Abstract
Most of the bone, cartilage, and connective tissue of the craniofacial region arise from cephalic neural crest cells. Presumably, patterning differences in crest cells are a result of regional action of transcription factors within the developing pharyngeal arches. The basic helix–loop– helix transcription factor dHAND/HAND2 is expressed throughout much of the neural crest-derived mesenchyme of the pharyngeal arches, suggesting that it plays a crucial role in craniofacial development. However, targeted inactivation of the dHAND gene results in embryonic lethality by E10.5 due to vascular defects, preventing further analysis of the role of dHAND in cephalic neural crest cell development. In order to examine putative roles of dHAND during later stages of embryogenesis, we have used a transgenic lineage marker approach, in which a portion of the dHAND upstream region containing an enhancer that directs dHAND expression to the pharyngeal arches is used to drive Cre recombinase expression. By crossing these dHAND-Cre transgenic mice with R26R mice, we can follow the fate of cells that expressed dHAND at any time during development by examining β-galactosidase activity. We show that dHAND is first expressed in postmigratory cephalic neural crest cells within the pharyngeal arches. In older embryos, β-galactosidase-labeled cells are observed in most of the neural crest-derived lower jaw skeleton and surrounding connective tissues. However, labeled cells only contribute to substructures within the middle ear, indicating that our transgene is not globally expressed in cephalic neural crest cells within the pharyngeal arches. Moreover, dHAND-Cre mice will provide a valuable tool for tissue-specific inactivation of gene expression in multiple tissue types of neural crest origin.
Keywords: Cephalic neural crest, Transgenic mouse, Facial development, dHAND/HAND2, Cre recombinase, Neural crest enhancer
Introduction
Among events in skeletogenesis, craniofacial morphogenesis is perhaps the most intriguing, partially due to the origin of the bone, cartilage, and connective tissue that composes the face and neck. While bone and cartilage throughout the body and skull vault are of mesodermal origin (Noden, 1983; Jiang et al., 2002), those in the facial region are derived from cephalic neural crest cells (Bronner-Fraser, 1995; Le Douarin, 1982; Noden, 1988). These cells arise along the lateral edge of the neural folds and begin a ventral migration around the time of neural tube closure, undergoing an epithelial-to-mesenchymal transformation during the process (Le Douarin et al., 1993; Serbedzija et al., 1992). They eventually come to reside in the pharyngeal arches, transient structures on the ventral aspect of the embryo, in which crest-derived ectomesenchymal cells continue to proliferate and differentiate, eventually giving rise to adult craniofacial structures.
Numerous signaling molecules direct the patterning and development of cephalic neural crest cells and their derivatives (Francis-West et al., 1998; Mina, 2001; Wilkie and Morriss-Kay, 2001). These signals include Wnt1 and Wnt3a [expansion of premigratory neural crest cells prior to migration (Ikeya et al., 1997)], AP-2 [migration of crest cells to the arches (Schorle et al., 1996; Zhang et al., 1996)], and members of the Distal-less gene family [postmigratory patterning and differentiation (for reviews, see Kraus and Lufkin, 1999; Merlo et al., 2000)]. It is during this last postmigratory time period that soluble factors emanating from the overlying epithelium trigger signaling cascades within crest-derived ectomesenchymal cells that subsequently provide patterning and differentiation information necessary for development of bone and cartilage structures of the face and neck (Clouthier et al., 2000; Thomas and Sharpe, 1998; Tucker et al., 1999). These epithelial expression domains appear to be tightly controlled, thus providing much of the spatial specificity in transcription factor expression in the underlying mesenchyme. However, it is also believed that crest cells of specific lineages may also be programmed to express or respond to specific signals (Dorsky et al., 1998).
Two factors expressed in the pharyngeal mesenchyme are the basic helix–loop– helix transcription factors dHAND/HAND2 and eHAND/HAND1 (Cserjesi et al., 1995; Hollenberg et al., 1995; Srivastava et al., 1995). The genes encoding these factors are expressed in numerous locations within the embryo, including postmigratory crest cells in the arches. Their expression domains partially overlap, with dHAND expression observed throughout the mesenchyme of mandibular arch 1 and arches 2– 6, whereas eHAND expression is confined to the ventral one-third of the mandibular arch one and arch two. While their expression patterns suggest that they might play crucial roles in craniofacial morphogenesis, targeted inactivation of the dHAND (Srivastava et al., 1997; Thomas et al., 1998) and eHAND (Firulli et al., 1998; Riley et al., 1998) genes in mice results in early embryonic lethality, thus preventing further analysis of their roles in cephalic neural crest cell development. However, some inferences on the role of dHAND can be made from studies in endothelin-A (ETA) receptor-deficient embryos, in which dHAND expression within the arch mesenchyme is almost completely down-regulated (Clouthier et al., 2000). embryos are born with severe craniofacial defects, including absence or malformation of the mandible, Meckel’s cartilage, malleus, incus, and tympanic ring (Clouthier et al., 1998). While expression of at least five genes, including dHAND and eHAND, is disrupted in the arch mesenchyme of embryos (Clouthier et al., 2000; and D.E.C., unpublished data), the dHAND expression domain encompasses most of the expression domains of other affected genes, suggesting that dHAND may play a dominant role in development of craniofacial bones and cartilages.
Recently, we have shown using an 11-kb fragment of the dHAND upstream region driving transgenic expression of lacZ that dHAND expression is observed in facial bones and cartilage until at least E16.75 (Charité et al., 2001). These included portions of the mandible and Meckel’s cartilage, as well as the tongue musculature and connective tissue of the lower jaw. To further examine potential roles of dHAND during craniofacial development, we have undertaken a lineage analysis of cells that express dHAND at any time during development. To accomplish this, we created transgenic mice in which a portion of the dHAND upstream region containing elements necessary for expression in the pharyngeal arch mesenchyme was used to drive Cre recombinase expression. By crossing dHAND-Cre transgenic mice with mice from the Cre reporter line R26R (Soriano, 1999), we determined the fate of dHAND-lineage cells by staining for β-galactosidase activity and then compared the staining with both staining in dHAND-lacZ transgenic embryos and endogenous dHAND expression as judged by in situ hybridization. We find that, during craniofacial morphogenesis, Cre expression in dHAND-Cre;R26R embryos is first detected in neural crest-derived pharyngeal arch mesenchyme at E9.5, matching both the expression of the dHAND-lacZ transgene and endogenous dHAND expression. In older dHAND-Cre;R26R embryos, labeled cells become restricted to a limited number of neural crest-derived structures, including Meckel’s cartilage, mandibular bone, dental pulp of mandibular teeth, and the mesenchyme surrounding the developing submandibular gland buds. Most striking is the restriction of staining observed in the middle ear ossicles, with only the manubrium of the malleus containing labeled cells. No cells are observed in the body of the malleus, incus, or stapes. In dHAND-lacZ embryos, a similar distribution of labeled cells is observed in most structures, though apparent differences in fated vs active expression exist within craniofacial cartilages and mandibular teeth. We observe very little endogenous dHAND expression in older embryos, with the primary exception being around the mandibular incisors. Together, our findings illustrate both the specificity and complexity of dHAND expression and function.
Materials and methods
Transgenic constructs
The plasmid pOG231 contains a CMV promoter, a synthetic intron, the Cre cDNA, a nuclear localization signal (NLS), and a SV40 polyadenylation signal (O’Gorman et al., 1997). To create the dHAND-Cre transgene, pOG231 was digested with XhoI, treated with Klenow enzyme to produce blunt ends, and then digested with EcoRI. This scheme liberated a 1.8-kb fragment containing the synthetic intron, Cre-NLS, and polyadenylation sequences. This fragment was then subcloned into pSP73 (Promega) cut with EcoRI and EcoRV, resulting in the plasmid pSP73-Cre-NLS. This plasmid was then digested with KpnI, treated with T4 DNA polymerase to produce blunt ends, followed by digestion with XhoI and gel purification. To prepare the promoter insert, pBSdHAND-lacZ (Charité et al., 2001), containing 11 kb of the dHAND upstream region, was cut with SfiI (3′ to the dHAND transcription initiation site), treated with T4 DNA polymerase to produce blunt ends, followed by restriction with XhoI (7.4 kb upstream of the SfiI site). The isolated 7.4-kb SfiI (b)–XhoI fragment was then subcloned into pSP73-Cre-NLS [KpnI (b)–XhoI]. The resulting plasmid, pdHAND-Cre, was verified by restriction digest and partial sequencing. The final 9.2-kb construct was isolated by restriction enzyme digest with SalI. The fragment was isolated on a 0.8% ultra-pure agarose gel (FMC), purified by perchlorate elution, and quantified (Comerford et al., 1995).
Production of transgenic mice
Construction and characterization of the −11-kb dHAND-lacZ transgenic mice has previously been described (Charité et al., 2001; McFadden et al., 2000). To construct dHAND-Cre transgenic mice, the purified dHAND-Cre construct was injected at 3 ng/μl into C57BL6/J × SJL F2 fertilized one-cell eggs, and two-cell embryos were transferred to pseudopregnant females as previously described (Clouthier et al., 1997). Initial genotyping of putative founders was performed by dot blot hybridization. Five founder mice were identified, with transgene copy number ranging from 1 to 10 copies per cell. Founder mice were bred to mice from the Cre reporter mouse strain R26R (strain 129/Sv-Gtrosa26tm/Sor; The Jackson Laboratory, Bar Harbor, ME) (Soriano, 1999). Embryos were collected at E10.5 and analyzed for β-galactosidase (β-gal) staining to evaluate the fidelity and robustness of Cre expression. Three lines were established that produced compound hemizygous pups with β-gal activity. While all three lines showed similar expression patterns, embryos from lines 7-1 and 7-2 had the most robust staining. Therefore, these lines were used for the current study.
Genotyping of dHAND-Cre transgenic mice and embryos
Genotyping was performed by PCR using genomic DNA isolated from a small tail biopsy or embryonic yolk sac. The presence of the dHAND-Cre transgene was determined by using the Cre primers 5′-GGACATGTTCAGGGATCGCCAGGCG-3′ and 5′-GCATAACCAGTGAAACAGCATTGCTG-3′ as previously described (Kisanuki et al., 2001). The presence of the R26R allele was determined by using primers 5′-GCGAAGAGTTTGTCCTCAACC-3′ (R1295) and 5′-AAAGTCGCTCTGAGTTGTTAT-3′ (R26RF) as previously described (Soriano, 1999). The presence of the dHAND-lacZ transgene was determined by using the lacZ primers 5′-GACACCAGACCAACTGGTAATGG-3′ and 5′-GCATCGAGCTGGGTAATAAGCG-3′. Bands were visualized on 1% agarose gels.
β-Galactosidase staining
For whole-mount staining [embryonic day (E) 8.5 to E11.5], embryos were collected and fixed for 1 h in 4% paraformaldehyde on ice. Embryos were then rinsed in lacZ rinse buffer (0.2 M sodium phosphate, pH 7.3, 2 mM magnesium chloride, 0.02% NP-40, 0.01% sodium deoxycholate) twice for 10 min and then placed in lacZ staining buffer (lacZ rinse buffer plus 5 mM potassium ferricyanide, 5 mM potassium ferrocyanide, and 1 mg/ml X-gal) and incubated overnight in the dark at room temperature. Embryos were rinsed in PBS, postfixed in 10% neutral-buffered formalin for 4 h, rinsed in PBS, and stored in 70% ethanol. Embryos were analyzed and photographed on an Olympus SZX12 stereomicroscope fitted with a DP11 digital camera. To observe β-gal staining at the cellular level, some of these stained embryos were dehydrated through graded ethanols and Cedarwood oil (Polysciences) and then embedded in paraffin and sectioned at 12 microns on a Lecia RM2135 microtome. Sections were collected on positively charged Superfrost Plus microscope slides (Fisher Scientific), partially rehydrated, counterstained with nuclear fast red, dehydrated, and coverslipped with DPX mounting media (BDH). Analysis of sections was performed by using a Nikon E600 microscope with an attached SPOT RT digital camera.
For staining of older embryos (E13.5–E18.5), embryos were isolated into PBS and immediately snap frozen in OCT on a dry ice/95% ethanol bath. Embryos were sectioned at 14–18 microns on a CM1900 cryostat (Leica). Sections were collected on Superfrost Plus slides, air dried for 2 h, fixed in 0.5% gluteraldehyde at room temperature for 30 min, washed three times for 20 min each in lacZ rinse buffer, and incubated overnight in lacZ staining solution at 37°C. After rinsing in PBS, slides were counterstained in nuclear fast red (E8.5–E15.5) or eosin (E18.5) and cover-slipped with PBX mounting media. Analysis and photography was performed as described above.
In situ hybridization analysis
Sectional in situ hybridization analysis was performed as previously described (Shelton et al., 2000) by using a 35S-UTP-labeled riboprobe against dHAND (Srivastava et al., 1997), with the hybridization carried out at 70°C in hybridization buffer containing 0.75 M NaCl. Sections were exposed for 2–3 weeks before developing, fixing, and counterstaining with Richard Allen hematoxylin 2 (VWR). Whole-mount in situ hybridization analysis was performed as previously described by using a digoxigenin-labeled riboprobe against dHAND (Clouthier et al., 2000). Following staining for alkaline phosphatase activity, embryos were photographed, embedded in paraffin, and sectioned at 16 microns. Sections were not counterstained. Photography was performed by using Nomarski DIC optics.
Results
Creation of dHAND-Cre transgenic mice
We initially set out to create a mouse strain in which we could follow the fate of dHAND-expressing cells throughout embryonic development. To accomplish this, we constructed a transgene in which a 7.4-kb fragment of the dHAND upstream region, which contains enhancer elements necessary for both cardiac and pharyngeal arch expression of dHAND (Charité et al., 2001; McFadden et al., 2000), was used to drive expression of Cre recombinase (Fig. 1). Microinjection of this construct resulted in five transgenic founder mice. To assess expression of the transgene, founder animals were bred with mice from the R26R Cre reporter line (Soriano, 1999). Expression of Cre in R26R mice results in activation of a lacZ gene and thus robust β-galactosidase (β-gal) staining, providing an indelible marker of cells that have expressed dHAND at any time during development. E10.5 dHAND-Cre;R26R embryos were collected from each line and assayed by β-gal staining. Three lines showed staining in the heart and pharyngeal arches in a dHAND-specific manner (data not shown); two of these (7-1 and 7-2) were subsequently used in the analyses described below. Findings were identical in both lines, though overall intensity of expression was less in the 7-2 line.
Fig. 1.
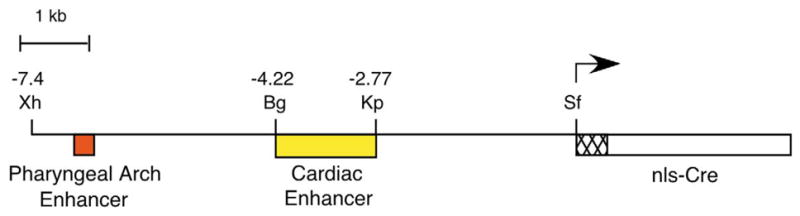
Diagram of the dHAND-Cre transgene. A 7.4-kb fragment of the dHAND upstream region, containing enhancers required for pharyngeal arch and cardiac expression, was fused to a Cre cDNA containing nuclear localization and SV40 polyadenylation sequences. A synthetic intron resides in the 5′ end of the Cre cDNA to aid transgene expression. Bg, BglII; Kp, KpnI; Sf, SfiI; Xh, XhoI.
dHAND-Cre expression during early embryogenesis
We initially examined β-gal staining in early dHAND-Cre;R26R embryos. Since β-gal staining could represent either active or past dHAND enhancer activity, we also examined β-gal staining in −11.0-kb dHAND-lacZ transgenic mouse embryos and compared that with the endogenous dHAND expression pattern. Though our dHAND-lacZ transgenic mouse line contained an additional 3.6 kb of upstream promoter region, both dHAND-lacZ and dHAND-Cre transgenic strains contained the same pharyngeal arch enhancer. Further, in our previous studies, −11.0 kb dHAND-lacZ and −7.4 kb dHAND-lacZ embryos had identical β-gal staining patterns in the developing pharyngeal arches (Charité et al., 2001).
Staining was first observed in dHAND-Cre;R26R embryos at E7.75 in the cardiac crescent (data not shown), matching the endogenous dHAND expression pattern (Srivastava et al., 1997). By E8.5, expression was observed in the lateral plate mesoderm and common ventricle of the heart but not in the pharyngeal arches, a finding recapitulated in dHAND-lacZ embryos (data not shown) and matching the endogenous dHAND expression pattern (Charité et al., 2000; Srivastava et al., 1997). However, the staining in the lateral plate mesoderm of dHAND-Cre;R26R was more restricted than the endogenous dHAND expression pattern. By E9.5, labeled cells in dHAND-Cre;R26R embryos were observed within much of the neural crest-derived mesenchyme of mandibular arch 1, with scattered cells also observed in the medial mesenchyme of arch 2 (Fig. 2A). Labeled cells were not observed in the overlying arch epithelium (Fig. 2A′). This staining pattern was identical to that observed in dHAND-lacZ embryos, though labeled cells were primarily confined to arch one (Fig. 2B and B′). β-gal staining in both of these transgene lines closely matched the endogenous dHAND expression pattern (Fig. 2C and C′), suggesting that the arch staining in dHAND-Cre;R26R embryos represented active dHAND expression.
Fig. 2.
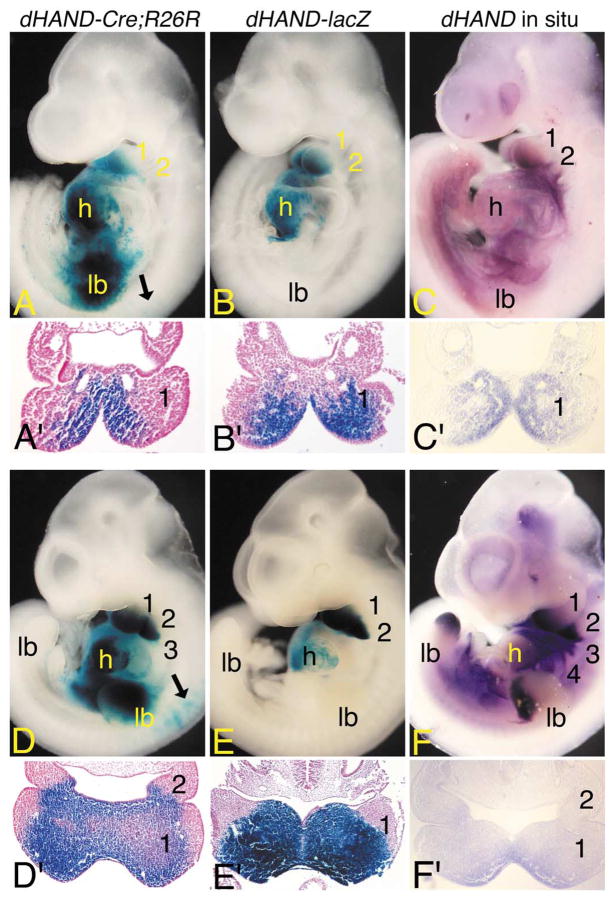
Early dHAND expression domains in dHAND-Cre; R26R embryos. (A–F) Lateral (A–F) and transverse (A′–F′) views of dHAND-Cre;R26R (A, A′; D, D′) and dHAND-lacZ (B, B′; E, E′) embryos stained in whole mount for β-gal activity and wild type embryos (C, C′; F, F′) after whole-mount in situ hybridization analysis of dHAND expression. Transverse sections of β-gal-stained embryos were counterstained with nuclear fast red and photographed by using brightfield optics. Transverse sections of whole-mount in situ embryos were photographed by using Nomarski DIC optics. (A) In E9.5 dHAND-Cre; R26R embryos, labeled cells are observed in the first mandibular (1) and second (2) arches, common heart ventricle (h), and forelimb bud (lb). Few scattered cells are observed in the early dorsal root ganglion (black arrow). (A′) A transverse section through the first arch of the embryo shown in (A) illustrates that stained cells are confined to the arch mesenchyme in a dHAND-specific pattern. (B) In E9.5 dHAND-lacZ embryos, staining is observed in the mandibular arch and common heart ventricle. (B′) A transverse section through the first arch of the embryo shown in (B) shows that labeled cells are confined to the arch mesenchyme. (C) In E9.5 wild type embryos, dHAND expression is observed in the first mandibular and second arches and in the future right ventricle of the heart (obscured by the left ventricle). (C′) As observed in (A′) and (B′), dHAND expression is confined to the arch mesenchyme. (D) By E10.5, labeled cells in dHAND-Cre;R26R embryos are observed in arches 1–3, the ventricle, and the limb bud. Scattered cells are also observed in the atrium. While not visible in this view, stained cells are also observed in arch 4. Labeled cells are also observed in the cervical dorsal root ganglion (black arrows). (D′) In a transverse section through the first and second arches, staining is observed in the arch mesenchyme, with the intensity of staining highest in the periphery of the arch. (E) In E10.5 dHAND-lacZ embryos, labeled cells are observed in arches one and two and in the ventricles of the heart. Scattered cells are also present in the atria. (E′) As observed in dHAND-Cre;R26R embryos, staining within the mesenchyme of the first arch is strongest at the periphery. While not shown due to a slight difference in the angle of section, the second arch staining in dHAND-lacZ embryos is comparable with that of dHAND-Cre;R26R embryos. (F) dHAND expression in E10.5 wild type embryos is observed in arches 1– 4, distal limb buds, and right ventricle of the heart. (F′) A section through the first and second arch again illustrates staining throughout the medial half of the arch mesenchyme, with the strongest staining observed along the periphery of the arch.
β-gal staining in E9.5 dHAND-Cre;R26R embryos was also observed in the neural crest-derived cervical dorsal root ganglia (DRG) (black arrow in Fig. 2A), a site that is not stained in dHAND-lacZ embryos (Fig. 2B) and that does not show any endogenous expression (Fig. 2C). These findings, along with the fact that the degree of labeling in the DRGs differed in different dHAND-Cre;R26R lines examined, suggest that the DRG staining is artifactual. Labeled cells in dHAND-Cre;R26R embryos were also present in the foregut and forelimb buds. Expression in the forelimb bud was unusually strong for the embryonic age of the embryo in comparison with the endogenous limb expression pattern of dHAND (Fig. 2C; and Charité et al., 2000; Fernandez-Teran et al., 2000; Srivastava et al., 1995). Further, while the endogenous dHAND gene is expressed in the posterior limb bud, labeled cells in E9.5 dHAND-Cre;R26R embryos were observed in the anterior portion of the forelimb. There was no apparent labeling of limb bud mesenchyme in dHAND-lacZ embryos (Fig. 2B).
By E10.5, the more caudal arches in dHAND-Cre;R26R embryos also contained labeled cells; other sites of expression remained unchanged (Fig. 2D). Within the arches, labeled cells appeared denser around the arch periphery (Fig. 2D′), resembling the pattern previously observed for neural crest cells (Chai et al, 2000). With the exceptions noted at E9.5, the overall expression pattern still resembled β-gal staining in dHAND-lacZ embryos (Fig. 2E and E′) and endogenous dHAND expression (Fig. 2F and F′), though very few labeled cells were observed in arches 3– 6 of dHAND-lacZ embryos. These findings suggest that the arch staining in dHAND-Cre;R26R embryos was still a result of active dHAND expression. While staining in locations other than the arches or derivatives continued throughout development in dHAND-Cre;R26R embryos, the majority of the present study focuses on findings during craniofacial development.
Fate of cells from a dHAND lineage during orofacial development
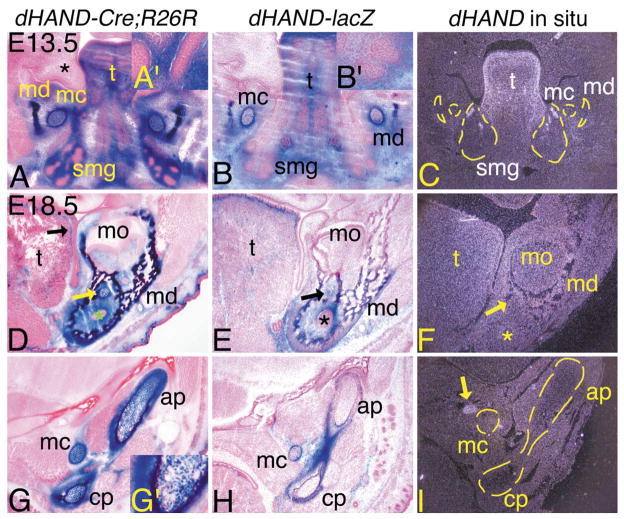
Using dHAND-lacZ transgenic mice, we have previously shown that dHAND is expressed in subsets of cells during orofacial development (Charité et al., 2001). To more directly determine the participation of dHAND lineage cells in this process, we examined β-gal staining in E13.5 and E18.5 dHAND-Cre;R26R embryos and compared that to both β-gal staining in dHAND-lacZ embryos and endogenous dHAND expression in wild type embryos. In E13.5 dHAND-Cre;R26R and dHAND-lacZ embryos, Meckel’s cartilage and surrounding perichondrium, as well as mandibular bone and surrounding connective tissue, all neural crest derivatives, were composed of labeled cells (Fig. 3A and B). The mesenchyme surrounding the buds and ducts of the developing submandibular gland (pseudoglandular stage), a neural crest derivative (Jaskoll et al., 2002), was also composed of β-gal-stained cells (see insets, Fig. 3A′ and B′). The ductal epithelium did not contain labeled cells. Mesodermal derivatives also contained labeled cells, including the lamina propria of the tongue. One notable difference was that Meckel’s cartilage in dHAND-lacZ embryos was composed of both labeled and unlabeled cells, compared with almost complete contribution of labeled cells in dHAND-Cre;R26R embryos. Endogenous dHAND gene expression was observed in the lamina propria of the tongue and in the submandibular gland mesenchyme, though the latter appeared to be more restricted than observed in dHAND-lacZ embryos (Fig. 3C). Expression in the early mandibular bone and Meckel’s cartilage was virtually undetectable. The other major site of labeled cells in E13.5 dHAND-Cre;R26R embryos was in the developing scapula, in which labeled cells composed the entire coracoid process and acromion of the scapula, but were mixed with unlabeled cells in the scapula blade (data not shown). The significance of this scapular staining is unclear, as endogenous dHAND expression was not observed in these structures at E13.5 (data not shown).
Fig. 3.
Contribution of dHAND lineage cells to arch-derived structures of the jaw of dHAND-Cre;R26R embryos. Transverse (A–C) and frontal (D–I) sections through the heads of dHAND-Cre;R26R (A, D, G), dHAND-lacZ (B, E, H), and wild type (C, F, I) embryos. Frozen sections from dHAND-Cre;R26R and dHAND-lacZ embryos were stained for β-gal activity and counterstained with nuclear fast red (E13.5) or eosin (E18.5). Paraffin sections from wild type embryos were probed with a 35S-labeled riboprobe against dHAND. After developing, sections were counterstained with Harris hematoxylin and photographed under darkfield conditions. To aid in the visualization of specific structures, yellow lines have been added to some of the darkfield panels. (A–C) Transverse sections through the lower jaw of E13.5 embryos. (A) Labeled cells in dHAND-Cre;R26R embryos are observed in structures derived from the first pharyngeal arch, including mandibular bone (md), Meckel’s cartilage (mc), tongue (t), and surrounding mesenchyme/connective tissue. Abundant labeled cells are also observed in the mesenchyme of the developing submandibular glands (smg), but not in the glandular epithelium (see inset, A′). The mesenchyme of the upper jaw (future maxilla; *) and oral cavity epithelium do not contain labeled cells. (B) Labeled cells in dHAND-lacZ transgenic embryos are observed in a similar pattern to those in dHAND-Cre;R26R embryos, though labeled cells in Meckel’s cartilage are mixed with unlabeled cells. (C) Endogenous dHAND expression is observed in the lamina propria of the tongue and in the mesenchyme surrounding the proximal ducts of the submandibular gland. Very little expression is observed in the mandible or Meckel’s cartilage. (D–I) Frontal sections through the mandible of E18.5 embryos. (D) β-gal-stained cells in dHAND-Cre;R26R embryos are present in the connective tissue of the tongue (t), with highest staining in the lamina propria (black arrow). Almost all cells in the mandible, Meckel’s cartilage (yellow arrow), and surrounding mesenchyme are also labeled. (E) Labeled cells in dHAND-lacZ embryos are again observed in a similar pattern to those in dHAND-Cre;R26R embryos. However, a mixture of labeled and unlabeled cells composes the mandibular bone and Meckel’s cartilage. (F) Faint dHAND expression is observed in the mandibular bone. Very little expression is observed in Meckel’s cartilage. The asterisks in (D–F) mark the enamel organ of the left incisor. (G) In a more proximal section to that shown in (D–F), labeled cells in dHAND-Cre;R26R embryos are observed in Meckel’s cartilage and in the articular (ap) and coronoid (cp) processes of the mandible and surrounding perichondrium. However, fewer labeled cells are observed near the junction with the ossified mandible, an area undergoing endochondral ossification and composed of hypertrophic chondrocytes (see inset, G′). (H) Labeled cells are mixed with unlabeled cells in Meckel’s cartilage, but are not observed in the anterior and coronoid processes of the mandible. The perichondrium surrounding the process is composed almost solely of labeled cells. (I) Endogenous dHAND expression is not observed in the mandibular processes or in Meckel’s cartilage, though expression is observed in the sympathetic ganglion (yellow arrow). Due to the angle of section, this structure is not observed in the dHAND-Cre-R26R or dHAND-lacZ embryo sections, though scattered labeled cells are present in the sympathetic ganglion of dHAND-Cre-R26R embryos (see Fig. 4). mo, molar.
By E18.5, the mandibular bone and surrounding lamina propria of dHAND-Cre;R26R embryos was composed of labeled cells along its entire proximodistal axis (Fig. 3D and G; and data not shown). Likewise, Meckel’s cartilage was also stained along its entire axis (Fig. 3D and G; and data not shown), as were the chondrocytes and surrounding perichondrium composing the anterior and lateral processes of the proximal mandible (Fig. 3G). The extent of cell labeling decreased near the junction of the processes with the mandibular bone, a zone composed of hypertrophic and presumably ossified chondrocytes (see inset, Fig. 3G′). Labeled cells were also observed throughout the tongue, with strongest staining again in the lamina propria (Fig. 3D), though the relative number of labeled cells appeared greater in the oral portion (distal two-thirds) than in the pharyngeal portion (proximal one-third) of the tongue (data not shown).
In dHAND-lacZ embryos, labeled cells were also observed throughout the proximodistal axis, though the relative presence of cells was higher in the caudal portion of the mandible along this axis (Fig. 3E). Labeled cells were also present in Meckel’s cartilage, though they again were mixed with unlabeled cells (Fig. 3E and H). Very few labeled cells were observed in the mandibular processes, though strong staining was observed in the perichondrium around both these processes and Meckel’s cartilage (Fig. 3E and H). Endogenous dHAND expression was only observed in scattered cells within the mandible (Fig. 3F and I). Expression was not observed in the perichondrium of the mandibular processes.
Contribution of dHAND lineage cells in middle ear and throat structures
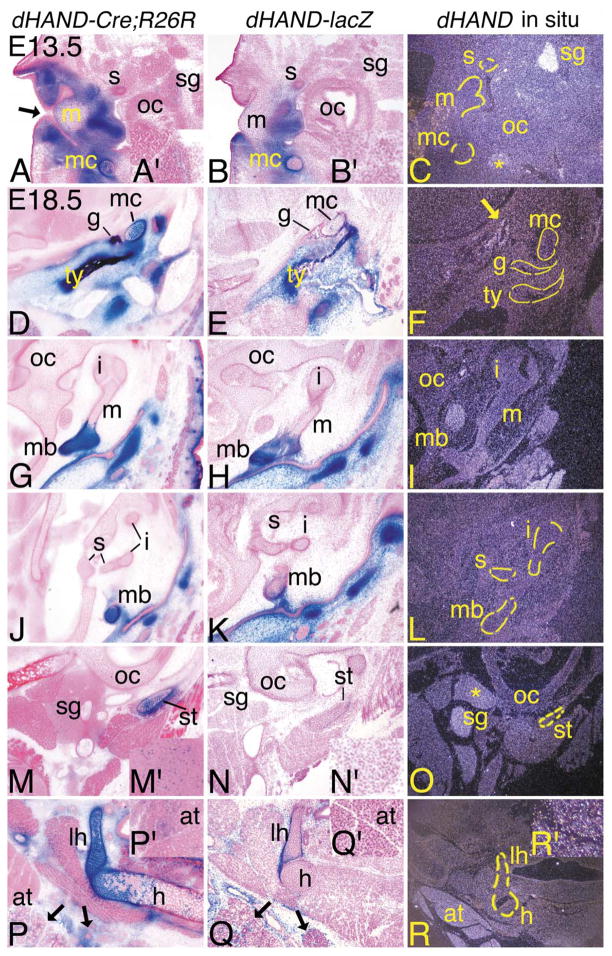
Much of the external and middle ear is composed of ectomesenchymal cells derived from the first and second arches (Fekete, 1999; Kontges and Lumsden, 1996; Mallo, 2001; Noden, 1988). In E13.5 dHAND-Cre;R26R embryos, most of the mesenchyme composing the connective tissue in the external region was composed of β-gal-positive cells (Fig. 4A), though the epithelium of the future external auditory meatus (derived from the first pharyngeal cleft) was unstained (arrow). In the middle ear, labeled cells were observed in Meckel’s cartilage as described above, though a strict delineation of labeled and unlabeled cells occurred at the junction with the cartilaginous precursor of the malleus, which was unlabeled (data not shown). However, labeled cells were observed in the distal malleus (Fig. 4A). Further, while most of the stapes was composed of unlabeled cells, the caudal stapes and surrounding perichondrium did contain a mixture of labeled and unlabeled cells (data not shown). We also observed labeled cells in the sympathetic ganglion, though there was extensive mixing with unlabeled cells (see inset, Fig. 4A′). Other sites of expression included the cervical dorsal root ganglia, superior cervical sympathetic trunk, and ventral primary ramus of the cervical nerve (data not shown). As observed for the sympathetic ganglion, contribution of labeled cells within these structures was scattered.
Fig. 4.
Distribution of dHAND progeny cells in the middle ear and throat during development of dHAND-Cre; R26R embryos. Transverse (A–C; E13.5) and frontal (D–R; E18.5) sections through the middle ear and throat of dHAND-Cre;R26R (A, D, G, J, M, P), dHAND-lacZ (B, E, H, K, N, Q), and wild type (C, F, I, L, O, R) embryos. Treatment of sections was performed as described in Fig. 3. (A–C) Sections through the middle ear of E13.5 embryos. (A) In dHAND-Cre;R26R embryos, stained cells are observed in the mesenchyme surrounding the epithelium of the external auditory meatus (black arrow), in Meckel’s cartilage (mc), and in the future manubrium of the malleus (m). Scattered labeled cells are also observed in the sympathetic ganglion (sg; see inset, A′). Labeled cells are not observed in the stapes (s), though are present in more proximal sections (see text). (B) Labeled cells in dHAND-lacZ embryos are also observed in the mesenchyme of the middle ear region, the manubrium of the malleus, and Meckel’s cartilage, though they are mixed with unlabeled cells in Meckel’s cartilage. No staining is observed in the stapes or sympathetic ganglion (see inset, B′). (C) Very little endogenous dHAND expression is observed in the middle ear. The exceptions are the sympathetic ganglion and the mandibular branch of the trigeminal ganglion (*). (D–R) Frontal sections through the heads of E18.5 embryos. (D) The tympanic (ty) and gonial (g) bones and Meckel’s cartilage are composed almost solely of labeled cells in dHAND-Cre;R26R embryos. The surrounding mesenchyme in this region of the head is also composed of labeled cells. (E) Labeled cells in dHAND-lacZ embryos are observed in the tympanic ring and gonial bone, though a small portion of unlabeled cells are also present. More significant mixing of labeled and unlabeled cells is observed in Meckel’s cartilage. (F) Little endogenous expression is observed in the tympanic and gonial bones and Meckel’s cartilage, though faint expression is found along some of the head vasculature (arrow). (G, H) In sections through the malleus, labeled cells in dHAND-Cre;R26R and dHAND-lacZ embryos are confined to the manubrium (mb) of the malleus and surrounding mesenchyme. (I) dHAND expression is not observed in any part of the malleus or incus (i) in wild type embryos. (J, K) β-gal-stained cells in dHAND-Cre;R26R and dHAND-lacZ embryos are not present in the incus or stapes (s). Labeled cells in the manubrium of dHAND-lacZ embryos are mixed with unstained cells. (L) dHAND expression is not observed in the incus, stapes, or manubrium of the malleus. (M) In sections through the distal portion of the styloid cartilage (st), labeled cells in dHAND-Cre;R26R embryos compose most of the styloid and are scattered in the sympathetic ganglion (see inset, M′). (N) In dHAND-lacZ embryos, labeled cells are present in the perichondrium of the styloid cartilage, but are mixed in the cartilage itself and are absent in the sympathetic ganglion (see inset, N′). (O) dHAND expression is observed in the sympathetic ganglion in the incus/stapes region, with weaker expression in the glossopharyngeal ganglion. (P) In the throat, β-gal-stained cells in dHAND-Cre;R26R embryos are observed in the lesser horn (lh) and body (h) of the hyoid. However, labeled cells are mixed with unlabeled cells in the body, possibly reflecting changes in staining in cartilage undergoing endochondral ossification, as few hypertrophic chondrocytes are stained. Labeled cells are also observed in the mesenchyme of the submandibular and sublingual glands (black arrows). Out of the photographic frame, adipose tissue also contains scattered labeled cells (inset, P′). (Q) As with other cartilages, labeled cells in dHAND-lacZ embryos compose the perichondrium of the lessor horns of hyoid but are mixed with a majority of unlabeled cells in the cartilage itself. Very few labeled cells are observed in the body of the hyoid. Labeled cells are observed in the mesenchyme of the submandibular and sublingual glands (arrows), but are absent in adipose tissue observed out of the photographic frame (inset, Q′). (R) dHAND expression is not observed in or around the hyoid cartilages or bones; however, adipose tissue at the base of the neck shows strong dHAND expression. Out of the plane of section, expression is also observed in the submandibular gland (inset, R′). oc, otic capsule.
A similar pattern of β-gal staining was observed in dHAND-lacZ embryos, though labeled cells in Meckel’s cartilage were mixed with unlabeled cells (Fig. 4B), while the sympathetic ganglion was composed solely of unlabeled cells (see inset, Fig. 4B′). This pattern of labeled cells was not necessarily reflected in the endogenous expression of dHAND, as very little expression was observed in the external or middle ear, though intense signal was present in the sympathetic ganglion and the mandibular branch of the trigeminal nerve (*) (Fig. 4C).
At E18.5, the tympanic ring and gonial bones, neural crest-derived structures formed by intramembranous ossification, were composed of labeled cells in dHAND-Cre; R26R embryos, as was the surrounding connective tissue and Meckel’s cartilage (Fig. 4D). Of the middle ear ossicles, only the manubrium of the malleus contained labeled cells; the cartilaginous precursors of the incus or stapes were unlabeled (Fig. 4G and J). Other elements composed of labeled cells included the distal portion of the styloid cartilage and cartilage associated with the pinna, with scattered cells also present in the sympathetic ganglion (Fig. 4M and 4M′). Labeled cells in E18.5 dHAND-lacZ embryos were observed in a similar pattern, with staining observed in the tympanic and gonial bones (Fig. 4E), Meckel’s cartilage (Fig. 4E), the manubrium of the malleus (Fig. 4H), and the styloid cartilage (Fig. 4N). However, as observed at E13.5, labeled cells in cartilage structures, while composing most of the perichondrium around the structures, were extensively mixed with unlabeled cells within the cartilage. This was most prominent in the styloid cartilage (Fig. 4N). Labeled cells were not present in the sympathetic ganglion (Fig. 4N). Endogenous dHAND expression was not widely observed, with virtually no expression observed in any of the middle ear structures examined (Fig. 4F, I, and L). However, as observed at E13.5, expression was observed in the sympathetic ganglion (Fig. 4O).
Labeled cells were also observed in the second and third arch derivatives of dHAND-Cre;R26R embryos. The hyoid bone of the throat is actually derived from three cartilaginous precursors derived from both arches. The lesser horns, a second arch derivative, were composed exclusively of labeled cells (Fig. 4P). However, the greater horns, a third arch derivative, were composed of both labeled and unlabeled cells (data not shown). The body of the hyoid, derived from both arches (Kontges and Lumsden, 1996), was also composed of a mixture of labeled and unlabeled cells in the cartilaginous portions (endochondral ossification begins in the middle of the body and spreads bilaterally). However, fewer hypertrophic chondrocytes and presumably ossified chondrocytes were labeled, similar to our observations in the mandibular processes. Scattered labeled cells were observed in the fourth arch-derived thyroid cartilage (data not shown). Labeled cells were also present in the mesenchyme composing the submandibular and sublingual glands (arrows in Fig. 4P) and in adipose tissue of the neck (see inset, Fig. 4P′).
In E18.5 dHAND-lacZ embryos, β-gal cells were observed in the perichondrium surrounding the lesser horns of the hyoid, with few labeled cells actually composing the lesser horns or hyoid body (Fig. 4Q). Labeled cells were also present in the submandibular and sublingual gland mesenchyme (arrow in Fig. 4Q), but were absent in adipose tissue (see inset, Fig. 4Q′). In contrast to this staining pattern, very little dHAND expression was detected by in situ hybridization in any hyoid structures (Fig. 4R), in stark contrast to the strong signal detected in adipose tissue. Expression was also detected in the submandibular and sublingual glands (see inset, Fig. 4R′).
Expression of dHAND during odontogenesis
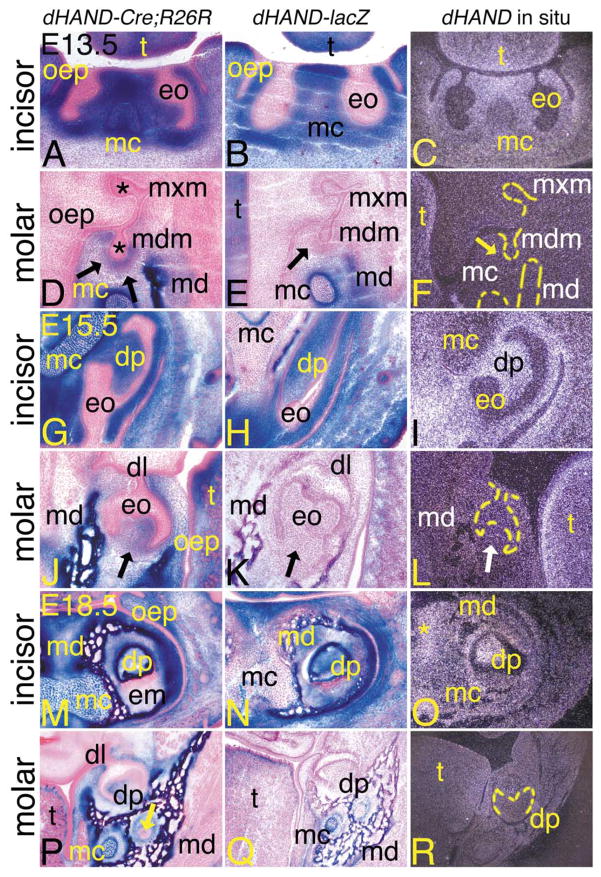
During odontogenesis, cephalic neural crest cell-derived ectomesenchyme contributes to the formation of odontoblasts (and consequently dentin matrix), dental pulp, cementum, and the periodontal ligaments of the incisors and molars (Chai et al., 2000). To examine potential roles of dHAND signaling in this process, we examined the extent of β-gal-stained cells at different stages of tooth development in dHAND-Cre;R26R embryos. As with our other analyses, we compared this staining with that observed in age-matched dHAND-lacZ embryos and with endogenous dHAND expression. At E13.5, tooth development is at the bud stage, which is characterized by proliferative areas (composed of neural crest-derived mesenchyme) surrounding the dental bud that ultimately forms the enamel organ. As shown in Fig. 5, these proliferative areas were composed solely of labeled cells around both mandibular incisor (Fig. 5A) and molar (Fig. 5D) buds of dHAND-Cre;R26R embryos. Labeled cells were not observed around the maxillary incisor and molar buds (Fig. 5D and data not shown). β-gal-positive cells in E13.5 dHAND-lacZ embryos were also observed in the neural crest-derived mesenchyme surrounding the enamel organs of mandibular incisors (Fig. 5B), but were absent around the mandibular molars (Fig. 5E). Likewise, endogenous dHAND expression was only observed in the mandibular incisor mesenchyme surrounding the enamel organs (Fig. 5C). Expression was not observed in Meckel’s cartilage (Fig. 5C) or around the mandibular molars (Fig. 5F). These findings thus indicate that the staining in the incisors of E13.5 dHAND-Cre;R26R embryos represents actual expression, while the staining around the molar buds represents fated expression.
Fig. 5.
Contribution of dHAND progeny cells during odontogenesis in dHAND-Cre;R26R embryos. Transverse (A–L) and frontal (M–R) sections through the heads of dHAND-Cre;R26R (A, D, G, J, M, P), dHAND-lacZ (B, E, H, K, N, Q), and wild type (C, F, I, L, O, R) embryos. Treatment of sections was performed as described in Fig. 3. (A–C) Transverse sections through the incisors of E13.5 embryos. (A, B) Labeled cells in dHAND-Cre;R26R and dHAND-lacZ embryos are apparent throughout the developing lower jaw, including in Meckel’s cartilage (mc) and mesenchyme destined to become dentin and pulp of the lower incisors. Labeled cells are not observed in the oral epithelium (oep) or developing enamel organ (eo). (C) dHAND expression in wild type embryos is observed in the tongue (t) and mesenchyme surrounding the incisor enamel organs. Less expression is observed in Meckel’s cartilage. (D–F) Transverse sections through the molar of E13.5 embryos, showing developing dentition of the molar at the late bud stage. (D) Labeled cells in dHAND-Cre;R26R embryos are observed in the mesenchyme surrounding the bud of the developing mandibular molar (mdm; arrows) and mandibular bone (md). Oral epithelium and developing enamel organs (*) of both maxillary (mxm) and mandibular molars are unlabeled. (E) Labeled cells in dHAND-lacZ embryos are not observed surrounding the mandibular molar buds (arrow). (F) Endogenous dHAND expression is also not observed surrounding the mandibular molar buds (arrow). (G–I) Transverse sections through the incisors of E15.5 embryos. The incisors have progressed to a cap stage, approaching the “bell” configuration. (G) The nonlabeled enamel organ in dHAND-Cre;R26R embryos partially encases the labeled dental papilla (dp) and appears irregular in shape due to the transverse angle of the section. The symphysis of Meckel’s cartilage is almost completely composed of labeled cells. (H) Labeled cells in dHAND-lacZ embryos also compose most of the dental pulp of the incisor, in contrast to the mixing of labeled and unlabeled cells in the symphysis of Meckel’s cartilage. (I) dHAND expression is observed in the dental pulp and surrounding mesenchyme. Expression is much weaker in Meckel’s cartilage symphysis. (J–L) Transverse sections through the molar of E15.5 embryos. (J) In the mandibular molar, also in the cap stage, labeled cells in dHAND-Cre; R26R embryos contribute to the dental papilla (arrow), though this area also contains unlabeled cells. (K) The dental papilla of mandibular molars from dHAND-lacZ embryos is composed solely of unlabeled cells (arrow). (L) dHAND expression is not observed in the papilla of the mandibular molars (white arrow), in contrast to the strong expression in the lamina propria of the tongue. (M–O) Frontal sections through the incisors of E18.5 embryos. (M) Labeled cells in dHAND-Cre;R26R embryos appear in the dental papilla of the mandibular incisors, mandibular bone (md), Meckel’s cartilage, and surrounding mesenchyme. Oral epithelium remains unlabeled, as does the developed enamel matrix (em). (N) A similar pattern of staining is observed in the incisors of dHAND-lacZ embryos, though few labeled cells are observed in Meckel’s cartilage. (O) dHAND expression is observed in the incisor dental pulp, surrounding bone, and mesenchyme. Very little expression is observed in Meckel’s cartilage. (P–R) Frontal sections through the molars of E18.5 embryos. (P) Labeled cells in dHAND-Cre;R26R embryos appear in the dental papilla, although extensive mixing with unlabeled cells has occurred. Labeled cells are also observed in the mandibular bone, Meckel’s cartilage, and surrounding connective tissue. The residual dental lamina (dl) can be seen connecting the unlabeled oral epithelium and enamel organ. Labeled cells are not observed in mandibular nerves (arrow). (Q) Labeled cells in dHAND-lacZ embryos are confined to the mandibular bone and Meckel’s cartilage. Labeled cells are not observed in the dental papilla of the molar. (R) A low level of dHAND expression is observed in the mandibular bone surrounding the molar of wild type embryos, but expression is not observed in the molar dental pulp.
By E15.5, odontogenesis has reached the cap stage, at which time the nonlabeled epithelial component of the developing tooth has begun to take the shape of the future tooth. Labeled cells comprising the dental papilla of both the mandibular molars and incisors were observed beneath the unlabeled epithelial cap in dHAND-Cre;R26R embryos (Fig. 5G and J). The relative number of unlabeled cells seemed to be higher in the molar papilla (arrow in Fig. 5G) than in the incisor papilla. The surrounding mandibular bone, symphysis of Meckel’s cartilage, and mesenchyme were also labeled. β-gal-positive cells were also observed in the dental papilla of the incisors of dHAND-lacZ embryos, though were mixed with unlabeled cells in the symphysis of Meckel’s cartilage (Fig. 5H). Labeled cells were again absent in the molar papilla (arrow in Fig. 5K). Endogenous dHAND expression was only observed in the incisor dental papilla and surrounding mesenchyme (Fig. 5I). As observed at E13.5, expression was absent in the dental papilla of lower molars (Fig. 5L) and Meckel’s cartilage (Fig. 5I), again suggesting differences in actual vs fated expression in the incisors and molars of dHAND-Cre;R26R embryos.
At E18.5, the distribution of dHAND progeny cells in the teeth of dHAND-Cre;R26R embryos was essentially unchanged. Dentin matrix, encompassing odontoblastic processes within dentinal tubules, showed strong staining in both developing mandibular incisors (Fig. 5M) and molars (Fig. 5P). Unlabeled cells were obvious in the dental papilla, likely reflecting the observed neural crest and nonneural crest contribution to the dental pulp (Chai et al., 2000). However, this mixing was once again more prominent in the molar pulp. The mandibular bone and surrounding mesenchyme were also composed of labeled cells. The entire enamel organ, including inner and outer enamel epithelial cells and the residual dental lamina, was unlabeled. Labeled cells were not observed in any part of the developing maxillary incisors (data not shown), though a few scattered cells were observed in the maxilla (data not shown).
In E18.5 dHAND-lacZ embryos, labeled cells were observed in the bone and mesenchyme surrounding the mandibular incisor dental papilla, with labeled cells in the symphysis of Meckel’s cartilage mixed with unlabeled cells (Fig. 5N). While β-gal-positive cells were present in the mandibular bone surrounding the lower molars, labeled cells were not present in the molar dental papilla (Fig. 5Q). This pattern closely reflected the endogenous dHAND expression pattern, with weak dHAND signal observed in the mandibular incisor dental papilla, distal mandibular bone, and surrounding mesenchyme (Fig. 5O). No signal was detected in the mandibular molar or surrounding mesenchyme (Fig. 5R) or in Meckel’s cartilage (Fig. 5O).
Discussion
Craniofacial development is regulated by multiple hierarchical signaling cascades that provide patterning and differentiation information to cephalic neural crest cells and their derivatives (Wilkie and Morriss-Kay, 2001). The transcription factor dHAND is likely involved in one or more of these cascades, based on previous studies of endogenous and transgenic expression patterns. We have shown here using fate analysis of dHAND progeny cells that dHAND likely plays crucial roles in development of neural crest-derived structures of the face, though based on our fate analysis and comparison with dHAND-lacZ expression and endogenous dHAND expression, the regulation of its expression is quite complex.
dHAND-Cre expression as a marker of current and past dHAND expression
One advantage of using Cre expression to examine gene expression patterns is the indelible nature of the staining, allowing observation of cell fate (Chai et al., 2000). These experiments also require cautious interpretation of resulting expression patterns, as transgenic expression constructs can lack elements necessary for complete gene expression or silencing. Indeed, analysis of the Mrf4/Myf5 locus reveals pharyngeal arch enhancer elements 88 kb upstream from the transcription start site (Carvajal et al., 2001). In this study, we have shown that labeled cells in dHAND-Cre;R26R embryos are observed in almost all cranial structures in which endogenous dHAND expression is observed (this study and Howard et al., 2000; Srivastava et al., 1995, 1997; Thomas et al., 1998). This includes the sympathetic ganglion, where dHAND-lacZ expression was not detected (this study, and Charité et al., 2001). While expression of the dHAND-lacZ transgene in the −11.0-kb line could be affected by transgene insertion effects, it is more likely that detection of dHAND-Cre expression is aided by the relative strength of the ROSA26 promoter. The ROSA26 gene is highly expressed in almost all embryonic and adult tissues (Soriano, 1997, 1999) and may therefore compensate for weak or partial enhancers, thus allowing weak Cre expression to be detected. This may be particularly true of the sympathetic ganglion expression, considering the scattered nature of staining in dHAND-Cre;R26R embryos vs the robust endogenous expression at E13.5. Using dHAND-lacZ deletion constructs, McFadden et al. (2000) demonstrated that, while the entire cardiac enhancer for dHAND was not required for expression of lacZ in the heart, partial enhancers resulted in an overall attenuation of cardiac expression.
Labeled cells in dHAND-Cre;R26R embryos are observed in several locations that likely reflect ectopic expression or activation of enhancers within the dHAND upstream region, including the DRG and limb bud. Aberrant induction of transgene expression by local enhancers surrounding the integration site is well documented. In our previous dHAND promoter study, limb bud expression was occasionally observed in F0 transgenic embryos carrying different deletion constructs of the dHAND promoter (Charité et al., 2001). However, embryos generated with the 7.4-kb fragment consistently showed the same abnormal limb bud pattern as we have observed in our dHAND-Cre embryos (J.C. and E.N.O. unpublished data). It is possible that the limb bud represents a permissive environment for ectopic transgene expression, or that an enhancer within the −7.4-kb upstream region acts in a promiscuous manner in the absence of the full dHAND upstream region.
Potential roles of dHAND during facial development
By comparing β-gal staining in dHAND-Cre;R26R and dHAND-lacZ embryos, we can begin to integrate the temporal expression of endogenous dHAND expression with overall dHAND progeny cell fate. In this study, we found labeled cells throughout the first pharyngeal arch of E10.5 dHAND-Cre;R26R and dHAND-lacZ embryos, with staining in E18.5 dHAND-Cre;R26R and dHAND-lacZ embryos observed in numerous neural crest-derived structures, including the mandible and Meckel’s cartilage. It is slightly surprising that β-gal staining in E18.5 dHAND-lacZ transgenic mice, while weaker, is similar to that observed in E18.5 dHAND-Cre;R26R embryos. This likely does not reflect aberrant activity of the dHAND pharyngeal arch enhancer, as dHAND-specific expression of the dHAND-lacZ transgene along arch arteries three and four and their derivatives is correctly downregulated by E13.5 (unpublished observations). Rather, these current findings suggest that dHAND likely has multiple roles in facial development. The first probably occurs during patterning of neural crest cells and their derivatives that will eventually form the facial skeleton. It is at this time (E9.5–E10.5) that β-gal staining was first observed in the arches of both dHAND-Cre;R26R and dHAND-lacZ embryos, coinciding with the endogenous expression pattern of dHAND. Further, while dHAND−/− embryos die by E10.5, disruption of mandibular development is already apparent (Thomas et al., 1998). Thus, it appears that dHAND function is crucial for the early development of postmigratory neural crest cells.
The second role appears to be later in jaw development, during which the dHAND-lacZ transgene is expressed in structures undergoing osteogenesis and chondrogenesis. In chondrogenic structures, labeled cells in E18.5 dHAND-Cre;R26R embryos are observed throughout the cartilage, whereas labeled cells in E18.5 dHAND-lacZ embryos are mainly confined to the perichondrium surrounding the cartilages. This suggests a limited role for dHAND in late chondrogenesis, such as possibly maintaining the proliferative environment surrounding developing cartilages in the jaw and throat. Past and active expression of the dHAND transgenes appears more equal in osteogenic structures in E18.5 embryos. Numerous genes and signaling pathways are involved in osteogenesis (Karsenty and Wagner, 2002); based on our current findings, it will be interesting to examine dHAND expression in mice lacking these genes to determine potential roles for dHAND in signaling pathways crucial for osteogenesis.
Positively defining the roles of dHAND, particularly in E18.5 embryos, is difficult due to the lack of correlation between endogenous dHAND expression and expression of a dHAND-lacZ transgene. Promoter–lacZ transgenic constructs or lacZ gene cassettes used to inactivate genes are routinely used to define gene expression patterns. However, we have shown here that β-gal staining is present even when endogenous gene expression is below the level of detection. This could reflect the relative stability of the lacZ message compared with that of the dHAND message. Further, while the half-life of dHAND protein is not known, in vitro experiments have shown that nuclear-localized β-galactosidase enzyme maintains 100% activity for at least 10 h (Tsuneoka and Mekada, 1992) and has a half-life of at least 20–30 h in cultured cells (Bachmair et al., 1986; Gonda et al., 1989). Because of this stability, it is quite likely that a low level of gene expression would result in a much larger pool of β-gal enzyme, thus making detection much easier compared with detection of mRNAs by in situ hybridization analysis. More study of this point needs to be conducted to fully understand the basis of these observations, and suggests that some caution should be taken when using β-gal staining to define temporal patterns of gene expression.
dHAND expression within subsets of cephalic neural crest cells
We have shown in this study that dHAND progeny cells compose most of the bone, cartilage, and connective tissue of the lower jaw, suggesting a significant role for dHAND in cephalic neural crest cell development. However, while neural crest cells contribute to the cartilaginous precursors of malleus, incus, and stapes of the middle ear (Le Douarin, 1982; Noden, 1983, 1988), labeled cells in dHAND-Cre; R26R embryos are only observed in the manubrium of the malleus. This could indicate that one or more additional enhancers are required in addition to the arch enhancer in our construct to fully recapitulate the full dHAND expression domain within the arches. It is also possible that, while early dHAND expression appears quite broad, dHAND action within the neural crest derived ectomesenchyme is highly compartmentalized. Fate map analysis of chick neural crest cells illustrates that the malleus is derived from crest cells of different midbrain/hindbrain origins (Kontges and Lumsden, 1996). This presumably reflects both the origin and final spatial distribution of these cells within the arches, as recent studies suggest that neural crest cells are patterned by both signals in the neural tube and environmental signals encountered during migration into the arches (Trainor and Krumlauf, 2000; Trainor et al., 2002). It is possible that our results reflect a similar mixed contribution in mouse embryos, with dHAND (or at least individual dHAND enhancers) participating in the development of neural crest cells from specific midbrain/hindbrain origins or those residing in specific arch subregions.
dHAND expression during odontogenesis
Another aspect of facial development that was uncovered in our fate analysis was the difference in dHAND expression between incisors and molars. During early development, dHAND-progeny cells in dHAND-Cre;R26R embryos are observed in both mandibular incisor and molar mesenchyme and later, dental papilla, though actual gene expression is only observed in cells in the incisor region. As mentioned above, evidence of past expression (such as β-gal staining in dHAND-Cre;R26R embryos in the absence of current gene expression) in developing structures does not necessarily imply a direct function in structure formation. Further, neither incisor nor molar defects are observed in embryos, in which expression of both dHAND and eHAND is disrupted (Clouthier et al., 2000), suggesting that loss of these two transcription factors alone does not disrupt odontogenesis. However, as dHAND progeny cells are observed in both neural crest-derived incisor and molar dental papilla, it is tempting to speculate that dHAND provides early patterning information to crest cells that later populate the dental papilla of the mandible and that it has redundant functions with other mandibular arch genes. Studies of multigene knockout embryos will be required to better address this question.
In later development, dHAND expression remains in the mesenchyme and dental papilla of the incisors, but not in the molars. We cannot exclude the possibility that lack of molar expression is due to absence of one or more as yet unidentified arch enhancers required to fully recapitulate the endogenous dHAND expression pattern. However, distinct signaling pathways within the mandible are known to pattern the dentition within the maxilla and mandible (Ferguson et al., 2000; Thomas et al., 2000). It is possible that dHAND represents an “incisor-specific” gene, possibly playing roles in both late tooth development and positioning of the incisors within the mandible. Analysis of dHAND expression in mouse mutants with defects in incisor and/or molar development should help clarify dHAND’s role in odontogenesis.
dHAND-Cre mice as a tool to investigate developmental processes
Our present results demonstrate the power and utility of using Cre-based fate mapping studies to understand possible developmental functions of specific genes. More specifically, dHAND-Cre transgenic mice provide the opportunity to follow the fate of dHAND lineage neural crest and nonneural crest cells in different mouse mutant lines. Further, these mice should be useful in concert with conditional knockout mouse lines in dissecting the basis of multiple developmental and disease processes.
Acknowledgments
We thank Bo Mason, Jennifer Beavin, Tim Morgan, Shelly Dixon, Jian Xie, and Cassandra Evans for technical assistance, and James E.T. Richardson for helpful discussions. M.Y. is an investigator of the Howard Hughes Medical Institute. D.E.C. is a recipient of a Career Development Award from the NIDCR/NIH. This work was supported in part by grants from the National Institutes of Health (to D.E.C. and E.N.O.), the Kentucky Excellence in Education Research Trust Fund (to D.E.C.), and the Donald W. Reynolds Clinical Cardiovascular Research Center, Dallas, TX (to E.N.O.).
References
- Bachmair A, Finley D, Varshavsky A. In vivo half-life of a protein is a function of its amino-terminal residue. Science. 1986;234:179–186. doi: 10.1126/science.3018930. [DOI] [PubMed] [Google Scholar]
- Bronner-Fraser M. Origins and developmental potential of the neural crest. Exp Cell Res. 1995;218:405–417. doi: 10.1006/excr.1995.1173. [DOI] [PubMed] [Google Scholar]
- Carvajal JJ, Cox D, Summerbell D, Rigby PWJ. A BAC transgenic analysis of the Mrf4/Myf5 locus reveals interdigitated elements that control activation and maintenance of gene expression during muscle development. Development. 2001;128:1857–1868. doi: 10.1242/dev.128.10.1857. [DOI] [PubMed] [Google Scholar]
- Chai Y, Jiang X, Ito Y, Bringas P, Han J, Rositch DH, Soriano P, McMahon AP, Sucov HM. Fate of the mammalian cranial neural crest during tooth and mandibular morphogenesis. Development. 2000;127:1671–1679. doi: 10.1242/dev.127.8.1671. [DOI] [PubMed] [Google Scholar]
- Charité J, McFadden DG, Merlo GR, Levi G, Clouthier DE, Yanagisawa M, Richardson JA, Olson EN. Role of Dlx6 in regulation of an endothelin-1-dependent, dHAND branchial arch enhancer. Genes Dev. 2001;15:3039–3049. doi: 10.1101/gad.931701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charité J, McFadden DG, Olson EN. The bHLH transcription factor dHAND controls Sonic hedgehog expression and establishment of the zone of polarizing activity during limb development. Development. 2000;127:2461–2470. doi: 10.1242/dev.127.11.2461. [DOI] [PubMed] [Google Scholar]
- Clouthier DE, Comerford SA, Hammer RE. Hepatic fibrosis, glomerulosclerosis, and a lipodystrophy-like syndrome in PEPCK-TGF-β1 transgenic mice. J Clin Invest. 1997;100:2697–2713. doi: 10.1172/JCI119815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clouthier DE, Hosoda K, Richardson JA, Williams SC, Yanagisawa H, Kuwaki T, Kumada M, Hammer RE, Yanagisawa M. Cranial and cardiac neural crest defects in endothelin-A receptor-deficient mice. Development. 1998;125:813–824. doi: 10.1242/dev.125.5.813. [DOI] [PubMed] [Google Scholar]
- Clouthier DE, Williams SC, Yanagisawa H, Wieduwilt M, Richardson JA, Yanagisawa M. Signaling pathways crucial for craniofacial development revealed by endothelin-A receptor-deficient mice. Dev Biol. 2000;217:10–24. doi: 10.1006/dbio.1999.9527. [DOI] [PubMed] [Google Scholar]
- Comerford SA, Maika SD, Laimins LA, Messing A, Elsasser HP, Hammer RE. E6 and E7 expression from the HPV 18 LCR: development of genital hyperplasia and neoplasia in transgenic mice. Oncogene. 1995;10:587–597. [PubMed] [Google Scholar]
- Cserjesi P, Brown D, Lyons GE, Olson EN. Expression of the novel basic helix–loop– helix gene eHAND in neural crest derivatives and extraembryonic membranes during mouse development. Dev Biol. 1995;170:664–678. doi: 10.1006/dbio.1995.1245. [DOI] [PubMed] [Google Scholar]
- Dorsky RI, Moon RT, Raible DW. Control of neural crest cell fate by the Wnt signalling pathway. Nature. 1998;396:370–373. doi: 10.1038/24620. [DOI] [PubMed] [Google Scholar]
- Fekete DM. Development of the vertebrate ear: insights from knockouts and mutants. Trends Neurosci. 1999;22:263–269. doi: 10.1016/s0166-2236(98)01366-6. [DOI] [PubMed] [Google Scholar]
- Ferguson CA, Tucker AS, Sharpe PT. Temporospatial cell interactions regulating mandibular and maxillary arch patterning. Development. 2000;127:403–412. doi: 10.1242/dev.127.2.403. [DOI] [PubMed] [Google Scholar]
- Fernandez-Teran M, Piedra ME, Kathiriya IS, Srivastava D, Rodriguez-Rey JC, MAR Role of dHAND in the anterior–posterior polarization of the limb bud: implications for the Sonic hedgehog pathway. Development. 2000;127:2133–2142. doi: 10.1242/dev.127.10.2133. [DOI] [PubMed] [Google Scholar]
- Firulli AB, McFadden DG, Lin Q, Srivastava D, Olson EN. Heart and extra-embryonic mesodermal defects in mouse embryos lacking the bHLH transcription factor Hand1. Nat Genet. 1998;18:266–270. doi: 10.1038/ng0398-266. [DOI] [PubMed] [Google Scholar]
- Francis-West P, Ladher R, Barlow A, Graveson A. Signalling interactions during facial development. Mech Dev. 1998;75:3–28. doi: 10.1016/s0925-4773(98)00082-3. [DOI] [PubMed] [Google Scholar]
- Gonda DK, Bachmair A, Wünning I, Tobias JW, Lane WS, Varshavsky A. Universality and structure of the N-end rule. J Biol Chem. 1989;264:16700–16712. [PubMed] [Google Scholar]
- Hollenberg SM, Sternglanz R, Cheng PF, Weintraub H. Identification of a new family of tissue-specific basic helix-loop-helix proteins with a two-hybrid screen. Mol Cell Biol. 1995;15:3813–3822. doi: 10.1128/mcb.15.7.3813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard MJ, Stanke M, Schneider C, Wu X, Rohrer H. The transcription factor dHAND is a downstream effector of BMPs in sympathetic neuron specification. Development. 2000;127:4073–4081. doi: 10.1242/dev.127.18.4073. [DOI] [PubMed] [Google Scholar]
- Ikeya M, Lee SMK, Johnson JE, McMahon AP, Takada S. Wnt signalling required for expansion of neural crest and CNS progenitors. Nature. 1997;389:966–970. doi: 10.1038/40146. [DOI] [PubMed] [Google Scholar]
- Jaskoll T, Zhou YM, Chai Y, Makarenkova HP, Collinson JM, West JD, Hajihosseini MK, Lee J, Melnick M. Embryonic submandibular gland morphogenesis: stage-specific protein localization of FGFs, BMPs, Pax6 and Pax9 in normal mice and abnormal SMG phenotypes in FgfR2-IIIc(+/Delta), BMP7(−/−) and Pax6(−/−) mice. Cells Tissues Organs. 2002;170:83–98. doi: 10.1159/000046183. [DOI] [PubMed] [Google Scholar]
- Jiang X, Iseki S, Maxson RE, Sucov HM, Morriss-Kay GM. Tissue origins and interactions in the mammalian skull vault. Dev Biol. 2002;241:106–116. doi: 10.1006/dbio.2001.0487. [DOI] [PubMed] [Google Scholar]
- Karsenty G, Wagner EF. Reaching a genetic and molecular understanding of skeletal development. Dev Cell. 2002;2:389–406. doi: 10.1016/s1534-5807(02)00157-0. [DOI] [PubMed] [Google Scholar]
- Kisanuki YY, Hammer RE, Miyazaki J, Williams SC, Richardson JA, Yanagisawa M. Tie-2Cre transgenic mice: a new model for endothelial cell- lineage analysis in vivo. Dev Biol. 2001;230:230–242. doi: 10.1006/dbio.2000.0106. [DOI] [PubMed] [Google Scholar]
- Kontges G, Lumsden A. Rhombencephalic neural crest segmentation is preserved throughout craniofacial ontogeny. Development. 1996;122:3229–3242. doi: 10.1242/dev.122.10.3229. [DOI] [PubMed] [Google Scholar]
- Kraus P, Lufkin T. Mammalian Dlx homeobox gene control of craniofacial and inner ear morphogenesis. J Cell Biochem Suppl. 1999;32–33:133–140. doi: 10.1002/(sici)1097-4644(1999)75:32+<133::aid-jcb16>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- Le Douarin NM. The Neural Crest. Cambridge Univ. Press; Cambridge: 1982. [Google Scholar]
- Le Douarin NM, Ziller C, Couly GF. Patterning of neural crest derivatives in the avian embryo: in vivo and in vitro studies. Dev Biol. 1993;159:24–49. doi: 10.1006/dbio.1993.1219. [DOI] [PubMed] [Google Scholar]
- Mallo M. Formation of the middle ear: recent progress on the developmental and molecular mechanisms. Dev Biol. 2001;231:410–419. doi: 10.1006/dbio.2001.0154. [DOI] [PubMed] [Google Scholar]
- McFadden DG, Charité J, Richardson JA, Srivastava D, Firulli AB, Olson EN. A GATA-dependent right ventricular enhancer controls dHAND transcription in the developing heart. Development. 2000;127:5331–5341. doi: 10.1242/dev.127.24.5331. [DOI] [PubMed] [Google Scholar]
- Merlo GR, Zerega B, Paleari L, Trombino S, Mantero S, Levi G. Multiple functions of Dlx genes. Int J Dev Biol. 2000;44:619–626. [PubMed] [Google Scholar]
- Mina M. Regulation of mandibular growth and morphogenesis. Crit Rev Oral Biol Med. 2001;12:276–300. doi: 10.1177/10454411010120040101. [DOI] [PubMed] [Google Scholar]
- Noden DM. The role of the neural crest in patterning of avian cranial skeletal, connective, and muscle tissues. Dev Biol. 1983;96:144–165. doi: 10.1016/0012-1606(83)90318-4. [DOI] [PubMed] [Google Scholar]
- Noden DM. Interactions and fates of avian craniofacial mesenchyme. Development. 1988;103:121–140. doi: 10.1242/dev.103.Supplement.121. [DOI] [PubMed] [Google Scholar]
- O’Gorman S, Dagenais NA, Qian M, Marchuk Y. Protamine-Cre recombinase transgenes efficiently recombine target sequences in the male germ line of mice, but not in embryonic stem cells. Proc Natl Acad Sci USA. 1997;94:14602–14607. doi: 10.1073/pnas.94.26.14602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley P, Anson-Cartwright L, Cross JC. The Hand1 bHLH transcription factor is essential for placentation and cardiac morphogenesis. Nat Genet. 1998;18:271–275. doi: 10.1038/ng0398-271. [DOI] [PubMed] [Google Scholar]
- Schorle H, Meier P, Buchert M, Jaenisch R, Mitchell PJ. Transcription factor AP-2 essential for cranial closure and craniofacial development. Nature. 1996;381:235–238. doi: 10.1038/381235a0. [DOI] [PubMed] [Google Scholar]
- Serbedzija GN, Bronner-Fraser M, Fraser SE. Vital dye analysis of cranial neural crest cell migration in the mouse embryo. Development. 1992;116:297–307. doi: 10.1242/dev.116.2.297. [DOI] [PubMed] [Google Scholar]
- Shelton JM, Lee MH, Richardson JA, SP Microsomal triglyceride transfer protein expression during mouse development. J Lipid Res. 2000;41:532–537. [PubMed] [Google Scholar]
- Soriano P. The PDGFα receptor is required for neural crest cell development and for normal patterning of the somites. Development. 1997;124:2691–2700. doi: 10.1242/dev.124.14.2691. [DOI] [PubMed] [Google Scholar]
- Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet. 1999;21:70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- Srivastava D, Cserjesi P, Olson EN. A subclass of bHLH proteins required for cardiac morphogenesis. Science. 1995;270:1995–1999. doi: 10.1126/science.270.5244.1995. [DOI] [PubMed] [Google Scholar]
- Srivastava D, Thomas T, Lin Q, Kirby ML, Brown D, Olson EN. Regulation of cardiac mesodermal and neural crest development by the bHLH transcription factor, dHAND. Nat Genet. 1997;16:154–160. doi: 10.1038/ng0697-154. [DOI] [PubMed] [Google Scholar]
- Thomas BL, Liu JK, Rubenstein JLR, Sharpe PT. Independent regulation of Dlx2 expression in the epithelium and mesenchyme of the first branchial arch. Development. 2000;127:217–224. doi: 10.1242/dev.127.2.217. [DOI] [PubMed] [Google Scholar]
- Thomas BL, Sharpe PT. Patterning of the murine dentition by homeobox genes. Eur J Oral Sci. 1998;106 (Suppl 1):48–54. doi: 10.1111/j.1600-0722.1998.tb02153.x. [DOI] [PubMed] [Google Scholar]
- Thomas T, Kurihara H, Yamagishi H, Kurihara Y, Yazaki Y, Olson EN, Srivastava D. A signaling cascade involving endothelin-1, dHAND and Msx1 regulates development of neural-crest-derived branchial arch mesenchyme. Development. 1998;125:3005–3014. doi: 10.1242/dev.125.16.3005. [DOI] [PubMed] [Google Scholar]
- Trainor P, Krumlauf R. Plasticity in mouse neural crest cells reveals a new patterning role for cranial mesoderm. Nat Cell Biol. 2000;2:96–102. doi: 10.1038/35000051. [DOI] [PubMed] [Google Scholar]
- Trainor PA, Ariza-McNaughton L, Krumlauf R. Role of the isthmus and FGFs in resolving the paradox of neural Crest plasticity and prepatterning. Science. 2002;295:1288–1291. doi: 10.1126/science.1064540. [DOI] [PubMed] [Google Scholar]
- Tsuneoka M, Mekada E. Degradation of a nuclear-localized protein in mammalian COS cells, using Escherichia coli β-galactosidase as a model protein. J Biol Chem. 1992;267:9107–9111. [PubMed] [Google Scholar]
- Tucker SA, Yamada G, Grigoriou M, Pachnis V, Sharpe PT. Fgf-8 determines rostral-caudal polarity in the first branchial arch. Development. 1999;126:51–61. doi: 10.1242/dev.126.1.51. [DOI] [PubMed] [Google Scholar]
- Wilkie AO, Morriss-Kay GM. Genetics of craniofacial development and malformation. Nat Rev Genet. 2001;2:458–468. doi: 10.1038/35076601. [DOI] [PubMed] [Google Scholar]
- Zhang J, Hagopian-Donaldson S, Serbedzija G, Elsemore J, Plehn-Dujowich D, McMahon AP, Flavell RA, Williams T. Neural tube, skeletal and body wall defects in mice lacking transcription factor AP-2. Nature. 1996;381:238–241. doi: 10.1038/381238a0. [DOI] [PubMed] [Google Scholar]