Introduction
In the Western world, cardiovascular disease, along with diabetes mellitus and its complications, are leading causes of death. A number of important risk factors have been associated with the virtual pandemic of these killers. These include smoking, sedentary lifestyle, high body mass index, hypertension, and so forth. Nonetheless, many individuals who develop cardiovascular and/or metabolic disease do not have these risk factors. Thus, it is clear that as yet unrecognized and underappreciated factors must be considered in the genesis of these disorders. In his monumental volume Stress, Hans Selye (1907-1982) observed that stress to the organism, in essentially any of its forms -- dietary, environmental, disease, and others -- could result in cellular, hormonal, and related damage, with the body mounting a response he termed the “General-Adaptation-Syndrome” (Selye, 1950). Writing years before the nuances of biochemical and molecular mechanisms were established, Selye envisioned an orchestrated biological defensive response to such challenges. This concept is of special relevance to the developing fetus, as during the course of gestation a number of stresses to the mother are known to affect placental, as well as embryonic/fetal development, many with life-long consequences.
Along this line, a factor that has received increasing attention is the idea of “programming” during fetal life, often as a consequence of maternal stress. Special features of antenatal programming include: critical periods of vulnerability, failure or unsatisfactory completion of specific developmental milestones, association with functional defects, the permanent nature of such sequelae, and so forth (Barker, 1989a; 1989b; 1992; 1994; 1995a; 1995b; 1998; 2004; Barker et al, 1995; Nijland et al, 2008). The concept of the developmental origins of adult health and disease, first articulated by David J.P. Barker in the mid-1980s, “... suggested that poor nutrition in early life increases susceptibility to the effects of an affluent diet” (Barker & Osmond, 1986a). In a related analysis, these authors noted the high correlation of cerebrovascular accidents in the 1970s, to increased infant mortality six decades earlier during the years 1911-1914 (Barker & Osmond, 1987). In addition, in men born from 1911 to 1930, Barker and his group have shown an inverse correlation between the weights both at birth and at 1 year of age to coronary artery disease as adults (Barker et al, 1989a; 1989b). A subsequent study disclosed a similar trend with birthweight among women (Osmond et al, 1993).
Since this concept first was proposed, supporting evidence has been provided by a series of epidemiologic studies from a number of countries and cultures. These include studies correlating adult mortality from acute myocardial infarction with high infant mortality rates in a given population, follow-up studies correlating adult hypertension, coronary artery disease, and type II diabetes with low birth weight, the relation of increased mortality from coronary artery disease to low weight at 1 yr of age, and the relation of both newborn ponderal index [weight (g) x 102/crown-heel length (cm)2] and placental-to-fetal weight ratio to hypertension in the adult (see Barker, 2003, and Barker et al, 1995 for review). These associations are independent of adult life style risk factors (Barker et al, 1993). Among a number of other maternal stress-induced sequelae are those of immune dysfunction (Götz et al, 2007; Merlot et al, 2008), cortisol secretion later in life (Reynolds et al, 2007; Tu et al, 2007), increased incidence of schizophrenia (Hoek et al, 1996; Hulshoff et al, 2000; Susser & Lin, 1992), and many others (Ham & Tronick, 2006). More recently, numerous studies in experimental animals have demonstrated a relation between intrauterine fetal stress, particularly that of maternal food deprivation and/or emotional stress, and adult disease (Gluckman et al, 2008; Green & Hanson, 2004; Hanson & Gluckman, 2005; Jansson & Powell, 2007). Among the major known intrauterine stresses about which the effects on subsequent adult health are largely unknown, are maternal hypoxia and dietary imbalance.
The placenta, a fetomaternal organ joining mother and offspring during pregnancy in mammals, serves as an endocrine organ in the “maternal-placental-fetal” complex, in addition to its role in the exchange of respiratory gases, a multitude of nutrients, an immunologic barrier, and other functions. As has been recognized for many years, compromised placental function can have both short- and long-term consequences for the developing conceptus. In the present review, we examine the current state of knowledge of placental gene expression responses to maternal stress such as hypoxia, protein deficiency, and caloric excess. For the most part these studies are in rodents, however, when applicable, we also review those studies of placental gene expression in the human. (We will not review thoroughly the field of antenatal origins of adult health and disease, nor the role of environmental toxins in developmental disorders, as these topics have been reviewed in extenso elsewhere). Importantly, beyond mere description, we attempt to place these gene expression changes into a framework of the biochemical pathways and molecular mechanisms, by which stresses to the maternal organism can result in alterations of great biologic and epigenetic importance to the developing embryo and fetus. Finally, we consider important issues for future investigation, i.e., questions that probe the limits of our understanding.
Why Study Gene Expression in the Placenta?
Successful placental development is crucial for optimal growth, maturation, and survival of the embryo/fetus. The placenta not only nurtures the fetus, but protects it from harmful waste products by acting as an excretory route, and also presents an immunologic barrier between the maternal and fetal circulatory beds. Although the nucleus of every cell in the body carries a complete set of DNA, these cells differ in function with placental and embryological development consisting of an elegantly orchestrated switching of genes on and off in the transition from single fertilized cell to fully formed placenta and fetus. Deviation in the normal gene expression pattern may lead to altered placental phenotype, as well as a modified phenotype of the conceptus. This is evidenced by the numerous lethal embryonic null mutants secondary to placental failure. The mouse has been employed as a useful model of placental development. While the mouse placenta is not identical to its human counterpart, many studies have shown that similar cell lineages are largely conserved, and similar genes direct placental development in both species. Placental cell lineages derive from trophoectoderm precursors. The mural trophoectoderm differentiates into primary trophoblast giant cells, while the polar trophoectoderm gives rise to the extraembryonic ectoderm and the ectoplacental cone. In many mammals, including the mouse, the extraembryonic ectoderm forms the chorion that fuses with the allantois, an outgrowth of extraembryonic mesoderm, at around embryonic day 8 (E8) to form the placental labyrinthine layer. The spongiotrophoblast layer of the murine placenta derives from ectoplacental precursor cells and forms the middle layer of the placenta, also known as the junctional zone. The outermost placental cells are the trophoblast giant cell layer. In addition to the primary trophoblast cells derived from the mural trophoectoderm, secondary trophoblast giant cells are derived from the spongiotrophoblast. Later in placental development, around E12.5, glycogen-filled trophoblast cells appear in the spongiotrophoblast layer. Although their function is unclear, these cells express several important gene products, and migrate into the decidua later in pregnancy. Several reviews have detailed placental cell lineages, and some of the genes involved in their differentiation (Cross, 2005; Simmons & Cross, 2005).
Recent studies reveal some of the fundamental mechanisms underlying placental development (Cross et al, 2003; Daoud et al, 2005; Gheorghe et al, 2006; Hemberger, 2007; Sood et al, 2006; Tanaka et al, 2000). Numerous genes are required for proper development of the placenta, and their number has increased greatly, in part, due to the discovery of numerous lethal embryonic null mutants secondary to placental failure (Adams et al, 2000; Schorpp-Kistner et al, 1999; Schreiber et al, 2000; Yamamoto et al, 1998). For example, the disruption of many genes, including growth factors, transcription factors, extracellular matrix proteins, and proteins involved in cell signaling, leads to embryonic lethality secondary to placental failure (Rossant & Cross, 2001). In human trophoblast in vitro, several gene classes are strongly up- and down-regulated in the course of differentiation (Aronow et al, 2001). Another study compared differentially expressed genes between the murine placenta and the embryo itself at E12.5 (Tanaka et al, 2000). Microarray analysis has provided insights into aspects of the genetic mechanisms of development, cell growth both normal and abnormal, responses to stress, and numerous other processes (Chu et al, 1998; Gasa et al, 2004; Iyer et al, 1999).
To What Extent is Placental Gene Expression Altered During Gestation?
The molecular basis of placental development remains incompletely understood. Recent studies have begun to shed some light on this process, and numerous genes have been demonstrated to be essential for the proper differentiation of placental cell lineages and fetal survival. Unfortunately, the details of the interactions and effects of these genes are unclear. In an effort to understand this process at a more fundamental level, we examined gene expression patterns in the developing murine placenta at days E10.5 E12.5, E15.5, and E17.5, testing the hypothesis that from E10.5 until E17.5, numerous placental genes are up- or down-regulated to a significant degree, and that specific functional groups of genes are regulated at the different developmental ages (Gheorghe et al, 2006). To examine gene clustering and functional analysis of pathways, we focused on those genes most highly regulated by development. At E10.5, several functional categories were over-represented, including genes involved in angiogenesis and blood vessel development, morphogenesis, and organogenesis, and genes involved in lipid metabolism and transport. At E12.5, over-represented gene categories were involved in cell cycle control and RNA binding proteins. At E15.5, notably over-represented were genes involved in cellular transport and cell growth and maintenance. At E17.5, we noted the up-regulation of an over-abundance of genes involved in the regulation of transcription and numerous proteins that localize to the nucleus.
This study identified several subsets of genes highly regulated during placental development. Clustering according to their expression patterns, suggests that at crucial times during placental ontogeny particular subsets of diverse genes are induced or repressed in concert (Fig. 1). Genes up-regulated early in placental development clearly underlie the rapid tissue growth, cell proliferation, and vascular development occurring during this period. At E10.5, genes involved in several key processes were strongly up-regulated. These include angiogenesis, lipid metabolism, and cell cycle regulation. Genes such as ELK3, c-fos induced growth factor, plasminogen (the precursor to angiostatin), serine (or cysteine) proteinase inhibitor clade F member 1, all are involved in blood vessel morphogenesis, and were up-regulated at E10.5. At both E10.5 and E12.5 cyclin, D1, cyclin E2, cyclin C, MAD 2, pleiotrophin, BRCA 2, which are involved in cell cycle control also are up-regulated strongly. At E12.5, ribosomal genes were notably up-regulated, as were several genes involved in lipid transport and metabolism including: apolipoprotein B, apolipoprotein C-II, lysophospholipase, microsomal triglyceride transfer protein and adiponectin receptor 1. As must be evident, lipid transport and metabolism are important for the proper fetal development (Shekhawat et al, 2003) and disruption of lipid transporters leads to embryonic lethality (Farese et al, 1996; Gimeno et al, 2003). Previous null mutant experiments have identified several of these genes as embryonic lethal, further confirming their importance in development. For instance, Cops2 mutants died soon after implantation (Lykke-Anderson et al, 2003), Pten mutants died at E9.5 secondary to placental failure (Yamamoto et al, 1998), and connexin 43 mutants died shortly after birth, due to cardiac and vascular abnormalities (Reaume et al, 1995).
Figure 1.
Over-represented gene functional classes throughout mouse placental development. Listed are the major groups of upregulated placental genes expressed from Embryonic day 10.5 to 17.5.
Genes such as growth hormone releasing hormone prolactin-like protein I, secretin, and chorionic somatomammotropin hormone 2 also were upregulated during the course of placental development. Recent studies in the sheep suggest that growth hormone releasing hormone regulates the expression of both placental growth hormone and lactogen (Lacroix et al, 2002). Insulin-like growth factor II (IGF-II) and Insulin-like growth factor binding protein 2, genes have been shown to be expressed in the placenta (Zollers et al, 2001), were up-regulated from E10.5 to E12.5. Previous studies have shown that after E12.5 IGF-II is mainly produced by trophoblast glycogen cells (Redline et al, 1993); however, in our study it was up-regulated at E12.5 and later (Gheorghe et al, 2006). IGF-II also appears to have key functions in placental transport and permeability (Sibley et al, 2004). A number of prolactin-like proteins have been shown to be regulated with development, such as: prolactin-like protein C 1, prolactin-like protein F, prolactin-like protein I, prolactin-like protein K. The prolactin gene family in the mouse has at least 26 identified members (Wiemers et al, 2003), and several studies have shown that in the placenta this gene family performs key reproductive and regulatory functions (Ain et al, 2003; 2004). In the near-term human placenta, mRNA for a number of factors associated with angiogenesis (vascular endothelial growth factor and annexin V) and homeostasis (plasminogen activator factor, thrombomodulin, and others) are widely distributed (Chinni et al, 2008). Circulating fetal fibrocytes, and perhaps other cells, also play a role in the development of the placenta and the umbilical arteries and vein (Kim et al, 2008).
To What Extent is Placental Gene Expression Altered by Maternal Hypoxia?
As noted above, a number of stressors can lead to altered placental and fetal growth and development. Of great importance in this regard, is the less than optimal supply of oxygen (O2), e.g., hypoxia. Hypoxia has been identified as a major stressor in development, and is believed to be a contributing cause to placental pathology such as that associated with preeclampsia (Austgulen et al, 2004; Challier & Uzan, 2003). Hypoxia can lead to low birth weight and intrauterine growth restriction (IUGR) and disease of the newborn such as persistent pulmonary hypertension (Zamudio, 2003). Little is known, however, about the adaptive mechanisms involved in the placental responses to suboptimal oxygen availability. Several studies have attempted to harness the power of microarray and proteomic analysis to elucidate responses to hypoxia in cultured human cytotrophoblasts (Hoang et al, 2001), and in rat embryos and placentas (Huang et al, 2004). As with other cell types, oxygen is a critical regulator of the normal trophoblast cell development, which undergo differentiation and/or proliferation in response to varying O2 concentrations. Acting through aryl receptor nuclear transporter and hypoxia-inducible factor 1α (HIF-1α), oxygen regulates placental cell phenotypes and gene expression (Adelman et al, 2000). Hypoxic stress can lead to placental cell death and dysregulation of vasculogenesis, which negatively affects the development of the placental vascular bed (Kingdom et al, 2000). In response to exposure to high altitude, long-term hypoxia, the ovine placentomes undergo significant structural changes (Penninga & Longo, 1998), and the vasculature displays significant increases in capillary density, vessel tortuosity, and a decrease in diffusion distance from maternal to fetal blood (Krebs et al, 1997). The human placenta also shows significant morphologic and morphometric changes in response to high altitude hypoxia (Zhang et al, 2002). Nonetheless, essentially nothing is known about the molecular basis of these changes.
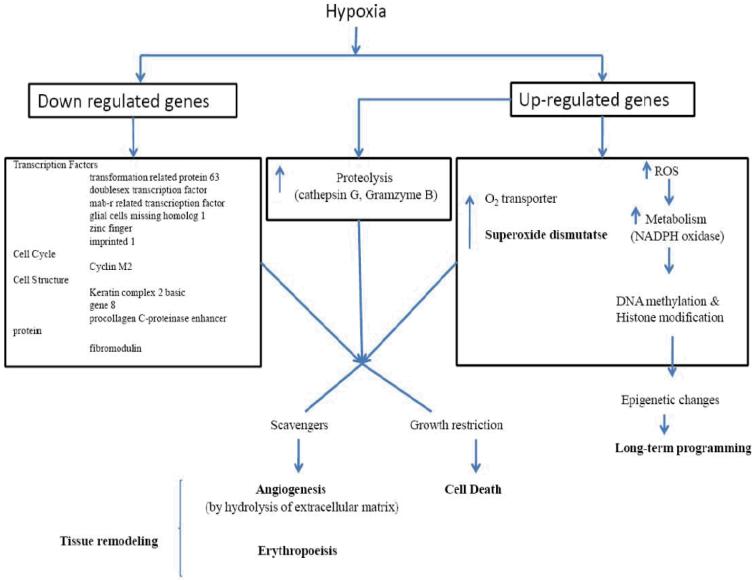
To understand in greater detail the role of hypoxia in placental gene expression, we tested the hypothesis that hypoxia-induced altered placental morphology is accompanied by significant changes in expression profiles. We compared gene expression levels between the normal murine placenta at E17.5, and that from dams exposed to 0.5 atmosphere hypoxia (10.5% O2) for 48 hours from E15.5 to E17.5. In response to this stress, some of the most highly up-regulated genes were those related to metabolism (alpha-keto reductase family-1 member 7, mitochondrial solute carrying protein, acetyl-coenzyme A synthetase 2, and NADPH oxidase 4), oxygen transport (erythroid associated factor, hemoglobin Y beta-like embryonic chain, erythrocyte protein band 4.2), proteolysis (cathepsin G, kallikrein 4, dipeptidase 1, serine protease 32), cell death (Bcl-like 2, perforin 1, glutathione peroxidase 1), and metabolism of reactive oxygen species (Gheorghe et al, 2007). Of particular note, several genes related to DNA methylation and epigenetic control were up-regulated (DNA methyltransferase 3B, methyl-CpG binding domain protein 1, RNA binding motif protein 3). Of the chromosomal distribution of genes up-regulated by hypoxia, we noted an over-abundance of genes from chromosome 14 (9.5% of the regulated genes as compared to 3.5% of genes on the array). Many of these correspond to the granzyme family of proteases.
Several of these genes have been noted previously to be regulated by hypoxia and/or involved in angiogenesis and metabolic responses. For instance, NAPDH oxidase 4 has been identified as a potential O2 sensor and regulator of HIF-1α (Zhu et al, 2002). Ferrocheloelastase is involved in heme metabolism, and has been shown to be up-regulated by hypoxia (Liu et al, 2004). Aminolevulinic acid synthase 2, glutathione peroxidase 1, and peroxiredoxin 2 are involved in the metabolism of reactive oxygen species; their activity, and expression levels have been identified as regulated by hypoxia (Abu-Farha et al, 2005; Mysore, 2005). Glycophorin A is involved in erythroid differentiation, and is regulated by erythropoietin (Gubin et al, 1999). Lactotransferrin is an antioxidant involved in iron metabolism and in scavenging of free radicals under hypoxic conditions (Morris et al, 1995). As noted above, hypoxia upregulated several members of the granzyme gene family. These are serine proteases expressed by lymphocytes, and thought to be involved in T-cell-mediated cytotoxicity. Granzymes are released along with perforin by natural killer (NK) cells and cytotoxic T lymphocytes, and trigger apoptosis in target cells through several mechanisms. A number of studies have demonstrated placental expression of granzymes, and they may play a broader role in placental development (Allen et al, 1998; Hirst et al, 2001). We also observed an up-regulation of perforin, which suggests that in response to hypoxia a greater number of NK cells, which have been shown to mediate a number of important functions, invade the placenta (Parham, 2004). Thus, the data suggest that granzymes not only play a role in normal placental development, but also are involved in hypoxia-mediated responses.
Again, of particular note in our study was the significant up-regulation of genes involved in DNA methylation and epigenetic control: DNA methyltransferase 3b and methyl CpG binding domain protein 1 (Gheorghe et al, 2007). As noted below, epigenetic modification is an important mechanism of gene expression regulation that does not involve modification of the DNA sequence, but rather DNA methylation and histone modification. These changes in expression patterns may be of importance in development, genomic imprinting, and the development of cancer (Turek-Plewa & Jagodzinski, 2005). The generation of reactive oxygen species (ROS) and the manipulation of glutathione metabolism have been shown to regulate a number of cellular processes, including DNA methylation (Fratelli et al, 2005). As depicted in Fig. 2, this suggests complex genetic regulation that commences with hypoxia and leads to alternations that result in long-term programming. In turn, this activates the DNA methylation machinery, and ultimately leads to long-term changes in the organism unrelated to modification of the nucleotide sequence. A key to unraveling this mechanism will be the identification of the targets for altered methylation and subsequent long-term down-regulation of transcription.
Figure 2.
Proposed mechanism of hypoxia-mediated epigenetic changes with long-term programming.
Among the down-regulated genes, most notable were several transcription factors (transformation-related protein 63, doublesex- and mab-3-related transcription factor 1, glial cells missing homolog 1, zinc finger, imprinted 1), cell cycle (cyclin M2), cell structure (keratin complex 2 basic, gene 8, procollagen C-proteinase enhancer protein, fibromodulin). We have presented a list of the genes up- or down-regulated and their known function, and verified the regulation of several of these genes using real time PCR (Gheorghe et al, 2007). In response to hypoxia, placental morphology also was altered significantly e.g. having a greater vascular density and containing many more red blood cells.
Several other studies have examined gene expression changes in response to hypoxia at the global level. One study catalogued the hypoxic-induced responses in the rat embryo to hypoxic exposure for both 24 hours and 11 days. Glycolysis-related genes, calcium homeostasis-related genes, and inflammatory genes (particularly as related to oxidative stress) were up-regulated, while cell growth-related genes were down-regulated (Huang et al2004). Other studies also have examined human trophoblast responses in-vitro to low oxygen tension. Observed were up-regulation of antioxidants (superoxide dismutase) and glycolysis-related genes, and the upregulation of glutathione-S-transferase (Nelson et al, 2003; Roh et al, 2005). In addition, gene expression changes in placentas from pregnancies complicated by pre-eclampsia and IUGR have been catalogued. In particular, H4 histone was down-regulated in women with severe pre-eclampsia (Chen et al, 2006; Soleymanloo et al, 2005), and up-regulation of glutathione-S-transferase was observed in human placentas from women at high altitude (3,100m) (Chen et al, 2006; Roh et al, 2005). These studies have highlighted the diverse manner in which placental cells respond to hypoxic stress.
In the placenta of patients that experienced chronic hypoxic ischemia, mRNA levels of both leptin and insulin-like growth factor (IGF)-1 were upregulated significantly (Trollmann et al, 2007). In near-term placental explants cultured in 1% O2, expression of the tumor suppressor protein p53, that promotes cell cycle arrest or apoptosis, was significantly elevated (Heazell et al, 2008). Exposure of human placental villous explants to 3% O2 for 48 h, resulted in significant increase in endoglin, a co-receptor for transforming growth factor (TGF-β3) pathway (Yinon et al, 2008). In the placentae of patients with severe preeclampsia at 31±2 weeks gestation, mRNA of the antioxidant protein glutathione reductase was reduced significantly, while that for thioredoxin peroxidase was increased (Vanderlelie et al, 2008). This suggests that oxidative stress may play a key role in the pathophysiology of the placentae in cases of preeclampsia.
In summary, the murine placenta appears to respond to hypoxia through several adaptive mechanisms. These include up-regulation of genes associated with erythropoiesis, increases in heme and iron metabolism, and in genes involved in proteolysis and peptidolysis. These varied responses suggest that the placenta responds by increasing its oxygen carrying capacity, increasing metabolic and antioxidant responses, and initiating tissue growth, turnover, and remodeling. Studies in both the human (Roh et al, 2005) and sheep (Krebs et al, 1997; Penninga & Longo, 1998) have demonstrated that the placenta undergoes multiple morphological and genetic changes in response to prolonged hypoxia. Hypoxic insults secondary to preeclampsia, maternal smoking, or exposure to high altitude can contribute to placental insufficiency and may lead to intrauterine growth restriction (Jones & Fox, 1980; Levi & Nelson, 2000; Reshetnikova et al, 1994; Spira et al, 1997). The exact mechanisms of these changes are not understood. Extracellular matrix remodeling, the modulation of apoptosis, altered cellular metabolism, and epigenetic changes all appear to be crucial steps in the physiological adaptations of the placenta to hypoxia. It is hoped that these, and other studies, will provide insights at the molecular level into these mechanisms and important clinical problems.
To What Extent is Placental Gene Expression Altered by Maternal Protein Restriction?
Several lines of evidence demonstrate that nutritional deprivation of the pregnant mother may have deleterious consequences for the progeny. For instance, a shocking “experiment” in humans was that during World War II of the “Hunger Winter” in Amsterdam and Western Holland from November 1944 until the Allied victory in May 1945. This tragedy provides useful lessons on the effects of caloric restriction/malnutrition on fetal development and disease prevalence in adulthood. During this seven month period, the caloric ration fell from ∼2400 to 400-800 calories per day, less than 25% of the recommended intake for adults. Although children, and to some extent pregnant and lactating women, received extra rations during the early part of this disastrous famine, they too suffered severe dietary deficiency (Roseboom et al, 2001a). In essence, upon reaching adulthood, the infants that were small at birth had significantly greater prevalence of cardiovascular disease, type II diabetes (Kyle & Prichard, 2006; Painter et al, 2005a; Roseboom et al, 2001a; 2001b; Stein & Susser, 1975; Stein et al, 2004), and mood and personality disorders (Godfrey, 1998). Those fetuses exposed to maternal caloric restriction in mid-gestation had a much greater incidence of pulmonary disease, including bronchitis (Lopuhaa et al, 2000), and renal disease as evidenced by microalbuminuria (Painter et al, 2005b). Females who were conceived during the famine also had a much higher prevalence of obesity as adults (Ravelli et al, 1999), and both males and females showed atherogenic lipid profiles (Roseboom et al, 2000). Concommently during WWII, the people of St. Petersburg and surrounding area of Russia were subjected to severe dietary restrictions due to interdiction of food supplies by the German army. The children born under these conditions were not only small for gestational age, but also developed health problems later in life (Neugebauer et al, 1999). However, records of the long-term sequelae of these individuals are not as clear as those in Holland (Lind, 1984; Ravelli et al, 1976). Importantly, the mechanisms of these in utero “programming” effects are unknown.
Epidemiologic data on the role of maternal nutrition in determining the long-term health of offspring derives largely from the studies of Barker and colleagues (Barker, 1995b; 2003; Barker & Clark, 1997; Barker & Osmond, 1986a; 1986b). Studies in several countries have correlated maternal dietary deficiencies that result in the newborn infant being small for gestational age, or growth restricted, with the prevalence of cardiovascular disease (Barker & Osmond, 1986b; Barker et al, 1989a), type 2 diabetes (Hales et al, 1991; Ravelli et al, 1998), and numerous other conditions in the adult. Maternal nutritional deprivation may influence placental growth and morphology, alter the hormonal milieu of the developing fetus, and cause subsequent cardiovascular, hormonal and behavioral consequences in the adult (Barker, 1992; 1994; 2003; Barker & Osmond, 1986a; 1986b; Barker et al, 1989; Gluckman et al, 2008).
The epidemiologic observations made in human subjects have been confirmed in animal models, and have led to speculation regarding the cellular mechanisms of changes in the placenta, and their effects on the developing fetus (Armitage et al, 2004; Hoet & Hanson, 1999). An important question is the extent to which these observed effects result from an overall caloric restriction, as opposed to a qualitative component in the diet that triggers the responses. Evidence from animal studies points to protein deprivation as a major factor in these defects. For example in the rat, the growth reducing effects of a low calorie diet can be reversed only by a dietary increase in protein levels, while vitamin supplements and caloric increases through carbohydrates did not reverse the effects observed (Hsueh et al, 1967). Other studies have revealed that dietary amino acid balance is a key mediator of some of the cardiovascular and metabolic effects observed in response to protein deprivation (Boujendar, 2003). Overall, the several studies indicate that nutritional deprivation, and protein restriction in particular, can have immediate deleterious effects on the placenta and the fetus, and may result in long-term sequelae that extend into adulthood.
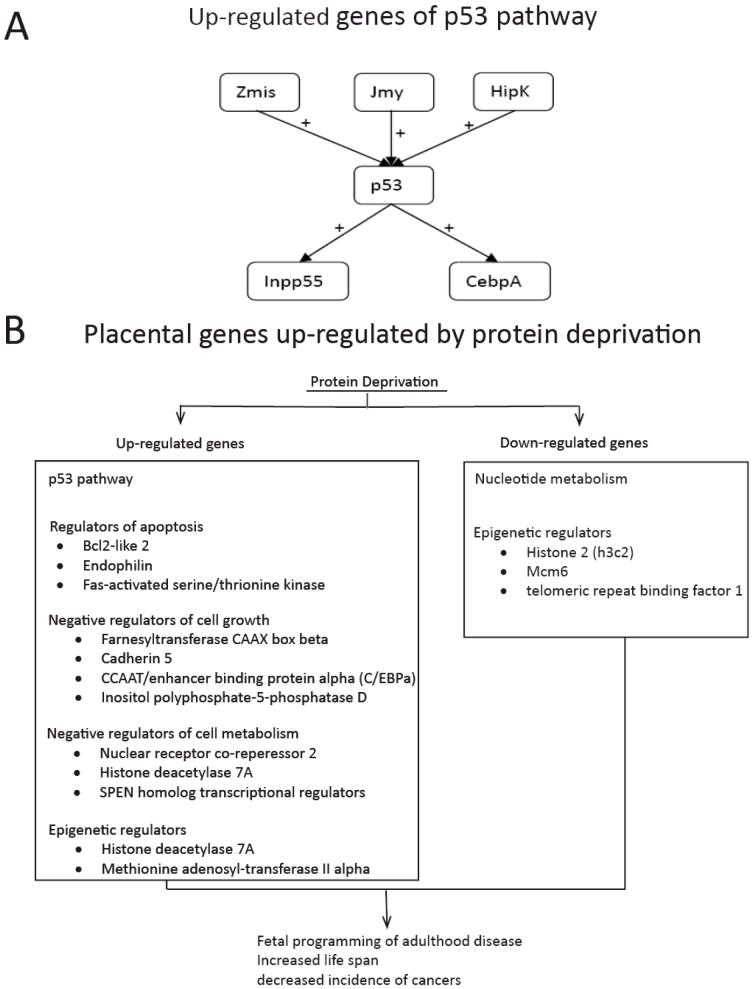
In a recent study in the mouse, we tested the hypothesis that moderate maternal protein deprivation would alter gene expression patterns in the placenta. We compared gene expression levels between normal placentas at E17.5, and those from pregnancies in which the mothers were exposed to seven days of protein deprivation (i.e., 10% protein by weight versus the 20% of normal chow) from E10.5 to E17.5. Of particular note, a number of genes involved in the p53 oncogene pathway were up-regulated. In addition to p53 itself, its positive regulators Zmis, Jmy, and Hipk2, as well as genes activated by p53 (Inpp5d, Cebpa), were induced (Figure 3A). These p53 pathway proteins are important regulators of cell growth and proliferation. This pathway serves as a G1 checkpoint, and arrests growth and/or induces apoptosis in response to cellular damage. Mutations in the p53 gene have been implicated in a number of cancers and other pathological processes (Ryan et al, 2001). Hipk2, an upstream regulator of p53, activates its transcriptional activity and pro-apoptotic activities through phosphorylation at Ser 46 (Hoffman et al, 2002). Cebpa, is a transcription factor induced by p53, and mediates some of the downstream effects of p53 activation (Yoon & Smart, 2004). Among the gene ontology classes most over-represented in the up-regulated group, we noted the mitogen-activated protein kinase pathway, regulators of apoptosis (Bcl2-like 2, p53, endophilin, Fas-activated serine/threonine kinase), negative regulators of cell growth (farnesyltransferase CAAX box beta, cadherin 5, CCAAT/enhancer binding protein (C/EBP) alpha, inositol polyphosphate-5-phosphatase D, p53), and negative regulators of cellular metabolism (nuclear receptor co-repressor 2, histone deacetylase 7A, SPEN homolog, transcriptional regulator). Acting in concert, activation of these genes could result in growth restriction during pregnancy (Fig. 3B). Among down-regulated genes, particularly striking were those related to nucleotide metabolism. For selected genes, we confirmed these results using qRT-PCR.
Figure 3.
A: p53 related genes up-regulated by protein restriction in the mouse placenta. + stands for induced
B: Proposed mechanisms of protein restriction induced long-term changes in gene expression.
Another potentially important finding, is that protein deprivation altered the expression of several genes involved in DNA methylation, histone acetylation, and epigenetic regulation of gene expression. The expression levels of histone deacetylase 7A and methionine adenosyltransferase II, alpha were elevated several fold. Histone acetylation triggers changes in chromatin structure, and regulates transcriptional availability of genes. In turn, histone deacetylation increases histone affinity for DNA, thereby repressing transcription (Bulger, 2005). Methionine adenosyltransferase II alpha synthesizes AdoMet the direct precursor used for DNA methylation by methyltransferases (Mao et al, 1998). Histone 2 (h3c2) is down-regulated, along with Mcm6 and telomeric repeat binding factor 1. These proteins contribute to DNA replication, stability, and structure (O’Connor et al, 2004; Yu et al, 2004). In the placenta of patients with preeclampsia, phosphorylation of extracellular signal-regulated kinase1/2 was significantly less frequent in the invasive trophoblasts, as compared to control (Moon et al, 2008). In another study, in placentas of preeclamptic patients, contrary to expectations, polymorphisms of several enzymes associated with oxidative stress (copper/zinc superoxide dismutase, manganese superoxide dismutase, glutathione-S-transferase, and others) did not differ from controls (Zhang et al, 2008). In contrast, in the placentas of patients with HELLP (Hemolysis, Elevated Liver Enzymes, Low Platelets) syndrome, genes encoding vascular endothelial growth factor receptor, leptin, and several other proteins, were up-regulated, as compared with the placenta of both normal control patients and those with preeclampsia (Buimer et al, 2008).
Because the various tissues and organ systems undergo critical, often brief, periods of growth and development during fetal life (Winick & Noble, 1966), “programming” as a consequence of maternal stress should not be unexpected, with insults to the developing organism having consequences later in life’s course. Studies in ruminants also have demonstrated that under-nutrition can have profound consequences for the fetus. In sheep, restricted maternal nutrition in early to mid-gestation was associated with an increase in placental weight, an increase in crown-rump length, and lower fetal to placental weight ratios (Heasman et al, 1998). Maternal under-nutrition also altered cardiovascular homeostatic regulation by the renin-angiotensin system, and exposed the lambs to higher levels of glucocorticoids (Edwards et al, 1999), and development of hypertension (Dodic et al, 2001). Protein restriction in bovines also resulted in an increase in placental weight and altered placental morphology (Perry et al, 1999).
Studies in rodents have shown similar effects. In rats, maternal protein restriction triggers hypertension in the pups in adulthood (Langley & Jackson, 1994), probably by augmentation of the pups’ renin-angiotensin system. In the spontaneously hypertensive rat placenta, several proteins, angiotensin receptor type I and inducible nitric oxide synthase (NOS), were up-regulated, while angiotensin converting enzyme and peroxisome proliferator-activated receptors alpha and gamma were downregulated (Raso et al, 2008). An alteration of placental glucocorticoid (GC) metabolism also was observed in placentae of rats fed a protein restricted diet, namely the activity of 11β-hydoxysteroid dehydrogenase that metabolizes glucocorticoids. This placental enzyme, which normally protects the pups from maternal glucocorticoid excess, was reduced in protein restricted rats (Langley-Evans et al, 1996), thus exposing the fetus to abnormally high GC concentrations. Elevated circulating cortisol concentrations, with modified responsiveness of the hypothalamic-pituitary-adrenal axis, and elevated mean arterial blood pressure with increased left ventricular wall thickness and mass, also were observed in guinea pigs in which the dam received only 70% of normal chow during either the first or second half of gestation. Some of these changes persisted in the F2 generation (Bertram et al, 2008). Another hormonal alteration in nutritionally deprived rat pups, was an increase in somatostatin expression in the periventricular nucleus. This led to much lower levels of growth hormone, and had deleterious effects on the growth of the pups post-partum (Huizinga et al, 2000). Fetal undernourishment also led to neuronal sequelae. The facial motor nucleus in pups was under-developed, resulting in decrease in the ability of pups to suckle and chew (Perez-Torrero et al, 2001). These observations also may relate to the epidemiologic findings, noted above, that abnormal antenatal nutrition may be associated with the development of schizophrenia and other mental illness.
In several animal models, in addition to the potential deleterious effects referenced above, a positive aspect of nutritional deprivation in the adult is that of prolonged lifespan and reduced cancer rates. A proposed mechanism for these benefits is that nutritional restriction without severe malnutrition inhibits cellular proliferation and induces apoptosis. This effect has been shown in mice lacking p53, in which -/- and +/- mutants have lowered spontaneous cancer rates when fed a calorically reduced, but otherwise complete, diet (Hursting et al, 2004). In the adult and aging animal, nutritional restriction has been shown to have beneficial effects that increased life span (Nikolich-Zugich & Messaoudi, 2005). A different picture has emerged in the fetus, however. As discussed above, caloric and protein deprivation have been shown to trigger fetal programming of adult disease, and lead to an increased prevalence of metabolic disorders in adulthood (Barker, 1995b; 1998; Barker & Clark, 1997). In the developing fetus, numerous animal studies have shown negative long-term effects of caloric and protein deprivation on the cardiovascular, renal, nervous system and metabolism (for review see McMillan & Robinson, 2005). A different form of nutritional compromise, that of placental restriction in sheep by removal of the endometrial caruncles in the nonpregnant ewe prior to mating, alters the expression of a number of genes associated with adipogenesis in adipose tissue of the fetus (Duffield et al, 2008). These findings emphasize the interrelation of placental development and its gene expression, to development of the fetus and its repertoire of gene expression.
To What Extent is Placental Gene Expression Altered by Maternal Caloric Excess?
Because maternal obesity poses an increased risk to the fetus during pregnancy, and has long-term consequences for the progeny, we tested the hypothesis that maternal caloric excess effects growth-related gene expression changes in the placenta. We fed female C57BL/65 mice a hypercaloric diet (20% fat, 38% sugar) or standard chow for six weeks prior to mating and throughout pregnancy. Near-term (E18), the dams were euthanized. We measured gene expression changes in the placenta, and performed pathway analysis on regulated genes.
Maternal overfeeding was associated with a two-fold increase in body fat mass, with several genes related to obesity, diabetes, DNA methylation, and the transforming growth factor-beta (TGF-β) pathway being differentially expressed (Poston et al, in preparation). The TGF-β superfamily comprises ∼30 growth and differential factors, including several TGF-βs, activins, inhibins, and other growth and cell cycle control factors (Goumans & Mummery, 2000; Kitisin et al, 2007; Massague et al, 2000; Roberts & Mishra, 2005; Roberts & Wakefield, 2003). Thus, our findings may have important implications for placental growth and epigenetic regulation. In other studies in mice, the chow was supplemented with methyl supplements (Wolff et al, 1998) or folic acid, vitamin B-12, choline, and betaine to enhance metabolism of cellular methyl donors (S-adenosylmethionine) (Waterland & Jirtle, 2003). These interventions resulted in altered coat color phenotype with concomitant increase in DNA methylation at the Avy locus. Conversely, in mice fed a methyl-donor-deficient diet that lacked folic acid, vitamin B-12, and choline the imprinted Igf2 gene was down-regulated with altered DNA methylation (Waterland et al, 2006a). Human studies also have demonstrated effects in the placenta on maternal dietary supplementation (Rush et al, 1984).
What is the Role Epigenetics in Placental Gene Expression?
During the course of life and reproduction, cells store information that has been handed down from their ancestors, and that will be transmitted to their descendents. For the most part, this “memory” is encoded in the sequence of nucleic acids that comprise the DNA of the genome, the genotype or entire compliment of genes that provides the stability and accurate heritability from generation to generation. Much traditional research has explored the combined effects of genetics and the environment in germline mutations of the coding and promoter regions of genes. In addition, cells can inherit and transmit information that is not part of the genomic sequence. This “epigenetic” [from Greek, upon, over, or beyond conventional genetic], cellular memory involves the heritable transmission of gene expression patterns that persist through cell division, but do not involve an alteration in DNA sequence. Epigenetic processes act in a cell specific, temporally-regulated manner to direct development, differentiation, organogensis, and related processes. Some have compared epigenetic mechanisms to the software to orchestrate and/or modulate the DNA hardware. One major class of epigenetic mechanisms termed “cytoplasmic”, is determined by cis-acting factors associated with DNA methylation and/or histone modification by acetylation/methylation/phosphorylation. DNA with accompanying histones are packaged in nucleosomes, the core of which contains an octamere of histone proteins. Four basic forms of histones (H2A, H2B, H3, and H4, as well as minor variants), are encircled by 146 base pairs of DNA (Finch et al, 1977); a fifth histone, H1, serves as a linker protein (Bernstein et al, 2007). The histone modifications noted above, and DNA methylation, confer a great increase in the regulatory capacity of each nucleosome, allowing specific functions such as DNA repair and gene activation to be modulated in the appropriate manner (Sarma & Reinberg, 2005). Enzymes critically associated with these nucleosomal modifications include: DNA methyltransferases (DMT), histone acetyltransferases (HAT), histone methyltransferase (HMT), histone deacetylases (HDAC), histone demethylases (HDM), and others (Dodd et al, 2007; Klose et al, 2006). It is by these nucleosomal modifications, with their influence on proximate genes, that genes may be regulated to affect phenotype by activity, chromatin structure, dosage compensation, and epigenetic memory, without changes in the nucleic acid code per se (Martin & Zhang, 2005; Wolffe & Matzke, 1999).
Epigenetic changes play a key role in normal cellular function, as well as the development and differentiation of various cell types (Drake & Walker, 2004; Monk, 1998; Rahnama et al, 2006; Reik, 2007). Examples include X-chromosome inactivation in female mammals, and genomic imprinting in which one parental allele is altered resulting in parent-of-origin, or random modification of gene transcription (Willard et al, 1993). The epigenetic state can be disrupted by maternal environmental influences such as hypoxia, protein deprivation, caloric excess, and so forth which alter DNA methylation or modify histones. Also importantly, a wide variety of environmental toxins, including low dose radiation and psychological stress, have been demonstrated to be important in epigenetic mechanisms (Dolinoy et al, 2007; Feinberg, 2007; Hertz-Piccioto et al, 2008; Jirtle & Skinner, 2007; Pryce et al, 2002; Szyf et al, 2007). Increasingly, epigenetic changes are being recognized to be of importance in ageing, and the development of cancer and other diseases. Despite the general understanding that DNA and/or histone modifications constitute a major factor in the pathogenesis of epigenesis, little is known of the molecular mechanisms whereby these chemical reactions/changes are regulated, and/or how they are transmitted between generations (Bird, 2007).
From an historical context, epigenetics has several facets. For the pioneer Edinburgh geneticist Conrad Hal Waddington (1905-1975), who coined the term, epigenetics was the study of how phenotypes arise from genotypes during development (Waddington, 1939; 1940; 1942; 1957). Epigenetics later was defined as heritable changes in gene expression not due to any alteration in DNA sequence (Holliday, 1987). In the mid-1970s, the concept of covalent chemical DNA modifications, including methylation was proposed to account for this phenomenon (Holliday & Pugh, 1975; Riggs, 1975). More recently, others have defined epigenesis as the study of mitotically and/or meiotically heritable changes in gene function without a change in DNA sequence (Dolinoy et al, 2007; Russo et al, 1996). As defined by Adrian Bird, epigenetics is “the structural adaptation of chromosomal regions so as to register, signal or perpetuate altered activity states” (Bird, 2007). The latter a definition focuses on chromosomes and genes, including those aspects such as DNA repair, cell-cycle phases, and those stable changes maintained from generation to generation.
In terms of perspective, epigenetics has a history that antedates understanding the genome and its regulation. The French biologist/zoologist and comparative anatomist who contributed greatly to classification of life forms, Jean-Baptiste Pierre Antoine de Mont, Chevalier de Lamarck (1774-1829), noted that organisms may inherit traits acquired during their parent’s lifetime. Although discredited by many, in part, because of his view that simple life forms arose from dead matter by spontaneous generation to become more complex as they were transformed into new species, Lamarck held that organisms inherit characteristics acquired during their parent’s lifetimes, evolving in a constant process of striving toward greater complexity and “perfection” (Lamarck, 1801; 1809; 1815-1822). Lamarck’s “First Law ...” stated that a change in the environment alters the needs of organisms in that environment, resulting in a change in behavior. Such behavioral alteration leads to greater or lesser use of a given structure with resultant increase or decrease in the size of that structure or organ. His “Second Law” stated that all such changes were heritable as a result (For instance, that a giraffe’s neck elongated as it ate from the highest leaves on a tree, and this feature would be seen in the next generation) (Haig, 2006; West-Eberhard, 2007). Of course, neo-Lamarckian biologists soundly reject such an idea. In more contemporary times, the views of the Russian biologist-agronomist, Trofim Denisovich Lysenko (1898-1976), gained considerable press for the agricultural “revolution” he promoted, in concert with Soviet collectivization policies. In essence, Lysenko held that acquired characteristics of a plant (or other organism) could be inherited by succeeding generations. An ideological-political creation, Lysenkoism held the study of classic genetics to be “bourgeois” or “fascist” pseudoscience. Lysenkoism invoked by biological determinists, as with the eugenics and scientific racism adopted by social constructivists, may be seen as the extremes to which political dogma can use science in promoting its propaganda (Soyfer, 2001).
As noted, environmental influences may have profound effects on gene regulation. This is clearly evident in the cells of multicellular organisms; although being genetically homogeneous, they are structurally and functionally heterogenesis. Many of these differences in gene expression arise during development and are retained through mitosis. Such stable, epigenetic changes, although heritable in the short term, are not a consequence of DNA mutation. Rather, as recent studies are demonstrating, to a great degree epigenesis appears to be a consequence of DNA methylation and histone modification. The term “epigenomics” has been applied to the study of altered chromatin structure, such as complex folding, altered nucleosome configuration, and related phenomena (Murrell et al, 2005). Importantly, several lines of evidence indicate that, in addition to maternal to fetal transfer, epigenetic modifications may be inherited across generations (Anway et al, 2005; 2006; Crews et al, 2007; Lane et al, 2003; Morgan et al, 1999; Pembray et al, 2006; Rakyan et al, 2003).
What are the Roles of DNA Methylation and Histone Modification in Placental Gene Expression?
As noted above, both DNA methylation and histone modification play important roles in development. These changes also may be important aspects of the ageing process and the development of cancer. In this review, we concentrate on the epigenetic influences of maternal diet, hypoxia, and related stress as observed in the placenta and fetal tissues, and that may have long-term consequences in the fetal origins of adult health and disease.
As with most phenomena of biology and life, the molecular mechanisms whereby genes are repressed or active in a stable manner are exceedingly complex. The best studied of these epigenetic modifications is that of DNA methylation, which first was suggested in 1975 by two groups (Holliday & Pugh, 1975; Riggs, 1975). This post-replication, covalent methylation occurs predominantly in repetitive genomic regions, on the 5′-carbon of cysteine residues that are followed by a guanine residue, i.e., “CpG methylation” which induces gene repression or “a silent chromatin state”, (The “p” in CpG refers to the phosphodiester bond between cytosine and guanine). Occurring at or around promoter regions, forming CpG islands, this can occur directly by inhibiting the binding of specific transcription factors, and indirectly by recruiting methyl-CPG binding proteins, with their associated repressive chromatin-remodeling activities (Razin & Riggs, 1980). A seeming paradox in this scenario, is that methylation of some specific DNA sequences may permit expression of neighboring genes. Both intrinsic factors and environmental/nutritional factors can determine the activity of methyltransferases upon which DNA methylation are dependent (Bestor, 2000). Because of the requirement for a high DNA synthesis rate during both gametogenesis and early embryogenesis, considerable activity in DNA methylation/demethylation patterning occurs during both this period of development, a time during which the cells are vulnerable to abnormal environmental factors. Nonetheless, nuances of molecular regulation of DNA methylation and histone modification during embryogenesis are beyond the scope of this synopsis, and a number of reviews on this topic are available (Bird, 2007c; Jaenisch, 1997; Jaenisch & Bird, 2003; Jones & Takai, 2001; Santos et al, 2002).
During the course of mammalian development, a wave of DNA demethylation occurs during cleavage, followed by genome-wide de novo methylation following implantation (Jaenish, 1997). Although the male genome is widely demethylated shortly after fertilization (Mayer et al, 2000; Oswald, 2000), the maternal genome is only partly demethylated with subsequent cleavage divisions (Li, 2002). In the gastrulating embryo, the extent of methylation is high, decreasing in various tissues during the course of differentiation (Ehrlich et al, 1982). For the developing embryo and fetus, the methylation/demethylation patterns while being of great significance, are enormously complex.
Additionally, gene expression is determined by the biochemical organization of the histones in the nucleosomes around which the DNA is wrapped. Several post-translational covalent modifications occur on the amino acids that constitute the histone N-terminal tails that modify their interaction with DNA and/or other nuclear proteins. Acetylation, methylation, phosphorylation and/or ubiquitination alone, or in combination play a key role in the regulation by repression or expression of contiguous genes (Jenuwein & Allis, 2001; Strahl & Allis, 2000; Turner, 2000). Again, the regulation of histone modification by acetylation and/or methylation is highly complex, has been shown to be specific for essentially every cell type, and may act with DNA methylation to constitute a system of cellular memory (Bird, 2007). The combination of the several epigenetic modifications of genes as well as non-coding sequences, the so-called “epigenome” or “epigenotype”, determine the extent to which a given gene is maintained repressed or active, and influences the phenotype at birth.
What is the Role of MicroRNAs in Placental Gene Expression?
MicroRNAs (miRNA) have emerged as important players in DNA methylation and post-transcriptional gene regulation (Lujambio et al, 2007; Saito et al, 2006). These are subtypes of small, non-coding RNA, which are 21-25 nucleotides in length. These miRNAs are capable of base pairing with mRNA, and fine-tuning gene expression during development and differentiation, by suppressing their expression in sequence specific manner. Following the discovery of first miRNA “lin4” in 1993, as a small temporal RNA (Lee et al, 1993), there has been enormous growth in this family, and identification of their targets. Although miRNAs are similar to small interfering RNA (siRNA) in their generation pathway and molecular characteristics, unlike siRNA, miRNA this does not degrade the target mRNA. Rather, they target the 3′ untranslated regions of mRNAs with which they share partial sequence complementarily, thereby silencing post-transcriptional gene translation. In this way, the biological system increases or decreases miRNA production to up- or down-regulate gene expression according to the developmental need, producing desired morphologic and physiological changes. Moreover, placental miRNA (miR-141, miR-149, miR-229-5p, and miR135b) are secreted in maternal plasma, and their concentration decreases significantly after parturition (Chim et al, 2008). This suggests that placental miRNA, in addition to regulating gene expression in placenta, may be playing an important role in maternal conditions with obscure etiology, such as preeclampsia or related hypertensive disorders. Studies reveal differential expression of miRNA (miR-210 and miR-182) in placenta from patients with preeclampsia and with small for gestational age newborn infants (Pineles et al, 2007). As must be evident, additional studies will be vital to examine and understand the complexity of placental genetic regulation, and their contribution to fetal and maternal health and disease.
What are the Human Correlates?
In several human population studies, it has been reported that the nutritional state of individuals may have phenotypic consequences for their grandchildren (Kaati et al, 2002; Lumley, 1992). An example of the role of diet in progeny DNA methylation status and phenotype is evident in patients with hyper-homocysteinemia (Ingrosso et al, 2003). This disorder is characterized by excess cellular adenosylhomocysteine, a potent inhibitor of S-adenosylmethionine-dependent methyltransferases. This suggests the possibility of significantly altered DNA methylation. In these patients, dietary supplementation with folate restored global methylation levels, as well as that of the imprinted IGF2-H19 locus (Ingrosso et al, 2003). Several earlier studies have indicated the developmental importance of folic acid as a dietary factory in utero, and the manner in which it modulates disease risks later in life (Torrens et al, 2006). It remains to be determined whether, as in the case of hyper-homocysteinaemia, these phenotypic effects occur through altered DNA methylation (McKay et al, 2004).
An optimal uterine environment has been shown to be essential for establishment and maintenance of embryonic epigenetic patterns (Vickaryous & Whitelaw, 2005). Because embryo culture and manipulation are employed in contemporary assisted reproductive technologies (ART), the question arises as to the extent to which ART or related procedures alter DNA methylation patterns, thereby inducing epigenetic changes in the developing organism (Brar et al, 2001; Feil, 2006; Khosla et al, 2001a; 2001b; Vickaryous & Whitelaw, 2005). Normally DNA methylation is confined to only one of the two parent alleles, thus imprinted gene loci allow minor alterations to be detected. An issue of great importance is the extent to which the chemical composition of culture medium, the duration of culture, or other factors, play a role in effecting changes in DNA methylation or histone modification (Doherty et al, 2000; Khosla et al, 2001a; 2001b; Mann et al, 2004; Young et al, 2001).
An additional consideration of importance, is the role of environmental toxins in producing alterations in the nucleosome with epigenetic consequences. An obvious example from mid-twentieth century is the ingestion of the estrogen-receptor agonist diethylstilbestrol (DES) by women in an attempt to reduce the risk of spontaneous abortion. This was followed by vaginal clear cell carcinoma (Swan, 2000), and altered limb development in the first generation, and deafness in the second generation (Stoll et al, 2003). Anticancer drugs and other environmental compounds may alter expression of specific genes, as well as the stress-related chaperone protein heat shock protein (HSP)-90, which may play a role in histone modification (Feil, 2006; Rutherford & Lindquist, 1998). A host of environmental contaminants including endocrine-disrupting chemicals are now known to demonstrate epigenetic effects on the germ line, and promote disease across several generations (Crews et al, 2007).
In humans, a number of factors, genetic and epigenetic, can influence placental/fetal growth, development, and long-term sequelae. Several hypotheses have been proposed to account for these phenomena. The “thrifty genotype” hypothesis, proposes the existence of genes that influence birthweight, and determining whether an infant will experience intrauterine growth restriction (Ong & Dunger, 2000; Prentice et al, 2005; Stöger, 2008). The “thrifty phenotype” hypothesis postulates that impairment of nutritional supply in early life results in permanent changes in tissue/organ function to conserve glucose, and prioritize development of the brain, heart, and other vital organs (Hales & Barker, 2001). A third hypothesis proposes that epigenetic alterations in gene expression, in the absence of altered DNA sequence, can be heritable, and may be reversible (Holness & Sugden, 2006). A challenge for our future is to develop strategies to negate the long-term consequences of these molecular alterations. A related issue of consequence is the epigenetic basis of dysregulation of gene expression in cancer. Rather than isolated instances, this may be a major factor in the seemingly increasing and intractable pandemic of this classes of diseases (for instance see Esteller, 2008; Gil-Yam et al, 2008; Palii & Robertson, 2007). Recognizing the importance of these vital issues, a recent National Institutes of Health initiative, as part of its “Roadmap” program, seeks applications to study the “Epigenomics of Human Health and Disease” (RFA-RM-08-017).
What are the Overall Perspectives, and Critical Questions to be Explored?
For the long-term well-being of an individual, both placental and fetal growth are essential. Thus, one would anticipate that profound inhibition of cellular growth at key time points during development would have grave long-term consequences for the embryo/fetus. This suggests that the timing of the treatment is a key determinant in the effect on the organism. Since their development, cDNA and oligo microarrays have proven to be powerful tools in the elucidation of gene expression patterns and discovery. In addition to examining cellular processes at the global gene expression level, these instruments have allowed analysis of numerous facets of normal growth and differentiation, as well as that occurring as a consequence of stress or malignant transformation. Of particular value, such studies allow analysis of gene expression by functional classes, as an aid in understanding pathways of cell metabolism, proliferation, senescence, and death.
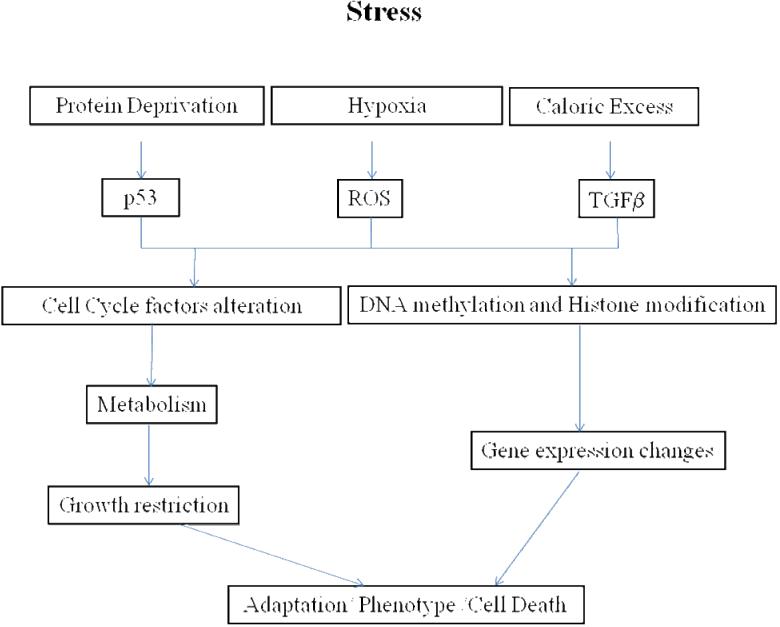
As with most tissues and organ systems, placental development and its response to stress remains a poorly understood process. Placental malfunction or failure accounts for numerous instances of fetal mortality (Cross et al, 2003), and may play an important role in the genesis of intrauterine growth restriction (Kingdom et al, 2000), as well as some maternal disease (Newstead et al, 2007). Numerous genes have been shown to be essential for placental function as an organ of respiratory gas and nutrient exchange, hormonal synthesis, immune function, and so forth. As is evident, maternal stresses, whether hypoxia, protein deprivation, caloric excess, or other, can result in profound alterations in placental gene expression patterns, and their consequences for growth, differentiation, and metabolism. Figure 4 presents in summary fashion established and potential pathways by which stress to the mother, whether hypoxia, protein deprivation, caloric excess, or others, can trigger changes in DNA methylation patterns and/or histone modification to effect alterations in the patterns of gene expression in the placenta and/or fetal organs.
Figure 4.
Proposed model of stress-induced long-term epigenetic changes in gene expression, adaptation, and phenotype.
Far from presenting a complete picture, the present review depicts but a fraction of what we need to know to understand more completely the molecular regulation of placental growth and development, and to lessen the ravages of placental dysfunction. A major challenge for the future will be to identify those portions of the genome particularly vulnerable to epigenetic modification which underlie states of health and diseases, and to understand the molecular mechanisms by which these changes occur. A few of the most obvious questions follow. How are the demonstrated gene expression profiles regulated? In terms of stress, what determines the individual patterns of expression, as opposed to the up- or down-regulation of those genes common to all stressors? What are the developmental stages/times of vulnerability to environmental, nutritional, or other stress? What environmental factors alter the epigenome in a deleterious manner, and what are their dose-response relations? What are the mechanisms by which DNA methylation and/or histone acetylation/methylation are regulated? To what extent do patterns of gene expression alterations in the placenta influence gene expression in the several fetal tissues/organs? To what extent can we use the findings of gene expression responses to stress, to gain an understanding of the phenomenon of epigenesis and its various manifestations. What is the role of epigenesis in normal development, and in the etiology of disease? How is it that epigenetic changes evident at the molecular level during embryonic/fetal life, do not become manifest in the adult organism for many years or decades? What is the relative importance of epigenetic, as opposed to genetic, changes for long-term sequelae? To what extent can we develop systems using molecular signatures/adducts to detect invidious interactions in early life? To what extent can an understanding of these issues provide us effective means to contain or counteract their influence and consequences? Can epigenetic biomarkers be identified that will allow disease detection at an early stage?
These are but a few of the vital questions that must be addressed in our pursuit to improve the lives and well being of mothers and infants, and the latter’s life as an adult. As biomedical scientists dedicated to betterment of the human condition, can we do less?
Summary
Successful placental development is crucial for optimal growth, development, maturation, and survival of the embryo/fetus into adulthood. Numerous epidemiologic and experimental studies demonstrate the profound influence of intrauterine environment on life, and the diseases to which one is subject as an adult. For the most part, these invidious influences, whether maternal hypoxia, protein or caloric deficiency or excess, and others, represent types of maternal stress. In the present review, we examine certain aspects of gene expression in the placenta as a consequence of maternal stressors. To examine these issues in a controlled manner, and in a species in which the genome has been sequenced, most of these reported studies have been performed in the mouse. Although each individual maternal stress is characterized by up- or down-regulation of specific genes in the placenta, functional analysis reveals some patterns of gene expression common to the several forms of stress. Of critical importance, these genes include those involved in DNA methylation and histone modification, cell cycle regulation, and related global pathways of great relevance to epigenesis and the developmental origins of adult health and disease.
Acknowledgement
We thank Brenda Kreutzer and Jimin Suh for their assistance in the preparation of this manuscript. This work was supported, in part, by USPHS grant HD-03807 to LDL.
REFERENCES
- Abu-Farha M, Niles J, Willmore WG. Erythroid-specific 5-aminolevulinate synthase protein is stabilized by low oxygen and proteasomal inhibition. Biochem Cell Biol. 2005;83:620–630. doi: 10.1139/o05-045. [DOI] [PubMed] [Google Scholar]
- Adams RH, Porras A, Alonso G, Jones M, Vintersten K, Panelli S, Valladares A, Perez L, Klein R, Nebreda AR. Essential role of p38alpha MAP kinase in placental but not embryonic cardiovascular development. Mol Cell. 2000;6:109–116. [PubMed] [Google Scholar]
- Adelman DM, Gertsenstein M, Nagy A, Simon MC, Maltepe E. Placental cell fates are regulated in vivo by HIF-mediated hypoxia responses. Genes Dev. 2000;14:3191–3203. doi: 10.1101/gad.853700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ain R, Dai G, Dunmore JH, Godwin AR, Soares MJ. A prolactin family paralog regulates reproductive adaptations to a physiological stressor. Proc Natl Acad Sci USA. 2004;101:16543–16548. doi: 10.1073/pnas.0406185101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ain R, Tash JS, Soares MJ. Prolactin-like protein-A is a functional modulator of natural killer cells at the maternal-fetal interface. Mol Cell Endocrinol. 2003;204:65–74. doi: 10.1016/s0303-7207(03)00125-4. [DOI] [PubMed] [Google Scholar]
- Allen MP, Nilsen-Hamilton M. Granzymes D, E, F, and G are regulated through pregnancy and by IL-2 and IL-15 in granulated metrial gland cells. J Immunol. 1998;161:2772–2779. [PubMed] [Google Scholar]
- Anway MD, Cupp AS, Uzumcu M, Skinner MK. Epigenetic transgenerational actions of endocrine disruptors and male fertility. Science. 2005;308:1466–1469. doi: 10.1126/science.1108190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anway MD, Skinner MK. Epigenetic transgenerational actions of endocrine disruptors. Endocrinology. 2006;147:S43–S49. doi: 10.1210/en.2005-1058. [DOI] [PubMed] [Google Scholar]
- Armitage JA, Khan IY, Taylor PD, Nathanielsz PW, Poston L. Developmental programming of the metabolic syndrome by maternal nutritional imbalance: how strong is the evidence from experimental models in mammals? J Physiol (Lond.) 2004;561:355–377. doi: 10.1113/jphysiol.2004.072009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aronow BJ, Richardson BD, Handwerger S. Microarray analysis of trophoblast differentiation: gene expression reprogramming in key gene function categories. Physiol Genomics. 2001;6:105–116. doi: 10.1152/physiolgenomics.2001.6.2.105. [DOI] [PubMed] [Google Scholar]
- Austgulen R, Isaksen CV, Chedwick L, Romundstad P, Vatten L, Craven C. Pre-eclampsia: associated with increased syncytial apoptosis when the infant is small-for-gestational-age. J Reprod Immunol. 2004;61:39–50. doi: 10.1016/j.jri.2003.10.001. [DOI] [PubMed] [Google Scholar]
- Barker DJP. Fetal and Infant Origins of Adult Disease. BMJ Publishing; London: 1992. [Google Scholar]
- Barker DJ. Fetal origins of coronary heart disease. BMJ. 1995a;311:171–174. doi: 10.1136/bmj.311.6998.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker DJ. Intrauterine programming of adult disease. Mol Med Today. 1995b;1:418–423. doi: 10.1016/s1357-4310(95)90793-9. [DOI] [PubMed] [Google Scholar]
- Barker DJ. In utero programming of chronic disease. Clin Sci (Lond) 1998;95:115–128. [PubMed] [Google Scholar]
- Barker D. The midwife, the coincidence, and the hypothesis. Brit Med J. 2003;327:1428–1430. doi: 10.1136/bmj.327.7429.1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker DJ. The developmental origins of chronic adult disease. Acta Paediatr Suppl. 2004;93:26–33. doi: 10.1111/j.1651-2227.2004.tb00236.x. [DOI] [PubMed] [Google Scholar]
- Barker DJP. Fetal origins of adult disease. In: Polin RA, Fox WW, Abman SH, editors. Fetal and Neonatal Physiology. 3rd Ed. Vol. 1. Saunders; Philadelphia, PA: 2004. pp. 160–165. [Google Scholar]
- Barker DJP. Mothers, Babies, and Health in Later Life. Churchill Livingstone; Edinburgh: 1994. [Google Scholar]
- Barker DJP. Mothers, Babies, and Health in Later Life. 2nd Ed. Churchill Livingstone; Edinburgh: 1998. [Google Scholar]
- Barker DJ, Clark PM. Rev Reprod. Vol. 2. 1997. Fetal undernutrition and disease in later life; pp. 105–112. [DOI] [PubMed] [Google Scholar]
- Barker DJ, Gluckman PD, Godfrey KM, Harding JE, Owens JA, Robinson JS. Fetal nutrition and cardiovascular disease in adult life. Lancet. 1993;341:938–941. doi: 10.1016/0140-6736(93)91224-a. [DOI] [PubMed] [Google Scholar]
- Barker DJ, Osmond C. Diet and coronary heart disease in England and Wales during and after the Second World War. J Epidemiol Community Health. 1986a;40:37–44. doi: 10.1136/jech.40.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker DJ, Osmond C. Infant mortality, childhood nutrition, and ischaemic heart disease in England and Wales. Lancet. 1986b;1:1077–1081. doi: 10.1016/s0140-6736(86)91340-1. [DOI] [PubMed] [Google Scholar]
- Barker DJP, Osmond C. Death rates from stroke in England and Wales predicted from past maternal mortality. BMJ. 1987;295:83–86. doi: 10.1136/bmj.295.6590.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker DJ, Osmond C, Golding J, Kuh D, Wadsworth ME. Growth in utero, blood pressure in childhood and adult life, and mortality from cardiovascular disease. BMJ. 1989a;298:564–567. doi: 10.1136/bmj.298.6673.564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker DJ, Winter PD, Osmond C, Margetts B, Simmonds SJ. Weight in infancy and death from ischaemic heart disease. Lancet. 1989b;2:577–580. doi: 10.1016/s0140-6736(89)90710-1. [DOI] [PubMed] [Google Scholar]
- Barker DJP, Sultant HY, Hanson MA, Rodeck CH, Spencer JAD, editors. Fetus and Neonate. Physiology and Clinical Applications. Vol. III. Growth. Cambridge Univ. Press; Cambridge: 1995. Fetal programming of human disease; pp. 255–276. [Google Scholar]
- Bernstein BE, Meissner A, Lander ES. The mammalian epigenome. Cell. 2007;128:669–681. doi: 10.1016/j.cell.2007.01.033. [DOI] [PubMed] [Google Scholar]
- Bertram C, Khan O, Ohri S, Phillips DI, Matthews SG, Hanson MA. Transgenerational effects of prenatal nutrient restriction on cardiovascular and hypothalamic-pituitary-adrenal function. J Physiol (Lond) 2008;586.8:2217–2229. doi: 10.1113/jphysiol.2007.147967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bestor TH. The DNA methyltransferases of mammals. Hum Mol Genet. 2000;9:2395–2402. doi: 10.1093/hmg/9.16.2395. [DOI] [PubMed] [Google Scholar]
- Bird A. Perceptions of epigenetics. Nature. 2007;447:396–398. doi: 10.1038/nature05913. [DOI] [PubMed] [Google Scholar]
- Boujendar S, Arany E, Hill D, Remacle C, Reusens B. Taurine supplementation of a low protein diet fed to rat dams normalizes the vascularization of the fetal endocrine pancreas. J Nutr. 2003;133:2820–2825. doi: 10.1093/jn/133.9.2820. [DOI] [PubMed] [Google Scholar]
- Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA, Lee TI, Levine SS, Wernig M, Tajonar A, Ray MK, Bell GW, Otte AP, Vidal M, Gifford DK, Young RA, Jaenisch R. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441:349–353. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- Brar AK, Handwerger S, Kessler CA, Aronow BJ. Gene induction and categorical reprogramming during in vitro human endometrial fibroblast decidualization. Physiol Genomics. 2001;7:135–148. doi: 10.1152/physiolgenomics.00061.2001. [DOI] [PubMed] [Google Scholar]
- Buimer M, Kijser R, Jebbink JM, Wehkamp D, van Kampen AHC, Boer K, van der Post JAM, Ris-Stalpers C. Seven placental transcripts characterize HELLP-syndrome. Placenta. 2008;29:444–453. doi: 10.1016/j.placenta.2008.02.007. [DOI] [PubMed] [Google Scholar]
- Bulger M. Hyperacetylated chromatin domains: lessons from heterochromatin. J Biol Chem. 2005;280:21689–21692. doi: 10.1074/jbc.R500004200. [DOI] [PubMed] [Google Scholar]
- Canli T, Qiu M, Omura K, Congdon E, Haas BW, Amin Z, Herrmann MJ, Constable RT, Lesch KP. Neural correlates of epigenesis. Proc Natl Acad Sci USA. 2006;103:16033–16038. doi: 10.1073/pnas.0601674103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cetin I, Foidart JM, Miozzo M, Raun T, Jansson T, Tsatsaris V, Reik W, Cross J, Hauguel-de-Mouson S, Illsley N, Kingdom J, Huppertz B. Fetal growth restriction: a workshop report. Placenta. 2004;25:753–757. doi: 10.1016/j.placenta.2004.02.004. [DOI] [PubMed] [Google Scholar]
- Challier JC, Uzan S. Le placenta humain et ses pathologies: l’oxygène en question. Med Sci (Paris) 2003;19:1111–1120. doi: 10.1051/medsci/200319111111. [DOI] [PubMed] [Google Scholar]
- Chen CP, Wang KG, Chen CY, Yu C, Chuang HC, Chen H. Altered placental syncytin and its receptor ASCT2 expression in placental development and pre-eclampsia. BJOG. 2006;113:152–158. doi: 10.1111/j.1471-0528.2005.00843.x. [DOI] [PubMed] [Google Scholar]
- Chim SS, Shing TK, Hung EC, Leung TY, Lau TK, Chiu RW, Lo YM. Detection and characterization of placental microRNAs in maternal plasma. Clin Chem. 2008;54:482–490. doi: 10.1373/clinchem.2007.097972. [DOI] [PubMed] [Google Scholar]
- Chinni E, Colaizzo D, Margaglione M, Rubini C, D’Ambrosio RL, Giuliani F, Di Vagno G, Grandone E. Correlation between factors involved in the local haemostasis and angiogenesis in full term human placenta. Thrombosis Res. 2008;122:376–382. doi: 10.1016/j.thromres.2007.12.016. [DOI] [PubMed] [Google Scholar]
- Chu S, DeRisi J, Eisen M, Mulholland J, Botstein D, Brown PO, Herskowitz I. The transcriptional program of sporulation in budding yeast. Science. 1998;282:699–705. doi: 10.1126/science.282.5389.699. [DOI] [PubMed] [Google Scholar]
- Collins FS, Barker AD. Mapping the cancer genome. Pinpointing the genes involved in cancer will help chart a new course across the complex landscape of human malignancies. Sci Am. 2007;296:50–57. [PubMed] [Google Scholar]
- Cooney CA, Dave AA, Wolff GL. Maternal methyl supplements in mice affect epigenetic variation and DNA methylation of offspring. J Nut. 2002;132:2393S–2400S. doi: 10.1093/jn/132.8.2393S. [DOI] [PubMed] [Google Scholar]
- Crews D, Gore AC, Hsu TS, Dangleben NL, Spinetta M, Schallert T, Anway MD, Skinner MK. Transgenerational epigenetic imprints on mate preference. Proc Natl Acad Sci USA. 2007;104:5942–5946. doi: 10.1073/pnas.0610410104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross JC. How to make a placenta: mechanisms of trophoblast cell differentiation in mice--a review. Placenta. 2005;26(Suppl A):S3–S9. doi: 10.1016/j.placenta.2005.01.015. [DOI] [PubMed] [Google Scholar]
- Cross JC, Baczyk D, Dobric N, Hemberger M, Hughes M, Simmons DG, Yamamoto H, Kingdom JC. Genes, development and evolution of the placenta. Placenta. 2003;24:123–130. doi: 10.1053/plac.2002.0887. [DOI] [PubMed] [Google Scholar]
- Daoud G, Amyot M, Rassart E, Masse A, Simoneau L, Lafond J. ERK1/2 and p38 regulate trophoblasts differentiation in human term placenta. J Physiol (Lond) 2005;566.2:409–423. doi: 10.1113/jphysiol.2005.089326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodd IB, Micheelsen MA, Sneppen K, Thon G. Theoretical analysis of epigenetic cell memory by nucleosome modification. Cell. 2007;129:813–822. doi: 10.1016/j.cell.2007.02.053. [DOI] [PubMed] [Google Scholar]
- Dodic M, Baird R, Hantzis V, Koukoulas I, Moritz K, Peers A, Wintour EM. Organs/systems potentially involved in one model of programmed hypertension in sheep. Clin Exp Pharmacol Physiol. 2001;28:952–956. doi: 10.1046/j.1440-1681.2001.03556.x. [DOI] [PubMed] [Google Scholar]
- Doherty AS, Mann MR, Tremblay KD, Bartolomei MS, Schultz RM. Differential effects of culture on imprinted H19 expression in the pre-implantation embryo. Biol Reprod. 2000;62:1526–1535. doi: 10.1095/biolreprod62.6.1526. [DOI] [PubMed] [Google Scholar]
- Dolinoy DC, Weidman JR, Jirtle RL. Epigenetic gene regulation: Linking early developmental environment to adult disease. Reprod Toxicol. 2007;23:297–307. doi: 10.1016/j.reprotox.2006.08.012. [DOI] [PubMed] [Google Scholar]
- Drake AJ, Walker BR. The intergenerational effects of fetal programming: non-genomic mechanisms for the inheritance of low birth weight and cardiovascular risk. J Endocrinol. 2004;180:1–16. doi: 10.1677/joe.0.1800001. [DOI] [PubMed] [Google Scholar]
- Duffield JA, Vuocolo T, Tellam R, Yuen BS, Muhlhausler BS, McMillen IC. Placental restriction of fetal growth decreases IGF1 and leptin mRNA expression in the perirenal adipose tissue of late gestation fetal sheep. Am J Physiol Regul Integr Comp Physiol. 2008;294:R1413–R1419. doi: 10.1152/ajpregu.00787.2007. [DOI] [PubMed] [Google Scholar]
- Edwards LJ, Simonetta G, Owens JA, Robinson JS, McMillen IC. Restriction of placental and fetal growth in sheep alters fetal blood pressure responses to angiotensin II and captopril. J Physiol (Lond) 1999;515.3:897–904. doi: 10.1111/j.1469-7793.1999.897ab.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrlich M, Gama-Sosa MA, Huang LH, Midgett RM, Kuo KC, McCune RA, Gehrke C. Amount and distribution of 5-methylcytosine in human DNA from different types of tissues of cells. Nucleic Acids Res. 1982;24:2709–2721. doi: 10.1093/nar/10.8.2709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteller M. Cancer epigenomics: DNA methylomes and histone-modification maps. Nat Rev Genet. 2007;8:286–298. doi: 10.1038/nrg2005. [DOI] [PubMed] [Google Scholar]
- Esteller M. Molecular origins of cancer. Epigenetics in cancer. N Engl J Med. 2008;358:1148–1159. doi: 10.1056/NEJMra072067. [DOI] [PubMed] [Google Scholar]
- Farese RV, Jr., Cases S, Ruland SL, Kayden HJ, Wong JS, Young SG, Hamilton RL. A novel function for apolipoprotein B: lipoprotein synthesis in the yolk sac is critical for maternal-fetal lipid transport in mice. J Lipid Res. 1996;37:347–360. [PubMed] [Google Scholar]
- Feil R. Environmental and nutritional effects on the epigenetic regulation of genes. Mutation Res. 2006;600:46–57. doi: 10.1016/j.mrfmmm.2006.05.029. [DOI] [PubMed] [Google Scholar]
- Feinberg AP. Phenotypic plasticity and the epigenetics of human disease. Nature. 2007;447:433–440. doi: 10.1038/nature05919. [DOI] [PubMed] [Google Scholar]
- Finch JT, Lutter LC, Rhodes D, Brown RS, Rushton B, Levitt M, Klug A. Structure of nucleosome core particles of chromatin. Nature. 1977;269:29–36. doi: 10.1038/269029a0. [DOI] [PubMed] [Google Scholar]
- Fratelli M, Goodwin LO, Ørom UA, Lombardi S, Tonelli R, Mengozzi M, Ghezzi P. Gene expression profiling reveals a signaling role of glutathione in redox regulation. Proc Natl Acad Sci USA. 2005;102:13998–14003. doi: 10.1073/pnas.0504398102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gal-Yam EN, Saito Y, Egger G, Jones PA. Cancer epigenetics: modifications, screening, and therapy. Annu Rev Med. 2008;59:267–280. doi: 10.1146/annurev.med.59.061606.095816. [DOI] [PubMed] [Google Scholar]
- Gasa R, Mrejen C, Leachman N, Otten M, Barnes M, Wang J, Chakrabarti S, Mirmira R, German M. Proendocrine genes coordinate the pancreatic islet differentiation program in vitro. Proc Natl Acad Sci USA. 2004;101:13245–13250. doi: 10.1073/pnas.0405301101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gheorghe CP, Mohan S, Oberg K, Longo LD. Gene expression patterns in the hypoxic murine placenta: a role in epigenesis? Reprod Sci. 2007;14:223–233. doi: 10.1177/1933719107302860. [DOI] [PubMed] [Google Scholar]
- Gheorghe C, Mohan S, Longo LD. Gene expression patterns in the developing murine placenta. J Soc Gynecol Invest. 2006;13:256–262. doi: 10.1016/j.jsgi.2006.02.007. [DOI] [PubMed] [Google Scholar]
- Gheorghe CP, Holweger J, Mohan S, Longo LD. Maternal protein deprivation and gene expression patterns in the murine placenta. Physiol Genomics. Submitted. [Google Scholar]
- Gimeno RE, Hirsch DJ, Punreddy S, Sun Y, Ortegon AM, Wu H, Daniels T, Stricker-Krongrad A, Lodish HF, Stahl A. Targeted deletion of fatty acid transport protein-4 results in early embryonic lethality. J Biol Chem. 2003;278:49512–49516. doi: 10.1074/jbc.M309759200. [DOI] [PubMed] [Google Scholar]
- Gluckman PD, Hanson MA, Cooper C, Thornburg KL. Effect of in utero and early-life conditions on adult health and disease. N Engl J Med. 2008;359:61–73. doi: 10.1056/NEJMra0708473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godfrey KM. Maternal regulation of fetal development and health in adult life. Eur J Obstet Gynecol Reprod Biol. 1998;78:141–150. doi: 10.1016/s0301-2115(98)00060-8. [DOI] [PubMed] [Google Scholar]
- Götz AA, Wittlinger S, Stefanski V. Maternal social stress during pregnancy alters immune function and immune cell numbers in adult male Long-Evans rat offspring during stressful life-events. J Neuroimmunol. 2007;185:95–102. doi: 10.1016/j.jneuroim.2007.01.019. [DOI] [PubMed] [Google Scholar]
- Goumans MJ, Mummery C. Funcitonal analysis of the TGFβ receptor/Smad pathway through gene ablation in mice. Int J Dev Biol. 2000;44:253–265. [PubMed] [Google Scholar]
- Green LR, Hanson MA. Programming of the fetal circulation. In: Polin RA, Fox WW, Abman SH, editors. Fetal and Neonatal Physiology. 3rd Ed. Vol. 1. Saunders; Philadelphia, PA: 2004. pp. 727–732. [Google Scholar]
- Gubin AN, Njoroge JM, Bouffard GG, Miller JL. Gene expression in proliferating human erythroid cells. Genomics. 1999;59:168–177. doi: 10.1006/geno.1999.5855. [DOI] [PubMed] [Google Scholar]
- Haig D. Weismann Rules! Epigenetics and the Lamarckian temptation. Biol Philosophy. 2007;22:415–428. [Google Scholar]
- Hales CN, Barker DJ. The thrifty phenotype hypothesis. Br Med Bull. 2001;60:5–20. doi: 10.1093/bmb/60.1.5. [DOI] [PubMed] [Google Scholar]
- Hales CN, Barker DJ, Clark PM, Cox LJ, Fall C, Osmond C, Winter PD. Fetal and infant growth and impaired glucose tolerance at age 64. BMJ. 1991;303:1019–1022. doi: 10.1136/bmj.303.6809.1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ham J, Tronick E. Infant resilience to the stress of the still-face. Infant and maternal psychophysiology are related. Ann NY Acad Sci. 2006;1094:297–301. doi: 10.1196/annals.1376.038. [DOI] [PubMed] [Google Scholar]
- Hanson MA, Gluckman PD. Developmental processes and the induction of cardiovascular function: conceptual aspects. J Physiol (Lond) 2005;565.1:27–34. doi: 10.1113/jphysiol.2004.082339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heasman L, Clarke L, Firth K, Stephenson T, Symonds ME. Influence of restricted maternal nutrition in early to mid gestation on placental and fetal development at term in sheep. Pediatr Res. 1998;44:546–551. doi: 10.1203/00006450-199810000-00013. [DOI] [PubMed] [Google Scholar]
- Heazel AEP, Lacey HA, Jones CJP, Huppertz B, Baker PN, Crocker IP. Effects of oxygen on cell turnover and expression of regulators of apoptosis in human placental trophoblast. Placenta. 2008;29:175–186. doi: 10.1016/j.placenta.2007.11.002. [DOI] [PubMed] [Google Scholar]
- Hemberger M. Epigenetic landscape required for placental development. Cell Mol Life Sci. 2007;64:2422–2436. doi: 10.1007/s00018-007-7113-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertz-Picciotto I, Park H-Y, Dostal M, Kocan A, Trnovec T, Sram R. Prenatal exposures to persistent and non-persistent organic compounds and effects on immune system development. Basic Clin Pharmacol Toxicol. 2008;102:146–154. doi: 10.1111/j.1742-7843.2007.00190.x. [DOI] [PubMed] [Google Scholar]
- Hirst CE, Buzza MS, Sutton VR, Trapani JA, Loveland KL, Bird PI. Perforin-independent expression of granzyme B and proteinase inhibitor 9 in human testis and placenta suggests a role for granzyme B-mediated proteolysis in reproduction. Mol Hum Reprod. 2001;7:1133–1142. doi: 10.1093/molehr/7.12.1133. [DOI] [PubMed] [Google Scholar]
- Hoang VM, Foulk R, Clauser K, Burlingame A, Gibson BW, Fusher SJ. Functional proteomics: examining the effects of hypoxia on the cytotrophoblast protein repertoire. Biochemistry. 2001;40:4077–4086. doi: 10.1021/bi0023910. [DOI] [PubMed] [Google Scholar]
- Hoek HW, Susser E, Buck KA, Lumey LH, Lin SP, Gorman JM. Schizoid personality disorder after prenatal exposure to famine. Am J Psychiatry. 1996;153:1637–1639. doi: 10.1176/ajp.153.12.1637. [DOI] [PubMed] [Google Scholar]
- Hoet JJ, Hanson MA. Intrauterine nutrition: its importance during critical periods for cardiovascular and endocrine development. J Physiol (Lond.) 1999;514:617–627. doi: 10.1111/j.1469-7793.1999.617ad.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann TG, Moller A, Sirma H, Zentgraf H, Taya Y, Droge W, Will H, Schmitz ML. Regulation of p53 activity by its interaction with homeodomain-interacting protein kinase-2. Nature Cell Biol. 2002;4:1–10. doi: 10.1038/ncb715. [DOI] [PubMed] [Google Scholar]
- Holliday R. The inheritance of epigenetic defects. Science. 1987;238:163–170. doi: 10.1126/science.3310230. [DOI] [PubMed] [Google Scholar]
- Holliday R, Pugh JE. DNA modification mechanisms and gene activity during development. Science. 1975;187:226–232. [PubMed] [Google Scholar]
- Holness MJ, Sugden MC. Epigenetic regulation of metabolism in children born small for gestational age. Curr Opin Clin Nutr Metab Care. 2006;9:482–488. doi: 10.1097/01.mco.0000232912.69236.e0. [DOI] [PubMed] [Google Scholar]
- Hsueh AM, Agustin CE, Chow BF. Growth of young rats after differential manipulation of maternal diet. J Nutr. 1967;91:195–200. doi: 10.1093/jn/91.2.195. [DOI] [PubMed] [Google Scholar]
- Huang ST, Vo KC, Lyell DJ, Faessen GH, Tulac S, Tibshirani R, Giaccia AJ, Giudice LC. Developmental response to hypoxia. FASEB J. 2004;18:1348–1365. doi: 10.1096/fj.03-1377com. [DOI] [PubMed] [Google Scholar]
- Huizinga CT, Oudejans CB, Steiner RA, Clifton DK, Delemarre-van de Waal HA. Effects of intrauterine and early postnatal growth restriction on hypothalamic somatostatin gene expression in the rat. Pediatr Res. 2000;48:815–820. doi: 10.1203/00006450-200012000-00019. [DOI] [PubMed] [Google Scholar]
- Hulshoff HE, Hoek HW, Susser E, Brown AS, Dingemans A, Schnack HG, van Haren NEM, Ramos LMP, Gispen-de Wied CC, Kahn RS. Prenatal exposure to famine and brain morphology in schizophrenia. Am J Psychiatry. 2000;157:1170–1172. doi: 10.1176/appi.ajp.157.7.1170. [DOI] [PubMed] [Google Scholar]
- Hursting SD, Lavigne JA, Berrigan D, Donehower LA, Davis BJ, Phang JM, Barrett JC, Perkins SN. Diet-gene interactions in p53-deficient mice: insulin-like growth factor-1 as a mechanistic target. J Nutr. 2004;134:2482S–2486S. doi: 10.1093/jn/134.9.2482S. [DOI] [PubMed] [Google Scholar]
- Ingrosso D, Cimmino A, Perna AF, Masella L, De Santo NG, De Bonis ML, Vacca M, D’Esposito M, D’Urso M, Galletti P, Zappia V. Folate treatment and unbalanced methylation and changes of allelic expression by hyper-homocysteinaemia in patients with uraemia. Lancet. 2003;17:1693–1699. doi: 10.1016/S0140-6736(03)13372-7. [DOI] [PubMed] [Google Scholar]
- Iyer VR, Eisen MB, Ross DT, Schuler G, Moore T, Lee JC, Trent JM, Staudt LM, Hudson J, Jr, Boguski MS, Lashkari D, Shalon D, Botstein D, Brown PO. The transcriptional program in the response of human fibroblasts to serum. Science. 1999;283:83–87. doi: 10.1126/science.283.5398.83. [DOI] [PubMed] [Google Scholar]
- Jablonka E, Lamb MJ. Epigenetic Inheritance and Evolution: The Lamarkian Dimension. Oxford Univ. Press; Oxford: 1995. [Google Scholar]
- Jablonka E, Lamb MJ. The changing concept of epigenetics. Ann NY Acad Sci. 2002;981:82–96. doi: 10.1111/j.1749-6632.2002.tb04913.x. [DOI] [PubMed] [Google Scholar]
- Jaenisch R. DNA methylation and imprinting: Why bother? Trends Genet. 1997;13:323–329. doi: 10.1016/s0168-9525(97)01180-3. [DOI] [PubMed] [Google Scholar]
- Jaenisch R, Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nature Genetics (Suppl) 2003;33:245–254. doi: 10.1038/ng1089. [DOI] [PubMed] [Google Scholar]
- Jansson T, Powell TL. Role of the placenta in fetal programming: underlying mechanisms and potential interventional approaches. Clin Sci. 2007;113:1–13. doi: 10.1042/CS20060339. [DOI] [PubMed] [Google Scholar]
- Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1079. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- Jirtle RL, Skinner MK. Environmental epigenomics and disease susceptibility. Nat Rev Genet. 2007;8:253–262. doi: 10.1038/nrg2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones CJ, Fox H. An ultrastructural and ultrahistochemical study of the human placenta in maternal pre-eclampsia. Placenta. 1980;1:61–76. doi: 10.1016/s0143-4004(80)80016-6. [DOI] [PubMed] [Google Scholar]
- Jones PA, Martienssen R. A blueprint for a Human Epigenome Project: the AACR Human Epigenome Workshop. Cancer Res. 2005;65:11241–11246. doi: 10.1158/0008-5472.CAN-05-3865. [DOI] [PubMed] [Google Scholar]
- Jones PA, Takai D. The role of DNA methylation in mammalian epigenetics. Science. 2001;293:1068–1070. doi: 10.1126/science.1063852. [DOI] [PubMed] [Google Scholar]
- Kaati G, Bygren LO, Edvinsson S. Cardiovascular and diabetes mortality determined by nutrition during parents’ and grandparents’ slow growth period. Eur J Hum Genet. 2002;10:682–688. doi: 10.1038/sj.ejhg.5200859. [DOI] [PubMed] [Google Scholar]
- Khosla S, Dean W, Reik W, Feil R. Culture of pre-implantation embryos and its long-term effects on gene expression and phenotype. Hum Reprod Update. 2001a;7:419–427. doi: 10.1093/humupd/7.4.419. [DOI] [PubMed] [Google Scholar]
- Khosla S, Dean W, Brown D, Reik R, Feil R. Culture of pre-implantation mouse embryos affects fetal development and the expression of imprinted genes. Biol Reprod. 2001b;64:918–926. doi: 10.1095/biolreprod64.3.918. [DOI] [PubMed] [Google Scholar]
- Kim JS, Romero R, Tarca A, LaJeunesse C, Han YM, Kim MJ, Suh YL, Draghici S, Mittal P, Gotsch F, Kusanovic JP, Hassan S, Kim CJ. Gene expression profiling demonstrates a novel role for fetal fibrocytes and the umbilical vessels in human fetoplacental development. J Cell Mol Med. doi: 10.1111/j.1582-4934.2008.00284.x. doi:10.1111/j.1582-4934.2008.00284.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingdom J, Huppertz B, Seaward G, Kaufmann P. Development of the placental villous tree and its consequences for fetal growth. Eur J Obstet Gynecol Reprod Biol. 2000;92:35–43. doi: 10.1016/s0301-2115(00)00423-1. [DOI] [PubMed] [Google Scholar]
- Kitisin K, Saha T, Blake T, Golestaneh N, Deng M, Kim C, Tang Y, Shetty K, Mishra B, Mishra L. TGFβ signaling in development. 2007:1–5. doi: 10.1126/stke.3992007cm1. stke.org/cgi/content/full/2007/399/cm1. [DOI] [PubMed]
- Klose RJ, Kallin EM, Zhang Y. JmjC-domain-containing proteins and histone demethylation. Nature Rev Genetics. 2006;7:715–727. doi: 10.1038/nrg1945. [DOI] [PubMed] [Google Scholar]
- Krebs C, Longo LD, Leiser R. Term ovine placental vasculature: comparison of sea level and high altitude conditions by corrosion cast and histomorphometry. Placenta. 1997;18:43–51. doi: 10.1016/s0143-4004(97)90070-9. [DOI] [PubMed] [Google Scholar]
- Kyle UG, Pichard C. The dutch Famine of 1944-1945 : A pathophsiological model of long-term consequences of wasting disease. Curr Opin Clin Nutrition Metabolic Care. 2006;9:388–394. doi: 10.1097/01.mco.0000232898.74415.42. [DOI] [PubMed] [Google Scholar]
- Lacroix MC, Bolifraud P, Durieux D, Pauloin A, Vidaud M, Kann G. Placental growth hormone and lactogen production by perifused ovine placental explants: regulation by growth hormone-releasing hormone and glucose. Biol Reprod. 2002;66:555–561. doi: 10.1095/biolreprod66.3.555. [DOI] [PubMed] [Google Scholar]
- Lamarck JBPA, de Monet . Système des aminaux sans vertèbris. Chez l’anteur; Paris: p. 1801. GM 215.5. [Google Scholar]
- Lamarck JBPA, de Monet . Philosophie zoologicque. Vol. 2. J.B. Baillère; Paris: p. 1809. GM 216. [Google Scholar]
- Lamarck JBPA, de Monet . Historoire naturelle des animaux sans vertèbris. Vol. 7. Verdière; Paris: pp. 1815–1822. GM 316. [Google Scholar]
- Lane N, Dean W, Erhardt S, Hajkova P, Surani A, Walter J, Reik W. Resistance of IAPs to methylation reprogramming may provide a mechanism for epigenetic inheritance in the mouse. Genesis. 2003;35:88–93. doi: 10.1002/gene.10168. [DOI] [PubMed] [Google Scholar]
- Langley SC, Jackson AA. Increased systolic blood pressure in adult rats induced by fetal exposure to maternal low protein diets. Clin Sci (Lond) 86:217–222. doi: 10.1042/cs0860217. discussion 121, 1994. [DOI] [PubMed] [Google Scholar]
- Langley-Evans SC, Phillips GJ, Benediktsson R, Gardner DS, Edwards CR, Jackson AA, Seckl JR. Protein intake in pregnancy, placental glucocorticoid metabolism and the programming of hypertension in the rat. Placenta. 1996;17:169–172. doi: 10.1016/s0143-4004(96)80010-5. [DOI] [PubMed] [Google Scholar]
- Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- Lee TI, Jenner RG, Boyer LA, Guenther MG, Levine SS, Kumar RM, Chevalier B, Johnstone SE, Cole MF, Isono K, Koseki H, Fuchikami T, Abe K, Murray HL, Zucker JP, Yuan B, Bell GW, Herbolsheimer E, Hannett NM, Sun K, Odom DT, Otte AP, Volkert TL, Bartel DP, Melton DA, Gifford DK, Jaenisch R, Young RA. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell. 2006;125:301–313. doi: 10.1016/j.cell.2006.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy R, Nelson DM. To be, or not to be, that is the question. Apoptosis in human trophoblast. Placenta. 2000;21:1–13. doi: 10.1053/plac.1999.0450. [DOI] [PubMed] [Google Scholar]
- Li M, Liu RH, Jiao LH, Wang WH. Synergetic effects of epidermal growth factor and estradiol on cytoplasmic maturation of porcine oocytes. Zygote. 2002;10:349–354. doi: 10.1017/s0967199402004094. [DOI] [PubMed] [Google Scholar]
- Lillycrop KA, Phillips ES, Torrens C, Hanson MA, Jackson AA, Burdge GC. Feeding pregnant rats a protein-restricted diet persistently alters the methylation of specific cytosines in the hepatic PPARalpha promoter of the offspring. Bri J Nutr. 2008;11:1–5. doi: 10.1017/S0007114507894438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lind T. Would more calories per day keep low birthweight at bay? Lancet. 1984;1:501–502. doi: 10.1016/s0140-6736(84)92860-5. [DOI] [PubMed] [Google Scholar]
- Liu YL, Ang SO, Weigent DA, Prchal JT, Bloomer JR. Regulation of ferrochelatase gene expression by hypoxia. Life Sci. 2004;75:2035–2043. doi: 10.1016/j.lfs.2004.03.027. [DOI] [PubMed] [Google Scholar]
- Longo LD, Pearce WJ. Invited Review: Fetal cerebrovascular acclimatization responses to high altitude, long-term hypoxia: A model for prenatal programming of adult disease? Am J Physiol Regul Integrative Comp Physiol. 2005;288:R16–R24. doi: 10.1152/ajpregu.00462.2004. [DOI] [PubMed] [Google Scholar]
- Lopuhaa CE, Roseboom TJ, Osmond C, Barker DJ, Ravelli AC, Bleker OP, van der Zee JS, van der Meulen JH. Atopy, lung function, and obstructive airways disease after prenatal exposure to famine. Thorax. 2000;55:555–561. doi: 10.1136/thorax.55.7.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lujambio A, Ropero S, Ballestar E, Fraga MF, Cerrato C, Setién F, Casado S, Suarez-Gauthier A, Sanchez-Cespedes M, Git A, Spiteri I, Das PP, Caldas C, Miska E, Esteller M. Genetic unmasking of an epigenetically silenced microRNA in human cancer cells. Cancer Res. 2007;67:1424–1429. doi: 10.1158/0008-5472.CAN-06-4218. [DOI] [PubMed] [Google Scholar]
- Lumey LH. Decreased birthweights in infants after maternal in utero exposure to the Dutch famine of 1944-1945. Paediatr Pernat Epidemiol. 1992;6:240–253. doi: 10.1111/j.1365-3016.1992.tb00764.x. [DOI] [PubMed] [Google Scholar]
- Lykke-Andersen K, Schaefer L, Menon S, Deng XW, Miller JB, Wei N. Disruption of the COP9 signalosome Csn2 subunit in mice causes deficient cell proliferation, accumulation of p53 and cyclin E, and early embryonic death. Mol Cell Biol. 2003;23:6790–6797. doi: 10.1128/MCB.23.19.6790-6797.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann MR, Lee SS, Doherty AS, Verona RI, Nolen LD, Schultz RM, Bartolomei MS. Selective loss of imprinting in the placenta following pre-implantation development in culture. Development. 2004;131:3727–3735. doi: 10.1242/dev.01241. [DOI] [PubMed] [Google Scholar]
- Mao Z, Liu S, Cai J, Huang ZZ, Lu SC. Cloning and functional characterization of the 5′-flanking region of human methionine adenosyltransferase 2A gene. Biochem Biophys Res Commun. 1998;248:479–484. doi: 10.1006/bbrc.1998.8965. [DOI] [PubMed] [Google Scholar]
- Martin C, Zhang Y. The diverse functions of histone lysine methylation. Nat Rev Mol Cell Biol. 2005;6:838–849. doi: 10.1038/nrm1761. [DOI] [PubMed] [Google Scholar]
- Massague J, lain SW, Lo RS. TGFβ signaling in growth control, cancer, and heritable disorders. Cell. 2000;103:295–309. doi: 10.1016/s0092-8674(00)00121-5. [DOI] [PubMed] [Google Scholar]
- Mayer W, Niveleau A, Walter J, Fundele R, Haaf T. Demethylation of the zygotic paternal genome. Nature. 2000;403:501–502. doi: 10.1038/35000656. [DOI] [PubMed] [Google Scholar]
- McKay JA, Williams EA, Mathers JC. Folate and DNA methylation during in utero development and aging. Biochem Soc Trans. 2004;32:1006–1007. doi: 10.1042/BST0321006. [DOI] [PubMed] [Google Scholar]
- McMillen IC, Robinson JS. Developmental origins of the metabolic syndrome: prediction, plasticity, and programming. Physiol Rev. 2005;85:571–633. doi: 10.1152/physrev.00053.2003. [DOI] [PubMed] [Google Scholar]
- Merlot E, Couret D, Otten W. Prenatal stress, fetal imprinting and immunity. Brain Behav Immun. 2008;22:42–51. doi: 10.1016/j.bbi.2007.05.007. [DOI] [PubMed] [Google Scholar]
- Monk M. Genomic imprinting. Genes Dev. 1988;2:921–925. doi: 10.1101/gad.2.8.921. [DOI] [PubMed] [Google Scholar]
- Moon KC, Park JS, Norwitz ER, Kim DI, Oh KJ, Park C-W, Jun JK, Syn HC. Expression of extracellular signal-regulated kinase1/2 and p38 mitogen-activated protein kinase in the invasive trophoblasts at the human placental bed. Placenta. 2008;29:391–395. doi: 10.1016/j.placenta.2008.02.001. [DOI] [PubMed] [Google Scholar]
- Morgan HD, Sutherland HGE, Martin DIK, Whitelaw E. Epigenetic inheritance at the agouti locus in the mouse. Nat Genet. 1999;23:314–318. doi: 10.1038/15490. [DOI] [PubMed] [Google Scholar]
- Morris CJ, Earl JR, Trenam CW, Blake DR. Reactive oxygen species and iron--a dangerous partnership in inflammation. Int J Biochem Cell Biol. 1995;27:109–122. doi: 10.1016/1357-2725(94)00084-o. [DOI] [PubMed] [Google Scholar]
- Murrell A, Rakyan VK, Beck S. From genome to epigenome. Hum Mole Genet. 2005;14(Suppl 1):R3–R10. doi: 10.1093/hmg/ddi110. [DOI] [PubMed] [Google Scholar]
- Mysore TB, Shinkel TA, Collins J, Salvaris EJ, Fisicaro N, Murray-Segal LJ, Johnson LE, Lepore DA, Walters SN, Stokes R, Chandra AP, O’Connell PJ, d’Apice AJ, Cowan PJ. Overexpression of glutathione peroxidase with two isoforms of superoxide dismutase protects mouse islets from oxidative injury and improves islet graft function. Diabetes. 2005;54:2109–2116. doi: 10.2337/diabetes.54.7.2109. [DOI] [PubMed] [Google Scholar]
- Nanney DL. Epigenetic control systems. Proc Natl Acad Sci USA. 1958;44:712–717. doi: 10.1073/pnas.44.7.712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson DM, Smith SD, Furesz TC, et al. Hypoxia reduces expression and function of system A amino acid transporters in cultured term human trophoblasts. Am J Physiol Cell Physiol. 2003;284:C310–C315. doi: 10.1152/ajpcell.00253.2002. [DOI] [PubMed] [Google Scholar]
- Neugebauer R, Hoek HW, Susser E. Prenatal exposure to wartime famine and development of antisocial personality disorder in early adulthood. JAMA. 1999;282:455–462. doi: 10.1001/jama.282.5.455. [DOI] [PubMed] [Google Scholar]
- Newstead J, von Dadelszen P, MaGee LA. Preeclampsia and future cardiovascular risk. Expert Rev Cardiovasc Ther. 2007;5:283–294. doi: 10.1586/14779072.5.2.283. [DOI] [PubMed] [Google Scholar]
- Nijland MJ, Ford SP, Nathanielsz PW. Prenatal origins of adult disease. Curr Opin Obstet Gynecol. 2008;20:132–138. doi: 10.1097/GCO.0b013e3282f76753. [DOI] [PubMed] [Google Scholar]
- Nikolich-Zugich J, Messaoudi I. Mice and flies and monkeys too: caloric restriction rejuvenates the aging immune system of non-human primates. Exp Gerontol. 2005;40:884–893. doi: 10.1016/j.exger.2005.06.007. [DOI] [PubMed] [Google Scholar]
- O’Connor MS, Safari A, Liu D, Qin J, Songyang Z. The human Rap1 protein complex and modulation of telomere length. J Biol Chem. 2004;279:28585–28591. doi: 10.1074/jbc.M312913200. [DOI] [PubMed] [Google Scholar]
- Ong KK, Dunger DB. Thrifty genotypes and phenotypes in the pathogenesis of type 2 diabetes mellitus. J Pediatr Endocrinol Metab. 2000;13(Suppl 6):1419–1424. doi: 10.1515/jpem-2000-s616. [DOI] [PubMed] [Google Scholar]
- Osmond C, Barker DJ, Winter PD, Fall CH, Simmonds SJ. Early growth and death from cardiovascular disease in women. Brit Med J. 1993;307:1519–1524. doi: 10.1136/bmj.307.6918.1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oswald J, Engemann S, Lane N, Mayer W, Olek A, Fundele R, Dean W, Reik W, Walter J. Active demethylation of the paternal genome in the mouse zygote. Curr Biol. 2000;10:475–478. doi: 10.1016/s0960-9822(00)00448-6. [DOI] [PubMed] [Google Scholar]
- Painter RC, Roseboom TJ, Bleker OP. Prenatal exposure to the Dutch famine and disease in later life: An overview. Reprod Toxicol. 2005a;20:345–352. doi: 10.1016/j.reprotox.2005.04.005. [DOI] [PubMed] [Google Scholar]
- Painter RC, Roseboom TJ, van Montfrans GA, Bossuyt PM, Krediet RT, Osmond C, Barker DJ, Bleker OP. Microalbuminuria in adults after prenatal exposure to the Dutch famine. J Am Soc Nephrol. 2005b;16:189–194. doi: 10.1681/ASN.2004060474. [DOI] [PubMed] [Google Scholar]
- Palii SS, Robertson KD. Epigenetic control of tumor suppression. Crit Rev Eukaryot Gene Expr. 2007;17:295–316. doi: 10.1615/critreveukargeneexpr.v17.i4.40. [DOI] [PubMed] [Google Scholar]
- Parham P. NK cells and trophoblasts: partners in pregnancy. J Exp Med. 2004;200:951–955. doi: 10.1084/jem.20041783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parodi A, Neasham D, Vineis P. Environment, population, and biology. A short history of modern epidemiology. Perspect Biol Med. 2006;49:357–368. doi: 10.1353/pbm.2006.0044. [DOI] [PubMed] [Google Scholar]
- Pembrey ME. Time to take epigenetic inheritance seriously. Eur J Hum Genet. 2002;10:669–671. doi: 10.1038/sj.ejhg.5200901. [DOI] [PubMed] [Google Scholar]
- Pembrey ME, Bygren LO, Kaati G, Edvinsson S, Northstone K, Sjöström M, Golding J, ALSPAC Study Team Sex-specific, male-line transgenerational responses in humans. Eur J Hum Genet. 2006;14:159–166. doi: 10.1038/sj.ejhg.5201538. [DOI] [PubMed] [Google Scholar]
- Penninga L, Longo LD. Ovine placentome morphology: effect of high altitude, long-term hypoxia. Placenta. 1998;19:187–193. doi: 10.1016/s0143-4004(98)90008-x. [DOI] [PubMed] [Google Scholar]
- Perez-Torrero E, Torrero C, Salas M. Effects of perinatal undernourishment on neuronal development of the facial motor nucleus in the rat. Brain Res. 2001;905:54–62. doi: 10.1016/s0006-8993(01)02500-8. [DOI] [PubMed] [Google Scholar]
- Perry VE, Norman ST, Owen JA, Daniel RC, Phillips N. Low dietary protein during early pregnancy alters bovine placental development. Anim Reprod Sci. 1999;55:13–21. doi: 10.1016/s0378-4320(98)00157-2. [DOI] [PubMed] [Google Scholar]
- Pineles BL, Romero R, Montenegro D, Tarca AL, Han YM, Kim YM, Draghici S, Espinoza J, Kusanovic JP, Mittal P, Hassan SS, Kim CJ. Distinct subsets of microRNAs are expressed differentially in the human placentas of patients with preeclampsia. Am J Obstet Gynecol. 2007;196:261.e1–e6. doi: 10.1016/j.ajog.2007.01.008. [DOI] [PubMed] [Google Scholar]
- Poston L, Taylor P, Gheorghe CP, Longo LD. Maternal caloric excess: placental TGFβ and DNA methylation pathway gene expression changes and epigenesis. In preparation. [Google Scholar]
- Prentice AM, Rayco-Solon P, Moore SE. Insights from the developing world: Thrifty genotypes and thrifty phenotypes. Proc Nutr Soc. 2005;64:153–161. doi: 10.1079/pns2005421. [DOI] [PubMed] [Google Scholar]
- Pryce CR, Ruedi-Bettschen D, Dettling AC, Feldon J. Early life stress: Long-term physiological impact in rodents and primates. News Physiol Sci. 2002;17:150–155. doi: 10.1152/nips.01367.2001. [DOI] [PubMed] [Google Scholar]
- Rahnama F, Shafiei F, Gluckman PD, Mitchell MD, Lobie PE. Epigenetic regulation of human trophoblastic cell migration and invasion. Endocrinology. 2006;147:5275–5283. doi: 10.1210/en.2006-0288. [DOI] [PubMed] [Google Scholar]
- Rakyan VK, Chong S, Champ ME, Cuthbert PC, Morgan HD, Luu KV, Whitelaw E. Transgenerational inheritance of epigenetic states at the murine AxinFu allele occurs after maternal and paternal transmission. Proc Natl Acad Sci USA. 2003;100:2538–2543. doi: 10.1073/pnas.0436776100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raso GM, Bianco G, Esposito E, Iacono A, Meli R, Autore G. Evaluation of placental protein modifications in normotensive and spontaneously hypertensive rats. Placenta. 2008;29:429–435. doi: 10.1016/j.placenta.2008.02.002. [DOI] [PubMed] [Google Scholar]
- Ravelli GP, Stein ZA, Susser MW. Obesity in young men after famine exposure in utero and early infancy. N Engl J Med. 1976;295:349–353. doi: 10.1056/NEJM197608122950701. [DOI] [PubMed] [Google Scholar]
- Ravelli AC, van der Meulen JH, Michels RP, Osmond C, Barker DJ, Hales CN, Bleker OP. Glucose tolerance in adults after prenatal exposure to famine. Lancet. 1998;351:173–177. doi: 10.1016/s0140-6736(97)07244-9. [DOI] [PubMed] [Google Scholar]
- Ravelli ACJ, van der Meulen JHP, Osmond C, Barker DJP, Bleker OP. Obesity at the age of 50 y in men and women exposed to famine prenatally. Am J Clin Nutr. 1999;70:811–816. doi: 10.1093/ajcn/70.5.811. [DOI] [PubMed] [Google Scholar]
- Razin A, Riggs AD. DNA methylation and gene function. Science. 1980;210:604–610. doi: 10.1126/science.6254144. [DOI] [PubMed] [Google Scholar]
- Reaume AG, de Sousa PA, Kulkarni S, Langille BL, Zhu D, Davies TC, Juneja SC, Kidder GM, Rossant J. Cardiac malformation in neonatal mice lacking connexin 43. Science. 1995;267:1831–1834. doi: 10.1126/science.7892609. [DOI] [PubMed] [Google Scholar]
- Redline RW, Chernicky CL, Tan HQ, Ilan J, Ilan J. Differential expression of insulin-like growth factor-II in specific regions of the late (post day 9.5) murine placenta. Mol Reprod Dev. 1993;36:121–129. doi: 10.1002/mrd.1080360202. [DOI] [PubMed] [Google Scholar]
- Reik W. Stability and flexibility of epigenetic gene regulation in mammalian development. Nature. 2007;447:425–432. doi: 10.1038/nature05918. [DOI] [PubMed] [Google Scholar]
- Reshetnikova OS, Burton GJ, Milovanov AP. Effects of hypobaric hypoxia on the fetoplacental unit: the morphometric diffusing capacity of the villous membrane at high altitude. Am J Obstet Gynecol. 1994;171:1560–1565. doi: 10.1016/0002-9378(94)90402-2. [DOI] [PubMed] [Google Scholar]
- Reynolds RM, Godfrey KM, Barker M, Osmond C, Phillips DIW. Stress responsiveness in adult life: influence of mother’s diet in late pregnancy. J Clin Endocrin Metab. 2007;92:2208–2210. doi: 10.1210/jc.2007-0071. [DOI] [PubMed] [Google Scholar]
- Riggs AD. X inactivation, differentiation, and DNA methylation. Cytogenet Cell Genet. 1975;14:9–25. doi: 10.1159/000130315. [DOI] [PubMed] [Google Scholar]
- Roberts A, Mishra L. Role of TGF-β in stem cells and cancer. Oncogene. 2005;24:5667. doi: 10.1038/sj.onc.1208924. [DOI] [PubMed] [Google Scholar]
- Roberts AB, Wakefield LM. The two faces of transforming growth factor β in carcinogenesis. Proc Natl Acad Sci USA. 2003;100:8621–8623. doi: 10.1073/pnas.1633291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roh CR, Budhraja V, Kim HS, Nelson DM, Sadovsky Y. Microarray-based identification of differentially expressed genes in hypoxic term human trophoblasts and in placental villi of pregnancies with growth restricted fetuses. Placenta. 2005;26:319–328. doi: 10.1016/j.placenta.2004.06.013. [DOI] [PubMed] [Google Scholar]
- Roseboom TJ, van der Meulen JH, Ravelli AC, Osmond C, Barker DJ, Bleker OP. Effects of prenatal exposure to the Dutch famine on adult disease in later life: an overview. Mol Cell Endocrinol. 2001a;185:93–98. doi: 10.1016/s0303-7207(01)00721-3. [DOI] [PubMed] [Google Scholar]
- Roseboom TJ, van der Meulen JH, Ravelli AC, Osmond C, Barker DJ, Bleker OP. Effects of prenatal exposure to the Dutch famine on adult disease in later life: an overview. Twin Res. 2001b;4:293–298. doi: 10.1375/1369052012605. [DOI] [PubMed] [Google Scholar]
- Roseboom TJ, van der Meulen JH, Osmond C, Barker DJ, Ravelli AC, Bleker OP. Plasma lipid profiles in adults after prenatal exposure to the Dutch famine. Am J Clin Nutr. 2000;72:1101–1106. doi: 10.1093/ajcn/72.5.1101. [DOI] [PubMed] [Google Scholar]
- Rossant J, Cross JC. Placental development: lessons from mouse mutants. Nat Rev Genet. 2001;2:538–548. doi: 10.1038/35080570. [DOI] [PubMed] [Google Scholar]
- Rush D, Kristal A, Navarro C, Chauhan P, Blanc W, Naeye R, Susser MW. The effects of dietary supplementation during pregnancy on placental morphology, pathology, and histomorphometry. Am J Clin Nutr. 1984;39:863–871. doi: 10.1093/ajcn/39.6.863. [DOI] [PubMed] [Google Scholar]
- Russo VEA, Martienssen RA, Riggs AD, editors. Epigenetic Mechanisms of Gene Regulation. Cold Spring Harbor Laboratory Press; Woodbury, NY: 1996. [Google Scholar]
- Rutherford SL, Lindquist S. Hsp90 as a capacitor for morphological evolution. Nature. 1998;396:336–342. doi: 10.1038/24550. [DOI] [PubMed] [Google Scholar]
- Ryan KM, Phillips AC, Vousden KH. Regulation and function of the p53 tumor suppressor protein. Curr Opin Cell Biol. 2001;13:332–337. doi: 10.1016/s0955-0674(00)00216-7. [DOI] [PubMed] [Google Scholar]
- Sadovsky Y, Wyatt SM, Collins L, Elchalal U, Kraus FT, Nelson DM. The use of needle biopsy for assessment of placental gene expression. Am J Obstet Gynecol. 2006;194:1137–1144. doi: 10.1016/j.ajog.2005.12.021. [DOI] [PubMed] [Google Scholar]
- Saito Y, Liang G, Egger G, Friedman JM, Chuang JC, Coetzee GA, Jones PA. Specific activation of microRNA-127 with downregulation of the proto-oncogene BCL6 by chromatin-modifying drugs in human cancer cells. Cancer Cell. 2006;9:435–443. doi: 10.1016/j.ccr.2006.04.020. [DOI] [PubMed] [Google Scholar]
- Santos F, Hendrich B, Reik W, Dean W. Dynamic reprogramming of DNA methylation in the early mouse embryo. Dev Biol. 2002;241:172–182. doi: 10.1006/dbio.2001.0501. [DOI] [PubMed] [Google Scholar]
- Sarma K, Reinberg D. Histone variants meet their match. Nat Rev Mol Cell Biol. 2005;6:139–149. doi: 10.1038/nrm1567. [DOI] [PubMed] [Google Scholar]
- Schmidt I. Metabolic diseases: the environment determines the odds, even for genes. News Physiol Sci. 2002;17:115–121. doi: 10.1152/nips.01380.2001. [DOI] [PubMed] [Google Scholar]
- Schorpp-Kistner M, Wang ZQ, Angel P, Wagner EF. JunB is essential for mammalian placentation. EMBO J. 1999;18:934–948. doi: 10.1093/emboj/18.4.934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber J, Riethmacher-Sonnenberg E, Riethmacher D, Tuerk EE, Enderich J, Bosl MR, Wegner M. Placental failure in mice lacking the mammalian homolog of glial cells missing, GCMa. Mol Cell Biol. 2000;20:2466–2474. doi: 10.1128/mcb.20.7.2466-2474.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selye H. A treatise based on the concepts of the GENERAL-ADAPTATION-SYNDROME and the DISEASES OF ADAPTATION. Acta Inc.; Montreal: 1950. The physiology and patheology of exposure to STRESS. [Google Scholar]
- Shekhawat P, Bennett MJ, Sadovsky Y, Nelson DM, Rakheja D, Strauss AW. Human placenta metabolizes fatty acids: implications for fetal fatty acid oxidation disorders and maternal liver diseases. Am J Physiol Endocrinol Metab. 2003;284:E1098–1105. doi: 10.1152/ajpendo.00481.2002. [DOI] [PubMed] [Google Scholar]
- Sibley CP, Coan PM, Ferguson-Smith AC, Dean W, Hughes J, Smith P, Reik W, Burton GJ, Fowden AL, Constancia M. Placental-specific insulin-like growth factor 2 (Igf2) regulates the diffusional exchange characteristics of the mouse placenta. Proc Natl Acad Sci USA. 2004;101:8204–8208. doi: 10.1073/pnas.0402508101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons DG, Cross JC. Determinants of trophoblast lineage and cell subtype specification in the mouse placenta. Dev Biol. 2005;284:12–24. doi: 10.1016/j.ydbio.2005.05.010. [DOI] [PubMed] [Google Scholar]
- Sitras V, Paulssen RH, Grønaas H, Vårtun Å, Acharya G. Gene expression profile in labouring and non-labouring human placenta near term. MHR-Basic Science of Reprod Med. 2008;14:61–65. doi: 10.1093/molehr/gam083. [DOI] [PubMed] [Google Scholar]
- Soleymanlou N, Jurisica I, Nevo O, Ietta F, Zhang X, Zamudio S, Post M, Caniggia I. Molecular evidence of placental hypoxia in preeclampsia. J Clin Endocrinol Metab. 2005;90:4299–4308. doi: 10.1210/jc.2005-0078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sood R, Zehnder JL, Druzin ML, Brown PO. Gene expression patterns in human placenta. PNAS. 2006;103:5478–5483. doi: 10.1073/pnas.0508035103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soyfer VN. The consequences of political dictatorhsip for Russian science. Nature Rev Genetics. 2001;2:723–729. doi: 10.1038/35088598. [DOI] [PubMed] [Google Scholar]
- Spira A, Philippe E, Spira N, Dreyfus J, Schwartz D. Smoking during pregnancy and placental pathology. Biomedicine. 1977;27:266–270. [PubMed] [Google Scholar]
- Stein AD, Zybert PA, van de Bor M, Lumey LH. Intrauterine famine exposure and body proportions at birth: the Dutch Hunger Winter. Int J Epidemiol. 2004;33:831–836. doi: 10.1093/ije/dyh083. [DOI] [PubMed] [Google Scholar]
- Stein Z, Susser M. The Dutch famine, 1944-1945, and the reproductive process. II. Interrelations of caloric rations and six indices at birth. Pediatr Res. 1975;9:76–83. doi: 10.1203/00006450-197502000-00004. [DOI] [PubMed] [Google Scholar]
- Stöger R. The thrifty epigenotype: An acquired and heritable predisposition for obesity and diabetes? Bioessays. 2008;30:156–166. doi: 10.1002/bies.20700. [DOI] [PubMed] [Google Scholar]
- Stoll C, Alembik Y, Dott B. Limb reduction defects in the first generation and deafness in the second generation of intrauterine exposed fetuses to diethylstilbe strol. Annales de genetique. 2003;46:459–465. doi: 10.1016/s0003-3995(03)00031-5. [DOI] [PubMed] [Google Scholar]
- Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- Susser ES, Lin SP. Schizophrenia after prenatal exposure to the Dutch Hunger Winter of 1944-1945. Arch Gen Psychiatry. 1992;49:983–988. doi: 10.1001/archpsyc.1992.01820120071010. [DOI] [PubMed] [Google Scholar]
- Swan SH. Intrauterine exposure to diethylstilbestrol: Long-term effects in humans. APMIS. 2000;108:793–804. doi: 10.1111/j.1600-0463.2000.tb00001.x. [DOI] [PubMed] [Google Scholar]
- Szyf M, Weaver I, Meaney M. Maternal care, the epigenome and phenotypic differences in behavior. Reprod Toxicol. 2007;24:9–19. doi: 10.1016/j.reprotox.2007.05.001. [DOI] [PubMed] [Google Scholar]
- Tanaka TS, Jaradat SA, Lim MK, Kargul GJ, Wang X, Grahovac MJ, Pantano S, Sano Y, Piao Y, Nagaraja R, Doi H, Wood WH, 3rd, Becker KG, Ko MS. Genome-wide expression profiling of mid-gestation placenta and embryo using a 15,000 mouse developmental cDNA microarray. Proc Natl Acad Sci USA. 2000;97:9127–9132. doi: 10.1073/pnas.97.16.9127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torrens C, Brawley L, Anthony FW, Dance CS, Dunn R, Jackson AA, Poston L, Hanson MA. Folate supplementation during pregnancy improves offspring cardiovascular dysfunction induced by protein restriction. Hypertension. 2006;47:982–987. doi: 10.1161/01.HYP.0000215580.43711.d1. [DOI] [PubMed] [Google Scholar]
- Trollman R, Klingmüller K, Schild RL, Rascher W, Dötsch J. Differential gene expression of somatotrophic and growth factors in response to in vivo hypoxia in human placenta. Am J Obstet Gynecol. 2007;197:601.e1–e6. doi: 10.1016/j.ajog.2007.04.008. [DOI] [PubMed] [Google Scholar]
- Tu MT, Grunau RE, Petrie-Thomas J, Haley DW, Weinberg J, Whitfield MF. Maternal stress and behavior modulate relationships between neonatal stress, attention, and basal cortisol at 8 months in preterm infants. Dev Psychobiol. 2007;49:150–164. doi: 10.1002/dev.20204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turek-Plewa J, Jagodzinski PP. The role of mammalian DNA methyltransferases in the regulation of gene expression. Cell Mol Biol Lett. 2005;10:631–647. [PubMed] [Google Scholar]
- Turner BM. Histone acetylation and an epigenetic code. Bioessays. 2000;22:836–845. doi: 10.1002/1521-1878(200009)22:9<836::AID-BIES9>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- Vadlamudi S, Kalhan SC, Patel MS. Persistence of metabolic consequences in the progeny of rats fed a HC formula in their early postnatal life. Am J Physiol. 1995;269:E731–E738. doi: 10.1152/ajpendo.1995.269.4.E731. [DOI] [PubMed] [Google Scholar]
- Vanderlelie J, Gude N, Perkins AV. Antioxidant gene expression in preeclamptic placentae: A preliminary investigation. Placenta. 2008;29:519–522. doi: 10.1016/j.placenta.2008.02.016. [DOI] [PubMed] [Google Scholar]
- Vickaryous N, Whitelaw E. The role of early embryonic environment on epigenotype and phenotype. Reprod Fertil Dev. 2005;17:335–340. doi: 10.1071/rd04133. [DOI] [PubMed] [Google Scholar]
- Waddington CH. Preliminary notes on the development of the wings in normal and mutant strains of drosophila. Proc Natl Acad Sci USA. 1939;25:299–307. doi: 10.1073/pnas.25.7.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waddington CH. Organisers and Genes. Cambridge University Press; Cambridge: 1940. [Google Scholar]
- Waddington CH. The epigenotype. Endeavour. 1942;1:18–20. [Google Scholar]
- Waddington CH. The Strategy of the Genes. Allen & Unwin; London: 1957. [Google Scholar]
- Waterland RA, Jirtle RL. Transposable elements: targets for early nutritional effects on epigenetic gene regulation. Mol Cell Biol. 2003;23:5293–5300. doi: 10.1128/MCB.23.15.5293-5300.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterland RA, Dolinoy DC, Lin J-R, Smith CA, Shi X, Tahiliani K. Maternal methyl supplements increase offspring DNA methylation at Axin (fused) Genesis. 2006a;44:401–406. doi: 10.1002/dvg.20230. [DOI] [PubMed] [Google Scholar]
- Waterland RA, Lin J-R, Smith CA, Jirtle RL. Post-weaning diet affects genomic imprinting at the insulin-like growth factor 2 (Igf2) locus. Hum Mol Genet. 2006b;15:705–716. doi: 10.1093/hmg/ddi484. [DOI] [PubMed] [Google Scholar]
- Watkins AJ, Wilkins A, Cunningham C, Perry VH, Seet MJ, Osmond C, Eckert JJ, Torrens C, Cagampang FRA, Cleal J, Gray WP, Hanson MA, Fleming TP. Low protein diet fed exclusively during mouse oocyte maturation leads to behavioural and cardiovascular abnormalities in offspring. J Physiol (Lond) 2008;586.8:2231–2244. doi: 10.1113/jphysiol.2007.149229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver ICG, Champagne FA, Brown SE, Dymov S, Sharma S, Meaney MJ, Szyf M. Reversal of maternal programming of stress responses in adult offspring through methyl supplementation: Altering epigenetic marking later in life. J Neurosci. 2005;25:11045–11054. doi: 10.1523/JNEUROSCI.3652-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wehby GL, Murray JC. The effects of prenatal use of folic acid and other dietary supplements on early child development. Matern Child Health J. 2008;12:180–187. doi: 10.1007/s10995-007-0230-3. [DOI] [PubMed] [Google Scholar]
- West-Eberhard MJ. Dancing with DNA and flirting with the ghost of Lamarck. Biol Philosophy. 2007;22:439–451. [Google Scholar]
- Wiemers DO, Shao LJ, Ain R, Dai G, Soares MJ. The mouse prolactin gene family locus. Endocrinology. 2003;44:313–325. doi: 10.1210/en.2002-220724. [DOI] [PubMed] [Google Scholar]
- Willard HF, Brown CJ, Carrel L, Hendrich B, Miller AP. Epigenetic and chromosomal control of gene expression: Molecular and genetic analysis of X chromosome inactivation. Cold Spring Harb Symp Quant Biol. 1993;58:315–322. doi: 10.1101/sqb.1993.058.01.037. [DOI] [PubMed] [Google Scholar]
- Winick M, Noble A. Quantitative changes in ribonucleic acids and protein during normal growth of rat placenta. Nature. 1966;212:34–35. doi: 10.1038/212034a0. [DOI] [PubMed] [Google Scholar]
- Wolffe GL, Kodell RL, Moore SR, Cooney CA. Maternal epigenetics and methyl supplements affect agouti gene expression in Avy/a mice. FASEB J. 1998;12:949–957. [PubMed] [Google Scholar]
- Wolffe AP, Matzke MA. Epigenetics: regulation through repression. Science. 1999;286:481–486. doi: 10.1126/science.286.5439.481. [DOI] [PubMed] [Google Scholar]
- Yamamoto H, Flannery ML, Kupriyanov S, Pearce J, McKercher SR, Henkel GW, Maki RA, Werb Z, Oshima RG. Defective trophoblast function in mice with a targeted mutation of Ets2. Genes Dev. 1998;12:1315–1326. doi: 10.1101/gad.12.9.1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yinon Y, Nevo O, Xu J, Many A, Rolfo A, Todros T, Post M, Caniggia I. Severe intrauterine growth restriction pregnancies have increased placental endoglin levels. Hypoxic regulation via transforming growth factor-β3. Am J Pathol. 2008;172:77–85. doi: 10.2353/ajpath.2008.070640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon K, Smart RC. C/EBPalpha is a DNA damage-inducible p53-regulated mediator of the G1 checkpoint in keratinocytes. Mol Cell Biol. 2004;24:10650–10660. doi: 10.1128/MCB.24.24.10650-10660.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young LE, Fernandes K, McEvoy TG, Butterwith SC, Gutierrez CG, Carolan C, Broadbent PJ, Robinson JJ, Wilmut I, Sinclair KD. Epigenetic changes in IGF2R is associated with fetal overgrowth after sheep embryo culture. Nature Genet. 2001;27:153–154. doi: 10.1038/84769. [DOI] [PubMed] [Google Scholar]
- Yu Z, Feng D, Liang C. Pairwise interactions of the six human MCM protein subunits. J Mol Biol. 2004;340:1197–1206. doi: 10.1016/j.jmb.2004.05.024. [DOI] [PubMed] [Google Scholar]
- Zamudio S. The placenta at high altitude. High Alt Med Biol. 2003;4:171–191. doi: 10.1089/152702903322022785. [DOI] [PubMed] [Google Scholar]
- Zhang EG, Burton GJ, Smith SK, Charnock-Jones DS. Placental vessel adaptation during gestation and to high altitude: changes in diameter and perivascular cell coverage. Placenta. 2002;23:751–762. doi: 10.1016/s0143-4004(02)90856-8. [DOI] [PubMed] [Google Scholar]
- Zhang J, Masiocchi M, Lewis D, Sun W, Liu A, Wang Y. Placental anti-oxidant gene polymorphisms, enzyme activity, and oxidative stress in preeclampsia. Placenta. 2008;29:439–443. doi: 10.1016/j.placenta.2008.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H, Jackson T, Bunn HF. Detecting and responding to hypoxia. Nephrol Dial Transplant. 2002;17(Suppl 1):3–7. doi: 10.1093/ndt/17.suppl_1.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zollers WG, Jr, Babischkin JS, Pepe GJ, Albrecht ED. Developmental regulation of placental insulin-like growth factor (IGF)-II and IGF-binding protein-1 and -2 messenger RNA expression during primate pregnancy. Biol Reprod. 2001;65:1208–1214. doi: 10.1095/biolreprod65.4.1208. [DOI] [PubMed] [Google Scholar]