Abstract
The Ets family of transcription factors is composed of more than 30 members. One of its members, Elf3, is expressed in virtually all epithelial cells as well as in many tumors, including breast tumors. Several studies observed that the promoter of the type II TGF-β receptor gene (TβR-II) is strongly stimulated by Elf3 via two adjacent Elf3 binding sites, A-site and B-site. Here we report the 2.2 Å resolution crystal structure of a mouse Elf3 C-terminal fragment, containing the DNA-binding Ets domain, in complex with the B-site of mouse type II TGF-β receptor promoter DNA (mTβR-IIDNA). Elf3 contacts the core GGAA motif of the B-site from major groove similar to that of known Ets proteins. However, unlike other Ets proteins, Elf3 also contacts sequences of the A-site from the minor groove of the DNA. DNA binding experiments and cell-based transcription studies indicate that minor groove interaction by Arg349 located in the Ets domain is important for Elf3 function. Equally interesting, previous studies have shown that the C-terminal region of Elf3, which flanks the Ets domain, is required for Elf3 binding to DNA. In this study, we determined that Elf3 amino acid residues within this flanking region, including Trp361, are important for the structural integrity of the protein as well as for the Efl3 DNA binding and transactivation activity.
Keywords: Elf3, Ets domain, protein-DNA complex, type II TGF-beta receptor, crystal structure
Introduction
E74-like factor-3 (Elf3) is an Ets transcription factor family member involved in the expression of at least 10 genes1. Elf3 works with other transcription factors to achieve specificity and to regulate genes involved in inflammation, differentiation, tumorigenesis, and metastasis2; 3; 4; 5; 6; 7; 8; 9; 10; 11. Elf3 has been identified in a wide range of epithelial carcinoma cells, and it is aberrantly expressed in cancers of the lung and breast12. In recent years, studies in several cell culture model systems have shown that the promoter of the type II TGF-β-receptor (TβR-II) gene is transactivated by Elf3. The TβR-II gene behaves as a tumor suppressor gene in many contexts, and it is expressed in nearly all cell types. The TβR-II gene is strongly stimulated by Elf3 via two adjacent Elf3 binding sites (A-site and B-site) in differentiated cells derived from mouse F9 embryonal carcinoma cells13; 14. Moreover, the forced expression of Elf3 in Hs578t breast cancer cells dramatically elevates the expression of TβR-II and decreases the tumorigenicity of these cells4. Given that both the A-site and the B-site are required for full promoter activity and the DNA binding domain of Elf3 appears to form a ternary complex in vitro with DNA containing both the A-sited and the B-site, it is likely that the stoichiometry of binding for Elf3 to this promoter is 2:113; 14.
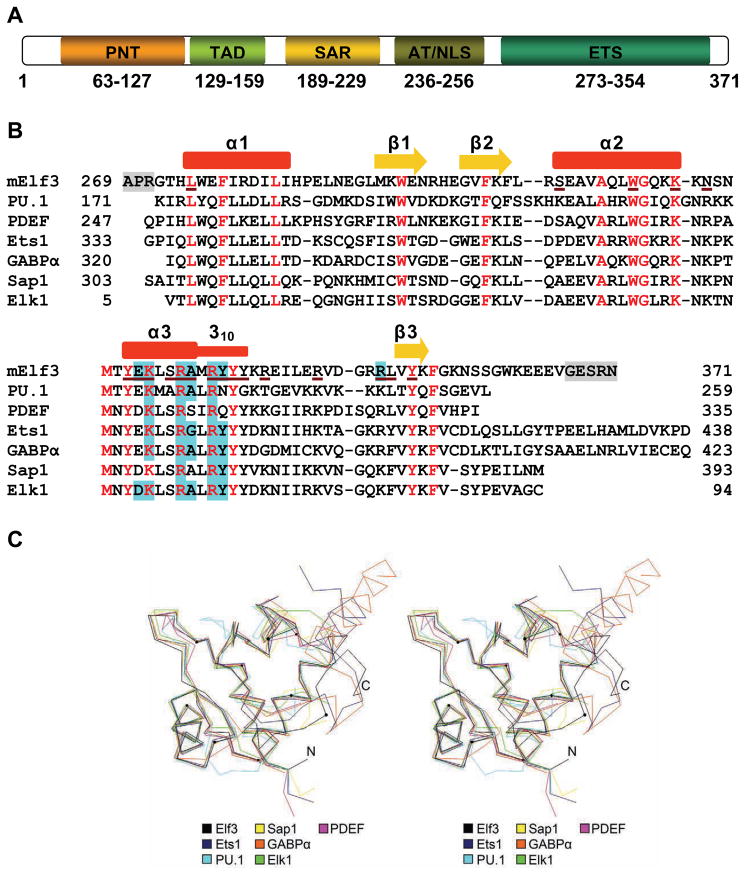
Elf3 is composed of five defined domains: a pointed domain, a transactivation domain (TAD), a serine and aspartic acid-rich (SAR) domain, an AT-hook domain, and an Ets domain (Figure 2A)1. The C-terminal Ets domain is a conserved DNA binding domain approximately 85 amino acids in length that is shared among the members of the Ets family15; 16; 17. Ets domains bind specifically to a core GGAA/T motif of DNA often referred to as an Ets-binding site (EBS)18; 19; 20. Structural analysis of Ets proteins reveals topological similarities in interactions with DNA21; however, the structural basis for the contribution of DNA sequences flanking the EBS is not well understood. To achieve a deeper understanding of the structural basis of the Ets domain binding to mouse TβR-II (mTβR-II) promoter DNA, we initiated structural studies of the mouse Elf3 Ets domain in complex with a mTβR-II promoter DNA containing a B-site and half of an A-site (Figure 1A)1; 13; 14. X-ray analysis revealed Elf3·DNA base interactions with the B-site GGAA core motif in the DNA major groove. In addition, interactions were found in the major and minor grooves of the 5′-TGTTT-3′ region of DNA. DNA binding experiments and cell-based transcription studies confirmed the importance of minor groove interactions for function of Elf3. Furthermore, the structural analysis and functional studies described here indicate that residues (355–361) C-terminal to Ets domain are necessary for the proper folding and function of Elf3. These findings provide a structural explanation for earlier work, which demonstrated that this region is required for in vivo binding of Elf3 to the promoter of the TβR-II gene1.
Figure 2.
Elf3 domain structure and sequence alignment. (A) Schematic diagram showing the domains of Elf3. (B) Structure-based sequence alignment of the DNA-bound Ets domains of Elf3, PDEF (1yo5), PU.1 (1pue), Sap1 (1k60), Elk1 (1dux), Ets1 (1k79), and GABPα (1awc). The secondary structure elements of Elf3 are indicated above the corresponding sequence. Helices are depicted by red rectangles and β strands as yellow arrows. The α3 helix terminates with a distorted 310 helix. Elf3 residues missing from the structure are highlighted in grey. Conserved residues are shown in red. The Elf3 residues interacting with DNA are underlined with brown color. Amino acid residues of Ets domains in direct contact with DNA bases are highlighted in cyan. (C) A stereoview of the three-dimensional alignment of Ets domains in the crystal structures of DNA-bound Elf3, PDEF, PU.1, Sap1, Elk1, Ets1, and GABPα. The α-carbon traces are shown as color-coded lines. The α-carbon position of every tenth amino acid residue of Elf3, starting from residue 280, is marked by a small circle.
Figure 1.
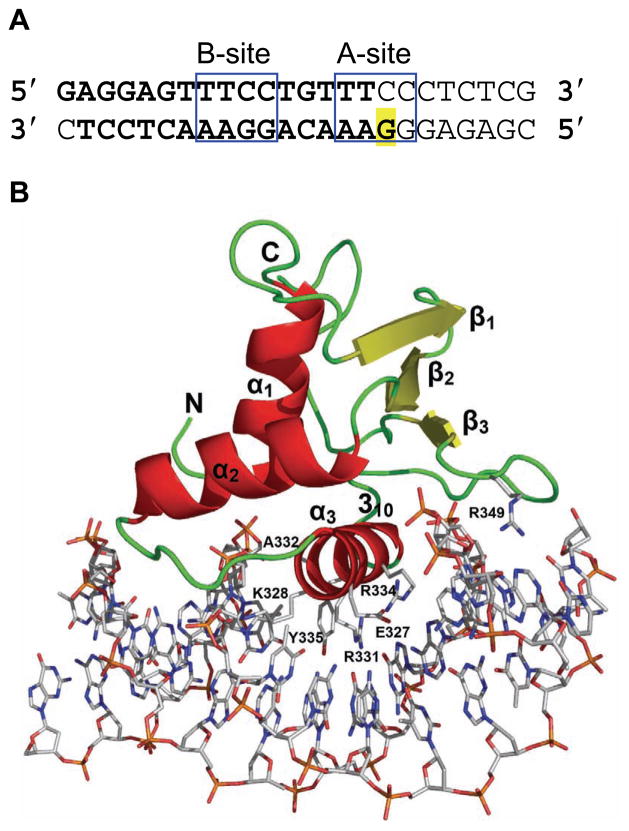
Overall view of mElf3269–371 bound to a B-site of mTβR-IIDNA. (A) mTβR-IIDNA sequences containing A and B Elf3 binding sites. The core regions of A- and B-sites are boxed. The DNA fragment used for crystallization is shown in bold. Highlighted G is replaced by C to enhance the crystallization36. (B) The structure of the mElf3269–371·mTβR-IIDNA complex. The protein is drawn as a ribbon diagram and DNA is represented as a stick diagram. The amino acid residues involved in direct DNA base interactions are also drawn in stick form.
Results and Discussion
Overall structure
We have determined the structure of the Ets domain-containing C-terminal part of the mouse Elf3 in complex with the B-site of mTβRII promoter DNA (Figure 1A). The mElf3269–371·mTβR-II dsDNA complex revealed protein interactions with the DNA bases and backbone both in the major and minor grooves (Figures 1B, 2B and 3). The Elf3 Ets domain displays an α/β architecture with three helices packed against three antiparallel β-strands similar to the ‘winged helix-turn-helix’ (wHTH) topology found in other Ets proteins22; 23; 24; 25. Superimpositions of Ets domain Cα positions in Elf3 and the crystal structures of PDEF (1yo5)21, PU.1 (1pue)17, Sap1 (1k6o)26, Elk1 (1dux)27, Ets1 (1k79)28, and GABPα (1awc)22 result in root mean square (rms) deviations of less than 0.97 Å (Figure 2C). The Elf3 Ets domain is embedded in mTβR-II dsDNA mainly through recognition helix 3 contacts (Figures 1B and 3B). The N-terminal part of helix 3 is an α-helix while the C-terminal part is a distorted 310-helix (Figures 1B and 2B). Similar observations were seen in this highly conserved tyrosine rich helix region of previous Ets domain crystal structures, including Sap126, GABPα22 and Ets128. The α1 helix makes only one DNA backbone contact through the Leu275 main chain, which is a typical feature of DNA recognition by Ets domains. Residues in the loop connecting helix 3 and β3 strand, part of the α2-helix, and the β3 strand are also involved in the protein·DNA interaction (Figures 1B and 2B). In addition, several water molecules mediate protein·DNA interactions (Figure 3A).
Figure 3.
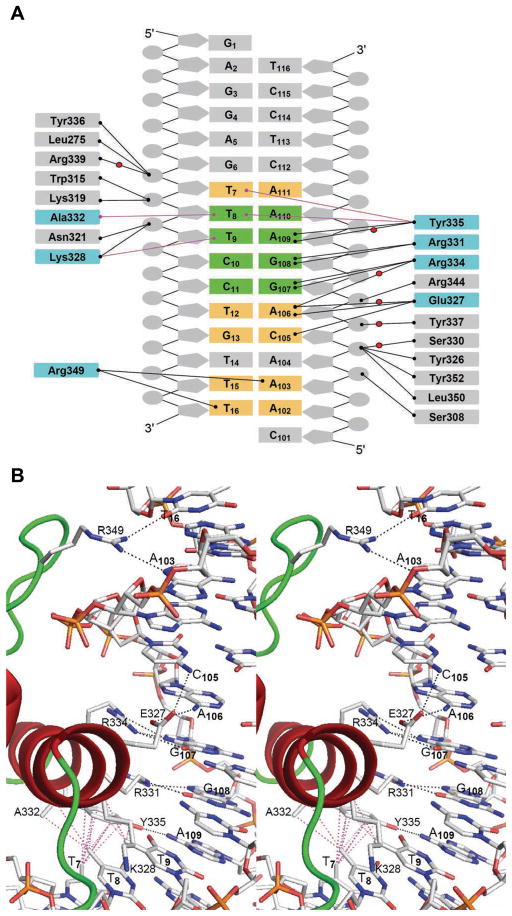
A summary of the Elf3·mTβR-IIDNA interactions. (A) A schematic representation of protein·DNA interactions. Hydrogen bonds are shown with black lines and van der Waals contacts are shown with magenta lines. Water molecules are represented by red balls. The DNA base pairs in the B-site core region are highlighted in green and DNA base pairs outside the core region that interact with the protein are highlighted in yellow. The Elf3 sequences involved in contacts with DNA bases are highlighted in cyan. (B and C) A stereoview of protein·DNA interface showing the details of Elf3’s direct interactions with (B) DNA bases and (C) DNA backbone. The hydrogen bonds are drawn with black dashed lines and the van der Waals interactions are drawn with dashed magenta lines.
Interactions with DNA bases in EBS core motif
The α3 helix protrudes into the major groove and forms a number of specific interactions with the bases of the 5′-G107G108A109 A110-3′ core sequence (Figure 3B). Arg331 and Arg334 form double hydrogen bonds with the bases of G108 and G107, respectively, which is characteristic for all Ets domains (Figure 2B). Tyr335 interacts with the base of A109 via direct and water-mediated hydrogen bonds. Similar observations were made in other Ets·DNA complex structures, except for PU.1 and PDEF having asparagine and glutamine instead of tyrosine, respectively (Figure 2B). Elf3 specificity for the core motif is also enhanced by van der Waals interactions, one between the methyl group of T9 and the side chain of Lys328, and another between the methyl group of T8 and the side chains of Ala332 and Tyr335. The van der Waals interactions with the base corresponding to T8 in the structure of Elf3 are absent in structures of Sap1 and PDEF since their DNAs contain a GGAT core instead of GGAA. Overall, Elf3 forms EBS core interactions similar to most of reported Ets domains (Figure 2B). However, as we discuss below, sequences flanking the EBS core also contribute to Elf3 specificity for the B-site on the mTβR-II promoter.
Interactions with DNA bases in EBS core flanking sequences
Elf3 forms several important interactions with DNA bases outside the EBS core (Figure 3). The methyl group of T7 is involved in van der Waals interactions with the aromatic portion of Tyr335. Glu327 forms a water-mediated hydrogen bond with A106. Glu327 also forms direct hydrogen bonds with the bases of A106 and C105, however the observed donor to acceptor distances of 3.6 Å are higher than the average value of 2.9 Å, indicating somewhat weaker interactions. This glutamic acid residue is substituted with aspartic acid in some Ets proteins (Figure 2B) where it does not interact with DNA bases with the exception of the corresponding Asp58 of Elk127. However, the specific interactions for the Glu327 of Elf3 and the Asp58 of Elk1 are different: Glu327 interacts with DNA bases outside the EBS core, while Asp58 interacts with bases in the EBS core27. The side chain of Arg349 invades the minor groove and forms contacts with both strands of DNA via hydrogen bonds with the T16 and A103 bases. The Arg349 is conserved in most of the Ets domains, except SAP-1, PU.1, and Elk-1 which have a lysine in this position. This unique feature of the arginine residue was not observed in other reported Ets·DNA complex structures, except that some resemblance is seen in the structure of the PDEF·DNA complex. But unlike the Arg349 of Elf3, the Arg326 of PDEF makes only water-mediated base interactions in the minor groove21.
Interactions with the DNA backbone
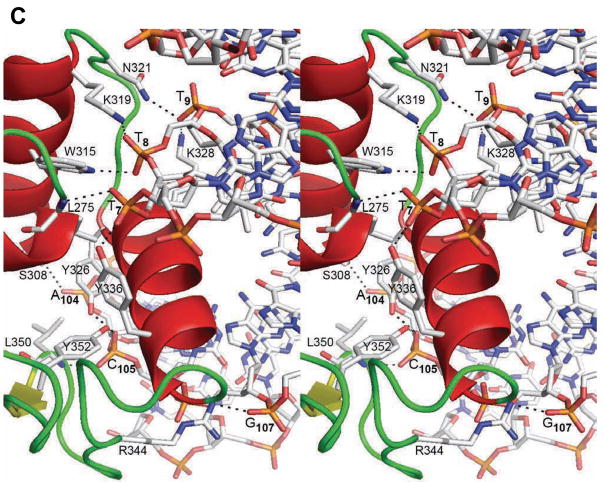
The Elf3 Ets domain makes a number of hydrogen bonds with the DNA sugar phosphate backbone in two areas, T7T8T9 and A104C105A106G107 (Figures 3A and 3C). In the T7T8T9 area, T8 is involved in hydrogen bonds with side chains of Trp315 and Lys319 of the α2 helix. The hydroxyl group of Tyr336 of the 310 helix and the backbone amide of Leu275 of the α1 helix form direct hydrogen bonds with the T7 phosphate oxygens from major and minor grooves, respectively. The Arg339 is involved only in water-mediated hydrogen bond with T7. The side chains of Asn321 of loop 4 and invariant Lys328 of α3 helix interact with the phosphate oxygen of T9 from the major groove side.
In the A104C105A106G107 area, the sugar phosphate backbone forms hydrogen bonds with the C-terminal amino acid residues of the Elf3 Ets domain. Interestingly, the conserved residue Ser330 is involved in water-mediated contacts with C105, while Ser308 displays a direct hydrogen bond with the A104 phosphate. However, none of the serine residues of Ets domains in other Ets proteins, except for Ser308 of PDEF, form DNA contacts21. The hydroxyl groups of Tyr326 and Tyr352 interact with the C105 phosphate in the major groove, and the backbone amide of Leu350 interacts with the C105 in the minor groove. The Arg344, which is substituted with lysine in other Ets domains (Figure 2B), forms hydrogen bonds with G107 phosphate. In addition, the highly conserved Tyr337 forms water-mediated hydrogen bond with the phosphate of A106.
The role of C-terminal residues
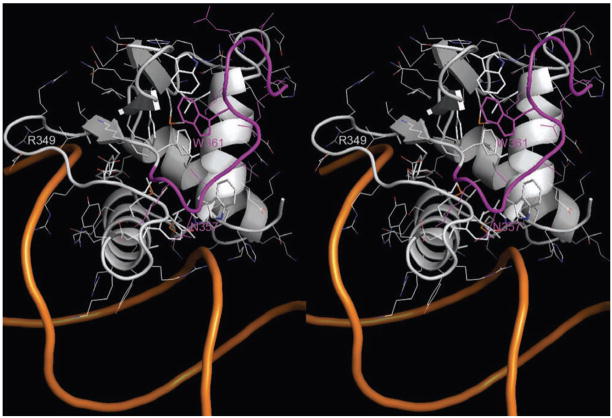
Contrary to the well conserved Ets domain sequences, sequences located N- and C-terminal to the Ets domain of different Ets proteins do not exhibit similarity. However, these sequences play an important role in the regulation of Ets domain DNA binding activity. For example, in Ets1 these amino acid sequences are folded as α-helices and form an inhibitory module29; 30, while in GABPα C-terminal residues are involved in formation of the complex with GABPβ22. Comparison of Ets protein structures and the folding of C-terminal residues are shown in Figure 2C. C-terminal residues are folded as random coils in Elf3, Sap1 and Elk1, while they form two α-helices in Ets1 and GABPα. Interestingly, corresponding C-terminal residues are missing in PU.1 and PDEF, but their Ets domains are well folded and bound to DNA. This raises the possibility that the C-terminal residues are dispensable for the stability and DNA binding of other Ets proteins. Our earlier studies revealed that removal of the last eight amino acids (364–371) of Elf3 reduced its ability to activate transcription from the mTβR-II −108/+56(B/A) promoter/reporter construct by only 20%. However, further deletion of 17 amino acids 355–371 completely abolished the DNA binding and transactivating capacities of Elf31. The current structure provides detailed insight for the interaction of Elf3 C-terminal residues (355–371) with the Ets domain (Figure 4). Residues 362 to 371 do not participate in stable interactions with the Ets domain and have high flexibility according to high temperature factors for 362–366 and absence of density for 367–371. This is consistent with the above mentioned deletion studies1. Among the amino acid residues 355–361, Trp361 appears to be the most extensively interacting residue. Its side chain covers and apparently completes the hydrophobic core formed between the β-sheet and three helices of the Ets domain (Figure 4). The van der Waals interactions of the Trp361 side chain with side chains of Trp295 and Phe354, and α-carbon of Gly301 anchors the C-terminal tail to the Ets domain, which in turn reduces the solvent exposure of the hydrophobic pocket. Another noticeable interaction is a hydrogen bond between the side chain of Asn357 and the main chain of Arg339.
Figure 4.
Location of Elf3 C-terminal. The structure of mElf3·mTβR-IIDNA is drawn as cartoons. The side-chains of Ets domain hydrophobic core residues and Trp361 are shown as sticks and the side chains of other residues are shown as lines. The Elf3 C-terminal residues 355–366 are in magenta color and the rest of residues are colored by atom type: the carbon, nitrogen and oxygen atoms are in grey, blue and red, respectively.
EMSA studies of minor groove interactions
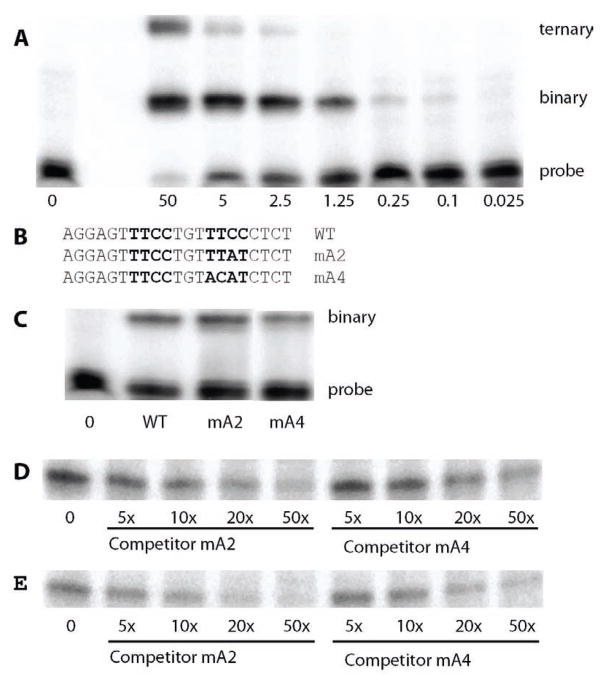
The above described Elf3·DNA interactions indicate that Elf3 bound to the B site also makes contact with thymine and adenine residues (T16 and A103) in the adjacent A site (Figures 1 and 3B). To test whether base pairs in the A site contribute to the binding of Elf3 to the B site, EMSA was performed with recombinant Elf3269–371 and a DNA probe based on the sequence surrounding these sites. Since Elf3 contains an autoinhibitory domain(s) that minimizes binding of the full length protein in vitro, a truncated version, Elf3269–371 (which includes the DNA binding domain and C-terminus of the protein) was used for all EMSA reported here. In an EMSA with increasing amounts of recombinant Elf3, we observed a slow migrating complex, which is likely to be a ternary complex composed of two molecules of Elf3 bound to the DNA probe, and a faster migrating complex, which is likely to be a binary complex composed of one molecule of Elf3 and the DNA probe (Figure 5A). The intensities diminished with decreasing protein concentration, and the slower migrating complex was only visible at 2.5 μM Elf3, while the greatest drop in intensity of the slower migrating complex occurred between 2.5 and 0.25 μM.
Figure 5.
Binding of Elf3 to the TGFβ-RII EBS B-site in the presence of A-site mutations. EMSA was performed with recombinant Elf3269–371 and 32P-labeled probes based on the TGFβR-II EBS sequence. Bands produced by binary and, where present, ternary complexes as well as the free probe are shown. (A) The wild-type probe was incubated with the indicated amount (μM) of Elf3269–371. (B) The sequence of the wild-type (WT) probe is shown with the core of Ets-binding sites B and A in bold. This sequence is compared to those of mA2, which has two modified base pairs in the A site, and mA4, which has four motified base pairs in the A site. (C) Incubations included 2 μM of Elf3269–371 and either the WT, mA2 or mA4 probe. As a control, the WT probe was incubated without Elf3 (0). It is evident in part A of this figure that 2 μM of Elf3269–371 generates little or no ternary complex. Either probe mA2 (D) or mA4 (E) was incubated with 2 μM Elf3269–371 and 5-fold to 50-fold excess of unlabelled competitor probe mA2 or mA4 was added as indicated. The band produced by the binary complex is shown.
To examine contributions of the T16 and A103 residues of the A site to Elf3 binding to the B site, two probes were used with modified A site sequences (Figure 5B). Both probes disrupted the A site by altering two residues in the 3′ half of the site, so that Elf3 can only be bound to the B site and not to the A site. However, one of the probes (mA2) left the thymine residues of the A site intact, while the other (mA4) also replaced two thymine residues. For the next set of EMSAs, 2 μM recombinant Elf3 was chosen since it only generates a binary complex and because at this concentration of Elf3 changes in DNA affinity should be more readily detected. Under these conditions, the mA2 probe does not produce a ternary complex in an EMSA, even when Elf3 concentrations were high enough for the wild-type probe to produce a ternary complex (data not shown). However, the mA2 probe gave a band for the binary complex of similar intensity to that with the wild-type probe (containing both A and B EBS), while the mA4 probe gave a less intense band (Figure 5C). This suggests that the residues in the A site contribute to the affinity of the B site for Elf3.
The relative affinities of the mA2 and mA4 DNA probes for Elf3 were tested further by comparing the abilities of the unlabeled oligonucleotides to compete with the labeled probes for binding of Elf3. In an EMSA using labeled mA2 and 5-fold to 50-fold excess of the unlabeled DNAs, the mA2 oligonucleotide decreased the intensity of the binary complex to a greater degree than did the mA4 oligonucleotide (Figure 5D) for any given concentration of unlabeled oligonucleotide. Similarly, mA2 also had the greater effect in an EMSA using the labeled mA4 probe (Figure 5E). This strongly supports the model that thymine residues in the Elf3-binding A site, which is retained in mA2, contribute to the binding of Elf3 to the B site.
Mutational and functional analysis of Elf3 binding to the TGF-β receptor II promoter
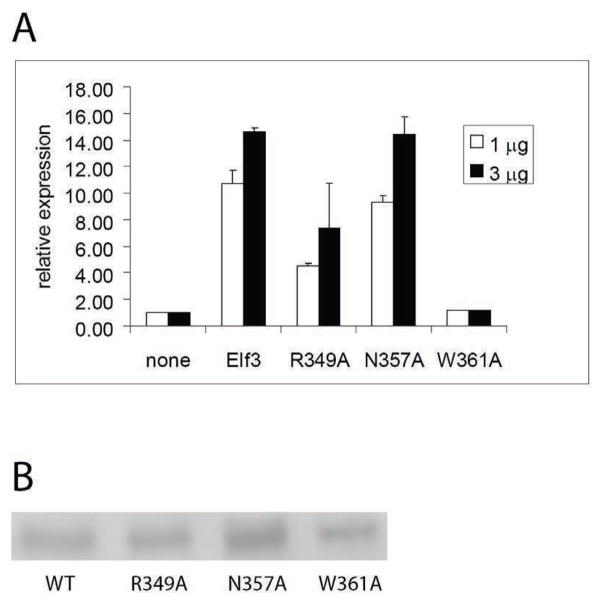
Our structural analysis of Elf3 binding to EBS in the mTβR-II promoter was also used to make predictions concerning the contributions of several amino acid residues of Elf3 to its DNA binding; specifically: Arg349, Asn357, and Trp361. Functional cell-based transcription studies were performed to evaluate the effects of Elf3 point mutations R349A, N357A, or W361A. Full-length versions of the flag-tagged Elf3 point mutant expression vectors were constructed and transiently transfected into F9-differentiated cells along with the mTGFβ-RII (−108/+56) promoter/reporter construct. Elf3 N357A was as effective as wild-type Elf3 in stimulating the promoter/reporter construct, while 1 or 3 μg of Elf3 R349A consistently stimulated the promoter/reporter construct to a lesser degree, and Elf3 W361A had no stimulatory effect (Figure 6A). To determine the expression levels of Elf3 and its mutant forms, we examined their expression in 293T cells, since their expression in transiently transfected F9-differentiated cells was too low to be detected by western blot analysis (data not shown). Western blot analysis using nuclear extracts prepared from 293T cells transfected with these expression vectors showed expression of Elf3 R349A at similar levels to that of the wild-type Elf3 as well as expression of Elf3 N357A at somewhat higher levels (Figure 6B). Interestingly, in contrast to mElf3269–371W361A, which is not expressed at easily detectable levels (data not shown), full-length Elf3W361A was readily observed although it was present at a lower concentration than the wild-type Elf3. From these results, we conclude that the hydrogen bond contributed by Asn357 in the C-terminal tail is functionally insignificant under the conditions of our studies. In contrast, the minor groove contacts made by Arg349 are significant, but not critical, to the transcriptional activity of Elf3. Even more importantly, Trp361 in the C-terminal tail is essential to the transcriptional activity of Elf3 and probably to its structural integrity.
Figure 6.
Transcriptional activity of Elf3 and its point mutants. (A) F9-differentiated cells were transiently transfected with the promoter/reporter construct mTGFβ-RII (−108/+56) and 1 or 3 μg of an expression plasmid for flag-tagged Elf3 with either the wild-type (WT) or indicated point mutant sequence. CAT reporter gene activity was measured and normalized as described in Materials and Methods. Independent clones of plasmids for the N357A and W361A mutants gave similar results and this experiment was repeated twice to verify the intermediate effects of the R349A clone. (B) Western blot analysis was performed using cell nuclear extracts from 293T cells transfected with the expression plasmids used in part (A) and an antibody against the N-terminal flag tag to visualize relative expression levels.
Conclusions
Ets family members were defined on the basis of the having the DNA binding Ets domain. Consequently, upon activation they are supposed to bind to promoter sequences having the same GGAA/T core motif. However, many genes contain GGAA/T sequences, plus different Ets factors are present simultaneously in many cell types. This indicates that the binding of each Ets protein to its regulatory sites at the required times must be tightly regulated. Most of the Ets factors, including Elf3, are produced in an inert autoinhibited state, and in order to bind DNA they have to be activated. Nevertheless, once activated, each Ets factor requires additional parameters for recognition of the correct EBS. In the latter case, promoter sequences other than the core EBS may play a crucial role in binding site preference by facilitating cooperative binding of two or more factors. For example Ets1 and Runx1 cooperatively bind to closely positioned sites on TCRβ31 and many other promoters.31; 32 Interestingly, the palindromic EBS on the stromelysin-1 and p53 promoters also facilitates the cooperative binding of Ets1 molecules which results in synergistic activation of gene expression33; 34.
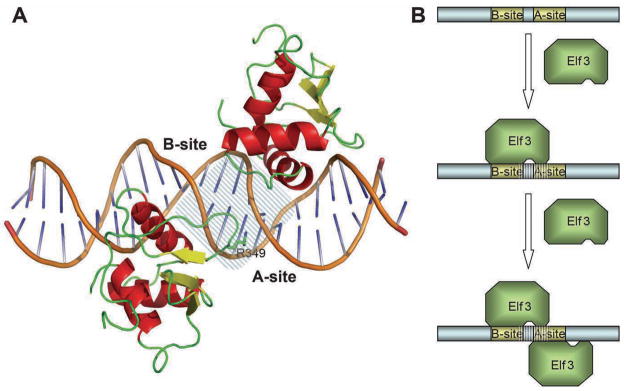
The binding of Elf3 to both the A- and the B-site is necessary for the synergistic activation of the mTβR-II gene in F9-differentiated cells13; 14; 35. Disruption of either site drastically reduces the promoter activity of this gene. This synergistic effect could occur by one or more mechanisms, including cooperative interactions that promote the binding of Elf3 to its two overlapping EBS. Importantly, we have determined previously that Elf3 exhibits higher affinity for the B-site than the A-site14. Based on the findings described in this study, we propose that the higher affinity for the B-site is due to DNA sequence differences outside the EBS core, including the R349 recognition site (TT for B-site vs. TC for A-site, Figure 1A). Our crystallographic analysis of mElf3269–371·mTβR-II dsDNA reveals that Elf3 interacts with the DNA bases from the major groove of the B-site introducing 26° bent and with bases from the minor groove of the A-site. Importantly, the B-site interaction introduces local conformational changes in half of the A-site (Figure 7A), which may enhance the binding affinity of the A-site for Elf3. Hence, it is tempting to speculate that binding of Elf3 to the two EBS of the mTβR-II promoter occurs by a two step mechanism: first, Elf3 binds the B-site, and then it binds to the structurally modified A-site (Figure 7B). Finally, we wish to stress that the findings described in this report and our previous studies1 argue strongly that regions flanking the Ets domain are essential for DNA binding both in vitro and in vivo.
Figure 7.
Elf3 binding to the TβR-II promoter. (A) A cartoon representation of the model of Elf3 bound to the A- and B-sites of the TβR-II promoter. The DNA fragment shared by both Elf3 molecules is marked by dashed lines (B) A schematic representation of the two-step mechanism of Elf3 binding to the TβR-II promoter. The first Elf3 molecule binds to the high-affinity B-site and induces conformational changes in the A-site (dashed lines). The conformational changes in the A-site allow binding of a second Elf3 molecule to the major groove of the A-site.
Materials and Methods
Structure determination and refinement
Preparations of mElf3 protein containing amino acid residues 269–371 (mElf3269–371), its complex with mTβR-II dsDNA, crystallization, cryoprotection of crystals, X-ray diffraction data collection and processing are described elsewhere36. The structure was determined by the molecular replacement method starting with the coordinates of Ets1 (PDB entry 1gvj). The major manual rebuilding of the initial model was performed with TURBO-FRODO software. The model was refined at 2.2 Å resolution to an Rcryst of 23.4% and an Rfree of 26.2%. CNS version 1.137 was used for all crystallographic computing. Application of zonal scaling38 and bulk solvent correction improved the quality of electron density maps. The final model contains amino acid residues 272 to 366, 16-mer mTβR-II dsDNA, and 54 water molecules per asymmetric unit. The electron density was not clear for the three N-terminal and five C-terminal amino acid residues and these residues were not included in the model. The Ramachandran plot demonstrates that of the non-glycine residues, 87.2% lie in the most favored regions and the other 12.8% in additionally allowed regions. The final refinement statistics are provided in Table 1. The figures containing molecular structures were drawn with PyMOL39. The distances used for the delineation of hydrogen bonds and van der Waals contacts are 3.4 Å and 4.2 Å, respectively.
Table 1.
Data collection and refinement statistics.
| Data collection | |
|---|---|
| Space group | P212121 |
| Cell dimensions: a, b, c (Å) | 42.66, 52.00, 99.78 |
| Resolution (Å)* | 50-2.2 (2.24-2.2) |
| Unique reflections | 11575 |
| Rmerge (%)* | 6.4 (39.0) |
| I/σ(I) | 53.5 (9.9) |
| Completeness (%) | 97.6 (97.5) |
| Redundancy | 5.8 (6.0) |
| Temperature (K) | 100 |
| Mosaicity (°) | 0.66–1.01 |
| Refinement | |
| Resolution (Å) | 40-2.2 |
| No. reflections | 11365 |
| Rwork/Rfree | 0.234/0.262 |
| No. atoms/B-factors (Å2) | |
| Protein | 819/43.3 |
| DNA | 650/47.6 |
| Water | 54/43.6 |
| R.m.s. deviations | |
| Bond lengths (Å) | 0.006 |
| Bond angles (°) | 1.06 |
| Ramachandran plot | |
| Favored (%) | 87.2 |
| Allowed (%) | 12.8 |
Values in parentheses are for the last shell.
Plasmid constructs for mutational analysis
The expression vectors used for flag-tagged full length Elf3 (in the FNpcDNA3 parent vector) have been described previously13. Expression vectors for Elf3 with particular amino acid modifications were created by site-directed mutagenesis using the QuickChange™ Site-directed Mutagenesis kit (Strategene). Newly created restriction endonuclease cleavage sites introduced into the expression vector along with each alteration were used for screening. The resulting plasmids were sequenced to verify that the products contained the desired alterations. Primers used for site-directed mutagenesis are listed here with the modified bases underlined.
R349A, 5′-GGAACGGGTGGATGGCCGTGCACTCGTCTACAAG-3′;
N357A, 5′-GTTTGGCAAGGCCTCTAGTGGCTGGAAGGAAGAAGAGG-3′;
W361A, 5′-GCAAGAACTCTAGTGGCGCCAAGGAAGAAGAGGTTGGAGAG-3′.
Promoter/reporter assays
Promoter/reporter assays were performed as described previously13. Briefly, F9 cells were grown in DME medium containing 10% fetal bovine serum, differentiated with 5 μM retinoic acid, and transfected using the calcium phosphate method. The mTβR-II(−108/+56) promoter/reporter construct was introduced at 15 μg/100-mm dish, and the resulting chloramphenicol acetyltransferase expression was normalized to correct for differences in transfection efficiency using the β-galactosidase activity of an internal control.
Nuclear extracts and western blot analysis
Nuclear extracts were prepared from 293T cells with the NE-PER kit (Pierce, Thermo Scientific) and western blot analysis was performed as described previously40. In this analysis, the M2 antiflag antibody (Sigma) was used to detect the flag-tagged proteins produced by expression vectors transiently transfected into the 293T cells 24–48 hours before preparation of the nuclear extracts.
Electrophoretic Mobility Shift Analysis (EMSA)
EMSA for Elf3 binding was performed as previously described41 with the following modifications. For labeling probes (sequences given in Figure 5B), the nucleotide sequence AGC was added to the 5′ end of both the sense and antisense oligonucleotides. In place of poly(dI-dC), a version of the mTβR-II Ets-binding region with mutated Ets sites was used to reduce non-specific binding (5′-TAGCTGGCGAGAAGTTTGAATTCACCCTCTCGGCGCGCTA-3′). The 0.5X TGE running buffer was used (12.5 mM Tris, 95 mM Glycine, 0.5 mM EDTA, pH 8.5). Recombinant Elf3269–371 for EMSA was from the protein preparation used for crystallization36.
Acknowledgments
We thank J. Lovelace and G.E. Borgstahl for maintenance and management of the Eppley Institute’s X-ray Crystallography facility; D.G. Vassylyev for the zonal scaling instruction files. Primers were synthesized in the Eppley Institute’s Molecular Biology Core facility. Both Eppley Institute’s facilities are supported by the Cancer Center Support Grant P30CA036727. This work is supported by the UNMC Eppley Cancer Center Pilot Project LB595, by NIGMS grant R01GM082923 to THT, and by the Nebraska Department of Health and Human Services grant LB506 (2009-32) to AR. This work is also based upon research conducted at the Northeastern Collaborative Access Team beamlines of the Advanced Photon Source, supported by award RR-15301 from the National Center for Research Resources at the National Institutes of Health. Use of the Advanced Photon Source is supported by the U.S. Department of Energy, Office of Basic Energy Sciences, under Contract No. DE-AC02-06CH11357.
Footnotes
Accession numbers
Atomic coordinates and structure factors have been deposited in the Protein Data Bank with accession number 3jtg.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kopp JL, Wilder PJ, Desler M, Kinarsky L, Rizzino A. Different domains of the transcription factor ELF3 are required in a promoter-specific manner and multiple domains control its binding to DNA. J Biol Chem. 2007;282:3027–41. doi: 10.1074/jbc.M609907200. [DOI] [PubMed] [Google Scholar]
- 2.Asada S, Choi Y, Yamada M, Wang SC, Hung MC, Qin J, Uesugi M. External control of Her2 expression and cancer cell growth by targeting a Ras-linked coactivator. Proc Natl Acad Sci U S A. 2002;99:12747–52. doi: 10.1073/pnas.202162199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Eckel KL, Tentler JJ, Cappetta GJ, Diamond SE, Gutierrez-Hartmann A. The epithelial-specific ETS transcription factor ESX/ESE-1/Elf-3 modulates breast cancer-associated gene expression. DNA Cell Biol. 2003;22:79–94. doi: 10.1089/104454903321515896. [DOI] [PubMed] [Google Scholar]
- 4.Chang J, Lee C, Hahm KB, Yi Y, Choi SG, Kim SJ. Over-expression of ERT(ESX/ESE-1/ELF3), an ets-related transcription factor, induces endogenous TGF-beta type II receptor expression and restores the TGF-beta signaling pathway in Hs578t human breast cancer cells. Oncogene. 2000;19:151–4. doi: 10.1038/sj.onc.1203252. [DOI] [PubMed] [Google Scholar]
- 5.Rudders S, Gaspar J, Madore R, Voland C, Grall F, Patel A, Pellacani A, Perrella MA, Libermann TA, Oettgen P. ESE-1 is a novel transcriptional mediator of inflammation that interacts with NF-kappa B to regulate the inducible nitric-oxide synthase gene. J Biol Chem. 2001;276:3302–9. doi: 10.1074/jbc.M006507200. [DOI] [PubMed] [Google Scholar]
- 6.Feldman RJ, Sementchenko VI, Watson DK. The epithelial-specific Ets factors occupy a unique position in defining epithelial proliferation, differentiation and carcinogenesis. Anticancer Res. 2003;23:2125–31. [PubMed] [Google Scholar]
- 7.Andreoli JM, Jang SI, Chung E, Coticchia CM, Steinert PM, Markova NG. The expression of a novel, epithelium-specific ets transcription factor is restricted to the most differentiated layers in the epidermis. Nucleic Acids Res. 1997;25:4287–95. doi: 10.1093/nar/25.21.4287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brembeck FH, Opitz OG, Libermann TA, Rustgi AK. Dual function of the epithelial specific ets transcription factor, ELF3, in modulating differentiation. Oncogene. 2000;19:1941–9. doi: 10.1038/sj.onc.1203441. [DOI] [PubMed] [Google Scholar]
- 9.Cabral A, Fischer DF, Vermeij WP, Backendorf C. Distinct functional interactions of human Skn-1 isoforms with Ese-1 during keratinocyte terminal differentiation. J Biol Chem. 2003;278:17792–9. doi: 10.1074/jbc.M300508200. [DOI] [PubMed] [Google Scholar]
- 10.Grall FT, Prall WC, Wei W, Gu X, Cho JY, Choy BK, Zerbini LF, Inan MS, Goldring SR, Gravallese EM, Goldring MB, Oettgen P, Libermann TA. The Ets transcription factor ESE-1 mediates induction of the COX-2 gene by LPS in monocytes. Febs J. 2005;272:1676–87. doi: 10.1111/j.1742-4658.2005.04592.x. [DOI] [PubMed] [Google Scholar]
- 11.Kwon JH, Keates S, Simeonidis S, Grall F, Libermann TA, Keates AC. ESE-1, an enterocyte-specific Ets transcription factor, regulates MIP-3alpha gene expression in Caco-2 human colonic epithelial cells. J Biol Chem. 2003;278:875–84. doi: 10.1074/jbc.M208241200. [DOI] [PubMed] [Google Scholar]
- 12.Chang CH, Scott GK, Kuo WL, Xiong X, Suzdaltseva Y, Park JW, Sayre P, Erny K, Collins C, Gray JW, Benz CC. ESX: a structurally unique Ets overexpressed early during human breast tumorigenesis. Oncogene. 1997;14:1617–22. doi: 10.1038/sj.onc.1200978. [DOI] [PubMed] [Google Scholar]
- 13.Kim JH, Wilder PJ, Hou J, Nowling T, Rizzino A. Activation of the murine type II transforming growth factor-beta receptor gene: up-regulation and function of the transcription factor Elf-3/Ert/Esx/Ese-1. J Biol Chem. 2002;277:17520–30. doi: 10.1074/jbc.M110434200. [DOI] [PubMed] [Google Scholar]
- 14.Kopp JL, Wilder PJ, Desler M, Kim JH, Hou J, Nowling T, Rizzino A. Unique and selective effects of five Ets family members, Elf3, Ets1, Ets2, PEA3, and PU.1, on the promoter of the type II transforming growth factor-beta receptor gene. J Biol Chem. 2004;279:19407–20. doi: 10.1074/jbc.M314115200. [DOI] [PubMed] [Google Scholar]
- 15.Ely KR, Kodandapani R. Ankyrin(g) ETS domains to DNA. Nat Struct Biol. 1998;5:255–9. doi: 10.1038/nsb0498-255. [DOI] [PubMed] [Google Scholar]
- 16.Seth A, Watson DK. ETS transcription factors and their emerging roles in human cancer. Eur J Cancer. 2005;41:2462–78. doi: 10.1016/j.ejca.2005.08.013. [DOI] [PubMed] [Google Scholar]
- 17.Kodandapani R, Pio F, Ni CZ, Piccialli G, Klemsz M, McKercher S, Maki RA, Ely KR. A new pattern for helix-turn-helix recognition revealed by the PU.1 ETS-domain-DNA complex. Nature. 1996;380:456–60. doi: 10.1038/380456a0. [DOI] [PubMed] [Google Scholar]
- 18.Garvie CW, Wolberger C. Recognition of specific DNA sequences. Mol Cell. 2001;8:937–46. doi: 10.1016/s1097-2765(01)00392-6. [DOI] [PubMed] [Google Scholar]
- 19.Piper DE, Batchelor AH, Chang CP, Cleary ML, Wolberger C. Structure of a HoxB1-Pbx1 heterodimer bound to DNA: role of the hexapeptide and a fourth homeodomain helix in complex formation. Cell. 1999;96:587–97. doi: 10.1016/s0092-8674(00)80662-5. [DOI] [PubMed] [Google Scholar]
- 20.Seth A, Robinson L, Thompson DM, Watson DK, Papas TS. Transactivation of GATA-1 promoter with ETS1, ETS2 and ERGB/Hu-FLI-1 proteins: stabilization of the ETS1 protein binding on GATA-1 promoter sequences by monoclonal antibody. Oncogene. 1993;8:1783–90. [PubMed] [Google Scholar]
- 21.Wang Y, Feng L, Said M, Balderman S, Fayazi Z, Liu Y, Ghosh D, Gulick AM. Analysis of the 2.0 A crystal structure of the protein-DNA complex of the human PDEF Ets domain bound to the prostate specific antigen regulatory site. Biochemistry. 2005;44:7095–106. doi: 10.1021/bi047352t. [DOI] [PubMed] [Google Scholar]
- 22.Batchelor AH, Piper DE, de la Brousse FC, McKnight SL, Wolberger C. The structure of GABPalpha/beta: an ETS domain- ankyrin repeat heterodimer bound to DNA. Science. 1998;279:1037–41. doi: 10.1126/science.279.5353.1037. [DOI] [PubMed] [Google Scholar]
- 23.Donaldson LW, Petersen JM, Graves BJ, McIntosh LP. Solution structure of the ETS domain from murine Ets-1: a winged helix-turn-helix DNA binding motif. Embo J. 1996;15:125–34. [PMC free article] [PubMed] [Google Scholar]
- 24.Liang H, Mao X, Olejniczak ET, Nettesheim DG, Yu L, Meadows RP, Thompson CB, Fesik SW. Solution structure of the ets domain of Fli-1 when bound to DNA. Nat Struct Biol. 1994;1:871–5. doi: 10.1038/nsb1294-871. [DOI] [PubMed] [Google Scholar]
- 25.Werner MH, Clore GM, Fisher CL, Fisher RJ, Trinh L, Shiloach J, Gronenborn AM. Correction of the NMR structure of the ETS1/DNA complex. J Biomol NMR. 1997;10:317–28. doi: 10.1023/a:1018399711996. [DOI] [PubMed] [Google Scholar]
- 26.Mo Y, Ho W, Johnston K, Marmorstein R. Crystal structure of a ternary SAP-1/SRF/c-fos SRE DNA complex. J Mol Biol. 2001;314:495–506. doi: 10.1006/jmbi.2001.5138. [DOI] [PubMed] [Google Scholar]
- 27.Mo Y, Vaessen B, Johnston K, Marmorstein R. Structure of the elk-1-DNA complex reveals how DNA-distal residues affect ETS domain recognition of DNA. Nat Struct Biol. 2000;7:292–7. doi: 10.1038/74055. [DOI] [PubMed] [Google Scholar]
- 28.Garvie CW, Hagman J, Wolberger C. Structural studies of Ets-1/Pax5 complex formation on DNA. Mol Cell. 2001;8:1267–76. doi: 10.1016/s1097-2765(01)00410-5. [DOI] [PubMed] [Google Scholar]
- 29.Garvie CW, Pufall MA, Graves BJ, Wolberger C. Structural analysis of the autoinhibition of Ets-1 and its role in protein partnerships. J Biol Chem. 2002;277:45529–36. doi: 10.1074/jbc.M206327200. [DOI] [PubMed] [Google Scholar]
- 30.Lee GM, Donaldson LW, Pufall MA, Kang HS, Pot I, Graves BJ, McIntosh LP. The structural and dynamic basis of Ets-1 DNA binding autoinhibition. J Biol Chem. 2005;280:7088–99. doi: 10.1074/jbc.M410722200. [DOI] [PubMed] [Google Scholar]
- 31.Hollenhorst PC, Shah AA, Hopkins C, Graves BJ. Genome-wide analyses reveal properties of redundant and specific promoter occupancy within the ETS gene family. Genes Dev. 2007;21:1882–94. doi: 10.1101/gad.1561707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wotton D, Ghysdael J, Wang S, Speck NA, Owen MJ. Cooperative binding of Ets-1 and core binding factor to DNA. Mol Cell Biol. 1994;14:840–50. doi: 10.1128/mcb.14.1.840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Baillat D, Laitem C, Leprivier G, Margerin C, Aumercier M. Ets-1 binds cooperatively to the palindromic Ets-binding sites in the p53 promoter. Biochem Biophys Res Commun. 2009;378:213–7. doi: 10.1016/j.bbrc.2008.11.035. [DOI] [PubMed] [Google Scholar]
- 34.Baillat D, Begue A, Stehelin D, Aumercier M. ETS-1 transcription factor binds cooperatively to the palindromic head to head ETS-binding sites of the stromelysin-1 promoter by counteracting autoinhibition. J Biol Chem. 2002;277:29386–98. doi: 10.1074/jbc.M200088200. [DOI] [PubMed] [Google Scholar]
- 35.Choi SG, Yi Y, Kim YS, Kato M, Chang J, Chung HW, Hahm KB, Yang HK, Rhee HH, Bang YJ, Kim SJ. A novel ets-related transcription factor, ERT/ESX/ESE-1, regulates expression of the transforming growth factor-beta type II receptor. J Biol Chem. 1998;273:110–7. doi: 10.1074/jbc.273.1.110. [DOI] [PubMed] [Google Scholar]
- 36.Agarkar VB, Babayeva ND, Rizzino A, Tahirov TH. Preliminary crystallographic analysis of mouse Elf3 C-terminal domain in complex with type II TGF-β receptor promoter DNA. Acta Crystallogr Sect F. 2009 doi: 10.1107/S1744309109038007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brünger AT, AP, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simmon T, Warren GL. Crystallography and NMR system (CNS): a new software system for macromolecular structure determination. Acta Crystallogr Sect D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 38.Vassylyev DG, Vassylyeva MN, Perederina A, Tahirov TH, Artsimovitch I. Structural basis for transcription elongation by bacterial RNA polymerase. Nature. 2007;448:157–62. doi: 10.1038/nature05932. [DOI] [PubMed] [Google Scholar]
- 39.DeLano WL. The PyMOL Molecular Graphics System DeLano Scientific. San Carlos; CA, USA: 2002. http://www.pymol.org. [Google Scholar]
- 40.Boer B, Kopp J, Mallanna S, Desler M, Chakravarthy H, Wilder PJ, Bernadt C, Rizzino A. Elevating the levels of Sox2 in embryonal carcinoma cells and embryonic stem cells inhibits the expression of Sox2: Oct-3/4 target genes. Nucleic Acids Res. 2007;35:1773–86. doi: 10.1093/nar/gkm059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wilder PJ, Bernadt CT, Kim JH, Rizzino A. Stimulation of the murine type II transforming growth factor-beta receptor promoter by the transcription factor Egr-1. Mol Reprod Dev. 2002;63:282–90. doi: 10.1002/mrd.10165. [DOI] [PubMed] [Google Scholar]