SUMMARY
Recent findings indicate that mammalian Sonic hedgehog (Shh) signal transduction occurs within primary cilia, although the cell biological mechanisms underlying both Shh signaling and ciliogenesis have not been fully elucidated. We show that an uncharacterized TBC domain-containing protein, Broad-minded (Bromi), is required for high-level Shh responses in the mouse neural tube. We find that Bromi controls ciliary morphology and proper Gli2 localization within the cilium. Using a zebrafish model, we further show that Bromi is required for proper association between the ciliary membrane and axoneme. Bromi physically interacts with cell cycle-related kinase (CCRK), whose Chlamydomonas homolog regulates flagellar length. Biochemical and genetic interaction data indicate that Bromi promotes CCRK stability and function. We propose that Bromi and CCRK control the structure of the primary cilium by coordinating assembly of the axoneme and ciliary membrane, allowing Gli proteins to be properly activated in response to Shh signaling.
INTRODUCTION
Signaling by the Hedgehog (Hh) family of secreted ligands is used in vertebrate embryonic development to control cellular identity, proliferation, differentiation, and survival (Ingham, 2008; Varjosalo and Taipale, 2008). Shh acts as a morphogen in specifying cell fates within the developing neural tube; graded concentrations of the ligand direct cells to adopt distinct identities along the dorsal-ventral axis (Dessaud et al., 2008; Ericson et al., 1997). How the quantitative information, represented by Shh ligand concentration, is accurately transduced by the signaling pathway to control gene expression patterns is an area of active research.
The molecular mechanism of Hh signaling transduction is understood best in Drosophila. In fruit flies, Hh ligands bind to their cell surface receptor, Patched (Ptc). This binding prevents Ptc from inhibiting a second transmembrane protein, Smoothened (Smo), allowing the Hh pathway to be activated (Jia and Jiang, 2006). Smo controls the activity of the downstream transcription factor Cubitus interruptus (Ci) through a cytoplasmic complex containing the atypical kinesin Costal-2 (Cos2), Fused (Fu) kinase, and Ci (Lum et al., 2003; Ruel et al., 2003). In addition, Suppressor of fused (Su(fu)) regulates the nuclear localization of Ci, although Su(fu) in Drosophila is dispensable for Hh signaling on its own (Methot and Basler, 2000). Ci acts both as a transcriptional activator and repressor of Hh target genes, depending on its proteolysis, which is regulated by the pathway (Methot and Basler, 1999).
The molecular basis of Hedgehog signaling in mammals is similar to that in Drosophila, although there are important differences. In mice, Hh ligands signal via Ptc and Smo homologs to control three Ci homologs, Gli1, Gli2, and Gli3. However, the functional significance of Fu and Su(fu) homologs in Hh signaling appears to have changed during evolution (Svard et al., 2006; Varjosalo et al., 2006; Wilson et al., 2009) and, although Cos2 homologs regulate Hh signaling in mammals and Drosophila, their biochemical functions may differ (Endoh-Yamagami et al., 2009; Liem et al., 2009). The role of intraflagellar transport and primary cilia in mammalian Hh signaling represents another key difference (Eggenschwiler and Anderson, 2007). During mouse development, many cell types generate primary cilia, including those responding to Hh signals. Mutations disrupting different aspects of ciliogenesis have distinct effects on the Hh pathway, interfering with steps downstream of Shh and Patched (Caspary et al., 2007; Huangfu and Anderson, 2005; Huangfu et al., 2003; Tran et al., 2008). Complementing the genetic data are cell biological studies showing that several key components of the mammalian Hh pathway, Patched1 (Ptch1), Smo, Gli2, Gli3, and Suppressor of Fused, localize to primary cilia (Corbit et al., 2005; Haycraft et al., 2005; Rohatgi et al., 2007). Despite these findings, how the regulatory cascade proceeds within a ciliary context remains obscure.
As with Hedgehog signaling, our understanding of ciliogenesis has seen significant advances, although many questions remain. The assembly of these structures is clinically important as they are involved in a group of human disorders, the so-called “ciliopathies”, such as polycystic kidney disease and Bardet-Biedl syndrome (Quinlan et al., 2008). Much of what we understand about ciliogenesis comes from research on the green algae Chlamydomonas reinhardtii. During interphase, Chlamydomonas cells generate a pair of motile flagella using strategies very similar to those acting in metazoan ciliogenesis. Studies in Chlamydomonas have highlighted the importance of intraflagellar transport (IFT), which allows bidirectional microtubule-based movement of cargo to and from the tips of growing cilia and flagella (Pedersen and Rosenbaum, 2008).
While the composition and movement of IFT protein complexes and the structure of axonemes have been studied in detail, other aspects of ciliogenesis are less clear. One example is how the lengths of cilia and flagella are controlled. The effects on flagellar length and growth rate after amputation of a single flagellum have led investigators to propose that Chlamydomonas actively monitor and regulate flagellar length (Rosenbaum et al., 1969). More recently, genetic and RNAi studies have identified several key regulators of the processes (Wilson et al., 2008). Among these are three proteins, one kinase related to cyclin-dependent kinases, LF2p, and two proteins of unknown function, LF1p and LF3p, which comprise a cytoplasmic Length Regulatory Complex (LRC; Barsel et al., 1988; Tam et al., 2007).
In this study, we show that an uncharacterized factor, Broad-minded (Bromi), links activity of the Shh signaling pathway to the coordinated assembly of the ciliary axoneme and membrane. In mouse bromi mutants, the Shh pathway can be properly activated by low and intermediate levels of signals, but high-level responses are blocked. Primary cilia on bromi mutant neural progenitors are abnormally shaped such that the axonemes appeared curled with Gli2 mislocalizing to their centers. A similar effect on ciliary morphology is seen in zebrafish bromi morphants. Ultrastructural analysis revealed detachment and massive expansion of the ciliary membrane away from the axoneme, indicating that proper assembly and association between the two require Bromi function. We find that Bromi physically interacts with cell cycle-related kinase (CCRK), a mammalian homolog of the LRC component LF2p, and that knockdown of ccrk in zebrafish results in curled cilia like those seen in bromi morphants. We observe a genetic interaction between bromi and ccrk, indicating that the two proteins function in a common mechanism, and our biochemical data suggest that Bromi-CCRK complex formation promotes CCRK stability. We propose that Bromi and CCRK act together to regulate ciliary membrane and axonemal growth and that the close association between these compartments ensures that Gli proteins can efficiently mediate responses to high levels of Shh signals.
RESULTS
A recessive mutation, broad-minded (bromi), was identified in a genetic screen for genes controlling mid-gestation development in the mouse (Zohn et al., 2005). bromi mutants exhibit exencephaly, poorly developed eyes, and preaxial polydactyly (Figure S1A-E). The cephalic ventral midline furrow, which can be seen exencephalic mouse mutants with normal neural patterning, was absent in bromi mutants, suggesting ventral neural fates are not properly specified (Figure S1D). These features resembled those of mutants with defects in Hedgehog signaling.
Bromi is required for appropriate responses to Shh signals
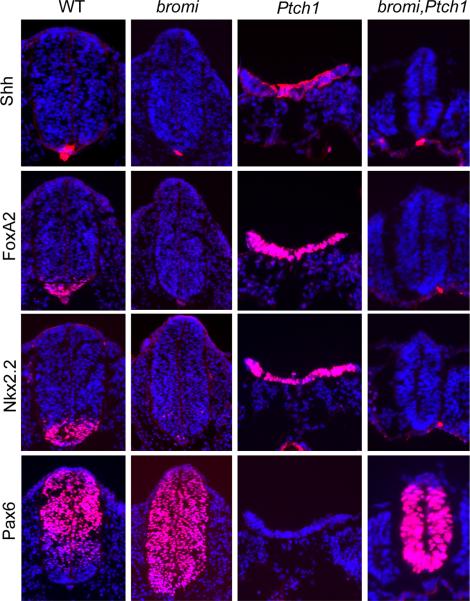
To investigate Shh signaling in bromi mutants, we assayed dorsal-ventral neural tube patterning, as this process is tightly controlled by the Shh morphogen (Dessaud et al., 2008). In the E10.5 bromi mutant neural tube, FoxA2+/Shh+ floor plate cells, specified by high levels of Shh signaling, were absent, Nkx2.2+ V3 interneuron progenitors (p3) were observed in the ventral midline and were variably reduced in number, and HB9+ motor neuron progenitors (pMN), specified by intermediate levels of Shh, were expanded ventrally along the midline (Figure 1A). Pax6 expression, which is normally repressed by high-level Shh signaling, expanded ventrally in bromi neural tubes. Expression of Pax7, which is sensitive to the lowest levels of Shh signaling, was unaffected in bromi mutants. We monitored the expression of an independent reporter of Shh signaling activity, Ptch1-LacZ (Goodrich et al., 1997). In bromi,Ptch1-LacZ/+ animals, staining for β-galactosidase activity was diminished in ventral neural progenitors compared with Ptch1-LacZ/+ controls, while the dorsal border of the expression domain was not appreciably affected (Figure 1B). These results suggested that bromi neural progenitors respond to low and intermediate levels of Shh signaling, but responses to high levels are blocked.
Figure 1. Neural patterning in bromi mutants.
(A) Spinal neural tubes of E10.5 wild-type (WT) and bromi mutants immunostained for markers of neural identity (in red, DAPI in blue). bromi mutant neural tubes lacked the floor plate, marked by Shh and FoxA2, although the notochord (arrowheads) expressed Shh normally. Nkx2.2+ p3 cells were inappropriately positioned in the mutant ventral midline and reduced in number. The Pax6+ and HB9+ (motor neuron) domains, which are supported by intermediate but not high levels of Shh, ectopically expanded into the most ventral regions in the mutants. The ventral limit of the Pax7+ domain, set by low levels of Shh signaling, was not affected in bromi mutants. (B) X-gal staining of E10.5 neural tubes from wild-type and bromi embryos heterozygous for the Ptch1-LacZ insertion. Ptch1-LacZ is expressed in a ventral-to-dorsal gradient in response to Shh. Expression in the ventral one-third of the bromi mutant neural tubes (n=3) was consistently weaker compared to that of wild-type.
bromi regulates the Shh pathway downstream of Ptch1 and Rab23
We next asked whether bromi can suppress the phenotype caused by loss of Ptch1. Ptch1 represses Hh target genes in the absence of ligand (Varjosalo and Taipale, 2008). Disruption of Ptch1 strongly activates the Hh pathway, resulting in potent ventralization of neural identity (Goodrich et al., 1997). In contrast to Ptch1 mutants, patterning in bromi,Ptch1 double mutant neural tubes was dorsalized as in bromi single mutants. The double mutant neural tubes failed to express Shh and FoxA2 and they maintained ventral Pax6 expression (Figure 2). Consistent with our hypothesis that the bromi mutation blocks responses to high but not low levels of Shh signaling, neural progenitors in the double mutants ectopically expressed Nkx6.1, a marker sensitive to both high and low levels of Shh signaling (Figure S2A). Moreover, bromi,Ptch1 double mutants exhibited ectopic Ptch1-LacZ activity throughout most of the embryo, but it was significantly weaker than that observed in Ptch1 single mutants (Figure S2B).
Figure 2. bromi is epistatic to Ptch1.
Spinal neural tubes from E9.5 wild-type (WT), bromi, Ptch1, and bromi,Ptch1 mutant embryos immunostained for markers of neural fate. In Ptch1 mutants, Shh, FoxA2, and Nkx2.2 were broadly expressed throughout the open neural tubes and Pax6 expression was absent. The Ptch1 mutant phenotype was strongly suppressed in bromi,Ptch1 double mutants, whose closed neural tubes were small, yet showed similar expression patterns as bromi single mutants.
We next investigated the relationship between bromi and Rab23. In mice, disruption of Rab23 causes activation of the Shh pathway and cell-autonomous ventralization of neural identity (Eggenschwiler et al., 2001). This effect occurs independently of Shh and Smo, but it is efficiently suppressed in Rab23,Gli2 double mutants, indicating that Gli2 activity is unrestrained in Rab23 mutants (Eggenschwiler et al., 2006). In contrast to Rab23opb2 mutants, bromi,Rab23opb2 double mutants lacked a Shh+/FoxA2+ floor plate and their neural patterning was identical to that of bromi single mutants (Figure S2C). These data suggest that bromi acts at a step downstream of Shh, Ptch1, Smo, and Rab23 and is required for Gli2 to potently activate Shh targets.
Gli activator function is impaired in bromi mutants
If the effects of bromi on cell identity are through Shh signaling, increasing pathway activity at a step further downstream should rescue the bromi patterning phenotype. Gli3 plays a dual role as an activator and repressor of Shh target genes. In the presence or absence of Shh signals, Gli3 is converted into a transcriptional activator, Gli3(Act), or it is processed into a repressor form, Gli3(Rep), respectively (Wang et al., 2000). A Gli3 null mutation (Gli3xt) causes modestly elevated pathway activity in the neural tube due to the loss of Gli3(Rep) (Persson et al., 2002). We found that increasing Shh pathway activity in bromi mutants by disrupting Gli3 (Figure 3A) was sufficient to restore ventral neural patterning, including specification of the floor plate, in bromi,Gli3xt double mutants. Thus, bromi neural progenitors are capable of adopting the most ventral of neural cell identities provided the Shh pathway is sufficiently activated.
Figure 3. Patterning in bromi,Gli3xt and bromi,Gli2 double mutants.
(A) FoxA2, Nkx2.2, and HB9 immunostaining of E10.5 wild-type (WT), bromi, Gli3xt, and bromi,Gli3xt double mutant spinal neural tubes. Expression of these markers was normal in Gli3xtmutants, but disruption of Gli3 restored floor plate specification in bromi,Gli3xt double mutants. (B) FoxA2, Nkx2.2, and HB9 immunostaining of E10.5 wild-type (WT), bromi, Gli2, and bromi,Gli2 double mutant spinal neural tubes. Ventral neural patterning is similar in bromi and Gli2 single mutants. bromi,Gli2 double mutant neural tubes lacked Nkx2.2+ cells, whereas they were always detected, albeit in reduced numbers, in either single mutant.
The bromi neural patterning phenotype was similar to that of Gli2 null mutants (Matise et al., 1998). However, whereas floor plate and V3 progenitors are lost entirely in Gli2,Gli3xt double mutants (Bai et al., 2004), these cell types were rescued in bromi,Gli3xt mutants, indicating Gli2 retains some function in bromi mutants (Figure 3A). Consistent with this hypothesis, disruption of Gli2 in bromi mutants weakly exacerbated the bromi phenotype; Nkx2.2+ p3 progenitors were not observed in any of 3 bromi,Gli2 double mutants, whereas Nkx2.2+ cells were observed in all 6 Gli2 and bromi single mutants analyzed (21± 8 and 16 ± 5 cells/section, respectively; Figure 3B).
Western blotting of bromi and wild-type extracts showed that the amount of processed Gli3, Gli3(Rep), was comparable between genotypes, although a greater amount of unprocessed full-length Gli3 was observed in the mutant samples (Figure S3A). A similar phenomenon has been observed in the ciliogenesis mutant ftm (Vierkotten et al., 2007). Disruption of Gli3(Rep) activity may explain the polydactylous phenotype of bromi mutants. However, because the expression patterns of Shh- and Gli3-dependent genes in the bromi mutant limbs were not dramatically affected (Figure S1F), such an effect on Gli3(Rep) function would be subtle. As steady-state Gli2 levels appeared normal in bromi mutants (Figure S3B), Bromi more likely regulates Gli2 activity rather than its synthesis or stability. The data suggest that the activator functions of Gli2 and Gli3 are less potent in the bromi mutant neural tube and that disruption of Gli3(Rep) can compensate for this defect.
bromi encodes C6orf170, an uncharacterized TBC domain-containing protein
We identified the bromi gene using meiotic recombinant mapping (Figure 4A). Sequencing of genes in the interval revealed an A-to-G transition in the splice acceptor site of exon 11 in the uncharacterized gene C6orf170 (D630037F22Rik). Mutant transcripts make use of a cryptic splice acceptor in exon 11 that shifts the reading frame and results in a nonsense codon immediately downstream (Figure 4B and 4C). We raised polyclonal antisera against Bromi and performed western blotting. A species of the predicted size (~140kDa) was observed in wild-type, but not bromi mutant, embryonic extracts (Figure 4D). A gene-trap insertion in C6orf170, RRF165, failed to complement bromi, showing the bromi phenotype in bromi/RRF165 transheterozygotes (Figure S4A), and RRF165 homozygotes showed a patterning phenotype identical to that of bromi mutants (Figure S4B). Thus, bromi disrupts C6orf170 and appears to be a null allele.
Figure 4. Positional cloning of bromi.
(A) bromi was mapped to a 2.3 Mb region on mouse chromosome 10 by analysis of recombinants (recombination events, out of 219 opportunities scored for each marker, are listed). This interval contained 6 known and predicted genes, which were fully sequenced in bromi and parental (C57BL/6J) strains. (B) An A-to-G splice acceptor mutation at the intron 10/exon 11 junction of C6orf170 was identified in bromi samples (trace shown in C) and sequencing of RTPCR products showed that bromi mutants use a cryptic splice acceptor site 13 base pairs downstream. (D) Western blotting of wild-type (WT) and bromi mutant embryonic extracts with polyclonal antisera against the Bromi protein.
Bromi homologs are restricted to chordates and are predicted to contain a TBC (Tre-2, Bub2 and Cdc16) domain near their C-termini. Proteins containing TBC domains typically function as GTPase activating proteins (GAPs) for Rab GTPases, functioning in membrane trafficking (Grosshans et al., 2006). Sequence comparison of the Bromi TBC domain with those of known RabGAPs (data not shown) revealed that Bromi lacks two key residues, an arginine and a glutamine in the catalytic finger motifs, essential for RabGAP activity (Pan et al., 2006). Neither Bromi nor its TBC domain showed an interaction with any among a panel of 60 GTP-locked Rab proteins (Figure S4D) in a yeast two-hybrid assay previously used to identify RabGAPs (Itoh et al., 2006). As Bromi functions in ciliogenesis (see below), we tested whether it co-immunoprecipitates with FLAG-tagged Rabs 8a, 17, and 23, which have been implicated in the control of ciliogenesis (Yoshimura et al., 2007), but found no evidence for such interactions (Figure S4E-F). Collectively, these data suggest that Bromi does not function as a classical RabGAP.
Bromi controls primary cilia morphology in the neural tube
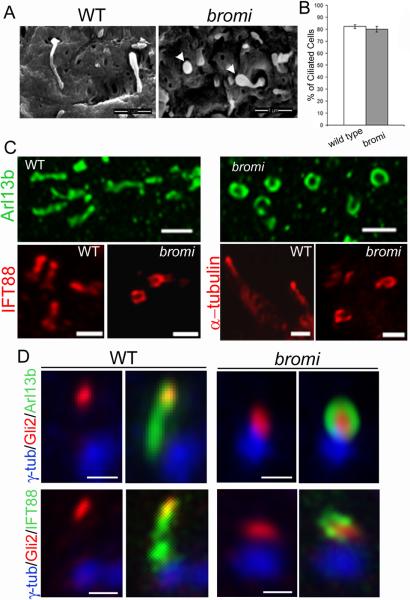
Because primary cilia are thought to play a key role in mouse Hh signal transduction, we investigated ciliogenesis in bromi mutants by scanning electron microscopy (SEM) and confocal microscopy. Cilia of the wild-type neuroepithelium were short and straight, projecting apically into the neural tube lumen (Figure 5A). In contrast, bromi neuroepithelial cilia, which were generated at the same frequency as wild-type (Figure 5B), appeared bulbous or spherical (Figure 5A). Staining with Arl13b, IFT88, and acetylated α-tubulin antibodies revealed structures resembling open rings when viewed from the apical surface (Figures 5C and 5D). We suggest that these ring-shaped structures represent curled axonemes enveloped by dilated ciliary membranes, although it is also possible that Arl13b, IFT88, and acetylated α-tubulin localize away from axonemes, adjacent to the dilated ciliary membranes. As the genetic data suggested that the function of the Gli activators is impaired in bromi mutants, we examined the localization of Gli2 within cilia (Figure 5D). In wild-type samples, Gli2 staining was concentrated at cilia tips and colocalized with Arl13b and IFT88. In contrast, Gli2 staining was largely concentrated in the centers of the bromi mutant cilia, encircled by rings of Arl13b and IFT88 staining. Co-labeling with gamma tubulin antibodies indicated that Gli2 localized within the mutant cilia rather than to the basal bodies.
Figure 5. Cilia defects in bromi mutants.
(A) Neuroepithelial cilia from E10.5 wild-type (WT) and bromi mutant neural tubes analyzed by scanning electron microscopy. bromi mutant cilia showed a swollen or bulbous morphology (arrowheads). (B) Cilia frequency in the wild-type and bromi mutant neuroepithelium. (C) Confocal images of neural tube cilia stained with Arl13b (upper panels), IFT88 and acetylated α-tubulin (lower panels). (D) Neural tube cilia from E10.5 wild-type (WT) and bromi mutant embryos immunostained for γ-tubulin/Arl13b/ Gli2 or γ-tubulin/IFT88/Gli2. Scale bars are 1 μm in A and C and 0.5 μm in D. Error bars in B reflect +/− standard error of the mean.
Bromi functions in zebrafish ciliogenesis
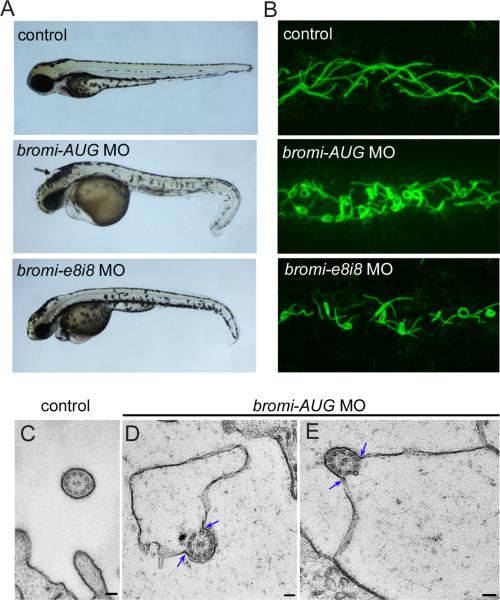
Cilia from neural progenitors are short and therefore difficult to study with high spatial resolution. For this reason, we further investigated Bromi's role in ciliogenesis using another system, zebrafish kidney tubules, where longer cilia could be better visualized. We investigated the function of the zebrafish bromi homolog (si:dkey-233p4.1) using morpholinos directed against the start site (bromi-AUG) and a splice site (bromi-e8i8) of the gene. Injection of either morpholino resulted in morphants that were viable during embryonic and early larval stages, but exhibited curvature of the body axis and hydrocephalus (Figure 6A). RT-PCR confirmed that splicing of zebrafish bromi was disrupted by the bromi-e8i8 morpholino (Figure S5A). We analyzed the cilia from distal kidney tubules, which are long and easily imaged by confocal microscopy using antibodies against acetylated α-tubulin (Figure 6B). Interestingly, the cilia seen in the majority of bromi-AUG and bromi-e8i8 morphants (20/31 and 8/11, respectively) exhibited pronounced curling, often forming rings. This ciliary phenotype resembled that seen in the bromi mouse mutant neuroepithelium, suggesting that Bromi plays a qualitatively similar function in controlling axonemal shape in both systems. To gain a better understanding of the ultrastructure of these cilia, we performed transmission electron microscopy (TEM). The cilia of bromi morphants showed a striking defect; their ciliary membranes were detached from axonemes along one side and dramatically expanded within the kidney tubule lumens (Figure 6D and 6E). The large space between the expanded membranes and axonemes appeared empty. In contrast, ciliary membranes were tightly associated along the periphery of the ciliary axonemes in control fish (Figure 6C). The axonemes of the morphant cilia showed the characteristic structure of 9 doublet microtubules surrounding a pair of central microtubules, indicating that morphant axonemes were largely intact. These defects indicate that coordinated assembly of the ciliary membrane and the axoneme is lost in the bromi morphants.
Figure 6. bromi regulates the association between ciliary membranes and axonemes.
(A) 72-hpf zebrafish embryos injected with morpholinos against the start site (bromi-AUG) and exon 8/intron 8 splice site (bromi-e8i8) of the zebrafish bromi homolog resulted in abnormal tail curvature. bromi-AUG MO fish exhibited hydrocephalus (arrow), while this was less frequent in bromi-e8i8 MO fish. (B) Confocal microscopy of acetylated alpha-tubulin-stained cilia in the posterior (distal early/late) regions of the zebrafish kidney tubules. Ciliary morphology in the anterior (proximal convoluted/straight) regions of the tubules was not appreciably affected in the morphants. (C-E) Posterior pronephric cilia visualized by TEM showing 9+2 axonemal structure in 48 hpf control and bromi-AUG MO fish. The membranes of control cilia were tightly associated with axonemes (C), whereas they detached along one side of the axonemes (blue arrows) and dramatically expanded within the tubule lumens of bromi-AUG MO fish (D, E). Scale bars are 100 nm.
Cell cycle-related kinase interacts with Bromi and controls vertebrate ciliogenesis
To gain insight into the molecular mechanism by which Bromi controls ciliogenesis, we searched for Bromi-associated proteins using tandem affinity purification and mass spectrometry. We identified a specific band of approximately 40 kDa on silver stained gels in the immunoprecipitates. Mass spectrometry identified this protein as cell cycle-related kinase (CCRK/PNQALRE). Using the Rheoswitch inducible system (New England Biolabs), we found that endogenous CCRK and Bromi co-immunoprecipitated with HA-tagged Bromi and CCRK, respectively (Figure 7A). We investigated the functional significance of this interaction by assaying CCRK levels in bromi mutants. In comparison to wild-type, endogenous CCRK in mutant embryos and fibroblasts was reduced by 58±12% and 51+15% of controls, respectively (Figure 7B). In addition, CCRK levels were increased (160±16%) in NIH3T3 cells induced to express HA-Bromi compared to uninduced controls. These data suggest that Bromi promotes CCRK stability and function.
Figure 7. Cell cycle-related kinase interacts with Bromi and acts in vertebrate ciliogenesis.
(A) HA-tagged Bromi or HA-tagged CCRK, expressed in NIH3T3 fibroblasts by Rheoswitch ligand (RSL1)-mediated induction, co-immunoprecipitated endogenous CCRK and Bromi, respectively. (B) CCRK protein levels in wild-type vs. bromi mutant embryos and embryonic fibroblasts and in uninduced vs. RSL1-induced Bromi-HA expressing NIH3T3 cells. RSL1 treatment of parental Rheoswitch cells had no effect on CCRK levels (data not shown). Anti-β-tubulin was used as a loading control. (C) Injection of morpholinos (4 ng) directed against the start site (ccrk-AUG) and exon 3/intron 3 splice site (ccrk-e3i3) of the zebrafish ccrk homolog resulted in kinking of the body axis and abnormal tail curvature. (D) Confocal microscopy of acetylated alpha-tubulin-stained cilia in the posterior (distal early/late) regions of the kidney tubules. (E) Percentage of animals exhibiting curled cilia. Injection of low doses of bromi-AUG or ccrk-e3i3 morpholinos (1ng or 2ng, respectively) had little effect on their own, whereas co-injection of both morpholinos at these doses had a synergistic effect. Values in parentheses represent numbers of animals scored.
The biological functions of CCRK are not well understood. CCRK has been suggested to function as a cyclin-dependent kinase (CDK)-activating kinase (CAK) (Liu et al., 2004), but other data have called this into question (Wohlbold et al., 2006). In addition, CCRK is the vertebrate homolog of the long flagella 2 protein (LF2p) of Chlamydomonas reinhardtii (Tam et al., 2007). LF2p, together with LF1p and LF3p, is a component of the Length Regulatory Complex (LRC), which controls the length of the flagellum (Barsel et al., 1988; Tam et al., 2007). LF2p is the only LRC component sharing homology with proteins outside of Chlamydomonas, but it is unknown whether such homologs function in ciliogenesis. We investigated this possibility by disrupting CCRK function using morpholinos directed against the single zebrafish homolog, zgc:101530/ccrk. Two morpholinos, one directed at the start site (ccrk-AUG) and another against a splice donor (ccrk-e3i3), caused similar phenotypes (Fig. 7C). RTPCR confirmed that splicing was largely disrupted by the splice site morpholino (Figure S5B). The ccrk morphant fish displayed a kinked body axis and curvature defects similar to bromi morphant fish (Figure 7C) and in most start site and splice site morphants (n= 11/16 and 18/27, respectively), the cilia in the distal kidney tubules appeared tightly curled as in bromi morphants (Figure 7D). Curling of the kidney tubule cilia was quite sensitive to morpholino dosage as injection of 2-fold lower doses of either ccrk-e3i3 or bromi-AUG morpholinos dramatically reduced the frequency of zebrafish with curled cilia (Figure 7E). However, injection of embryos with the same low doses of both morpholinos together had a strong, synergistic effect (Figure 7E). These findings, together with the biochemical data, indicate that Bromi and CCRK act together to regulate ciliary assembly through a common mechanism.
In Chlamydomonas, components of the LRC such as LF2p also show punctate cytoplasmic, rather than flagellar, localization (Tam et al., 2007). We found that epitope-tagged CCRK and Bromi also showed cytoplasmic localization in NIH3T3 cells, although it appeared diffuse rather than punctate. Importantly, neither Bromi nor CCRK showed discrete localization within the cilium (Figure S6). In addition, CCRK showed nuclear staining, consistent with a previous report (Liu et al., 2004).
DISCUSSION
Here, we show that mouse Broad-minded (Bromi), a previously uncharacterized TBC domain-containing protein, is required for proper cellular responses to the highest levels of Shh signaling in the neural tube. The genetic data indicate that this factor acts in Shh signal-receiving cells while the cell biological experiments indicate that it regulates the structure of the primary cilium and coordinates the growth of ciliary membranes with that of the axoneme. We find that the vertebrate cell cycle-related kinase (CCRK) interacts with, and is stabilized by, Bromi and that both proteins play similar roles in the assembly of cilia.
Bromi and the primary cilium
Bromi controls ciliogenesis, yet its mutant ciliary phenotype appears distinct from that of other mutants previously described. While Bromi regulates the manner in which cilia are assembled, it is not required for their generation in the embryo. Rather, Bromi appears to control axonemal shape and the close apposition of axonemes with ciliary membranes. IFT proteins and electron-dense material accumulate between the membranes and axonemes of mutants with defective retrograde IFT leading to mild, symmetric expansion of ciliary/flagellar membranes around axonemes without affecting axonemal shape (Hou et al., 2004; Perkins et al., 1986; Qin et al., 2001; Tran et al., 2008). In contrast, disruption of bromi in zebrafish results in a more dramatic expansion of the ciliary membrane. This expansion is limited to one side of the cilium and the space between the membrane and axoneme is devoid of ectopic material. Moreover, IFT88 does not accumulate at the distal ends of bromi mutant cilia as it does in mouse, C. elegans and Chlamydomonas mutants with retrograde IFT defects. Thus, the expansion of the ciliary membrane away from the axoneme that we observe more likely results from defective axonemal/membrane growth control rather than membrane displacement resulting from accumulation of IFT or structural proteins in the cilium.
The ciliary membrane does not arise from evagination of the plasma membrane but, rather, it is a specialized membrane with unique lipid and protein composition produced from specific Golgi-derived vesicles (Chailley and Boisvieux-Ulrich, 1985; Pazour and Bloodgood, 2008). With the exception of specialized cilia (e.g., photoreceptors), flagellar and ciliary membranes remain tightly associated with axonemes during their growth (Pazour and Bloodgood, 2008). This suggests that the deposition and growth of ciliary membranes are coordinated with growth of the axoneme. However, work on Trypanosome flagella indicate that flagellar membranes can form without axonemes, and vice versa, when IFT proteins or TbLRTP are disrupted, respectively (Davidge et al., 2006; Morgan et al., 2005). Thus, at least in Trypanosomes, biogenesis of flagellar membranes and axonemes are separable under experimental conditions.
Bromi may control cilia assembly through several mechanisms. Its primary role could be in proper assembly of the axonemal structure, which, in turn, controls the growth of the ciliary membrane. However, because the axonemal structure of zebrafish bromi morphant cilia appeared intact, whereas ciliary membranes showed a dramatic defect, we suggest that the curling of their axonemes results from stress placed on the axoneme by the asymmetric growth of the ciliary membrane along one side. Alternatively, Bromi may directly control the addition of newly synthesized ciliary membranes from within the cell. Although many of the details of ciliary membrane growth remain obscure, Rab8a, IFT20, and Bardet-Biedl Syndrome (BBS) proteins have been shown to function in transporting Golgi-derived membranes to the cilium (Follit et al., 2006; Nachury et al., 2007). Bromi may function to control the delivery of membrane to the cilium or coordinate this process with axonemal growth through membrane trafficking. Our data suggest this function of Bromi is due to its regulation of CCRK function rather than through RabGAP activity of its TBC domain.
CCRK links Bromi to ciliary assembly
One clue as to the cellular function of Bromi is its physical association with CCRK. Like Bromi, CCRK localizes to the cytoplasm and in zebrafish its disruption results in curling of ciliary axonemes. CCRK is particularly interesting because it is the closest vertebrate homolog to the long flagella 2 (LF2p) protein of Chlamydomonas. Together with two novel proteins, LF1p and LF3p, LF2p is a component of the Length Regulatory Complex. Although the biochemical function of the LRC remains elusive, mutations in any one of these components cause flagella to be unusually long or short, and of unequal length within the same cell (Barsel et al., 1988; Tam et al., 2007). Homologs of LF1p and LF3p have not been identified in other species and a role for LF2p homologs outside of Chlamydomonas in ciliogenesis has not been described (Tam et al., 2003). It is unclear whether LF1p and LF3p regulate or potentiate LF2p activity and, if so, whether vertebrate Bromi performs an analogous function with CCRK.
Our work suggests that the Bromi/CCRK complex helps to stabilize CCRK in the cell, but Bromi may provide additional functions such as recruiting CCRK substrates. One of the few CCRK substrates identified thus far is MRK (also known as ICK), a member of the MAK/MOK/MRK subfamily of MAP kinases (Fu et al., 2006). Interestingly, this subfamily shares considerable homology with Chlamydomonas LF4p, C. elegans DYF-5, and Leishmania mexicana LmxMPK9, which negatively regulate flagellelar and ciliary length (Bengs et al., 2005; Berman et al., 2003; Burghoorn et al., 2007). The Chlamydomonas lf4 mutant phenotype, like that of lf2, perturbs flagellar length. However, null alleles of these genes show distinct phenotypic features and genetic interactions, suggesting that LF2p and LF4p have related, yet separable, cell biological functions (Asleson and Lefebvre, 1998; Berman et al., 2003; Tam et al., 2007). Identification of additional substrates for these kinases should clarify their functions with respect to the assembly of cilia and flagella.
Bromi and Shh signaling
In mice, the primary cilium appears to have diverse and complex functions in regulating the Shh signaling pathway. In mouse mutants with ciliary defects, the specific manner in which the output of the pathway is disrupted depends on the way in which ciliogenesis is affected. Mutations that disrupt ciliogenesis altogether (e.g., Ift172, Ift88, and Kif3a null mutants), have two effects on Shh signaling; while Gli activators lose their abilities to activate Shh target genes, diminished processing of Gli3 to form the Gli3 repressor leads to weak derepression of Shh target genes (Huangfu and Anderson, 2005; Huangfu et al., 2003). Disruption of Arl13b results in constitutive, intermediate-level activity of Shh pathway throughout the neural tube (Caspary et al., 2007). These mutants generate cilia but they are short and exhibit open axonemal B tubules. Disruption of THM1/aln results in ball-shaped cilia with defects in retrograde IFT and constitutive, high-level pathway activity in the neural tube (Tran et al., 2008). In contrast to these mutants, the Shh pathway in bromi mutants retains appropriate responses to all but the highest levels of Shh signaling in the neural tube. This phenotype is most similar to mouse mutants with hypomorphic C2cd3 or IFT88 function (Hoover et al., 2008; Huangfu et al., 2003). The phenotype of bromi,Ptch1 double mutants indicates that loss of bromi places an upper boundary on the extent to which the pathway may be activated but, at lower levels of Shh signaling, responses to graded Shh signals are intact. The suppression of the Rab23 phenotype in bromi,Rab23 double mutants indicates that Bromi acts downstream in the Shh pathway and that strong, constitutive activation of Gli2 in the absence of Rab23 relies on Bromi function.
The notion that cilia merely provide a permissive environment where Hedgehog signaling occurs has been challenged with the characterization of mouse ciliogenesis mutants with complex Shh responses (Caspary et al., 2007; Eggenschwiler and Anderson, 2007; Tran et al., 2008). The localization of Gli proteins to cilia is regulated, at least in part, by activity of the Hedgehog pathway (Chen et al., 2009; Endoh-Yamagami et al., 2009) and it appears that the precise level of Gli activity in response to a given level of stimulus depends on the way in which ciliary transport and structure are affected in various mutants. In bromi neuroepithelial cells, Gli2 fails to colocalize with Arl13b and Ift88 at cilia tips. We propose that Ift88, Arl13b, and acetylated α- tubulin label the axonemes within the mutant cilia and that Gli2 localizes interior to, and distinct from, the curled axonemes. This dissociation may attenuate the efficacy of the Glis, possibly by affecting their flux into and out of cilia (Kim et. al. 2009) or by preventing efficient interactions between Gli proteins and their upstream regulators.
EXPERIMENTAL PROCEDURES
Mouse strains and genotyping
The Ptch1tm1Mps allele, obtained from M. Scott, is null for Ptch1 and harbors a LacZ insertion driven by the Ptch1 regulatory elements (Goodrich et al., 1997). Rab23opb2, Gli2tm1Alj and Gli3xt/J are null alleles obtained from K. Anderson, A. Joyner, and the Jackson Laboratories, respectively, and were genotyped as described (Eggenschwiler et al., 2001; Hui and Joyner, 1993; Matise et al., 1998). bromi mutants were identified using flanking SSLP markers and an assay based on a DdeI RFLP created by the A/G mutation in the splice acceptor site of bromi/C6orf170 exon 11. RRF165 is a LacZ gene-trap insertion within intron 4 of C6orf170 of mouse embryonic stem cells obtained from BayGenomics. The insertion site and splicing events were confirmed by genomic and RT-PCR. RRF165/+ mouse lines were generated by blastocyst injection using standard approaches. RRF165/+ mice were backcrossed to C3Heb/FeJ and were genotyped using primers in LacZ and SSLP markers. All animal work was conducted according to protocols approved by the Institutional Animal Care and Use Committee (IACUC) at Princeton University.
Mapping and cloning of the bromi locus
bromi was isolated in a recessive N-ethyl-N-nitrosourea mutagenesis screen (on a C57BL/6J background) and mapped using MIT SSLP polymorphic markers. For fine mapping, we generated additional polymorphic markers (information available upon request). Recombinant chromosomes defined the 2.3 Mb critical region which contains six known genes. Sequencing of genomic DNA (exons and exon-intron junctions) of all six genes from bromi mutant and its parental strain revealed a single change, an A to G transition, in the intron 10/exon 11 splice acceptor site of D630037F22Rik (C6orf170). The full-length transcript sequence (accession# NM_001033385) was confirmed using a SMART RACE cDNA amplification kit (Clontech).
Immunohistochemistry
Immunohistochemical analysis of neural tubes was carried out on fixed frozen sections as described (Eggenschwiler and Anderson, 2000). Mouse monoclonal antibodies against HB9/MNR2, Nkx2.2, Nkx6.1, FoxA2/Hnf3β, Pax7, Pax6, and Shh were obtained from the Developmental Studies Hybridoma Bank. Additional antibodies included rabbit anti-Pax6 (Covance Research Products), and anti-Olig2 (R&D Systems). Cy2- and Cy-3-conjugated secondary antibodies were purchased from Jackson ImmunoResearch. Sections were counterstained with DAPI (Sigma-Aldrich). Images were obtained using a fluorescent microscope (E800, Nikon) with CCD camera (Princeton Instruments), using MetaMorph software (Universal Imaging Corporation). Image intensity and contrast were adjusted using Photoshop 7.0 software (Adobe). In all epistasis experiments, at least 5 single mutants and 3 to 4 double mutants per genotype were analyzed.
X-gal staining, skeletal preparation, and in situ hybridization
Embryos were prepared for X-gal staining to monitor β-galactosidase activity as described (Nagy et al., 2003). Embryos of different genotypes stained to monitor Ptch1-LacZ activity were treated identically in the same batch. The weak, ubiquitous expression of C6orf170/bromi in the embryo made it difficult to unambiguously assay by in situ hybridization. Therefore, C6orf170 expression was assayed by X-gal staining of RRF165/+ (gene-trap) embryos. Non-transgenic (+/+) embryos were treated identically to control for potential nonspecific staining. Limb skeletons were stained with alcian blue as described (Nagy et al., 2003). Whole mount in situ hybridization was performed on E10.5 embryos using Gli1 (Hui et al., 1994), Hoxd11 (Dolle et al., 1989), and Hoxd13 (Dolle et al., 1989) ribroprobes as previously described (Eggenschwiler and Anderson, 2000).
Analysis of neural tube cilia
Neural tube cilia were imaged from sections (4 embryos per genotype) stained with anti-Arl13b (Caspary et al., 2007), anti-IFT88 (gift from G. Pazour), anti-acetylated α-tubulin (Sigma), anti-γ-tubulin (Sigma), and anti-Gli2 (Cho et al., 2008) antisera followed by Cy2-, Cy3-, and Cy5-labeled secondary anibodies (Jackson ImmunoResearch). Cilia were imaged by confocal microscopy using a Leica TCS SP5 confocal microscope. Z-stack projections were subjected to deconvolution using Volocity software (Improvision/Perkin Elmer). Ciliogenesis frequency in the neural tube was assayed from specimens prepared in open-book format labeled with anti-Arl13b for cilia and Alexa-488 phalloidin (Molecular Probes) to identify the apical contours of individual cells.
Electron microscopy
To image neuroepithelial cilia by scanning electron microscopy, E10.5 embryos were dissected in PBS to expose the neural tube apical surface, fixed overnight with 2.5% glutaraldehyde in 200 mM sodium cacodylate buffer at 4°C. Fixed embryos were postfixed in 1% osmium tetroxide for 30 minutes, washed with sodium cacodylate buffer, and dehydrated in ethanol. Embryos were air dried after serial tetramethylsilane washes and sputter-coating with gold-palladium. Cilia from 3 mutants and 3 controls were visualized using a Phillips XLS30 scanning electron microscope.
Transmission electron microscopy was performed on bromi-AUG morpholino-injected (n=2) and control (n=2) 48 hours post fertilization (hpf) zebrafish embryos. Embryos were dissected and fixed overnight (1.5% glutaraldehyde, 1% paraformaldehyde, 3% sucrose, 0.1% tannic acid, 70mM NaPO4, pH7.2). Subsequently, embryos were washed three times in 0.1M phosphate buffer (pH7.4), postfixed in 1% osmium tetroxide with potassium ferrocyanide, and washed further in phosphate buffer. The fixed samples were then dehydrated and embedded in epon resin (EM Sciences). The posterior regions of kidney tubules adjacent to the cloaca were cut using a Leica UC-6 ultramicrotome and examined by using a Leo 912AB transmission electron microscope.
Analysis of bromi and ccrk in zebrafish
Morpholinos (MO) were designed against zebrafish bromi (si:dkey-233p4.1) or ccrk (zgc:101530) translation start site and splice donor sites. The following antisense morpholino sequences were synthesized 5’-CATCCTCGGTGGAGAACTGAGACAT-3’ for bromi translational start site (bromi-AUG MO), 5’-AAATGCCACATACTTGTTTGAGGAG-3’ for bromi exon 8/intron 8 splice site (bromi-e8i8 MO), 5’-GGCCAAGAATACTATACTG GTCCAT-3’ for ccrk translational start site (ccrk-AUG MO) and 5’-ACACGGGAACACTAGC TCACCCTAT-3’ for ccrk exon 3/intron 3 splice site (ccrk-e3i3 MO) (Gene Tools). Morpholinos were injected as described (Schottenfeld et al., 2007). In pilot experiments, varying doses of morpholinos were injected to determine the lowest dose resulting in <50% phenotypic animals (2ng for bromi-AUG MO, 12ng for bromi-e8i8 MO, and 4ng for ccrk-AUG MO and ccrk-e3i3 MO). RT-PCR using RNA from each splice donor site MO-injected embryos was used to assay the efficacy of bromi and ccrk knockdown with primers flanking the exon 8/intron 8 junction for bromi-e8i8 MO and exon 3/intron 3 junction for ccrk-e3i3 MO. Gadph RT-PCR was used as an internal control. MO-injected 27hpf embryos were fixed in Dent's fixative at 4°C overnight. Fixed embryos were gradually rehydrated with PBDT (1X PBS, 0.1% Tween, 1% DMSO) and blocked in 10% normal goat serum/PBDT. Embryos were stained with anti-acetylated α-tubulin antibodies and Cy2-conjugated secondary antibodies. Cilia in the proximal and distal regions of the pronephric tubules were visualized by confocal microscopy.
Generation of polyclonal Bromi antibody
D630037F22Rik/C6orf170 mouse cDNA sequence, encoding the N-terminal 227 amino acids, was PCR-amplified and cloned in the expression vector pET28A (Novagen). Recombinant N- and C-terminally His-tagged Bromi fragments were produced in BL21(DE3)pLysS competent cells as recommended by the manufacturer (Stratagene) and purified under denaturing conditions (8 M urea) using TALON metal affinity resin (Clontech). Purified fusion proteins were used to produce antisera in New Zealand White rabbits at Princeton University. Antisera from independent rabbits gave similar results.
Tandem affinity purification and mass spectrometry
To identify the Bromi interacting proteins, the Rheoswitch inducible expression system was used following manufacturer's instruction (New England Biolabs). A construct containing HA and FLAG tandem epitope tags at the C-terminus of mouse Bromi in pNEBRx1 plasmid (New England Biolabs) was generated and used to produce stable NIH3T3 cell lines expressing HA/FLAG tandem tagged Bromi upon induction by Rheoswitch ligand (RSL1). To isolate HA/FLAG-Bromi complexes, the cells were induced for 24 hrs with 500 nM RSL1, and lysed using NP40 lysis buffer (50 mM Tris-HCl, pH7.4, 150 mM NaCl, 10% glycerol, 1 mM EDTA, 0.5% NP40) supplemented with 1 mM PMSF, and Complete™ Protease inhibitor cocktail (Roche). Cell extracts were incubated with EZview™ Red anti-FLAG® M2 beads (Sigma-Aldrich) overnight at 4 °C, washed with NP40 lysis buffer, and complexes were eluted with 3X FLAG peptides (Sigma-Aldrich). Eluates were subjected to a second round of affinity purification using anti-HA agarose beads (Sigma-Aldrich). Complexes were eluted a second time with 2X HA peptides (Genscript). Final eluates were run on silver-stained gels, together with a negative control for nonspecific binding, and were separately analyzed in-house by LC/MS/MS using a hybrid LTQ-ORBITRAP mass spectrometer (Thermo Fisher) interfaced with a Eksigent NanoLC-2D system (Eksigent Technologies, Inc.). The resulting MS/MS data were used to search a mouse database using Bioworks Browser software (Thermo Fisher).
Immunoblotting
Embryos (E10.5), primary embryonic fibroblasts, and NIH3T3 cells expressing HA-tagged Bromi using the Rheoswitch induction system (New England Biolabs) were homogenized/lysed in RIPA buffer supplemented with 0.5 mM PMSF and Complete™ Protease inhibitor cocktail (Roche). Extracts were quantified with Coomassie Plus Assay Reagent (Pierce). Equal quantities of protein extracts were analyzed by western blotting using rabbit anti-Bromi, chicken anti-mouse GLI3 (Bulgakov et al., 2004), guinea pig anti-mouse Gli2 (Cho et al., 2008), rabbit anti-CCRK (Wohlbold et al., 2006), mouse β-tubulin (Sigma) and rabbit anti-HSP60 (abcam) antibodies. Quantitation of band intensity was performed using NIH ImageJ.
Immunoprecipitation
HA-Bromi or HA-CCRK NIH3T3 Rheoswitch cells were treated with 500 nM RSL1 or vehicle (DMSO) and lysed with a modified RIPA buffer (50 mM Tris pH 7.4, 150 mM NaCl, 1% NP-40, 0.25% deoxycholic acid, 1 mM EDTA, 1 mM NaF) supplemented with 0.5 mM PMSF, and Complete™ Protease inhibitor cocktail (Roche). Anti-HA Ab-agarose beads were added to the extracts, and incubated with gentle rotation for 3-5 hrs at 4°C followed by washing with modified RIPA buffer. Beads were mixed with SDS-sample buffer, boiled, and spun. Western blotting was performed on supernatants and total extracts using anti-Bromi and anti-CCRK antibodies. Immunoprecipitation assays were similarly performed on NIH3T3 cells cotransfected with HA-Bromi plus FLAG-tagged mouse Rab8a, Rab17, and Rab23 (Itoh et al., 2006) or V5 epitope-tagged CCRK constructs using anti-HA Ab-agarose beads and blotted with anti-FLAG (M2, Sigma) or anti-V5 antibodies (abcam) antibodies.
Statistical analysis
Statistical significance was investigated with nonparametric Mann-Whitney U tests using Statplus software (AnalystSoft). Significance was separately confirmed using Student's t-tests (two-tailed, equal variance).
Supplementary Material
ACKNOWLEDGEMENTS
We thank members of K. Anderson and L. Niswander's laboratories for help with the mutagenesis screen. We thank K. Anderson, M. Scott, A. Joyner, T. Caspary, G. Pazour, R. Fisher, X. Sun and BayGenomics, for contributing mouse lines, cell lines, and reagents. We thank J. Goodhouse, M. Bisher, J. Levorse, S. Kyin, Y. Lin, and J. Schottenfeld for technical assistance and R. Burdine and T. Caspary for comments on the manuscript. Monoclonal antibodies against Shh, FoxA2/Hnf3β, Nkx2.2, Nkx6.1, HB9/Mnr2, Pax6, and Pax7 were obtained from the Developmental Studies Hybridoma Bank developed under the auspices of the NICHD and maintained by The University of Iowa, Department of Biological Sciences, Iowa City, IA 52242. This work was supported by the New Jersey Commission on Spinal Cord Research (NJCSCR) grant 07-3069-SCR-E-0 and a US National Institutes of Health grant, R01 HD050761. H.W.K. was supported by a postdoctoral fellowship from the NJCSCR.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
SUPPLEMENTAL MATERIALS
Six supplemental figures and figure legends, and Supplemental Experimental Procedures.
REFERENCES
- Asleson CM, Lefebvre PA. Genetic analysis of flagellar length control in Chlamydomonas reinhardtii: a new long-flagella locus and extragenic suppressor mutations. Genetics. 1998;148:693–702. doi: 10.1093/genetics/148.2.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai CB, Stephen D, Joyner AL. All mouse ventral spinal cord patterning by hedgehog is Gli dependent and involves an activator function of Gli3. Dev. Cell. 2004;6:103–115. doi: 10.1016/s1534-5807(03)00394-0. [DOI] [PubMed] [Google Scholar]
- Barsel SE, Wexler DE, Lefebvre PA. Genetic analysis of long-flagella mutants of Chlamydomonas reinhardtii. Genetics. 1988;118:637–648. doi: 10.1093/genetics/118.4.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bengs F, Scholz A, Kuhn D, Wiese M. LmxMPK9, a mitogen-activated protein kinase homologue affects flagellar length in Leishmania mexicana. Mol Microbiol. 2005;55:1606–15. doi: 10.1111/j.1365-2958.2005.04498.x. [DOI] [PubMed] [Google Scholar]
- Berman SA, Wilson NF, Haas NA, Lefebvre PA. A novel MAP kinase regulates flagellar length in Chlamydomonas. Curr Biol. 2003;13:1145–9. doi: 10.1016/s0960-9822(03)00415-9. [DOI] [PubMed] [Google Scholar]
- Bulgakov OV, Eggenschwiler JT, Hong DH, Anderson KV, Li T. FKBP8 is a negative regulator of mouse sonic hedgehog signaling in neural tissues. Development. 2004;131:2149–59. doi: 10.1242/dev.01122. [DOI] [PubMed] [Google Scholar]
- Burghoorn J, Dekkers MP, Rademakers S, de Jong T, Willemsen R, Jansen G. Mutation of the MAP kinase DYF-5 affects docking and undocking of kinesin-2 motors and reduces their speed in the cilia of Caenorhabditis elegans. Proc Natl Acad Sci U S A. 2007;104:7157–62. doi: 10.1073/pnas.0606974104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caspary T, Larkins CE, Anderson KV. The graded response to Sonic Hedgehog depends on cilia architecture. Dev. Cell. 2007;12:767–778. doi: 10.1016/j.devcel.2007.03.004. [DOI] [PubMed] [Google Scholar]
- Chailley B, Boisvieux-Ulrich E. Detection of plasma membrane cholesterol by filipin during microvillogenesis and ciliogenesis in quail oviduct. J. Histochem. Cytochem. 1985;33:1–10. doi: 10.1177/33.1.3965567. [DOI] [PubMed] [Google Scholar]
- Chen MH, Wilson CW, Li YJ, Law KK, Lu CS, Gacayan R, Zhang X, Hui CC, Chuang PT. Cilium-independent regulation of Gli protein function by Sufu in Hedgehog signaling is evolutionarily conserved. Genes Dev. 2009;23:1910–1928. doi: 10.1101/gad.1794109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho A, Ko HW, Eggenschwiler JT. FKBP8 cell-autonomously controls neural tube patterning through a Gli2- and Kif3a-dependent mechanism. Dev. Biol. 2008;321:27–39. doi: 10.1016/j.ydbio.2008.05.558. [DOI] [PubMed] [Google Scholar]
- Corbit KC, Aanstad P, Singla V, Norman AR, Stainier DY, Reiter JF. Vertebrate Smoothened functions at the primary cilium. Nature. 2005;437:1018–1021. doi: 10.1038/nature04117. [DOI] [PubMed] [Google Scholar]
- Davidge JA, Chambers E, Dickinson HA, Towers K, Ginger ML, McKean PG, Gull K. Trypanosome IFT mutants provide insight into the motor location for mobility of the flagella connector and flagellar membrane formation. J. Cell Sci. 2006;119:3935–3943. doi: 10.1242/jcs.03203. [DOI] [PubMed] [Google Scholar]
- Dessaud E, McMahon AP, Briscoe J. Pattern formation in the vertebrate neural tube: a sonic hedgehog morphogen-regulated transcriptional network. Development. 2008;135:2489–2503. doi: 10.1242/dev.009324. [DOI] [PubMed] [Google Scholar]
- Dolle P, Izpisua-Belmonte JC, Falkenstein H, Renucci A, Duboule D. Coordinate expression of the murine Hox-5 complex homoeobox-containing genes during limb pattern formation. Nature. 1989;342:767–72. doi: 10.1038/342767a0. [DOI] [PubMed] [Google Scholar]
- Eggenschwiler JT, Anderson KV. Dorsal and lateral fates in the mouse neural tube require the cell-autonomous activity of the open brain gene. Dev. Biol. 2000;227:648–660. doi: 10.1006/dbio.2000.9918. [DOI] [PubMed] [Google Scholar]
- Eggenschwiler JT, Espinoza E, Anderson KV. Rab23 is an essential negative regulator of the mouse Sonic hedgehog signalling pathway. Nature. 2001;412:194–8. doi: 10.1038/35084089. [DOI] [PubMed] [Google Scholar]
- Eggenschwiler JT, Anderson KV. Cilia and developmental signaling. Annu. Rev. Cell Dev. Biol. 2007;23:345–373. doi: 10.1146/annurev.cellbio.23.090506.123249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggenschwiler JT, Bulgakov OV, Qin J, Li T, Anderson KV. Mouse Rab23 regulates hedgehog signaling from smoothened to Gli proteins. Dev. Biol. 2006;290:1–12. doi: 10.1016/j.ydbio.2005.09.022. [DOI] [PubMed] [Google Scholar]
- Endoh-Yamagami S, Evangelista M, Wilson D, Wen X, Theunissen JW, Phamluong K, Davis M, Scales SJ, Solloway MJ, de Sauvage FJ, Peterson AS. The mammalian Cos2 homolog Kif7 plays an essential role in modulating Hh signal transduction during development. Curr. Biol. 2009;19:1320–1326. doi: 10.1016/j.cub.2009.06.046. [DOI] [PubMed] [Google Scholar]
- Ericson J, Rashbass P, Schedl A, Brenner-Morton S, Kawakami A, van Heyningen V, Jessell TM, Briscoe J. Pax6 controls progenitor cell identity and neuronal fate in response to graded Shh signaling. Cell. 1997;90:169–180. doi: 10.1016/s0092-8674(00)80323-2. [DOI] [PubMed] [Google Scholar]
- Follit JA, Tuft RA, Fogarty KE, Pazour GJ. The intraflagellar transport protein IFT20 is associated with the Golgi complex and is required for cilia assembly. Mol. Biol. Cell. 2006;17:3781–3792. doi: 10.1091/mbc.E06-02-0133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Z, Larson KA, Chitta RK, Parker SA, Turk BE, Lawrence MW, Kaldis P, Galaktionov K, Cohn SM, Shabanowitz J, Hunt DF, Sturgill TW. Identification of yin-yang regulators and a phosphorylation consensus for male germ cell-associated kinase (MAK)-related kinase. Mol Cell Biol. 2006;26:8639–54. doi: 10.1128/MCB.00816-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodrich LV, Milenkovic L, Higgins KM, Scott MP. Altered neural cell fates and medulloblastoma in mouse patched mutants. Science. 1997;277:1109–1113. doi: 10.1126/science.277.5329.1109. [DOI] [PubMed] [Google Scholar]
- Grosshans BL, Ortiz D, Novick P. Rabs and their effectors: achieving specificity in membrane traffic. Proc. Natl. Acad. Sci. U S A. 2006;103:11821–11827. doi: 10.1073/pnas.0601617103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haycraft CJ, Banizs B, Aydin-Son Y, Zhang Q, Michaud EJ, Yoder BK. Gli2 and Gli3 localize to cilia and require the intraflagellar transport protein polaris for processing and function. PLoS Genet. 2005;1:e53. doi: 10.1371/journal.pgen.0010053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoover AN, Wynkoop A, Zeng H, Jia J, Niswander LA, Liu A. C2cd3 is required for cilia formation and Hedgehog signaling in mouse. Development. 2008;135:4049–4058. doi: 10.1242/dev.029835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Y, Pazour GJ, Witman GB. A dynein light intermediate chain, D1bLIC, is required for retrograde intraflagellar transport. Mol. Biol. Cell. 2004;15:4382–4394. doi: 10.1091/mbc.E04-05-0377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huangfu D, Anderson KV. Cilia and Hedgehog responsiveness in the mouse. Proc. Natl. Acad. Sci. U S A. 2005;102:11325–11330. doi: 10.1073/pnas.0505328102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huangfu D, Liu A, Rakeman AS, Murcia NS, Niswander L, Anderson KV. Hedgehog signalling in the mouse requires intraflagellar transport proteins. Nature. 2003;426:83–87. doi: 10.1038/nature02061. [DOI] [PubMed] [Google Scholar]
- Hui CC, Joyner AL. A mouse model of greig cephalopolysyndactyly syndrome: the extra-toesJ mutation contains an intragenic deletion of the Gli3 gene. Nat. Genet. 1993;3:241–6. doi: 10.1038/ng0393-241. [DOI] [PubMed] [Google Scholar]
- Hui CC, Slusarski D, Platt KA, Holmgren R, Joyner AL. Expression of three mouse homologs of the Drosophila segment polarity gene cubitus interruptus, Gli, Gli-2, and Gli-3, in ectoderm- and mesoderm-derived tissues suggests multiple roles during postimplantation development. Dev. Biol. 1994;162:402–13. doi: 10.1006/dbio.1994.1097. [DOI] [PubMed] [Google Scholar]
- Ingham PW. Hedgehog signalling. Curr. Biol. 2008;18:R238–241. doi: 10.1016/j.cub.2008.01.050. [DOI] [PubMed] [Google Scholar]
- Itoh T, Satoh M, Kanno E, Fukuda M. Screening for target Rabs of TBC (Tre-2/Bub2/Cdc16) domain-containing proteins based on their Rab-binding activity. Genes Cells. 2006;11:1023–1037. doi: 10.1111/j.1365-2443.2006.00997.x. [DOI] [PubMed] [Google Scholar]
- Jia J, Jiang J. Decoding the Hedgehog signal in animal development. Cell. Mol. Life Sci. 2006;63:1249–1265. doi: 10.1007/s00018-005-5519-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Kato M, Beachy PA. Gli2 trafficking links Hedgehog-dependent activation of Smoothened in the primary cilium to transcriptional activation in the nucleus. Proc Natl Acad Sci U S A. 2009 doi: 10.1073/pnas.0912180106. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liem KF, Jr., He M, Ocbina PJ, Anderson KV. Mouse Kif7/Costal2 is a cilia-associated protein that regulates Sonic hedgehog signaling. Proc. Natl. Acad. Sci. U S A. 2009;106:13377–13382. doi: 10.1073/pnas.0906944106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Wu C, Galaktionov K. p42, a novel cyclin-dependent kinase-activating kinase in mammalian cells. J. Biol. Chem. 2004;279:4507–4514. doi: 10.1074/jbc.M309995200. [DOI] [PubMed] [Google Scholar]
- Lum L, Zhang C, Oh S, Mann RK, von Kessler DP, Taipale J, Weis-Garcia F, Gong R, Wang B, Beachy PA. Hedgehog signal transduction via Smoothened association with a cytoplasmic complex scaffolded by the atypical kinesin, Costal-2. Mol. Cell. 2003;12:1261–1274. doi: 10.1016/s1097-2765(03)00426-x. [DOI] [PubMed] [Google Scholar]
- Matise MP, Epstein DJ, Park HL, Platt KA, Joyner AL. Gli2 is required for induction of floor plate and adjacent cells, but not most ventral neurons in the mouse central nervous system. Development. 1998;125:2759–2770. doi: 10.1242/dev.125.15.2759. [DOI] [PubMed] [Google Scholar]
- Methot N, Basler K. Hedgehog controls limb development by regulating the activities of distinct transcriptional activator and repressor forms of Cubitus interruptus. Cell. 1999;96:819–831. doi: 10.1016/s0092-8674(00)80592-9. [DOI] [PubMed] [Google Scholar]
- Methot N, Basler K. Suppressor of fused opposes hedgehog signal transduction by impeding nuclear accumulation of the activator form of Cubitus interruptus. Development. 2000;127:4001–4010. doi: 10.1242/dev.127.18.4001. [DOI] [PubMed] [Google Scholar]
- Morgan GW, Denny PW, Vaughan S, Goulding D, Jeffries TR, Smith DF, Gull K, Field MC. An evolutionarily conserved coiled-coil protein implicated in polycystic kidney disease is involved in basal body duplication and flagellar biogenesis in Trypanosoma brucei. Mol. Cell Biol. 2005;25:3774–3783. doi: 10.1128/MCB.25.9.3774-3783.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nachury MV, Loktev AV, Zhang Q, Westlake CJ, Peranen J, Merdes A, Slusarski DC, Scheller RH, Bazan JF, Sheffield VC, Jackson PK. A core complex of BBS proteins cooperates with the GTPase Rab8 to promote ciliary membrane biogenesis. Cell. 2007;129:1201–1213. doi: 10.1016/j.cell.2007.03.053. [DOI] [PubMed] [Google Scholar]
- Nagy A, Gertenstein M, Vintersten K, Behringer R. Manipulating the Mouse Embryo, A Laboratory Manual. Cold Spring Harbor Press; Cold Spring Harbor, New York: 2003. [Google Scholar]
- Pan X, Eathiraj S, Munson M, Lambright DG. TBC-domain GAPs for Rab GTPases accelerate GTP hydrolysis by a dual-finger mechanism. Nature. 2006;442:303–306. doi: 10.1038/nature04847. [DOI] [PubMed] [Google Scholar]
- Pazour GJ, Bloodgood RA. Targeting proteins to the ciliary membrane. Curr. Top. Dev. Biol. 2008;85:115–149. doi: 10.1016/S0070-2153(08)00805-3. [DOI] [PubMed] [Google Scholar]
- Pedersen LB, Rosenbaum JL. Intraflagellar transport (IFT) role in ciliary assembly, resorption and signalling. Curr .Top. Dev. Biol. 2008;85:23–61. doi: 10.1016/S0070-2153(08)00802-8. [DOI] [PubMed] [Google Scholar]
- Perkins LA, Hedgecock EM, Thomson JN, Culotti JG. Mutant sensory cilia in the nematode Caenorhabditis elegans. Dev. Biol. 1986;117:456–487. doi: 10.1016/0012-1606(86)90314-3. [DOI] [PubMed] [Google Scholar]
- Persson M, Stamataki D, te Welscher P, Andersson E, Bose J, Ruther U, Ericson J, Briscoe J. Dorsal-ventral patterning of the spinal cord requires Gli3 transcriptional repressor activity. Genes Dev. 2002;16:2865–2878. doi: 10.1101/gad.243402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin H, Rosenbaum JL, Barr MM. An autosomal recessive polycystic kidney disease gene homolog is involved in intraflagellar transport in C. elegans ciliated sensory neurons. Curr. Biol. 2001;11:457–461. doi: 10.1016/s0960-9822(01)00122-1. [DOI] [PubMed] [Google Scholar]
- Quinlan RJ, Tobin JL, Beales PL. Modeling ciliopathies: Primary cilia in development and disease. Curr. Top. Dev. Biol. 2008;84:249–310. doi: 10.1016/S0070-2153(08)00605-4. [DOI] [PubMed] [Google Scholar]
- Rohatgi R, Milenkovic L, Scott MP. Patched1 regulates hedgehog signaling at the primary cilium. Science. 2007;317:372–376. doi: 10.1126/science.1139740. [DOI] [PubMed] [Google Scholar]
- Rosenbaum JL, Moulder JE, Ringo DL. Flagellar elongation and shortening in Chlamydomonas. The use of cycloheximide and colchicine to study the synthesis and assembly of flagellar proteins. J. Cell Biol. 1969;41:600–619. doi: 10.1083/jcb.41.2.600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruel L, Rodriguez R, Gallet A, Lavenant-Staccini L, Therond PP. Stability and association of Smoothened, Costal2 and Fused with Cubitus interruptus are regulated by Hedgehog. Nat. Cell Biol. 2003;5:907–913. doi: 10.1038/ncb1052. [DOI] [PubMed] [Google Scholar]
- Schottenfeld J, Sullivan-Brown J, Burdine RD. Zebrafish curly up encodes a Pkd2 ortholog that restricts left-side-specific expression of southpaw. Development. 2007;134:1605–1615. doi: 10.1242/dev.02827. [DOI] [PubMed] [Google Scholar]
- Svard J, Heby-Henricson K, Persson-Lek M, Rozell B, Lauth M, Bergstrom A, Ericson J, Toftgard R, Teglund S. Genetic elimination of Suppressor of fused reveals an essential repressor function in the mammalian Hedgehog signaling pathway. Dev. Cell. 2006;10:187–197. doi: 10.1016/j.devcel.2005.12.013. [DOI] [PubMed] [Google Scholar]
- Tam LW, Dentler WL, Lefebvre PA. Defective flagellar assembly and length regulation in LF3 null mutants in Chlamydomonas. J. Cell Biol. 2003;163:597–607. doi: 10.1083/jcb.200307143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam LW, Wilson NF, Lefebvre PA. A CDK-related kinase regulates the length and assembly of flagella in Chlamydomonas. J. Cell Biol. 2007;176:819–829. doi: 10.1083/jcb.200610022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran PV, Haycraft CJ, Besschetnova TY, Turbe-Doan A, Stottmann RW, Herron BJ, Chesebro AL, Qiu H, Scherz PJ, Shah JV, et al. THM1 negatively modulates mouse sonic hedgehog signal transduction and affects retrograde intraflagellar transport in cilia. Nat. Genet. 2008;40:403–410. doi: 10.1038/ng.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varjosalo M, Li SP, Taipale J. Divergence of hedgehog signal transduction mechanism between Drosophila and mammals. Dev. Cell. 2006;10:177–186. doi: 10.1016/j.devcel.2005.12.014. [DOI] [PubMed] [Google Scholar]
- Varjosalo M, Taipale J. Hedgehog: functions and mechanisms. Genes Dev. 2008;22:2454–2472. doi: 10.1101/gad.1693608. [DOI] [PubMed] [Google Scholar]
- Vierkotten J, Dildrop R, Peters T, Wang B, Ruther U. Ftm is a novel basal body protein of cilia involved in Shh signalling. Development. 2007;134:2569–2577. doi: 10.1242/dev.003715. [DOI] [PubMed] [Google Scholar]
- Wang B, Fallon JF, Beachy PA. Hedgehog-regulated processing of Gli3 produces an anterior/posterior repressor gradient in the developing vertebrate limb. Cell. 2000;100:423–434. doi: 10.1016/s0092-8674(00)80678-9. [DOI] [PubMed] [Google Scholar]
- Wilson CW, Nguyen CT, Chen MH, Yang JH, Gacayan R, Huang J, Chen JN, Chuang PT. Fused has evolved divergent roles in vertebrate Hedgehog signalling and motile ciliogenesis. Nature. 2009;459:98–102. doi: 10.1038/nature07883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson NF, Iyer JK, Buchheim JA, Meek W. Regulation of flagellar length in Chlamydomonas. Semin. Cell Dev. Biol. 2008;19:494–501. doi: 10.1016/j.semcdb.2008.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wohlbold L, Larochelle S, Liao JC, Livshits G, Singer J, Shokat KM, Fisher RP. The cyclin-dependent kinase (CDK) family member PNQALRE/CCRK supports cell proliferation but has no intrinsic CDK-activating kinase (CAK) activity. Cell Cycle. 2006;5:546–554. doi: 10.4161/cc.5.5.2541. [DOI] [PubMed] [Google Scholar]
- Yoshimura S, Egerer J, Fuchs E, Haas AK, Barr FA. Functional dissection of Rab GTPases involved in primary cilium formation. J. Cell Biol. 2007;178:363–369. doi: 10.1083/jcb.200703047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zohn IE, Anderson KV, Niswander L. Using genomewide mutagenesis screens to identify the genes required for neural tube closure in the mouse. Birth Defects Res. A. Clin. Mol. Teratol. 2005;73:583–590. doi: 10.1002/bdra.20164. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.