Abstract
Some xanthine analogues, including 1,3,7-trimethylxanthine (caffeine) and 1,3-dimethylxanthine (theophylline), have been shown to exert anticancer activities in both cell culture and animal models. The present study focused on the relationship of structure and activity of 50 different caffeine analogues in preventing epidermal growth factor (EGF)-induced malignant transformation of mouse epidermal JB6 promotion-sensitive (P+) Cl41 (JB6 P+) cells. Results indicated that the inhibition of cell transformation by the 1,3,7-trialkylxanthines depends on the number of carbons at the alkyl groups R1 and R3, but not R7. Notably, 1-ethyl-3-hexylxanthine (xanthine 70) was the most effective compound for inhibiting EGF-induced neoplastic transformation among the 50 xanthine analogues tested. The 50% inhibition of cell transformation (ICT50) value for xanthine 70 was 48- or 75-fold less than the ICT50 value of caffeine or theophylline, respectively. Further study revealed that xanthine 70 (5–40 μM) dose dependently inhibited EGF-induced transactivation of activator protein 1 (AP-1), whereas theophylline or caffeine (up to 500 μM) had no effect on AP-1 activity. In addition, xanthine 70 (10 μM) inhibited 12-O-tetradecanoylphorbol-13-acetate- or H-Ras-induced neoplastic transformation in JB6 P+ cells by 78.2 or 62.0%, respectively. Collectively, these results indicated that the number of carbons at R1 and R3 is important for the antitumor-promoting activity of the trialkylxanthines and xanthine 70 might be a promising anticancer agent.
Introduction
Nitrogen-containing compounds from terrestrial and marine organisms have been intensively investigated over the last few years for their potential as drug candidates and anticancer agents (1–5). These compounds include 1,3,7-trimethylxanthine or caffeine, a natural methylxanthine present in coffee and tea, and its various analogues, referred to as 1,3,7-trialkylxanthines or xanthines. Previous studies revealed that caffeine inhibits the development of tumors induced by various carcinogens in numerous organs including skin (6), lung (7), stomach (8) and liver (9). Oral administration of caffeine was also shown to substantially inhibit UVB-induced carcinogenesis in SKH-1 mice (10). A recent study further revealed that caffeine inhibited 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone-induced lung tumor progression from adenoma to adenocarcinoma (7). Caffeine was shown to suppress the proliferation of various cancer and transformed cell lines including human neuroblastoma cells, human pancreatic adenocarcinoma cells, human A549 lung adenocarcinoma cells and mouse epidermal JB6 promotion-sensitive (P+) cells (11–14). Tumor promoter-induced neoplastic transformation was also inhibited by caffeine treatment (11). 1,3-Dimethylxanthine (theophylline) and 1-methyl-3-propyl-7-butylxanthine (xanthine 77) were proposed to be potential anticancer drugs especially in combination with other chemotherapeutic drugs (15–18). Some xanthine analogues containing methyl groups were also reported to enhance the killing of p53-deficient cells (19).
The JB6 cell system of clonal genetic variants, including those that are P+ or promotion resistant (P−), facilitates the study of genetic susceptibility to transformation and promotion/progression at the molecular level (20–23). The JB6 P+, P− and transformed variants are a series of cell lines representing ‘earlier-to-later’ stages of preneoplastic-to-neoplastic progression (20–23). JB6 P+ cells transform when stimulated with epidermal growth factor (EGF), 12-O-tetradecanoylphorbol-13-acetate (TPA) or H-Ras, resulting in the formation of colonies in soft agar. The transformation is thought to involve the activation of activator protein 1 (AP-1), which regulates the transcription of various genes related to inflammation, proliferation and metastasis (21,24). The purpose of the present study was to investigate the potential chemopreventive activities of 50 different 1,3,7-trialkylxanthines, which resemble caffeine in their structures (Figure 1A) but differ in the length of the alkyl side chains. Here we report that the number of carbons at R1 or R3 is important for the antitumor-promoting activity of trialkylxanthines and that xanthine 70 may be a potential anticancer agent.
Materials and methods
Reagents
Caffeine, xanthine, theobromine, theophylline, 1-methylxanthine, 3-methylxanthine, 7-methylxanthine, 1,7-dimethxanthine and TPA were purchased from Sigma (St Louis, MO). Other xanthines were synthesized as described previously (25–28). All compounds were analyzed for carbon, hydrogen and nitrogen and the analytical results were within ±0.4% of theoretical values and data from high-resolution mass spectral analysis showed that all compounds were of high purity as described previously (26,29). Xanthines were dissolved in dimethyl sulfoxide. Eagle's minimum essential medium (MEM), basal medium Eagle, gentamicin, L-glutamine and EGF were purchased from Gibco BRL (Carlsbad, CA). Fetal bovine serum (FBS) was obtained from Gemini Bio-Products (Calabasas, CA). G418 and the luciferase assay substrate were purchased from Promega (Madison, WI).
Cell culture
The JB6 P+ and H-Ras-transformed JB6 mouse epidermal cell lines were cultured in monolayers in MEM containing 5% FBS, 2 mM L-glutamine and 25 μg/ml gentamicin at 37°C in a 5% CO2 incubator. The JB6 mouse epidermal cell line was stably transfected with an AP-1 luciferase reporter plasmid and maintained in MEM supplemented with 5% FBS containing 200 μg/ml G418.
Anchorage-independent transformation assay
The effect of xanthines on EGF- or TPA-induced cell transformation was investigated using JB6 P+ cells. Cells (8 × 103) were exposed to EGF or TPA, with or without xanthines (0.005–3 mM), in 1 ml of 0.33% basal medium Eagle agar containing 10% FBS and overlaid with 3.5 ml of 0.5% basal medium Eagle agar containing 10% FBS. The cultures were maintained in a 37°C, 5% CO2 incubator for 2–3 weeks. Cell colonies were then scored using the LEICA DM IRB inverted research microscope (Leica Mikroskopie und Systeme GmbH, Germany) and the Image-Pro Plus software program (v.3) (Media Cybernetics, Silver Spring, MD). The effect of the compounds on transformation of JB6 P+ cells is expressed as a percent inhibition of cell transformation compared with EGF- or TPA-stimulated cells as a positive control.
Luciferase assay for AP-1 transactivation
Confluent monolayers of JB6 P+ cells stably transfected with an AP-1 luciferase reporter plasmid were trypsinized and 8 × 103 viable cells suspended in 100 μl of 5% FBS–MEM were added to each well of a 96-well plate. Plates were incubated at 37°C in a humidified atmosphere of 5% CO2. When cells reached 80–90% confluence, they were starved by culturing in 0.1% FBS–MEM for another 24 h. The cells were treated or not treated with selected xanthine analogues at increasing concentrations (125, 250 or 500 μM) for 1 h and then they were exposed to 10 ng/ml EGF for 24 h. After treatment, cells were disrupted with 100 μl of lysis buffer [0.1 mM potassium phosphate buffer (pH 7.8), 1% Triton X-100, 1 mM dithiothreitol and 2 mM ethylenediaminetetraacetic acid and the luciferase activity was measured using a luminometer (Luinoskan Ascent, Labsystems, MD).
Statistical methods
A 50% inhibition of cell transformation (ICT50) was calculated using a linear or a quadratic regression. Data regarding inhibition of cell transformation were calculated as the percentage of positive (EGF-stimulated only) control. Five points around the ICT50 were used for regression analysis. Multiple linear regression was used for estimating the effect of the number of carbon atoms of each alkyl chain in each group on the ICT50. Significant differences between the positive control and the effect of each xanthine, as determined from the anchorage-independent soft agar assay or luciferase assay, were determined using the Mann–Whitney U-test. Each data point is shown as the mean ± SE or mean ± SD. The R version 2.5.1, Matlab 7.4.0 and SAS 9.1.3 computer programs on the Sun Fi−re v440 computer at the University of Minnesota Supercomputer Institute were used for analysis.
Results
Inhibitory effects of various xanthines on EGF-induced neoplastic transformation
As indicated earlier, the JB6 P+ cell system is a well-developed model for studying the tumor promotion process and potential of antitumor-promoting agents. We determined whether trialkyl-substituted xanthines (Table I) could have an inhibitory effect on EGF-induced neoplastic transformation of JB6 P+ cells. Results indicated that various trialkyl-substituted xanthines inhibited EGF-induced neoplastic transformation of JB6 P+ cells (Table II). Interestingly, the potency of inhibition of cell transformation increased as the number of carbons in the alkyl chain at R1 and R3 increased (Table III). For example, xanthine 105, xanthine 58, xanthine 62 and xanthine 50 all have identical groups at R1 (hexyl) and R7 (hydrogen). However, as the number of carbons at group 3 in these xanthines increased from zero to four, the potency for inhibition of cell transformation by each respective xanthine increased as well. Specifically, the number of carbons for xanthine 105 at R3 was zero with an ICT50 of 0.39 mM; the number of carbons for xanthine 58 at R3 was one with an ICT50 of 0.36 mM; the number of carbons for xanthine 62 at R3 was two with an ICT50 of 0.05 mM and the number of carbons for xanthine 50 at R3 was four with an ICT50 of 0.03 mM. Therefore, the number of carbons in the alkyl chain of R1 and R3, but not R7 (Table III), is associated with the ability of the respective xanthine to inhibit cell transformation.
Table I.
Chemical structure of 1,3,7-substituted alkylxanthines
| No. | Compound name | Radical 1 | Radical 3 | Radical 7 |
| 1 | Xanthine | H | H | H |
| 2 | 1-Methylxanthine | Me | H | H |
| 3 | 3-Methylxanthine | H | Me | H |
| 4 | 7-Methylxanthine | H | H | Me |
| 5 | 1,3-Dimethylxanthine (theophylline) | Me | Me | H |
| 6 | 1,7-Dimethylxanthine (paraxanthine) | Me | H | Me |
| 7 | 3,7-Dimethylxanthine (theobromine) | H | Me | Me |
| 8 | 1,3,7-Trimethylxanthine (caffeine) | Me | Me | Me |
| 9 | 1-Ethylxanthine (Xt 101) | Et | H | H |
| 10 | 3-Ethylxanthine (NY 457) | H | Et | H |
| 11 | 1-Ethyl-3-methylxanthine (Xt 054) | Et | Me | H |
| 12 | 1,3-Ethylxanthine (Xt 023) | Et | Et | H |
| 13 | 1-Propylxanthine (Xt 102) | n-Pro | H | H |
| 14 | 3-Propylxanthine (enprofylline) | H | n-Pro | H |
| 15 | 1-Propyl-3-methylxanthine (Xt 055) | n-Pro | Me | H |
| 16 | 3-Propyl-7-methylxanthine (Xt 016) | H | n-Pro | Me |
| 17 | 1,7-Dimethyl-3-propylxanthine (Xt 017) | Me | n-Pro | Me |
| 18 | 1-Propyl-3-ethylxanthine (Xt 059) | n-Pro | Et | H |
| 19 | 1-Ethyl-3-propylxanthine (Xt 015) | Et | n-Pro | H |
| 20 | 3-Propyl-7-ethylxanthine (Xt 018) | H | n-Pro | Et |
| 21 | 1-Methyl-3-propyl-7-ethylxanthine (Xt 074) | Me | n-Pro | Et |
| 22 | 1-Ethyl-3-propyl-7-methylxanthine (Xt 071) | Et | n-Pro | Me |
| 23 | 1,3-Dipropylxanthine (Xt 043) | n-Pro | n-Pro | H |
| 24 | 3,7-Dipropylxanthine (Xt 019) | H | n-Pro | n-Pro |
| 25 | 1,3-Dipropylxanthine (Xt 072) | n-Pro | n-Pro | Me |
| 26 | 1-Methyl-3,7-dipropylxanthine (Xt 075) | Me | n-Pro | n-Pro |
| 27 | 1-Butylxanthine (Xt 103) | n-Bu | H | H |
| 28 | 3-Butylxanthine (NY 466) | H | n-Bu | H |
| 29 | 1-Butyl-3-methylxanthine (Xt 056) | n-Bu | Me | H |
| 30 | 1-Methyl-3-butylxanthine (Xt 011) | Me | n-Bu | H |
| 31 | 1-Butyl-3-ethylxanthine (Xt 060) | n-Bu | Et | H |
| 32 | 1-Ethyl-3-butylxanthine (Xt 031) | Et | n-Bu | H |
| 33 | 1-Propyl-3-butylxanthine (Xt 047) | n-Pro | n-Bu | H |
| 34 | 1-Butyl-3-propyl-7-methylxanthine (Xt 073) | n-Bu | n-Pro | Me |
| 35 | 1-Methyl-3-propyl-7-butylxanthine (Xt 077) | Me | n-Pro | n-Bu |
| 36 | 1,3-Dibutylxanthine (Xt 048) | n-Bu | n-Bu | H |
| 37 | 1-Pentylxanthine (Xt 104) | n-Pent | H | H |
| 38 | 1-Pentyl-3-methylxanthine (Xt 057) | n-Pent | Me | H |
| 39 | 1-Pentyl-3-ethylxanthine (Xt 061) | n-Pent | Et | H |
| 40 | 1-Ethyl-3-pentylxanthine (Xt 066) | Et | n-Pent | H |
| 41 | 1-Pentyl-3-propyl-7-methylxanthine (Xt 078) | n-Pent | n-Pro | Me |
| 42 | 1-Pentyl-3-butylxanthine (Xt 049) | n-Pent | n-Bu | H |
| 43 | 1-Hexylxanthine (Xt 105) | n-Hex | H | H |
| 44 | 3-Hexylxanthine (Xt 040) | H | n-Hex | H |
| 45 | 1-Hexyl-3-methylxanthine (Xt 058) | n-Hex | Me | H |
| 46 | 1-Methyl-3-hexylxanthine (Xt 069) | Me | n-Hex | H |
| 47 | 1-Hexyl-3-ethylxanthine (Xt 062) | n-Hex | Et | H |
| 48 | 1-Ethyl-3-hexylxanthine (Xt 070) | Et | n-Hex | H |
| 49 | 1-Hexyl-3-propyl-7-methylxanthine (Xt 079) | n-Hex | n-Pro | Me |
| 50 | 1-Hexyl-3-butylxanthine (Xt 050) | n-Hex | n-Bu | H |
Each compound tested is listed in the first column. The compounds are placed in order of increasing number of carbons from radical 1 to radical 7. The columns labeled group 1, 3 or 7 are based on the structure shown in Figure 1 and indicate hydrogen (H), methyl (Me), ethyl (Et), propyl (n-Pro), butyl (n-Bu), pentyl (n-Pent) and hexyl (n-Hex).
Table II.
The inhibitory activity of the 1,3,7-substituted alkylxanthines on EGF-induced neoplastic transformation in JB6 P+ cells
| No. | Compound name | Concentration of compound in agar/% of colonies relatively untreated control | ICT50 mM | ||||
| 0.05 mM | 0.1 mM | 0.2 mM | 0.4 mM | 0.8 mM | |||
| 1 | Xanthine | 93.90 ± 6.67 | 96.09 ± 11.54 | 84.61 ± 9.46 | 54.75 ± 5.72 | 6.24 ± 1.33 | 0.45 |
| 2 | 1-Methylxanthine | 97.70 ± 8.11 | 94.57 ± 6.93 | 82.65 ± 7.03 | 60.37 ± 4.97 | 23.96 ± 3.15 | 0.53 |
| 3 | 3-Methylxanthine | 96.49 ± 5.84 | 84.89 ± 5.60 | 84.86 ± 4.68 | 53.17 ± 3.69 | 15.70 ± 1.79 | 0.47 |
| 0.5 mM | 1 mM | 2 mM | 2.5 mM | 3 mM | |||
| 4 | 7-Methylxanthine | 85.59 ± 34.59 | 2.96 ± 2.00 | 1.42 ± 0.45 | 1.79 ± 0.97 | 1.37 ± 0.46 | 1.18 |
| 0.125 mM | 0.250 mM | 0.5 mM | 1.0 mM | 1.5 mM | |||
| 5 | 1,3-Dimethylxanthine (theophylline) | 111.32 ± 12.96 | 85.00 ± 11.55 | 51.35 ± 6.13 | 20.97 ± 3.44 | 11.69 ± 2.36 | 0.75 |
| 0.5 mM | 1 mM | 2 mM | 2.5 mM | 3 mM | |||
| 6 | 1,7-Dimethylxanthine (paraxanthine) | 89.67 ± 31.28 | 46.01 ± 21.14 | 1.33 ± 0.45 | 0.82 ± 0.04 | 1.34 ± 0.50 | 1.27 |
| 0.05 mM | 0.1 mM | 0.2 mM | 0.4 mM | 0.8 mM | |||
| 7 | 3,7-Dimethylxanthine (theobromine) | 123.80 ± 3.65 | 92.67 ± 4.23 | 81.17 ± 4.05 | 48.99 ± 2.22 | 15.93 ± 0.75 | 0.47 |
| 8 | 1,3,7-Trimethylxanthine (caffeine) | 100.24 ± 3.88 | 97.67 ± 4.97 | 92.86 ± 5.09 | 78.92 ± 4.22 | 1.93 ± 0.28 | 0.48 |
| 9 | 1-Ethylxanthine (Xt 101) | 151.60 ± 19.78 | 119.83 ± 17.38 | 85.85 ± 14.20 | 57.16 ± 7.29 | 8.41 ± 0.76 | 0.49 |
| 0.05 mM | 0.1 mM | 0.2 mM | 0.3 mM | 0.4 mM | |||
| 10 | 3-Ethylxanthine (NY 457) | 129.15 ± 11.49 | 106.49 ± 8.37 | 97.15 ± 4.16 | 82.65 ± 4.25 | 73.40 ± 5.38 | 0.51 |
| 0.02 mM | 0.04 mM | 0.08 mM | 0.2 mM | 0.4 mM | |||
| 11 | 1-Ethyl-3-methylxanthine (Xt 054) | 111.19 ± 10.89 | 106.55 ± 8.54 | 84.92 ± 9.46 | 42.24 ± 4.10 | 21.93 ± 3.15 | 0.24 |
| 0.05 mM | 0.1 mM | 0.2 mM | 0.4 mM | 0.8 mM | |||
| 12 | 1,3-Diethylxanthine (Xt 023) | 100.30 ± 16.38 | 93.62 ± 16.90 | 50.54 ± 10.48 | 39.40 ± 8.45 | 13.41 ± 1.68 | 0.38 |
| 13 | 1-Propylxanthine (Xt 102) | 121.72 ± 17.02 | 62.27 ± 5.21 | 42.76 ± 5.33 | 21.38 ± 2.66 | 10.69 ± 1.33 | 0.43 |
| 0.05 mM | 0.1 mM | 0.2 mM | 0.4 mM | 0.8 mM | |||
| 14 | 3-Propylxanthine (enprofylline) | 80.52 ± 8.15 | 60.59 ± 8.11 | 66.70 ± 5.49 | 49.05 ± 9.29 | 17.48 ± 4.42 | 0.37 |
| 15 | 1-Propyl-3-methylxanthine (Xt 055) | 108.71 ± 8.48 | 100.70 ± 10.17 | 97.74 ± 6.19 | 61.49 ± 2.31 | 1.47 ± 1.74 | 0.47 |
| 0.5 mM | 1 mM | 2 mM | 2.5 mM | 3 mM | |||
| 16 | 3-Propyl-7-methylxanthine (Xt 016) | 54.95 ± 3.54 | 8.22 ± 1.61 | 3.25 ± 0.38 | 2.42 ± 0.37 | 2.24 ± 0.19 | 0.56 |
| 0.05 mM | 0.1 mM | 0.2 mM | 0.4 mM | 0.8 mM | |||
| 17 | 1,7-Dimethyl-3-propylxanthine (Xt 017) | 82.19 ± 7.22 | 53.78 ± 4.57 | 27.48 ± 2.93 | 6.69 ± 0.78 | 4.13 ± 0.54 | 0.20 |
| 0.02 mM | 0.04 mM | 0.08 mM | 0.2 mM | 0.4 mM | |||
| 18 | 1-Propyl-3-ethylxanthine (Xt 059) | 101.16 ± 5.78 | 81.10 ± 6.62 | 71.97 ± 8.07 | 28.37 ± 3.38 | 6.25 ± 0.78 | 0.12 |
| 0.05 mM | 0.1 mM | 0.2 mM | 0.4 mM | 0.8 mM | |||
| 19 | 1-Ethyl-3-propylxanthine (Xt 015) | 95.92 ± 5.89 | 71.29 ± 5.03 | 64.97 ± 7.14 | 41.85 ± 3.59 | 14.68 ± 0.93 | 0.38 |
| 20 | 3-Propyl-7-ethylxanthine (Xt 018) | 64.76 ± 8.71 | 58.31 ± 9.65 | 36.11 ± 8.07 | 15.09 ± 4.31 | 9.13 ± 1.53 | 0.16 |
| 21 | 1-Methyl-3-propyl-7-ethylxanthine (Xt 074) | 84.03 ± 5.68 | 69.10 ± 6.01 | 40.03 ± 3.77 | 11.88 ± 1.24 | 4.63 ± 0.85 | 0.25 |
| 22 | 1-Ethyl-3-propyl-7-methylxanthine (Xt 071) | 108.40 ± 22.55 | 87.18 ± 10.72 | 20.61 ± 8.04 | 12.11 ± 2.07 | 11.13 ± 1.72 | 0.30 |
| 0.02 mM | 0.04 mM | 0.08 mM | 0.2 mM | 0.4 mM | |||
| 23 | 1,3-Dipropylxanthine (Xt 043) | 83.28 ± 8.23 | 75.56 ± 7.54 | 45.58 ± 6.02 | 10.35 ± 1.44 | 5.86 ± 0.97 | 0.13 |
| 0.05 mM | 0.1 mM | 0.2 mM | 0.4 mM | 0.8 mM | |||
| 24 | 3,7-Dipropylxanthine (Xt 019) | 115.60 ± 5.65 | 88.33 ± 4.65 | 52.31 ± 4.87 | 9.97 ± 1.45 | 4.48 ± 0.68 | 0.33 |
| 25 | 1,3-Dipropylxanthine (Xt 072) | 105.58 ± 15.58 | 83.52 ± 24.57 | 27.89 ± 4.56 | 27.54 ± 9.93 | 14.95 ± 3.45 | 0.32 |
| 26 | 1-Methyl-3,7-dipropylxanthine (Xt 075) | 95.18 ± 7.97 | 74.91 ± 6.19 | 51.52 ± 4.82 | 13.63 ± 1.68 | 4.84 ± 0.75 | 0.30 |
| 0.04 mM | 0.2 mM | 0.4 mM | 0.6 mM | 0.8 mM | |||
| 27 | 1-Butylxanthine (Xt 103) | 179.88 ± 14.33 | 156.71 ± 13.76 | 100.70 ± 9.54 | 57.02 ± 6.84 | 34.41 ± 2.33 | 0.68 |
| 0.05 mM | 0.1 mM | 0.2 mM | 0.4 mM | 0.8 mM | |||
| 28 | 3-Butylxanthine (NY 466) | 111.11 ± 7.71 | 85.74 ± 9.72 | 69.02 ± 5.42 | 32.50 ± 5.05 | 5.85 ± 0.67 | 0.29 |
| 29 | 1-Butyl-3-methylxanthine (Xt 056) | 96.54 ± 8.16 | 81.13 ± 5.89 | 46.48 ± 4.37 | 11.67 ± 1.45 | 4.11 ± 0.33 | 0.30 |
| 0.02 mM | 0.04 mM | 0.08 mM | 0.2 mM | 0.4 mM | |||
| 30 | 1-Methyl-3-butylxanthine (Xt 011) | 89.54 ± 6.13 | 76.49 ± 8.83 | 75.21 ± 6.52 | 30.18 ± 3.96 | 16.46 ± 2.18 | 0.18 |
| 0.005 mM | 0.01 mM | 0.02 mM | 0.04 mM | 0.05 mM | |||
| 31 | 1-Butyl-3-ethylxanthine (Xt 060) | 105.56 ± 4.23 | 101.22 ± 5.46 | 97.38 ± 8.02 | 69.72 ± 5.48 | 54.35 ± 4.53 | 0.05 |
| 0.02 mM | 0.04 mM | 0.08 mM | 0.2 mM | 0.4 mM | |||
| 32 | 1-Ethyl-3-butylxanthine (Xt 031) | 61.71 ± 4.95 | 50.36 ± 3.45 | 17.41 ± 2.72 | 4.24 ± 0.27 | 4.32 ± 0.41 | 0.05 |
| 33 | 1-Propyl-3-butylxanthine (Xt 047) | 93.76 ± 9.15 | 73.28 ± 5.58 | 37.86 ± 4.45 | 5.50 ± 1.01 | 4.49 ± 0.63 | 0.12 |
| 0.05 mM | 0.1 mM | 0.2 mM | 0.4 mM | 0.8 mM | |||
| 34 | 1-Butyl-3-propyl-7-methylxanthine (Xt 073) | 51.20 ± 5.39 | 26.31 ± 3.68 | 9.26 ± 1.99 | 3.79 ± 0.17 | 5.10 ± 0.74 | 0.05 |
| 0.05 mM | 0.1 mM | 0.2 mM | 0.3 mM | 0.4 mM | |||
| 35 | 1-Methyl-3-propyl-7-butylxanthine (Xt 077) | 83.40 ± 5.11 | 36.74 ± 2.62 | 4.14 ± 0.40 | 3.60 ± 0.26 | 3.35 ± 0.15 | 0.13 |
| 0.02 mM | 0.04 mM | 0.08 mM | 0.2 mM | 0.4 mM | |||
| 36 | 1,3-Dibutylxanthine (Xt 048) | 107.24 ± 10.92 | 84.39 ± 8.50 | 45.38 ± 5.13 | 7.08 ± 1.51 | 8.11 ± 1.70 | 0.15 |
| 0.05 mM | 0.1 mM | 0.2 mM | 0.4 mM | 0.8 mM | |||
| 37 | 1-Pentylxanthine (Xt 104) | 121.73 ± 14.79 | 119.54 ± 18.21 | 96.79 ± 10.64 | 69.26 ± 12.95 | 17.51 ± 2.98 | 0.56 |
| 0.02 mM | 0.04 mM | 0.08 mM | 0.2 mM | 0.4 mM | |||
| 38 | 1-Pentyl-3-methylxanthine (Xt 057) | 72.30 ± 6.38 | 69.41 ± 7.35 | 35.33 ± 1.93 | 7.39 ± 1.51 | 4.99 ± 0.65 | 0.10 |
| 0.005 mM | 0.01 mM | 0.02 mM | 0.04 mM | 0.05 mM | |||
| 39 | 1-Pentyl-3-ethylxanthine (Xt 061) | 92.14 ± 5.87 | 92.34 ± 7.19 | 74.11 ± 6.32 | 64.23 ± 5.24 | 55.70 ± 3.80 | 0.05 |
| 0.05 mM | 0.1 mM | 0.2 mM | 0.4 mM | 0.8 mM | |||
| 40 | 1-Ethyl-3-pentylxanthine (Xt 066) | 44.80 ± 2.76 | 18.26 ± 1.43 | 5.61 ± 0.68 | 5.20 ± 0.55 | 5.59 ± 0.66 | 0.15 |
| 0.02 mM | 0.03 mM | 0.04 mM | 0.05 mM | 0.1 mM | |||
| 41 | 1-Pentyl-3-propyl-7-methylxanthine (Xt 078) | 76.22 ± 12.06 | 62.22 ± 9.33 | 43.47 ± 9.08 | 27.83 ± 4.45 | 23.82 ± 2.58 | 0.04 |
| 0.005 mM | 0.01 mM | 0.02 mM | 0.04 mM | 0.05 mM | |||
| 42 | 1-Pentyl-3-butylxanthine (Xt 049) | 98.91 ± 7.68 | 83.33 ± 6.90 | 74.80 ± 4.22 | 31.88 ± 3.51 | 29.54 ± 2.93 | 0.03 |
| 0.05 mM | 0.1 mM | 0.2 mM | 0.4 mM | 0.8 mM | |||
| 43 | 1-Hexylxanthine (Xt 105) | 121.59 ± 13.15 | 117.87 ± 13.39 | 73.30 ± 9.01 | 14.20 ± 2.95 | 8.33 ± 1.51 | 0.39 |
| 0.01 mM | 0.02 mM | 0.04 mM | 0.06 mM | 0.08 mM | |||
| 44 | 3-Hexylxanthine (Xt 040) | 114.66 ± 3.05 | 96.63 ± 3.47 | 49.69 ± 3.78 | 31.29 ± 2.17 | 14.95 ± 0.95 | 0.05 |
| 0.04 mM | 0.1 mM | 0.4 mM | 0.6 mM | 0.8 mM | |||
| 45 | 1-Hexyl-3-methylxanthine (Xt 058) | 115.21 ± 18.06 | 36.53 ± 8.65 | 29.59 ± 7.37 | 22.87 ± 5.35 | 24.52 ± 4.95 | 0.36 |
| 0.005 mM | 0.01 mM | 0.02 mM | 0.04 mM | 0.05 mM | |||
| 46 | 1-Methyl-3-hexylxanthine (Xt 069) | 83.41 ± 5.69 | 73.10 ± 3.99 | 37.82 ± 4.33 | 11.70 ± 1.85 | 11.09 ± 1.92 | 0.02 |
| 0.05 mM | 0.1 mM | 0.2 mM | 0.4 mM | 0.8 mM | |||
| 47 | 1-Hexyl-3-ethylxanthine (Xt 062) | 57.03 ± 4.86 | 32.44 ± 3.10 | 5.94 ± 0.68 | 7.02 ± 1.01 | 5.57 ± 0.62 | 0.05 |
| 0.005 mM | 0.01 mM | 0.02 mM | 0.04 mM | 0.05 mM | |||
| 48 | 1-Ethyl-3-hexylxanthine (Xt 070) | 66.93 ± 5.64 | 43.87 ± 2.51 | 16.01 ± 2.35 | 4.08 ± 0.52 | 3.38 ± 0.32 | 0.01 |
| 0.02 mM | 0.03 mM | 0.04 mM | 0.05 mM | 0.1 mM | |||
| 49 | 1-Hexyl-3-propyl-7-methylxanthine (Xt 079) | 60.70 ± 5.46 | 44.24 ± 7.90 | 40.55 ± 1.97 | 24.09 ± 3.41 | 18.88 ± 2.14 | 0.03 |
| 0.02 mM | 0.04 mM | 0.08 mM | 0.2 mM | 0.4 mM | |||
| 50 | 1-Hexyl-3-butylxanthine (Xt 050) | 59.79 ± 5.41 | 16.90 ± 2.36 | 5.88 ± 0.75 | 6.31 ± 0.84 | 5.10 ± 0.73 | 0.03 |
The compounds are placed in order of increasing number of carbons from radical 1 to radical 7. Each data point is represented as the mean ± SE from 12 measurements of the number of colonies in soft agar (anchorage-independent assay). The numbers are indicated as the percentage of colonies relative to the untreated control for each concentration of each compound. Inhibition of cell transformation is represented as the ICT50 (last column, bold), which indicates 50% inhibition compared with the EGF-only treated control.
Table III.
Multiple regression results for calculating ICT50
| Regression equation | No. of cases | Radical | P level of beta |
| ICT50 = 0.72546 − 0.06318 × R1 − 0.11207 × R3 − 0.01340 × R7 | 50 | R1 beta = −0.06 | P < 0.0001 |
| R3 beta = −0.11 | P < 0.0001 | ||
| R7 beta = −0.01 | P = 0.6384 |
Significance level of the contributions of groups (R1 and R3) is highlighted in bold (last two columns).
In a comparison of xanthine 62 with xanthine 70 or xanthine 105 with xanthine 40, the analysis suggested that a longer carbon chain at R3 may be more important than at R1. For example, xanthine 62 has six carbons at R1 and two carbons at R3 with an ICT50 of 0.05 mM. Xanthine 70 has two carbons at R1 and six carbons at R3 with an ICT50 of 0.01 mM. Another example is the comparison between xanthine 105 and xanthine 40. Xanthine 105 has six carbons of R1 and no carbons at R3 with an ICT50 of 0.39 mM. Xanthine 40 has no carbons at R1 and six carbons at R3 with an ICT50 of 0.05 mM. Thus, these results support the idea that a longer carbon chain at R3 is more important than at R1.
Caffeine (Figure 1B) has been shown to exert anticancer activities in both cell culture and animal models (10,11,30,31). Our previous studies have shown that caffeine inhibited cell proliferation (12,32) and EGF-induced neoplastic transformation (11). In the present study, 1-ethyl-3-hexylxanthine (xanthine 70, Figure 1C) was the most effective of the 50 xanthine analogues tested in inhibiting EGF-induced neoplastic transformation (Table II). The ICT50 value of xanthine 70 (with six carbons at group 3) was 48-fold less than caffeine (one carbon at group 3). These results suggested that the length of the alkyl chain of the respective xanthine plays a key role in the inhibition of neoplastic transformation and that xanthine 70 might be an effective antitumor-promoting agent.
Fig. 1.
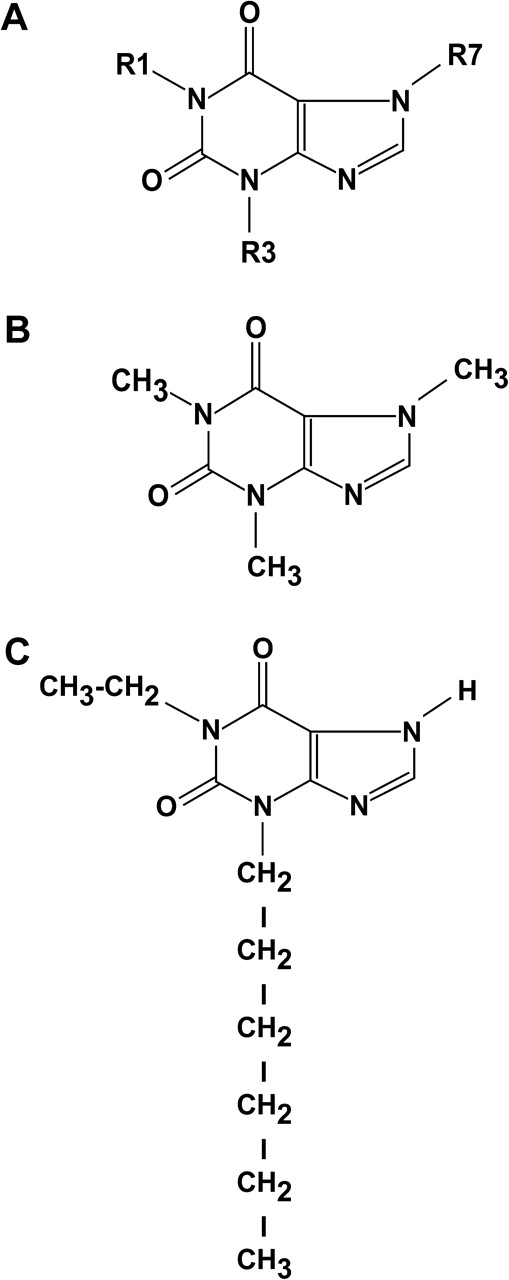
The chemical structure of the 1,3,7-trialkylxanthines, caffeine and xanthine 70. (A) The basic structure of the 1,3,7-trialkylxanthines. (B) Chemical structure of caffeine. Caffeine has R1 = R3 = R7 = methyl. (C) Chemical structures of xanthine 70. Xanthine 70 has R1 = ethyl, R3 = hexyl, R7 = hydrogen.
Effect of some selected xanthine analogues on EGF-induced AP-1 activation
AP-1 activation was shown previously to be required for neoplastic transformation in JB6 cells (21,24). Xanthines 49, 50, 79 and 70 each have different groups at position 1, 3 or 7. The most effective xanthines for inhibiting cell transformation included xanthines 79 and 50, each of which has a hexyl group at position 1, and xanthine 70, which has a hexyl group at position 3. Xanthine 79 has a methyl group at position 7 and xanthine 50 has only a hydrogen group at position 7, but a butyl group at position 3. In this case, the compound that has a group with a one carbon alkyl chain at position 7 seemed to be more effective in inhibiting cell transformation compared with a compound that has a group with a one carbon alkyl chain at position 3. To determine whether the inhibition of transformation by these xanthine analogues involves the suppression of AP-1 activity, we measured AP-1 transactivation in JB6 cells stably transfected with an AP-1 luciferase reporter plasmid. Consistent with the results above, xanthines 40, 50, 69, 70 or 79 inhibited EGF-induced transactivation of AP-1 at concentrations 125, 250 or 500 μM in a dose-dependent manner, whereas caffeine did not (Figure 2A). Theobromine exerted an inhibitory effect at a concentration of 250 and 500 μM, whereas theophylline and xanthines 58 and 78 were only effective at 500 μM (Figure 2A). These results indicated that xanthines 40, 50, 69, 70 or 79 were substantially more effective than caffeine or theophylline in suppressing AP-1 transactivation in JB6 P+ cells, which may account for their antitumor-promoting activity. For example, xanthine 70 at 125 μM was significantly more effective for inhibition of EGF-induced transactivation of AP-1 than caffeine (P < 0.05) or theophylline (P < 0.05) as indicated by the Mann–Whitney U-test (Figure 2B). In addition, we found that xanthine 70 at 5 μM significantly inhibited EGF-induced transactivation of AP-1 in JB6 P+ cells (Figure 2C), suggesting that AP-1 might be primarily involved in xanthine 70-mediated inhibition of cell transformation.
Fig. 2.
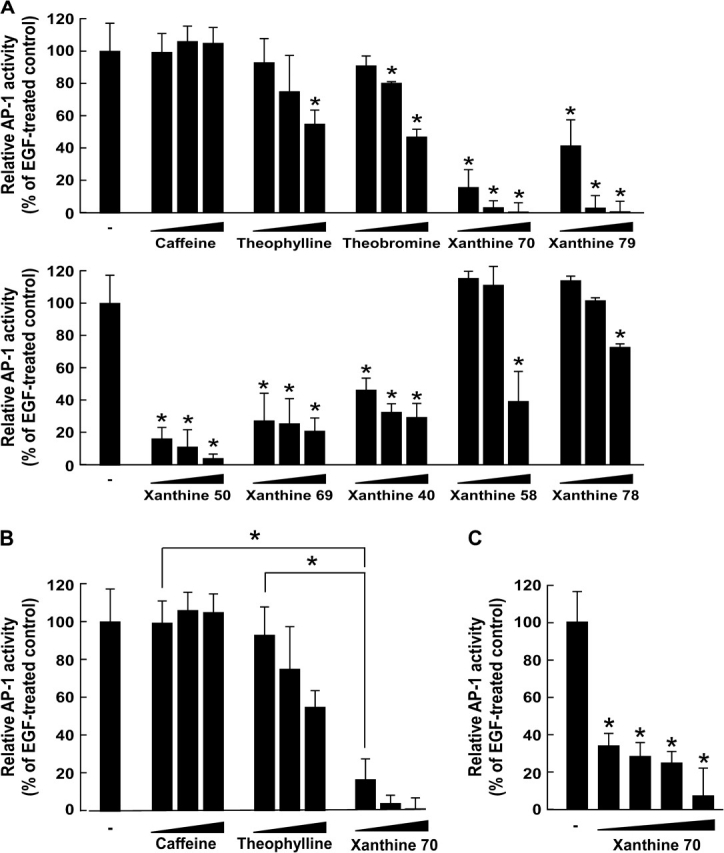
The effects of some selected xanthine analogues on EGF-induced AP-1 transactivation. (A) Effect of caffeine, theophylline, theobromine, xanthine 70, xanthine 79, xanthine 50, xanthine 69, xanthine 40, xanthine 58 or xanthine 78 on EGF-induced AP-1 transactivation in JB6 P+ cells. For the luciferase assay, JB6 cells stably transfected with an AP-1 luciferase reporter plasmid were cultured as described in Materials and methods. The cells were starved in 0.1% FBS–MEM and then treated or not treated with selected xanthine analogues at increasing concentrations (125, 250 or 500 μM) for 1 h and then they were exposed to 10 ng/ml EGF for 24 h. Luciferase activity was then assayed and AP-1 activity is expressed as the percent inhibition by each xanthine relative to cells treated with only EGF. Data are represented as mean ± SD of AP-1 luciferase activity calculated from three independent experiments. (B) Effect of caffeine, theophylline and xanthine 70 on EGF-induced AP-1 transactivation in JB6 P+ cells. Xanthine 70 at 125 μM was significantly more effective in inhibition of EGF-induced transactivation of AP-1 than caffeine or theophylline. Significant differences were determined by the Mann–Whitney U-test. The asterisk (*) indicates a significant difference (P < 0.05) between the EGF-treated control cells and cells treated with each respective xanthine. (C) Effect of xanthine 70 on EGF-induced AP-1 transactivation in JB6 P+ cells. For the luciferase assay, JB6 cells stably transfected with an AP-1 luciferase reporter plasmid were cultured as described in Materials and methods. The cells were starved in 0.1% FBS–MEM and then treated or not treated with selected xanthine analogues at increasing concentrations (5, 10, 20 or 40 μM) for 1 h and then they were exposed to 10 ng/ml EGF for 24 h. Luciferase activity was then assayed and AP-1 activity is expressed as the percent inhibition by each xanthine relative to cells treated with only EGF.
Xanthine 70 inhibits TPA- or H-Ras-induced neoplastic transformation of JB6 P+ cells
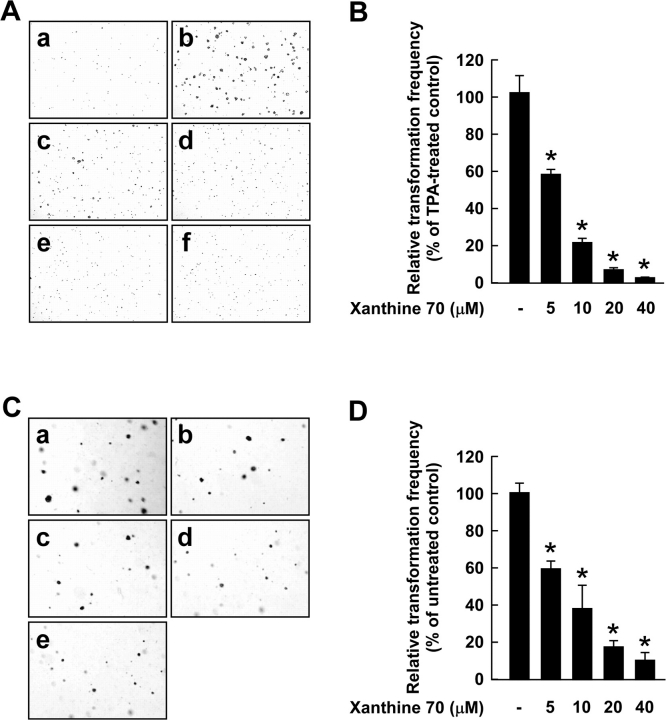
Our previous studies showed that TPA induces anchorage-independent growth in JB6 P+ cells (21,24). Various human cancers exhibit mutations and overexpression of ras genes and one of the most frequent events in carcinogenesis is the uncontrolled activation of the Ras signaling pathway (33). Theophylline, when used in combination with cisplatin to treat ovarian carcinoma cells or granulosa cells transformed with SV40 and the ras oncogene, could efficiently inhibit the proliferation of these cells (16). We examined the potential inhibitory effect of xanthine 70 on TPA-induced cell transformation. Based on the number of cell colonies, xanthine 70 at a concentration of 10 μM inhibited TPA-induced neoplastic transformation by 78.2% (P < 0.0001) (Figure 3A and B). Caffeine or theophylline (up to 80 μM) did not affect TPA-induced neoplastic transformation (data not shown). Xanthine 70 (0–40 μM) inhibited H-Ras-induced neoplastic transformation in a dose-dependent manner, and xanthine 70 at a concentration of 10 μM inhibited H-Ras-induced neoplastic transformation by 62.0% (P < 0.0001) (Figure 3C and D). These results supported the idea that xanthine 70 is a potent inhibitor of neoplastic transformation.
Fig. 3.
Effects of xanthine 70 on TPA- or H-Ras-induced neoplastic transformation in JB6 P+ cells. (A) Xanthine 70 inhibits TPA-induced cell transformation. JB6 P+ cells were treated as described in the Materials and methods and colonies were counted 14 days later. Panels indicate (a) untreated control, (b) treatment with TPA alone, (c) TPA and 5 μM xanthine 70, (d) TPA and 10 μM xanthine 70, (e) TPA and 20 μM xanthine 70 or (f) TPA and 40 μM xanthine 70. Cell colonies were counted under a microscope with the aid of the Image-Pro Plus software program (v.3). (B) The effects of xanthine 70 on cell transformation of JB6 P+ cells are presented as a percent inhibition of cell transformation compared with TPA-stimulated cells. (C) Xanthine 70 inhibits H-Ras-induced cell transformation. H-Ras-transformed JB6 cells were treated as described in Materials and methods and colonies were counted 14 days later. Panels indicate (a) untreated control, (b) 5 μM xanthine 70, (c) 10 μM xanthine 70, (d) 20 μM xanthine 70 or (e) 40 μM xanthine 70. (D) The cell colonies were counted under a microscope with the aid of the Image-Pro Plus software program (v.3). The effects of xanthine 70 on H-Ras-induced cell transformation of JB6 P+ cells are presented as a percent inhibition of cell transformation compared with untreated cells. For (B) and (D), data are expressed as mean ± SD. Significant differences were determined by the Mann–Whitney U-test and the asterisk (*) indicates a significant (P < 0.0001) difference between the respective positive control and effect of various concentrations of xanthine 70.
Discussion
A serving of instant, filtered or percolated coffee contains 60–115 mg caffeine (34,35) and the human plasma concentration of caffeine obtainable is ∼10–30 μM (36). A recent study has shown that caffeine (500 μM) can suppress EGF- or TPA-induced malignant cell transformation (11). However, these results suggested that the caffeine concentrations required to exert these effects may be difficult to achieve by drinking only a few cups of coffee each day. Therefore, developing more potent analogues of caffeine with fewer side effects may provide a feasible means of achieving an effective physiologic concentration. In this study, we found that xanthine derivatives inhibited cell transformation and that the potency increased as the number of carbons increased at group 1 or 3, but not 7. Notably, a longer carbon chain at R3 may be more important than at R1 for the inhibition of neoplastic transformation. Our results also suggested that xanthine 70 may be a potential anticancer agent.
All alkylxanthines that were investigated in the present study should have similar characteristics for absorption and cell permeation, which would be important for physiologic involvement in intracellular processes. Xanthines of this group have molecular weights ranging from 152 (xanthine) to 292 (xanthine 50 or 79), which is suitable for good absorption and permeation (37). Studies have shown that the number of NH and OH groups is associated with cellular absorption and permeability. In the present study, all 50 compounds had a similar number of OH or NH groups even though the number of carbons of R1, R3 or R7 was variable. Thus, the 50 compounds most probably had similar absorption and permeability characteristics. An increase in hydrophobicity of alkylxanthine derivatives may enhance the biological activity of the xanthine molecule and be associated with the inhibition of cell transformation. The addition of a single methyl group that can occupy a receptor pocket has been shown to improve binding by ∼0.7 kcal/mol (38). Thus, one might assume that alkylxanthine derivatives with groups containing a larger number of carbons might interact more strongly with hydrophobic protein-binding pockets.
In general, our findings indicated that increasing the number of carbons at the alkyl chain of the alkylxanthine groups appears to be associated with enhancing the ability of the respective xanthine to inhibit cell transformation. Previous studies (28,29) have shown an inhibitory effect of trialkyl-substituted xanthines on the activity of cyclic adenosine 3′,5′-monophosphate-phosphodiesterase that depends on the number of carbons at R1 and R3. Substitution in a carbon chain at R7 was shown to affect some activities (29). For example, the relaxant action in the tracheal muscle and chronortopic action in the right atrium were decreased by alkylation of alkylxanthines at the 7 position (29). However, in most cases, changes in the length of the chain at R7 of alkylxanthines did not influence activities other than cell viability (25–29). Importantly, in the present study, we used xanthines with a small variation in the number of carbons at group 7 (zero to four), whereas the number of carbons at R1 or R3 varied from zero to six. Also, in the present study, the xanthines contained a hydrogen or methyl group at position 7, whereas in R1 and R3, only the number of carbons was different.
AP-1 is an inducible eukaryotic transcription factor comprised of proteins of the fos and jun oncogene families and plays a key role in ‘preneoplastic-to-neoplastic’ transformation and proliferation (21,24). Furthermore, activation of AP-1 induces the expression of target genes such as cyclooxygenase-2, fos, cyclin D1, matrix metalloproteinases, urokinase-type plasminogen activator and vascular endothelial growth factor, which are involved in many disease processes such as neoplastic transformation, inflammation, tumor progression, metastasis and angiogenesis (39). A diverse variety of stimuli induce AP-1 binding to the DNA of various genes that govern cellular processes such as transformation and proliferation (40). Many reports have suggested that AP-1 activity is a critical factor for tumor promoter-induced cell transformation in JB6 P+ cells (20,21,40–42). EGF, TPA and H-Ras are commonly used experimental stimuli used to activate AP-1 and induce cellular transformation in many different cell types and animal models (20,21,40). Previous studies (40,43) indicated that AP-1 plays a critical role in tumor promotion through a comparison of the JB6 P+ and P− cells. Caffeine was shown to slightly attenuate EGF-induced AP-1 activation at 1 mM (11). We found that xanthine 70 dose dependently suppressed AP-1 activation. Nomura et al. (11) suggested that AP-1 activation might not be substantially involved in caffeine's inhibition of cell transformation. Some transcription factors such as p53, nuclear factor-κB or NFAT are also related to the transformation of JB6 P+ cells. We do not exclude the possibility that xanthine analogues, depending on structure, can affect other transcription factors. However, the concentration at which xanthine 70 inhibited cell transformation was similar to the effective concentration at which it inhibited AP-1 activity, suggesting that AP-1 might be primarily involved in xanthine 70-mediated inhibition of cell transformation.
Taken together, the biological activities of the xanthine analogues suggest the importance of the number of carbons at R1 or R3 in the relationship of the structure of xanthine analogues and their antitumor-promoting activities. Our findings lead us to conclude that the ability of 1,3,7-trialkylxanthines to inhibit cell transformation increases with the increase in the number of carbons from 7 to 10 at groups 1 and 3 as follows: n-heptyl → n-octyl → n-nonyl → n-decyl. Xanthine 70 shows promise as a possible antitumor-promoting agent. Further study will be needed to understand the molecular basis for the chemopreventive properties of xanthine 70 and other analogues, which will be critical in the design of more effective agents to prevent or treat cancer.
Funding
The Hormel Foundation; National Institutes of Health grants (CA120388, CA111536, CA88961, CA81064); BioGreen 21 Program, Rural Development Administration, Republic of Korea (20070301-034-042-007-02-00).
Acknowledgments
Conflict of Interest Statement: None declared.
Glossary
Abbreviations
- AP-1
activator protein 1
- EGF
epidermal growth factor
- FBS
fetal bovine serum
- ICT50
50% inhibition of cell transformation
- MEM
minimum essential medium
- P+
promotion sensitive
- TPA
12-O-tetradecanoylphorbol-13-acetate
References
- 1.Epifano F, et al. Chemistry and pharmacology of oxyprenylated secondary plant metabolites. Phytochemistry. 2007;68:939–953. doi: 10.1016/j.phytochem.2007.01.019. [DOI] [PubMed] [Google Scholar]
- 2.Bode AM, et al. Molecular and cellular targets. Mol. Carcinog. 2006;45:422–430. doi: 10.1002/mc.20222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fedorov SN, et al. Evaluation of cancer-preventive activity and structure-activity relationships of 3-demethylubiquinone Q2, isolated from the ascidian Aplidium glabrum, and its synthetic analogs. Pharm. Res. 2006;23:70–81. doi: 10.1007/s11095-005-8813-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Somei M, et al. Simple indole alkaloids and those with a non-rearranged monoterpenoid unit. Nat. Prod. Rep. 2005;22:73–103. doi: 10.1039/b316241a. [DOI] [PubMed] [Google Scholar]
- 5.Fedorov SN, et al. Marine alkaloid polycarpine and its synthetic derivative dimethylpolycarpine induce apoptosis in JB6 cells through p53- and caspase 3-dependent pathways. Pharm. Res. 2004;21:2307–2319. doi: 10.1007/s11095-004-7683-5. [DOI] [PubMed] [Google Scholar]
- 6.Huang MT, et al. Effects of tea, decaffeinated tea, and caffeine on UVB light-induced complete carcinogenesis in SKH-1 mice: demonstration of caffeine as a biologically important constituent of tea. Cancer Res. 1997;57:2623–2629. [PubMed] [Google Scholar]
- 7.Lu G, et al. Inhibition of adenoma progression to adenocarcinoma in a 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone-induced lung tumorigenesis model in A/J mice by tea polyphenols and caffeine. Cancer Res. 2006;66:11494–11501. doi: 10.1158/0008-5472.CAN-06-1497. [DOI] [PubMed] [Google Scholar]
- 8.Nishikawa A, et al. Effects of caffeine on glandular stomach carcinogenesis induced in rats by N-methyl-N′-nitro-N-nitrosoguanidine and sodium chloride. Food Chem. Toxicol. 1995;33:21–26. doi: 10.1016/0278-6915(95)80243-6. [DOI] [PubMed] [Google Scholar]
- 9.Hosaka S, et al. Suppressive effect of caffeine on the development of hepatic tumors induced by 2-acetylaminofluorene in ACI rats. Gann. 1984;75:1058–1061. [PubMed] [Google Scholar]
- 10.Lou YR, et al. Effects of oral administration of tea, decaffeinated tea, and caffeine on the formation and growth of tumors in high-risk SKH-1 mice previously treated with ultraviolet B light. Nutr. Cancer. 1999;33:146–153. doi: 10.1207/S15327914NC330205. [DOI] [PubMed] [Google Scholar]
- 11.Nomura M, et al. Inhibition of epidermal growth factor-induced cell transformation and Akt activation by caffeine. Mol. Carcinog. 2005;44:67–76. doi: 10.1002/mc.20120. [DOI] [PubMed] [Google Scholar]
- 12.He Z, et al. Induction of apoptosis by caffeine is mediated by the p53, Bax, and caspase 3 pathways. Cancer Res. 2003;63:4396–4401. [PubMed] [Google Scholar]
- 13.Jang MH, et al. Caffeine induces apoptosis in human neuroblastoma cell line SK-N-MC. J. Korean Med. Sci. 2002;17:674–678. doi: 10.3346/jkms.2002.17.5.674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Qi W, et al. Caffeine induces TP53-independent G(1)-phase arrest and apoptosis in human lung tumor cells in a dose-dependent manner. Radiat. Res. 2002;157:166–174. doi: 10.1667/0033-7587(2002)157[0166:citigp]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 15.Hirsh L, et al. Phosphodiesterase inhibitors as anti-cancer drugs. Biochem. Pharmacol. 2004;68:981–988. doi: 10.1016/j.bcp.2004.05.026. [DOI] [PubMed] [Google Scholar]
- 16.Yoshida Y, et al. Theophylline and cisplatin synergize in down regulation of BCL-2 induction of apoptosis in human granulosa cells transformed by a mutated p53 (p53 val135) and Ha-ras oncogene. Int. J. Oncol. 2000;17:227–235. doi: 10.3892/ijo.17.2.227. [DOI] [PubMed] [Google Scholar]
- 17.Sadzuka Y, et al. Effects of 1-methyl-3-propyl-7-butylxanthine (MPBX) on idarubicin-induced antitumor activity and bone marrow suppression. Jpn. J. Cancer Res. 2000;91:651–657. doi: 10.1111/j.1349-7006.2000.tb00995.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sadzuka Y, et al. 1-Methyl-3-propyl-7-butylxanthine, a novel biochemical modulator, enhances therapeutic efficacy of adriamycin. Jpn. J. Cancer Res. 1998;89:228–233. doi: 10.1111/j.1349-7006.1998.tb00553.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jiang X, et al. Structure-activity relationships for G2 checkpoint inhibition by caffeine analogs. Int. J. Oncol. 2000;16:971–978. doi: 10.3892/ijo.16.5.971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bernstein LR, et al. AP1/jun function is differentially induced in promotion-sensitive and resistant JB6 cells. Science. 1989;244:566–569. doi: 10.1126/science.2541502. [DOI] [PubMed] [Google Scholar]
- 21.Dong Z, et al. Blocking of tumor promoter-induced AP-1 activity inhibits induced transformation in JB6 mouse epidermal cells. Proc. Natl Acad. Sci. USA. 1994;91:609–613. doi: 10.1073/pnas.91.2.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang C, et al. Shortage of mitogen-activated protein kinase is responsible for resistance to AP-1 transactivation and transformation in mouse JB6 cells. Proc. Natl Acad. Sci. USA. 1998;95:156–161. doi: 10.1073/pnas.95.1.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dong Z, et al. Harvesting cells under anchorage-independent cell transformation conditions for biochemical analyses. Sci. STKE. 2002;2002:PL7. doi: 10.1126/stke.2002.130.pl7. [DOI] [PubMed] [Google Scholar]
- 24.Huang C, et al. Blocking activator protein-1 activity, but not activating retinoic acid response element, is required for the antitumor promotion effect of retinoic acid. Proc. Natl Acad. Sci. USA. 1997;94:5826–5830. doi: 10.1073/pnas.94.11.5826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miyamoto KI, et al. Selective bronchodilators from 1-(5′-oxohexyl) xanthines. J. Pharm. Pharmacol. 1992;44:888–892. doi: 10.1111/j.2042-7158.1992.tb03230.x. [DOI] [PubMed] [Google Scholar]
- 26.Miyamoto K, et al. Bronchodilator activity of xanthine derivatives substituted with functional groups at the 1- or 7-position. J. Med. Chem. 1993;36:1380–1386. doi: 10.1021/jm00062a010. [DOI] [PubMed] [Google Scholar]
- 27.Miyamoto K, et al. Selective tracheal relaxation and phosphodiesterase-IV inhibition by xanthine derivatives. Eur. J. Pharmacol. 1994;267:317–322. doi: 10.1016/0922-4106(94)90156-2. [DOI] [PubMed] [Google Scholar]
- 28.Sanae F, et al. Structure-activity relationships of alkylxanthines: alkyl chain elongation at the N1- or N7-position decreases cardiotonic activity in the isolated guinea pig heart. Jpn. J. Pharmacol. 1995;69:75–82. doi: 10.1254/jjp.69.75. [DOI] [PubMed] [Google Scholar]
- 29.Sakai R, et al. Effects of alkyl substitutions of xanthine skeleton on bronchodilation. J. Med. Chem. 1992;35:4039–4044. doi: 10.1021/jm00100a008. [DOI] [PubMed] [Google Scholar]
- 30.Levi-Schaffer F, et al. Xanthines inhibit 3T3 fibroblast proliferation. Skin Pharmacol. 1991;4:286–290. doi: 10.1159/000210963. [DOI] [PubMed] [Google Scholar]
- 31.Belman S, et al. Phorbol-12-myristate-13-acetate effect on cyclic adenosine 3′,5′-monophosphate levels in mouse skin and inhibition of phorbol-myristate-acetate-promoted tumorigenesis by theophylline. Cancer Res. 1974;34:3446–3455. [PubMed] [Google Scholar]
- 32.Hashimoto T, et al. Caffeine inhibits cell proliferation by G0/G1 phase arrest in JB6 cells. Cancer Res. 2004;64:3344–3349. doi: 10.1158/0008-5472.can-03-3453. [DOI] [PubMed] [Google Scholar]
- 33.Zachos G, et al. Expression of ras proto-oncogenes: regulation and implications in the development of human tumors. Crit. Rev. Oncol. Hematol. 1997;26:65–75. doi: 10.1016/s1040-8428(97)00013-9. [DOI] [PubMed] [Google Scholar]
- 34.Barone JJ, et al. Caffeine consumption. Food Chem. Toxicol. 1996;34:119–129. doi: 10.1016/0278-6915(95)00093-3. [DOI] [PubMed] [Google Scholar]
- 35.Lelo A, et al. Assessment of caffeine exposure: caffeine content of beverages, caffeine intake, and plasma concentrations of methylxanthines. Clin. Pharmacol. Ther. 1986;39:54–59. doi: 10.1038/clpt.1986.10. [DOI] [PubMed] [Google Scholar]
- 36.MAFF. Survey of Caffeine and Other Methylxanthines in Energy Drinks and Other Caffeine-Containing Products. London: MAFF Publications; 1997. Vol. Food Surveillance Information Sheet No. 103. [Google Scholar]
- 37.Lipinski CA, et al. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. Rev. 2001;46:3–26. doi: 10.1016/s0169-409x(00)00129-0. [DOI] [PubMed] [Google Scholar]
- 38.Andrews PR, et al. Functional group contributions to drug-receptor interactions. J. Med. Chem. 1984;27:1648–1657. doi: 10.1021/jm00378a021. [DOI] [PubMed] [Google Scholar]
- 39.Lee KW, et al. The roles of polyphenols in cancer chemoprevention. Biofactors. 2006;26:105–121. doi: 10.1002/biof.5520260202. [DOI] [PubMed] [Google Scholar]
- 40.Angel P, et al. The role of Jun, Fos and the AP-1 complex in cell-proliferation and transformation. Biochim. Biophys. Acta. 1991;1072:129–157. doi: 10.1016/0304-419x(91)90011-9. [DOI] [PubMed] [Google Scholar]
- 41.Dong Z, et al. Effects of tea polyphenols on the signal transduction pathways. Adv. Exp. Med. Biol. 2001;492:55–67. doi: 10.1007/978-1-4615-1283-7_6. [DOI] [PubMed] [Google Scholar]
- 42.Bode AM, et al. Cancer prevention by food factors through targeting signal transduction pathways. Nutrition. 2004;20:89–94. doi: 10.1016/j.nut.2003.09.016. [DOI] [PubMed] [Google Scholar]
- 43.Sun Y, et al. No point mutation of Ha-ras or p53 genes expressed in preneoplastic-to-neoplastic progression as modeled in mouse JB6 cell variants. Mol. Carcinog. 1993;8:49–57. doi: 10.1002/mc.2940080111. [DOI] [PubMed] [Google Scholar]