Abstract
Protein degradation in the 20S proteasome is regulated in eukaryotes by the 19S ATPase complex and in archaea by the homologous PAN ATPase ring complex. Subunits of these hexameric ATPases contain on their C-termini a conserved hydrophobic-tyrosine-X (HbYX) motif that docks into pockets in the 20S to stimulate the opening of a gated substrate entry channel. Here, we report the crystal structure of the archaeal 20S proteasome in complex with the C-terminus of the archaeal proteasome regulatory ATPase, PAN. This structure defines the detailed interactions between the critical C-terminal HbYX motif and the 20S α-subunits and indicates that the intersubunit pocket in the 20S undergoes an induced-fit conformational change on binding of the HbYX motif. This structure together with related mutagenesis data suggest how in eukaryotes certain proteasomal ATPases bind to specific pockets in an asymmetrical manner to regulate gate opening.
Keywords: cryoelectron microscopy, proteasomal ATPase, proteasome, protein degradation, X-ray crystallography
Introduction
The 26S proteasome is an ATP-dependent proteolytic complex that catalyses most of the protein degradation in eukaryotic cells (Glickman and Ciechanover, 2002; Goldberg, 2005). In the ubiquitin-proteasome pathway, this large complex is the site for hydrolysis of proteins targeted for degradation by ubiquitination. It is composed of a hollow 20S proteasome core particle, capped at each end by a 19S regulatory particle (Voges et al, 1999). The barrel-shaped 20S proteasome contains four stacked heptameric rings, with two central β-rings flanked by two outer α-rings, forming three continuous chambers. The proteolytic active sites sequestered in the middle chamber (Lowe et al, 1995; Groll et al, 1997; Unno et al, 2002) and protein substrates therefore have to be translocated into this central chamber for degradation. The out wall of the 20S particle is tightly packed and does not allow for entry of substrate. Instead, there is a narrow channel in the centre of the outer α-rings, which allows passage only of unfolded polypeptides. The N-termini residues of α-subunits interact with one another and function as a gate that is normally closed in the free 20S proteasome preventing nonspecific substrate entry into the degradation chamber (Groll et al, 2000). In the 26S proteasome, opening of this gate and stimulation of substrate proteolysis are regulated by the hexameric ATPases ring in the 19S particles. Although archaea lack ubiquitin and the lid components of the 19S complex, it has a simpler yet homologous 20S proteasome and a proteasomal regulatory ATPases complex called PAN that is homologous to the 19S ATPases (Benaroudj and Goldberg, 2000; Benaroudj et al, 2003). Similar to the 19S regulatory particle, PAN binds protein substrates, unfolds them, and promotes their translocation into 20S for degradation. The homo-hexameric PAN ATPases and the hetero-hexameric 19S ATPases activate their corresponding 20S proteasome by inducing gate opening and do so by similar mechanisms (Smith et al, 2007).
The proteasomal regulatory ATPases, both PAN from archaea and three of the six Rpt subunits from eukaryotes, contain a conserved hydrophobic-tyrosine-X (HbYX) motif in their C-termini that is required for ATP-dependent gate opening (Smith et al, 2007). In fact, seven-residue peptides corresponding to PAN's C-terminal sequence or the HbYX-containing C-terminal sequences of the 19S ATPases (Rpt2 and Rpt5) by themselves can cause 20S gate opening (Smith et al, 2007; Gillette et al, 2008). We recently showed by single particle cryoelectron microscopy (cryoEM) that the C-terminus of PAN ATPase binds to the α-rings of 20S in the pocket between the adjacent α-subunits (named ‘intersubunit pocket') and induces a rigid body rotation in the α-subunits (Rabl et al, 2008). This α-subunit rotation relocates a reverse turn loop and stabilizes the open-gate conformation in the 20S proteasome. Unfortunately, efforts to understand further the atomic details of gate opening by C-termini of PAN has been frustrated by the failures of many laboratories to crystallize and solve the structure of 20S in complex with this mobile C-terminal region of PAN.
A relocation of this reverse turn loop in the α-subunits is also critical in the mechanism of gate opening by the 11S proteasome activator, PA26, a homolog of mammalian PA28α, β, and γ complexes. The exact physiological roles of these 11S complexes remain uncertain, but they associate with one or both ends of the 20S proteasome, enhance gate opening, and can promote the degradation of small peptides. The mechanism of gate opening by the proteasomal activator was first demonstrated by Hill and colleagues who solved the structure of PA26 (a homo-heptameric complex from Trypanosoma brucei) bound to the 20S from yeast (Saccharomyces cerevisiae) (Whitby et al, 2000; Forster et al, 2003) and archaea (Thermoplasma acidophilum) (Forster et al, 2005). In these complexes, the C-termini of PA26 bind to the 20S α-ring in the same intersubunit pocket where PAN's C-termini bind, even though PA26 and other 11S regulators lack an HbYX motif in their C-termini. The binding of PA26's C-termini is essential for complex formation but does not induce gate opening. Instead, an activation loop in PA26 directly interacts with the reverse turn loop in the 20S α-subunit and relocates it outwards away from the central pore stabilizing the open-gate conformation. Thus, PA26 requires two separate interactions to induce gate opening. In contrast, the proteasomal ATPases require only one type of interaction to induce gate opening—the binding of the conserved C-terminal HbYX motif by itself is sufficient to do so (Smith et al, 2007).
While the proteasomal ATPases contain the C-terminal HbYX motif, PA26 lacks any such conserved motif and is not homologous to PAN's C-terminus (Forster et al, 2005). Although the C-termini of PAN and PA26 bind to the same pockets in the 20S α-ring, their interactions with the residues in this pocket must be very different as the C-terminal HbYX motif from PAN induces a conformational change in the α-ring, whereas the C-termini from PA26 does not. The interactions between the C-terminus of PA26 and the intersubunit pocket of 20S were resolved in atomic details by X-ray crystallography (Forster et al, 2005), but the mechanisms how PAN interacts with the same pocket and induces different actions remains elusive, despite intensive efforts. In this study, we investigated the atomic details of how the conserved C-terminal HbYX motif of PAN interacts with the intersubunit pockets of the 20S proteasome and to understand how it induces gate opening.
Efforts to study the interactions of PAN or the 19S ATPases with the 20S have proven difficult largely because the ATPases complex has six subunits, whereas there are seven intersubunit pockets in the 20S's α-ring. Therefore, we took a different approach by creating an artificial hybrid proteasomal activator based on the heptameric PA26 structure, in which the activation loop was eliminated by site-directed mutagenesis to disable PA26's ability to induce gate opening. We then replaced the last eight residues of PA26's C-terminus with the C-terminus of PAN, which contains the critical HbYX motif. This heptameric hybrid activator forms a stable complex with the 20S proteasome by capping its both ends and opens the 20S gate and such effects depend on the presence of the C-terminal HbYX motif. Using both single particle cryoEM and X-ray crystallography, we determined the structure of the 20S proteasome from T. acidophilum in complex with our hybrid activator. This structure reveals how the conserved HbYX motif interacts specifically with the intersubunit pocket of the 20S proteasome and induces gate opening. In addition, this structure clarifies how the proteasomal ATPases regulate the functions of the 20S proteasome and provides new insights into how the eukaryotic 19S ATPases, which are composed of six distinct but homologous subunits (Rpt1–6) associate with and induce gate opening in the eukaryotic 20S proteasome, whose outer ring contains seven distinct α-subunits and thus seven different intersubunit pockets.
Results
Hybrid proteasomal activator PA26/PAN
The atomic structure of the T. brucei PA26 in complex with the T. acidophilum 20S proteasome (PDB ID: 1YA7; Forster et al, 2005) revealed how PA26's C-termini bind to each of the 20S's intersubunit pockets and how its seven activation loops interact with the seven reverse turn loops of the 20S α-subunits so as to maintain the gate in the open state (Figure 1A and B). As PAN is a homo-hexamer whose C-termini alone can induce gate opening in the 20S proteasome without any activation loop (Smith et al, 2007; Rabl et al, 2008), PAN must function by a different mechanism. To facilitate analysis of its structure, we designed, expressed, and purified from Escherichia coli an artificial seven-fold symmetric hybrid proteasomal activator. In this hybrid complex, the PA26 has an alanine mutation in its activation loop, which by itself prevents proteasome activation. Moreover, the last eight residues of its C-terminus were replaced with the last seven residues of PAN's C-terminus plus two glycine residues in between as a linker (Figure 1C). These additions restore the ability of this complex to activate the 20S proteasome. Such a hybrid proteasomal activator was designed to allow a stable interaction between the conserved HbYX motif and the 20S intersubunit pockets that would facilitate high-resolution structural studies of the proteasomal ATPases' C-termini bound to the 20S intersubunit pockets in a manner that triggers gate opening in an HbYX-dependent manner.
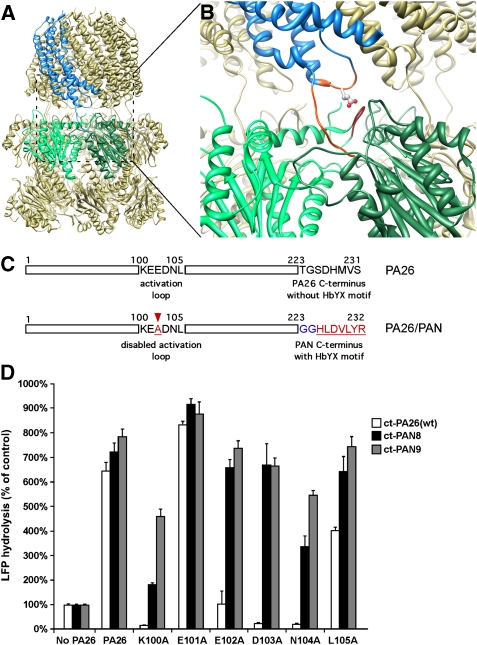
Figure 1.
Hybrid proteasomal activator PA26/PAN. (A) Atomic structure of wild-type 20S–PA26 complex (PDB ID: 1YA7, Forster et al, 2005). Two neighbouring α-subunits are coloured in light and dark green. The PA26 subunit that binds to this intersubunit pocket is coloured in blue. (B) An enlarged view of (A) shows interactions of the PA26 activation loop (orange) with the 20S reverse turn loop and the PA26 C-terminus (orange) with the 20S intersubunit pocket. The residue indicated by stick is E102. (C) Schematics shows the sequence of PA26 activation loop and its C-terminus (upper) and the design of a hybrid PA26/PAN activator (lower). (D) 20S (0.7 μg of wild-type (wt) T. acidophilum 20S) and LFP are incubated with the wt PA26 (ct-Pa26) and PA26/PAN hybrid complex with one (ct-PAN8) or two (ct-PAN9) glycine residues in the linker. The stimulation of gate opening was measured by the increase of LFP hydrolysis over the control without any activator. The values are mean±standard deviation from at least three independent measurements.
A mutagenesis study of PA28α, the mammalian homolog of PA26, showed that the single amino acid mutation in the activation loop completely inactivates its gate-opening function (Zhang et al, 1998). Although there is no sequence homology in the activation loops of the different PA26 homologs, the crystal structures of 20S–PA26 (Forster et al, 2005) revealed the location of this activation loop, which is formed by residues between Lys100 and Asp103 (Figure 1B). These residues are shown to interact directly with the reverse turn loop in the 20S α-subunit. To confirm that these residues are required for gate opening by PA26, we introduced single and triple alanine replacements for the residues between Lys100 and Leu105. Both negative staining EM and size exclusion chromatography indicated that all PA26 mutants with a single alanine mutation in its activation loop assembled into a ring. However, the triple alanine mutation did not assemble into a ring and did not form stable complex with the 20S (data not shown), and therefore was excluded for any further studies.
By measuring the hydrolysis of the quenched nine-residue fluorogenic substrates LFP (Smith et al, 2007) by T. acidophilum 20S proteasome in the presence of the PA26 activation loop mutants, we confirmed that the PA26 mutant with a single alanine replacement for Lys100, Glu102, Asp103, or Asn104 cannot induce gate opening in the 20S (Figure 1D). We also used negative staining EM to examine whether these PA26 mutants could still form a stable complex with the 20S proteasome. We found very few if any complexes between the PA26 activation domain mutants and the 20S, except for Glu101Ala and Leu105Ala mutants. Under these same conditions, we observed many complexes formed by the wild-type PA26 and the 20S. Thus, these mutations in the key residues of PA26's activation loop impair their binding to the 20S proteasome, and PA26's C-termini alone are not sufficient to mediate complex formation between PA26 and the 20S particle. Thus, the activation loop appear to be important in determining somehow the affinity of the 20S–PA26 complex, perhaps to maintain the PA26 heptamer in the appropriate conformation or to directly provide binding energy.
In the crystal structure of PA26 (Forster et al, 2005), its C-termini are extended away from the main body (Figure 1B). Therefore, replacing its C-terminal residues with a different sequence should not affect its folding or heptameric state. We therefore generated hybrid activators by replacing the last eight residues of the PA26 activation loop mutants with the last seven residues of PAN, HLDVLYR (Figure 1C). Short peptides containing these seven C-terminal residues by themselves stimulate 20S gate opening (Smith et al, 2007; Rabl et al, 2008). We also inserted one (PAN8) or two (PAN9) glycine residues in between the PA26 sequence and the C-terminus of PAN to introduce some flexibility into the C-termini of these hybrid complexes. In the presence of T. acidophilum 20S proteasomes, all these hybrid activators stimulated the hydrolysis of LFP by the 20S to the same extent as the wild-type PA26 (Figure 1D). Thus, replacing PA26's C-termini with PAN's restored the gate-opening function of the PA26, even though its activation loops had been inactivated. In addition, by negative staining EM, we observed that all hybrid activators stably associate with the 20S proteasome (data not shown). When the C-termini of the wild-type PA26 and the two PA26 activation loop mutants (Glu101Ala and Leu105Ala) were replaced with PAN's C-terminus, these mutants have similar stimulations of the 20S gate opening as the wild-type PA26. However, such gate opening may not follow the HbYX-dependent mechanism, as their activation loops are still functional.
Among all these hybrid activators, we chose the PA26 with an E102A mutation and a PAN9 in its C-terminus (named PA26E102A−PAN9) for structure determination. To further verify this construct, we replaced its penultimate tyrosine with a phenylalanine. A similar Tyr to Phe mutation in PAN prevents PAN from associating with the 20S and opening its gate. As expected, this same mutation in PA26E102A−PAN9 also prevented opening of the 20S gate (Table 1) and its association with the 20S (data not shown). This result confirms that our symmetry matched hybrid activators require an intact HbYX motif and that function in gate opening in a similar manner as PAN's C-termini.
Table 1.
The penultimate tyrosine is required for hybrid PA26E102A−PAN9 to activate archaeal 20S proteasome
| LFP hydrolysis | s.d. | |
|---|---|---|
| 20S | 100 | 3.14 |
| 20S+PAN | 370 | 26.5 |
| 20S+PA26E102A−PAN9 | 878 | 69.4 |
| 20S+PA26E102A−PAN9(Y231F) | 74.8 | 7.0 |
| PAN and PA26E102A−PAN9 are used as controls. All measurements were normalized against the 20S alone without activators. Standard deviations were calculated from the results of three independent measurements. | ||
Structure of the 20S–PA26E102A−PAN9 complex by single particle cryoEM
If the hybrid activator stimulates gate opening by the same mechanism as PAN (and not by the PA26 mechanism involving the activation loop), it should induce a conformational change in the 20S α-ring that resembles the conformational change observed in our prior studies (Rabl et al, 2008). To confirm this prediction, we used single particle cryoEM to determine the structure of the complex of the hybrid activator PA26E102A−PAN9 with the archaeal 20S proteasome. Visual inspection of the raw images of frozen hydrated specimens (Supplementary Figure 1A) reveals three types of particles: 20S alone and 20S with one end or both ends capped with a hybrid PA26E102A−PAN9 activator. Only particles of side views were manually selected to ensure that all the particles selected were doubly capped by the PA26E102A−PAN9 activators. Images of ∼17 000 double-capped 20S–PA26E102A−PAN9 complexes were used to calculate the three-dimensional (3D) reconstruction. The final 3D reconstruction of this complex with D7 symmetry was calculated at nominal resolutions of ∼10 Å (using FSC=0.143 criterion (Rosenthal and Henderson, 2003)). The resolution of the 20S core particle is ∼7.5 Å, which is significantly better than that of the entire complex as a whole (Supplementary Figure 1B). The secondary structural features, such as α-helices and β-sheets, are resolved in the 3D reconstructions (Figure 2A and B) confirming the nominal resolutions. The difference in the resolutions from different parts of the 3D reconstruction implies that there may be a small degree of mobility of the PA26E102A−PAN9 activator bound on top of the 20S, probably because of the glycine linker.
Figure 2.
Structure of 20S–PA26E102A−PAN9 complex by single particle cryoEM. (A) Side views of a 3D density map. (B) Cut in half and look from inside out of 20S–PA26E102A−PAN9 complex with the atomic structure (determined in this study) docked. (C) Top view of the 20S α-ring shows an open gate and the density of the C-termini of PAN bound in the intersubunit pockets. (D) An enlarged view of the α-ring pockets.
The architecture of the 20S–PA26E102A−PAN9 complex (Figure 2A) is similar to that of the wild-type 20S–PA26 complex. However, when we docked the entire atomic structure of wild-type (PDB ID: 1YA7; Forster et al, 2005) into our cryoEM density map, only the double β-rings in the middle of the 20S fit nicely into the cryoEM density map without any modification (Supplementary Figure 1C). We generated a pseudo atomic model of the 20S–PA26E102A−PAN9 complex (without the C-terminus of the hybrid complex) by docking separately the atomic structures of (1) the T. acidophilum 20S's β-rings (double β-ring as one rigid body), (2) the α-subunit (each α-subunit as an individual rigid body), and (3) PA26 (as a single rigid body) into the 3D density map. Comparisons of this pseudo atomic model with that of wild-type 20S–PA26 revealed a conformational change in the 20S α-ring, in which each α-subunit rotated about ∼7° pivoting around the central channel (Supplementary Figure 1C). This rotation also causes the intersubunit pockets to undergo a rotational shift about 4° around the seven-fold symmetry axis, and this axial rotation of the intersubunit pockets in turn resulted in an axial rotation of the hybrid activator. Such a rotation of the PA26 body was not observed in the crystal structure of the wild-type 20S–PA26 complex (Forster et al, 2005), and thus it must be a result of the HbYX-dependent mechanism of gate opening. Moreover, as expected, the gate of the 20S in the complex was open (Figure 2C). The conformational change in the α-ring and the open-gate conformation of 20S are consistent with our prior study, which elucidated the conformational changes in the α-ring caused by binding of the C-terminal peptides of PAN (Rabl et al, 2008). Therefore, the hybrid PA26E102A−PAN9 induces α-subunit conformational changes and gate opening in a similar HbYX-dependent manner as do the proteasomal ATPases. Even though the densities of PAN's C-termini in the 20S–PA26E102A−PAN9 complex show better features (Figure 2D) than those of free HbYX-containing peptides in our previous reconstruction (Rabl et al, 2008), the resolution was still not sufficient to derive atomic details about the interactions between the C-terminal HbYX motif and the 20S intersubunit pocket.
Structure of the 20S–PA26E102A−PAN9 complex by X-ray crystallography
We initially crystallized 20S–PA26E102A−PAN9 complex under a number of different buffer conditions. For structure determination, we chose crystals grown under conditions that most closely resembled the experimental assay conditions, in which we collected our single particle cryoEM data and measured gate opening by the LFP assay. The crystals under this specific buffer condition have a space group of P4222, with its two-fold crystallographic axis along one of the seven two-fold symmetry axes of the complex. The structure of the 20S–PA26E102A−PAN9 complex was solved using molecular replacement with the structure of wild-type T20S–PA26 complex (PDB ID: 1YA7; Forster et al, 2005) as the starting template. Non-crystallographic seven-fold symmetry was applied during the structure refinement. The final structure was refined to a resolution of 4.0 Å, with R and free R factor being 0.249 and 0.284. The complete crystallographic statistics are shown in Supplementary Table I.
A simulated-annealing composite omit electron density map was calculated from the final refinement along with regular difference maps. In all these maps, the main chains of both the 20S proteasome and the PA26E102A−PAN9 (except two loops, 128–135 and 162–171 of PA26) can be traced unambiguously with many of the bulky residue side chains well resolved. A portion of the representative electron density map of the 20S is shown in Figure 3A. Importantly, the structure of PAN's C-terminus in the intersubunit pocket is well defined (Figure 3B). We also docked this structure as a rigid body into the 3D density map of this complex determined by single particle cryoEM. The X-ray crystallographic structure of the 20S–PA26E102A−PAN9 fits very well into the cryoEM 3D density map (Figure 2), suggesting that the 20S–PA26E102A−PAN9 complex in crystals assumed the native conformation found in solution by single particle cryoEM.
Figure 3.
Structure of 20S–PA26E102A−PAN9 complex by X-ray crystallography. (A) Stereo views of electron density map (omit map) of a fragment of 20S core particle show the overall quality of the density map. (B) Stereo view of the omit map of PAN's C-terminus in the pocket between neighbouring 20S α-subunits. The numbering of residues follows the sequence of PA26.
As was shown by single particle cryoEM, the overall architecture of the 20S–PA26E102A−PAN9 complex is similar to that of the wild type (Supplementary Figure 2A). By superimposing our crystal structure with that of the wild-type 20S–PA26 complex, the overall root-mean-square-deviation (RMSD) between a total of 4480 Cα atoms of the entire complex is 1.75 Å, covering all but seven PA26 C-terminal residues in the complex. However, when the two structures were aligned using only the Cα atoms of the central β-rings, the β-ring has the lowest RMSD of only 0.65 Å, whereas that of the α-ring is 2.51 Å (see below) and of the heptamer activator is 3.03 Å. Therefore, the RMSD increases progressively from the central plane of the complex (between the two β-rings) to the distal end (the top of the PA26) along the seven-fold axis. Thus, though the β-ring is basically unchanged after the binding of PA26E102A−PAN9 to the 20S, the α-rings together with the heptameric activator undergo conformational changes with respect to the β-ring (see below).
Interactions between the HbYX motif and 20S intersubunit pocket
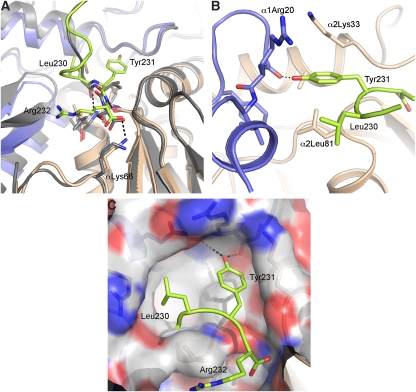
The crystal structure of the 20S–PA26E102A−PAN9 revealed in atomic detail the interactions between the conserved C-terminal HbYX motif of the proteasomal ATPases and the 20S proteasome intersubunit pocket. Figure 3B shows the omit map (contoured at 2σ level) of the C-terminal tail of the hybrid PA26E102A−PAN9 complex. The side chain densities of the terminal arginine and the penultimate tyrosine are well defined (Figure 3B). The structure of this C-terminus bound to the intersubunit pocket differs significantly from that of the wild-type PA26 bound to the same pocket (Figure 4A). The main chain of the last seven residues forms a single-turn α-helical structure (Figure 4A, green). In contrast, the C-terminus of the PA26 does not have such a helical conformation (Figure 4A, grey).
Figure 4.
Interactions between PAN's C-terminus and the 20S intersubunit pocket. (A) Comparison of C-termini's conformations of PA26 (grey) and PAN (green) in the intersubunit pocket. The α-subunits on the right of the same pocket was aligned and superimposed. (B) The position of the tyrosine residue of the C-terminal HbYX motif. (C) Position of the C-terminal HbYX motif in the intersubunit pocket that is represented as space filling. Positively charged residues are coloured in blue, and negatively charged residues are coloured in red. The white grey are hydrophobic regions.
More specifically, the carboxyl group of the terminal arginine forms a salt bridge with the ɛ-NH2 group of the Lys66 in the 20S α-subunit (∼2.5 Å, Figure 4A), whereas the side chain of this arginine residue does not specifically interact with any residue of the pocket. This result helps account for the previous demonstration by mutagenesis that Lys66 is critical for the binding and gate opening by both types of proteasomal activators, the PAN ATPases and the PA26 (Forster et al, 2005; Smith et al, 2007). Moreover, this observation is consistent with the finding that only the carboxyl group of the terminal residue (X) in the HbYX motif is critical for gate opening. For example, if terminal arginine is replaced by an alanine, the capacity to induce gate opening is maintained, but derivatizing the carboxyl group of this terminal residue impairs gate opening by the HbYX motif (Smith et al, 2007).
The side chain of the most critical and most conserved residue in the HbYX motif, the penultimate tyrosine, forms specific interactions with a number of residues within the intersubunit pocket. Its hydroxyl group on the aromatic ring forms a hydrogen bond with the main chain carbonyl group of Gly19 near the reverse turn loop of the α-subunit (Figure 4B). This hydrogen bond appears to be critical for gate opening because replacement of this tyrosine residue with a phenylalanine, which also has an aromatic ring but lacks the hydroxyl group, abolishes PAN's binding to the 20S and the stimulation of gate opening (Table 1; Smith et al, 2007). In addition to this hydrogen bond, the two positively charged residues, Arg20 from one α-subunit and Lys33 from the neighbouring α-subunit, further stabilize the position and orientation of this penultimate tyrosine by forming a cation–π interaction (Figure 4B). Although the Arg20 is conserved as a positively charged residue (arginine or lysine) in both archaeal and eukaryotic proteasomes, the Lys33 is less conserved in eukaryotic α-subunits. Finally, the first residue of the HbYX motif, leucine, forms a hydrophobic interaction with the hydrophobic patch (Leu21, val24, Leu81, and val82) in the intersubunit pocket (Figure 4C).
The conformational change in the intersubunit pocket induced by binding of the HbYX motif
Upon binding of PAN's C-termini to the intersubunit pocket formed by two neighbouring α-subunits, the pocket becomes smaller (Figure 4A). When one α-subunit from the 20S–PA26E102A−PAN9 (the one on the right side in Figure 4A) is superimposed on one from the 20S, the neighbouring α-subunit of the 20S–PA26E102A−PAN9 (the one on the left side of the pocket) is ∼4 Å closer (measured from the movement of Glu177), thus forming a tighter pocket around the HbYX motif. Within this tighter pocket, PAN's C-terminus interacts with both sides of the pocket. Thus, the binding pocket seems to undergo an induced-fit conformational change. In contrast, the pocket changes very subtly, if at all, upon binding of PA26's C-terminus (grey in Figure 4A).
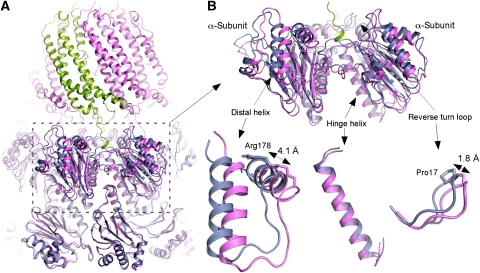
When β-rings of the 20S–PA26E102A−PAN9 and 20S are aligned and superimposed together, the α-subunit of 20S–PA26E102A−PAN9 appears rotated about 7° from the α-subunit of 20S. This rotation is hinged around the helix located in the wedge between the neighbouring β-subunits and connecting to the loop that forms the narrow substrate entry channel (Figure 5). Two residues in this channel, Gly127 and Gly128, have the smallest shift of all the residues in the subunit (0.3 and 0.4 Å respectively). This hinge helix (residues 81–104) has the smallest RMSD of only 0.7 Å (Figure 5B). The distal helix (residues 168–180) has a largest RMSD of 3.2 Å, and the Cα atom of Arg178 moved ∼4.1 Å. The Cα atom of Pro17 in the reverse turn loop moved ∼1.8 Å. Similarly, when β-rings of the 20S–PA26E102A−PAN9 and wild-type 20S–PA26 (PDB 1YA7) are superimposed, RMSD of the hinge helix is 0.7 Å, of the distal helix is 3.3 Å, and the Arg178 moved 5.2 Å. However, the Pro17 moved only ∼0.9 Å (Supplementary Figure 2B).
Figure 5.
Comparison of 20S–PA26E102A−PAN9 and wild-type 20S. (A) The atomic structures of 20S–PA26E102A−PAN9 complex (pink and green) and 20S (PDB ID: 1PMA; blue) are superimposed. All Cα atoms of β-rings were used to align the two structures. (B) An enlarged view of adjacent α-subunits. The rotation of α-subunit is also illustrated in the enlarged fragments. The two distal helices linked by a flexible loop has the largest movement with Arg178 moved 4.1 Å. The hinge helix has the smallest RMSD of only 0.7 Å. The reverse turn loop (Pro17) moved ∼1.8 Å.
As every pocket in the 20S α-ring contains a C-terminus of PAN, the combined conformational changes in each intersubunit pocket results in a symmetrical rotation of the α-ring relative to the β-rings and around the seven-fold symmetry axis. To accommodate the shift in each intersubunit pocket in the 20S, the PA26E102A−PAN9 rotates as a rigid body around the seven-fold axis.
Binding of ATPases to the eukaryotic 20S proteasome
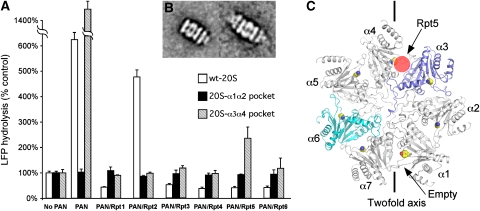
In the eukaryotic 26S proteasome, three (or in some species, four) of the six 19S ATPases, Rpt2, Rpt3, and Rpt5, have the complete HbYX motif in their C-termini. However, only the C-terminal peptides of Rpt2 and Rpt5 can stimulate gate opening in the mammalian and yeast 20S (Smith et al, 2007; Gillette et al, 2008) with Rpt5 being more potent than Rpt2 (Gillette et al, 2008). These data suggest that Rpt2 and Rpt5 each bind to a specific intersubunit pocket in the eukaryotic 20S α-ring to stimulate gate opening. Although the binding of the Rpt3-derived peptide by itself does not stimulate gate opening, in the 26S proteasome the Rpt3's HbYX motif does appear to contribute to gating (Smith et al, 2007; Gillette et al, 2008). To determine which Rpt subunit's C-terminus binds to which intersubunit pocket, we generated seven different T. acidophilum 20S mutants. In each of these mutants, a number of residues within the pocket were mutated to mimic each specific intersubunit pocket in the yeast 20S proteasome (Supplement Table II). The residues mutated were the ones shown in our crystal structure as ones that appear to interact with the HbYX motif (Supplementary Figure 3). The resulting 20S mutants would therefore be expected to contain seven identical intersubunit pockets that each mimics one of the seven pockets in the yeast 20S proteasome. These mutants were expressed and purified from E. coli, but only the mutants that mimic the pockets between subunits α1 and α2 and between subunit α3 and α4 produced intact 20S proteasomes, as examined by negative stain EM and size exclusion chromatography (data not shown). PAN stimulated gate opening very well in the 20S proteasome mutant containing the predicted α3–α4 pocket but showed no activity in the mutant containing the mimic of the α1–α2 pocket (Figure 6A).
Figure 6.
Binding of yeast proteasomal ATPases to the yeast 20S proteasome. (A) Wild-type T. acidophilum 20S (wt-20S), 20S with α1–α2 pocket mutations (20S-α1α2 pocket) and with α3–α4 pocket mutations (20S-α3α4 pocket) and the substrate LFP are incubated with the wild-type PAN and PAN mutants that have their C-termini replace by seven residues of yeast Rpt1–6. The stimulation of gate opening was measured by the increase of LFP hydrolysis over the control without any activator. The values are the mean±standard deviation from at least three independent measurements. (B) Averages of 20S with α3–α4 pocket mutations single (left) and double (right) capped with the PANRpt5 mutant. (C) Top view of yeast 20S proteasome α-ring, with each α-subunit marked. The side chains of lysine residues within the pocket, which is required to bind the C-termini of Rpt, are shown.
In addition, we expressed and purified six mutants of the PAN ATPases, in which PAN's seven C-terminal residues were replaced by the corresponding C-terminal residues of one of the yeast ATPases, Rpt1–Rpt6. The six different PAN mutants we generated thus contained six identical C-termini that were identical to those of one of the yeast Rpt's. We then tested which PAN mutants could stimulate gate opening of which 20S intersubunit pocket mutants by measuring the stimulation of LFP hydrolysis (Figure 6A). The wild-type T. acidophilum 20S and PAN were used as controls. The wild-type PAN and the PANRpt5 stimulated gate opening of the α3–α4 20S pocket mutant. None of the PANRpt mutants or wild-type PAN could stimulate gate opening of 20S α1–α2 pocket mutant, and PANRpt2 stimulates gate opening of wild-type 20S, but not the α3–α4 pocket mutant. Furthermore, by negative staining EM, only PANRpt5 was found to bind to one or both ends of the 20S α3–α4 pocket mutant (Figure 6B), whereas none of the PANRpts formed a complex with the α1–α2 pocket mutant. Our previous work showed that a Leu-to-Phe mutation in PAN's HbYX motif, though preserving the hydrophobic character, reduced the stimulation of gate opening significantly to only 20% of the wild-type PAN (Smith et al, 2007). Possibly, PANRpt5 and PANRpt3 did not stimulate T. acidophilum 20S gate opening, because the hydrophobic residue in these two hybrid PAN is phenylalanine.
These findings together argue strongly that the eukaryotic ATPases' C-termini bind very specifically to individual 20S pockets. Specifically, they strongly suggest that the pocket between subunit α3 and α4 is the binding site for Rpt5 (Figure 6C) that stimulates gate opening in the 20S. This result is consistent with the observation of Gillette et al (2008) that yeast Rpt5 cross-linked to the 20S's α4 subunit (which is present also in the pocket between α4 and α5). However, the negative data do not suggest which Rpt binds to the α1–α2 pocket.
Discussion
In the eukaryotic 26S proteasome, a key function of the six 19S ATPases is to open the gate in the 20S proteasome to allow substrate entry. The conserved HbYX motif in the C-terminus of the ATPases is a critical element in stimulating the 20S gate opening, especially for Rpt2 and Rpt5. The structural studies described here revealed that binding of PAN's C-terminus with the HbYX motif causes an induced-fit conformational change in the 20S intersubunit pocket and a rotation of the α-subunit. In addition, these structures identify at atomic resolution the critical residues in the 20S proteasome that interact with the HbYX motif and allow it to regulate gate opening in the 20S.
Solving high-resolution structure of the 20S proteasome in complex with the proteasomal ATPases has been a difficult task (Cheng, 2009). Most likely, the symmetry mismatches between the heptameric 20S proteasome and the hexameric proteasomal PAN ATPases is a primary cause of multiple conformations of the 20S–PAN complex (Smith et al, 2005). Although the structure of parts of PAN subunit has recently been determined (Djuranovic et al, 2009; Zhang et al, 2009), its interaction with the 20S has remained elusive. In the hybrid proteasomal activator, PA26E102A−PAN9, we used the PA26 as a scaffold for the C-terminal HbYX domain of PAN to promote a stable complex with the 20S that is amenable to crystallization. As shown here, this hybrid causes gate opening by the same mechanism as the proteasomal ATPase, rather than by that of PA26, and induces the same conformational change in the 20S α-subunits as are induced by the C-terminal peptides of PAN (Rabl et al, 2008).
One obvious concern is whether the conformation of the last seven C-terminal residues of PA26E102A−PAN9 actually mimics the conformation of PAN's C-termini when bound to the 20S intersubunit pockets. As peptides corresponding to the C-termini of PAN can autonomously induce gate opening, it is likely that attaching this sequence to the main body of PA26 does not alter its conformational properties from that in PAN. To ensure sufficient flexibility for the C-terminal HbYX domain in its interaction with residues in the intersubunit pocket, two glycine residues were inserted between the C-terminal residues of PAN and the body of PA26. This construct appeared to allow sufficient flexibility because another hybrid complex with four glycine residues in the linker (PA26E102A−PAN11) behaved the same, biochemically, as the one with two-glycine linker (PA26E102A−PAN9). The crystal structure of 20S–PA26E102A−PAN11 has the same conformation of its C-terminus as the one with only two-glycine linker, except with a less ordered conformation only in the linker region (data not shown). We also confirmed that even with the symmetrical binding of PA26E102A−PAN9 to the 20S proteasome, the intact HbYX motif is still required for gate opening and complex formation as shown by mutating the critical penultimate tyrosine. These observations make it likely that the conformation of the C-terminus in the hybrid is very similar to the native conformation of PAN's C-terminus when bound to the 20S intersubunit pocket.
This study does not resolve the symmetry mismatch problem, that is how the six C-termini on the PAN complex can dock into the seven pockets in 20S α-ring and activate gate opening. Our earlier work has pointed out that it is not necessary for the proteasomal ATPases to engage most intersubunit pockets simultaneously and is even unlikely to do so (Smith et al, 2005; Rabl et al, 2008). In fact, the specialized and redundant roles of Rpt2 and Rpt5 make such symmetric engagement highly unlikely. Although the use of a seven-fold symmetric PA26 as a scaffold offers clear advantage for structure determination, the rotation of the hybrid activator PA26E102A−PAN9 around the seven-fold axis could be an artefact caused by this symmetrical binding to all intersubunit pockets. However, the interactions showed here between the HbYX motif and the residues in the intersubunit pocket and the induced-fit conformational change within the pocket are clear findings that are independent of the symmetrical binding.
The major difference between PA26's C-terminus and PAN's HbYX motif is the penultimate tyrosine residue that does not exist in any of the PA26/PA28 family members. As it is absolutely essential for PAN's activation of the 20S, it is not surprising that this tyrosine residue forms multiple specific interactions with the intersubunit pocket that cannot be achieved by any other residues, even a phenylalanine. Several PAN family members and some of the eukaryotic 19S ATPases do not have a tyrosine in the penultimate residue. Thus, the main difference between 19S C-termini with or without an HbYX motif is the presence or absence of the penultimate tyrosine, which probably determines the conformation of the C-terminus bound to the intersubunit pocket. Many observations make it likely that the eukaryotic 19S ATPases subunits with HbYX motifs bind to the 20S in a similar conformation and open the 20S gate by a similar mechanism (Smith et al, 2005; Rabl et al, 2008). Our current findings suggest that whenever a proteasomal activator with the HbYX motif (ATPases or PA200) binds to the archaeal or eukaryotic 20S, the C-terminus with the HbYX motif assumes the structure of PAN's C-terminus and the intersubunit pocket undergoes an induced-fit conformational change that the intersubunit becomes smaller. In contrast, the C-termini lacking the HbYX motif may assume a structure resembling that of PA26's C-terminus. Consequently, the pocket would not undergo significant conformational change and would not move or rotate significantly.
Interactions of specific ATPases' C-termini with the individual intersubunit pockets
In the eukaryotic 20S, the N-terminus of each α-subunit contributes differently to the formation of the gate, but the N-terminus of α3 forms the scaffold of the gate (Groll et al, 2000). This subunit has a long N-terminus that spans across the entire pore in the middle of the α-ring and forms a number of interactions with residues in the N-termini of the other α-subunits (Figure 6C), and deletion of α3's N-terminus leads to a constitutively open gate (Groll et al, 2000). Binding of an ATPase C-terminal HbYX motif to the α3–α4 pocket should induce the conformational changes that move the reverse turn loop of α3-subunit away from the central pore, which should have the largest effects on disrupting the closed gate, more than moving the N-termini of any other α-subunits (Figure 6C). Therefore, docking of C-termini into this pocket should be more critical in opening the gate than binding to any of the other pockets.
Although the 19S Rpt2, Rpt3, and Rpt5 subunits all contain the canonical HbYX motif in their C-termini, only the peptides corresponding to the Rpt2 and Rpt5 C-termini can stimulate gate opening of the mammalian and yeast 20S, with Rpt5 being more potent than Rpt2 (Smith et al, 2007; Gillette et al, 2008). Therefore, Rpt2 and Rpt5's C-termini each binds to a specific intersubunit pockets to destabilize the closed gate. Our findings indicate that the C-terminus of Rpt5 most likely binds to the α3–α4 pocket. This conclusion is consistent with the previous observation that the C-terminal peptide of Rpt5 cross-linked preferentially to subunit α4 (Gillette et al, 2008), which could indicate Rpt5 docks into either α3–α4 or α4–α5 pockets. As the long N-terminus of α3 subunit serves as the backbone of the eukaryotic gate, the α3–α4 pocket is most likely to be a critical one in gate opening. Our data suggest the C-terminus of Rpt5 binds the α3–α4 pocket rather than the α4–α5 pocket.
Gate opening in the 26S proteasome is an ATP-dependent process but other factors influence the extent or duration of the open gate. Recently, three groups have found that ubiquitinated proteins further enhance gate opening beyond that seen with ATP alone (Bech-Otschir et al, 2009; Li and Demartino, 2009; Peth et al, 2009). All these findings and considerations emphasize the necessity of ordering the different ATPase subunits in definite positions during proteasome biogenesis. Several recent studies not only identified a number of factors that promote the precise assembly of the 19S ATPase ring, but also identified three sub-complexes of Rpt subunits that function as precursors in its assembly process (Besche et al, 2009; Funakoshi et al, 2009; Park et al, 2009; Roelofs et al, 2009; Saeki et al, 2009). Mutation of the extreme C-terminal residues prevented assembly of the ATPases on the 20S α-ring, resulting in the buildup of precursors in the cell, presumable because they were no longer able to associate with the 20S intersubunit pockets. Each of these precursors contained two ATPase subunits. It is likely that the specific interactions between the neighbouring Rpt subunits in these sub-complexes (Rpn1–Rpt2–Rpt1–Hsm3, Rpt4–Rpt5–Nas2, and Nas6–Rpt3–Rpt6–Rpn14) are maintained in the formation of the Rpt ring on the 19S base. The adjacent Rpt subunits of each three sub-complexes are likely held together through interaction of their N-termini coiled-coil domains, which requires a cis-proline, found in Rpt2, Rpt3, and Rpt5, to pair with a residue in trans-conformation in the neighbouring subunit (Zhang et al, 2009). On the basis of these precursors the likely order of the ATPases around the ring is Rpt1-2-6-3-4-5, as noted by Forster et al (2009). The data we present here suggest that Rpt5 binds to the α3–α4 pocket (Figure 6C). It is noteworthy that this proposed model is contradictory to the model proposed by Forster et al (2009), in which the Rpt2 binds to α3–α4 and Rpt5 binds to α6–α7.
Materials and methods
Molecular cloning, protein expression, and purification
Cloning of T20S, PAN plasmids was described earlier (Smith et al, 2007). Briefly, the α-, β-subunit genes of T. acidophilum 20S proteasome were sub-cloned in pRSET-A plasmid, a His6-tag attached to β-subunit C-terminus to facilitate protein purification. The Methanocaldococcus jannaschii PAN bearing M74A mutation was sub-cloned into the same vector but without the tag. T. brucei PA26 gene was ab initio synthesized along with an N-terminal His6-tag, and sub-cloned in NcoI, XhoI cutting sites of pET-15b plasmid.
T20S pRSET-A construct was transformed into BL21(DE3) cells. The transformant was inoculated into LB and grown to OD600 0.6–1.0. 0.1 mM IPTG was added to the culture to induce protein expression, and the culture was incubated at 37°C for another 4 h. Cells were resuspended in Lysis buffer (50 mM NaH2PO4 pH 8.0, 300 mM NaCl; 30 ml/l culture), French pressed, and spun at 20 000 r.p.m., 4°C for 20 min. Lysate supernatant was heat precipitated by adding 4 volume of boiling lysis buffer, and kept at 75°C for 15 min, and spun at 40 000 r.p.m., 4°C for 35 min. The protein was purified on an ÄKTA Purifier system (GE Healthcare) by Ni–NTA affinity chromatography on HiTrap Chelating column (GE Healthcare) and gel filtration on Superose 6 column (GE Healthcare). Gel filtration fractions were pooled and concentrated in Amicon ultrafiltration tube (Millipore) to 5–15 mg/ml in 20 mM Tris pH 8.0, 50 mM NaCl, 0.1 mM EDTA.
PANM74A pRSET-A construct was transformed into BL21(DE3) pLysS competent cells. Protein expression and cell lysis were performed the same way as 20S proteasome. Lysate supernatant was heat precipitated at 60°C for 15 min, spun to remove pellet, then heated to 80°C for 15 min, and spun again. Protein was purified by ion-exchange chromatography on a HiTrap Q XL column (GE Healthcare), and gel filtration on Superose 6 column. Protein was concentrated to 5–15 mg/ml in buffer containing 20 mM Tris pH 8.0, 50 mM NaCl, 0.1 mM EDTA.
PA26 pET-15b construct was transformed into BL21(DE3) competent cells. Protein expression and cell lysis were performed the same way as above. Protein was purified by Ni–NTA affinity chromatography and gel filtration. PA26 and mutants were concentrated to 10–20 mg/ml in buffer containing 20 mM Tris pH 8.0, 50 mM NaCl, 0.1 mM EDTA.
LFP activity assay
The sequence and hydrolysis of LFP, which is a fluorogenic nona-peptide substrate, were described earlier (Smith et al, 2007). In a typical assay, 0.7 μg of T20S proteasome was added in 200 μl LFP buffer (50 mM Tris pH 8.5, 1 mM DTT, 10 mM MgCl2, 5% glycerol, and 10 μM LFP), 1.3 μg PA26, or 2.1 μg PAN protein supplemented at a molar ratio 7:1 to 20S, and 10 μM ATPγS included if PAN presents. Reactions were prepared on ice and kept at room temperature after the first measurement. Fluorescence 322/398 nm was measured on Safire microtiter plate reader (Tecan). For each condition three independent measurements were performed.
CryoEM and image processing
CryoEM grids were prepared as described before (Rabl et al, 2008). Grids of frozen hydrated samples were imaged using a Tecnai TF20 electron microscope equipped with a field emission gun (FEI Company, USA) and operated at an acceleration voltage of 200 kV. Images were recorded at a nominal magnification of × 62 000 using a Gatan 4K × 4K charge-coupled-device camera by an automated data acquisition procedure, Leginon (Suloway et al, 2005). Defocus is ranged from −4 to −1 μm. Particle picking and contrast transfer function (CTF) determination were performed as described before (Rabl et al, 2008). FREALIGN (Grigorieff, 2007) was used for the refinement of Euler angles, in plane x–y shift of all particles, as well as for the CTF correction and 3D reconstruction. Wild-type archaeal 20S proteasome (PDB ID: 1PMA) was filtered to a resolution of 40 Å to serve as an initial model. A D7 symmetry was applied. During the refinement, a total of ∼17 000 double-capped 20S–PA26E102A−PAN9 particles were used to calculate two different 3D reconstructions of this complex. One 3D reconstruction was calculated with a small soft-edge mask that includes only the 20S proteasome and the other one was calculated with the same parameters, but a larger mask to include the entire complex, the 20S in the middle and the two PA26E102A−PAN9 complexes capped at the each end. The only difference between these two 3D reconstructions is that the first one does not have the densities of the activators, whereas the second one does. The densities of the 20S proteasome in both 3D reconstructions are identical. The 3D reconstruction without the densities of the activators is used as the reference for the next cycle of refinement to improve the accuracy of the geometrical parameters. The resolutions of both 3D reconstructions were estimated using Fourier Shell Correlation curve. Both 0.143 criteria (Rosenthal and Henderson, 2003) as well as commonly used 0.5 criteria were used for estimating the nominal resolution (Supplementary Figure 1B). The surface rendering of the cryoEM density map was produced with the program Chimera (Pettersen et al, 2004).
Crystallization and structure determination
Purified T20S was mixed with an excess of PA26E102A−PAN9 at a total protein concentration 5.0 mg/ml. The molar ratio was optimized to 1:7 so that ∼80% T20S were double capped by PA26E102A−PAN9. High-throughput screening of crystallization conditions resulted in several hits using Classic, PEG, and AmSO4 kits (Qiagen), a total of 288 conditions. Bigger crystals grew in up-scaled and refined conditions of these hits from hanging drops of 2 μl protein mixed with 2 μl reservoir solution and equilibrated against 0.5 ml reservoir solution. The best crystals from condition 0.1 M MES pH 6.8, 20–25% PEG 1500 showed a perfect bipyramid shape with size 0.1–0.2 mm. Several such crystals, some soaked shortly in crystallization solution supplemented with 5–10% PEG 400, were flash-frozen in liquid nitrogen before mounting on a goniometer in a cold nitrogen stream. Data were collected on beamline BL8.3.1 at Advanced Light Source synchrotron. The best data set reaching 4.0 Å resolution was collected from a single crystal. Diffraction patterns were integrated by Mosflm (Leslie, 1992), and data reduction processed using the CCP4 program package (No.4 CCP, 1994).
The structure was determined by molecular replacement using AMORE (Navaza, 1994). The T20S–PA26 complex structure (PDB ID: 1YA7) with the last eight residues of PA26 omitted was used as the template. The molecular replacement solution first went through rigid-body refinement and simulated-annealing molecular dynamics in CCP4; the difference electron density maps showed an obvious density for PAN's C-terminal peptide in the 20S intersubunit pocket. Several cycles of model building using program O (Jones et al, 1991) and refinement using CNS program package (Brunger et al, 1998) until R/R-free factors did not decrease. Throughout all refinement procedures, seven-fold non-crystallographic symmetry was applied. The insufficient high-resolution data for Wilson plot may limit the accuracy of B-factor.
Accession codes
Protein Data Bank (PDB): coordinate of the crystal structure has been deposited with accession code . Electron Microscopy Data Bank (EMDB): the density map of single particle cryoEM 3D reconstruction has been deposited with accession code 5130.
Supplementary Material
Supplementary Material
Review Process File
Acknowledgments
We thank Bridget Carragher and Clint Potter (Scripps Research Institute) for allowing us to use their electron microscope for data collection. CryoEM data were collected at the National Resource for Automated Molecular Microscopy (NRAMM), which is supported by the NIH through the National Center for Research Resources' P41 program (PR17573). X-ray diffractions were collected using Beamline 5.0.2 and 8.3.1 at Advanced Light Source. This study has been supported in part by a grant from NIH (R01GM082893) and a grant from UCSF Program for Breakthrough Biomedical Research (Opportunity Award in Basic Science) to YC and by grants from NIGMS (R01GM51923-13), the Multiple Myeloma Foundation and Johnson & Johnson Foundation to ALG, who is a senior fellow of the Ellison Foundation. DMS was supported by a fellowship from the Medical Foundation.
Footnotes
The authors declare that they have no conflict of interest.
References
- Bech-Otschir D, Helfrich A, Enenkel C, Consiglieri G, Seeger M, Holzhutter HG, Dahlmann B, Kloetzel PM (2009) Polyubiquitin substrates allosterically activate their own degradation by the 26S proteasome. Nat Struct Mol Biol 16: 219–225 [DOI] [PubMed] [Google Scholar]
- Benaroudj N, Goldberg AL (2000) PAN, the proteasome-activating nucleotidase from archaebacteria, is a protein-unfolding molecular chaperone. Nat Cell Biol 2: 833–839 [DOI] [PubMed] [Google Scholar]
- Benaroudj N, Zwickl P, Seemuller E, Baumeister W, Goldberg AL (2003) ATP hydrolysis by the proteasome regulatory complex PAN serves multiple functions in protein degradation. Mol Cell 11: 69–78 [DOI] [PubMed] [Google Scholar]
- Besche HC, Peth A, Goldberg AL (2009) Getting to first base in proteasome assembly. Cell 138: 25–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL (1998) Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr D 54: 905–921 [DOI] [PubMed] [Google Scholar]
- Cheng Y (2009) Toward an atomic model of the 26S proteasome. Curr Opin Struct Biol 19: 203–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djuranovic S, Hartmann MD, Habeck M, Ursinus A, Zwickl P, Martin J, Lupas AN, Zeth K (2009) Structure and activity of the N-terminal substrate recognition domains in proteasomal ATPases. Mol Cell 34: 580–590 [DOI] [PubMed] [Google Scholar]
- Forster A, Masters EI, Whitby FG, Robinson H, Hill CP (2005) The 1.9 A structure of a proteasome-11S activator complex and implications for proteasome-PAN/PA700 interactions. Mol Cell 18: 589–599 [DOI] [PubMed] [Google Scholar]
- Forster A, Whitby FG, Hill CP (2003) The pore of activated 20S proteasomes has an ordered 7-fold symmetric conformation. EMBO J 22: 4356–4364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forster F, Lasker K, Beck F, Nickell S, Sali A, Baumeister W (2009) An atomic model AAA-ATPase/20S core particle sub-complex of the 26S proteasome. Biochem Biophys Res Commun 388: 228–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funakoshi M, Tomko RJ Jr, Kobayashi H, Hochstrasser M (2009) Multiple assembly chaperones govern biogenesis of the proteasome regulatory particle base. Cell 137: 887–899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillette TG, Kumar B, Thompson D, Slaughter CA, DeMartino GN (2008) Differential roles of the COOH termini of AAA subunits of PA700 (19 S regulator) in asymmetric assembly and activation of the 26 S proteasome. J Biol Chem 283: 31813–31822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glickman MH, Ciechanover A (2002) The ubiquitin-proteasome proteolytic pathway: destruction for the sake of construction. Physiol Rev 82: 373–428 [DOI] [PubMed] [Google Scholar]
- Goldberg AL (2005) Nobel committee tags ubiquitin for distinction. Neuron 45: 339–344 [DOI] [PubMed] [Google Scholar]
- Grigorieff N (2007) FREALIGN: high-resolution refinement of single particle structures. J Struct Biol 157: 117–125 [DOI] [PubMed] [Google Scholar]
- Groll M, Bajorek M, Kohler A, Moroder L, Rubin DM, Huber R, Glickman MH, Finley D (2000) A gated channel into the proteasome core particle. Nat Struct Biol 7: 1062–1067 [DOI] [PubMed] [Google Scholar]
- Groll M, Ditzel L, Lowe J, Stock D, Bochtler M, Bartunik HD, Huber R (1997) Structure of 20S proteasome from yeast at 2.4 A resolution. Nature 386: 463–471 [DOI] [PubMed] [Google Scholar]
- Jones TA, Zou J-Y, Cowan SW, Kjeldgaard M (1991) Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A 47: 110–119 [DOI] [PubMed] [Google Scholar]
- Leslie AG (1992) Joint CCP4 and ESF-EACMB Newsletter on Protein Crystallography, vol. 26 Daresbury Laboratory: Warrington, UK [Google Scholar]
- Li X, Demartino GN (2009) Variably modulated gating of the 26S proteasome by ATP and polyubiquitin. Biochem J 421: 397–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe J, Stock D, Jap B, Zwickl P, Baumeister W, Huber R (1995) Crystal structure of the 20S proteasome from the archaeon T. acidophilum at 3.4 A resolution. Science 268: 533–539 [DOI] [PubMed] [Google Scholar]
- Navaza J (1994) AMoRe: an automated package for molecular replacement. Acta Crystallogr A 50: 157–163 [Google Scholar]
- No.4 CCP (1994) The CCP4 suite: program for protein crystallography. Acta Crystallogr D 50: 760–763 [DOI] [PubMed] [Google Scholar]
- Park S, Roelofs J, Kim W, Robert J, Schmidt M, Gygi SP, Finley D (2009) Hexameric assembly of the proteasomal ATPases is templated through their C termini. Nature 459: 866–870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peth A, Besche HC, Goldberg AL (2009) Ubiquitinated proteins activate teh proteasome by binding to USP14/UBP6 which cause 20S gate opening. Mol Cell (doi:10.1016/j.molcel.2009.11.015) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE (2004) UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem 25: 1605–1612 [DOI] [PubMed] [Google Scholar]
- Rabl J, Smith DM, Yu Y, Chang SC, Goldberg AL, Cheng Y (2008) Mechanism of gate opening in the 20S proteasome by the proteasomal ATPases. Mol Cell 30: 360–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roelofs J, Park S, Haas W, Tian G, McAllister FE, Huo Y, Lee BH, Zhang F, Shi Y, Gygi SP, Finley D (2009) Chaperone-mediated pathway of proteasome regulatory particle assembly. Nature 459: 861–865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenthal PB, Henderson R (2003) Optimal determination of particle orientation, absolute hand, and contrast loss in single-particle electron cryomicroscopy. J Mol Biol 333: 721–745 [DOI] [PubMed] [Google Scholar]
- Saeki Y, Toh EA, Kudo T, Kawamura H, Tanaka K (2009) Multiple proteasome-interacting proteins assist the assembly of the yeast 19S regulatory particle. Cell 137: 900–913 [DOI] [PubMed] [Google Scholar]
- Smith DM, Chang SC, Park S, Finley D, Cheng Y, Goldberg AL (2007) Docking of the Proteasomal ATPases' carboxyl termini in the 20S proteasome's alpha ring opens the gate for substrate entry. Mol Cell 27: 731–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DM, Kafri G, Cheng Y, Ng D, Walz T, Goldberg AL (2005) ATP binding to PAN or the 26S ATPases causes association with the 20S proteasome, gate opening, and translocation of unfolded proteins. Mol Cell 20: 687–698 [DOI] [PubMed] [Google Scholar]
- Suloway C, Pulokas J, Fellmann D, Cheng A, Guerra F, Quispe J, Stagg S, Potter CS, Carragher B (2005) Automated molecular microscopy: the new Leginon system. J Struct Biol 151: 41–60 [DOI] [PubMed] [Google Scholar]
- Unno M, Mizushima T, Morimoto Y, Tomisugi Y, Tanaka K, Yasuoka N, Tsukihara T (2002) The structure of the mammalian 20S proteasome at 2.75 A resolution. Structure 10: 609–618 [DOI] [PubMed] [Google Scholar]
- Voges D, Zwickl P, Baumeister W (1999) The 26S proteasome: a molecular machine designed for controlled proteolysis. Annu Rev Biochem 68: 1015–1068 [DOI] [PubMed] [Google Scholar]
- Whitby FG, Masters EI, Kramer L, Knowlton JR, Yao Y, Wang CC, Hill CP (2000) Structural basis for the activation of 20S proteasomes by 11S regulators. Nature 408: 115–120 [DOI] [PubMed] [Google Scholar]
- Zhang F, Hu M, Tian G, Zhang P, Finley D, Jeffrey PD, Shi Y (2009) Structural insights into the regulatory particle of the proteasome from Methanocaldococcus jannaschii. Mol Cell 34: 473–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Clawson A, Realini C, Jensen CC, Knowlton JR, Hill CP, Rechsteiner M (1998) Identification of an activation region in the proteasome activator REGalpha. Proc Natl Acad Sci USA 95: 2807–2811 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material
Review Process File