson Laboratories (Bar Harbor, ME). ROSA26-EGFP mice (CD1 background strain) express EGFP under control of a bacterial artificial chromosome containing the ROSA26 locus and have been described earlier (8), and wild-type CD1 (strain code 022) mice were purchased from Charles River Laboratories (Wilmington, MA). Transgenic mice were bred and maintained at the DLAM under specific pathogen-free conditions. Eight week-old virgin female mice were used for all experiments. Two mice representing each transgenic and corresponding background strain were analyzed using all techniques described below, one set for each of two experiments.
Tissue collection
Blood was obtained by cardiac puncture. Thymus, heart, lung, liver, spleen, kidney, brain and bone marrow were obtained by dissection.
Whole animal (in vivo) and organ (ex vivo) imaging
For in vivo imaging of whole animals, mice were anesthetized and then shaved from the neck to the lower torso in order to allow for optimal visualization of fluorescence without interference from fur. Animals were then placed in the IVIS Imaging System 200 and analyzed for fluorescence based on the manufacturer’s recommendations (Xenogen, Alameda, CA). For organ (ex vivo) imaging, fresh organs were placed on 10 cm plates and analyzed for fluorescence using the IVIS system (Xenogen). EGFP was excited at 488 nm (filter range 445 to 490 nm) and detected at 510 nm. Data was collected as photons/sec/cm2 using Living Image software v2.50 (Xenogen).
DNA extraction and real-time PCR amplification
Twenty-five mg of each tissue was frozen at −80°C immediately after organ imaging, with the exception of spleen, from which 10 mg was obtained. After subsequent thawing of tissue, genomic DNA was extracted from all samples using the QIAamp DNA Mini Kit, as recommended by the manufacturer (Qiagen, Valencia, CA). A 75 base pair region of the gfp transgene was amplified as previously described (11) using the following primers and dual-labeled fluorescent probe: forward primer; 5′-ACTACAACAGCCACAACGTCTATATCA-3′, reverse primer; 5′-GGCGGATCTTGAAGTTCACC-3′ and Taqman probe; 5′-FAM-CCGACAAGCAGAAGAACGGCATCA-TAMRA-3′, where FAM is 6-carboxyfluorescein and TAMRA is 6-carboxytetramethylrhodamine. As an internal control for the concentration of genomic DNA, a sequence of the mouse apolipoprotein b (apob) gene was amplified as previously described (11) using the following primers and dual-labeled fluorescent probe: forward primer; 5′-CGTGGGCTCCAGCATTCTA-3′, reverse primer; 5′-TCACCAGTCATTTCTGCCTTTG-3′ and Taqman probe; 5′-FAM-CCTTGAGCAGTGCCCGACCATTC-TAMRA-3′. Real-time PCR was performed for gfp and apob gene sequences using an ABI 7900 Sequence Detection System with SDS v2.2 software (Applied Biosystems, Foster City, CA). All PCR experiments were performed in triplicate, with results reported as median values. All results are expressed as pg of GFP DNA in 2500 pg genomic DNA, as determined by PCR amplification of the apob sequence.
Flow cytometry (FCM)
Erythrocytes were removed from whole blood by lysis (2.075% NH4Cl, 0.25% NaHCO3) and washed twice with FCM buffer (PBS with 1% BSA and 0.5% sodium azide). PBMC were stained with PE-conjugated rat anti-mouse CD45 (BD Pharmingen, San Jose, CA). Other tissues were dissected into small pieces in FCM buffer and homogenized. The homogenates were filtered through a 40 μm nylon cell strainer (BD Falcon, Bedford, MA) to remove debris. Nucleated cells from blood and organs were washed twice in FCM buffer and re-suspended in FCM buffer with 1 μg/mL 7-AAD (BD Pharmingen) to stain dead cells. FCM analysis was conducted using a FACS Calibur (Becton Dickinson, San Jose, CA). 7-AAD, PE and EGFP were excited at 488 nm and measured at 647 nm, 575 nm and 510 nm, respectively. 7-AAD positive cells were excluded by gating, and PE or EGFP cells were included based on FSC and SSC (see Supplemental Figure 1). List mode data were obtained from at least 30,000 cells. FlowJo version 8.5.3 (Tree Star Inc., Ashland, OR) and Summit version 4.3 (Dako Colorado, Inc., Fort Collins, CO) were used to analyze FCM data.
Statistics
Student’s t test was used to compare signal (EGFP) to background (wild-type) noise ratios from in and ex vivo imaging analyses and quantities of the gfp transgene amplified by PCR from organs of the two transgenic mouse strains.
RESULTS
In vivo imaging
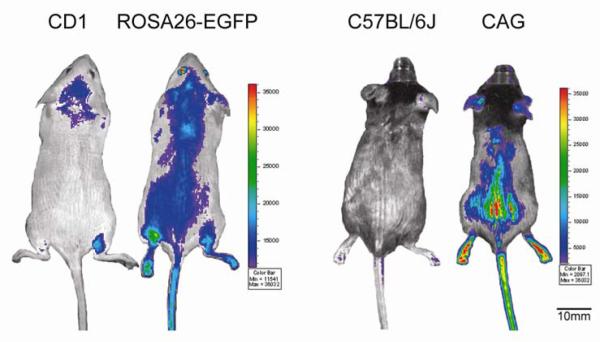
Whole body imaging of ROSA26-EGFP and CAG transgenic mice primarily depends upon coat color. In wild-type CD1 and transgenic ROSA26-EGFP mice, which have white fur, nearly the whole unshaven body of both wild type and transgenic strains fluoresces due to autofluorescence (light reflection) of the fur. Following shaving from neck to lower torso, only the non-shaved parts of the wild-type animal fluoresce, while the entire transgenic animal fluoresces whether shaved or not (Figure 1). Eyes, ears, legs and tail yield bright signals in the transgenic animal but no signal in the wild-type animal, as these areas have little or no fur (Figure 1).
Figure 1.
Comparison of EGFP gene expression represented by whole-body fluorescent intensity of ROSA26-EGFP and CAG transgenic mice and their corresponding wild-type background strains (to determine non-specific fluorescence) as measured by in vivo imaging. CD1 and ROSA26-EGFP mice shown were used in Experiment 2; C57BL/6J and CAG mice shown were used in Experiment 1. Mice have been shaved from the neck to the lower torso. Fluorescent intensity is recorded as photons/sec/cm2, and the color of the signal represents the amount of EGFP protein present.
This is in contrast to wild-type C57BL/6J and CAG transgenic mice, which have a black coat color. In these animals, the black fur does not autofluoresce but rather absorbs light. Therefore, neither the shaved or unshaved parts of the wild-type animal yield fluorescent signals (Figure 1). In the transgenic strain, only those areas that have been shaved or those areas that do not possess fur (eyes, ears, legs and tail) yield fluorescent signals (Figure 1).
Ex vivo imaging
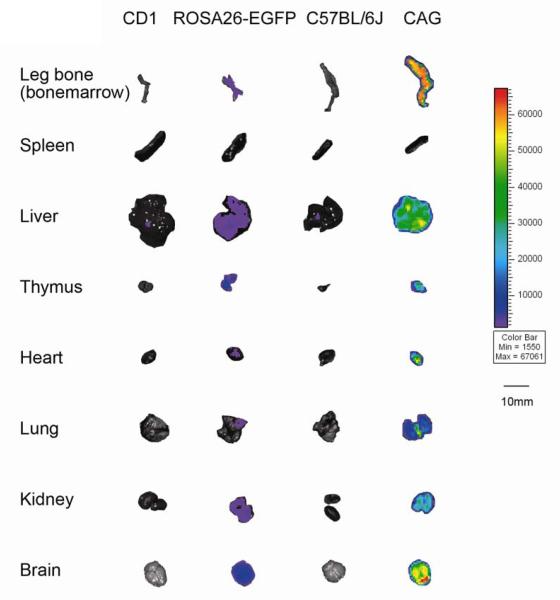
Figure 2 shows the relative fluorescent intensities of internal organs dissected from wild-type CD1 and transgenic ROSA26-EGFP mice, and from wild-type C57BL/6J and transgenic CAG mice. With the exception of spleen, all organs from CAG mice are brightly fluorescent, in contrast to the lower level of fluorescence seen from organs dissected from ROSA26-EGFP mice. While the organs from both wild-type strains have comparable background fluorescence, organs from CAG mice are on average an order of magnitude brighter than organs from ROSA26-EGFP mice. The corresponding quantities of emitted photons from each organ from both stains of transgenic mice shown in Figure 2 are presented in Table 1.
Figure 2.
Comparison of EGFP expression from intact organs of ROSA26-EGFP and CAG transgenic mice and their corresponding wild-type background strains (to determine non-specific fluorescence) as measured by ex vivo imaging immediately after sacrifice. Organs represent animals from Experiment 2. Fluorescent intensity is recorded as photons/sec/cm2, and the color of the signal represents the amount of EGFP protein present.
Table 1.
Fluorescent intensity of intact organs as determined by in vivo imaging
| Experiment 1 |
||||||||
|---|---|---|---|---|---|---|---|---|
| Strain | Leg bone | Spleen | Liver | Thymus | Heart | Lung | Kidney | Brain |
| ROSA26- EGFP |
- | 7.91×108 | 3.67×109 | - | 5.88×108 | 7.20×108 | 2.79×109 | 3.75×109 |
| CD1 (wild- type) |
- | 4.63×108 | 1.70×109 | - | 2.02×108 | 2.88×108 | 6.80×108 | 1.27×109 |
| Ratio of ROSA26- EGFP to CD1 |
- | 1.71 | 2.16 | - | 2.91 | 2.49 | 4.10 | 2.96 |
| CAG | - | 6.80×108 | 3.69×1010 | - | 1.00×1010 | 8.62×109 | 1.53×1010 | 4.28×1010 |
| C57BL/6J (wild-type) |
- | 1.43×108 | 2.57×109 | - | 1.31×108 | 3.81×108 | 7.57×108 | 9.56×108 |
| Ratio of CAG to C57BL/6J |
- | 4.76 | 14.39 | - | 76.65 | 22.60 | 20.18 | 44.72 |
| Experiment 2 |
||||||||
|---|---|---|---|---|---|---|---|---|
| Strain | Leg Bone | Spleen | Liver | Thymus | Heart | Lung | Kidney | Brain |
| ROSA26- EGFP |
6.96×108 | 9.00×107 | 3.57×109 | 1.49×109 | 2.91×108 | 9.37×108 | 1.63×109 | 4.34×109 |
| CD1 (wild- type) |
3.09×108 | 3.93×108 | 2.46×109 | 1.74×108 | 7.01×107 | 3.56×108 | 5.42×108 | 7.05×108 |
| Ratio of ROSA26- EGFP to CD1 |
2.25 | 0.23 | 1.45 | 8.56 | 4.15 | 2.63 | 3.01 | 6.16 |
| CAG | 3.84×1010 | 8.71×106 | 6.35×1010 | 4.20×109 | 5.63×109 | 1.15×1010 | 1.15×1010 | 3.55×1010 |
| C57BL/6J (wild-type) |
4.40×108 | 6.29×107 | 1.60×109 | 5.25×107 | 9.18×107 | 4.82×108 | 4.82×108 | 7.05×108 |
| Ratio of CAG to C57BL/6J |
87.27 | 0.14 | 39.69 | 80.15 | 61.33 | 23.86 | 23.86 | 50.35 |
Each row represents organs from a single mouse
Results expressed in photons/sec/cm2
CAG: C57BL/6-Tg(ACTB-EGFP)10sb/J
From ROSA26-EGFP mice, brain, liver and kidney emit the highest fluorescence, while heart, lung and spleen emit approximately 50% less fluorescence (Table 1). Thymus and bone from ROSA26-EGFP emit fluorescence comparable to brain, liver and kidney, although these organs were dissected and imaged from only one mouse. From CAG mice, brain, liver and bone emit the greatest fluorescence, while kidney, heart, lung and thymus emit approximately 50% less fluorescence. Spleen emits the lowest fluorescence from CAG mice. The ratio of transgenic to background fluorescence of organs from ROSA26-EGFP mice has an average of 3.20, with a range of 0.23 (spleen) to 8.56 (thymus). In contrast, the ratio of transgenic to background fluorescence in organs from CAG mice has an average of 39.3, with a range of 0.14 (spleen) to 87.27 (bone)(p<0.0001). Overall, there is a higher level of emitted fluorescence and signal to noise ratio from organs of CAG mice compared to organs of ROSA26-EGFP mice. With the exception of spleen, the fluorescent intensity of organs from a CAG animal is approximately 1 order of magnitude brighter than organs from a ROSA26-EGFP animal. Therefore, the signal-to-noise ratio is higher in CAG animals compared to ROSA26-EGFP animals.
PCR amplification
The results of PCR amplification of various organs from wild-type and transgenic animals are shown in Table 2. An equal amount of genomic DNA based on PCR amplification of the ApoB gene sequence from each tissue was added to each well for PCR amplification of the gfp transgene. With the exception of heart tissue in Experiment 1 and blood in Experiment 2, a greater quantity of transgene from CAG mice was detected compared to that from ROSA26-EGFP mice (p=0.04). In both experiments, DNA extracted from heart and kidney tissue was most efficiently amplified and DNA extracted from peripheral blood was least efficiently amplified. In all cases, there was no amplification of the transgenic sequence from wild-type DNA.
Table 2.
PCR amplification of gfp transgene
| Experiment 1 |
||||||||
|---|---|---|---|---|---|---|---|---|
| Strain | Blood | Spleen | Liver | Thymus | Heart | Lung | Kidney | Brain |
| ROSA26- EGFP |
- | 814 | 849 | - | 4421 | 660 | 1530 | 698 |
| CD1 (wild- type) |
- | 0 | 0 | - | 0 | 0 | 0 | 0 |
| CAG | - | 1295 | 1774 | - | 3150 | 2121 | 3934 | 1644 |
| C57BL/6J (wild-type) |
- | 0 | 0 | - | 0 | 0 | 0 | 0 |
| Experiment 2 |
||||||||
|---|---|---|---|---|---|---|---|---|
| Strain | Blood | Spleen | Liver | Thymus | Heart | Lung | Kidney | Brain |
| ROSA26- EGFP |
376 | 983 | 463 | 646 | 2713 | 298 | 2451 | 793 |
| CD1 (wild- type) |
0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| CAG | 295 | 2640 | 1390 | 830 | 4539 | 595 | 7265 | 1341 |
| C57BL/6J (wild-type) |
0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
Each row represents organs from a single mouse
Results expressed as pg of GFP DNA in 2500 pg genomic DNA
CAG: C57BL/6-Tg(ACTB-EGFP)10sb/J
Flow cytometry
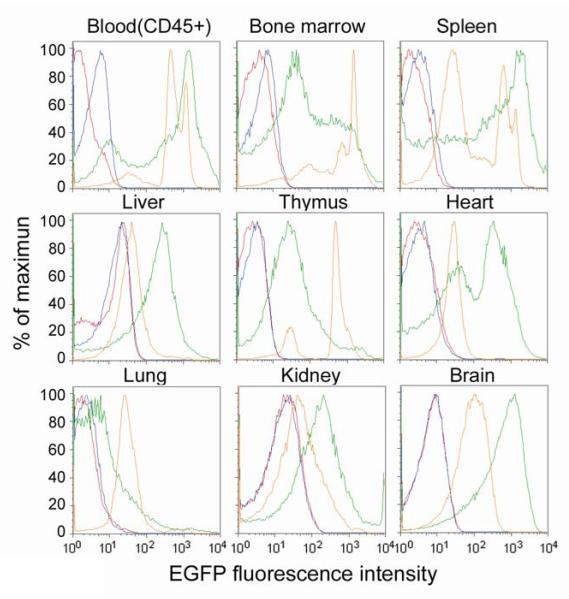
Figure 3 shows the results of the flow cytometric analysis of single cell suspensions from various organs of wild-type and transgenic animals. Table 3 shows mean and median EGFP fluorescent intensities as well as CVs among all organs studied from the transgenic strains and their corresponding wild-type strains. Overall, the fluorescence of ROSA26-EGFP cells is more uniform than CAG cells. For example, the fluorescence of ROSA26-EGFP heart cells is represented by a single, narrow fluorescent peak, while the peaks representing the fluorescence of CAG heart cells are much broader with cell populations that exhibit highly variable fluorescent intensities (Figure 3). Furthermore, the CV in fluorescence intensity is higher in CAG compared to ROSA26-EGFP mice (Table 3). This can specifically be seen in bone marrow and thymus, where the CV from Experiment 1 for CAG mice is 247.8 and 352.1, respectively, while the CV from in ROSA26-EGFP is only 97.4 and 79.7, respectively. Similar results were obtained in Experiment 2. An exception is kidney, where the CV from Experiment 1 in CAG mice is 195.2, while in ROSA26-EGFP mice it is 315.3, although this difference was not seen in Experiment 2.
Figure 3.
Comparison of EGFP expression in live (i.e. 7-AAD negative) CD45+ blood cells and live cells isolated from various organs of ROSA26-EGFP and CAG transgenic mice and their corresponding wild-type controls (to determine non-specific fluorescence) as measured by FCM immediately after sacrifice. Organs represent animals from Experiment 2. Blue: CD1 wild-type mice; yellow: ROSA26-EGFP transgenic mice; red: C57BL/6J wild-type mice; green: CAG transgenic mice.
Table 3.
Mean and median fluorescent intensities and CV of wild-type and transgenic organs
| Organ | Experiment | Mean | Median | CV | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CD1 | ROSA26- EGFP |
C57BL/6J | CAG | CD1 | ROSA26- EGFP |
C57BL/6J | CAG | CD1 | ROSA26- EGFP |
C57BL/6J | CAG | ||
| Blood (CD45+) |
1 | 7.4 | 890.8 | 4.7 | 1281.1 | 6.0 | 626.4 | 4.1 | 1000.0 | 76.3 | 91.2 | 67.6 | 106.9 |
| 2 | 5.1 | 742.4 | 4.7 | 1179.1 | 4.5 | 604.3 | 3.9 | 649.4 | 66.1 | 56.9 | 70.7 | 130.7 | |
| Bone marrow | 1 | 4.4 | 879.4 | 3.7 | 405.1 | 3.3 | 697.8 | 2.6 | 24.6 | 85.0 | 97.4 | 110.4 | 247.8 |
| 2 | 5.2 | 1037.5 | 3.9 | 449.1 | 4.1 | 805.8 | 2.6 | 42.2 | 188.6 | 98.0 | 91.5 | 226.1 | |
| Spleen | 1 | 3.4 | 357.9 | 2.4 | 1016.6 | 2.6 | 72.3 | 1.7 | 421.7 | 89.8 | 129.8 | 89.1 | 135.4 |
| 2 | 3.3 | 405.1 | 2.6 | 832.2 | 2.4 | 96.5 | 7.8 | 120.0 | 88.7 | 130.3 | 86.9 | 172.6 | |
| Liver | 1 | 31.2 | 46.8 | 18.5 | 54.6 | 27.4 | 34.0 | 19.1 | 27.4 | 117.6 | 194.6 | 87.4 | 483.6 |
| 2 | 14.6 | 63.5 | 11.9 | 282.2 | 11.6 | 36.5 | 5.2 | 124.1 | 91.2 | 234.4 | 111.9 | 215.8 | |
| Thymus | 1 | 3.7 | 478.8 | 2.7 | 179.5 | 2.8 | 453.2 | 2.1 | 13.8 | 90.1 | 79.7 | 76.9 | 352.1 |
| 2 | 3.3 | 534.9 | 3.0 | 211.8 | 2.8 | 504.8 | 2.3 | 28.4 | 71.0 | 78.2 | 75.1 | 315.2 | |
| Heart | 1 | 17.5 | 121.6 | 20.1 | 383.7 | 15.4 | 37.9 | 18.4 | 23.7 | 79.2 | 287.3 | 72.0 | 355.5 |
| 2 | 4.1 | 32.3 | 3.9 | 321.0 | 2.7 | 26.4 | 2.3 | 89.8 | 112.4 | 171.9 | 122.6 | 183.4 | |
| Lung | 1 | 4.8 | 74.6 | 2.8 | 427.7 | 2.7 | 32.8 | 1.8 | 14.3 | 130.5 | 355.6 | 138.1 | 287.2 |
| 2 | 2.5 | 47.5 | 2.2 | 37.1 | 1.8 | 26.4 | 1.4 | 2.6 | 113.7 | 265.3 | 127.6 | 828.9 | |
| Kidney | 1 | 14.6 | 138.9 | 13.4 | 1320.8 | 6.3 | 37.9 | 6.3 | 205.4 | 121.0 | 315.3 | 120.8 | 195.2 |
| 2 | 17.0 | 92.7 | 17.3 | 663.2 | 10.4 | 32.8 | 10.4 | 138.2 | 118.2 | 233.1 | 118.3 | 242.6 | |
| Brain | 1 | 19.2 | 94.2 | 22.2 | 937.2 | 16.0 | 75.0 | 17.2 | 805.8 | 68.6 | 82.8 | 80.5 | 66.9 |
| 2 | 7.8 | 107.0 | 7.7 | 1087.7 | 6.5 | 80.6 | 6.3 | 897.7 | 75.5 | 94.4 | 79.7 | 81.8 |
CAG:C57BL/6-Tg(ACTB-EGFP)10sb/J
Table 4 shows the percentage of cells from various organs of transgenic mice with fluorescent intensities greater than background fluorescence from their corresponding wild-type strains. Overall, a greater proportion of cells from ROSA26-EGFP organs were positive for EGFP than cells from CAG organs. For example, approximately 98% and 85% of CD45+ blood cells from ROSA26-EGFP animals and CAG animals, respectively, were EGFP positive. A difference between the transgenic strains was also observed with spleen, thymus, lung and bone marrow cells. One exception is kidney, where only approximately 20% of ROSA26-EGFP cells were positive for EGFP, while approximately 60% of CAG cells were EGFP positive. Brian and liver also had a higher proportion of EGFP+ cells in CAG than ROSA26-EGFP transgenic strains. Interestingly, the wild-type fluorescence of cells from liver, kidney and brain in both transgenic strains is an order of magnitude higher than that of spleen, thymus, blood and bone marrow (20 compared to 2 EGFP intensity)(Figure 3)
Table 4.
Percentage of transgenic cells exhibiting fluorescence above threshold*
| Organ | Experiment | ROSA26-EGFP | CAG |
|---|---|---|---|
| Blood (CD45+) | 1 | 98.9 | 91.1 |
| 2 | 97.0 | 82.4 | |
| Bone marrow | 1 | 97.1 | 64.8 |
| 2 | 90.5 | 67.9 | |
| Spleen | 1 | 99.0 | 69.9 |
| 2 | 88.0 | 62.2 | |
| Liver | 1 | 3.3 | 15.1 |
| 2 | 31.6 | 64.0 | |
| Thymus | 1 | 99.2 | 60.8 |
| 2 | 97.9 | 75.4 | |
| Heart | 1 | 27.4 | 26.4 |
| 2 | 64.4 | 63.5 | |
| Lung | 1 | 73.5 | 45.1 |
| 2 | 84.1 | 14.9 | |
| Kidney | 1 | 17.8 | 61.1 |
| 2 | 23.0 | 55.0 | |
| Brain | 1 | 84.5 | 98.0 |
| 2 | 93.9 | 99.0 |
Value above which 1% of wild-type cells are considered fluorescent
CAG:C57BL/6-Tg(ACTB-EGFP)10sb/J
Fluorescence of the main peak representing EGFP cells is higher in blood, spleen, liver, heart, kidney and brain from CAG mice than ROSA26-EGFP mice, while it is higher in bone marrow, thymus and lung from ROSA26-EGFP mice (Figure 3). However, CAG mice have a greater proportion of cells with the brightest EGFP intensity than those of ROSA26-EGFP mice (Figure 3). This is true in all organs, although this difference is most pronounced in spleen, heart and brain.
In most tissues from CAG mice, there are a considerable proportion of transgenic cells that exhibit “dull” fluorescence (i.e. < 102 EGFP intensity), which are not distinguishable from wild-type cells (Figure 3). Bone marrow, spleen and heart from CAG mice have a significant proportion of cells with dull fluorescence, and the nearly all lung cells from this transgenic strain exhibit dull fluorescence. In ROSA26-EGFP mice, only spleen has more than one distinct population of fluorescent cells, although blood, bone marrow and thymus have a detectable subpopulation of cells with dull fluorescence (Figure 3).
DISCUSSION
The purpose of these experiments was to determine the most useful transgenic animal model and analytical technique to apply to studies of fetal cell microchimerism. The low frequency of fetal cells that are retained in maternal tissues following pregnancy necessitates the most efficient and powerful system to analyze these cells, with a high signal to background noise ratio. Therefore, we compared two transgenic strains of EGFP+ mice using three different analytical techniques at whole body, organ and cellular levels. While the results demonstrated that neither transgenic strain is preferable to the other for all techniques studied, we show instead that each strain possesses characteristics that makes it useful under specific experimental circumstances, such as the analytical tool being used or the organs of interest to be analyzed. Therefore, an investigator must carefully select the transgenic strain that is most suited to the experimental design in order to obtain the most consistent and reproducible data. However, imaging of fluorescent cells in live animals is currently limited to labeled cells in blood vessels or in regions of known location (i.e. area of injection of cells)(12). Therefore, due to the existing status of this imaging technology and the purely quantitative nature of PCR amplification, flow cytometry provides the best overall approach for the quantitative and qualitative evaluation of rare transgenic microchimeric fetal cells.
For in vivo imaging, whole body fluorescence is brighter in CAG (black) mice compared to ROSA26-EGFP (white) mice after shaving fur. Due to reflection and absorption of the excitation light used for imaging in animals with white and black coat colors, respectively, all animals regardless of color should be shaved to minimize false positive or negative results and to create uniform circumstances for the visualization of real fluorescent signals. Shaving will also increase the likelihood of visualizing transgenic, microchimeric fetal cell populations in a wild-type mother.
Using ex vivo imaging, while each organ autofluoresces resulting in baseline photon count differences between organs, all organs are brighter in CAG mice compared to ROSA26-EGFP mice (with the exception of spleen). There are several possible reasons for the differences seen in fluorescent intensity of organs between the two strains. These include differences in transgene expression inherent to each strain or due to unique promoters, or due to different numbers of transgene copies incorporated into each strain. With respect to spleen, photon counts are higher in wild-type mice compared to transgenic mice, and spleen is the only organ in which fluorescence is brighter in ROSA26-EGFP compared to CAG mice. The reasons for these differences are unclear. However, since the spleen contains a large and perhaps variable number of cells from the erythroid lineage with reduced or absent transgene expression compared to other nucleated cells, fluorescent emission would be similarly affected. One solution may be the incorporation of spectral deconvolution into the methodology described here, leading to improvement of imaging quality through the highly specific discrimination between overlapping fluorescent signals (i.e. real and autofluorescence)(10).
Because both CAG and ROSA26-EGFP mice have the same EGFP transgene sequence, the same pair of primers and fluorescently labeled probe was used for amplifying the target sequence by real-time PCR. The transgene was amplified in all organs analyzed. PCR amplification occurs at an earlier cycle threshold in some organs (such as kidney and heart) than other organs (such as thymus and blood). The quantity of the PCR product of CAG mice was 1.5 to 3 fold higher than ROSA26-EGFP with only one exception, blood. The reasons for the organ to organ differences are unclear. However, the copy number of the transgene may be higher in CAG mice than in ROSA26-EGFP. There may also be different insertion sites of the two transgenes. PCR has higher specificity than fluorescent imaging, as PCR of the EGFP sequence is always negative in all organs from wild type mice.
Overall, a greater proportion of cells from ROSA26-EGFP organs were positive for EGFP than cells from CAG organs as determined by FCM, particularly in blood, bone marrow, spleen and thymus. This is agreement with the results of Giel-Moloney et al. (8), who showed that greater than 90% of B220+ splenocytes, CD4+ and CD8+ thymocytes and CD11b+ bone marrow myeloid cells from ROSA26-EGFP bone marrow transplant recipients were EGFP positive over background strain levels. This was in contrast to levels in CAG recipients, where these levels were ~90%, 50% and 65%, respectively (8). However, our results showed that the highest level of transgene expression in these organs, represented by maximum EGFP intensity above background fluorescence, occurs in cells from CAG mice. In addition, organs from CAG mice were brighter in peak fluorescence compared to those from ROSA26-EGFP in blood, spleen, liver, heart, kidney and brain. Interestingly, peak fluorescence of bone marrow, thymus and lung tissue is higher in ROSA26-EGFP mice than CAG. The reasons for these organ differences are unclear. In addition, there appear to be differences in fluorescence intensity from animal to animal, as demonstrated by differences in EGFP intensity of cells from bone marrow. These differences could be due to technical variation, or could be due to biological factors, such as differences in transgene expression that are influenced by parental imprinting (13). Nevertheless, blood cells from ROSA26-EGFP mice exhibit fairly uniform EGFP expression, which may make this model useful for studies of hematopoietic chimerism. Conversely, cells from CAG organs exhibit a wider range of transgene expression than ROSA26-EGFP mice, from very bright to very dim. There was also a considerable number of cells that showed no EGFP expression in CAG mice. Overall, the expression level of EGFP in ROSA26-EGFP is much more consistent between each organ than CAG mice, even though the peak fluorescence levels were lower in ROSA26-EGFP mice.
As with blood, the level of fluorescence of liver, heart, lung and kidney from ROSA26-EGFP mice is highly consistent, but the fluorescence is lower than that in CAG mice. Among cells with variable transgene expression in ROSA26-EGFP (e.g. those from spleen and bone marrow), it is not possible to distinguish between transgenic cells with a low level of transgene expression and wild-type cells. Overall, the CAG mouse model is useful when experiments require brighter cells, whereas ROSA26-EGFP is more appropriate when uniform or ubiquitous expression is more important than brightness. Further characterization of both EGFP positive and negative cell types following breeding of transgenic males and wild type females will be necessary to continue to contribute to the understanding of the dynamics of maternal-fetal cell trafficking. This includes an assessment of the phenotype of fluorescently negative cells from specific transgenic organs, such as lung, to determine why a relatively high subset does not fluoresce. It is possible that the difference in fluorescent intensity among transgenic cell types in various organs is due to EGFP promoter specificity. Nevertheless, the aim of the current study was to subjectively assess multiple strains and methodologies for the detection of fetal transgenic DNA sequences or their corresponding RNA transcripts and protein products to more fully understand the biology of fetal maternal cell trafficking. Additional experiments are undoubtedly necessary using combined analytical approaches (e.g. flow cytometry and immunohistochemistry) to continue to understand this phenomenon. These experiments also include the assessment of bright, far-red fluorescent proteins, such as the far-red mutant TurboFP635 (14), and DsRed mouse strains that are currently commercially available, such as B6.Cg-Tg(ACTB-Bgeo,-DsRed*MST)1Nagy/J. These new and highly innovative vectors may allow for improved sensitivity and specificity due to better tissue penetration, increased signal to noise ratios through the reduction of autofluorescence. However, for the current study we selected mouse strains that are transgenic for green fluorescent protein as these are the animals for which we have the most experience and which have been most commonly used as animal models of fetal cell microchimerism.
The aim of this study was to determine the potential usefulness of the newly created ROSA26-EGFP transgenic mouse strain compared to the widely used strain of mice transgenic for EGFP, CAG. The very low frequency of microchimeric cells in the host organs means that the selection of an optimal transgenic mouse strain and methods of transgene and/or transgenic cell detection are essential for the acquisition of consistent and reproducible data. We show here that in vivo imaging, PCR and FCM possess differences in sensitivity and specificity between strains of transgenic mice. Our results suggest that sensitivity and specificity of these techniques are mutually exclusive when used for rare cell detection. For higher specificity, the brighter peak fluorescence intensity of cells from CAG mice give this model a definitive advantage to distinguish true signals from background. In contrast, ROSA26-EGFP mice have the advantage of sensitivity, as the majority of transgenic cells have ubiquitous EGFP expression. Regarding methodology, while cellular and subcellular resolution using in vivo imaging techniques is possible, reports to date have only described its use in the detection of labeled cells in blood vessels or areas of known location (e.g. region of injection of cells)(12), not for cells of unknown location deep in solid tissue. While further development of this imaging technology may allow for the localization of rare fetal microchimeric cells, currently the ability of flow cytometry to provide for highly sensitive quantitative and qualitative evaluations of rare transgenic cells suggests it is the most versatile and useful method for studies of fetal cell microchimerism and should be considered as the primary tool for these investigations.
Supplementary Material
Supplemental Figure 1. Gating strategy for FCM. First, gating of FSC and SSC is used to exclude cells based on abnormal shape (top row). Then, for blood (left column) dead cells are excluded based on 7-AAD staining (X axis) and CD45 negative cells are excluded based on lack of CD45-PE staining (Y axis)(middle row). For all other tissues (right column), dead cells are excluded based on 7-AAD staining (X axis)(middle row). Finally, gated cells are then analyzed for EGFP fluorescence intensity (bottom row). Cells shown are from ROSA26-EGFP tissue used in Experiment 2.
ACKNOWLEDGMENTS
This study was supported by NIH grant R01 HD049469-01 (to DWB). YF was supported by a grant from Kanzawa Medical Research Foundation in Nagano, Japan.
REFERENCES
- 1.Ariga H, Ohto H, Busch MP, Imamura S, Watson R, Reed W, Lee TH. Kinetics of fetal cellular and cell-free DNA in the maternal circulation during and after pregnancy: implications for noninvasive prenatal diagnosis. Transfusion. 2001;41:1524–1530. doi: 10.1046/j.1537-2995.2001.41121524.x. [DOI] [PubMed] [Google Scholar]
- 2.Bianchi DW, Zickwolf GK, Weil GJ, Sylvester S, DeMaria MA. Male fetal progenitor cells persist in maternal blood for as long as 27 years postpartum. Proc Natl Acad Sci USA. 1996;93:705–708. doi: 10.1073/pnas.93.2.705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liégeois A, Escourrou J, Ouvre E, Charreire J. Microchimerism: a stable state of low-ratio proliferation of allogeneic bone marrow. Transplant Proc. 1977;9:273–276. [PubMed] [Google Scholar]
- 4.Liégeois A, Gaillard MC, Ouvre E, Lewin D. Microchimerism in pregnant mice. Transplant Proc. 1981;13:1250–1252. [PubMed] [Google Scholar]
- 5.Khosrotehrani K, Johnson KL, Guégan S, Stroh H, Bianchi DW. Natural history of fetal cell microchimerism during and following murine pregnancy. J Reprod Immunol. 2005;66:1–12. doi: 10.1016/j.jri.2005.02.001. [DOI] [PubMed] [Google Scholar]
- 6.Khosrotehrani K, Reyes RR, Johnson KL, Freeman RB, Salomon RN, Peter I, Stroh H, Guégan S, Bianchi DW. Fetal cells participate over time in the response to specific types of murine maternal hepatic injury. Hum Reprod. 2007;22:654–661. doi: 10.1093/humrep/del426. [DOI] [PubMed] [Google Scholar]
- 7.Okabe M, Ikawa M, Kominami K, Nakanishi T, Nishimune Y. ‘Green mice’ as a source of ubiquitous green cells. FEBS Lett. 1997;407:313–319. doi: 10.1016/s0014-5793(97)00313-x. [DOI] [PubMed] [Google Scholar]
- 8.Giel-Maloney M, Krause DS, Chen G, Van Etten RA, Leiter AB. Ubiquitous and uniform in vivo fluorescence in ROSA26-EGFP BAC transgenic mice. Genesis. 2007;45:83–89. doi: 10.1002/dvg.20269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Contag PR, Olomu IN, Stevenson DK, Contag CH. Bioluminescent indicators in living mammals. Nat Med. 1998;4:245–247. doi: 10.1038/nm0298-245. [DOI] [PubMed] [Google Scholar]
- 10.Wessels JT, Busse AC, Mahrt J, Dullin C, Grabbe E, Mueller GA. In vivo imaging in experimental preclinical tumor research--a review. Cytometry A. 2007;71:542–549. doi: 10.1002/cyto.a.20419. [DOI] [PubMed] [Google Scholar]
- 11.Pan D, Gunther R, Duan W, Wendell S, Kaemmerer W, Kafri T, Verma IM, Whitley CB. Biodistribution and toxicity studies of VSVG-pseudotyped lentiviral vector after intravenous administration in mice with the observation of in vivo transduction of bone marrow. Mol Ther. 2002;6:19–29. doi: 10.1006/mthe.2002.0630. [DOI] [PubMed] [Google Scholar]
- 12.Yamauchi K, Yang M, Hayashi K, Jiang P, Yamamoto N, Tsuchiya H, Tomita K, Moossa AR, Bouvet M, Hoffman RM. Imaging of nucleolar dynamics during the cell cycle of cancer cells in live mice. Cell Cycle. 2007;6:2706–2708. doi: 10.4161/cc.6.21.4861. [DOI] [PubMed] [Google Scholar]
- 13.Pries JI, Downes M, Oates NA, Rasko JE, Whitelaw E. Sensitive flow cytometric analysis reveals a novel type of parent-of-origin effect in the mouse genome. Curr Biol. 2003;13:955–959. doi: 10.1016/s0960-9822(03)00335-x. [DOI] [PubMed] [Google Scholar]
- 14.Shcherbo D, Merzlyak EM, Chepurnykh TV, Fradkov AF, Ermakova GV, Solovieva EA, Lukyanov KA, Bogdanova EA, Zaraisky AG, Lukyanov S, Chudakov DM. Bright far-red fluorescent protein for whole-body imaging. Nat Methods. 2007;4:741–746. doi: 10.1038/nmeth1083. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1. Gating strategy for FCM. First, gating of FSC and SSC is used to exclude cells based on abnormal shape (top row). Then, for blood (left column) dead cells are excluded based on 7-AAD staining (X axis) and CD45 negative cells are excluded based on lack of CD45-PE staining (Y axis)(middle row). For all other tissues (right column), dead cells are excluded based on 7-AAD staining (X axis)(middle row). Finally, gated cells are then analyzed for EGFP fluorescence intensity (bottom row). Cells shown are from ROSA26-EGFP tissue used in Experiment 2.