Abstract
Levels of the peptide hormone hepcidin negatively correlate with systemic iron status and are increased in disorders in which iron metabolism is secondarily dysregulated, such as the anemia of chronic disease. Consequently, the ability to measure hepcidin in the clinical setting may have diagnostic value for a broad range of indications. We describe a novel quantitative MALDI-TOF mass spectrometry assay for hepcidin in human urine which involves, i) direct enrichment from minute volumes (5 μL) of minimally treated urine on the surface of a functionalized chip, ii) quantification by the use of a stable isotope labeled internal standard and iii) analysis by MALDI-TOF. Performance features include: a wide linear range (1–1000 nM; LOQ 2.5 nM), high accuracy (90–110% recovery) and precision (intra-day CV 12.11%; inter-day CV 13.21%), and a strong correlation upon inter-laboratory cross validation with an existing immunoassay. The assay is simple, accurate, efficient, and the high throughput performance features of the assay make large-scale clinical research studies feasible.
Keywords: hepcidin, iron, urine, mass spectrometry, MALDI-TOF
Introduction
Hepcidin is a small peptide hormone produced by the liver that functions as the master regulator of iron metabolism in mammalian systems. Hepcidin is produced predominantly by hepatocytes as an 84-amino acid (84-aa) precursor, that is processed intracellularly into a 64-aa propeptide and then cleaved extracellularly to yield a 25-aa (HepC-25, 2789.4 Da) mature, bioactive form1–3. Hepcidin acts to regulate iron metabolism by binding directly to the transmembrane iron exporter, ferroportin, on the surface of intestinal enterocytes and macrophages, causing its internalization and degradation4. In doing so, hepcidin acts as a negative regulator of iron efflux from the intestine and iron release from recycling macrophages5; consequently, high hepcidin levels serve to down regulate systemic iron stores and limit macrophage recycling of iron derived from senescent red blood cells.
Since its initial description in the year 2000, several hundred reports have appeared in the literature regarding the role of hepcidin in health and disease2, 3. In uncomplicated iron deficiency, serum or urinary hepcidin is very low or undetectable6, whereas in transfusional iron overload hepcidin is increased. Dysregulation of hepcidin expression has been linked with a number of disorders, including the anemia of inflammation/chronic disease7–11, and the anemia of renal failure, where hepcidin levels are inappropriately elevated. On the other hand, in most forms of hereditary hemochromatosis, hepcidin levels are inappropriately reduced for systemic iron stores, with the degree of reduction roughly correlating with the severity of the disease12–15.
Despite the obvious significance of hepcidin and its potential as a clinical indicator of iron status, no commercial assay is presently available. Several immunochemical and mass spectrometric methods have been described, yet each is methodologically suboptimal due to limited accuracy, applicability for routine diagnostics, or utility for large clinical research studies. For example, common antibody-based methods have not been shown to discriminate between biologically active HepC-25 and biologically inactive, shorter 22 and 20 amino acid forms that are variably abundant in urine or serum16, 17. Previously described SELDI-TOF methods are challenged by low mass resolution, low throughput, and in certain cases suboptimal quantitative accuracy18–20. SELDI measurements, which are collected in linear mode and lack of isotopic resolution, are susceptible to detection interferences which impair accurate quantification using peak area data. Furthermore, the use of internal standards with characteristics which differ significantly from the analyte, such as an artificially truncated hepcidin with distinctive biophysical properties, is unsuitable for accurate quantification especially when selective enrichment protocols are applied21. Published LC-MS/MS methods offer high sensitivity, and, with the use of a stable isotope internal standard, high accuracy, but are generally restricted by low throughput workflows22, 23. A recently described method involving off-line WCX magnetic bead-based enrichment prior to traditional dried droplet spotting and MALDI-TOF analysis benefits from isotopic resolution and enhanced accuracy compared with SELDI, however, the high throughput capacity of the assay and it’s applicability in serum or plasma were not demonstrated24.
The methodological complexities and restrictions of existing methods limit their potential use in large scale clinical applications, which are often resource intensive, demand high sample throughput, and, in certain cases, may be constrained by limited sample volumes. To this end, we have focused upon the major limitations of existing methods and have made several key advances towards stream-lining sample preparation and analysis in the context of the MALDI-TOF MS platform. Specifically, we have coupled selective “on-chip” enrichment with matrix application through the use of a novel functionalized matrix pre-spotted MALDI chip, thus eliminating the need for separate offline enrichment and matrix addition steps. Using a stable isotope labeled synthetic hepcidin internal standard we measured linear range, analytical sensitivity, accuracy, and precision in the course of assay validation. The assay is simple and scalable and has the capacity to process several hundred samples per run, which should make large-scale analysis of clinical research studies feasible. We demonstrate proof-of-concept and its high-throughput potential by cross validation against an immunoassay method, processing 60 samples in duplicate in a single run.
Experimental Section
Urine samples were collected from subjects suspected to have inherited disorders of iron metabolism and their immediate family members. All subjects were recruited and informed consent was obtained under the auspices of a human subject’s research protocol approved by the Children’s Hospital Boston Committee on Clinical Investigation. Clean catch urine samples were collected in the early morning, and, when possible, after fasting. Samples collected nationally were shipped priority overnight at 4°C while those collected internationally were shipped on replenished dry ice (−20°C). Overnight incubation at +4°C had no observable effects on samples. Upon receipt, urine samples were centrifuged at 2800 g for 10 minutes and the supernatant was aliquoted into 1.5 mL microtubes and then frozen at −80°C. Samples collected locally were immediately processed and stored frozen at −80°C.
All solvents were HPLC grade purity (Burdick and Jackson). Synthetic hepcidin (HepC-25) was synthesized by Bachem (Torrance, CA) and stable isotope labeled synthetic HepC-25(Lys15N213C6)2 was synthesized by Sigma-Genosys (Woodlands, TX). Concentrations of peptide stocks in water were determined in duplicate by amino acid analysis (Molecular Biology Core Facilities at Dana Farber Cancer Institute, Boston, MA). All commercial/synthetic peptide solutions were stored at −20°C; working stocks were kept at +4°C under argon. Mass Spec Turbo Chips (384-spot format; 600 μm) and finishing solution were obtained from QIAGEN (Hilden, Germany). MALDI-TOF experiments were carried out on an Applied Biosystems/MDS SCIEX 4800 instrument maintained at the Blais Proteomics Center and Molecular Biology Core Facilities at Dana Farber Cancer Institute.
The assay was based on the detection and quantification of endogenous hepcidin relative to that of a stable isotope labeled hepcidin internal standard added to samples at a known concentration. The HepC-25(Lys15N213C6)2 and HepC-25 synthetic peptides exhibited indistinguishable MS/MS CID fragmentation as determined by MALDI-TOF/TOF experiments. Each hepcidin form exhibited linear detection over the concentration range investigated with the sole distinguishing feature being the observed difference in [M+H]+ (monoisotopic masses: HepC-25 2788.2 Da, HepC-25(Lys15N213C6)2 2804.3 Da).
Urine samples stored at −80°C were thawed on ice and vortexed briefly. TFA (5%) was added 1:5 (10 μL) to the urine samples (50 μL) to normalize the pH (pH 2–3). Internal standard in water was added at 1/20 total sample volume (3 μL in 57 μL urine). Samples were mixed and centrifuged at 2800 g for 10 min. +4°C in an Eppendorf 5402 microcentrifuge. In situ hepcidin enrichment was performed by incubating treated urine (5 μL) on the Mass Spec Turbo Chip spot surface for 20 min. under controlled environmental conditions (20°C, 50% relative humidity). Following incubation, urine was removed and three brief (10 sec) washes (5 μL) were performed using finishing solution prior to MALDI-TOF MS analysis.
A standard peptide mixture deposited onto designated spots was used for external chip calibrations. Positive ion reflector mode MS spectra are the average of 1000 shots. Raw data files were exported and analyzed using Data Explorer software (Applied Biosystems v.4.9, Foster City, CA). Offline analysis and graphing was performed using Excel (Microsoft Office 2007, Redmond, WA) and Origin (OriginLab v.7.0, Springfield, MA). Spectra were analyzed manually and relative intensities were determined by obtaining the baseline corrected average of the three most abundant isotopic peaks for each HepC-25 and HepC-25(Lys15N213C6)2 peak series. The same isotope peaks were selected for analysis throughout experiments to ensure accuracy and consistency. The following equation was used to determine relative HepC-25 concentration from peak area data and known internal standard concentration:
HepC-25 is regular (light) hepcidin peptide and I.S. is the stable isotope labeled internal standard (heavy) synthetic peptide (HepC-25(Lys15N213C6)2).
Cross validation of the described MALDI-TOF method against an existing immunoassay method16, 17 was performed by independent analysis of a collection of clinical urine samples (n=60). Data were normalized for urine creatinine determined by standard methods25 and reported as ng hepcidin/mg creatinine. In MALDI-TOF experiments, twice thawed urine samples (47 μL) were transferred to wells of a 96-well polypropylene microplate (Greiner bio-one, Frickenhausen, Germany) containing 10 μL 5% TFA and 3 μL HepC-25(Lys15N213C6)2 internal standard (100 nM final). Samples were mixed and the plate was flooded with Argon, sealed, and spun in a refrigerated centrifuge 2000 g for 10 min at +4°C. The soluble fraction (50 μL) was transferred to a pre-chilled 96-well microplate for spotting onto the MALDI chip.
Results and Discussion
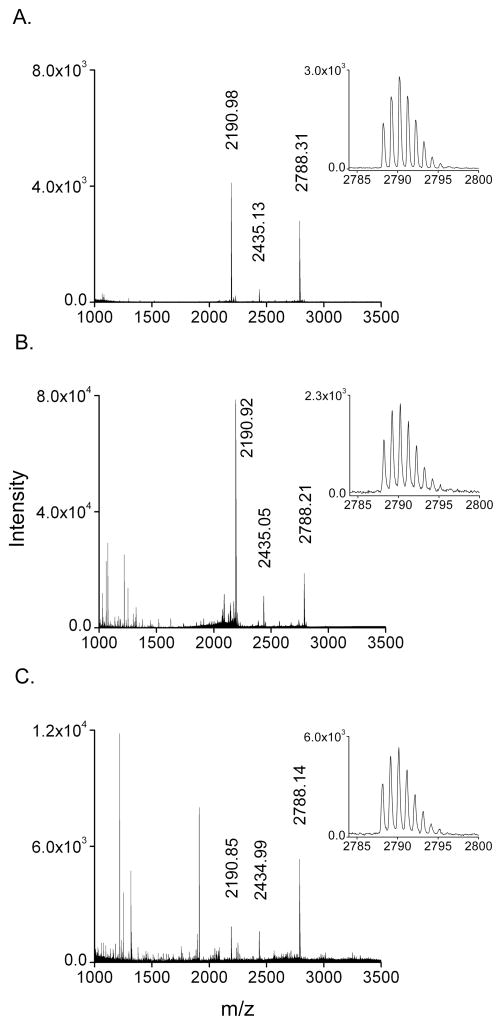
Figure 1 illustrates a powerful feature of the described method, the selective enrichment of hepcidin from a minute amount of minimally treated urine following a simple on-chip incubation and wash sequence. This direct enrichment approach is a departure from weak-cation exchange (WCX) extraction of hepcidin from urine or serum prior to MS analysis, a method which requires large volume sample input and results in lower throughput, additional sample handling and thus additional potential sample losses16, 20, 21, 23. The functionalized MALDI chip contains pre-deposited matrix spots prepared by vacuum sublimation onto an ultra-hydrophobic surface of extremely low wettability. Incubation of 5 μL droplets on these matrix spots (0.3 mm2) permits the simultaneous concentration and enrichment of hepcidin in the fine structured top layers of the spots, while the homogeneous matrix formulation dramatically increases MS sensitivity 26.
Figure 1.
A. All hepcidin forms (HepC-25 2788.31; HepC-22 2435.13; HepC-20 2190.98) can be detected in urine using off-line weak-cation exchange (WCX) enrichment (20-fold) upstream of traditional dried droplet deposition on a stainless steel MALDI plate. Comparisons are drawn from the first three averaged isotope peaks representing HepC-25 peak characteristics (inset) (peak area = 22969, signal/noise = 1361, resolution = 8569). B. The identical off-line WCX enrichment coupled with analysis using the functionalized MALDI chip shows similar S/N and peak resolution, and a >8-fold increase in peak area (194583; the inset is 1/10th dilution shown for uniform comparison). C. Direct analysis of a minute volume (5 μL) of minimally treated urine directly on the Turbo chip surface (without offline enrichment) shows comparable peak area (46850) and resolution (8500) as Figure 1A with a ~2-fold decrease in signal/noise. Monoisotopic masses are labeled.
In order to compare differences in detection and sensitivity, we first performed off-line WCX enrichment and examined performance when combined with either traditional dried droplet matrix preparation on a stainless steel substrate or vacuum sublimated spot analysis via the functionalized chip. We then compared these results with enrichment and analysis of urine by direct application on the functionalized chip. The results showed that, compared to dried droplet preparations, the functionalized chip preparations provided slight increases in signal-to-noise and peak resolution and a significant enhancement of HepC-25 peptide ion intensities (>8-fold) (Figure 1A, 1B). Prior investigations of sublimated matrix performance compared to dried droplet spots documented similar observed increases in peptide ion intensities, which arise from more efficient ionization and higher ion survival yields, a consequence of homogeneous matrix crystals and lower internal energies due to collisional cooling 26
Analysis of urine by direct on-chip enrichment showed the presence of HepC-25 signal with peak area and resolution similar to those from the 20-fold WCX enrichment analyzed with the dried droplet technique (Figure 1C, 1A). A significant difference, however, was the total input volume of sample; 100 μL urine for WCX (Fig. 1A and B) versus 5 μL urine for the functionalized chip (Figure 1C). Direct on-chip selective enrichment from urine arises from the partitioning of Hepcidin into the fine structured top layers of matrix, which excludes larger peptides, proteins, and contaminants that are ultimately removed through subsequent washes.
An additional feature of note involves the change in the apparent distribution of HepC peptide forms when comparing WCX with direct enrichment (compare panels 1A–1C). The cysteine disulfide bridged forms of the HepC-20 and HepC-22 peptides are more basic (pI 11.2) with respect to HepC-25 (pI 10.0), which may translate to an increase in WCX affinity and a bias in the detection of the two strongly cationic peptides (HepC-20 and HepC-22) over HepC-25. Further investigations regarding the nature of the enrichment may help clarify this observation.
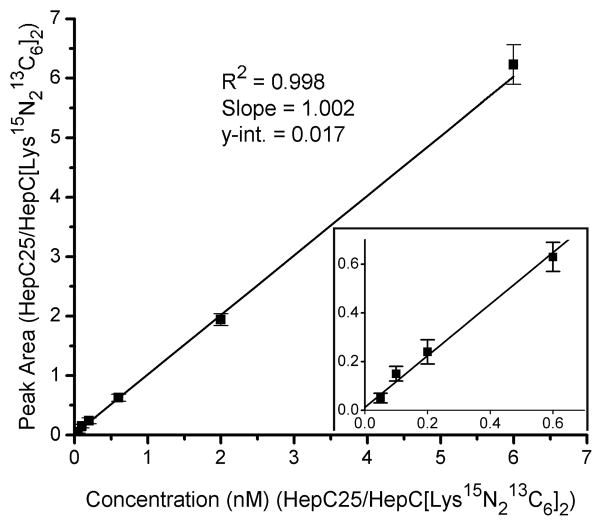
Using blank urine samples, we observed linearity over the span of three orders of magnitude (1–1000 nM; 5–5000 fmol/spot). This wide linear range may be attributed to the combination of efficient enrichment of hepcidin from urine, efficient ionization performance of the chip, and use of the stable isotope labeled internal standard, which exhibits an accurate wide linear response in relation to HepC-25. In validation experiments, a series of six dilutions of HepC-25 peptide (1–300 nM final) spiked into blank urine and two blanks (± I.S.) were analyzed (n=6) and HepC-25/HepC-25(Lys15N213C6)2 peak area (mean and S.D.) was plotted against concentration ratio (Figure 2). Mean, % recovery, and % CV values for four of the dilutions are displayed in Supplemental Table 1. The limit of detection (LOD) was 1 nM, and the limit of quantification (LOQ) was 2.5 nM in these experiments.
Figure 2.
Linearity of HepC-25 quantification using a urine sample without detectable hepcidin, a single internal standard concentration (50nM), and a series of HepC-25 synthetic peptide calibrants (2.5 – 300 nM). The ratio of average peak area of HepC25/HepC([Lys15N213C6]2) is plotted against the ratio of known HepC25/HepC([Lys15N213C6]2) concentrations (nM), and the resulting relationship is fitted with a linear regression. The slope (1.002) and y-intercept (0.017) indicate the absence of significant matrix effects or proportional error in the urine background.
In spike and recovery experiments, slight deviations in accuracy were observed with additions of large (300 nM; 98%) and intermediate (60 nM; 109%) amounts of HepC-25 compared with low (10 nM; 101%) amounts (Supplemental Table 1). Salt concentrations in urine, typically in the hundred millimolar range, can fluctuate widely based on metabolic and hydration status. Interestingly, addition of up to 1M NaCl had only modest effects on measured HepC-25, (measurement difference of 13.63 nM or 12% and compared with control sample). Long term assay precision measurements of three urine samples representing the high, mid, and low sectors of the linear range showed intra-day, inter-spot CV values ranging from 5–23% between the three samples, with an average CV 12.11% (Supplemental Table 1). The largest inter-spot CV was associated with samples near the low end of the detection range. This general trend was also observed for inter-day CV values which ranged from 8–18% with an inter-sample average CV 13.21%. Repeated (3 consecutive days) freeze/thaw treatment had no significant effects on measured hepcidin levels (Supplemental Figure 1B). Following the three day freeze/thaw cycle the HepC-25 signal was robust and the oxidized methionine species of HepC-25 appeared in very low abundance (~5%) which was also attributed to the use of argon to ensure a protective atmosphere (Supplemental Figures 1C and 3).
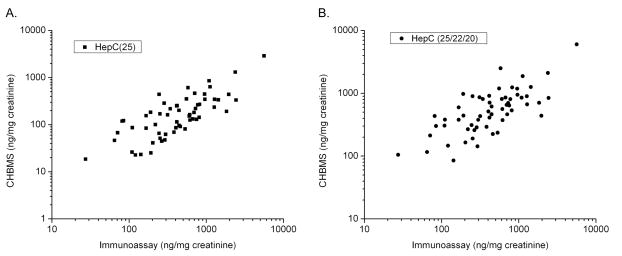
After internally validating the assay (heretofore referred to as the Children’s Hospital Boston mass spectrometry (CHBMS) hepcidin assay), we sought to cross validate the assay against a previously reported hepcidin immunoassay 16, 17. A large cohort of clinical research samples were collected under described procedures and twice-thawed aliquots were subjected to both immunoassay and CHBMS analyses (60 samples, analyzed in duplicate). Log-log comparison of CHBMS HepC-25 values with immunoassay values demonstrated a definitive correlation (Figure 3) with Deming regression slope = 0.446 (± 0.033), intercept (y-int.) = −38.9 (± 35.3), and Spearman correlation r = 0.733. While the correlation is good considering the distinct testing platforms, the slope <1 indicates the presence of a proportional systematic offset. The offset between the assays may result from signal interferences in the urine background or from differences in HepC-25/HepC-22/HepC-20 detection specificities.
Figure 3.
A. Correlation between HepC-25 levels in the urine sample set measured by the CHBMS method plotted against values determined using the immunoassay method, normalized for urine creatinine (ng hepcidin/mg creatinine) and plotted in log scale. B. HepC-25 and estimates of HepC-22 and HepC-20 based on summation of peak area are plotted in log scale against the immunoassay method.
An advantage of the described MALDI-TOF method is the ability to accurately measure HepC-25 by the use of the HepC-25(Lys15N213C6)2 internal standard while also capturing peak intensity data for HepC-22 and HepC-20. Assuming that HepC-22 and -20 have similar enrichment and ionization efficiencies, we extracted the average peak intensities of HepC-22 and HepC-20, when present, and performed a summed average of these in addition to HepC-25 and then calculated an estimated total hepcidin concentration based on the I.S. average intensity (Figure 3B). Log-log comparison of these data resulted in Deming regression slope = 0.997 (± 0.091), and intercept (y-int) = 55.1 (± 98.2), and Spearman correlation r = 0.824. The slope indicates a correction in the previously observed offset.
Further information is required to clarify these results, including knowledge of the relative specificities of the immunoassay antibody for the different hepcidin forms, as well as CHBMS MS-based quantification of the HepC-20 and HepC-22 peptides using the appropriate internal standards. Moreover, the performance of either assay in conjunction with clinical data, including additional iron markers (e.g. ferritin, hemoglobin, transferrin saturation, etc.), will help to further establish the clinical validity of each assay and may reveal interesting and important differences with regard to diagnostic potential.
Conclusions
The assay presented here offers several key advantages over previously reported methods. The novel on-chip enrichment method eliminates exogenous enrichment procedures and consumes approximately 20–100 fold lower sample volumes than existing methods 21–24. The vacuum sublimated matrix spots offer efficient hepcidin enrichment while avoiding the requirement for offline matrix preparation and spotting, thereby supporting a high throughput workflow 27. The ultra-fine homogeneous matrix composition provides a significant enhancement in sensitivity and spot-spot reproducibility compared with traditional dried droplet preparations 27. The wide dynamic detection range reported here (2.5–300 nM) exceeds that of previously reported methods19–21, 23, 24, 28, 29. Accuracy measurements were within 10% of expected values, consistent with the use of the stable isotope labeled hepcidin internal standard and overall better than those obtained using alternative internal standards 22–24.
A significant advancement in this work is the design of an accurate and precise high throughput assay which enables automated on-line sample preparation, rapid detection using large format (384/1536 spot) MALDI chips, and computational quantitative analysis using dedicated peak-picking software30. We are optimizing methods for hepcidin analysis in urine and plasma, and plan to perform cross validation against immunoassay and MS-based assays to assess accuracy and consistency in sample measurement and to compare results in the context of additional iron status markers. The high throughput MALDI-TOF format is amenable to reference laboratories and should make the analysis of large patient cohorts, as in clinical trials, feasible at a relatively low cost.
Supplementary Material
S2 Supplemental Table 1.
Assay validation figures of merit.
S3 Supplemental Figure 1.
Preanalytical factors. A. Sequence of urine treatment prior to MALDI-TOF analysis, B. Consecutive freeze/thaw treatments, C. Spectra following third day of freeze/thaw treatment, D. Influence of I.S. on HepC peak intensity.
S4 Supplemental Figure 2.
Representative set of spectra from urine cross validation. Inset: magnified view showing isotopic peak series for HepC-25 and the internal standard
S5-7 Supplemental Figure 3.
Representative set of spectra demonstrating methionine oxidation and the concurrent appearance of metastable decay peaks originating from neutral loss of the sulfoxide ion (CH3SOH).
Supplemental Figure 1. Assessment of pre-analytical factors impacting assay performance. A. The sequence of urine treatment prior to direct on-chip enrichment and MALDI-TOF analysis. Method 1 – centrifugation, acidification, internal standard addition, Method 2 – acidification, centrifugation, I.S. addition, Method 3 – I.S. addition, acidification, centrifugation, Method 4 – no centrifugation step. Shown are samples from a single urine sample analyzed in duplicate, errors bars are S.D. B. The effects of successive freeze/thaw cycles on HepC-25 measurement assessed using five urine samples tested over a three day period, values are normalized for urine creatinine. C. Representative spectra showing a sample with and without internal standard following the third day of freeze/thaw treatment. D. Assessment of the potential influence of internal standard on HepC-25 ion suppression. Five urine samples were analyzed with and without added internal standard and peak area of HepC-25 was plotted.
Supplemental Figure 2. Representative spectra from urine sample cross validation. A. Spectrum of sample in the absence of I.S. B. Magnified section of the spectrum detailing the HepC-25 peak isotope series; note the absence of methionine oxidation (+16 m/z). C. Spectrum of sample with added I.S. (100 nM). D. Magnified section of the spectrum detailing the HepC-25 and HepC-25(Lys15N213C6)2 peak isotope series.
Supplemental Figure 3. Previous reports have noted potential assay interference caused by hepcidin methionine oxidation, likely arising from handling and manipulation or urine under oxidative conditions during the enrichment phase of hepcidin analysis 1–3. All described methods to date involve incubation/agitation (typically ≥30 min at r.t.) of urine at neutral pH with solid phase binding substrate (WCX or IMAC SELDI or IEX resin) prior to elution and analysis. We observed nominal oxidation in our experiments as the described method involves minimal handling (time and sample transfers) and thus minimal sample exposure to oxidative conditions (see text and Supplemental Figures 1, 2). Nonetheless, in order to investigate the effects of potential oxidation, we incubated a normal urine sample (Figure 3 panel a) with 0.6% H2O2 to induce oxidation prior to WCX enrichment and analysis (Figure 3 panel B). Interestingly, we observed the appearance of metastable decay peaks (Δ64 m/z (peaks 2740.45 and 2757.56) in the oxidized sample representing an apparent loss of the sulfoxide (CH3SOH) from oxidized methionine, which arises from post source decay of oxidized methionine in the mass spectrometer. Although we anticipate no confounding issues regarding oxidation in our studies, the observation of the intense metastable decay ions is useful as a diagnostic (or correction factor) for methods involving extended sample handling and enrichment under oxidative conditions, or in the assessment of clinical samples that may have been handled under less than ideal conditions.
Table 1.
Linearity, accuracy, and precision measurements for assay validation.
| Linearity | ||||
| Concentration (nM) | 300 | 100 | 30 | 10 |
| Mean (nM) n=6 | 311.42 | 97.22 | 31.38 | 10.10 |
| Accuracy (% Recovery) n=6 | 103.81 | 97.22 | 104.6 | 101 |
| Precision (% CV) n=6 | 5.30 | 5.21 | 10.05 | 13.17 |
| Dilution Linearity | Sample 1 | Dilution 1 | Dilution 2 | Dilution 3 |
| Concentration (nM) | 101.12 | 33.71 | 16.85 | 8.43 |
| Mean (nM) | 94.50 | 34.11 | 16.80 | 7.59 |
| Accuracy (% Recovery) | 93.45 | 101.19 | 99.70 | 90.04 |
| Accuracy | ||||
| Sample Recovery | Sample 1 | Recovery 1 | Recovery 2 | Recovery 3 |
| Mean (nM) | 40.90 | 50.96 | 106.45 | 334.94 |
| HepC-25 Added (nM) | 10 | 60 | 300 | |
| Recovered (nM) | 10.06 | 65.55 | 294.04 | |
| Recovery (%) | 101 | 109 | 98 | |
| Analyte Interference | Sample 1 | Interference 1 | Interference 2 | |
| Mean (nM) | 101.12 | 114.74 | 108.89 | |
| NaCl Added (M) | 0 | 0.25 | 1.0 | |
| Interference (nM) | 13.63 | 7.78 | ||
| Precision | ||||
| Inter-spot/Inter-day | Measurements | Sample 1 | Sample 2 | Sample 3 |
| Day 1 | Mean (nM) | 106.05 | 25.33 | 10.30 |
| Precision (%CV) | 5.75 | 11.28 | 20.47 | |
| n | 6 | 6 | 4 | |
| Day 2 | Mean (nM) | 110.77 | 30.23 | 11.97 |
| Precision (%CV) | 7.38 | 11.41 | 10.04 | |
| n | 6 | 6 | 6 | |
| Day 3 | Mean (nM) | 101.79 | 29.76 | 11.78 |
| Precision (%CV) | 9.43 | 12.42 | 23.63 | |
| n | 6 | 6 | 6 | |
| Total | Mean (nM) | 106.20 | 28.44 | 11.48 |
| Precision (%CV) | 7.92 | 13.65 | 18.38 | |
| n | 18 | 18 | 16 | |
Acknowledgments
The authors thank Dean Campagna for assistance with sample processing, and Mark Westerman from Intrinsic Life Sciences for making the hepcidin immunoassay available on a research basis. The authors also thank Jim Lee and Will Beavers for technical assistance and Mark Kellogg and Robert Everley for critical review of the manuscript. Synthetic HepC-25 was a generous gift from Ellis Neufeld, Division of Hematology/Oncology, and Children’s Hospital Boston. Dalila Bensaddek is acknowledged for experiments performed in the earliest phase of this project. This work was supported in part by the Children’s Hospital Boston Department of Pathology. D.S.A. is supported by NIH T32 HL076115. M.H.H. is supported by K12 HL087164. M.D.F. is supported by R01 DK080011.
Abbreviations
- MALDI-TOF
matrix-assisted laser desorption ionization/time of flight
- HepC-20/22/25
hepcidin 20, 22, and 25 amino acid forms
- MS
mass spectrometry
- SELDI-TOF
surface enhanced laser desorption ionization/time of flight
- LC-MS/MS
liquid chromatography tandem mass spectrometry
- HPLC
high performance liquid chromatography
- CHCA
alpha-cyano-4-hydroxy cinnamic acid
- TFA
trifluoroacetic acid
- WCX
weak cation exchange
- I.S
internal standard
- C.V
coefficient of variation
- CHBMS
Children’s Hospital Boston Mass Spectrometry hepcidin assay
Footnotes
Conflict of interest
U.R. and C.M. are employees of QIAGEN GmbH, Hilden, Germany. The other authors have no relevant conflicts to disclose.
References
- 1.Valore EV, Ganz T. Blood Cells Mol Dis. 2008;40:132–138. doi: 10.1016/j.bcmd.2007.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Krause A, Neitz S, Magert HJ, Schulz A, Forssmann WG, Schulz-Knappe P, Adermann K. FEBS Lett. 2000;480:147–150. doi: 10.1016/s0014-5793(00)01920-7. [DOI] [PubMed] [Google Scholar]
- 3.Park CH, Valore EV, Waring AJ, Ganz T. J Biol Chem. 2001;276:7806–7810. doi: 10.1074/jbc.M008922200. [DOI] [PubMed] [Google Scholar]
- 4.Nemeth E, Tuttle MS, Powelson J, Vaughn MB, Donovan A, Ward DM, Ganz T, Kaplan J. Science. 2004;306:2090–2093. doi: 10.1126/science.1104742. [DOI] [PubMed] [Google Scholar]
- 5.Ganz T. Best Pract Res Clin Haematol. 2005;18:171–182. doi: 10.1016/j.beha.2004.08.020. [DOI] [PubMed] [Google Scholar]
- 6.Nemeth E, Ganz T. Haematologica. 2006;91:727–732. [PubMed] [Google Scholar]
- 7.Weinstein DA, Roy CN, Fleming MD, Loda MF, Wolfsdorf JI, Andrews NC. Blood. 2002;100:3776–3781. doi: 10.1182/blood-2002-04-1260. [DOI] [PubMed] [Google Scholar]
- 8.Kearney SL, Nemeth E, Neufeld EJ, Thapa D, Ganz T, Weinstein DA, Cunningham MJ. Pediatr Blood Cancer. 2007;48:57–63. doi: 10.1002/pbc.20616. [DOI] [PubMed] [Google Scholar]
- 9.Nemeth E, Rivera S, Gabayan V, Keller C, Taudorf S, Pedersen BK, Ganz T. J Clin Invest. 2004;113:1271–1276. doi: 10.1172/JCI20945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee P, Peng H, Gelbart T, Beutler E. Proc Natl Acad Sci U S A. 2004;101:9263–9265. doi: 10.1073/pnas.0403108101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Roy CN, Custodio AO, de Graaf J, Schneider S, Akpan I, Montross LK, Sanchez M, Gaudino A, Hentze MW, Andrews NC, Muckenthaler MU. Nat Genet. 2004;36:481–485. doi: 10.1038/ng1350. [DOI] [PubMed] [Google Scholar]
- 12.Bridle KR, Frazer DM, Wilkins SJ, Dixon JL, Purdie DM, Crawford DH, Subramaniam VN, Powell LW, Anderson GJ, Ram GA. Lancet. 2003;361:669–673. doi: 10.1016/S0140-6736(03)12602-5. [DOI] [PubMed] [Google Scholar]
- 13.Drakesmith H, Schimanski LM, Ormerod E, Merryweather-Clarke AT, Viprakasit V, Edwards JP, Sweetland E, Bastin JM, Cowley D, Chinthammitr Y, Robson KJ, Townsend AR. Blood. 2005;106:1092–1097. doi: 10.1182/blood-2005-02-0561. [DOI] [PubMed] [Google Scholar]
- 14.Papanikolaou G, Samuels ME, Ludwig EH, MacDonald ML, Franchini PL, Dube MP, Andres L, MacFarlane J, Sakellaropoulos N, Politou M, Nemeth E, Thompson J, Risler JK, Zaborowska C, Babakaiff R, Radomski CC, Pape TD, Davidas O, Christakis J, Brissot P, Lockitch G, Ganz T, Hayden MR, Goldberg YP. Nat Genet. 2004;36:77–82. doi: 10.1038/ng1274. [DOI] [PubMed] [Google Scholar]
- 15.Huang FW, Pinkus JL, Pinkus GS, Fleming MD, Andrews NC. J Clin Invest. 2005;115:2187–2191. doi: 10.1172/JCI25049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nemeth E, Valore EV, Territo M, Schiller G, Lichtenstein A, Ganz T. Blood. 2003;101:2461–2463. doi: 10.1182/blood-2002-10-3235. [DOI] [PubMed] [Google Scholar]
- 17.Ganz T, Olbina G, Girelli D, Nemeth E, Westerman M. Blood. 2008;112:4292–4297. doi: 10.1182/blood-2008-02-139915. [DOI] [PubMed] [Google Scholar]
- 18.Kemna E, Tjalsma H, Laarakkers C, Nemeth E, Willems H, Swinkels D. Blood. 2005;106:3268–3270. doi: 10.1182/blood-2005-05-1873. [DOI] [PubMed] [Google Scholar]
- 19.Tomosugi N, Kawabata H, Wakatabe R, Higuchi M, Yamaya H, Umehara H, Ishikawa I. Blood. 2006;108:1381–1387. doi: 10.1182/blood-2005-10-4043. [DOI] [PubMed] [Google Scholar]
- 20.Kemna EH, Tjalsma H, Podust VN, Swinkels DW. Clin Chem. 2007;53:620–628. doi: 10.1373/clinchem.2006.079186. [DOI] [PubMed] [Google Scholar]
- 21.Swinkels DW, Girelli D, Laarakkers C, Kroot J, Campostrini N, Kemna EH, Tjalsma H. PLoS ONE. 2008;3:e2706. doi: 10.1371/journal.pone.0002706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Murphy AT, Witcher DR, Luan P, Wroblewski VJ. Blood. 2007 doi: 10.1182/blood-2006-11-057471. [DOI] [PubMed] [Google Scholar]
- 23.Bansal SS, Halket JM, Bomford A, Simpson RJ, Vasavda N, Thein SL, Hider RC. Anal Biochem. 2009;384:245–253. doi: 10.1016/j.ab.2008.09.045. [DOI] [PubMed] [Google Scholar]
- 24.Bansal SS, Halket JM, Fusova J, Bomford A, Simpson RJ, Vasavda N, Thein SL, Hider RC. Rapid Commun Mass Spectrom. 2009;23:1531–1542. doi: 10.1002/rcm.4033. [DOI] [PubMed] [Google Scholar]
- 25.Slot C. Scand J Clin Lab Invest. 1965;17:381–387. doi: 10.3109/00365516509077065. [DOI] [PubMed] [Google Scholar]
- 26.Poetsch A, Schlusener D, Florizone C, Eltis L, Menzel C, Rogner M, Steinert K, Roth U. J Biomol Tech. 2008;19:129–138. [PMC free article] [PubMed] [Google Scholar]
- 27.Jaskolla TW, Karas M, Roth U, Steinert K, Menzel C, Reihs K. J Am Soc Mass Spectrom. 2009 doi: 10.1016/j.jasms.2009.02.010. [DOI] [PubMed] [Google Scholar]
- 28.Ward DG, Roberts K, Stonelake P, Goon P, Zampronio CG, Martin A, Johnson PJ, Iqbal T, Tselepis C. Proteome Sci. 2008;6:28. doi: 10.1186/1477-5956-6-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Altamura S, Kiss J, Blattmann C, Gilles W, Muckenthaler MU. Biochimie. 2009 doi: 10.1016/j.biochi.2009.04.010. [DOI] [PubMed] [Google Scholar]
- 30.Renard BY, Kirchner M, Steen H, Steen JA, Hamprecht FA. BMC Bioinformatics. 2008;9:355. doi: 10.1186/1471-2105-9-355. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
S2 Supplemental Table 1.
Assay validation figures of merit.
S3 Supplemental Figure 1.
Preanalytical factors. A. Sequence of urine treatment prior to MALDI-TOF analysis, B. Consecutive freeze/thaw treatments, C. Spectra following third day of freeze/thaw treatment, D. Influence of I.S. on HepC peak intensity.
S4 Supplemental Figure 2.
Representative set of spectra from urine cross validation. Inset: magnified view showing isotopic peak series for HepC-25 and the internal standard
S5-7 Supplemental Figure 3.
Representative set of spectra demonstrating methionine oxidation and the concurrent appearance of metastable decay peaks originating from neutral loss of the sulfoxide ion (CH3SOH).
Supplemental Figure 1. Assessment of pre-analytical factors impacting assay performance. A. The sequence of urine treatment prior to direct on-chip enrichment and MALDI-TOF analysis. Method 1 – centrifugation, acidification, internal standard addition, Method 2 – acidification, centrifugation, I.S. addition, Method 3 – I.S. addition, acidification, centrifugation, Method 4 – no centrifugation step. Shown are samples from a single urine sample analyzed in duplicate, errors bars are S.D. B. The effects of successive freeze/thaw cycles on HepC-25 measurement assessed using five urine samples tested over a three day period, values are normalized for urine creatinine. C. Representative spectra showing a sample with and without internal standard following the third day of freeze/thaw treatment. D. Assessment of the potential influence of internal standard on HepC-25 ion suppression. Five urine samples were analyzed with and without added internal standard and peak area of HepC-25 was plotted.
Supplemental Figure 2. Representative spectra from urine sample cross validation. A. Spectrum of sample in the absence of I.S. B. Magnified section of the spectrum detailing the HepC-25 peak isotope series; note the absence of methionine oxidation (+16 m/z). C. Spectrum of sample with added I.S. (100 nM). D. Magnified section of the spectrum detailing the HepC-25 and HepC-25(Lys15N213C6)2 peak isotope series.
Supplemental Figure 3. Previous reports have noted potential assay interference caused by hepcidin methionine oxidation, likely arising from handling and manipulation or urine under oxidative conditions during the enrichment phase of hepcidin analysis 1–3. All described methods to date involve incubation/agitation (typically ≥30 min at r.t.) of urine at neutral pH with solid phase binding substrate (WCX or IMAC SELDI or IEX resin) prior to elution and analysis. We observed nominal oxidation in our experiments as the described method involves minimal handling (time and sample transfers) and thus minimal sample exposure to oxidative conditions (see text and Supplemental Figures 1, 2). Nonetheless, in order to investigate the effects of potential oxidation, we incubated a normal urine sample (Figure 3 panel a) with 0.6% H2O2 to induce oxidation prior to WCX enrichment and analysis (Figure 3 panel B). Interestingly, we observed the appearance of metastable decay peaks (Δ64 m/z (peaks 2740.45 and 2757.56) in the oxidized sample representing an apparent loss of the sulfoxide (CH3SOH) from oxidized methionine, which arises from post source decay of oxidized methionine in the mass spectrometer. Although we anticipate no confounding issues regarding oxidation in our studies, the observation of the intense metastable decay ions is useful as a diagnostic (or correction factor) for methods involving extended sample handling and enrichment under oxidative conditions, or in the assessment of clinical samples that may have been handled under less than ideal conditions.