Abstract
Fanconi Anemia (FA) is an inherited genomic instability disorder, caused by mutations in genes regulating replication-dependent removal of interstrand DNA crosslinks. The Fanconi Anemia pathway is thought to coordinate a complex mechanism that enlists elements of three classic DNA repair pathways, namely homologous recombination, nucleotide excision repair, and mutagenic translesion synthesis, in response to genotoxic insults. To this end, the Fanconi Anemia pathway employs a unique nuclear protein complex that ubiquitinates FANCD2 and FANCI, leading to formation of DNA repair structures. Lack of obvious enzymatic activities among most FA members has made it challenging to unravel its precise modus operandi. Here we review the current understanding of how the Fanconi Anemia pathway components participate in DNA repair and discuss the mechanisms that regulate this pathway to ensure timely, efficient, and correct restoration of chromosomal integrity.
Keywords: DNA repair, interstrand crosslink, cancer, homologous recombination, translesion synthesis, protein ubiquitination
INTRODUCTION
Fanconi Anemia (FA) is a rare autosomal recessive or X-linked genetic disease, which has become a very well appreciated model system for several biological processes, including DNA repair, cancer progression and protein ubiquitination (31, 35, 73, 120, 143, 146). Clinically, FA is strikingly heterogenous. Its hallmarks are bone marrow failure, congenital abnormalities including skeletal defects and hypopigmentation, and early onset of cancer (5, 6). The renal, cardiac, gastrointestinal, and reproductive systems can also be affected. This surprisingly wide range of clinical findings can be explained by the fact that FA is a chromosomal instability disorder, and cells from FA patients accumulate DNA damage at an increased rate. Unrepaired DNA damage can activate pro-apoptotic pathways, leading, for example, to depletion of hematopoietic stem cells, causing pancytopaenia. Alternatively, defective DNA repair in FA cells can lead to mutations and translocations that cause inactivation of cell cycle barriers and result in acute myeloblastic leukemia (AML) and other blood and solid tumors. These stochastic events are responsible for the paradoxical phenomenon that the same population of cells can either be depleted (in anemia) or hyper-represented (in cancer) (101). In fact, bone marrow failure appears earlier in development, on average at age 7, and is the major cause of death, occurring around the age of 16 (5, 6). In contrast, the median age of cancer onset is 14, representing a major hurdle for patients that received bone marrow transplant to treat the anemia.
The most striking cellular hallmark of FA is hyper-sensitivity to a class of DNA damaging agents that create DNA interstrand crosslinks (ICLs), such as mitomycin C (MMC) or diepoxybutane (DEB) (9, 51, 125). ICLs are very toxic lesions. These covalent links prevent DNA unwinding, thereby blocking both DNA replication and transcription. In fact, ICL-inducing agents are so toxic to dividing cells that they are used successfully as chemotherapeutic agents. Cellular toxicity and quantification of chromosomal abnormalities induced by these chemicals (Figure 1a) are also used for clinical diagnosis of the FA disease (10), highlighting the difficulties of treating cancers in FA patients by classic chemotherapy. Administration of ICL-inducing chemotherapy has serious side effects on FA patients, as their cells cannot repair crosslinks.
Figure 1.
The Fanconi Anemia pathway protects against genomic instability. (a) Loss of the FA pathway leads to chromosomal aberrations, with a specific increase in the frequency of radial chromosomes following induction of ICLs. Shown is a metaphase spread of a Fanconi Anemia cell. (b) Schematic representation of the FA protein complex: FANCM and FAAP24 recruit a large multisubunit ubiquitin ligase, termed the core complex, to DNA lesions. This structure is composed of subcomplexes (shown in different colors): FANCA-FANCG, FANCC-FANCE-FANCF and FANCB-FANCL-FAAP100. The core complex monoubiquitinates FANCD2 and FANCI, which then localize to DNA repair foci together with FANCD1, FANCJ, and FANCN.
ICLs are difficult to repair, and they affect both strands of the helix (126). Therefore, it seems that higher eukaryotes have evolved the FA pathway as a special means to deal with this class of DNA damage. As we will discuss in this review, the main function of the FA pathway seems to be the coordination of several distinct repair activities, belonging to different classic repair pathways including nucleotide excision repair (NER), translesion synthesis (TLS), and homologous recombination (HR), in order to remove crosslinks (103).
Given that it is activated not only by crosslink-inducing chemicals, but also by other DNA damaging agents, such as ultraviolet radiation (UV), ionizing radiation (IR), hydroxyurea, and even spontaneously during replication (42, 48, 66, 139), the FA pathway is likely to be involved in the replication-dependent repair of many types of lesions. But unlike ICLs, most of these lesions are also repairable by other pathways. Thus, loss of FA proteins only mildly or not at all affects survival rates in response to IR or UV (15, 42, 65, 103). However, the FA pathway is the major mechanism that can remove crosslinks efficiently, accounting for the hypersensitivity of FA patient cells to ICLs.
The source and identity of the DNA lesions that affect progenitor cells during development, leading to the clinical characteristics of FA, is not clearly defined. It is likely that the endogenous metabolism of certain cells might specifically create agents that cause ICLs or DNA-protein crosslinks. Lipid metabolism in the liver, for example, may generate endogenous crosslinking compounds, perhaps accounting for the increased liver tumors in FA patients (101, 112). Additionally, circulating crosslinking metabolites such as formaldehyde may account for the sensitivity of FA hematopoietic stem cells (122). DEB, used in the diagnostic test for FA, might mimic the effect of some endogenous crosslinking compounds.
GENETIC SYSTEMS EMPLOYED IN STUDYING FANCONI ANEMIA
The availability of powerful genetic systems has allowed FA to become a highly valued model. There are thirteen FA complementation groups identified so far (subtypes A, B, C, D1, D2, E, F, G, I, J, L, M, N) (36–38, 40, 66, 83, 84, 86, 87, 92, 93, 98, 121, 129, 130, 132, 144, 157). In particular, the use of patient-derived cell lines has been essential for accumulating the current knowledge of the FA pathway. These cells were employed for somatic hybridization and functional complementation, leading to the identification of many of the thirteen complementation groups known. Initially, genes known to be required for crosslink repair in yeast and Drosophila were investigated but were not found mutated in FA patients. Instead, the first FA genes were cloned by positional cloning and complementation with cDNA libraries (36–38, 87, 132). Isogenic patient-derived cell lines corrected with the respective FA gene could then be used to investigate cellular phenotypes long before RNA interference technologies became available.
More recently, use of chicken B-cell derived DT40 cell lines has greatly bolstered the field (135). These cells show a high degree of conservation of basic cellular processes with mammalian cells, and the relative ease of achieving multiple gene knockouts by recombination techniques has allowed researchers to perform epistasis analyses. Also, the development of assays that evaluate the rates of somatic hypermutation and gene conversion at the endogenous immunoglobulin (Ig) loci, processes that rely on basic DNA repair machineries, made DT40 cells invaluable to the DNA repair field. Although the pathway is only partially conserved in nematodes, C elegans is also becoming a prized genetic system for FA study in the context of the whole organism, owing to its simplicity and genetic malleability (163).
Finally, knockout mice have been obtained for many FA or FA-associated genes (1, 20, 22, 65, 78, 150, 153, 162). Although, at the cellular level these mice recapitulate the phenotypes of human FA patient cells, including sensitivity to crosslinking agents and increased chromosomal abnormalities, the clinical features of FA are only partially represented in the knockout mice. Most notably, these mice do not develop anemia, perhaps reflecting some differences in the metabolisms of hematopoietic progenitor cells in mice versus humans, that make mouse cells less exposed to damage than the human ones. However, the cancer predisposition is recapitulated in this system, especially in the FANCD2 subtype (65).
THE CLASSIC FANCONI ANEMIA GENES
As mentioned before, somatic cell genetics has astoundingly revealed thirteen complementation groups (Table 1). A few other gene products were found to be associated with the FA protein complexes and required for FA activation, so it is presumed that their inactivation would lead to FA; however, patients with such mutations have not yet been described. The majority of FA proteins form a complex with ubiquitin ligase activity, termed the FA core complex (Figure 1b). This complex is required for monoubiquitination of FANCD2 and FANCI in response to DNA lesions during replication. FANCD2 and FANCI thereafter localize to DNA repair foci together with FANCD1 (and presumably FANCJ and FANCN), and other repair proteins including Rad51 and PCNA.
Table 1.
The thirteen complementation groups of Fanconi Anemia
| FA genes | Prevalence | Chromosomal position | Protein size (kDa) | Activity |
|---|---|---|---|---|
| FANCA | 66% | 16q24.3 | 163 | Core complex member; required for FANCD2-I ubiquitination |
| FANCB | 2% | Xp22.31 | 95 | Core complex member; required for FANCD2-I ubiquitination |
| FANCC | 10% | 9q22.3 | 63 | Core complex member; required for FANCD2-I ubiquitination |
| FANCD1 | 2% | 13q12–13 | 380 | HR mediator; downstream of FANCD2-I ubiquitination |
| FANCD2 | 2% | 3q25.3 | 155, 162 | Ubiquitinated following DNA damage |
| FANCE | 2% | 6p21–22 | 60 | Core complex member; required for FANCD2-I ubiquitination; binds directly FANCD2 |
| FANCF | 2% | 11p15 | 42 | Core complex member; required for FANCD2-I ubiquitination |
| FANCG | 9% | 9p13 | 68 | Core complex member; required for FANCD2-I ubiquitination |
| FANCI | 2% | 15q25–26 | 140, 147 | Ubiquitinated following DNA damage |
| FANCJ | 2% | 17q22–24 | 140 | Helicase; downstream of FANCD2-I ubiquitination |
| FANCL | 0.2% | 2p16.1 | 43 | Core complex member; required for FANCD2-I ubiquitination; PHD domain, ubiquitin ligase activity |
| FANCM | 0.2% | 14q21.3 | 250 | Helicase; localizes the core complex to DNA; required for FANCD2-I ubiquitination |
| FANCN | 2% | 16p12.1 | 140 | FANCD1 interactor; downstream of FANCD2-I ubiquitination |
The Core Complex: A Large Multisubunit Ubiquitin Ligase
Most FA patients harbor mutations in the genes that encode the core complex, with a wide majority (more than 60%) being defective in FANCA. Consequently, this was one of the first FA genes to be cloned. Other core complex members include FANCB, FANCC, FANCE, FANCF, FANCG, FANCL, and FANCM. Two other genes, FAAP24 and FAAP100 encode proteins that were found to be part of the core complex and required for the activity of the FA pathway, but mutations in these genes have not yet been associated with the FA disease (23, 85). Importantly, FANCM and FAAP24 seem to be functionally distinct from the other core members, as they are not constitutively associated with the complex. Instead, they are required for recognizing DNA lesions or stalled replication forks and are likely to be recruiting the complex to these sites (23, 74). Thus, we will discuss them separately. Structurally, very few FA proteins bear recognizable domains, which has made it very difficult to functionally characterize them. Crystal structures of sections of FANCE and FANCF revealed extended helical regions (79, 106). Interaction studies have shown the presence of subcomplexes, but whether they are functionally relevant is not clear. For example, FANCA and FANCG form a stable subcomplex (47), as do FANCB, FANCL, and FAAP100 (90). Finally, FANCC, FANCE, and FANCF are part of a subcomplex of their own (53, 137). The core complex is stabilized by weaker interactions among several members of different subcomplexes. FANCL is an E3 ubiquitin ligase with a PHD domain that modifies FANCD2 and FANCI with a single ubiquitin moiety, employing UBE2T as an E2 (57, 88, 91). Mutations in UBE2T have not been found in FA patients to this date. FANCL seems to be the only member of the ubiquitin ligase complex present in lower multicellular eukaryotes (together with FANCD2 and FANCI), and it is able to ubiquitinate its substrates in vitro in the absence of any other complex members. This suggests that the function of the other core proteins is to regulate the recruitment of FANCL to FANCD2 and FANCI, and possibly to modulate the ligase activity (4, 163). The carboxy-terminal sequence of FANCE interacts directly with the N-terminal region of FANCD2 (53, 108). Monoubiquitination of FANCD2 is a convenient diagnostic test for FA, given that it requires the function of all upstream FA proteins in the core complex (128).
FANCM and FAAP24: targeting the core complex to DNA
FANCM and FAAP24 are members of the XPF endonuclease family, which normally operate as heterodimers (24). Generally in the case of this family, both members of the complex have a nuclease domain, required for dimerization, but only one member is enzymatically active, whereas the other is required for DNA binding. In the case of FANCM-FAAP24, neither nuclease domain is active; instead FANCM has a DEAH helicase domain that combines the translocation of Holliday junctions with fork reversal activities to regress replication forks in vitro (49, 50). FAAP24 is able to bind single stranded DNA junctions via its nuclease region (23). In the absence of FANCM and FAAP24, binding of the core complex to chromatin is reduced and FANCD2-I is not efficiently ubiquitinated (74). Thus, the prevailing model suggests that FAAP24-FANCM recognize stalled replication forks and recruit the core complex to ubiquitinate FANCD2 and FANCI (35). However, unlike the case with core complex genes such as FANCA or FANCG, knocking out FANCM only partially decreases FANCD2 ubiquitination (122a, 129a). Perhaps there are alternative mechanisms for localizing the ubiquitin ligase activity to its substrates on chromatin. Alternatively, the core complex might be able ubiquitinate FANCD2 to some extent, without being targeted to chromatin. Indeed, the core complex remains intact in the absence of FANCM, suggesting that FANCM-FAAP24 acts as a landing pad that does not affect the ubiquitin ligase activity of the complex, but rather its availability for substrate binding (74).
Recent experiments suggested that the role of FANCM is more complex than previously appreciated. An ATPase-inactivating mutation supports mono-ubiquitination, but not crosslinker resistance, arguing that FANCM acts in the FA pathway both upstream and downstream of FANCD2-I ubiquitination (129a, 158a). Finally, unlike other FA proteins, FANCM is also involved in supressing sister chromatid exchanges (detailed below), showing that it has broader functions, outside the FA pathway.
FANCD2 and FANCI: the substrates for ubiquitination
FANCD2 is mutated in 5% of FA patients, and together with FANCI (even less common among FA patients) forms a dynamic complex that moves in and out of the chromatin, depending on the status of its posttranslational modifications (48, 97, 130). The two proteins are similar in size and domain structure, and they are monoubiquitinated by FANCL at K561 and K523, respectively. Monoubiquitination is considered the essential step in FA activation. Other roles of the core complex, outside of FANCD2-I ubiquitination, cannot be excluded. Indeed, a FANCD2 K561R-ubiquitin fusion can complement the phenotypes of FANCD2 knockout DT40 cells (suggesting that the position of the ubiquitin is not important), but cannot complement core complex deficient cells (89).
Whereas FANCD2 ubiquitination is indispensable for the DNA repair process, FANCI ubiquitination is not essential, although it may enhance repair (69). Instead, as discussed below, FANCI provides a major regulatory mechanism of the pathway, through its ataxia telangiectasia and Rad3 related (ATR)-dependent phosphorylation. FANCI phosphorylation might be required for its localization to chromatin, independently of core complex activity. Consistent with this notion, deletion of FANCI in DT40 cells ablates FANCD2 ubiquitination, but this is restored upon complementing the cells with a FANCI ubiquitination site point mutant, arguing that features of FANCI other than its ubiquitination site are essential for FA activation (69).
Ubiquitination of FANCD2-I leads to its localization to chromatin foci. These foci are considered to be DNA repair structures because they contain repair factors such as Rad51, BRCA1, BRCA2, NBS1, PCNA, or γH2AX (14, 48, 68, 99, 139, 148). The mechanisms that mediate these transitions, as well as the function of ubiquitinated FANCD2-I in these foci, are currently unknown. FANCD2 and FANCI contain poorly characterized ARM repeats (a protein-protein interaction motif) and acidic EDGE domains (130), but the relevance of these regions is also not known. An EDGE domain mutant of FANCD2 is normally ubiquitinated and localizes to DNA repair foci, but does not support crosslink repair, suggesting that this region is involved in the repair process downstream or independent of the ubiquitination step. The C-terminal EDGE domain has been proposed to be a binding site for an (unidentified) protein in the pathway (97).
FANCD1, FANCJ, and FANCN: The Downstream Factors
Members of this group are not required for FANCD2-I ubiquitination, and appear to function downstream in the repair process, or even independently, in a parallel pathway. FANCJ, also known as BRIP1 or BACH1, is the second helicase in the FA pathway (156). FANCJ interacts with the BRCT domains of BRCA1 (18) and localizes to DNA repair structures containing BRCA2 and RPA (56, 80). The function of FANCJ in crosslink repair is not known, but its 5′ to 3′ helicase activity is likely used to remodel DNA structures, thus facilitating or activating repair (17). Alternatively, given that FANCJ can unwind D-loop structures in vitro (55), it may be involved in resolving Rad51 nucleofilaments to finish the HR reaction.
FANCD1, better known as BRCA2, is a recombination mediator that facilitates the formation of Rad51 nucleofilaments (134, 161). Next to BRCA1, BRCA2 is a well-described tumor suppressor found inactivated in inherited breast, ovarian, and other cancers. Our laboratory identified BRCA2 to be mutated in FANCD1 patients (66). Rad51 foci are absent in these cells (52). Moreover, BRCA2 colocalizes with ubiquitinated FANCD2, and this protein is important for BRCA2 focus formation, thus clearly connecting the FA pathway to HR (68, 148). However, BRCA2 may have additional functions, outside the FA pathway. Indeed, the activity of BRCA2 is essential for recombination, unlike the other FA genes’ products, which only mildly impact HR (100, 134). Consistent with this notion, FANCD1 patients have a more severe phenotype, with a high increase in the risk of blood and solid tumors. Additionally, disruption of murine BRCA2 leads to embryonic lethality, which contrasts the mild phenotypes of other FA knockout mice (7, 127).
FANCN (also termed PALB2) was initially identified as a BRCA2 interaction partner required for localization of BRCA2 to chromatin and consequently for its function in HR (158). Similar to other FA genes, such as FANCF, inactivation of FANCN has been identified in several cancers (21, 138, 141). The precise function of FANCN in promoting crosslink repair is not known, although it seems likely to involve its BRCA2-binding activity to promote HR.
The Evolution of FA Genes
In line with the fact that the FA pathway is required for protection of progenitor cells in the hematopoietic system, FA genes appeared relatively late in evolution (35, 112, 146, 163). Core complex members are only present in vertebrates. Interestingly, FANCD2 and FANCI were also identified in lower multicellular eukaryotes such as C. elegans. In worms, FANCD2 is required for ICL repair and was shown to be ubiquitinated in response to DNA damage, raising the intriguing possibility that ubiquitin ligases other than the core complex might be involved in FANCD2 ubiquitination (28, 163). BRCA2 is present in multicellular eukaryotes, but it is not found in yeast.
The helicases FANCJ and FANCM appear the earliest in evolution, having structural and functional homology to the bacterial helicase dinG and the archeal Hef, respectively (93, 98, 156). FANCJ and dinG share the same substrate preference in vitro (they require a pre-existing 5′ single-stranded tail) (56, 145). It seems likely that the ancient homologs of these helicases were co-opted in the FA pathway as they provided an enzymatic activity critical for ICL repair.
REPAIR OF INTERSTRAND CROSSLINKS
Models of ICL Repair
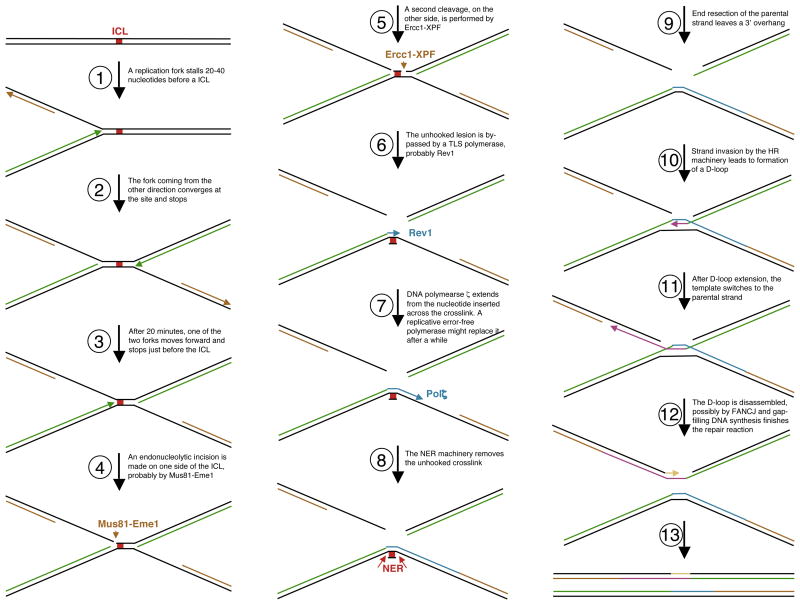
ICLs covalently link both strands of a DNA duplex, thereby blocking unwinding by DNA helicases and stopping the progression of the replication and transcription machineries. Therefore, ICLs pose an extreme danger to the cell: a single crosslink can kill a yeast cell if not repaired or bypassed (126). A single DNA repair pathway (in the classic sense) cannot deal with such a formidable structure. ICLs seem to be removed almost exclusively during DNA replication (2). Based on the biochemical and genetic characteristics of the pertinent enzymes, it has been proposed that ICL repair requires the coordinated and sequential activity of enzymes belonging to several different pathways (41, 101, 103, 146) (Figure 2).
Figure 2.
Speculative model for the coordinated use of multiple classic DNA repair pathways in replication-dependent repair of ICLs (adapted from ref. 119). Replication forks converging from different directions stall at an ICL site, 20–40 nucleotides before the lesion (steps 1 and 2). Subsequently, one fork moves forward and stops just before the lesion (step 3). Dual incision on each side of the ICL, likely performed by Mus81-Eme1 (the proximal –step 4) and Ercc1-XPF (the distal –step 5) unhook the ICL, which is then bypassed by a TLS polymerase, probably Rev1 (step 6). DNA polymerase ζ extends from this initial insertion (step 7), and the NER system removes the bypassed crosslink (step 8). The HR machinery then repairs the broken chromatid using the newly repaired sister as a template (steps 9–13). (It is also possible that a double Holliday junction is the relevant structure of the HR reaction –steps 11 and 12.)
Once DNA replication is blocked at a crosslink, a double strand break (DSB) is generated by the activity of endonucleases (34, 58). This event uncouples one sister chromatid from the other. Once the same strand is cleaved on the other side of the lesion (102), the crosslinked base can be unhooked and the remaining structure bypassed by specialized polymerases that are able to replicate through DNA lesions (103). The bypassed, unhooked crosslink resembles single nucleotide lesions and can be repaired by hydrolysis or nucleotide replacement. This allows the re-establishment of the replication fork, likely by HR machinery-mediated invasion of the repaired chromatid by the sister. Indeed, ICLs induce sister chromatid exchanges (SCEs), a quantifiable event which requires the HR machinery (101, 113). This suggests that HR repair is used at a step during the succession of events leading to crosslink removal. Thus, efficient crosslink repair is believed to require the coordinated efforts of three classic pathways: NER, TLS, and HR.
It is worth mentioning that a secondary, minor pathway, that is replication and recombination-independent, has been described in yeast. This mechanism employs endonucleolytic unhooking followed by TLS on one strand and a second round of incisions and DNA synthesis on the other strand (124, 126). A candidate nuclease for this alternative crosslink repair activity is the Pso2 (SNM1) nuclease (12, 39). This pathway is perhaps important for removal of ICLs in nonreplicating cells (126), and it may be hyperactive in FA pathway deficient cells.
A Biochemical System for ICL Repair
A well-defined biochemical setup, taking advantage of the Xenopus egg extract replication system, was recently developed by J. Walter and colleagues to study replication-dependent crosslink removal (119). Repair of a circular plasmid with a chemically engineered ICL takes place in a stepwise manner (Figure 2). Importantly, in this system, not one but two replication forks, coming from opposing directions, converge on a crosslink, with the 3′ ends of their leading strands arresting ~20 nucleotides before the lesion. After approximately 20 minutes, during which time the replication machinery is probably reconfigured, one of the two leading strands advances to within one nucleotide of the crosslink. This replication fork stops again for approximately 30 minutes. Dual incisions take place on one strand, on both sides of the crosslink. The unhooked lesion is bypassed, probably by translesion polymerases, in two steps: insertion of a nucleotide across from the template base that forms the ICL, followed by DNA polymerase ζ-dependent extension of the growing strand past the lesion. Finally, two fully repaired sister duplexes are formed. The HR and NER machineries are probably employed to repair the broken sister chromatid and to remove the adduct left hooked on the parental strand, respectively. Importantly, during the replication of this crosslinked plasmid, both the ATR and the FA pathways become activated, as measured by Chk1 phosphorylation and FANCD2 ubiquitination (119).
EPISTASIS PATHWAYS CONTROLLED BY FANCONI ANEMIA
The use of in vivo assays to quantify the efficiencies of different DNA repair mechanisms has allowed for the identification of defective repair activities in FA deficient cells. Of particular interest are homologous recombination, translesion synthesis, and nucleotide excision repair mechanisms, theoretically required for crosslink removal.
NER
The initial incisions that unhook the crosslink are likely performed by NER endonucleases, and this process does not appear to require other NER factors. Although endonucleases have not been found mutated in FA patients up to date, the phenotypes of some endonuclease mutants partly mimic FA phenotypes, strengthening the argument for their involvement in the FA pathway. Two structure-specific heterodimeric nucleases, Ercc1-XPF and Mus81-Eme1 seem to be involved in crosslink repair, given that cells deleted of these factors are exquisitely (the former) or moderately (the latter) sensitive to crosslinking agents (58, 70, 81, 102, 117). Unlike other NER members, Ercc1 deficient mice show hematopoietic defects similar to FA pathway deficient cells (117).
It is believed that the cleavage is achieved in a highly coordinated manner, with Mus81-Eme1 performing the initial incision 3′ of the lesion and Ercc1-XPF being responsible for the secondary 5′ cleavage (24, 35). As Mus81-Eme1-deficient cells are not hypersensitive to ICLs, it is possible that other endonucleases can perform this function in their absence. It is not known how this process is regulated or whether the FA pathway is involved, but XPF colocalizes with FANCA (131). Recent studies indicate that the Ercc1-XPF cleavage step is required for efficient recruitment of monoubiquitinated FANCD2 at the sites of crosslink repair (13).
HR
In some cases, ICLs induce the formation of radial chromosomes, which may represent fusions of chromosomes at crosslink sites, or aberrant pairing of nonhomologous chromosomes. It is likely that radials arise from the activity of repair mechanisms that do not employ homology-mediated recombination, such as end-joining of chromosome breaks (101). Indeed, FA cells exhibit higher rates of microdeletions and insertions, consistent with a state of hyperactive nonhomologous end joining (NHEJ) (109). Although SCE levels are not strongly affected in FA cells, the frequency of radial formation is greatly increased, suggesting that one function of the FA pathway is to correctly harness the HR machinery (77, 98, 103). Mutations in the FA core complex, as well as in FANCD2 or FANCI, reduce homologous recombination rates, consistent with the observation that activated FANCD2 is present in Rad51 foci (100, 103, 130, 159). HR proteins such as Rad51, XRCC2, or XRCC3 are known to be involved in crosslink repair (103, 136). Importantly, IR-induced Rad51 focus formation requires both FANCD2 and FANCD1 (BRCA2), and the three proteins colocalize (52, 134, 139, 148). Moreover, FA factors are epistatic to XRCC2 and XRCC3 in DT40 (61, 103, 152). Loss of BRCA2 has a much more severe HR phenotype compared with core complex mutants, suggesting that BRCA2 has additional functions in HR independent of the FA pathway (7, 127). Indeed, FA proteins are also required for single strand annealing (SSA), a pathway that is upregulated in BRCA2 deficient cells (82, 100, 134).
TLS
A role for the FA pathway in controlling mutation rates has also been revealed. FA patient cells deficient in the core complex have a lower frequency of substitutions and small deletions/insertions at endogenous sites, and DT40 cells with a deletion of the FANCC gene have reduced somatic hypermutation of the immunoglobulin loci (59, 60, 95, 103, 109, 110, 142). This suggests that TLS pathways, generally responsible for this type of point mutation, are negatively affected. Indeed, the genes encoding TLS polymerases Rev1 and ζ (Rev3/Rev7) are required for crosslink repair and epistatic to FANCC in DT40 cells (103, 105). Rev1 focus formation is partially dependent on the core complex in human cells, requiring the BRCT domain of Rev1, as opposed to its ubiquitin binding motifs, which are activated by PCNA ubiquitination (54, 95). Also, DNA polymerase ζ is necessary for completion of ICL repair in the Xenopus biochemical system (119).
Using a plasmid-based assay to quantify the frequency of point mutations, an unexpected feature of the FA pathway in controlling mutagenesis was recently described (95). Whereas FA patient-derived core complex mutants show reduced mutagenesis, surprisingly, loss of FANCD2 or FANCI has the opposite effect, leading to a significant increase in the mutation rate. Thus, the FA pathway might in fact be forked, with the FANCD2-I ubiquitination branch responsible for HR control and the core complex-dependent branch controlling TLS via Rev1. Moreover, there is crosstalk between the two branches, as FANCD2-I deficiency results in upregulation of TLS (95). However, DT40 cells bearing a disruption in the FANCD2 gene show a slight reduction in the number of point mutations at the endogenous IgV locus (159), suggesting differential involvement of FANCD2 in controlling mutagenesis.
It is important to note that most experiments addressing the role of FA proteins in HR and TLS were performed either in the absence of exogenous damage, or after induction of other types of non-ICL damage (UV, enzymatically-induced DSBs) (95, 100). Moreover, in DT40 cells, FA factors are required for point mutagenesis at the endogenous Ig locus (103, 159). Finally, FANCD2 ubiquitination is induced by a broad spectrum of DNA damaging agents (42, 48, 66, 139). Taken together, these observations suggest that the FA pathway has a more general role in DNA repair than the dedicated removal of DNA crosslinks. The FA pathway appears to be extremely plastic, capable of fine-tuning the repair process. It can direct removal on non-ICL lesions using either error-prone (via TLS) or error-free (via HR) repair. When it comes to crosslink repair, it must use both pathways, in succession. Accordingly, it was shown that cells lacking Rev3 or Rev7 do not form SCEs in response to crosslinks, suggesting that in the absence of TLS there is no HR (which creates SCEs), and thus confirming the model of a sequential activity of DNA repair pathways in crosslink removal (103).
Importantly, knockdown of TLS or HR factors does not affect FANCD2 ubiquitination, suggesting that they are downstream of FA activation. It is also of note that FA mutants have normal or even elevated NHEJ repair (99, 109).
REGULATION OF THE FANCONI ANEMIA PATHWAY
Probably reflecting the deleterious effects of unrestricted activation of the FA pathway, numerous mechanisms that control its activity have been described (Figure 3). The current understanding is that the pathway is off by default, but it is rapidly turned on when required. Upon completion of DNA repair, it is switched off again. Finally, the FA pathway seems to be restricted to S-phase, and several mechanisms ensure that it cannot be activated in mitosis. It is not known whether unscheduled activation of the FA pathway causes DNA damage. One possibility is that its ability to harness mutagenic repair mechanisms, such as DNA replication using TLS polymerases, must be kept in check. Also, the helicase activities associated with FA might be deleteriously acting on catenation intermediates during chromosome disjunction in mitosis. In the following, we will summarize the most relevant elements that modulate FA activity.
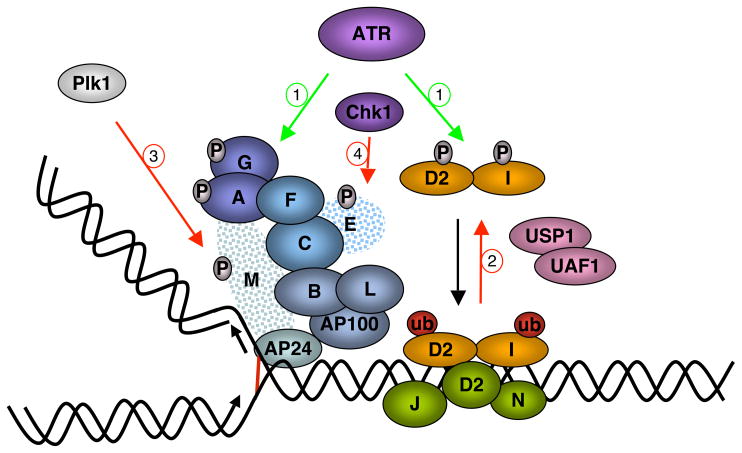
Figure 3.
Regulation of the FA pathway. Positive regulation is marked by green arrows, and negative control is represented by red arrows. (a) ATR initiates the pathway by phosphorylating FANCI, FANCD2, FANCA, and possibly FANCG. (b) The deubiquitinating enzyme USP1 and its partner and activator UAF1 remove ubiquitin from FANCD2 and FANCI, thus suppressing the pathway. (c) In mitosis, the Plk1 kinase phophorylates FANCM, creating a recognition motif for the ubiquitin ligase βTRCP. FANCM becomes multiubiquitinated and degraded by the proteasome. (d) Chk1-dependent phosphorylation of FANCE leads to its degradation.
Role of ATR in FA Activation
The main upstream regulator of the FA pathway is ATR (46). This kinase, responsible for coordinating the cellular response to DNA damage in S-phase, is required for efficient FANCD2 ubiquitination and focus formation (8, 62, 115). Cells from Seckel syndrome patients, defective in ATR, are similar to FA cells, showing crosslink hypersensitivity and increased chromosomal aberrations (8). ATR phosphorylates, directly or via its effector kinase Chk1, several components of the FA machinery, including subunits of the core complex such as FANCA and FANCE, and its substrates FANCD2 and FANCI (8, 62, 115, 118, 130, 149, 152).
The most relevant targets for ATR-mediated phosphorylation in this pathway seem to be FANCD2 and FANCI. Although FANCD2 phosphorylation was initially associated with FA activation (8), recently it has been inferred from studies in DT40 cells that FANCI phosphorylation plays a dominant regulatory role (69). ATR-dependent phosphorylation of at least six residues in FANCI is sufficient to induce FANCD2 ubiquitination and efficient repair, even in the absence of FANCD2 or core complex phosphorylation. It is believed that this modification can somehow recruit FANCD2-I to the core complex, or alternatively inhibit its deubiquitination (147). As mentioned above, experiments in DT40 cells showed that the relevant substrate of ubiquitination by the core complex is FANCD2 (69). Therefore, the FA pathway provides a novel example of in trans phosphorylation-ubiquitination cross-talk: phosphorylation of a factor is required for ubiquitination of its binding partner (147).
Besides acting on FANCD2 and FANCI, ATR also phosphorylates FANCA after induction of DNA crosslinks, and this event is required for the nuclear localization of FANCA and for efficient crosslink repair (27). FANCG is also phosphorylated to promote repair (118, 152).
Deubiquitination of FANCD2 by USP1-UAF1
A major mechanism for keeping the FA pathway in check is the removal of ubiquitin from FANCD2 and FANCI. This is achieved by the activity of the deubiquitinating enzyme USP1 (104, 130), which is also under several levels of control. USP1 is found in vivo in a constitutively active deubiquitinating complex, together with its partner UAF1 (USP1 Associated Factor 1). UAF1 is a WD40-domain protein that acts as an activator of USP1, stimulating its activity toward the substrate (25, 26). The USP1-UAF1 complex keeps in check FANCD2 ubiquitination under normal conditions (26, 67). Upon DNA damage, the activity of this complex is repressed by two mechanisms. First, transcription of the USP1 gene is turned off, and second, the USP1 protein present in the cells is quickly degraded by the proteasome, in a process that appears to be initiated by an endo-proteolytic cleavage (possibly autocleavage) at an internal GG motif of USP1. This exquisite regulation allows the buildup of excess ubiquitinated FANCD2 and FANCI.
FANCM Phosphorylation and Degradation
A novel level of control of the FA pathway, involving FANCM post-translational modifications, was recently described (71, 74, 157). The FANCM-FAAP24 complex is bound to chromatin during the cell cycle but can only recruit the core complex in S-phase, when FANCM phosphorylation is reduced (74). After completing replication, FANCM is hyperphosphorylated leading to the release of the core complex from chromatin. This coincides with loss of FANCD2 monoubiquitination. Several kinases seem to be involved in FANCM phosphorylation, and we recently identified the mitotic regulator polo-like kinase (Plk1) as one of them (71). FANCM has a consensus Plk1 binding site, which is required for its Plk1-mediated phosphorylation in mitosis. This event creates a phospho-degron in FANCM, recognized by the SCF-βTRCP ubiquitin ligase and leads to ubiquitination and subsequent proteasomal degradation of FANCM. This mechanism ensures that the core complex is not recruited to chromatin during mitosis. Expression of nondegradable mutants of FANCM allows chromatin binding of the core complex in mitosis, leading to formation of radial chromosomes, suggesting that FANCD2-I ubiquitination in mitosis is deleterious.
Thus, it seems that shutting off the FA pathway occurs by two distinct mechanisms: deubiquitination of FANCD2-I by USP1-UAF1 or blockage of ubiquitination by release of the core complex from chromatin.
Chk1-dependent FANCE Degradation
A Chk1-mediated regulatory mechanism that modulates the ubiquitin ligase activity of the core complex, rather than its localization to substrates, was also described (149). Two Chk1 consensus phosphorylation sites in FANCE are required for crosslink repair. Depletion of Chk1 or mutations in Chk1 phosphorylation sites of FANCE result in basal increase in FANCD2 ubiquitination, but no damage-induced modification. Surprisingly, FANCE phosphorylation by Chk1 ultimately leads to its proteasomal degradation, suggesting that this might be yet another mechanism for negatively regulating the FA pathway.
Modulating the Local Availability of the Factors Required for FA Activation
Another possible level of regulation is controlling the local concentrations of each of the factors involved in FA activation: the E3 ligase, the E2 conjugating enzyme, and the substrates. As mentioned above, the core complex is targeted to chromatin by FANCM-FAAP24 and released upon FANCM phosphorylation and degradation (71, 74). Experiments employing DT40 cells showed that after DNA damage, the FANCD2-I complex is initially loaded in chromatin independently of the core complex, even in the absence of ubiquitination (3). How FANCD2-I is targeted to chromatin is unknown. This process might involve FANCI phosphorylation by ATR, as described above (69). The E2 enzyme UBE2T is constitutively present on chromatin, its localization not being affected by cell cycle progression or induction of DNA damage (3).
HOW DOES THE FANCONI ANEMIA PATHWAY ACTIVATE DNA REPAIR?
An integrated model of DNA repair by FA proteins is currently only speculative, and many details are missing. The activation of the pathway occurs almost exclusively in S-phase, both constitutively and especially in response to DNA damage. The constitutive S-phase activation may represent a response to endogenous damage or to replication intermediates that require some form of processing or lead to replication fork stalling. As described above, ATR activation is considered the initial trigger for turning the FA pathway on (8, 69). Recruitment of the core complex to sites of DNA damage by FANCM-FAAP24 allows it to ubiquitinate FANCD2 and FANCI (74), which are probably targeted independently to chromatin (3), following ATR-mediated phosphorylation of FANCI. Interestingly, in DT40 cells, FANCM inactivation partly alleviated the crosslink toxicity of FANCC mutants (98), and acquired mutation of FANCM appears to reduce the severity of the FA phenotype in a FANCA patient. (129a and R. Meetei and J. de Winter, personal communication). These results suggest that FANCM channels the repair process into the FA pathway by creating a structure that can only be processed by the FA complex. If this is disrupted, the FA-dependent repair may be deleterious, leading to the high chromosomal aberrations observed in these mutants. If, however, FANCM is not present, repair may occur through another pathway leading to partial suppression of the deleterious events observed in core complex mutants (35). Alternatively, FANCM might normally inhibit another crosslink repair mechanism, leaving cells with no ICL defense pathway in the absence of the core complex. In this scenario, acquired mutation of FANCM may restore some crosslink repair.
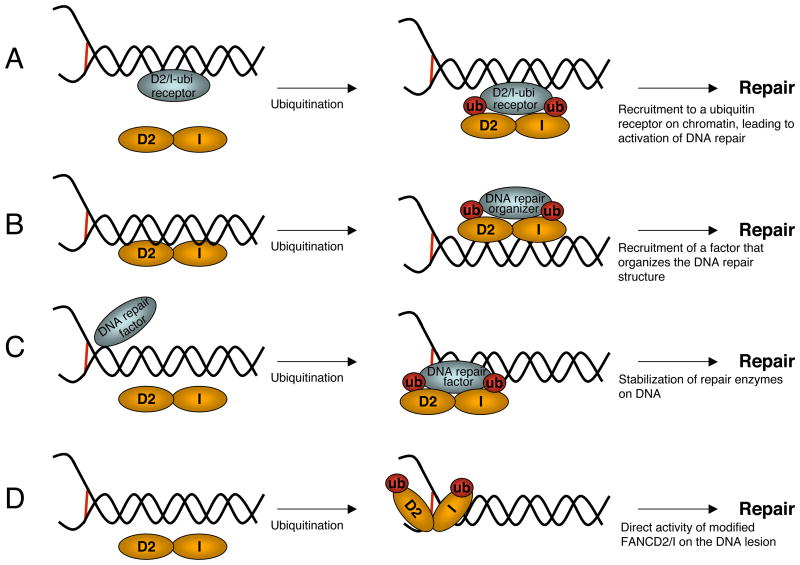
Once ubiquitinated, FANCD2-I complexes show redistribution to specific foci, where they colocalize with BRCA2, Rad51, and PCNA, among other proteins (Figure 4) (48, 68, 100, 139, 148). It is not clear how ubiquitination allows the formation of these complexes. Moreover, the function of FANCD2-I at these sites is not known. One model poses that so-called ubiquitin receptors are required to concentrate ubiquitinated FANCD2-I to foci, where the presence of this complex somehow activates DNA repair (Figure 4a) (64). Alternatively, ubiquitinated FANCD2-I might recruit another molecule that can, in turn, localize repair factors to the damage sites (Figure 4b), similarly to the proposed role for H2AX ubiquitination in DNA damage signaling (160). Finally, it is attractive to imagine that modified FANCD2-I can enhance the activity of repair enzymes, perhaps by localizing them to their substrate (Figure 4c), in analogy to PCNA ubiquitination-dependent recruitment of TLS polymerases (63, 96). Candidates for such factors include endonucleases, TLS polymerases, and HR factors, all of which localize to FANCD2 foci. Concordantly, recent experiments employing the Xenopus egg extract crosslink repair system showed that depletion of FANCD2 blocks lesion bypass, with the leading strand stalling immediately before the damaged template base (J. Walter, personal communication). This suggests that an essential factor for lesion bypass, perhaps a specific TLS polymerase, is recruited by FANCD2 during crosslink removal. Finally, ubiquitination of FANCD2-I may lead to structural modifications, allowing them to function in repair (Figure 4d), as described for the SUMOylation of the base excision repair enzyme thymine DNA glycosylase (11). Although FANCD2 and FANCI have no known enzymatic activity, they are capable of directly binding DNA structures in vitro (111) (P. Sung, personal communication), and it will be interesting to study how ubiquitination affects this property.
Figure 4.
Models for the function of FANCD2-I ubiquitination. (a) ubiquitination allows FANCD2-I binding to a nuclear receptor on chromatin, leading to its accumulation in foci and activation of DNA repair. (b) Ubiquitinated FANCD2-I recruit factors that organize the repair response and bind other repair enzymes. (c) Modified FANCD2-I complexes stabilize, activate, or enhance the enzymatic activities of repair factors. (d) Ubiquitination leads to a structural change in FANCD2-I that activates a cryptic DNA repair activity of the complex. These models are not mutually exclusive.
Another important question is the function of deubiquitination. Surprisingly, USP1 deletion confers ICL sensitivity and chromosomal aberrations in mouse and chicken cells (75, 107), suggesting that FANCD2 deubiquitination is an essential step in crosslink repair. A USP1 knockout mouse generated in our lab showed a phenotype strikingly reminiscent of FANCD2 mutant mice, including small size, infertility, and a reduction in the number of bone marrow hematopoietic stem cells (75). These results suggest that, rather than just turning the pathway off, USP1-mediated deubiquitination may be part of the repair process per se. Indeed, overexpression of free FANCD2 cannot alleviate the repair defect of USP1-deleted cells, arguing that it is not due to titrating out unmodified FANCD2. Concordantly, in USP1-deleted cells ubiquitinated FANCD2 is unable to localize to foci, even though it could accumulate on chromatin (75). Thus, USP1 may be required for correctly localizing FANCD2-I to DNA repair sites. Whether this function requires USP1-mediated deubiquitination of its substrates or some nonenzymatic activity of USP1 is not currently understood.
Finally, the activities of the downstream FA factors FANCJ, FANCN, and FANCD1 are possibly required for resolving repair intermediates. All three proteins are involved in HR; so it is conceivable that FA-dependent DNA repair employs a recombination-mediated event to complete the repair (156, 158, 161).
FUNCTIONS OF THE FANCONI ANEMIA PATHWAY OUTSIDE REPLICATION-DEPENDENT ICL REMOVAL
In addition to the major activity of the FA pathway in removing DNA lesions during replication, several of its components are involved in alternative mechanisms that promote genome stability (Table 2). It is not clear whether these processes are distinct from the FA pathway, simply representing harnessing of FA constituents by other pathways, or are coregulated and integrated as part of the FA-mediated response.
Table 2.
Functions of the FA proteins
| Function | FA members involved | Requires FANCD2/I ubiquitination? |
|---|---|---|
| Replication-coupled removal of ICLs (and probably of other DNA lesions) | All | Yes |
| ATM-dependent intra-S checkpoint | FANCD2 | No |
| ATR-dependent DNA damage checkpoint | FANCM | No |
| Suppression of SCEs | FANCM | No |
| Maintenance of G-rich DNA sequences | FANCJ | No |
| Cytokinesis | FANCD1 | No |
| Mediators of Homologous Recombination | FANCD1 | No |
| Handling reactive oxygen species | FANCC | No |
FANCD2 and the Intra-S Checkpoint
As part of the response to DSBs, the checkpoint kinase ataxia telangiectasia mutated (ATM) activates the intra-S checkpoint and arrests replication (76). FANCD2 was found to be phosphorylated by ATM at S222 (140). Although this modification is not important for FANCD2 ubiquitination, focus formation, or crosslink repair, it is essential for establishing the intra-S checkpoint. FANCD2 deficient cells expressing a FANCD2 phospho-mutant protein show radio-resistant DNA synthesis, failing to block replication after IR treatment. It is not known how FANCD2, once phosphorylated by ATM, acts as an effector of this checkpoint, but it is likely that this function is independent of its ubiquitination-mediated DNA repair role (140).
The activation of ATM is facilitated by the Mre11/Rad50/Nbs1 (MRN) complex, which acts as a sensor of DSBs (151). Accordingly, FANCD2 phosphorylation is absent in cells from ATLD or Nijmegen syndromes patients, which are defective in Nbs1 and Mre11, respectively (99). The MRN complex is also involved in crosslink repair, and it colocalizes with FANCD2 in chromatin foci. However, MRN and FANCD2 foci are formed independently, and probably represent separate mechanisms for removal of crosslinks (99).
FANCM in ATR/Chk1 Checkpoint Signaling
Recently, FANCM and FAAP24 were identified as members of a complex formed independently of the FA pathway, containing ATR and its effector HCLK2 (29). Cells depleted of FANCM or FAAP24 are unable to block mitotic entry after replication stress. It is not clear if FANCM-FAAP24 are sensors or effectors of the ATR-Chk1-mediated checkpoint. The helicase activity of FANCM is required for checkpoint signaling, suggesting that FANCM might be involved in remodeling stalled replication forks to facilitate the checkpoint signaling (29).
Involvement of FA Proteins in Suppressing SCEs
The yeast homologs of FANCM, Mph1 (in S. cerevisiae), and Fml (in S. pombe) are involved in suppressing sister chromatid exchanges by dissociating Rad51-induced D-loops, thereby channeling the recombination reaction into a noncrossover branch (116, 133). In DT40 cells, deletion of FANCM also leads to a fivefold increase in SCEs, a level significantly higher than the mild twofold increase of FANCC or FANCD2 mutants (122a). The function of FANCM in suppressing SCEs requires its helicase activity, which is not necessary for efficient FANCD2 ubiquitination (122a).
BLM is a helicase required for genomic stability and for suppressing SCEs (154). BLM and its cofactors, TopIIIα and RPA, are associated with the FA core complex (94). Intriguingly, BLM and ubiquitinated FANCD2 interact and colocalize in crosslink-induced foci (61, 114). BLM localization to these foci requires the FA proteins, whereas FANCD2 activation occurs normally in BLM mutants. The increased SCE of FA mutants in DT40 is epistatic to BLM, suggesting that BLM might be employed by the FA pathway to suppress SCEs (61, 122a).
Maintenance of Guanine-rich DNA Tracts by FANCJ
FANCJ and its C. elegans homolog, named dog-1, are involved in the maintenance of guanine-rich DNA sequences (155, 164). Such sequences, abundant in many genomes, can form G-quadruplexes and might represent sites of replication fork stalling. The FANCJ family of helicases can unwind these G4 structures in vitro; moreover, G-rich sequences in the genome are lost in patient-derived FANCJ mutant cells or in dog-1 worm mutants, probably due to deleterious excision-and-ligation events (86, 163, 164). Loss of G-rich regions was not found in FANCD2 mutants, suggesting that their maintenance by FANCJ is a function independent of the classic FA pathway (164).
Functions of FA Proteins in Mitosis
In addition to its involvement in HR, BRCA2 (FANCD1) is also involved in controlling cytokinesis (33). BRCA2 localizes to the central spindle and the cytokinetic midbody during cell division. In its absence, cytokinesis is delayed and often abortive, leading to formation of binucleate cells.
During anaphase, the separating chromatids are bridged by ultrafine DNA structures coated with BLM molecules (19). FANCD2 and FANCI were recently found to form foci on both sister chromatids, at each end of these bridges, in a FA core complex-dependent manner (19a, 98a). These FANCD2-I sister foci probably represent sites of unresolved replication or recombination intermediates, generated during late S-phase at chromosomal fragile sites. The FA pathway was also shown to prevent chromosome segregation defects (98a). FA pathway deficient cells exhibited cytokinesis failure and an increase percentage of binucleated cells (98a).
It will be important to address whether the recently reported FANCM phosphorylation and degradation (71) regulates the mitotic role of FANCD2-I.
Other Functions of FA Proteins
FANCG interacts with BRCA2, FANCD2, and XRCC3, forming a complex that was proposed to function in HR (152). The formation of this structure requires phosphorylation of FANCG by ATR but not the presence of the core complex. FANCC may be involved in JAK/STAT signaling and in regulating apoptotic responses (44). It has also been found to interact with GSTP1 and other cytoplasmic factors that control the levels of reactive oxygen species (30).
A CLINICAL PERSPECTIVE
Owing to the critical importance of the FA pathway in maintaining genome stability, there are currently great limitations in treating Fanconi Anemia (5, 6, 32). The initial clinical approach is to alleviate the anemia, which otherwise would be lethal. Heterologous bone marrow transplant, frequently from siblings, has been the classic approach. FA-corrected hematopoietic progenitors were recently derived from induced pluripotent stem cells of FA patients, that were produced from skin grafts, following genetic correction (119a). This raises the possibility that gene therapy strategies might be used successfully in the future. Once anemia is relieved, the sporadic cancers often represent impassible hurdles for FA patients. As their cells are extremely sensitive to crosslinking agents and mildly sensitive to DSBs, classic chemotherapy and radiotherapy for FA patients must be applied in lower doses, which minimizes their antitumor effects. Attempts have been made to generate chemotherapeutic agents that limit DNA damage and delay onset of cancer (165).
In many sporadic tumors, inactivation of the FA pathway has been observed in otherwise normal patients. Loss of heterozygosity and promoter methylation can inactivate the FA response in tumors (138). It is not clear what advantages FA disruption confers in transformed cells, but this DNA repair defect can be used for therapeutic purposes. Indeed, tumors having inactivated BRCA2 are responsive to chemotherapy such as cisplatin. The downside, however is that, probably owing to their lower accuracy of DNA repair, these cells accumulate secondary intragenic modifications that can lead to reversal of the BRCA2 mutation, allowing these cells to acquire crosslink resistance (43, 123).
Novel therapeutic approaches might also include targeting other DNA repair pathways that become essential for cellular survival in FA disrupted cancer cells. Indeed, it is believed that although inactivation of one DNA repair pathway might confer advantages to tumors, cancer cells may rely more heavily on other repair pathways. Therefore, inactivation of a second pathway will be deleterious for these cells, causing synthetic lethality. An RNA interference screen, knocking down a large collection of DNA repair genes, identified the ATM pathway to be synthetically lethal with FA (72). Therefore, ATM inhibitors might be used as potent antitumor agents for treating cancer patients with tumor-specific FA inactivation. A similar strategy for synthetic lethality is currently under investigation---namely, the use of base excision repair PARP1 inhibitors in the setting of HR-deficient breast and ovarian cancer (16, 45).
Although intricate details of crosslink repair and Fanconi Anemia mechanisms of action are still not available, recent advances employing a variety of genetic, biochemical, and cell biological systems have greatly improved our knowledge of these two intertwining fields. Owing to its clinical relevance to cancer, genetic instability, and hematopoietic differentiation defects, Fanconi Anemia is currently under great scrutiny, and more efficient treatment options will certainly become available in the near future. Finally, it is worth mentioning that many basic molecular circuits identified initially in FA research, such as consecutive monoubiquitination/deubiquitination cycles, were found to be conserved mechanisms employed by cells to relay DNA repair responses to effector molecules.
Summary points
Mutations in Fanconi Anemia genes cause a disorder characterized by bone marrow failure, developmental defects, and cancer proneness.
FA-deficient cells are hypersensitive to DNA crosslinking agents and show chromosomal aberrations, with a hallmark increase in radial chromosomes.
The FA pathway is activated during S-phase to remove crosslinks and possibly other types of DNA lesions.
The FA-dependent response to DNA damage involves monoubiquitination of FANCD2 and FANCI by a large multisubunit ubiquitin ligase, the FA core complex. Modified FANCD2-I localize to DNA repair foci, together with downstream FA factors.
Crosslink repair involves the coordinated activities of nucleotide excision repair, translesion synthesis, and homologous recombination.
Complex redundant mechanisms, employing ubiquitination, phosphorylation, and degradation signals, ensure the correct regulation of the FA pathway to promote effective and timely DNA repair.
Ubiquitinated FANCD2-I is likely to provide a signal that activates specific DNA repair factors.
Novel functions of FA members recently emerged, suggesting [**AU: change ok?**] that the pathway has a more general role in protecting the accuracy of the genetic information.
Future issues
What is the precise function of the FANCD2-I complex? Is it an enzyme involved in crosslink repair? Is it a docking site for a TLS polymerase? Does it coordinate NER, TLS, and HR? Does it help to stabilize the replication fork?
What are the factors that recognize ubiquitinated FANCD2-I and how are they regulated by this interaction?
What is the basis of ICL-specific sensitivity of FA mutants? The FA pathway is activated by many exogenous and endogenous stimuli, but FA mutants are hypersensitive only to ICL-inducing agents.
How is the localization of FANCD2-I regulated? Is its initial binding to chromatin achieved independently of the core complex? Is FANCI phosphorylation involved? Once ubiquitinated, is FANCD2-I mobilized to chromatin and is deubiquitination required for its removal from chromatin? Or does deubiquitination relocalize it to foci?
What are the alternative TLS polymerases involved in crosslink repair and how are they recruited to the lesion? Is PCNA ubiquitination involved?
Are there other substrates of the core complex? Are there other substrates of USP1?
Are there other human FA complementation groups? Their identification will help answer the questions above.
Acknowledgments
We would like to thank Johannes Walter, Patrizia Vinciguerra, Younghoon Kee, and Mahesh Madhavan for helpful discussions and comments, and Lisa Moreau for providing Figure 1a. We are grateful to our colleagues for sharing unpublished results. The work was supported by NIH grants RO1DK43889, RO1HL52725, PO1DK50654, U19A1067751, and 5PO1CA092584. GLM is supported by a postdoctoral fellowship from the International Human Frontiers Science Program Organization.
- FA
Fanconi Anemia
- Ubiquitination
reversible covalent post-translational of proteins with the ubiquitin polypeptide, involving a three step enzymatic cascade
- Pancytopaenia
a condition characterized by marked reduction in the number of blood cells
- ICL
Interstrand crosslink
- Translesion synthesis (TLS)
DNA polymerization on damaged templates; it employs specialized low fidelity polymerases able to tolerate DNA lesions, which frequently induce mutations
- Homologous recombination (HR)
exchange of genetic material between two homologous strands of DNA
- Nucleotide excision repair (NER)
Repair mechanism that removes bulky lesions from DNA by introducing single strand breaks to excise the damage
- UV
ultraviolet radiation
- IR
ionizing radiation, typically X-rays or gamma-rays
- Ataxia telangiectasia and Rad3 related (ATR)
a kinase activated by single stranded DNA, that directs the cell cycle arrest and stabilization of stalled replication forks in S-phase
- D-loop (displacement loop)
an HR-specific DNA structure, in which the two DNA strands are separated and a third strand is annealed
- Recombination mediators
a class of proteins that propel the formation of Rad51-coated single stranded DNA during the homologous recombination reaction
- Sister chromatid exchange (SCE)
reciprocal substitution of segments between two sister chromatids
- DSB
double strand break
- NHEJ
non-homologous end-joining
- Proteasome
proteolytic machinery that degrades polyubiquitinated substrates
- Polo-like kinase (Plk1)
an enzyme that governs cell cycle progression through mitosis, controlling among others, the activity of CDK-cyclin complexes and the mitotic spindle
- SUMOylation
post-translational modification with the ubiquitin-related modifier SUMO; it involves an enzymatic cascade similar to ubiquitination
- Ataxia telangiectasia mutated (ATM)
a kinase activated at double strand breaks, that initiates a signaling cascade controlling DNA repair and cell cycle arrest
Footnotes
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- 1.Agoulnik AI, Lu B, Zhu Q, Truong C, Ty MT, et al. A novel gene, Pog, is necessary for primordial germ cell proliferation in the mouse and underlies the germ cell deficient mutation, gcd. Hum Mol Genet. 2002;11:3047–53. doi: 10.1093/hmg/11.24.3047. [DOI] [PubMed] [Google Scholar]
- 2.Akkari YM, Bateman RL, Reifsteck CA, Olson SB, Grompe M. DNA replication is required to elicit cellular responses to psoralen-induced DNA interstrand cross-links. Mol Cell Biol. 2000;20:8283–89. doi: 10.1128/mcb.20.21.8283-8289.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alpi A, Langevin F, Mosedale G, Machida YJ, Dutta A, Patel KJ. UBE2T, the Fanconi anemia core complex, and FANCD2 are recruited independently to chromatin: a basis for the regulation of FANCD2 monoubiquitination. Mol Cell Biol. 2007;27:8421–30. doi: 10.1128/MCB.00504-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alpi AF, Pace PE, Babu MM, Patel KJ. Mechanistic insight into site-restricted monoubiquitination of FANCD2 by Ube2t, FANCL, and FANCI. Mol Cell. 2008;32:767–77. doi: 10.1016/j.molcel.2008.12.003. [DOI] [PubMed] [Google Scholar]
- 5.Alter BP. Fanconi’s anemia and malignancies. Am J Hematol. 1996;53:99–110. doi: 10.1002/(SICI)1096-8652(199610)53:2<99::AID-AJH7>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 6.Alter BP, Greene MH, Velazquez I, Rosenberg PS. Cancer in Fanconi anemia. Blood. 2003;101:2072. doi: 10.1182/blood-2002-11-3597. [DOI] [PubMed] [Google Scholar]
- 7.Alter BP, Rosenberg PS, Brody LC. Clinical and molecular features associated with biallelic mutations in FANCD1/BRCA2. J Med Genet. 2007;44:1–9. doi: 10.1136/jmg.2006.043257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Andreassen PR, D’Andrea AD, Taniguchi T. ATR couples FANCD2 monoubiquitination to the DNA-damage response. Genes Dev. 2004;18:1958–63. doi: 10.1101/gad.1196104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Auerbach AD. A test for Fanconi’s anemia. Blood. 1988;72:366–67. [PubMed] [Google Scholar]
- 10.Auerbach AD, Wolman SR. Susceptibility of Fanconi’s anaemia fibroblasts to chromosome damage by carcinogens. Nature. 1976;261:494–96. doi: 10.1038/261494a0. [DOI] [PubMed] [Google Scholar]
- 11.Baba D, Maita N, Jee JG, Uchimura Y, Saitoh H, et al. Crystal structure of thymine DNA glycosylase conjugated to SUMO-1. Nature. 2005;435:979–82. doi: 10.1038/nature03634. [DOI] [PubMed] [Google Scholar]
- 12.Barber LJ, Ward TA, Hartley JA, McHugh PJ. DNA interstrand cross-link repair in the Saccharomyces cerevisiae cell cycle: overlapping roles for PSO2 (SNM1) with MutS factors and EXO1 during S phase. Mol Cell Biol. 2005;25:2297–309. doi: 10.1128/MCB.25.6.2297-2309.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bhagwat N, Olsen AL, Wang A, Hanada K, Stuckert P, et al. XPF-ERCC1 participates in the Fanconi Anemia pathway of crosslink repair. 2009. submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bogliolo M, Lyakhovich A, Callen E, Castella M, Cappelli E, et al. Histone H2AX and Fanconi anemia FANCD2 function in the same pathway to maintain chromosome stability. EMBO J. 2007;26:1340–51. doi: 10.1038/sj.emboj.7601574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bridge WL, Vandenberg CJ, Franklin RJ, Hiom K. The BRIP1 helicase functions independently of BRCA1 in the Fanconi anemia pathway for DNA crosslink repair. Nat Genet. 2005;37:953–57. doi: 10.1038/ng1627. [DOI] [PubMed] [Google Scholar]
- 16.Bryant HE, Schultz N, Thomas HD, Parker KM, Flower D, et al. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature. 2005;434:913–17. doi: 10.1038/nature03443. [DOI] [PubMed] [Google Scholar]
- 17.Cantor S, Drapkin R, Zhang F, Lin Y, Han J, et al. The BRCA1-associated protein BACH1 is a DNA helicase targeted by clinically relevant inactivating mutations. Proc Natl Acad Sci USA. 2004;101:2357–62. doi: 10.1073/pnas.0308717101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cantor SB, Bell DW, Ganesan S, Kass EM, Drapkin R, et al. BACH1, a novel helicase-like protein, interacts directly with BRCA1 and contributes to its DNA repair function. Cell. 2001;105:149–60. doi: 10.1016/s0092-8674(01)00304-x. [DOI] [PubMed] [Google Scholar]
- 19.Chan KL, North PS, Hickson ID. BLM is required for faithful chromosome segregation and its localization defines a class of ultrafine anaphase bridges. EMBO J. 2007;26:3397–409. doi: 10.1038/sj.emboj.7601777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19a.Chan KL, Palmai-Pallag T, Ying S, Hickson ID. Replication stress induces sister-chromatid bridges at fragile site loci in mitosis. Nat Cell Biol. 2009;11:753–60. doi: 10.1038/ncb1882. [DOI] [PubMed] [Google Scholar]
- 20.Chen M, Tomkins DJ, Auerbach W, McKerlie C, Youssoufian H, et al. Inactivation of Fac in mice produces inducible chromosomal instability and reduced fertility reminiscent of Fanconi anaemia. Nat Genet. 1996;12:448–51. doi: 10.1038/ng0496-448. [DOI] [PubMed] [Google Scholar]
- 21.Chen P, Liang J, Wang Z, Zhou X, Chen L, et al. Association of common PALB2 polymorphisms with breast cancer risk: a case-control study. Clin Cancer Res. 2008;14:5931–37. doi: 10.1158/1078-0432.CCR-08-0429. [DOI] [PubMed] [Google Scholar]
- 22.Cheung AM, Elia A, Tsao MS, Done S, Wagner KU, et al. Brca2 deficiency does not impair mammary epithelium development but promotes mammary adenocarcinoma formation in p53(+/−) mutant mice. Cancer Res. 2004;64:1959–65. doi: 10.1158/0008-5472.can-03-2270. [DOI] [PubMed] [Google Scholar]
- 23.Ciccia A, Ling C, Coulthard R, Yan Z, Xue Y, et al. Identification of FAAP24, a Fanconi anemia core complex protein that interacts with FANCM. Mol Cell. 2007;25:331–43. doi: 10.1016/j.molcel.2007.01.003. [DOI] [PubMed] [Google Scholar]
- 24.Ciccia A, McDonald N, West SC. Structural and functional relationships of the XPF/MUS81 family of proteins. Annu Rev Biochem. 2008;77:259–87. doi: 10.1146/annurev.biochem.77.070306.102408. [DOI] [PubMed] [Google Scholar]
- 25.Cohn MA, Kee Y, Haas W, Gygi SP, D’Andrea AD. UAF1 is a subunit of multiple deubiquitinating enzyme complexes. J Biol Chem. 2009;284:5343–51. doi: 10.1074/jbc.M808430200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cohn MA, Kowal P, Yang K, Haas W, Huang TT, et al. A UAF1-containing multisubunit protein complex regulates the Fanconi anemia pathway. Mol Cell. 2007;28:786–97. doi: 10.1016/j.molcel.2007.09.031. [DOI] [PubMed] [Google Scholar]
- 27.Collins NB, Wilson JB, Bush T, Tomashevski A, Roberts KJ, et al. ATR-dependent phosphorylation of FANCA on serine 1449 after DNA damage is important for FA pathway function. Blood. 2009;113:2181–90. doi: 10.1182/blood-2008-05-154294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Collis SJ, Barber LJ, Ward JD, Martin JS, Boulton SJ. C. elegans FANCD2 responds to replication stress and functions in interstrand cross-link repair. DNA Repair (Amst) 2006;5:1398–406. doi: 10.1016/j.dnarep.2006.06.010. [DOI] [PubMed] [Google Scholar]
- 29.Collis SJ, Ciccia A, Deans AJ, Horejsi Z, Martin JS, et al. FANCM and FAAP24 function in ATR-mediated checkpoint signaling independently of the Fanconi anemia core complex. Mol Cell. 2008;32:313–24. doi: 10.1016/j.molcel.2008.10.014. [DOI] [PubMed] [Google Scholar]
- 30.Cumming RC, Lightfoot J, Beard K, Youssoufian H, O’Brien PJ, Buchwald M. Fanconi anemia group C protein prevents apoptosis in hematopoietic cells through redox regulation of GSTP1. Nat Med. 2001;7:814–20. doi: 10.1038/89937. [DOI] [PubMed] [Google Scholar]
- 31.D’Andrea AD, Grompe M. The Fanconi anaemia/BRCA pathway. Nat Rev Cancer. 2003;3:23–34. doi: 10.1038/nrc970. [DOI] [PubMed] [Google Scholar]
- 32.Dalle JH. HSCT for Fanconi anemia in children: factors that influence early and late results. Bone Marrow Transplant. 2008;42(Suppl 2):S51–53. doi: 10.1038/bmt.2008.284. [DOI] [PubMed] [Google Scholar]
- 33.Daniels MJ, Wang Y, Lee M, Venkitaraman AR. Abnormal cytokinesis in cells deficient in the breast cancer susceptibility protein BRCA2. Science. 2004;306:876–79. doi: 10.1126/science.1102574. [DOI] [PubMed] [Google Scholar]
- 34.De Silva IU, McHugh PJ, Clingen PH, Hartley JA. Defining the roles of nucleotide excision repair and recombination in the repair of DNA interstrand cross-links in mammalian cells. Mol Cell Biol. 2000;20:7980–90. doi: 10.1128/mcb.20.21.7980-7990.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.de Winter JP, Joenje H. The genetic and molecular basis of Fanconi anemia. Mutat Res: Fundam Mol Mech Mutagen. 2008 doi: 10.1016/j.mrfmmm.2008.11.004. [DOI] [PubMed] [Google Scholar]
- 36.de Winter JP, Leveille F, van Berkel CG, Rooimans MA, Van Der Weel L, et al. Isolation of a cDNA representing the Fanconi anemia complementation group E gene. Am J Hum Genet. 2000;67:1306–8. doi: 10.1016/s0002-9297(07)62959-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.de Winter JP, Rooimans MA, Van Der Weel L, van Berkel CG, Alon N, et al. The Fanconi anaemia gene FANCF encodes a novel protein with homology to ROM. Nat Genet. 2000;24:15–16. doi: 10.1038/71626. [DOI] [PubMed] [Google Scholar]
- 38.de Winter JP, Waisfisz Q, Rooimans MA, van Berkel CG, Bosnoyan-Collins L, et al. The Fanconi anaemia group G gene FANCG is identical with XRCC9. Nat Genet. 1998;20:281–83. doi: 10.1038/3093. [DOI] [PubMed] [Google Scholar]
- 39.Dominski Z. Nucleases of the metallo-beta-lactamase family and their role in DNA and RNA metabolism. Crit Rev Biochem Mol Biol. 2007;42:67–93. doi: 10.1080/10409230701279118. [DOI] [PubMed] [Google Scholar]
- 40.Dorsman JC, Levitus M, Rockx D, Rooimans MA, Oostra AB, et al. Identification of the Fanconi anemia complementation group I gene, FANCI. Cell Oncol. 2007;29:211–8. doi: 10.1155/2007/151968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dronkert ML, Kanaar R. Repair of DNA interstrand cross-links. Mutat Res. 2001;486:217–47. doi: 10.1016/s0921-8777(01)00092-1. [DOI] [PubMed] [Google Scholar]
- 42.Dunn J, Potter M, Rees A, Runger TM. Activation of the Fanconi anemia/BRCA pathway and recombination repair in the cellular response to solar UV light. Cancer Res. 2006;66:11140–47. doi: 10.1158/0008-5472.CAN-06-0563. [DOI] [PubMed] [Google Scholar]
- 43.Edwards SL, Brough R, Lord CJ, Natrajan R, Vatcheva R, et al. Resistance to therapy caused by intragenic deletion in BRCA2. Nature. 2008;451:1111–15. doi: 10.1038/nature06548. [DOI] [PubMed] [Google Scholar]
- 44.Fagerlie SR, Koretsky T, Torok-Storb B, Bagby GC. Impaired type I IFN-induced Jak/STAT signaling in FA-C cells and abnormal CD4+ Th cell subsets in Fancc−/− mice. J Immunol. 2004;173:3863–70. doi: 10.4049/jimmunol.173.6.3863. [DOI] [PubMed] [Google Scholar]
- 45.Farmer H, McCabe N, Lord CJ, Tutt AN, Johnson DA, et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434:917–21. doi: 10.1038/nature03445. [DOI] [PubMed] [Google Scholar]
- 46.Friedel AM, Pike BL, Gasser SM. ATR/Mec1: coordinating fork stability and repair. Curr Opin Cell Biol. 2009;21:237–44. doi: 10.1016/j.ceb.2009.01.017. [DOI] [PubMed] [Google Scholar]
- 47.Garcia-Higuera I, Kuang Y, Denham J, D’Andrea AD. The fanconi anemia proteins FANCA and FANCG stabilize each other and promote the nuclear accumulation of the Fanconi anemia complex. Blood. 2000;96:3224–30. [PubMed] [Google Scholar]
- 48.Garcia-Higuera I, Taniguchi T, Ganesan S, Meyn MS, Timmers C, et al. Interaction of the Fanconi anemia proteins and BRCA1 in a common pathway. Mol Cell. 2001;7:249–62. doi: 10.1016/s1097-2765(01)00173-3. [DOI] [PubMed] [Google Scholar]
- 49.Gari K, Decaillet C, Delannoy M, Wu L, Constantinou A. Remodeling of DNA replication structures by the branch point translocase FANCM. Proc Natl Acad Sci USA. 2008;105:16107–12. doi: 10.1073/pnas.0804777105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gari K, Decaillet C, Stasiak AZ, Stasiak A, Constantinou A. The Fanconi anemia protein FANCM can promote branch migration of Holliday junctions and replication forks. Mol Cell. 2008;29:141–48. doi: 10.1016/j.molcel.2007.11.032. [DOI] [PubMed] [Google Scholar]
- 51.German J, Schonberg S, Caskie S, Warburton D, Falk C, Ray JH. A test for Fanconi’s anemia. Blood. 1987;69:1637–41. [PubMed] [Google Scholar]
- 52.Godthelp BC, Wiegant WW, Waisfisz Q, Medhurst AL, Arwert F, et al. Inducibility of nuclear Rad51 foci after DNA damage distinguishes all Fanconi anemia complementation groups from D1/BRCA2. Mutat Res. 2006;594:39–48. doi: 10.1016/j.mrfmmm.2005.07.008. [DOI] [PubMed] [Google Scholar]
- 53.Gordon SM, Buchwald M. Fanconi anemia protein complex: mapping protein interactions in the yeast 2- and 3-hybrid systems. Blood. 2003;102:136–41. doi: 10.1182/blood-2002-11-3517. [DOI] [PubMed] [Google Scholar]
- 54.Guo C, Sonoda E, Tang TS, Parker JL, Bielen AB, et al. REV1 protein interacts with PCNA: significance of the REV1 BRCT domain in vitro and in vivo. Mol Cell. 2006;23:265–71. doi: 10.1016/j.molcel.2006.05.038. [DOI] [PubMed] [Google Scholar]
- 55.Gupta R, Sharma S, Sommers JA, Jin Z, Cantor SB, Brosh RM., Jr Analysis of the DNA substrate specificity of the human BACH1 helicase associated with breast cancer. J Biol Chem. 2005;280:25450–60. doi: 10.1074/jbc.M501995200. [DOI] [PubMed] [Google Scholar]
- 56.Gupta R, Sharma S, Sommers JA, Kenny MK, Cantor SB, Brosh RM., Jr FANCJ (BACH1) helicase forms DNA damage inducible foci with replication protein A and interacts physically and functionally with the single-stranded DNA-binding protein. Blood. 2007;110:2390–98. doi: 10.1182/blood-2006-11-057273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gurtan AM, Stuckert P, D’Andrea AD. The WD40 repeats of FANCL are required for Fanconi anemia core complex assembly. J Biol Chem. 2006;281:10896–905. doi: 10.1074/jbc.M511411200. [DOI] [PubMed] [Google Scholar]
- 58.Hanada K, Budzowska M, Modesti M, Maas A, Wyman C, et al. The structure-specific endonuclease Mus81-Eme1 promotes conversion of interstrand DNA crosslinks into double-strands breaks. EMBO J. 2006;25:4921–32. doi: 10.1038/sj.emboj.7601344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hinz JM, Nham PB, Salazar EP, Thompson LH. The Fanconi anemia pathway limits the severity of mutagenesis. DNA Repair (Amst) 2006;5:875–84. doi: 10.1016/j.dnarep.2006.05.039. [DOI] [PubMed] [Google Scholar]
- 60.Hinz JM, Nham PB, Urbin SS, Jones IM, Thompson LH. Disparate contributions of the Fanconi anemia pathway and homologous recombination in preventing spontaneous mutagenesis. Nucleic Acids Res. 2007;35:3733–40. doi: 10.1093/nar/gkm315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hirano S, Yamamoto K, Ishiai M, Yamazoe M, Seki M, et al. Functional relationships of FANCC to homologous recombination, translesion synthesis, and BLM. EMBO J. 2005;24:418–27. doi: 10.1038/sj.emboj.7600534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ho GP, Margossian S, Taniguchi T, D’Andrea AD. Phosphorylation of FANCD2 on two novel sites is required for mitomycin C resistance. Mol Cell Biol. 2006;26:7005–15. doi: 10.1128/MCB.02018-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hoege C, Pfander B, Moldovan GL, Pyrowolakis G, Jentsch S. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature. 2002;419:135–41. doi: 10.1038/nature00991. [DOI] [PubMed] [Google Scholar]
- 64.Hofmann K. Ubiquitin-binding domains and their role in the DNA damage response. DNA Repair (Amst) 2009;14:544–56. doi: 10.1016/j.dnarep.2009.01.003. [DOI] [PubMed] [Google Scholar]
- 65.Houghtaling S, Timmers C, Noll M, Finegold MJ, Jones SN, et al. Epithelial cancer in Fanconi anemia complementation group D2 (Fancd2) knockout mice. Genes Dev. 2003;17:2021–35. doi: 10.1101/gad.1103403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Howlett NG, Taniguchi T, Olson S, Cox B, Waisfisz Q, et al. Biallelic inactivation of BRCA2 in Fanconi anemia. Science. 2002;297:606–9. doi: 10.1126/science.1073834. [DOI] [PubMed] [Google Scholar]
- 67.Huang TT, Nijman SM, Mirchandani KD, Galardy PJ, Cohn MA, et al. Regulation of monoubiquitinated PCNA by DUB autocleavage. Nat Cell Biol. 2006;8:339–47. doi: 10.1038/ncb1378. [DOI] [PubMed] [Google Scholar]
- 68.Hussain S, Wilson JB, Medhurst AL, Hejna J, Witt E, et al. Direct interaction of FANCD2 with BRCA2 in DNA damage response pathways. Hum Mol Genet. 2004;13:1241–48. doi: 10.1093/hmg/ddh135. [DOI] [PubMed] [Google Scholar]
- 69.Ishiai M, Kitao H, Smogorzewska A, Tomida J, Kinomura A, et al. FANCI phosphorylation functions as a molecular switch to turn on the Fanconi anemia pathway. Nat Struct Mol Biol. 2008;15:1138–46. doi: 10.1038/nsmb.1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jaspers NG, Raams A, Silengo MC, Wijgers N, Niedernhofer LJ, et al. First reported patient with human ERCC1 deficiency has cerebro-oculo-facio-skeletal syndrome with a mild defect in nucleotide excision repair and severe developmental failure. Am J Hum Genet. 2007;80:457–66. doi: 10.1086/512486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kee Y, Kim JM, D’Andrea AD. Regulated degradation of FANCM in the Fanconi Anemia pathway during mitosis. Genes Dev. 2009;23:549–54. doi: 10.1101/gad.1761309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kennedy RD, Chen CC, Stuckert P, Archila EM, De la Vega MA, et al. Fanconi anemia pathway-deficient tumor cells are hypersensitive to inhibition of ataxia telangiectasia mutated. J Clin Invest. 2007;117:1440–49. doi: 10.1172/JCI31245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kennedy RD, D’Andrea AD. The Fanconi Anemia/BRCA pathway: new faces in the crowd. Genes Dev. 2005;19:2925–40. doi: 10.1101/gad.1370505. [DOI] [PubMed] [Google Scholar]
- 74.Kim JM, Kee Y, Gurtan A, D’Andrea AD. Cell cycle-dependent chromatin loading of the Fanconi anemia core complex by FANCM/FAAP24. Blood. 2008;111:5215–22. doi: 10.1182/blood-2007-09-113092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kim JM, Parmar K, Huang M, Weinstock DM, Ruit CA, et al. Inactivation of murine Usp1 results in genomic instability and a fanconi anemia phenotype. Dev Cell. 2009;16:314–20. doi: 10.1016/j.devcel.2009.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kitagawa R, Kastan MB. The ATM-dependent DNA damage signaling pathway. Cold Spring Harb Symp Quant Biol. 2005;70:99–109. doi: 10.1101/sqb.2005.70.002. [DOI] [PubMed] [Google Scholar]
- 77.Kook H. Fanconi anemia: current management. Hematology. 2005;10(Suppl 1):108–10. doi: 10.1080/10245330512331390096. [DOI] [PubMed] [Google Scholar]
- 78.Koomen M, Cheng NC, van de Vrugt HJ, Godthelp BC, Van Der Valk MA, et al. Reduced fertility and hypersensitivity to mitomycin C characterize Fancg/Xrcc9 null mice. Hum Mol Genet. 2002;11:273–81. doi: 10.1093/hmg/11.3.273. [DOI] [PubMed] [Google Scholar]
- 79.Kowal P, Gurtan AM, Stuckert P, D’Andrea AD, Ellenberger T. Structural determinants of human FANCF protein that function in the assembly of a DNA damage signaling complex. J Biol Chem. 2007;282:2047–55. doi: 10.1074/jbc.M608356200. [DOI] [PubMed] [Google Scholar]
- 80.Kumaraswamy E, Shiekhattar R. Activation of BRCA1/BRCA2-associated helicase BACH1 is required for timely progression through S phase. Mol Cell Biol. 2007;27:6733–41. doi: 10.1128/MCB.00961-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kuraoka I, Kobertz WR, Ariza RR, Biggerstaff M, Essigmann JM, Wood RD. Repair of an interstrand DNA cross-link initiated by ERCC1-XPF repair/recombination nuclease. J Biol Chem. 2000;275:26632–36. doi: 10.1074/jbc.C000337200. [DOI] [PubMed] [Google Scholar]
- 82.Larminat F, Germanier M, Papouli E, Defais M. Deficiency in BRCA2 leads to increase in nonconservative homologous recombination. Oncogene. 2002;21:5188–92. doi: 10.1038/sj.onc.1205659. [DOI] [PubMed] [Google Scholar]
- 83.Levitus M, Waisfisz Q, Godthelp BC, de Vries Y, Hussain S, et al. The DNA helicase BRIP1 is defective in Fanconi anemia complementation group J. Nat Genet. 2005;37:934–35. doi: 10.1038/ng1625. [DOI] [PubMed] [Google Scholar]
- 84.Levran O, Attwooll C, Henry RT, Milton KL, Neveling K, et al. The BRCA1-interacting helicase BRIP1 is deficient in Fanconi anemia. Nat Genet. 2005;37:931–33. doi: 10.1038/ng1624. [DOI] [PubMed] [Google Scholar]
- 85.Ling C, Ishiai M, Ali AM, Medhurst AL, Neveling K, et al. FAAP100 is essential for activation of the Fanconi anemia-associated DNA damage response pathway. EMBO J. 2007;26:2104–14. doi: 10.1038/sj.emboj.7601666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Litman R, Peng M, Jin Z, Zhang F, Zhang J, et al. BACH1 is critical for homologous recombination and appears to be the Fanconi anemia gene product FANCJ. Cancer Cell. 2005;8:255–65. doi: 10.1016/j.ccr.2005.08.004. [DOI] [PubMed] [Google Scholar]
- 87.Lo Ten Foe JR, Rooimans MA, Bosnoyan-Collins L, Alon N, Wijker M, et al. Expression cloning of a cDNA for the major Fanconi anaemia gene, FAA. Nat Genet. 1996;14:320–23. doi: 10.1038/ng1196-320. [DOI] [PubMed] [Google Scholar]
- 88.Machida YJ, Machida Y, Chen Y, Gurtan AM, Kupfer GM, et al. UBE2T is the E2 in the Fanconi anemia pathway and undergoes negative autoregulation. Mol Cell. 2006;23:589–96. doi: 10.1016/j.molcel.2006.06.024. [DOI] [PubMed] [Google Scholar]
- 89.Matsushita N, Kitao H, Ishiai M, Nagashima N, Hirano S, et al. A FancD2-monoubiquitin fusion reveals hidden functions of Fanconi anemia core complex in DNA repair. Mol Cell. 2005;19:841–47. doi: 10.1016/j.molcel.2005.08.018. [DOI] [PubMed] [Google Scholar]
- 90.Medhurst AL, Laghmani el H, Steltenpool J, Ferrer M, Fontaine C, et al. Evidence for subcomplexes in the Fanconi anemia pathway. Blood. 2006;108:2072–80. doi: 10.1182/blood-2005-11-008151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Meetei AR, de Winter JP, Medhurst AL, Wallisch M, Waisfisz Q, et al. A novel ubiquitin ligase is deficient in Fanconi anemia. Nat Genet. 2003;35:165–70. doi: 10.1038/ng1241. [DOI] [PubMed] [Google Scholar]
- 92.Meetei AR, Levitus M, Xue Y, Medhurst AL, Zwaan M, et al. X-linked inheritance of Fanconi anemia complementation group B. Nat Genet. 2004;36:1219–24. doi: 10.1038/ng1458. [DOI] [PubMed] [Google Scholar]
- 93.Meetei AR, Medhurst AL, Ling C, Xue Y, Singh TR, et al. A human ortholog of archaeal DNA repair protein Hef is defective in Fanconi anemia complementation group M. Nat Genet. 2005;37:958–63. doi: 10.1038/ng1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Meetei AR, Sechi S, Wallisch M, Yang D, Young MK, et al. A multiprotein nuclear complex connects Fanconi anemia and Bloom syndrome. Mol Cell Biol. 2003;23:3417–26. doi: 10.1128/MCB.23.10.3417-3426.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Mirchandani KD, McCaffrey RM, D’Andrea AD. The Fanconi anemia core complex is required for efficient point mutagenesis and Rev1 foci assembly. DNA Repair (Amst) 2008;7:902–11. doi: 10.1016/j.dnarep.2008.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Moldovan GL, Pfander B, Jentsch S. PCNA, the maestro of the replication fork. Cell. 2007;129:665–79. doi: 10.1016/j.cell.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 97.Montes de Oca R, Andreassen PR, Margossian SP, Gregory RC, Taniguchi T, et al. Regulated interaction of the Fanconi anemia protein, FANCD2, with chromatin. Blood. 2005;105:1003–9. doi: 10.1182/blood-2003-11-3997. [DOI] [PubMed] [Google Scholar]
- 98.Mosedale G, Niedzwiedz W, Alpi A, Perrina F, Pereira-Leal JB, et al. The vertebrate Hef ortholog is a component of the Fanconi anemia tumor-suppressor pathway. Nat Struct Mol Biol. 2005;12:763–71. doi: 10.1038/nsmb981. [DOI] [PubMed] [Google Scholar]
- 98a.Naim V, Rosselli F. The FANC pathway and BLM collaborate during mitosis to prevent micro-nucleation and chromosome abnormalities. Nat Cell Biol. 2009;11:761–68. doi: 10.1038/ncb1883. [DOI] [PubMed] [Google Scholar]
- 99.Nakanishi K, Taniguchi T, Ranganathan V, New HV, Moreau LA, et al. Interaction of FANCD2 and NBS1 in the DNA damage response. Nat Cell Biol. 2002;4:913–20. doi: 10.1038/ncb879. [DOI] [PubMed] [Google Scholar]
- 100.Nakanishi K, Yang YG, Pierce AJ, Taniguchi T, Digweed M, et al. Human Fanconi anemia monoubiquitination pathway promotes homologous DNA repair. Proc Natl Acad Sci USA. 2005;102:1110–15. doi: 10.1073/pnas.0407796102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Niedernhofer LJ, Lalai AS, Hoeijmakers JH. Fanconi anemia (cross)linked to DNA repair. Cell. 2005;123:1191–98. doi: 10.1016/j.cell.2005.12.009. [DOI] [PubMed] [Google Scholar]
- 102.Niedernhofer LJ, Odijk H, Budzowska M, van Drunen E, Maas A, et al. The structure-specific endonuclease Ercc1-Xpf is required to resolve DNA interstrand cross-link-induced double-strand breaks. Mol Cell Biol. 2004;24:5776–87. doi: 10.1128/MCB.24.13.5776-5787.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Niedzwiedz W, Mosedale G, Johnson M, Ong CY, Pace P, Patel KJ. The Fanconi anaemia gene FANCC promotes homologous recombination and error-prone DNA repair. Mol Cell. 2004;15:607–20. doi: 10.1016/j.molcel.2004.08.009. [DOI] [PubMed] [Google Scholar]
- 104.Nijman SM, Huang TT, Dirac AM, Brummelkamp TR, Kerkhoven RM, et al. The deubiquitinating enzyme USP1 regulates the Fanconi anemia pathway. Mol Cell. 2005;17:331–39. doi: 10.1016/j.molcel.2005.01.008. [DOI] [PubMed] [Google Scholar]
- 105.Nojima K, Hochegger H, Saberi A, Fukushima T, Kikuchi K, et al. Multiple repair pathways mediate tolerance to chemotherapeutic cross-linking agents in vertebrate cells. Cancer Res. 2005;65:11704–11. doi: 10.1158/0008-5472.CAN-05-1214. [DOI] [PubMed] [Google Scholar]
- 106.Nookala RK, Hussain S, Pellegrini L. Insights into Fanconi Anaemia from the structure of human FANCE. Nucleic Acids Res. 2007;35:1638–48. doi: 10.1093/nar/gkm033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Oestergaard VH, Langevin F, Kuiken HJ, Pace P, Niedzwiedz W, et al. Deubiquitination of FANCD2 is required for DNA crosslink repair. Mol Cell. 2007;28:798–809. doi: 10.1016/j.molcel.2007.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Pace P, Johnson M, Tan WM, Mosedale G, Sng C, et al. FANCE: the link between Fanconi anaemia complex assembly and activity. EMBO J. 2002;21:3414–23. doi: 10.1093/emboj/cdf355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Papadopoulo D, Guillouf C, Mohrenweiser H, Moustacchi E. Hypomutability in Fanconi anemia cells is associated with increased deletion frequency at the HPRT locus. Proc Natl Acad Sci USA. 1990;87:8383–87. doi: 10.1073/pnas.87.21.8383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Papadopoulo D, Guillouf C, Porfirio B, Moustacchi E. Decreased mutagenicity in Fanconi’s anemia lymphoblasts following treatment with photoactivated psoralens. Prog Clin Biol Res. 1990;340A:241–48. [PubMed] [Google Scholar]
- 111.Park WH, Margossian S, Horwitz AA, Simons AM, D’Andrea AD, Parvin JD. Direct DNA binding activity of the Fanconi anemia D2 protein. J Biol Chem. 2005;280:23593–98. doi: 10.1074/jbc.M503730200. [DOI] [PubMed] [Google Scholar]
- 112.Patel KJ, Joenje H. Fanconi anemia and DNA replication repair. DNA Repair (Amst) 2007;6:885–90. doi: 10.1016/j.dnarep.2007.02.002. [DOI] [PubMed] [Google Scholar]
- 113.Perry P, Evans HJ. Cytological detection of mutagen-carcinogen exposure by sister chromatid exchange. Nature. 1975;258:121–25. doi: 10.1038/258121a0. [DOI] [PubMed] [Google Scholar]
- 114.Pichierri P, Franchitto A, Rosselli F. BLM and the FANC proteins collaborate in a common pathway in response to stalled replication forks. EMBO J. 2004;23:3154–63. doi: 10.1038/sj.emboj.7600277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Pichierri P, Rosselli F. The DNA crosslink-induced S-phase checkpoint depends on ATR-CHK1 and ATR-NBS1-FANCD2 pathways. EMBO J. 2004;23:1178–87. doi: 10.1038/sj.emboj.7600113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Prakash R, Satory D, Dray E, Papusha A, Scheller J, et al. Yeast Mph1 helicase dissociates Rad51-made D-loops: implications for crossover control in mitotic recombination. Genes Dev. 2009;23:67–79. doi: 10.1101/gad.1737809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Prasher JM, Lalai AS, Heijmans-Antonissen C, Ploemacher RE, Hoeijmakers JH, et al. Reduced hematopoietic reserves in DNA interstrand crosslink repair-deficient Ercc1−/− mice. EMBO J. 2005;24:861–71. doi: 10.1038/sj.emboj.7600542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Qiao F, Mi J, Wilson JB, Zhi G, Bucheimer NR, et al. Phosphorylation of fanconi anemia (FA) complementation group G protein, FANCG, at serine 7 is important for function of the FA pathway. J Biol Chem. 2004;279:46035–45. doi: 10.1074/jbc.M408323200. [DOI] [PubMed] [Google Scholar]
- 119.Raschle M, Knipsheer P, Enoiu M, Angelov T, Sun J, et al. Mechanism of replication-coupled DNA interstrand crosslink repair. Cell. 2008;134:969–80. doi: 10.1016/j.cell.2008.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119a.Raya A, Rodriguez-Piza I, Guenechea G, Vassena R, Navarro S, et al. Disease-corrected haematopoietic progenitors from Fanconi anaemia induced pluripotent stem cells. Nature. 2009 doi: 10.1038/nature08129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Rego MA, Kolling FWT, Howlett NG. The Fanconi anemia protein interaction network: Casting a wide net. Mutat Res: Fundam Mol Mech Mutagen. 2008 doi: 10.1016/j.mrfmmm.2008.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Reid S, Schindler D, Hanenberg H, Barker K, Hanks S, et al. Biallelic mutations in PALB2 cause Fanconi anemia subtype FA-N and predispose to childhood cancer. Nat Genet. 2007;39:162–64. doi: 10.1038/ng1947. [DOI] [PubMed] [Google Scholar]
- 122.Ridpath JR, Nakamura A, Tano K, Luke AM, Sonoda E, et al. Cells deficient in the FANC/BRCA pathway are hypersensitive to plasma levels of formaldehyde. Cancer Res. 2007;67:11117–22. doi: 10.1158/0008-5472.CAN-07-3028. [DOI] [PubMed] [Google Scholar]
- 122a.Rosado IV, Niedzwiedz W, Alpi AF, Patel KJ. The Walker B motif in avian FANCM is required to limit sister chromatid exchanges but it is dispensable for DNA crosslink repair. Nucleic Acids Res. 2009 doi: 10.1093/nar/gkp365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Sakai W, Swisher EM, Karlan BY, Agarwal MK, Higgins J, et al. Secondary mutations as a mechanism of cisplatin resistance in BRCA2-mutated cancers. Nature. 2008;451:1116–20. doi: 10.1038/nature06633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Sarkar S, Davies AA, Ulrich HD, McHugh PJ. DNA interstrand crosslink repair during G1 involves nucleotide excision repair and DNA polymerase zeta. EMBO J. 2006;25:1285–94. doi: 10.1038/sj.emboj.7600993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Sasaki MS. Is Fanconi’s anaemia defective in a process essential to the repair of DNA cross links? Nature. 1975;257:501–3. doi: 10.1038/257501a0. [DOI] [PubMed] [Google Scholar]
- 126.Scharer OD. DNA interstrand crosslinks: natural and drug-induced DNA adducts that induce unique cellular responses. Chembiochem. 2005;6:27–32. doi: 10.1002/cbic.200400287. [DOI] [PubMed] [Google Scholar]
- 127.Sharan SK, Morimatsu M, Albrecht U, Lim DS, Regel E, et al. Embryonic lethality and radiation hypersensitivity mediated by Rad51 in mice lacking Brca2. Nature. 1997;386:804–10. doi: 10.1038/386804a0. [DOI] [PubMed] [Google Scholar]
- 128.Shimamura A, Montes de Oca R, Svenson JL, Haining N, Moreau LA, et al. A novel diagnostic screen for defects in the Fanconi anemia pathway. Blood. 2002;100:4649–54. doi: 10.1182/blood-2002-05-1399. [DOI] [PubMed] [Google Scholar]
- 129.Sims AE, Spiteri E, Sims RJ, 3rd, Arita AG, Lach FP, et al. FANCI is a second monoubiquitinated member of the Fanconi anemia pathway. Nat Struct Mol Biol. 2007;14:564–67. doi: 10.1038/nsmb1252. [DOI] [PubMed] [Google Scholar]
- 129a.Singh TR, Bakker ST, Agarwal S, Jansen M, Grassman E, et al. Impaired FANCD2 monoubiquitination and hypersensitivity to camptothecin uniquely characterize Fanconi Anemia complementation group M. Blood. 2009 doi: 10.1182/blood-2009-02-207811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Smogorzewska A, Matsuoka S, Vinciguerra P, McDonald ER, 3rd, Hurov KE, et al. Identification of the FANCI protein, a monoubiquitinated FANCD2 paralog required for DNA repair. Cell. 2007;129:289–301. doi: 10.1016/j.cell.2007.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Sridharan D, Brown M, Lambert WC, McMahon LW, Lambert MW. Nonerythroid alphaII spectrin is required for recruitment of FANCA and XPF to nuclear foci induced by DNA interstrand cross-links. J Cell Sci. 2003;116:823–35. doi: 10.1242/jcs.00294. [DOI] [PubMed] [Google Scholar]
- 132.Strathdee CA, Gavish H, Shannon WR, Buchwald M. Cloning of cDNAs for Fanconi’s anaemia by functional complementation. Nature. 1992;358:434. doi: 10.1038/358434a0. [DOI] [PubMed] [Google Scholar]
- 133.Sun W, Nandi S, Osman F, Ahn JS, Jakovleska J, et al. The FANCM ortholog Fml1 promotes recombination at stalled replication forks and limits crossing over during DNA double-strand break repair. Mol Cell. 2008;32:118–28. doi: 10.1016/j.molcel.2008.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Sung P, Klein H. Mechanism of homologous recombination: mediators and helicases take on regulatory functions. Nat Rev Mol Cell Biol. 2006;7:739–50. doi: 10.1038/nrm2008. [DOI] [PubMed] [Google Scholar]
- 135.Takata M, Kitao H, Ishiai M. Fanconi anemia: genetic analysis of a human disease using chicken system. Cytogenet Genome Res. 2007;117:346–51. doi: 10.1159/000103197. [DOI] [PubMed] [Google Scholar]
- 136.Takata M, Sasaki MS, Tachiiri S, Fukushima T, Sonoda E, et al. Chromosome instability and defective recombinational repair in knockout mutants of the five Rad51 paralogs. Mol Cell Biol. 2001;21:2858–66. doi: 10.1128/MCB.21.8.2858-2866.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Taniguchi T, D’Andrea AD. The Fanconi anemia protein, FANCE, promotes the nuclear accumulation of FANCC. Blood. 2002;100:2457–62. doi: 10.1182/blood-2002-03-0860. [DOI] [PubMed] [Google Scholar]
- 138.Taniguchi T, D’Andrea AD. Molecular pathogenesis of Fanconi anemia: recent progress. Blood. 2006;107:4223–33. doi: 10.1182/blood-2005-10-4240. [DOI] [PubMed] [Google Scholar]
- 139.Taniguchi T, Garcia-Higuera I, Andreassen PR, Gregory RC, Grompe M, D’Andrea AD. S-phase-specific interaction of the Fanconi anemia protein, FANCD2, with BRCA1 and RAD51. Blood. 2002;100:2414–20. doi: 10.1182/blood-2002-01-0278. [DOI] [PubMed] [Google Scholar]
- 140.Taniguchi T, Garcia-Higuera I, Xu B, Andreassen PR, Gregory RC, et al. Convergence of the fanconi anemia and ataxia telangiectasia signaling pathways. Cell. 2002;109:459–72. doi: 10.1016/s0092-8674(02)00747-x. [DOI] [PubMed] [Google Scholar]
- 141.Taniguchi T, Tischkowitz M, Ameziane N, Hodgson SV, Mathew CG, et al. Disruption of the Fanconi anemia-BRCA pathway in cisplatin-sensitive ovarian tumors. Nat Med. 2003;9:568–74. doi: 10.1038/nm852. [DOI] [PubMed] [Google Scholar]
- 142.Thompson LH, Hinz JM. Cellular and molecular consequences of defective Fanconi anemia proteins in replication-coupled DNA repair: mechanistic insights. Mutat Res. 2009 doi: 10.1016/j.mrfmmm.2009.02.003. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Thompson LH, Hinz JM, Yamada NA, Jones NJ. How Fanconi anemia proteins promote the four Rs: replication, recombination, repair, and recovery. Environ Mol Mutagen. 2005;45:128–42. doi: 10.1002/em.20109. [DOI] [PubMed] [Google Scholar]
- 144.Timmers C, Taniguchi T, Hejna J, Reifsteck C, Lucas L, et al. Positional cloning of a novel Fanconi anemia gene, FANCD2. Mol Cell. 2001;7:241–48. doi: 10.1016/s1097-2765(01)00172-1. [DOI] [PubMed] [Google Scholar]
- 145.Voloshin ON, Vanevski F, Khil PP, Camerini-Otero RD. Characterization of the DNA damage-inducible helicase DinG from Escherichia coli. J Biol Chem. 2003;278:28284–93. doi: 10.1074/jbc.M301188200. [DOI] [PubMed] [Google Scholar]
- 146.Wang W. Emergence of a DNA-damage response network consisting of Fanconi anaemia and BRCA proteins. Nat Rev Genet. 2007;8:735–48. doi: 10.1038/nrg2159. [DOI] [PubMed] [Google Scholar]
- 147.Wang W. A major switch for the Fanconi anemia DNA damage-response pathway. Nat Struct Mol Biol. 2008;15:1128–30. doi: 10.1038/nsmb1108-1128. [DOI] [PubMed] [Google Scholar]
- 148.Wang X, Andreassen PR, D’Andrea AD. Functional interaction of monoubiquitinated FANCD2 and BRCA2/FANCD1 in chromatin. Mol Cell Biol. 2004;24:5850–62. doi: 10.1128/MCB.24.13.5850-5862.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Wang X, Kennedy RD, Ray K, Stuckert P, Ellenberger T, D’Andrea AD. Chk1-mediated phosphorylation of FANCE is required for the Fanconi anemia/BRCA pathway. Mol Cell Biol. 2007;27:3098–108. doi: 10.1128/MCB.02357-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Whitney MA, Royle G, Low MJ, Kelly MA, Axthelm MK, et al. Germ cell defects and hematopoietic hypersensitivity to gamma-interferon in mice with a targeted disruption of the Fanconi anemia C gene. Blood. 1996;88:49–58. [PubMed] [Google Scholar]
- 151.Williams RS, Williams JS, Tainer JA. Mre11-Rad50-Nbs1 is a keystone complex connecting DNA repair machinery, double-strand break signaling, and the chromatin template. Biochem Cell Biol. 2007;85:509–20. doi: 10.1139/O07-069. [DOI] [PubMed] [Google Scholar]
- 152.Wilson JB, Yamamoto K, Marriott AS, Hussain S, Sung P, et al. FANCG promotes formation of a newly identified protein complex containing BRCA2, FANCD2 and XRCC3. Oncogene. 2008;27:3641–52. doi: 10.1038/sj.onc.1211034. [DOI] [PubMed] [Google Scholar]
- 153.Wong JC, Alon N, McKerlie C, Huang JR, Meyn MS, Buchwald M. Targeted disruption of exons 1 to 6 of the Fanconi Anemia group A gene leads to growth retardation, strain-specific microphthalmia, meiotic defects and primordial germ cell hypoplasia. Hum Mol Genet. 2003;12:2063–76. doi: 10.1093/hmg/ddg219. [DOI] [PubMed] [Google Scholar]
- 154.Wu L. Role of the BLM helicase in replication fork management. DNA Repair (Amst) 2007;6:936–44. doi: 10.1016/j.dnarep.2007.02.007. [DOI] [PubMed] [Google Scholar]
- 155.Wu Y, Shin-ya K, Brosh RM., Jr FANCJ helicase defective in Fanconia anemia and breast cancer unwinds G-quadruplex DNA to defend genomic stability. Mol Cell Biol. 2008;28:4116–28. doi: 10.1128/MCB.02210-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Wu Y, Suhasini AN, Brosh RM. Welcome the family of FANCJ-like helicases to the block of genome stability maintenance proteins. Cell Mol Life Sci. 2008;66:1209–22. doi: 10.1007/s00018-008-8580-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Xia B, Dorsman JC, Ameziane N, de Vries Y, Rooimans MA, et al. Fanconi anemia is associated with a defect in the BRCA2 partner PALB2. Nat Genet. 2007;39:159–61. doi: 10.1038/ng1942. [DOI] [PubMed] [Google Scholar]
- 158.Xia B, Sheng Q, Nakanishi K, Ohashi A, Wu J, et al. Control of BRCA2 cellular and clinical functions by a nuclear partner, PALB2. Mol Cell. 2006;22:719–29. doi: 10.1016/j.molcel.2006.05.022. [DOI] [PubMed] [Google Scholar]
- 158a.Xue Y, Li Y, Guo R, Ling C, Wang W. FANCM of the Fanconi anemia core complexis required for both monoubiquitination and DNA repair. Hum Mol Genet. 2008;17:1641–52. doi: 10.1093/hmg/ddn054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Yamamoto K, Hirano S, Ishiai M, Morishima K, Kitao H, et al. Fanconi anemia protein FANCD2 promotes immunoglobulin gene conversion and DNA repair through a mechanism related to homologous recombination. Mol Cell Biol. 2005;25:34–43. doi: 10.1128/MCB.25.1.34-43.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Yan J, Jetten AM. RAP80 and RNF8, key players in the recruitment of repair proteins to DNA damage sites. Cancer Lett. 2008;271:179–90. doi: 10.1016/j.canlet.2008.04.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Yang H, Jeffrey PD, Miller J, Kinnucan E, Sun Y, et al. BRCA2 function in DNA binding and recombination from a BRCA2-DSS1-ssDNA structure. Science. 2002;297:1837–48. doi: 10.1126/science.297.5588.1837. [DOI] [PubMed] [Google Scholar]
- 162.Yang Y, Kuang Y, Montes De Oca R, Hays T, Moreau L, et al. Targeted disruption of the murine Fanconi anemia gene, Fancg/Xrcc9. Blood. 2001;98:3435–40. doi: 10.1182/blood.v98.12.3435. [DOI] [PubMed] [Google Scholar]
- 163.Youds JL, Barber LJ, Boulton SJ. C. elegans: a model of Fanconi anemia and ICL repair. Mutat Res: Fundam Mol Mech Mutagen. 2008 doi: 10.1016/j.mrfmmm.2008.11.007. [DOI] [PubMed] [Google Scholar]
- 164.Youds JL, Barber LJ, Ward JD, Collis SJ, O’Neil NJ, et al. DOG-1 is the Caenorhabditis elegans BRIP1/FANCJ homologue and functions in interstrand cross-link repair. Mol Cell Biol. 2008;28:1470–79. doi: 10.1128/MCB.01641-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Zhang QS, Eaton L, Snyder ER, Houghtaling S, Mitchell JB, et al. Tempol protects against oxidative damage and delays epithelial tumor onset in Fanconi anemia mice. Cancer Res. 2008;68:1601–8. doi: 10.1158/0008-5472.CAN-07-5186. [DOI] [PubMed] [Google Scholar]