Abstract
Thymic nurse cells (TNCs) are epithelial cells in the thymic cortex that contain as many as fifty thymocytes within specialized cytoplasmic vacuoles. The function of this cell-in-cell interaction has created controversy since their discovery in 1980. Further, some skepticism exists about the idea that apoptotic thymocytes within the TNC complex result from negative selection, a process believed to occur exclusively within the medulla. In this report, we have microscopic evidence that defines a unique membranous environment wherein lipid raft aggregates around the αβTCR expressed on captured thymocytes and class II MHC molecules expressed on TNCs. Further, immunohistological examination of thymic sections show TNCs located within the cortico-medullary junction to express cytokeratins five and eight (K5 and K8), and the transcription factor Trp-63, the phenotype defined elsewhere as the thymic epithelial progenitor subset. Our results suggest that the microenvironment provided by TNCs plays an important role in thymocyte selection as well as the potential for TNCs to be involved in the maintenance of thymic epithelia.
Keywords: Thymic nurse cell, epithelial progenitor, membrane extensions, cytokeratins, heterotypic internalization, MHC restriction, Trp 63
INTRODUCTION
Although the internalization of a viable cell by another cell has been described for over a century, not until very recently has this phenomenon been accepted as scientifically plausible [1]. Although phagocytes, cells with the ability to take up dead or dying cells, have for a long time been a part of scientific dogma, it has been difficult to advance the idea that a viable cell can internalize another viable cell, and in some cases release the trapped cell from its intra-cytoplasmic space [1]. Thymic nurse cells (TNCs) were discovered in mice by Wekerle and Ketelson in 1980 [2; 3]. Their initial report described TNCs as keratin expressing cells containing several thymocytes completely enclosed within specialized cytoplasmic vacuoles. The number of thymocytes enclosed was reported to vary from about 7 to 50. TNCs were also shown to express both class I and class II MHC antigens on their cell surfaces as well as on the surfaces of the vacuoles surrounding internalized thymocytes. The expression of membrane class II MHC antigens is atypical for epithelial cells. The expression of class II MHC antigens is generally thought to be restricted to cells of the immune system. Typically, epithelial cells do not function within the immune system. Following their initial discovery in mice, TNCs were isolated from the thymus of fish, frogs, chicken, sheep, pigs, rats and humans [2; 3; 4; 5; 6; 7; 8]. Since then, the major focus of their study has been to determine the immunological function of the TNC/thymocyte interaction within the thymic cortex. Initial studies of TNCs were performed using freshly isolated cells [2; 3; 4; 5]. However, more than twenty years passed before new information was obtained about TNC function because, upon isolation, cytoplasmic thymocytes are released, which does not allow for the identification of the internalized subset. Further, once freshly isolated TNCs release their internalized thymocytes they do not retain the capacity to further internalize thymocytes, making functional studies of the interaction impossible.
Much information has been reported recently in support of TNCs ability to engulf another cell, as well as to define a role for this interaction in shaping the T cell repertoire [4; 9; 10; 11]. The most convincing evidence has been obtained from the generation of TNC lines that produce cells with the ability to internalize thymocytes in vitro [11; 12]. Upon addition of freshly isolated thymocytes to cells of the TNC lines, only αβTCRlowCD4+CD8+ cells were found to be bound and internalized [12]. TNCs were shown to selectively rescue a subset of triple positive thymocytes from apoptosis, and antibodies against MHC I and MHC II molecules prevented this rescue activity, suggesting that the rescue was a function of MHC driven selection [11]. The rescued population matured to the αβTCRhiCD69+ stage of development before being released from the TNC complex. In studies using the TNC-specific monoclonal antibody (mAb) pH91, which blocks the TNC/thymocyte interaction, it was demonstrated that this interaction is required for thymocyte viability during the triple positive stage of development [13]. In fetal thymic organ culture, the presence of pH91 reduced the viability of developing thymocytes by 80%, with the largest reduction found in the αβTCRhiCD69+ thymocyte subset [14].
Collectively, these data have been difficult to accept because they define a subset of epithelial cells in the thymic cortex that facilitate the MHC restriction process using a cell-in-cell activity, a not well-accepted biological phenomenon [1]. Further, these data suggest that both positive and negative selection can occur within TNC complexes, which reside in the thymic cortex. Current dogma insists that positive selection but not negative selection occurs in the cortex of the mouse thymus [15]. MHC restriction is defined as the positive selection of triple positive thymocytes (rescue from apoptosis), or negative selection (induction of apoptosis) that results from an interaction between the αβTCR on developing thymocytes and MHC molecules on antigen presenting cells (APC) [16; 17]. While it has been accepted that TNC cytoplasmic thymocytes undergo apoptosis, most reports suggest that cells of the thymic cortex are not functionally equipped to perform negative selection [15]. More specifically, it is believed that negative selection requires the expression of the AIRE protein, which has been reported to control the expression of tissue-restricted antigens (TRA) [18; 19; 20]. Both of these functions have been reported to be restricted to cells located within the medulla. However, very recently both AIRE and TRA expression was detected within the TNC complex [21]. These current findings along with the data presented here showing an αβTCR/MHC interaction within the TNC complex adds support to data suggesting that TNCs have the capacity to facilitate MHC restriction, both positive and negative selection. Further, we define the structures involved in thymocyte uptake and show that the initial internalization event results in the delivery of trapped thymocytes into specialized membrane spaces created as a result of extensive cytoplasmic membrane folding. These membrane spaces are external to the TNC cytoplasm but create the two dimensional illusion that trapped thymocytes are cytoplasmic. We propose that these unique membrane structures provide a microenvironment for the αβTCR/MHC interaction and easy release of positively selected thymocytes, while allowing the cytoplasmic uptake of negatively selected thymocytes. If this is correct, the only thymocyte subset that truly becomes cytoplasmic is apoptotic and destined for destruction through lysosomal fusion [22]. Finally, and most unexpectedly, in vitro and in vivo staining results show a subset of TNCs to express the thymic epithelial cell progenitor phenotype which has been shown to have the capacity to generate an entire functional thymus when transplanted under the kidney capsule [23].
MATERIALS AND METHODS
Isolation of TNCs and Thymocytes
C57BL/6 mice (Jackson Laboratory, Bar Harbor, ME) were dissected aseptically and the thymi were removed. Thymi were slightly disrupted with fine needles and subjected to enzymatic digestion in a solution of 0.015% collagenase D (Sigma Aldrich, St Louis, MO), 0.01% DNAse I (Sigma Aldrich), and 25 ml of trypsin (GIBCO, Carlsbad, CA) along with gentle agitation. The solution was changed every 10 minutes until the thymi were completely digested. The resulting cells were subjected to 1×g gradient separation in fetal bovine serum (Atlas Biological, Fort Collins, CO) at 4°C to enrich TNC numbers. Thymocytes were obtained by the mechanical disruption of thymi obtained from 4 to 6 week old C57BL/6 mice. Macrophage depletion was accomplished by negative sorting using CD11b Microbeads (Miltenyi Biotech, Auburn, CA)
Scanning Electron Microscopy
One million thymocytes were allowed to incubate with 1×105 TNCs from our temperature sensitive cell line, tsTNC-1 [24], at 37°C in Terisaki culture plates for 0 - 12 hours and transferred to microscope slides. Freshly isolated TNCs were allowed to incubate on microscope slides for 2 hours at 37°C. The samples were fixed with 3.2% gluteraldehyde in 0.1 M sodium cacodylate buffer pH 7.2 (Electron Microscopy Sciences, Hatsfield, PA), and stored at 4°C for 12 - 24 hours. The cells were then rinsed in distilled water, dehydrated in a graded series of ethanol (Electron Microscopy Sciences), rinsed twice in amyl acetate (Electron Microscopy Sciences) and critical point dried. The cells were then sputter coated with 6-10 nm of gold and observed in a Zeiss DSM 940 Scanning Electron Microscope. Secondary electron images were captured using the Spirit image acquisition system (version 1.07) at a 1024 × 1024 pixel resolution.
Transmission Electron Microscopy
For studies requiring co-incubation, 5×106 TNCs were incubated with 5×107 thymocytes for 0 - 20 hours in glass petri dishes at 37°C. Co-incubated cells and isolated TNCs were fixed in 0.1 M cacodylate, 2% gluteraldehyde, and 1% osmium tetroxide (Electron Microscopy Sciences), pH 7.4 at 4°C for 30 minutes. Cells were then dehydrated in ascending concentrations of acetone (Electron Microscopy Sciences). After dehydration, cells were embedded in Embed 812 (Electron Microscopy Sciences). Ultra thin sections were made on a LKB Ultrotome III and stained with uranyl acetate followed by lead citrate (Electron Microscopy Sciences). Cells were viewed on a Zeiss EM 902 Electron Microscope using a SIS MegaView III digital camera at a resolution of 1376 × 1032 pixels.
Video Microscopy
Phase contrast videography of 1×104 TNCs co-incubated with 2×106 thymocytes was viewed using a Nikon Diaphat Microscope with a Hoffman Modulation Contrast System. The microscope was attached to a Nikon CCD-72 camera. The samples were visualized on a Sony 19 inch color monitor coupled to a JVC VCR. Videography using a light microscope was observed using an I×70 Olympus microscope attached to a DP11 Olympus camera. Video images were captured in real time and immediately digitized. All video microscopy was performed at 37°C.
Thymic Sections
Thymi were dissected aseptically from C57BL/6 mice. Individual lobes were embedded in OTC medium (Richard Allan Scientific, Kalamazoo, MI). Thymic sections 7μm in thickness were made using a Leica CM1950 Cryostat. Sections were mounted on Bond-Rite microscope slides (Richard Allan Scientific) for immunostaining.
Immunostaining of TNCs and Thymic Sections
Isolated TNCs were deposited onto glass slides using a Thermo Scientific Shandon Cytospin 4. Thymic sections or isolated TNCs were fixed in 2% paraformaldehyde (Baker, Phillipsburg, PA) for 30 minutes followed by 3 washes with phosphate buffered saline (PBS) (GIBCO). Sections were blocked and permeabilized in 3% bovine serum albumin (BSA) (Fisher Scientific, Pittsburg, PA), 0.1% Triton-X (Fisher Scientific) in PBS. Samples were incubated with primary and secondary antibodies at 37°C for 1 hour each. Samples were mounted in ProLongGold antifade with DAPI (Molecular Probes, Carlsbad, CA). Images were acquired using the Zeiss LSM510 Confocal Microscope.
Primary antibodies used were as follows: rat anti-mouse pH91 monoclonal antibody (IgG2a) [13], cytokeratin 8 (K8) - TROMA-I (IgG2a) (Developmental Studies Hybridoma Bank, Iowa City, IA), chicken anti-mouse K8 polyclonal antibody (IgY) (Abcam, Cambridge, MA), goat anti-rabbit cytokeratin 5 (K5) polyclonal antibody PRB-160B (IgG) (Covance, Princeton, NJ), rabbit anti-goat ΔNp63 (N-16): sc-8609 (Santa Cruz Biotechnology, Santa Cruz, CA), FITC-conjugated anti-mouse MHC class II (Miltenyi Biotech), biotinylated anti-mouse αβTCR (BD Pharmingen, San Jose, CA), APC-conjugated CD4 (BD Pharmingen), PE-conjugated CD8 (BD Pharmingen), FITC-conjugated Thy 1.2 (BD Pharmingen), FITC-conjugated rat IgG2a isotype control (BD Pharmingen), and TRITC-conjugated rabbit IgG2a isotype control (BD Pharmingen). Lipid rafts were visualized using Alexa Fluor 647-conjugated cholera toxin subunit B (Invitrogen, Carlsbad, CA). Secondary antibodies used are as follows: FITC-conjugated mouse anti-rat IgG2a (BD Pharmingen), APC-conjugated donkey anti-chicken IgY (Jackson ImmunoResearch Laboratories, West Grove, PA), TRITC-conjugated goat anti-rabbit IgG (Jackson ImmunoResearch Laboratories), APC-conjugated donkey anti-rabbit IgG (Jackson ImmunoResearch Laboratories), TRITC-conjugated donkey anti-rabbit IgG (Jackson ImmunoResearch Laboratories), TRITC-conjugated donkey anti-goat IgG (Jackson ImmunoResearch Laboratories), TRITC-conjugated rabbit anti-goat IgG (Jackson ImmunoResearch Laboratories), and TRITC-conjugated streptavidin (BD Pharmingen).
RESULTS
Characterization of In Vitro and In Vivo TNCs
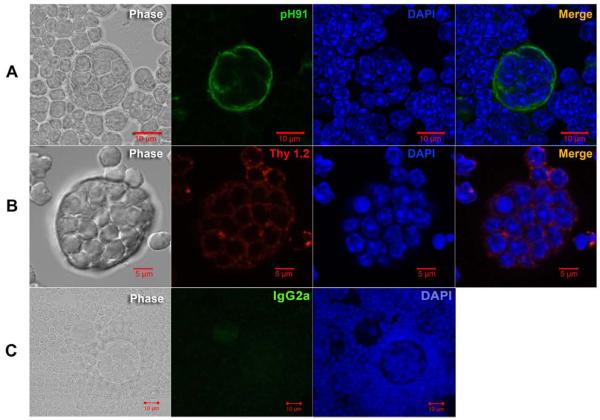
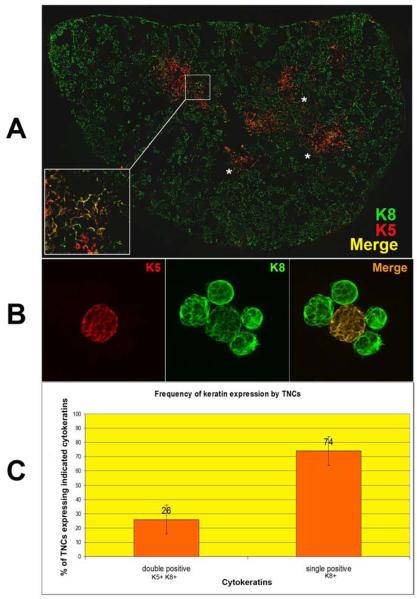
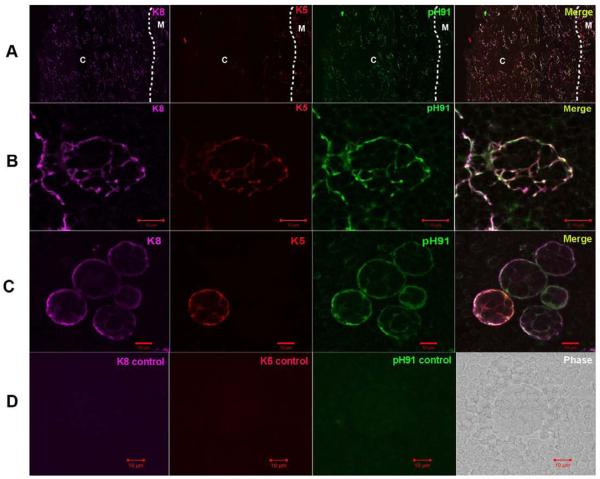
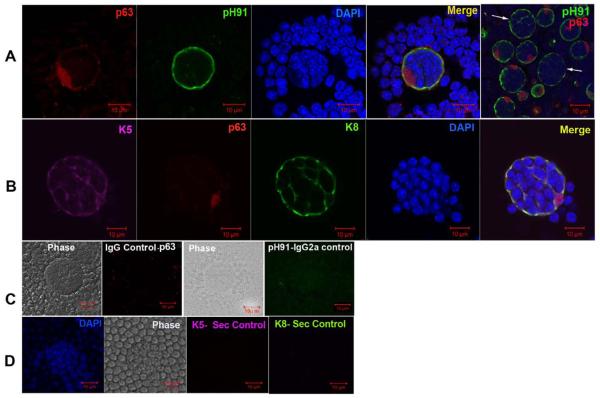
In Figure 1, the structure of freshly isolated thymic nurse cells is shown. The classical TNC structure is shown in the phase panels of Figures 1A, 1B and 1C. Other than their unique multi-cellular structure, TNCs can only be identified through staining with the TNC-specific mAb pH91. Figure 1A shows a freshly isolated TNC stained with pH91. DAPI and Thy 1.2 staining reveal the presence of internalized thymocytes (Figure 1B). When stained with antibodies (Abs) against cytokeratin 8 (K8) and cytokeratin 5 (K5), thymic sections show distinct cortical (K8) and medullary (K5) regions (Figure 2A). A small population of cells at the cortico-medullary junction is double positive for K5 and K8 (Figure 2A, inset and asterisks). Thymic epithelial cells within the cortico-medullary junction that express both K5 and K8 have been proposed to be thymic epithelial progenitor cells [25]. Enumeration of freshly isolated TNCs co-stained with Abs to K8 and K5 (Figure 2B) showed that 74% are K8+ single positive cells and 26% are K8+K5+ cells (Figure 2C). Cells with the multi-cellular structures of TNCs that expressed the K5 only phenotype were not detected. When freshly isolated TNCs were stained, a K8+K5+ subset was detected. The in vivo thymic location of TNCs was determined using pH91 staining of frozen thymic sections (Figure 3A). TNCs were found within the cortico-medullary junction and throughout the central cortex. All freshly isolated TNCs that exhibited the classical TNC morphology stained with pH91 whether they express K8+ only or K8+K5+ (Figure 3C) PH91+ cells also express the nuclear transcription factor Trp-63 (p63) (Figure 4A), which has also been shown to be associated with the epithelial stem cell phenotype [26]. The expression of p63, K8 and K5 in TNCs is consistent (Figure 4B) with the phenotype of thymic epithelial progenitors [25]. Not all TNCs express p63 (Figure 4A, arrows) suggesting that TNCs exist at different stages of development within the thymic cortex.
Figure 1.
PH91 identifies the TNC lymphostromal complex. (A) Phase image, pH91 staining (green), DAPI staining (blue), and overlay of a freshly isolated TNC. Scale bar represents 10 μm. (B) Phase image, Thy 1.2 staining (red), DAPI staining (blue), and overlay of a freshly isolated TNC. Scale bar represents 5 μm. (C) Panel shows phase image and isotype control for pH91 (IgG2a). Data is representative of three independent experiments.
Figure 2.
K8+K5+ cells exist at the cortico-medullary junction and are a subset of TNCs. Confocal analysis of keratin and pH91 staining of thymic sections and freshly isolated TNCs. (A) Thymic section stained with K5 (red) and K8 (green). Yellow regions (inset and asterisks) show double positive (K8+K5+) epithelial cells. Original magnification: 40X. (B) Freshly isolated TNCs stained with K5 (red) and K8 (green). The double positive TNC appears as yellow in the merge. Original magnification: 40X. (C) Frequency of K8+ and K8+K5+ populations in freshly isolated TNCs determined by manual counting of 1000 cytospun TNCs after immunostaining. No K5+ single positive cells with the TNC complex morphology were detected. Data for (A) and (B) is representative of three independent experiments.
Figure 3.
Expression of K8 and K5 by pH91-labeled TNCs. (A) Thymic sections stained with anti-K8 (magenta), anti-K5 (red), and pH91 (green) Abs. The cortex and medulla are indicated by “C” and “M”, respectively, and the cortico-medullary junction by a dotted line. The merge shows areas of co-localization. Original magnification: 40X. (B) Magnified area of section in panel A. The triple stained TNC exhibits a fenestrated structure. (C) Freshly isolated TNCs stained with anti-K8 (magenta), anti-K5 (red), and pH91 (green) Abs. Both K8+K5+ and K8+ only TNCs are visible. (D) Shows secondary Ab controls for K8, K5, and pH91 as well as a phage image. Data is representative of three independent experiments.
Figure 4.
Trp63 expression of freshly isolated TNCs. (A) Confocal micrograph of a TNC labeled with Ab against p63 (red) and pH91 (green). Merge shows cell surface staining with pH91, nuclear localization of p63, and DAPI stained nuclei. Last panel shows that not all TNCs (white arrows) express p63. (B) Confocal micrograph of a TNC labeled with Ab against p63 (red), K5 (magenta), and K8 (green) with DAPI stained nuclei (blue). Merge shows cell surface staining with pH91, nuclear localization of p63, and DAPI stained nuclei. (C) TNCs stained with secondary controls for p63 (IgG) and pH91 (IgG2a) with phase image. (D) TNCs stained with secondary controls for K5 and K8 showing phase image and DAPI stained nuclei. Data is representative of three independent experiments.
Thymocyte Internalization
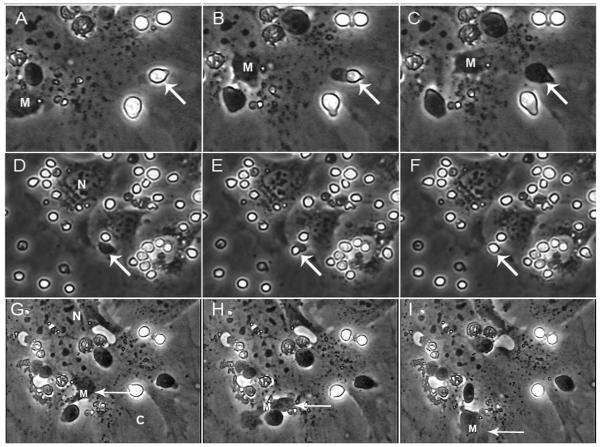
A controversy about the ability of TNCs to internalize thymocytes continues. In Figure 5, thymocytes were detected within a TNC complex after a 4 hours exposure of freshly isolated thymocytes to a monolayer of TNCs from the tsTNC-1 cell line. Using time-lapse video microscopy, we have captured the entire internalization process. Figure 5A-C shows a thymocyte (arrow), bound to the surface of a TNC (the thymocyte is phase bright outside and phase dark inside). The process of internalization begins with the development of a uropod on the internalizing thymocyte. As the process continues, the uropod remains visible and the internalizing thymocyte becomes phase dark as it enters the membrane of the targeted TNC. Also in Figure 5D-F (arrow), thymocyte release was detected. As the thymocyte is released, it becomes phase bright. Macrophages were also present and mobile within the TNC complex (Figure 5G-I).
Figure 5.
Thymic nurse cells facilitate the inward and outward migration of thymocytes. tsTNC-1 cells were co-incubated with freshly isolated thymocytes for 5 hours. Panels A-C show the stepwise internalization of a thymocyte (arrow). Panels D-F show the outward movement of a different thymocyte (arrow). The movement of a macrophage within a TNC complex is shown in panels G-I. The “N” indicates the location of the nucleus of the thymic nurse cell.
Thymocyte uptake into TNCs was also analyzed using scanning electron microscopy (SEM). Within one hour of adding tsTNC-1 cells and freshly isolated thymocytes to co-culture in Terisaki plates, TNCs display a highly ruffled membrane surface (Figure 6A and B), and a subset of thymocytes is found trapped within the ruffled extensions of membrane (Figure 6C and D, arrows). Thymic nurse cells trap thymocytes within specialized membrane folds which will eventually fuse leading to the capture of thymocytes (Figure 6D, E and F) with layers of membrane (Figure 6E and F). In Figure 6E, thymocytes were visible through fenestras created by overlapping membrane extensions (arrow). Figure 6F shows the lymphoepithelial complex after 10 hours in culture. Thymocytes are not visible after a 10 hours incubation period, but are visible within the complex using confocal microscopy (Figure 6G).
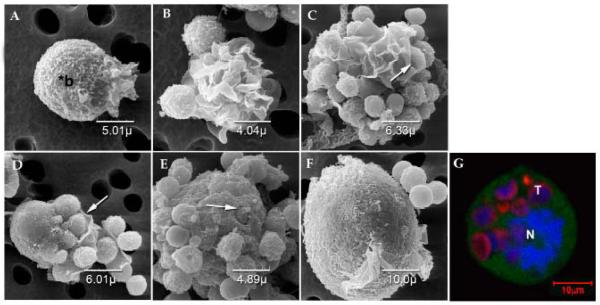
Figure 6.
SEM of tsTNC-1 cells co-incubated with thymocytes for up to 10 hours. (A) An individual TNC displaying membrane extensions polarized to one side of the cell. The body of the cell is indicated by “*b”. (Time of incubation = 0 hour) (B) TNCs co-incubated with thymocytes for 1 hours display larger areas of membrane extensions. (C) Thymocytes are trapped within the folds of the membrane extensions after 2 hours of incubation (arrow). (D) Membrane extensions of TNCs wrapped around the surface bound thymocytes (arrow). Cells were incubated for 2 hours. (E) After 4 hours of incubation, TNC no longer displays prolific membrane extensions but does contain internalized thymocytes. An arrow points to a partially internalized thymocyte. (F) Large TNC complex contains internalized thymocytes. (G) Confocal micrograph of tsTNC co-incubated with thymocytes for 10 hours is shown in inset. Prior to co-incubation the TNCs were labeled with CFDA (green) and the thymocytes were labeled with CFSE (red). The TNC nucleus (N) is visible as are internalized thymocytes (T).
When freshly isolated TNCs (Figures 7A and 7G) were exposed to tissue culture, multiple layers of membranes unfurl to reveal thymocytes trapped within a honeycomb cage-like structure containing fenestra through which trapped thymocytes were visible (Figure 7B and C). This membrane structure was similar to that detected in Figure 6E. With time (Figure 7D and E), thymocytes were detected within the ruffled membrane extensions similar to those described above in Figure 6C. Most trapped thymocytes were completely enclosed within membrane extensions (Figure 7D, inset), while other thymocytes were partially exposed within cocoon-like membrane structures (Figure 7E, inset). By 3 hours, all thymocytes were released and the ruffled membrane surface of the freshly isolated TNC remained visible (Figure 7F).
Figure 7.
SEM of TNCs isolated from murine thymus and incubated over a 3 hours time period. (A) Thirty minute after isolation, the cell exhibits a circular morphology and layers of membrane folds (see inset). (B) Membrane begins to unfold and wraps around released thymocytes (see inset) (60 minutes post isolation). (C) Membrane layer is completely unfolds exposing thymocytes located within the fenestra (inset) (90 minutes post isolation). (D) TNC membrane is completely unfolded (120 minutes post isolation). (E) (150 min) Thymocytes are then gradually released from the TNC. (F) No thymocytes are associated with the TNC. (180 min) (G) TEM image of a freshly isolated TNC corresponds to cell shown in panel A.
Both tsTNC-1 cells and freshly isolated TNCs were then analyzed using transmission electron microscopy (TEM) (Figure 8). Figure 8A shows a freshly isolated TNC complex. Thymocytes (Figure 8A, asterisk) appear to be trapped by the TNC membrane extension with structures that are similar to those observed in Figure 6E. The TEM micrographs also show membrane extensions equivalent to those detected in scanning studies. When co-cultured with thymocytes for 10 hours, tsTNC-1 cells contain cytoplasmic thymocytes and display membrane extensions (Figure 8B). Cytoplasmic thymocytes are clearly in various stages of apoptosis (Figure 8B, insets).
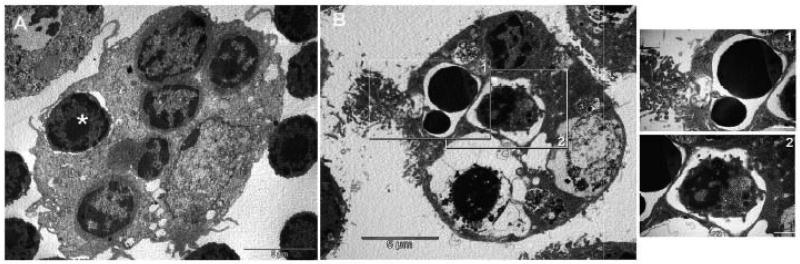
Figure 8.
Transmission electron micrographs of TNCs containing internalized thymocytes. (A) Freshly isolated TNC fixed and prepared for TEM analysis immediately following isolation. Membrane partially encloses a thymocyte (asterisk). (B) Thymocytes and tsTNC-1 cells were incubated for 10 hours and then analyzed by TEM. Thymocytes at different stages of apoptosis (insets 1 and 2) are visible within the cytoplasmic vacuoles.
The intertwining complex of membrane extensions are visible in both SEM and TEM micrographs (Figure 9A and B) and are localized to one side of the cell opposite to the nucleus (also see Figure 6G). Figure 9A shows the membrane extensions in an open configuration that creates a membrane maze that enters into the cytoplasm through classical cytoplasmic vacuoles (Figure 9A, panels 2 and 3). This network of membranes was detectable only in tsTNC-1 cultures to which thymocytes were added. Thymocytes are shown trapped within membrane extensions (Figure 9A, panels 1 and 3). In panel 3, the trapped thymocyte is located above a cytoplasmic vacuole. In the closed configuration, the network of membranes creates a cage-like structure (Figure 9B). Trapped thymocytes are visible within the cage (Figure 9B panel 1, inset). Similar structures are visible in TEM images of freshly isolated TNCs (Figure 9B, panel 2) and confocal images of TNCs in thymic sections (Figure (B, panel 3). Phase contrast video microscopy shows TNC cytoplasmic membrane asymmetry with respect to thymocyte contact (Figure 9C). Both membrane bound (phase bright) and thymocytes trapped within the cage (phase dark) (see circle in Figure 9C) are detectable. Also, the initial TNC interaction with thymocytes is shown to occur through contact with membrane extensions in an open configuration, which pull bound thymocytes to the TNC complex (Figure 9C, arrows).
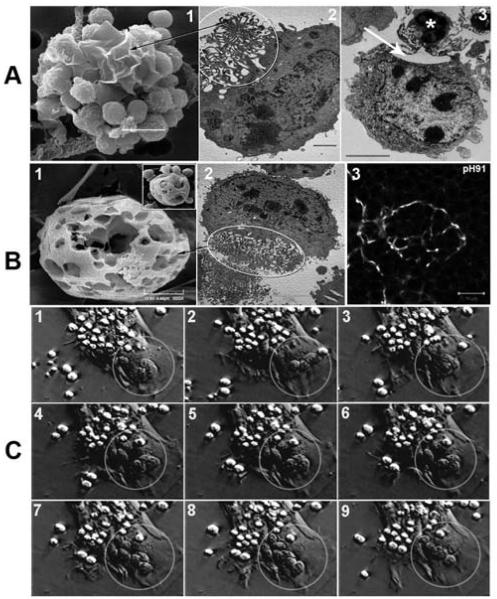
Figure 9.
Microscopic identification of membrane extensions and cage structure of TNCs during thymocyte binding and internalization. (A) Membrane extensions of TNCs visualized with SEM (panel 1) and TEM (panel 2). The circle in panel 2 corresponds to the membrane extensions seen in panel 1. TNC membrane extensions, equivalent to those in panels 1 and 2, are wrapped around a thymocyte (asterisk, panel 3) with a large cytoplasmic vesicle directly beneath (arrow). (B) The cage structure of TNCs visualized with SEM (panel 1), TEM (panel 2), and confocal microscopy of thymic section stained with pH91 (panel 3). The inset in panel 1 shows thymocytes residing within the cage like structure. The circle in panel 2 surrounds the cage as it appears in TEM. The arrow points to its SEM equivalent in panel 1. (C) Phase contrast video microscopy of TNC - thymocyte interaction. Thymocytes bound to the TNC surface are phase bright. Thymocytes trapped within the cage-like structure (circle, B and C) are phase dark. Thymocyte movement within cage was also detected. The arrow shows initial contact of thymocytes by membrane extensions from TNC. Membrane extensions pull thymocytes to the TNC complex.
MHC Restriction within TNCs
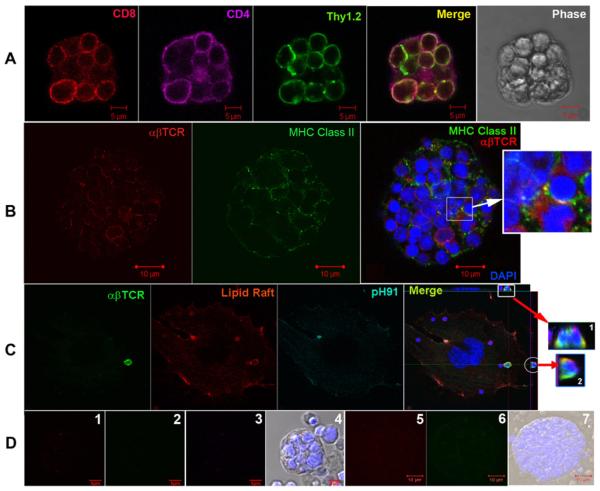
MHC class II expression on antigen presenting cells is required for the proper selection of thymocytes slated to become CD4+ T-cells. The negative selection of developing thymocytes has been reported to require an interaction with dendritic cells or macrophages within the medulla of the thymus [27]. Thymocytes within TNCs were stained with mAb against both CD4 and CD8 and analyzed using confocal microscopy (Figure 10A). Double positive thymocytes were detected within a complex which displayed the classical TNC phenotype. Freshly isolated TNCs containing trapped thymocytes (Figure 10B) were stained with mAbs against both the αβTCR and the MHC class II antigen. An interaction between the αβTCR and MHC class II molecules was detected (Figure 10B, inset). Also, lipid raft aggregation has been shown to occur during MHC restriction [28; 29]. Thymocytes that were co-cultured with tsTNC-1 cells showed lipid raft accumulation around cell surface αβTCR (Figure 10C).
Figure 10.
TNCs participate in the MHC restriction of thymocytes. (A) Freshly isolated TNCs were stained with mAbs against CD8 (red), CD4 (magenta), and Thy 1.2 (green). Merge panel shows captured thymocytes to be CD4+CD8+. (B) Freshly isolated TNC stained with mAbs to αβTCR (red) and MHC Class II (green). Thymocyte nuclei are visualized using DAPI staining (blue). Insets show co-localization of MHC and αβTCR (yellow). (C) Co-localization of thymocyte lipid raft with αβTCR. tsTNC-1 cells were co-incubated with freshly isolated thymocytes for 4 hours. Cells were stained with cholera toxin subunit B (red) for presence of lipid rafts and mAbs to αβTCR (green) and pH91 (magenta). Merge images (insets 1 and 2) are orthogonal projections that show the contact areas between αβTCR and lipid raft (yellow). (D) Panels 1, 2, and 3 show controls for CD8, CD4, and Thy 1, respectively. Panels 5 and 6 show mAb controls for αβTCR and MHC class II. Data is representative of three independent experiments.
DISCUSSION
TNCs as Thymic Epithelial Progenitors
In the studies presented here, we identify thymic nurse cells in the thymic cortex using the TNC-specific mAb-ph91 and mAbs to cytokeratins K8 and K5 (Figure 3). Traditionally, TNCs were only recognizable through their unique cell-in-cell structure. The multi-cellular nature of these complexes is clearly visible when isolated from the thymus (Figure 1). All TNCs, whether in culture or in vivo, were found to express the pH91-specific antigen (Figure 3) [13]. A subset of these pH91+ cells expressed the transcription factor p63 (Figure 4). Using freshly isolated cells, we also showed that nearly a quarter of TNCs are pH91+K8+K5+ cells (Figure 2). Our in vivo studies show that the K8+K5+ double positive cells with the unique structure of TNCs to be located in the cortico-medullary junction. The thymic epithelial progenitor cell type has been reported to reside in the cortico-medullary junction and to express both K8 and K5 cytokeratins and p63 [25; 26]. Several studies show thymic epithelial progenitor cells to be able to regenerate an entire functional thymus when transplanted under the kidney capsule [23; 30; 31]. Our data show a subset of TNCs located in the cortico-medullary junction express the reported characteristics of thymic epithelial progenitors.
The TNC/Thymocyte Interaction
TNCs were isolated and examined using scanning electron microscopy. Our results show enclosed thymocytes to be sequestered within membrane extensions (Figure 6). These membrane extensions were visible on the surface of TNCs, and some thymocytes were found trapped in specialized cocoon-like structures captured here for the first time (Figures 6E, 7C and 9B, panel 1). With time, multiple layers of membrane enclosed trapped thymocytes. This membrane overlay made it impossible to determine the following steps in the process using SEM. We continued to follow the process using transmission electron microscopy. The network of membrane extensions was detected protruding from one side of the TNC. It was only found in freshly isolated TNCs and tsTNC-1 cells exposed to thymocytes. A close examination of this unique membrane network revealed the area nearest the cytoplasm of the network to terminate with classical cytoplasmic vacuoles (Figure 9A and B). There were two types of membrane networks detected in our TEM results. One open, the ends of the membranes were open to the external microenvironment (Figure 9A, panel 2). The other closed, the ends were weaved together like straws of a basket (Figure 9B, panel 2). An analysis of the structures observed in Figures 7C and 9B suggested that the cage-like structures observed may have formed as a function of dovetailing or interlocking of the membrane extensions detected in Figure 6B as compared to Figure 6D. We propose that the interlocking of membrane extensions create the fenestrated cage seen in Figure 9B, panel 1. It is highly unlikely that these two unusual but similar membrane structures have different origins. If this is correct, the structure created would limit the outward movement of trapped thymocytes, as well as facilitate the thymocyte movement detected in our video studies (Figure 9C).
We evaluated the images of the thymocyte/TNC interaction using video microscopy with respect to the data collected in our EM studies. The fact that uropods consistently appear on the internalizing thymocyte indicates that thymocytes have a partial role in the internalization process. Once inside thymocytes are clearly mobile, as is the macrophage detected. A close examination of the macrophage's movement (Figure 5, panels G-I) shows it to squeeze through small spaces from and into larger ones. Using the information gathered from our EM studies, we believe the structural distortions displayed by the macrophage as it moved in the video resulted from its movement through fenestras detected in Figure 7C. A close analysis of the honeycomb cage-like structure, seen in Figure 9B, shows multiple external fenestras volumetrically sufficient to hold thymocytes, along with open internal spaces large enough to facilitate thymocyte movement. We believe that all of these structures are external to the cytoplasm of the TNC. Collectively, we propose these data to show that most thymocytes, as seen in classical micrographs (Figures 1 and 8, and [32]) of TNCs complexes, are external to cytoplasmic spaces. That is, the majority of trapped thymocytes dwell within the fenestrated cage-like structure where they are free to move (as seen in the video, Figure 9C). Thymocytes trapped in such a structure can escape the membrane complex without the complicated series of membrane fusion events required for external expulsion from classical cytoplasmic vacuoles. This could also explain the rapid release of thymocytes from the TNC complex upon isolation in vitro, as well as the existence of a rapidly moving macrophage within these spaces (which cannot occur within the cytoplasm of a cell). At the same time, we found the proximal side of the membrane network to terminate into classical cytoplasmic vacuoles. This makes possible for the movement of a selected subset of trapped thymocytes into cytoplasmic vacuoles. However, the selection process for the thymocyte subset chosen to enter cytoplasmic vacuoles remains unknown. On the other hand, previous studies have shown apoptotic thymocytes to be degraded through fusion with lysosomes within TNCs [22]. Thymocyte fusion with lysosomes (which are cytoplasmic organelles) requires the apoptotic thymocytes to be within cytoplasmic vacuoles. This suggests that minimally, the subset of thymocytes within the complex selected to undergo apoptosis must move into vacuoles within the cytoplasm to facilitate an interaction with lysosomes.
Does Negative Selection Occur Within the TNC Complex?
MHC restriction involves the selective removal of potentially autoreactive T cells through an interaction between the αβTCR and MHC molecules expressed on thymocytes and antigen presenting cells, respectively. The process is separated into activities that either select for thymocyte maturation (positive selection), or thymocyte deletion (negative selection through apoptosis). Low affinity interactions between the αβTCR and MHC molecules facilitate positive selection exclusively. All other interactions result in negative selection. Much data has been generated to show that these activities occur at different locations within the thymus [15; 33; 34; 35; 36]. Positive selection is believed to take place in the cortex, while negative selection has been shown to be restricted to the medulla. Recent reports conflict with this idea and suggest that negative selection can occur in the cortex as well [37; 38; 39; 40]. Until recently, it was believed that negative selection was restricted to cells of the medulla because they exclusively expressed AIRE and TRA, both of which have been proposed to be required for negative selection. However, a recent publication showed both AIRE and TRA expression to occur within the TNC complex [21]. Data collected in this study show, for the first time, a functional interaction between the αβTCR on thymocytes and MHC molecules expressed on vacuoles within the TNC complex, which activates the co-localization of lipid raft accumulation around the αβTCR. Lipid raft co-localization has been shown to result from αβTCR signaling during MHC restriction [41; 42]. The protein composition of these lipid micro-domains is different during positive versus negative selection [28]. It has been shown that distinct TCR lipid rafts are generated to control positive selection through the generation of ERK signaling modules [29; 43]. Negative selection is proposed to involve lipid rafts formed for the activation of JNK and p38 pathways [44].
So, what are the requirements for negative selection? Negative selection is the process that activates apoptosis during MHC restriction. Apoptotic as well as viable thymocytes are found within the TNC complex [11; 45]. MHC restriction reportedly occurs at the CD4+CD8+αβTCRlow stage of development [46]. Thymocytes within the TNC complex were shown to express CD4, CD8 and the αβTCR (Figure 10) [3]. CD4+CD8+αβTCRlow CD69− cells within the TNC complex have been shown to either die through apoptosis or to mature to the CD4+CD8+αβTCRhigh CD69+ stage of development [45]. In another study, using HY-TCR transgenic mice, isolated TNCs contained triple positive thymocytes exclusively [47]. The thymus of the female HY-TCR transgenic mouse represents a microenvironment for positive selection, while negative selection is restricted to the male thymi because HY is a male–specific antigen [48; 49]. Five times more viable thymocytes were found within TNCs isolated from the female transgenic mouse, with less than 4% apoptosis, while almost fifty percent of the thymocytes found in male TNCs were apoptotic, suggesting the increased apoptosis within the male TNCs is associated with negative selection. Negative selection requires a functional interaction between the αβTCR and MHC molecules, as was described to occur within TNCs in this study. Negative selection requires the expression of both AIRE and TRAs. TNCs were recently shown to express both [50]. Barring any other requirements, these data strengthen the possibility that negative selection occurs within the TNC complex.
ACKNOWLEDGEMENTS
We would like to thank Jorge Morales and Daniel Fimiarz for their excellent technical assistance. We are very grateful to Garnet Lewis and Rhembert Walker for their continued support. This work was supported by NIH-RCMI grant 5G12RR03060, NSF grant MCB-0412822and CUNY-RF grant 95202-02 01.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Overholtzer M, Brugge JS. The cell biology of cell-in-cell structures. Nat Rev Mol Cell Biol. 2008;9:796–809. doi: 10.1038/nrm2504. [DOI] [PubMed] [Google Scholar]
- 2.Wekerle H, Ketelsen UP. Thymic nurse cells--Ia-bearing epithelium involved in T-lymphocyte differentiation? Nature. 1980;283:402–4. doi: 10.1038/283402a0. [DOI] [PubMed] [Google Scholar]
- 3.Wekerle H, Ketelsen UP, Ernst M. Thymic nurse cells. Lymphoepithelial cell complexes in murine thymuses: morphological and serological characterization. J Exp Med. 1980;151:925–44. doi: 10.1084/jem.151.4.925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ritter MA, Sauvage CA, Cotmore SF. The human thymus microenvironment: in vivo identification of thymic nurse cells and other antigenically-distinct subpopulations of epithelial cells. Immunology. 1981;44:439–46. [PMC free article] [PubMed] [Google Scholar]
- 5.Boyd RL, Oberhuber G, Hala K, Wick G. Obese strain (OS) chickens with spontaneous autoimmune thyroiditis have a deficiency in thymic nurse cells. J Immunol. 1984;132:718–24. [PubMed] [Google Scholar]
- 6.Flano E, Alvarez F, Lopez-Fierro P, Razquin BE, Villena AJ, Zapata AG. In vitro and in situ characterization of fish thymic nurse cells. Dev Immunol. 1996;5:17–24. doi: 10.1155/1996/14738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Romano N, Taverne-Thiele AJ, Fanelli M, Baldassini MR, Abelli L, Mastrolia L, Van Muiswinkel WB, Rombout JH. Ontogeny of the thymus in a teleost fish, Cyprinus carpio L.: developing thymocytes in the epithelial microenvironment. Dev Comp Immunol. 1999;23:123–37. doi: 10.1016/s0145-305x(98)00053-6. [DOI] [PubMed] [Google Scholar]
- 8.Mohammad MG, Chilmonczyk S, Birch D, Aladaileh S, Raftos D, Joss J. Anatomy and cytology of the thymus in juvenile Australian lungfish, Neoceratodus forsteri. J Anat. 2007;211:784–97. doi: 10.1111/j.1469-7580.2007.00814.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Andrews P, Boyd RL, Shortman K. The limited immunocompetence of thymocytes within murine thymic nurse cells. Eur J Immunol. 1985;15:1043–8. doi: 10.1002/eji.1830151016. [DOI] [PubMed] [Google Scholar]
- 10.Kyewski BA, Kaplan HS. Lymphoepithelial interactions in the mouse thymus: phenotypic and kinetic studies on thymic nurse cells. J Immunol. 1982;128:2287–94. [PubMed] [Google Scholar]
- 11.Pezzano M, Li Y, Philp D, Omene C, Cantey M, Saunders G, Guyden JC. Thymic nurse cell rescue of early CD4+CD8+ thymocytes from apoptosis. Cell Mol Biol (Noisy-le-grand) 1995;41:1099–111. [PubMed] [Google Scholar]
- 12.Li Y, Pezzano M, Philp D, Reid V, Guyden J. Thymic nurse cells exclusively bind and internalize CD4+CD8+ thymocytes. Cell Immunol. 1992;140:495–506. doi: 10.1016/0008-8749(92)90214-a. [DOI] [PubMed] [Google Scholar]
- 13.Pezzano M, King KD, Philp DD, Adeyemi A, Gardiner B, Yang J, Samms M, Boto W, Guyden JC. A thymic nurse cell-specific monoclonal antibody. Cell Immunol. 1998;185:123–33. doi: 10.1006/cimm.1998.1279. [DOI] [PubMed] [Google Scholar]
- 14.Philp D, Pezzano M, Li Y, Omene C, Boto W, Guyden J. The binding, internalization, and release of thymocytes by thymic nurse cells. Cell Immunol. 1993;148:301–15. doi: 10.1006/cimm.1993.1114. [DOI] [PubMed] [Google Scholar]
- 15.Surh CD, Sprent J. T-cell apoptosis detected in situ during positive and negative selection in the thymus. Nature. 1994;372:100–3. doi: 10.1038/372100a0. [DOI] [PubMed] [Google Scholar]
- 16.Robey E, Fowlkes BJ. Selective events in T cell development. Annu Rev Immunol. 1994;12:675–705. doi: 10.1146/annurev.iy.12.040194.003331. [DOI] [PubMed] [Google Scholar]
- 17.von Boehmer H, Kisielow P. Self-nonself discrimination by T cells. Science. 1990;248:1369–73. doi: 10.1126/science.1972594. [DOI] [PubMed] [Google Scholar]
- 18.Peterson P, Org T, Rebane A. Transcriptional regulation by AIRE: molecular mechanisms of central tolerance. Nat Rev Immunol. 2008;8:948–57. doi: 10.1038/nri2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Su MA, Anderson MS. Aire: an update. Curr Opin Immunol. 2004;16:746–52. doi: 10.1016/j.coi.2004.09.009. [DOI] [PubMed] [Google Scholar]
- 20.Derbinski J, Schulte A, Kyewski B, Klein L. Promiscuous gene expression in medullary thymic epithelial cells mirrors the peripheral self. Nat Immunol. 2001;2:1032–9. doi: 10.1038/ni723. [DOI] [PubMed] [Google Scholar]
- 21.Hansenne I, Louis C, Martens H, Dorban G, Charlet-Renard C, Peterson P, Geenen V. Aire and Foxp3 expression in a particular microenvironment for T cell differentiation. Neuroimmunomodulation. 2009;16:35–44. doi: 10.1159/000179665. [DOI] [PubMed] [Google Scholar]
- 22.Samms M, Philp D, Emanus F, Osuji O, Pezzano M, Guyden JC. Lysosomal-mediated degradation of apoptotic thymocytes within thymic nurse cells. Cell Immunol. 1999;197:108–15. doi: 10.1006/cimm.1999.1559. [DOI] [PubMed] [Google Scholar]
- 23.Bennett AR, Farley A, Blair NF, Gordon J, Sharp L, Blackburn CC. Identification and characterization of thymic epithelial progenitor cells. Immunity. 2002;16:803–14. doi: 10.1016/s1074-7613(02)00321-7. [DOI] [PubMed] [Google Scholar]
- 24.Pezzano M, Li Y, Yang YM, Guyden J. The immortalization of thymic nurse cells by SV40 virus. Cell Immunol. 1991;133:434–45. doi: 10.1016/0008-8749(91)90116-s. [DOI] [PubMed] [Google Scholar]
- 25.Popa I, Zubkova I, Medvedovic M, Romantseva T, Mostowski H, Boyd R, Zaitseva M. Regeneration of the adult thymus is preceded by the expansion of K5+K8+ epithelial cell progenitors and by increased expression of Trp63, cMyc and Tcf3 transcription factors in the thymic stroma. Int Immunol. 2007;19:1249–60. doi: 10.1093/intimm/dxm092. [DOI] [PubMed] [Google Scholar]
- 26.Senoo M, Pinto F, Crum CP, McKeon F. p63 Is essential for the proliferative potential of stem cells in stratified epithelia. Cell. 2007;129:523–36. doi: 10.1016/j.cell.2007.02.045. [DOI] [PubMed] [Google Scholar]
- 27.Lorenz RG, Allen PM. Thymic cortical epithelial cells can present self-antigens in vivo. Nature. 1989;337:560–2. doi: 10.1038/337560a0. [DOI] [PubMed] [Google Scholar]
- 28.Werlen G, Hausmann B, Naeher D, Palmer E. Signaling life and death in the thymus: timing is everything. Science. 2003;299:1859–63. doi: 10.1126/science.1067833. [DOI] [PubMed] [Google Scholar]
- 29.Delgado P, Fernandez E, Dave V, Kappes D, Alarcon B. CD3delta couples T-cell receptor signalling to ERK activation and thymocyte positive selection. Nature. 2000;406:426–30. doi: 10.1038/35019102. [DOI] [PubMed] [Google Scholar]
- 30.Gill J, Malin M, Hollander GA, Boyd R. Generation of a complete thymic microenvironment by MTS24(+) thymic epithelial cells. Nat Immunol. 2002;3:635–42. doi: 10.1038/ni812. [DOI] [PubMed] [Google Scholar]
- 31.Rossi SW, Jenkinson WE, Anderson G, Jenkinson EJ. Clonal analysis reveals a common progenitor for thymic cortical and medullary epithelium. Nature. 2006;441:988–91. doi: 10.1038/nature04813. [DOI] [PubMed] [Google Scholar]
- 32.Brelinska R, Warchol JB. Thymic nurse cells: their functional ultrastructure. Microsc Res Tech. 1997;38:250–66. doi: 10.1002/(SICI)1097-0029(19970801)38:3<250::AID-JEMT6>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 33.Anderson G, Jenkinson EJ. Lymphostromal interactions in thymic development and function. Nat Rev Immunol. 2001;1:31–40. doi: 10.1038/35095500. [DOI] [PubMed] [Google Scholar]
- 34.Hogquist KA, Bevan MJ. The nature of the peptide/MHC ligand involved in positive selection. Semin Immunol. 1996;8:63–8. doi: 10.1006/smim.1996.0009. [DOI] [PubMed] [Google Scholar]
- 35.Boehm T, Scheu S, Pfeffer K, Bleul CC. Thymic medullary epithelial cell differentiation, thymocyte emigration, and the control of autoimmunity require lympho-epithelial cross talk via LTbetaR. J Exp Med. 2003;198:757–69. doi: 10.1084/jem.20030794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Anderson MS, Venanzi ES, Klein L, Chen Z, Berzins SP, Turley SJ, von Boehmer H, Bronson R, Dierich A, Benoist C, Mathis D. Projection of an immunological self shadow within the thymus by the aire protein. Science. 2002;298:1395–401. doi: 10.1126/science.1075958. [DOI] [PubMed] [Google Scholar]
- 37.McCaughtry TM, Baldwin TA, Wilken MS, Hogquist KA. Clonal deletion of thymocytes can occur in the cortex with no involvement of the medulla. J Exp Med. 2008;205:2575–84. doi: 10.1084/jem.20080866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Klein L, Kyewski B. Self-antigen presentation by thymic stromal cells: a subtle division of labor. Curr Opin Immunol. 2000;12:179–86. doi: 10.1016/s0952-7915(99)00069-2. [DOI] [PubMed] [Google Scholar]
- 39.Goldman KP, Park CS, Kim M, Matzinger P, Anderson CC. Thymic cortical epithelium induces self tolerance. Eur J Immunol. 2005;35:709–17. doi: 10.1002/eji.200425675. [DOI] [PubMed] [Google Scholar]
- 40.Spain LM, Berg LJ. Developmental regulation of thymocyte susceptibility to deletion by “self”-peptide. J Exp Med. 1992;176:213–23. doi: 10.1084/jem.176.1.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Janes PW, Ley SC, Magee AI, Kabouridis PS. The role of lipid rafts in T cell antigen receptor (TCR) signalling. Semin Immunol. 2000;12:23–34. doi: 10.1006/smim.2000.0204. [DOI] [PubMed] [Google Scholar]
- 42.Magee T, Pirinen N, Adler J, Pagakis SN, Parmryd I. Lipid rafts: cell surface platforms for T cell signaling. Biol Res. 2002;35:127–31. doi: 10.4067/s0716-97602002000200003. [DOI] [PubMed] [Google Scholar]
- 43.Fischer AM, Katayama CD, Pages G, Pouyssegur J, Hedrick SM. The role of erk1 and erk2 in multiple stages of T cell development. Immunity. 2005;23:431–43. doi: 10.1016/j.immuni.2005.08.013. [DOI] [PubMed] [Google Scholar]
- 44.Sohn SJ, Thompson J, Winoto A. Apoptosis during negative selection of autoreactive thymocytes. Curr Opin Immunol. 2007;19:510–5. doi: 10.1016/j.coi.2007.06.001. [DOI] [PubMed] [Google Scholar]
- 45.Pezzano M, Philp D, Stephenson S, Li Y, Reid V, Maitta R, Guyden JC. Positive selection by thymic nurse cells requires IL-1 beta and is associated with an increased Bcl-2 expression. Cell Immunol. 1996;169:174–84. doi: 10.1006/cimm.1996.0108. [DOI] [PubMed] [Google Scholar]
- 46.von Boehmer H. Developmental biology of T cells in T cell-receptor transgenic mice. Annu Rev Immunol. 1990;8:531–56. doi: 10.1146/annurev.iy.08.040190.002531. [DOI] [PubMed] [Google Scholar]
- 47.Martinez M, Samms M, Hendrix TM, Adeosun O, Pezzano M, Guyden JC. Thymic nurse cell multicellular complexes in HY-TCR transgenic mice demonstrate their association with MHC restriction. Exp Biol Med (Maywood) 2007;232:780–8. [PubMed] [Google Scholar]
- 48.Kisielow P, Bluthmann H, Staerz UD, Steinmetz M, von Boehmer H. Tolerance in T-cell-receptor transgenic mice involves deletion of nonmature CD4+8+ thymocytes. Nature. 1988;333:742–6. doi: 10.1038/333742a0. [DOI] [PubMed] [Google Scholar]
- 49.Teh HS, Kisielow P, Scott B, Kishi H, Uematsu Y, Bluthmann H, von Boehmer H. Thymic major histocompatibility complex antigens and the alpha beta T-cell receptor determine the CD4/CD8 phenotype of T cells. Nature. 1988;335:229–33. doi: 10.1038/335229a0. [DOI] [PubMed] [Google Scholar]
- 50.Zuklys S, Balciunaite G, Agarwal A, Fasler-Kan E, Palmer E, Hollander GA. Normal thymic architecture and negative selection are associated with Aire expression, the gene defective in the autoimmune-polyendocrinopathy-candidiasis-ectodermal dystrophy (APECED) J Immunol. 2000;165:1976–83. doi: 10.4049/jimmunol.165.4.1976. [DOI] [PubMed] [Google Scholar]