Abstract
Plasmodium falciparum is the pathogen responsible for over 90% of human deaths from malaria1. Therefore, it has been the focus of a considerable research initiative, involving the complete DNA sequencing of the genome2, large-scale expression analyses3,4, and protein characterization of its life-cycle stages5. The Plasmodium genome sequence is relatively distant from those of most other eukaryotes, with more than 60% of the 5,334 encoded proteins lacking any notable sequence similarity to other organisms2. To systematically elucidate functional relationships among these proteins, a large two-hybrid study has recently mapped a network of 2,846 interactions involving 1,312 proteins within Plasmodium6. This network adds to a growing collection of available interaction maps for a number of different organisms, and raises questions about whether the divergence of Plasmodium at the sequence level is reflected in the configuration of its protein network. Here we examine the degree of conservation between the Plasmodium protein network and those of model organisms. Although we find 29 highly connected protein complexes specific to the network of the pathogen, we find very little conservation with complexes observed in other organisms (three in yeast, none in the others). Overall, the patterns of protein interaction in Plasmodium, like its genome sequence, set it apart from other species.
With the recent accumulation of protein interaction maps in public databases, cross-species comparisons are becoming critical for analysing the large networks formed by these interactions to delineate protein function and evolution7. At a fundamental level, protein networks can be compared to identify ‘interologues’—that is, interactions that are conserved across species8. Beyond the individual comparison of interactions, methods such as PathBLAST (refs 9, 10) create a global alignment between protein networks to identify dense clusters of conserved interactions, suggestive of protein complexes. Such comparative approaches are important because they can tease conserved components of cellular machinery out of a highly connected network and increase overall confidence in the underlying interaction measurements.
We compared the protein–protein interaction network of Plasmodium reported by LaCount et al. (ref. 6) to protein networks for the budding yeast Saccharomyces cerevisiae11, the nematode worm Caenorhabditis elegans12, the fruitfly Drosophila melanogaster13 and the bacterial pathogen Helicobacter pylori14. Surprisingly, the pairwise alignment of these networks using PathBLAST (ref. 9) revealed that Plasmodium had only three conserved complexes with yeast (Fig. 1a–c), and no conserved complexes with any of the other organisms examined. However, yeast, fly and worm shared substantial numbers of conserved complexes with each other (Fig. 2a); for instance, yeast and fly had the highest degree of conservation with 61 conserved complexes.
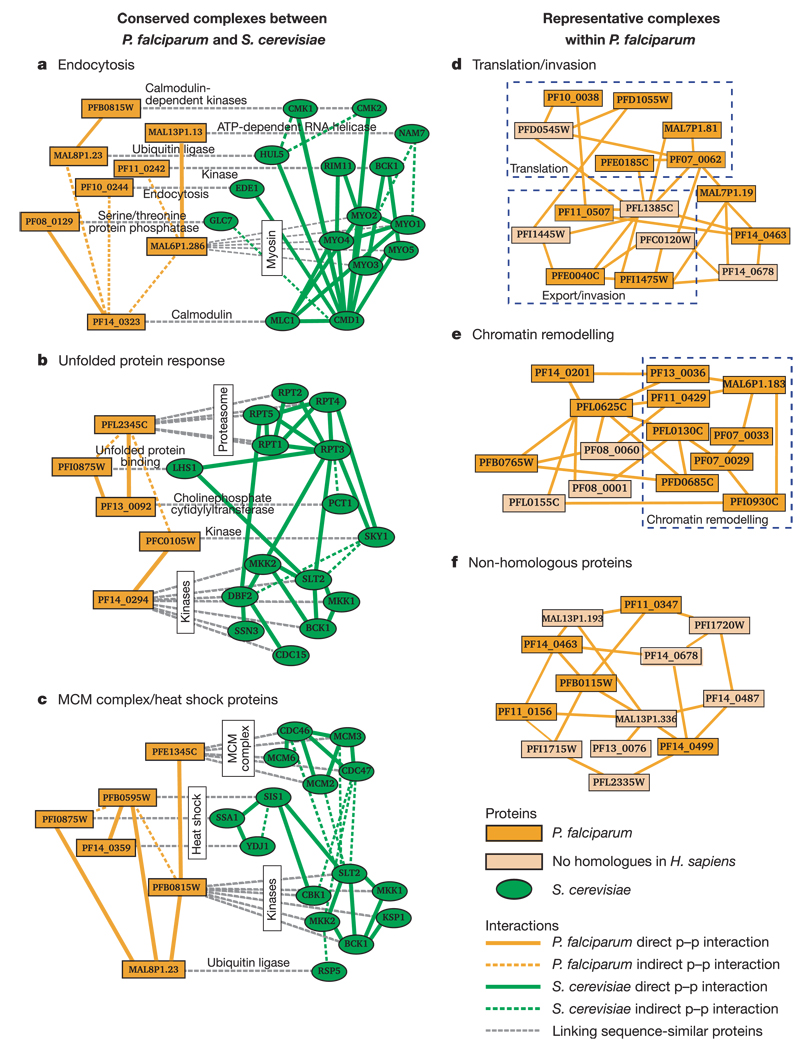
Figure 1. Conserved and distinct complexes within P. falciparum.
a–c, The three conserved complexes identified between P. falciparum and S. cerevisiae. Orange versus green nodes correspond to P. falciparum versus S. cerevisiae proteins. Solid links represent direct protein–protein (p–p) interactions, while dashed links represent indirect protein–protein interactions mediated by one other protein. Grey dashed lines connect sequence-similar proteins across the two species. MCM, minichromosome maintenance. d–f, Three representative complexes found within the P. falciparum network only (out of 29 total complexes; see Supplementary Table 7). Cream-coloured nodes denote Plasmodium proteins without human homologues (Homo sapiens; using a permissive BLAST E value threshold ≤1 × 10−2 to allow for distant homologues). All figures were drawn using the software Cytoscape (http://www.cytoscape.org).
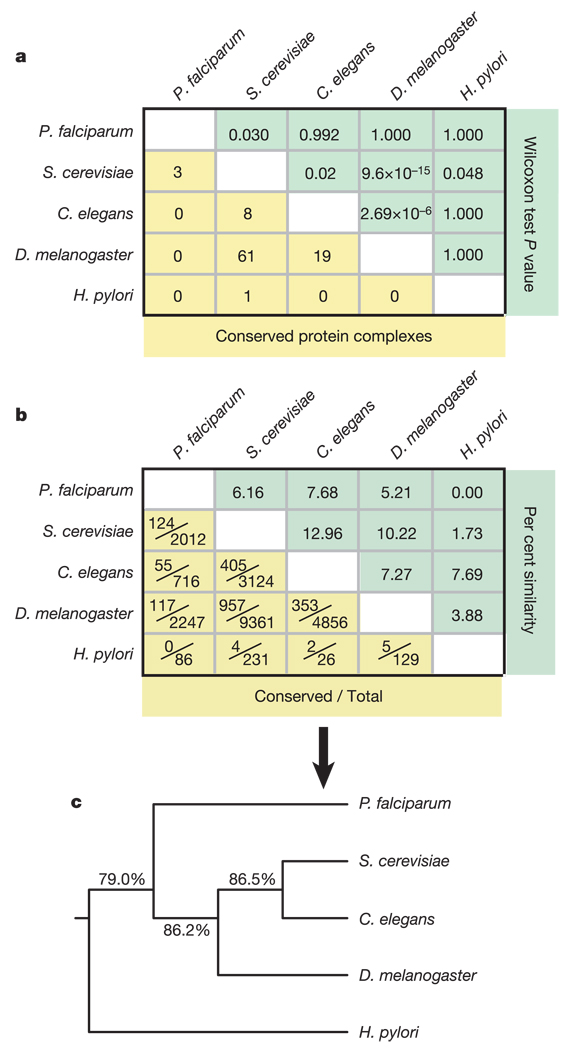
Figure 2. Network similarity across the five organisms.
a, The results of all pairwise PathBLAST comparisons. The number of conserved complexes is shown for each pair of species (yellow). The Wilcoxon rank-sum test P value (green) represents the significance of the distribution of all complex scores versus the distribution of complex scores found in equivalent random networks. b, The interaction-by-interaction similarity between networks is reported as both fractional values (yellow) and percentages (green). c, A phylogenetic tree constructed using these similarities. Percentages indicate the reproducibility of each branch during bootstrap analysis.
The relatively low similarity between the Plasmodium protein network and those of the other eukaryotes suggested that it encodes important functional differences worthy of further investigation. Alternatively, it was possible that differences in the number of complexes were related to network size. Thus, in addition to searching for conserved complexes, we investigated whether the observed similarities and differences were reflected in the probability of conservation of each protein interaction individually (Fig. 2b). For each pair of species, a protein–protein interaction was considered ‘conserved’ if both proteins had homologues that interacted in the opposite species (BLAST E value ≤1 × 10−4, normalized for genome size). A global pairwise similarity metric was then defined as the overall fraction of interactions that were conserved, restricted to proteins with at least one homologue in the opposite species.
Figure 2c expresses the pairwise interaction similarities as a phylogenetic tree drawn using the method of Kitsch15. This tree was relatively robust to sampling errors as determined by bootstrap analysis: 86.2% of trials placed Plasmodium as an outgroup relative to yeast, worm and fly. Among the three model eukaryotes, yeast and worm were closest on the basis of interaction similarity (Fig. 2b), while yeast and fly were closest on the basis of conserved complexes (Fig. 2a). This discrepancy was probably due to network size or coverage. Nonetheless, the particular phylogenetic placement of Plasmodium was consistent across both analyses, and also agrees with the accepted taxonomical relationships among these organisms as established by morphological and sequence comparisons2.
Another possibility for the low similarity of the Plasmodium protein network to the other species was that its interaction network had been measured predominantly among proteins expressed in the asexual stages of the parasite’s life cycle (see ref. 6). There are two ways in which this sampling could have affected network similarity. First, it was possible that a high (or low) level of messenger RNA expression increases (or decreases) the number of interactions identified for the corresponding proteins, and thus alters the topology of the Plasmodium network relative to the other species. However, as shown in Supplementary Fig. 1, we found no correlation between expression level in any stage and the number of protein interactions. Second, it was possible that proteins from asexual stages tended to have lower similarity across species than proteins from other stages of the Plasmodium life cycle. However, we found that the Plasmodium interaction set was enriched for proteins with homologues in the other species, and that the protein interaction networks from all five organisms were enriched for yeast homologues in particular (Table 1; Supplementary Table 1). Such enrichment was observed even in worm, for which baits were explicitly selected to be non-homologous to yeast12. This effect requires further study, but might indicate a bias of the yeast two-hybrid system in measuring interactions among yeast homologues, because all two-hybrid constructs must be expressible in the yeast cell.
Table 1.
Topological properties of protein interaction networks
| Organism | No. of interactions |
Proteins covered |
Average degree |
Average shortest path |
Average clustering coefficient* |
No. of yeast homologues† (P value) |
No. of single-species complexes |
|---|---|---|---|---|---|---|---|
| S. cerevisiae (DIP)‡ | 14,319 | 4,389 | 6.53 | 4.12 | 0.193 | – | 145 |
| S. cerevisiae (Uetz)‡ | 1,449 | 1,345 | 2.16 | 6.95 | 0.049 | – | 66 |
| P. falciparum | 2,846 | 1,312 | 4.35 | 4.20 | 0.032 | 286 (2 × 10−133) | 29 |
| D. melanogaster | 20,720 | 7,038 | 5.89 | 4.70 | 0.019 | 2,429 (2 × 10−205) | 296 |
| C. elegans | 3,926 | 2,718 | 2.89 | 5.10 | 0.031 | 673 (4 × 10−10) | 12 |
| H. pylori | 1,465 | 732 | 4.00 | 4.15 | 0.063 | 143 (1 × 10−2) | 21 |
The clustering coefficient measures local density of the network around a protein and is computed as previously described16.
Yeast homologues are determined using a conservative BLAST E value threshold of ≤1 × 10−10. The P values score the significance of enrichment for yeast homologues within the set of proteins covered by each interaction network, using the hypergeometric test. These enrichments are significant over a broad range of E value thresholds (data not shown).
Unlike other networks that are generated from single two-hybrid studies, the network of yeast interactions in the Database of Interacting Proteins (DIP)11 consists of many experiments and experimental types. A separate analysis is included considering only the data from a single two-hybrid screen by Uetz et al. (ref. 29).
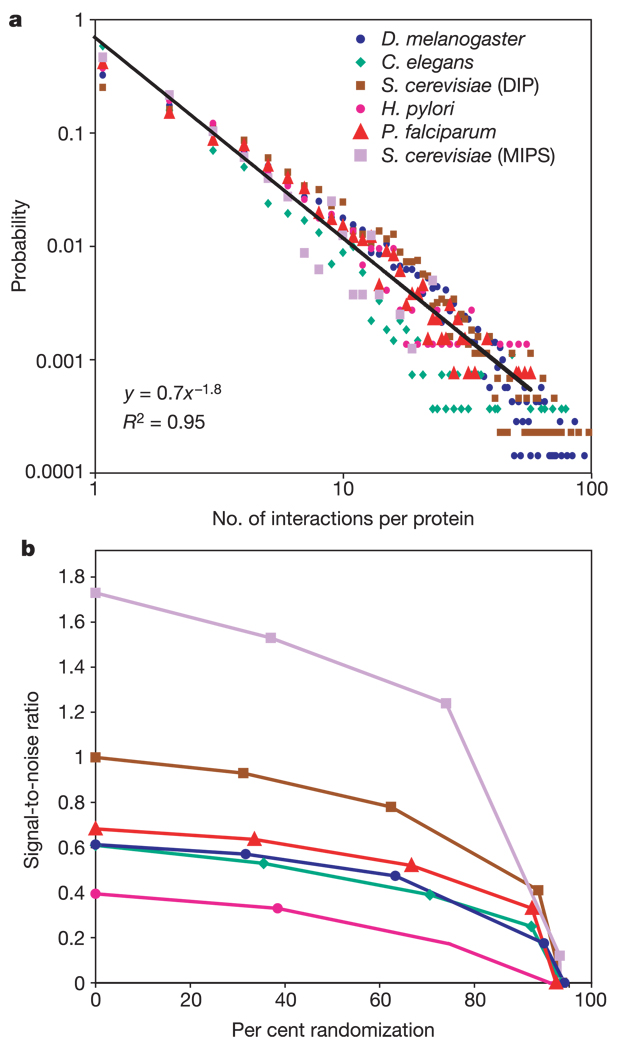
A final possibility for the low level of conservation between the Plasmodium protein network and those of the other organisms was that the Plasmodium network might have a substantially higher proportion of false-positive interactions relative to the networks of yeast, fly and worm. Lacking a ‘gold standard’ set of true interactions, we characterized the relative quality of the Plasmodium network by examining: (1) its global topological properties, and (2) the signal-to-noise ratio (SNR) of its protein complexes. Several common topological measures16 were computed on each network, including the average number of interactions per protein (average degree), the average shortest path length between proteins, and the average clustering coefficient (Table 1). The number of interactions per protein in the Plasmodium interaction network followed a scale-free distribution, similar to other networks (Fig. 3a). Moreover, the Plasmodium network was never the outlier in any of the various measurements, suggesting that its global organization was consistent with the others.
Figure 3. Number of interactions per protein and the signal-to-noise ratio of protein complexes.
a, Scale-free network behaviour is shown in a manner similar to ref. 16. The linear fit is for Plasmodium data only. b, Dependence of SNR on error rate. See the key in panel a for an explanation of the symbols. Each network was modified by randomly shuffling from 0–100% of its protein interactions to simulate the addition of false positives and negatives. The subsequent decrease in SNR, converging to SNR = 0 at 100% noise, validates that each network contains a substantial fraction of true positive interactions. The high-throughput S. cerevisiae data shown is from the network of yeast interactions in the Database of Interacting Proteins (DIP)11. In addition to the high-throughput networks in this study, a literature-curated network—S. cerevisiae physical interactions according to the Munich Information Center for Protein Sequences (MIPS)30—is provided as a positive control. Similar trends are observed for the average clustering coefficient and the per cent of interactions covered by established protein-domain interactions (Supplementary Table 3).
Next, we applied the PathBLAST procedure to identify dense interaction complexes within each organism independently. A total of 29 single-species complexes were identified for Plasmodium, three of which are shown in Fig. 1d–f. This number was the median of the range observed over the five species (Table 1). Single-species complexes were used to assess the overall quality of each network by computing their SNR, a standard measure of assessing data quality in information theory and signal processing17. The SNR was computed by comparing the scores of complexes identified in the observed versus random interaction data for each organism (see the Methods). Plasmodium, worm and fly had very similar SNR values (Fig. 3b), while the SNR of the yeast network was slightly higher, and that of the H. pylori network was slightly lower. The network distances of Plasmodium versus yeast, worm or fly do not appear to depend on SNR.
As the observed network differences could not be attributed to bias or error, we next examined the novel functional predictions suggested by the three conserved and 29 Plasmodium-specific complexes. The conserved protein complex shown in Fig. 1a predicts that the proteins PF10_0244 and MAL6P1.286 may have previously uncharacterized roles in endocytosis. The counterpart of PF10_0244 in the yeast network, Ede1, localizes to the cortical patch18 of the cell membrane at sites of polarized growth and seems to be involved in endocytosis19. Myo5 and Myo3, yeast counterparts of MAL6P1.286, are class I myosins that also localize to actin cortical patches20, where the calmodulin protein Cmd1 has been implicated in the uptake step of receptor-mediated endocytosis21. Taken together, this evidence suggests a role for this complex in calmodulin-mediated endocytosis. Calmodulin inhibitors have been shown to attenuate growth22 and chloroquine extrusion (thus effecting drug resistance)23 in malarial parasites, and endocytosis has recently been linked to the mechanism of anti-malarial drugs including chloroquine and artemisinin24. The proximity of calmodulin to the formation of endocytic vacuoles in Plasmodium provides for a discrete hypothesis linking endocytosis, drug resistance and drug mechanism-of-action.
Within the 29 Plasmodium-specific complexes, chromatin remodelling was a prominent function, as shown in Fig. 1e. This complex involves the chromatin-remodelling protein ISWI (MAL6P1.183) interacting with a nucleosome assembly protein (PFI0930C)25. The protein PF11_0429 has a PHD domain (for plant homeodomain), and PF07_0029 has an HSP90 domain (for heat shock protein of 90 kDa), both postulated to be involved in the remodelling process25,26. Together, these known functions suggest that other proteins in the complex, such as PF08_0060, PFB0765W and PFL0625C, also participate in chromatin remodelling. For instance, although PFL0625C is annotated as a translation initiation factor, its yeast homologue has been found in complex with histone acetyl-transferases27. Further analysis of other complexes shown in Fig. 1 is available in the Supplementary Information.
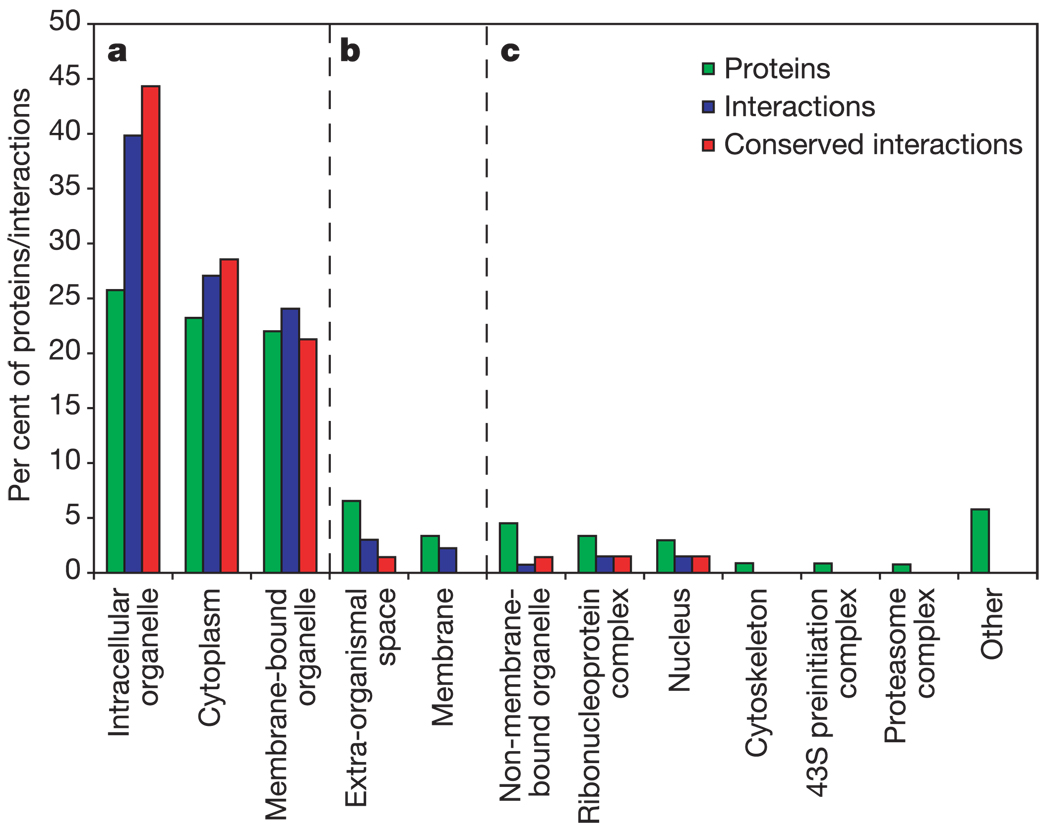
Several cellular components that we expected to be present, such as the proteasome, were missing from the set of complexes conserved between Plasmodium and the other species. To investigate this issue, we plotted the distributions of known functional annotations (according to Gene Ontology Cellular Component Level Three)28 among Plasmodium proteins, protein interactions and conserved interactions (Fig. 4 and Supplementary Fig. 2; note that a protein or an interaction can participate in multiple categories). Considerable proportions of all three data sets were associated with intracellular organelles, membrane-bound organelles or the cytoplasm (Fig. 4a). Other cellular components, such as the membrane and extra-organismal space, were represented among proteins and interactions but to a lesser extent among conserved interactions (Fig. 4b). Many membrane-associated components were also reported in the 29 Plasmodium-specific complexes, and are suggestive of machinery unique to this organism. Finally, components such as the proteasome and cytoskeleton were represented among proteins but were absent from the interaction set, and hence were not found as conserved interactions or complexes (Fig. 4c). Interactions among proteins in these components may have yet to be uncovered. These observations are reinforced by a complementary analysis of the functional distributions of yeast, worm and fly protein networks (Supplementary Fig. 2).
Figure 4. Functional roles within the Plasmodium protein network.
Functional annotations within the Plasmodium protein network associated with intracellular organelles, membrane-bound organelles or the cytoplasm (a), the membrane and extra-organismal space (b), and other components such as the proteasome and cytoskeleton (c). The histograms show the distribution of Gene Ontology Cellular Component assignments over all annotated Plasmodium proteins (green bars), protein interactions (blue bars) and conserved protein interactions (red bars). Interactions are considered ‘annotated’ if the interacting proteins share the same Gene Ontology category (these interactions are listed in Supplementary Table 6). For conserved interactions, the percentages in each category are cumulative over the three pairwise comparisons of Plasmodium versus the other three eukaryotes (yeast, fly or worm). Note that a protein or interaction can participate in multiple categories.
In summary, we have characterized conserved patterns of interaction between the protein network of Plasmodium falciparum and those of other species, and reported the specific network regions that are conserved. All of the examined networks contain dense complex-like structures of interactions, some of which are shared by yeast, worm and fly but not Plasmodium. These relationships are not clearly related to noise or bias in the Plasmodium interaction set. Some of the observed differences are almost certainly due to incomplete coverage in one or more networks: for instance, the present Plasmodium interaction set is focused on asexual life-cycle stages. Nevertheless, our comparison reflects the relative degree of similarity between the different networks. These differences are observed even when considering only those genes that are homologous across species.
It is generally expected that conserved genes will retain their functions and interactions. From this comparison, a different principle emerges: conservation of specific groups of related genes does not necessarily imply conservation of interaction among their encoded proteins. Further studies may distinguish the true differences from those related to network coverage and, ultimately, facilitate the discovery of new pharmaceuticals directed at the protein complexes unique to this parasite.
METHODS
Identification of conserved and species-specific complexes
Identification of protein complexes was performed using the PathBLAST family of network alignment tools, as previously described9. Briefly, these methods integrate protein interaction data from two species with protein sequence homology to generate an ‘aligned network’, in which each node represents a pair of homologous proteins (one from each organism; BLAST E value ≤1 × 10−10) and each link represents a conserved interaction. The network alignment is searched to identify high-scoring subnetworks, for which the score is based on the density of interactions within the subnetwork as well as confidence estimates for each protein interaction (see below). The search is then repeated over 100 random trials, in which the interactions of both species are arbitrarily reassigned while maintaining the same number of interactions per protein, resulting in a distribution of random subnetwork scores pooled over all trials. Dense subnetworks that score in the top fifth percentile of this random score distribution are considered significant and reported as ‘conserved complexes’. The search for ‘single-species complexes’ is identical to the search for conserved complexes, except that an individual protein network is searched instead of the network alignment. This process identifies dense subnetworks constrained by the interactions of one organism rather than two.
Interaction confidence scores
We estimated the probability that each measured protein interaction is true using a logistic regression model based on mRNA expression correlation, the network cluster coefficient, and the number of times the interaction had been experimentally observed. Further information on these confidence assignments is provided in the Supplementary Methods.
Phylogenetic tree construction
The Kitsch algorithm (provided by the PHYLIP package15) assumes the presence of an evolutionary clock and is based on pairwise distances between species. For each pair of species, an interaction between proteins a and b was considered ‘conserved’ if both proteins had sequence-similar counterparts a′ and b′ (BLAST E value ≤1 × 10−4) that interacted in the opposite species. A pairwise similarity between networks was computed as s 1,2 = (c 1 + c 2)/(t 1 + t 2), where c is the number of conserved interactions and t is the total number of interactions in species 1 or 2, respectively (with all interactions restricted to the set of proteins with homologues in the opposite species). Pairwise network distance was then defined as 1–s 1,2. The resulting phylogenetic tree shown in Fig. 2c is the consensus over 10,000 bootstrap simulations. Values of c and t for each network are listed in Supplementary Table 2.
Signal-to-noise ratio of protein complexes
SNR was computed for the single-species complexes as follows. The search for dense interaction complexes is initiated from each node (protein) and the highest scoring complex from each is reported (see the ‘Identification of conserved and species-specific complexes’ section of the Methods). This yields a distribution of complex scores over all nodes in the network. A score distribution is also generated for 100 randomized networks, which have an identical degree distribution as the original network. The SNR ratio is computed from these original and random score distributions (representing signal and noise, respectively) according to the standard formula17 using the root mean square (r.m.s.):
| (1) |
and where xi is the score of a complex and M is the total number of complexes.
Supplementary Material
Acknowledgements
We are indebted to M. Vignali, D. LaCount and S. Fields at the University of Washington, and B. Hughes and S. Sahasrabudhe at Prolexys, for providing us with advance access to the Plasmodium interaction data and for suggestions on our manuscript. We also thank E. Winzeler and J. Vinetz for advice on Plasmodium protein function, R. Sharan for help with the PathBLAST algorithm, and V. Bafna for assistance with the false-positive analysis. Finally, we acknowledge the following funding support: the National Science Foundation (S.S.); the National Institute of General Medical Sciences (T.I.); a David and Lucille Packard Fellowship award (T.I.); the Howard Hughes Medical Institute (T.S.); and Unilever (T.S.).
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Author Contributions S.S. and T.S. contributed equally to this work. All authors discussed the results and wrote the paper.
Reprints and permissions information is available at npg.nature.com/reprintsandpermissions. The authors declare no competing financial interests.
The authors declare no competing financial interests.
References
- 1.Miller LH, Baruch DI, Marsh K, Doumbo OK. The pathogenic basis of malaria. Nature. 2002;415:673–679. doi: 10.1038/415673a. [DOI] [PubMed] [Google Scholar]
- 2.Gardner MJ, et al. Genome sequence of the human malaria parasite Plasmodium falciparum. Nature. 2002;419:498–511. doi: 10.1038/nature01097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bozdech Z, et al. The transcriptome of the intraerythrocytic developmental cycle of Plasmodium falciparum. PLoS Biol. 2003;1:E5. doi: 10.1371/journal.pbio.0000005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Le Roch KG, et al. Discovery of gene function by expression profiling of the malaria parasite life cycle. Science. 2003;301:1503–1508. doi: 10.1126/science.1087025. [DOI] [PubMed] [Google Scholar]
- 5.Florens L, et al. A proteomic view of the Plasmodium falciparum life cycle. Nature. 2002;419:520–526. doi: 10.1038/nature01107. [DOI] [PubMed] [Google Scholar]
- 6.LaCount DJ, et al. A protein interaction network of the malaria parasite Plasmodium falciparum. Nature. doi: 10.1038/nature04104. doi:10.1038/nature04104 (this issue) [DOI] [PubMed] [Google Scholar]
- 7.Conant GC, Wagner A. Convergent evolution of gene circuits. Nature Genet. 2003;34:264–266. doi: 10.1038/ng1181. [DOI] [PubMed] [Google Scholar]
- 8.Yu H, et al. Annotation transfer between genomes: protein–protein interologs and protein–DNA regulogs. Genome Res. 2004;14:1107–1118. doi: 10.1101/gr.1774904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sharan R, et al. Conserved patterns of protein interaction in multiple species. Proc. Natl Acad. Sci. USA. 2005;102:1974–1979. doi: 10.1073/pnas.0409522102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kelley BP, et al. Conserved pathways within bacteria and yeast as revealed by global protein network alignment. Proc. Natl Acad. Sci. USA. 2003;100:11394–11399. doi: 10.1073/pnas.1534710100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xenarios I, et al. DIP, the Database of Interacting Proteins: a research tool for studying cellular networks of protein interactions. Nucleic Acids Res. 2002;30:303–305. doi: 10.1093/nar/30.1.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li S, et al. A map of the interactome network of the metazoan C. elegans. Science. 2004;303:540–543. doi: 10.1126/science.1091403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Giot L, et al. A protein interaction map of Drosophila melanogaster. Science. 2003;302:1727–1736. doi: 10.1126/science.1090289. [DOI] [PubMed] [Google Scholar]
- 14.Rain JC, et al. The protein–protein interaction map of Helicobacter pylori. Nature. 2001;409:211–215. doi: 10.1038/35051615. [DOI] [PubMed] [Google Scholar]
- 15.Felsenstein J. PHYLIP—phylogeny inference package (version 3.2) Cladistics. 1989;5:164–166. [Google Scholar]
- 16.Barabasi AL, Oltvai ZN. Network biology: understanding the cell’s functional organization. Nature Rev. Genet. 2004;5:101–113. doi: 10.1038/nrg1272. [DOI] [PubMed] [Google Scholar]
- 17.Shanmugam KS. Digital and Analog Communication Systems. New York: Wiley; 1979. [Google Scholar]
- 18.Gagny B, et al. A novel EH domain protein of Saccharomyces cerevisiae, Ede1p, involved in endocytosis. J. Cell Sci. 2000;113:3309–3319. doi: 10.1242/jcs.113.18.3309. [DOI] [PubMed] [Google Scholar]
- 19.Engqvist-Goldstein AE, Drubin DG. Actin assembly and endocytosis: from yeast to mammals. Annu. Rev. Cell Dev. Biol. 2003;19:287–332. doi: 10.1146/annurev.cellbio.19.111401.093127. [DOI] [PubMed] [Google Scholar]
- 20.Goodson HV, Anderson BL, Warrick HM, Pon LA, Spudich JA. Synthetic lethality screen identifies a novel yeast myosin I gene (MYO5): myosin I proteins are required for polarization of the actin cytoskeleton. J. Cell Biol. 1996;133:1277–1291. doi: 10.1083/jcb.133.6.1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Salisbury JL, Condeelis JS, Maihle NJ, Satir P. Calmodulin localization during capping and receptor-mediated endocytosis. Nature. 1981;294:163–166. doi: 10.1038/294163a0. [DOI] [PubMed] [Google Scholar]
- 22.Scheibel LW, et al. Calcium and calmodulin antagonists inhibit human malaria parasites (Plasmodium falciparum): implications for drug design. Proc. Natl Acad. Sci. USA. 1987;84:7310–7314. doi: 10.1073/pnas.84.20.7310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sanchez CP, McLean JE, Stein W, Lanzer M. Evidence for a substrate specific and inhibitable drug efflux system in chloroquine resistant Plasmodium falciparum strains. Biochemistry. 2004;43:16365–16373. doi: 10.1021/bi048241x. [DOI] [PubMed] [Google Scholar]
- 24.Hoppe HC, et al. Antimalarial quinolines and artemisinin inhibit endocytosis in Plasmodium falciparum. Antimicrob. Agents Chemother. 2004;48:2370–2378. doi: 10.1128/AAC.48.7.2370-2378.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Langst G, Becker PB. Nucleosome mobilization and positioning by ISWI-containing chromatin-remodeling factors. J. Cell Sci. 2001;114:2561–2568. doi: 10.1242/jcs.114.14.2561. [DOI] [PubMed] [Google Scholar]
- 26.Sollars V, et al. Evidence for an epigenetic mechanism by which Hsp90 acts as a capacitor for morphological evolution. Nature Genet. 2003;33:70–74. doi: 10.1038/ng1067. [DOI] [PubMed] [Google Scholar]
- 27.Gavin AC, et al. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature. 2002;415:141–147. doi: 10.1038/415141a. [DOI] [PubMed] [Google Scholar]
- 28.Ashburner M, et al. The Gene Ontology Consortium. Gene ontology: tool for the unification of biology. Nature Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Uetz P, et al. A comprehensive analysis of protein–protein interactions in Saccharomyces cerevisiae. Nature. 2000;403:623–627. doi: 10.1038/35001009. [DOI] [PubMed] [Google Scholar]
- 30.Mewes HW, et al. MIPS: analysis and annotation of proteins from whole genomes. Nucleic Acids Res. 2004;32:D41–D44. doi: 10.1093/nar/gkh092. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.