Abstract
In the majority of patients with acquired thrombotic thrombocytopenic purpura (TTP), antibodies are directed toward the spacer domain of ADAMTS13. We have previously shown that region Y658-Y665 is involved. We now show that replacement of R660, Y661, or Y665 with alanine in ADAMTS13 reduced/abolished the binding of 2 previously isolated human monoclonal antibodies and polyclonal antibodies derived from plasma of 6 patients with acquired TTP. We investigated whether these residues also influenced cleavage of short von Willebrand factor (VWF) fragment substrate VWF115. An ADAMTS13 variant (R660A/Y661A/Y665A, ADAMTS13-RYY) showed a 12-fold reduced catalytic efficiency (kcat/Km) arising from greatly reduced (> 25-fold) binding, demonstrated by surface plasmon resonance. The influence of these residue changes on full-length VWF was determined with denaturing and flow assays. ADAMTS13-RYY had reduced activity in both, with proteolysis of VWF unaffected by autoantibody. Binding of ADAMTS13-RYY mutant to VWF was, however, similar to normal. Our results demonstrate that residues within Y658-Y665 of the ADAMTS13 spacer domain that are targeted by autoantibodies in TTP directly interact with a complementary exosite (E1660-R1668) within the VWF A2 domain. Residues R660, Y661, and Y665 are critical for proteolysis of short VWF substrates, but wider domain interactions also make important contributions to cleavage of full-length VWF.
Introduction
Von Willebrand factor (VWF) is a key hemostatic glycoprotein involved in the adhesion of platelets to sites of vascular perturbation.1 During its biosynthesis in endothelial cells, VWF undergoes a number of posttranslational modifications that include the formation of intermolecular disulfide bonds between the carboxyl-terminal cysteine knot domains and the amino-terminal D3 domains.2,3 Current findings suggest that the resulting VWF polymers condense into tubular structures in the trans Golgi network and are subsequently packaged into Weibel-Palade bodies, rod-shaped subcellular organelles.4,5 Upon release of Weibel-Palade body contents, the VWF tubules rapidly unfold, and the fluid shear stress in the flowing blood induces the formation of ultra-large VWF (UL-VWF) strings on the surface of endothelial cells.6,7 Andre et al6 have shown that the appearance of platelet-decorated strings is a transient process in vivo. This transient nature is attributed to the rapid proteolysis of UL-VWF multimers by the metalloprotease ADAMTS13.8,9 In the absence of ADAMTS13, the rate at which platelet strings disappear from the endothelium is markedly reduced.
Thrombotic thrombocytopenic purpura (TTP) is a thrombotic microangiopathy characterized by hemolytic anemia, severe thrombocytopenia, and the presence of schistocytes in blood smears and is accompanied by a deficiency in ADAMTS13. ADAMTS13 is a large multidomain protein that consists of a propeptide, a catalytic metalloprotease domain, a disintegrin-like domain, a thrombospondin type I repeat (TSP), a cysteine-rich domain, a spacer domain, 7 additional TSP repeats, and 2 carboxyl-terminal CUB domains.10–12 In the majority of patients with TTP, inhibitory antibodies targeting ADAMTS13 have been found.13–16 In addition to inhibition of ADAMTS13 activity, anti-ADAMTS13 antibodies may also accelerate ADAMTS13 clearance.17 Epitope mapping studies have shown that the cysteine-rich/spacer domains contain the major binding site for human anti-ADAMTS13 antibodies.18–21 Additional epitopes for human anti-ADAMTS13 antibodies located outside the spacer domain have also been identified.18,22,23 In a previous study, we have shown that amino acid residues Y658-Y665 within the spacer domain comprise part of a core binding site for anti-ADAMTS13 antibodies.24
The ADAMTS13 metalloprotease domain cleaves the VWF A2 domain at the Y1605-M1606 scissile bond. The metalloprotease domain by itself cannot efficiently proteolyse VWF; the proximal disintegrin-like, TSP1, cysteine-rich, and spacer (DTCS) domains are all required for ADAMTS13 activity under static conditions.19,25–27 Comparison of the activities of carboxyl-terminally truncated ADAMTS13 variants toward short substrates of VWF and inhibition studies with the use of VWF A2-derived peptides led to the identification of VWF A2 domain sequence E1660-R1668 as a high-affinity exosite that interacts with the spacer domain.28,29 Additional orientation or repositioning of the protease over the cleavage site occurs by interaction of the disintegrin-like domain of ADAMTS13 with another exosite located closer to the cleavage site, involving residue D1614 in the VWF A2 domain.30 Apart from the proximal metalloprotease and DTCS domains, the TSP2-8 and CUB1-2 domains have recently been implicated in the binding of the D4-CK domains of globular VWF in the absence of flow.31,32 Together, these findings suggest a model in which the TSP2-8/CUB1-2 domains mediate initial binding of ADAMTS13 to VWF, and once ADAMTS13 is docked on VWF, multiple interactions between the MDTCS domains and VWF A2 domain residues ensure correct positioning of the active site for cleavage of the Y1605-M1606 scissile bond. For these multiple interactions to occur, VWF must change from its globular to an unfolded conformation, which enables ADAMTS13 to access the cleavage site.
In this study, we specifically explored spacer domain residues within region Y658-Y665 that are targeted by antibodies in acquired TTP. Because anti-ADAMTS13 antibodies often directly inhibit ADAMTS13 proteolysis of VWF, we also sought to characterize the role of these residues in ADAMTS13 function. We provide evidence for an interactive surface comprising R660, Y661, and Y665 in the spacer domain that is not only an important recognition site for anti-ADAMTS13 antibodies in TTP but also plays a role in binding to and cleavage of VWF.
Methods
Patients
Plasma samples from a panel of patients (designated I-VI) presenting with acute, acquired TTP were included in this study. Patients I and II have been described previously.22,23 Informed consent for these studies was obtained from the patients, and the protocol was approved by the Medical Ethical Committee of the University Medical Center Utrecht in accordance with the Declaration of Helsinki. ADAMTS13 activity levels in all plasma samples were less than 5%, as measured by use of the fluorogenic FRETS-VWF73 substrate assay kit (Peptides International). Patient samples were selected for high inhibitor titers to obtain a clear signal in immunoprecipitation analysis. Inhibitor titers were measured with the Technozym ADAMTS13 inhibitor enzyme-linked immunosorbent assay (ELISA; Technoclone: I = 54 U/mL, II = 128 U/mL, III = 220 U/mL, IV = 156 U/mL, V = 101 U/mL, and VI = 215 U/mL).
Construction and expression of recombinant ADAMTS13 MDTCS variants for epitope-mapping studies
Construction and expression of wild-type ADAMTS13 propeptide/metalloprotease/disintegrin-like/TSP1/cysteine-rich/spacer fragment (PMDTCS13, but termed MDTCS13 here), a hybrid fragment containing the spacer domain of ADAMTS1 (MDTCS1), a TSP2-8 fragment, and a CUB1-2 domain fragment in High Five insect cells has been described previously.20 Single alanine mutants within region Y658-Y665 (Y658A, R659A, R660A, Y661A, G662A, E663A, E664A, Y665A), and mutant combinations R660A/Y661A (RY1), R660A/Y665A (RY2), Y661A/Y665A (YY), and R660A/Y661A/Y665A (RYY) were introduced into the wild-type pMIB-V5/his6 MDTCS13 vector by use of Quik-Change polymerase chain reaction (PCR; Stratagene). Sequences of the sense and antisense oligonucleotide primers, containing the nucleotides to be changed with 15-bp flanking regions, are available upon request. All vectors were verified by sequencing. Epitope-mapping of anti-ADAMTS13 antibodies derived from patients with TTP was performed by immunoprecipitation, essentially as described in Luken et al.20 For each immunoprecipitation reaction 35 μL of patient plasma was used. Human monoclonal anti-spacer domain antibodies I-9 and II-1 have previously been described and were derived from immunoglobulin V gene phage display libraries created from B cells from patients with TTP.22,23
Construction and expression of full-length ADAMTS13 variants for functional analysis
For expression of full-length ADAMTS13 variants in HEK293T cells, the same mutations described previously were introduced into vectors pcDNA3.1-myc-his ADAMTS1333 (single-point mutants) and pcDNA3.1-V5-his ADAMTS1334 (combination mutations). Transient transfection of HEK293T cells was performed with linear polyethylenimine (Polysciences). Proteins were expressed in Optimem supplemented with Glutamax (Invitrogen). Medium was harvested after 4 days, concentrated with 100-kDa molecular weight cutoff Amicon spin columns (Millipore), and dialyzed into 20mM Tris-HCl, pH 7.8; 150mM NaCl; and 10mM benzamidine. Expression and secretion of ADAMTS13 was confirmed by Western blotting with an anti-metalloprotease antibody (Abcam), an anti-V5 antibody (Invitrogen), or an anti-myc antibody (Santa Cruz Biotechnology). For binding studies, the ADAMTS13 variants were purified as described previously by the use of Ni2+ HiTrap columns coupled to an ÄKTA FPLC (GE Healthcare, Bio-Sciences AB).30,33 ADAMTS13 concentration was determined with a previously developed ELISA with purified rabbit polyclonal antibodies against ADAMTS13.33,35 Maxisorp plates (NUNC) were coated with polyclonal antibodies depleted from TSP2-4 antibodies by affinity chromatography, whereas bound ADAMTS13 was detected with the polyclonal TSP2-4 antibodies labeled with biotin.35
Expression and purification of VWF115 and VWF106
The short VWF A2 domain fragment VWF115, spanning residues 1554 to 1668, was expressed in Rosetta Escherichia coli and resolubilized and purified as described.36 A VWF fragment comprising residues 1554 to 1659 that lacks the spacer binding site (VWF106) was amplified by PCR with site-specific primers and cloned into pET100/D-TOPO (Invitrogen), which fuses a his-tag and Xpress-epitope to the N-terminus of the protein, similar to VWF115. VWF106 was purified with the same method as used for VWF115. Purified protein fragments were dialyzed into 20mM Tris-HCl, pH 7.8 and 500mM NaCl and then quantified by BCA assay (Pierce Biotechnology).
Proteolysis of VWF115/VWF106 by ADAMTS13 and its variants
For determination of VWF115/VWF106 proteolysis by qualitative sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) analysis, 2nM of each ADAMTS13 variant was preincubated in 20mM Tris-HCl, pH 7.8; 150mM NaCl; and 5mM CaCl2 at 37°C for 45 minutes before addition of 6μM to 10μM substrate (as indicated for Figures 3–4). At different time points (0-3 hours) samples were taken, and 10mM EDTA (ethylenediaminetetraacetic acid) was added to stop the reaction. Samples were analyzed on 4% to 12% NuPAGE gels (Invitrogen) followed by staining with Imperial Protein Stain (Pierce). For quantitative analysis of VWF115 proteolysis, similar experiments were performed with 2nM ADAMTS13 variant and 2.5μM VWF115. Samples were analyzed by high-performance liquid chromatography (HPLC) analysis and catalytic efficiencies (kcat/Km) were calculated as described.36
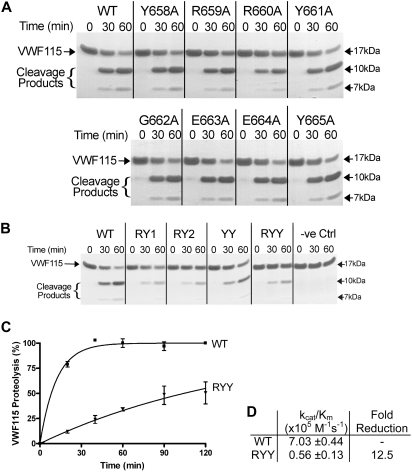
Figure 3.
Proteolytic cleavage of VWF115 by spacer domain ADAMTS13 variants with mutations in Y658-Y665. (A) Each single alanine variant (2nM each) in region Y658-Y665 or wild-type ADAMTS13 was incubated with 6μM VWF115 substrate at 37°C. After 0, 30, and 60 minutes samples were taken, and the reaction was stopped with EDTA and analyzed on SDS-PAGE. The VWF115 cleavage products of 10 and 7 kDa are indicated. (B) Similar reactions were set up by the use of ADAMTS13-RY1, RY2, YY, and RYY. Conditioned medium derived from nontransfected cells was included as a negative control (−ve Ctrl). (C) Cleavage products generated in activity assays with 2nM wild-type ADAMTS13 or ADAMTS13-RYY and 2μM VWF115 activity at the times indicated were quantified by HPLC. VWF115 proteolysis is depicted in percentages and mean values of 3 experiments, with error bars representing SD are shown. (D) From the graph in panel C, the catalytic efficiencies (kcat/Km) for VWF115 proteolysis were derived and mean values ± SD are shown in the table.
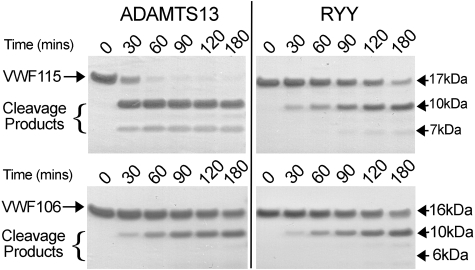
Figure 4.
Comparison of VWF115 and VWF106 proteolysis by ADAMTS13 and ADAMTS13-RYY. ADAMTS13 or ADAMTS13-RYY, 2nM, was incubated with 10μM VWF115 (top) or VWF106 (bottom) substrate at 37°C. At the time points indicated samples were taken, reactions stopped with EDTA, and analyzed on SDS-PAGE. Cleavage products of VWF115 (10 and 7 kDa) and VWF106 (10 and 6 kDa) are indicated.
Expression and purification of full-length recombinant VWF
Recombinant VWF was stably expressed by HEK293 cells and purified by the use of monoclonal antibody CLB-RAg20 as described previously.37 VWF multimers were eluted with 50mM HEPES, pH 7.4; 0.1M NaCl; and 1M KSCN. Fractions containing VWF were dialyzed against 50mM HEPES, pH 7.4; 150mM NaCl; and 50% (vol/vol) glycerol and stored at −20°C.
Binding of wild-type ADAMTS13 and ADAMTS13-RYY to VWF115, VWF106, and full-length VWF
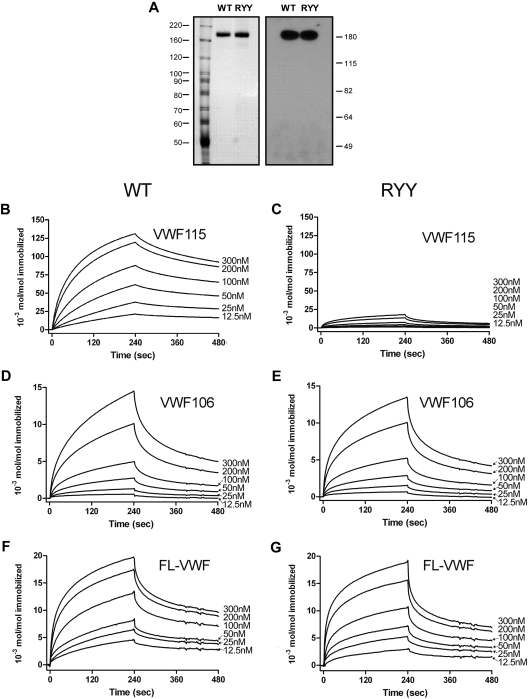
The binding of purified wild-type ADAMTS13 and ADAMTS13-RYY for VWF was determined by surface plasmon resonance (SPR) analysis on a BiaCORE2000 biosensor system (Biacore). We and others have previously established the validity of using SPR to study protein interactions between ADAMTS13 and VWF.32,36,38,39 Purified recombinant ADAMTS13 was dialyzed against 10mM HEPES, pH 7.4; 150mM NaCl; 0.0034mM EDTA; and 0.005% Tween20 and passed over a CM5 sensor chip at different concentrations (12.5-300nM) for 4 minutes at a flow of 20 μL/minute at 25°C. Dissociation of ADAMTS13 was then measured for 4 minutes. The chip was coated with either purified VWF115 substrate (∼ 163 fmol/mm2), purified recombinant VWF106 substrate (∼ 177 fmol/mm2), or purified recombinant full-length VWF (∼ 64 fmol/mm2), by use of the amine-coupling kit (Biacore). To enable comparison of results obtained with different sensor chips, the binding response was expressed as moles of ADAMTS13 bound per mole of VWF (fragment) immobilized on the chip, on the basis of the assumption that 1000 RU corresponds to 1 ng of bound protein per square millimeter. Regeneration of the sensorchip was performed with a buffer containing 50% ethylene glycol, 25mM lysine, and 0.5M NaCl, pH 7.4. Binding was corrected for background binding to a noncoated control channel.
Proteolysis of full-length recombinant VWF under denaturing conditions
Recombinant VWF was incubated with 1.5M guanidine-HCl at 37°C for 30 minutes. The denatured VWF was then diluted 10-fold to a final concentration of 40nM into 20mM Tris-HCl, pH 7.8; 150mM NaCl; 0.5% bovine serum albumin, and 5mM CaCl2 in which 3.5nM ADAMTS13 and its variants had been preincubated for 60 minutes. Monoclonal anti-spacer domain antibody II-1 or a control antibody was included at a concentration of 1.5μM during preincubation of ADAMTS13, as indicated in the figure legend. Samples were taken at sequential time points and quenched by adding loading buffer (10mM Tris; 2% SDS; 8M urea; and 1mM EDTA, pH 8.0). These were analyzed by 2% agarose gel electrophoresis and Western blotting.40,41
Proteolysis of newly secreted VWF on the surface of endothelial cells under flow
Endothelial cells were obtained from human umbilical veins and cultured as described previously.42 Secretion and formation of UL-VWF strings on the surface of endothelial cells was performed with a Wide Field Axiovert 200 microscope (Carl Zeiss Vision) and the Axiovision 4.7 software. Human umbilical vein endothelial cells were grown in disposable flow chambers (Flow-Through-Kit-μ-slide I-0.4 luer, 80 076; ibidi) in EGM2 medium (Invitrogen) until 100% confluency was reached. The assembled chamber was connected to a syringe pump to perfuse medium (Optimem; Invitrogen) containing 100μM histamine for 10 minutes, followed by the addition of freshly isolated washed platelets from 3 different donors (180 × 109/L) for 5 minutes in medium, and after that, medium containing ADAMTS13 or ADAMTS13-RYY was perfused for 5 minutes. ADAMTS13 had been preincubated for 30 minutes at 37°C in the presence or absence of monoclonal antibody II-1 (0.33μM). Assays were performed at a flow rate of 2.5 dyn/cm2.7 Images were collected at 10-second intervals for 5 minutes after the onset of perfusion with ADAMTS13 variants. UL-VWF released from endothelial cells formed strings with bound platelets, which were quantified by counting individual strings in 3 separate image fields.
Results
Critical amino acids in a conserved region of the spacer domain are involved in binding of anti-ADAMTS13 antibodies
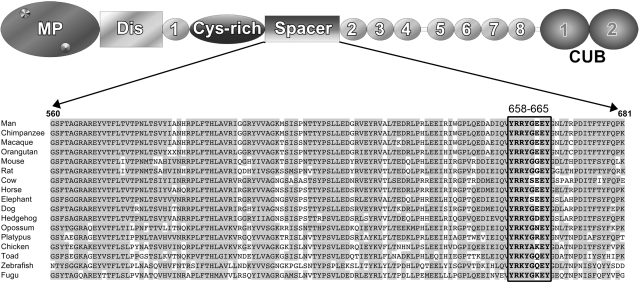
We have previously reported that exchange of region V657-G666 in ADAMTS13 for the corresponding region of ADAMTS1 abrogated binding of autoantibodies derived from 6 different TTP patients to ADAMTS13. Because residues V657 and G666 at either end of this region are conserved in the ADAMTS1 spacer domain, the observed effect is likely caused by the intervening residues Y658-Y665. Although the ADAMTS13 spacer domain is poorly conserved among ADAMTS family members, residues Y658-Y665 are highly conserved among species (Figure 1), with the exception of E663. To identify residues critical for antibody binding, we performed alanine-scanning mutagenesis of region Y658-Y665. We evaluated the binding of 2 previously isolated inhibitory human monoclonal antibodies, I-9 and II-1, both directed toward the spacer domain of ADAMTS13.22,23 Inspection of the reactivity of antibody I-9 and II-1 with MDCTS variants containing single alanine substitutions within region Y658-Y665 revealed that replacement of either R660, Y661, or Y665 completely abrogates the binding of these monoclonal antibodies to MDCTS (Figure 2A).
Figure 1.
Alignment of the ADAMTS13 spacer domain from different species. Above the sequence alignment a schematic domain structure of ADAMTS13 is depicted with the various different domains indicated; metalloprotease domain (MP), disintegrin-like (Dis), first thrombospondin type-I repeat (1), cysteine-rich domain (Cys-rich), spacer domain (Spacer), 2nd through 8th thrombospondin type-I repeats (2-8), and 2 CUB domains (CUB1-2). For the alignment of residues 560-681 of the ADAMTS13 spacer domain, predicted ADAMTS13 sequences from different species were derived from http://www.ensembl.org and the alignment was made with the ClustalW algorithm (http://www.ebi.ac.uk). Identical residues are in gray, and spacer domain residues Y658-Y665 are boxed.
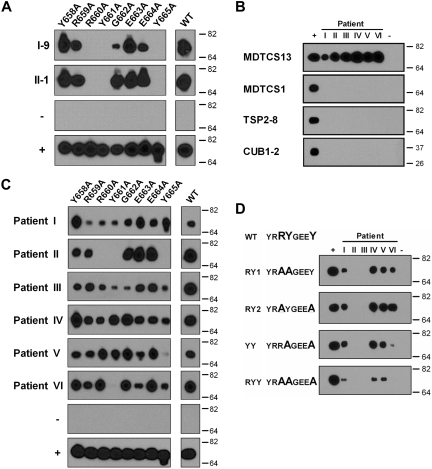
Figure 2.
Binding of single alanine variants in region Y658-Y665 of the spacer domain to antibodies from patients with acquired TTP. (A) Immunoprecipitation of single alanine variants in region Y658-Y665 of MDTCS with protein G Sepharose coupled with human monoclonal antibodies I-9 and II-1. (B) Reactivity of IgG present in 6 patient samples (I-VI) with recombinant ADAMTS13 proteins MDTCS13, MDTCS in which the spacer domain of ADAMTS13 was replaced by the spacer domain of ADAMTS1 (MDTCS1), TSP2-8, and CUB1-2. (C) Binding of patient-derived IgG (samples I-VI) to single alanine variants in region Y658-Y665 of MDTCS. (D) Reactivity of patient-derived IgG (samples I-VI) toward truncated ADAMTS13 variants containing multiple amino acid substitutions: MDTCS-RY1, MDTCS-RY2, MDTCS-YY, and MDTCS-RYY. Pull-down of ADAMTS13 proteins with anti-V5 antibody was included in all experiments as positive control (+), whereas in a monoclonal antipneumococcal antibody (subclass IgG1; panel A), or IgG derived from normal plasma was used as a negative control (−; panels B-D).
We subsequently explored the involvement of residues Y657-Y665 in the binding of polyclonal antibodies derived from plasma of 6 patients with acquired TTP (labeled I-VI in Figure 2B-D). First, to confirm that antibodies against the spacer domain were present in these patients, we compared the reactivity of these polyclonal antibodies with the wild-type MDTCS fragment and a recombinant protein in which the ADAMTS13 spacer domain was replaced by the spacer domain of ADAMTS1 (MDTCS1). Using this approach, we found that antibodies in the plasma of these patients bound to the MDTCS fragment but exhibited no reactivity with the MDTCS1 variant (Figure 2B). In addition, these polyclonal antibodies failed to react with isolated carboxyl-terminal TSP2-8 and CUB1-2 domain fragments (Figure 2B), thus excluding a major role for other antibody binding sites on ADAMTS13 that could confound the study of anti-spacer domain antibodies. Together, these analyses indicated that binding of polyclonal antibodies from all 6 TTP patients to ADAMTS13 is dependent on the presence of the spacer domain.
We then determined the reactivity of polyclonal antibodies present in plasma of patients with acquired TTP with the single alanine variants within region Y658-Y665 (Figure 2C). The antibodies present in 1 of the 6 patient samples (patient II) displayed an identical pattern of binding to the ADAMTS13 variants as the isolated human monoclonal antibodies, in that changing either residue R660, Y661, or Y665 abrogated antibody binding (compare Figure 2A and 2C, patient II). Replacement of Y661 also ablated the binding of antibodies present in plasma sample VI to MDTCS, whereas changing Y665 almost completely abolished binding of polyclonal antibodies present in plasma sample V. Binding of the polyclonal antibodies present in the other patient samples was more complex and was not completely abolished upon changing a single amino acid residue. Despite this, the other patient samples analyzed all showed reduced binding to certain single alanine variants. Overall, inspection of the patterns obtained for the different patient-derived immunoglobulin G (IgG) preparations suggested that particularly R660, Y661, and Y665 are important determinants for binding of anti-ADAMTS13 antibodies derived from TTP patient plasmas (Figure 2C).
We therefore generated MDTCS variants containing double alanine substitutions at R660, Y661, and Y665 (Figure 2D). Replacement of both R660 and Y661 by alanine (MDTCS13-RY1) abolished the binding of antibodies from 2 of 6 patients. Replacement of both R661 and Y665 (MDTCS13-RY2) yielded similar results. A strong reduction in reactivity of patient sample VI was observed when both Y661 and Y665 were replaced by alanine. Substitution of all 3 residues, R660, Y661, and Y665, to alanine (MDTCS13-RYY) revealed reduced (patients I, IV, and V) or even absent (patients II, III, VI) binding of antibodies of all patient samples analyzed, confirming that these residues are critical for optimal binding of patient derived antibodies to the spacer domain. Our findings thus suggest that R660, Y661, and Y665 are crucial residues providing an antigenic surface that is targeted by anti-ADAMTS13 antibodies that develop in patients with acquired TTP.
Proteolysis of VWF substrates by ADAMTS13 spacer domain variants
Because anti-ADAMTS13 autoantibodies in TTP patients frequently inhibit VWF proteolysis, we examined whether residues within region Y658-Y665 directly contribute to the function of ADAMTS13. We therefore introduced the single-point mutations in region Y658-Y665 into full-length ADAMTS13 to be able to perform functional analyses. All ADAMTS13 variants were expressed at similar level as full-length wild-type ADAMTS13, with the exception of variant Y658A, which was expressed at a modestly reduced level (data not shown). We evaluated the activity of the single alanine variants toward the previously described VWF115 substrate (VWF residues 1554-1668).36 Single alanine substitutions within region Y658-Y665 of ADAMTS13 (Y658A, R659A, R660A, Y661A, G662A, E663A, E664A, Y665A) were all able to proteolyze VWF115 (Figure 3A). Only ADAMTS13-R660A exhibited a minor reduction in activity, as evidenced by the amount of uncleaved VWF115 substrate after 1 hour compared with wild-type ADAMTS13. However, the double RY1 and RY2 and triple RYY variants all exhibited reduced proteolysis of VWF115, and, although less prominent, also using variant ADAMTS13-YY (Figure 3B). For quantitative assessment of the reduction in proteolytic activity caused by these substitutions, we analyzed the VWF115 cleavage products generated using ADAMTS13-RYY at different time points by HPLC (Figure 3C). From this analysis, the catalytic efficiency (kcat/Km) of ADAMTS13-RYY was found to be 0.56 plus or minus 0.13 × 105 M−1 s−1, 12.5-fold reduced compared with wild-type ADAMTS13 (Figure 3D).
Spacer domain residues R660, Y661, and Y665 interact with the previously identified exosite E1660-R1668 within the VWF A2 domain
The ADAMTS13 spacer domain mediates a high-affinity interaction with the VWF A2 domain, more precisely through an exosite comprising VWF residues E1660-R1668 that is contained within the VWF115 substrate.28 To study whether residues R660, Y661, and Y665 are involved in mediating the interaction of the ADAMTS13 spacer with VWF residues E1660-R1668, we examined the ability of ADAMTS13-RYY to cleave a shortened VWF A2 substrate that lacks residues E1660-R1668 (VWF106). Compared with the cleavage of VWF115, proteolysis of VWF106 by wild-type ADAMTS13 proceeded slowly (Figure 4). Processing of VWF106 by ADAMTS13-RYY proceeded with similar time course to that observed for wild-type ADAMTS13 (Figure 4). Proteolysis of VWF115 and VWF106 by ADAMTS13-RYY appeared equally efficient, that is, lack of the spacer binding region had no effect on proteolysis by ADAMTS13-RYY. These results indicate that residues R660, Y661, and Y665 contribute to an interactive surface within the spacer domain and that the function of this site is dependent upon residues E1660-R1668 in the VWF A2 domain.
To further confirm and extend the information on the ADAMTS13 R660-Y661-Y665 interaction with VWF residues E1660-R1668, we performed binding experiments using SPR. For this we used highly purified ADAMTS13 and ADAMTS13-RYY (see silver-stained gel and Western blot that confirm the fidelity of the quantitative ELISA in Figure 5A). The interaction of wild-type ADAMTS13 and ADAMTS13-RYY with immobilized VWF115 and VWF106 was studied. A dose-dependent increase in binding of wild-type ADAMTS13 to VWF115 was observed (Figure 5B-C). Conversely, ADAMTS13-RYY displayed strongly reduced binding to immobilized VWF115. Both ADAMTS13 and ADAMTS13-RYY displayed a similarly reduced binding to VWF106 (Figure 5D-E). After correction for the small differences in the amounts of VWF115 and VWF106 coated on the chip, we found that the binding of ADAMTS13-RYY to both VWF115 and VWF106 was similar. The binding between ADAMTS13 and VWF fragments appeared to be dependent upon multiple interactions between both proteins; accordingly, we could not fit the data to simple binding and dissociation models. In an attempt to quantitate the clear differences between wild-type and mutant ADAMTS13 in binding to VWF115, we plotted maximal binding response against ADAMTS13 concentration. From this, a greater than 25-fold difference in affinity was estimated. We then studied the binding of wild-type ADAMTS13 and ADAMTS13-RYY to full-length VWF. Again, we observed a dose-dependent binding of wild-type and mutant ADAMTS13 to full-length VWF (Figure 5F-G). However, when using this approach we detected no major difference between binding affinities of wild-type ADAMTS13 and ADAMTS13 RYY.
Figure 5.
Binding of wild-type ADAMTS13 and ADAMTS13-RYY to VWF studied with SPR. (A) Equal amounts of the ADAMTS13 preparations, as determined by ELISA, were used in binding experiments; see silver-stained SDS-PAGE gel (left), and Western blot with an anti-metalloprotease domain antibody (right). ADAMTS13-WT (B) or ADAMTS13-RYY (C), 12.5 to 300nM, was perfused over VWF115 immobilized to a CM5 sensor chip at a rate of 20 μL/minute for 240 seconds. Dissociation in the absence of ADAMTS13 was then studied for a further 240 seconds. (D-E) Experiments were conducted as in panels B and C but with immobilized VWF106. (F-G) Experiments were conducted as in panels B and C but with immobilized full-length VWF. In all graphs representative binding curves obtained with the use of 12.5, 25, 50, 100, 200, or 300nM wild-type ADAMTS13 or ADAMTS13-RYY are shown. Binding is represented as mol ADAMTS13/mol immobilized VWF (fragment).
Influence of ADAMTS13 spacer domain residues R660, Y661, and Y665 on cleavage of full-length VWF
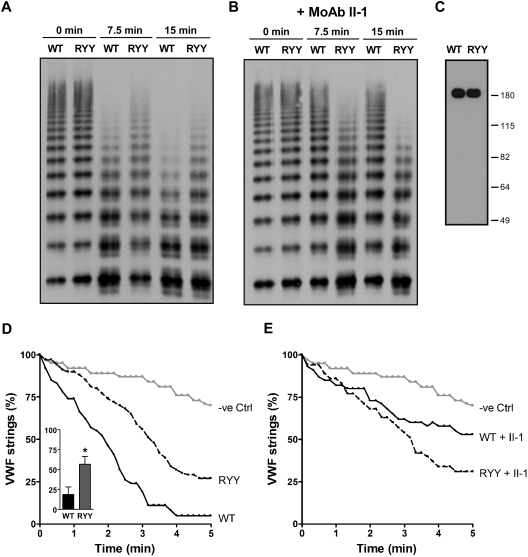
To further study the role of residues R660, Y661, and Y665 in ADAMTS13 function, we determined proteolytic activity of wild-type ADAMTS13 and ADAMTS13-RYY toward full-length recombinant VWF under denaturing conditions. A time-dependent loss of high-molecular-weight multimers was observed upon incubation of denatured VWF with wild-type ADAMTS13 (Figure 6A). Although ADAMTS13-RYY was still able to cleave VWF multimers, the efficiency of processing was reduced compared with ADAMTS13—already evident at 7.5 and 15 minutes. The ability of the patient-derived monoclonal antibody directed to the spacer domain, II-1, to inhibit ADAMTS13 cleavage of VWF is demonstrated in Figure 6B, which also shows an absence of inhibitory effect of this antibody on ADAMTS13-RYY activity, as predicted. Once again, it was confirmed with Western blotting that these differences in ADAMTS13 and ADAMTS13-RYY were not the result of concentration-dependent artifacts (Figure 6C).
Figure 6.
Proteolytic cleavage of full-length VWF by ADAMTS13 and ADAMTS13-RYY. (A) Processing activity of ADAMTS13 (WT) and ADAMTS13-RYY (RYY) toward denatured, multimeric recombinant VWF was determined essentially as described previously.30 Proteolysis of VWF multimers was determined at different time points (0, 7.5, and 15 minutes) and samples analyzed by Western blot. These experiments were performed in the presence of a control, monoclonal, antipneumococcal antibody (see Figure 2 legend), which does not affect proteolysis. Cleavage of full-length VWF results in a reduction in the largest VWF multimers and the appearance of characteristic triplet bands. (B) Experiments were conducted as in panel A, except the patient derived inhibitory monoclonal antibody II-1 directed to the spacer domain was used instead of the control antibody. (C) Western blot analysis of ADAMTS13 preparations in conditioned media. This demonstrates the fidelity of ELISA quantitation of these 2 proteins used in this figure. (D) Processing activity of ADAMTS13 (WT), ADAMTS13-RYY (RYY), and control medium (−ve Ctrl) under flow on the surface of endothelial cells. The number of UL-VWF strings after the addition of ADAMTS13 is expressed as a percentage of the number of UL-VWF strings that were present at the onset of the experiment. The number of UL-VWF strings was monitored for 5 minutes at 10-second intervals. A decline in the number of UL-VWF strings also was observed in the absence of ADAMTS13, reflecting spontaneous detachment of UL-VWF strings from the surface of endothelial cells (−ve Ctrl). The inset shows the percentage of VWF strings remaining at 2.5 minutes, when repeated (n = 3) experiments were analyzed. *P < .05 determined by Student paired t test. (E) Experiments conducted as in panel D but in the presence of monoclonal antibody II-1.
In vivo, processing of UL-VWF multimers most likely occurs on the surface of endothelial cells under flow.6–8 We therefore used a previously established assay to determine the activity of ADAMTS13-RYY on the surface of endothelial cells under a laminar flow of 2.5 dyn/cm2.7 Release of UL-VWF strings was provoked by stimulation of endothelial cells with 100μM histamine. After the addition of ADAMTS13, UL-VWF strings rapidly disappeared (Figure 6D). UL-VWF strings also disappeared in a control experiment in which control medium without ADAMTS13 was perfused, although at an appreciably slower rate. Comparison of ADAMTS13-RYY with the control revealed that ADAMTS13-RYY was still capable of processing of UL-VWF on the surface of endothelial cells, albeit at a slower rate than wild-type ADAMTS13. The rate of VWF proteolysis was dose dependently related to ADAMTS13 and ADAMTS13-RYY enzyme concentration (data not shown). Repeated determination of the percentage of VWF strings that remain at 2.5 minutes demonstrated a reproducible reduction in cleavage by ADAMTS13-RYY (see inset in Figure 6D). Preincubation of antibody II-1 with ADAMTS13 and ADAMTS13-RYY (in the same experiment) demonstrated inhibition of the activity of the former, but not the latter (Figure 6E).
Discussion
A major antigenic determinant for antibodies that develop in patients with acquired TTP resides within the spacer domain of ADAMTS13.18–21 In the current study, we show that ADAMTS13 residues R660, Y661, and Y665 are crucial for the binding of 2 previously isolated human monoclonal anti-ADAMTS13 antibodies designated I-9 and II-122,23 (Figure 2A). The reactivity of a panel of patient plasma–derived polyclonal antibodies confirms that R660, Y661, and Y665 also contribute to the binding of polyclonal anti-ADAMTS13 antibodies, with similar binding characteristics observed for patient sample II to those of monoclonal antibodies I-9 and II-1 (Figure 2C). It should be noted that antibody II-1 was isolated from peripheral B cells of patient II. The restricted epitope specificity observed for patient-derived IgG points to a largely oligoclonal response in this patient. For the other patients, a more heterogeneous pattern is observed; whereas mutation of R660, Y661, and Y665 abrogates the binding of antibodies from patients II, III, and VI, some antibodies of patients I, IV, and V are still able to bind, although the binding signal intensity obtained is reduced. This finding can be explained by a reduction in affinity of antibodies for the RYY variant, but it is more likely that a mixture of polyclonal antibodies is present in these patients.
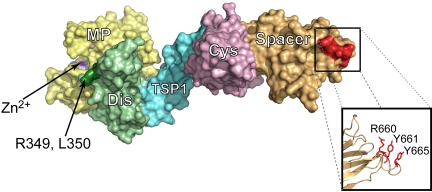
We hypothesized that as well as being an essential part of pathogenic antibody recognition, ADAMTS13 spacer domain residues R660, Y661, and Y665 also would form part of the binding surface recognized specifically by the VWF A2 domain in its high-affinity interaction with ADAMTS13. Experimental investigation of the functional role of these spacer domain residues in ADAMTS13 function is complicated potentially by several important experimental considerations. First, in past studies, the role of the ADAMTS13 spacer domain has mostly been derived from studies on carboxyl-terminally truncated ADAMTS13 variants, such as MDTCS. Such studies are potentially confounded by the finding that MDTCS often displays a greater catalytic efficiency than full-length ADAMTS13.21,27,43 The activity of truncated mutant MDTC, on the other hand, is approximately 25-fold reduced over MDTCS, but only approximately 10 times less active than full-length VWF.27 In this study, we have therefore introduced mutations into full-length ADAMTS13 rather than truncated fragments. A potential confounder in our mutagenesis approach is that the introduced point mutations may have resulted in structural changes in the ADAMTS13 spacer domain. Of note, however, is the recent crystal structure of the ADAMTS13 DTCS domains44 that was published during the preparation of this article. Residues R660, Y661, and Y665 align very well as a surface exposed cluster, available for ligand interactions (Figure 7).30,44 It therefore seems highly likely that there is an important role of these residues as a surface exposed exosite for VWF.
Figure 7.
Residues R660, Y661, and Y665 align as a cluster on the surface of ADAMTS13 MDTCS. The recent ADAMTS13 DTCS crystal structure (3GHM)44 and previously generated model of the ADAMTS13 metalloprotease domain30 were aligned to create a model of the ADAMTS13 MDTCS domains (PyMOL; DeLano Scientific). The metalloprotease domain is shown in yellow (MP) with the catalytic zinc ion indicated in magenta. The disintegrin-like domain (Dis) is in green and residues R349 and D350 that have previously been established as exosite for VWF are shown in dark green. The TSP1, cysteine-rich (Cys), and spacer domains are shown in blue, pink, and orange, respectively. Residues R660, Y661, and Y665 are highlighted by the box to show where they are located on the surface of MDTCS. The alternative representation of the side chains of these residues shows that they align in this structure to form a cluster that is surface exposed.
A second consideration is the need to unfold VWF to reveal binding sites and to expose the scissile bond. It has recently been shown that the isolated VWF A2 domain can unfold almost completely when force is applied at its amino- and carboxyl-terminus.45 Mechanical unfolding of this domain facilitates cleavage by ADAMTS13; without unfolding cleavage does not occur.45 The 3-dimensional structure of the VWF A2 domain has recently been determined at 1.9 Å resolution.46 The Y1605-M1606 bond is present in the middle of a β-sheet (β4), which is buried and inaccessible within the domain.46 In addition, tension induced unfolding is necessary to make its carboxyl-terminal sequence containing the E1660-R1668 alpha-helix available for binding.28 Results obtained with ADAMTS13 mutants are therefore highly dependent upon the specific VWF substrate used and whether they need unfolding for cleavage. Full-length substrates need to be unfolded with denaturants or flow,30,47,48 whereas in shorter substrates the binding and cleavage sites are exposed and available under physiologic conditions.28,29,36,49
Because of this complexity, we first used short VWF substrates, VWF106 and 115, to define the role that ADAMTS13 spacer domain interactions make to their cleavage. We wished to determine how specific binding of complementary domains influences substrate cleavage, without the need for denaturant- or shear-dependent unfolding. Here, we have shown that single alanine variants of the spacer domain of ADAMTS13 did not by themselves display reduced proteolysis of the VWF115 substrate, but the triple mutant, ADAMTS13-RYY, displays an approximately 12-fold reduced conversion of VWF115, but not of the shorter substrate VWF106. Reduced binding, greater than 25-fold, of ADAMTS13-RYY to VWF115 was demonstrated by SPR (Figure 5B), confirming that the substituted residues contribute to a primary binding site on ADAMTS13 for this fragment. Cleavage of VWF106 has previously been used by Gao et al27 to study the contribution of amino-terminal VWF A2 regions to proteolysis in the absence of the proposed high-affinity spacer interaction binding site. The similar activity toward VWF106 of WT-ADAMTS13 and ADAMTS13-RYY suggests that residues E1660-R1668, which are lacking in VWF106, interact directly with region Y658-Y665 of the ADAMTS13 spacer domain. This finding was again confirmed with direct binding studies, which revealed a strongly reduced, but yet residual, binding of both ADAMTS13 and ADAMTS13-RYY to VWF106 (Figure 5D-E). This similar binding of ADAMTS13 and ADAMTS13-RYY to VWF 106 confirms that R660, Y661, and Y665 do not participate in ADAMTS13 binding to VWF A2 1554 to 1659. In the 3-dimensional structure of the VWF A2 domain residues E1660 to R1668 are part of alpha-helixα6.46 It has been proposed that at high shear stress the adjacent vicinal cysteines C1669 and C1670 are dislocated from the hydrophobic core and causes rapid VWF A2 domain unfolding.46 VWF A2 domain unfolding leads to exposure of the binding site E1660-R1668, thereby allowing binding to residues Y658-Y665 of the spacer domain of ADAMTS13. Concurrent exposure of the Y1605-M1606 cleavage site, allows processing of by the protease domain once this is correctly positioned.27,30
To address the role and contribution of the ADAMTS13 spacer domain in interactions with full-length VWF, we used denaturing conditions to study processing of VWF multimers by the triple-mutant ADAMTS13-RYY (Figure 6A). Although there was a clear defect in VWF proteolysis under these conditions, the defect was perhaps less than was anticipated in view of the results with the VWF115 substrate. We therefore studied full-length VWF cleavage under physiologic shear stress on the surface of endothelial cells.7,8 Decreased activity of ADAMTS13-RYY also was observed under these conditions (Figure 6A). Newly released UL-VWF is highly hemostatically active in that it binds platelets very efficiently. Directly after its release, UL-VWF contains a larger number of VWF A2 domains that are in an “open” unfolded conformation50 and that allow for the high-affinity interaction with the spacer domain to take place. It appears that in the absence of the Y658-Y665 spacer/E1660-R1668 VWF A2 interaction, the binding of ADAMTS13 is less efficient but can, however, still take place.
These results with the full-length VWF substrate, studied under different conditions, therefore point to important functional roles of domain interactions other than ADAMTS13 spacer/VWF A2, as the full-length VWF substrate can still be effectively proteolyzed when these specific binding interactions are greatly reduced by mutagenesis. Moreover, direct binding of ADAMTS13-RYY to full-length VWF is similar to that observed for wild-type ADAMTS13 (Figure 5F-G), in contrast to the binding of ADAMTS13-RYY to VWF115. Evidence has been obtained for involvement of the carboxyl-terminal TSP2-8 and CUB domains in binding and processing of VWF.26,27,39,51 Very recently, 2 independent studies described a role for the carboxyl-terminal ADAMTS13 domains in the interaction with globular, folded VWF, mostly likely between TSP5-8 and the VWF D4-CK domains.31,32 Interestingly, inhibition of wild-type ADAMTS13 proteolysis by antibody II-1 at 1.5μM under flow is greater than the effect caused by changing residues R660, Y661, and Y665 of ADAMTS13 to alanine (ADAMTS13-RYY, compare Figure 6D-E). We anticipate that the antibody, because of its large size, may also shield other exosites required for binding of ADAMTS13 to VWF.
In summary, we have shown that specific ADAMTS13 spacer domain residues R660, Y661, and Y665 are the target for pathogenic antibodies that cause TTP. These same residues are part of the functional binding exosite that makes contact with the VWF A2 domain. Although we have shown that ADAMTS13 residues R660, Y661, and Y665 are critical determinants of molecular recognition of the VWF A2 domain, we also have shown that alternative binding interactions outside of the spacer/A2 domains also influence cleavage considerably.
Acknowledgments
We thank K. Gijzen (Sanquin-AMC Landsteiner Laboratory) for analyzing the processing of UL-VWF on the surface of endothelial cells.
This work was supported by grants from Landsteiner Foundation for Scientific Research (LSBR), the Netherlands Organization for Scientific Research (NWO), British Council Partnership in Science program (grant number PPS 851), The British Heart Foundation (Grant RG/02/008, now RG/06/007), and from the National Institute for Health Research (NIHR) Biomedical Research Center funding scheme.
Footnotes
Presented in part in abstract form at the 50th Annual Meeting of the American Society of Hematology, San Francisco, CA, December 8, 2008, and at the XXII Congress of the International Society on Thrombosis and Haemostasis, Boston, MA, July 15, 2009.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: W.P., J.T.B.C., and B.M.L. designed the study, performed experiments, analyzed results, and wrote the manuscript; R.F. provided patient samples and coordinated clinical aspects of the study; and J.V. and D.A.L. designed the study and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Dr Brenda M. Luken, Department of Haematology, Imperial College London, Hammersmith Hospital Campus, 5.S5 Commonwealth Bldg, Du Cane Rd, London, W12 0NN, UK; e-mail: b.luken@imperial.ac.uk.
References
- 1.Sadler JE. Von Willebrand factor, ADAMTS13, and thrombotic thrombocytopenic purpura. Blood. 2008;112(1):11–18. doi: 10.1182/blood-2008-02-078170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Voorberg J, Fontijn R, van Mourik JA, Pannekoek H. Domains involved in multimer assembly of von Willebrand factor (vWF): multimerization is independent of dimerization. EMBO J. 1990;9(3):797–803. doi: 10.1002/j.1460-2075.1990.tb08176.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Voorberg J, Fontijn R, Calafat J, Janssen H, van Mourik JA, Pannekoek H. Assembly and routing of von Willebrand factor variants: the requirements for disulfide-linked dimerization reside within the carboxy-terminal 151 amino acids. J Cell Biol. 1991;113(1):195–205. doi: 10.1083/jcb.113.1.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Michaux G, Abbitt KB, Collinson LM, Haberichter SL, Norman KE, Cutler DF. The physiological function of von Willebrand's factor depends on its tubular storage in endothelial Weibel-Palade bodies. Dev Cell. 2006;10(2):223–232. doi: 10.1016/j.devcel.2005.12.012. [DOI] [PubMed] [Google Scholar]
- 5.Huang RH, Wang Y, Roth R, et al. Assembly of Weibel-Palade body-like tubules from N-terminal domains of von Willebrand factor. Proc Natl Acad Sci U S A. 2008;105(2):482–487. doi: 10.1073/pnas.0710079105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.André P, Denis CV, Ware J, et al. Platelets adhere to and translocate on von Willebrand factor presented by endothelium in stimulated veins. Blood. 2000;96(10):3322–3328. [PubMed] [Google Scholar]
- 7.Dong JF, Moake JL, Nolasco L, et al. ADAMTS-13 rapidly cleaves newly secreted ultralarge von Willebrand factor multimers on the endothelial surface under flowing conditions. Blood. 2002;100(12):4033–4039. doi: 10.1182/blood-2002-05-1401. [DOI] [PubMed] [Google Scholar]
- 8.Motto DG, Chauhan AK, Zhu G, et al. Shigatoxin triggers thrombotic thrombocytopenic purpura in genetically susceptible ADAMTS13-deficient mice. J Clin Invest. 2005;115(10):2752–2761. doi: 10.1172/JCI26007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zheng XL, Sadler JE. Pathogenesis of thrombotic microangiopathies. Annu Rev Pathol. 2008;3:249–277. doi: 10.1146/annurev.pathmechdis.3.121806.154311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zheng X, Chung D, Takayama TK, Majerus EM, Sadler JE, Fujikawa K. Structure of von Willebrand factor-cleaving protease (ADAMTS13), a metalloprotease involved in thrombotic thrombocytopenic purpura. J Biol Chem. 2001;276(44):41059–41063. doi: 10.1074/jbc.C100515200. [DOI] [PubMed] [Google Scholar]
- 11.Levy GG, Nichols WC, Lian EC, et al. Mutations in a member of the ADAMTS gene family cause thrombotic thrombocytopenic purpura. Nature. 2001;413(6855):488–494. doi: 10.1038/35097008. [DOI] [PubMed] [Google Scholar]
- 12.Soejima K, Mimura N, Hirashima M, et al. A novel human metalloprotease synthesized in the liver and secreted into the blood: possibly, the von Willebrand factor-cleaving protease? J Biochem (Tokyo) 2001;130(4):475–480. doi: 10.1093/oxfordjournals.jbchem.a003009. [DOI] [PubMed] [Google Scholar]
- 13.Zheng XL, Kaufman RM, Goodnough LT, Sadler JE. Effect of plasma exchange on plasma ADAMTS13 metalloprotease activity, inhibitor level, and clinical outcome in patients with idiopathic and nonidiopathic thrombotic thrombocytopenic purpura. Blood. 2004;103(11):4043–4049. doi: 10.1182/blood-2003-11-4035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Peyvandi F, Ferrari S, Lavoretano S, Canciani MT, Mannucci PM. von Willebrand factor cleaving protease (ADAMTS-13) and ADAMTS-13 neutralizing autoantibodies in 100 patients with thrombotic thrombocytopenic purpura. Br J Haematol. 2004;127(4):433–439. doi: 10.1111/j.1365-2141.2004.05217.x. [DOI] [PubMed] [Google Scholar]
- 15.Ferrari S, Scheiflinger F, Rieger M, et al. Prognostic value of anti-ADAMTS 13 antibody features (Ig isotype, titer, and inhibitory effect) in a cohort of 35 adult French patients undergoing a first episode of thrombotic microangiopathy with undetectable ADAMTS 13 activity. Blood. 2007;109(7):2815–2822. doi: 10.1182/blood-2006-02-006064. [DOI] [PubMed] [Google Scholar]
- 16.Peyvandi F, Lavoretano S, Palla R, et al. ADAMTS13 and anti-ADAMTS13 antibodies as markers for recurrence of acquired thrombotic thrombocytopenic purpura during remission. Haematologica. 2008;93(2):232–239. doi: 10.3324/haematol.11739. [DOI] [PubMed] [Google Scholar]
- 17.Shelat SG, Smith P, Ai J, Zheng XL. Inhibitory autoantibodies against ADAMTS-13 in patients with thrombotic thrombocytopenic purpura bind ADAMTS-13 protease and may accelerate its clearance in vivo. J Thromb Haemost. 2006;4(8):1707–1717. doi: 10.1111/j.1538-7836.2006.02025.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Klaus C, Plaimauer B, Studt JD, et al. Epitope mapping of ADAMTS13 autoantibodies in acquired thrombotic thrombocytopenic purpura. Blood. 2004;103(12):4514–4519. doi: 10.1182/blood-2003-12-4165. [DOI] [PubMed] [Google Scholar]
- 19.Soejima K, Matsumoto M, Kokame K, et al. ADAMTS-13 cysteine-rich/spacer domains are functionally essential for von Willebrand factor cleavage. Blood. 2003;102(9):3232–3237. doi: 10.1182/blood-2003-03-0908. [DOI] [PubMed] [Google Scholar]
- 20.Luken BM, Turenhout EA, Hulstein JJ, Van Mourik JA, Fijnheer R, Voorberg J. The spacer domain of ADAMTS13 contains a major binding site for antibodies in patients with thrombotic thrombocytopenic purpura. Thromb Haemost. 2005;93(2):267–274. doi: 10.1160/TH04-05-0301. [DOI] [PubMed] [Google Scholar]
- 21.Zhou W, Dong L, Ginsburg D, Bouhassira EE, Tsai HM. Enzymatically active ADAMTS13 variants are not inhibited by anti-ADAMTS13 autoantibodies: a novel therapeutic strategy? J Biol Chem. 2005;280(48):39934–39941. doi: 10.1074/jbc.M504919200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Luken BM, Kaijen PH, Turenhout EA, et al. Multiple B-cell clones producing antibodies directed to the spacer and disintegrin/thrombospondin type-1 repeat 1 (TSP1) of ADAMTS13 in a patient with acquired thrombotic thrombocytopenic purpura. J Thromb Haemost. 2006;4(11):2355–2364. doi: 10.1111/j.1538-7836.2006.02164.x. [DOI] [PubMed] [Google Scholar]
- 23.Pos W, Luken BM, Hovinga JA, et al. VH1-69 germline encoded antibodies directed towards ADAMTS13 in patients with acquired thrombotic thrombocytopenic purpura. J Thromb Haemost. 2009;7(3):421–428. doi: 10.1111/j.1538-7836.2008.03250.x. [DOI] [PubMed] [Google Scholar]
- 24.Luken BM, Turenhout EA, Kaijen PH, et al. Amino acid regions 572-579 and 657-666 of the spacer domain of ADAMTS13 provide a common antigenic core required for binding of antibodies in patients with acquired TTP. Thromb Haemost. 2006;96(3):295–301. doi: 10.1160/TH06-03-0135. [DOI] [PubMed] [Google Scholar]
- 25.Zheng X, Nishio K, Majerus EM, Sadler JE. Cleavage of von Willebrand factor requires the spacer domain of the metalloprotease ADAMTS13. J Biol Chem. 2003;278(32):30136–30141. doi: 10.1074/jbc.M305331200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ai J, Smith P, Wang S, Zhang P, Zheng XL. The proximal carboxyl-terminal domains of ADAMTS13 determine substrate specificity and are all required for cleavage of von Willebrand factor. J Biol Chem. 2005;280(33):29428–29434. doi: 10.1074/jbc.M505513200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gao W, Anderson PJ, Sadler JE. Extensive contacts between ADAMTS13 exosites and von Willebrand factor domain A2 contribute to substrate specificity. Blood. 2008;112(5):1713–1719. doi: 10.1182/blood-2008-04-148759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gao W, Anderson PJ, Majerus EM, Tuley EA, Sadler JE. Exosite interactions contribute to tension-induced cleavage of von Willebrand factor by the antithrombotic ADAMTS13 metalloprotease. Proc Natl Acad Sci U S A. 2006;103(50):19099–19104. doi: 10.1073/pnas.0607264104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu JJ, Fujikawa K, McMullen BA, Chung DW. Characterization of a core binding site for ADAMTS-13 in the A2 domain of von Willebrand factor. Proc Natl Acad Sci U S A. 2006;103(49):18470–18474. doi: 10.1073/pnas.0609190103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.de Groot R, Bardhan A, Ramroop N, Lane DA, Crawley JT. Essential role of the disintegrin-like domain in ADAMTS13 function. Blood. 2009;113(22):5609–5616. doi: 10.1182/blood-2008-11-187914. [DOI] [PubMed] [Google Scholar]
- 31.Feys HB, Anderson PJ, Vanhoorelbeke K, Majerus EM, Sadler JE. Multi-step binding of ADAMTS13 to VWF. J Thromb Haemost. 2009;7(12):2088–2095. doi: 10.1111/j.1538-7836.2009.03620.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zanardelli S, Chion AC, Groot E, et al. A novel binding site for ADAMTS13 constitutively exposed on the surface of globular VWF. Blood. 2009;114(13):2819–2828. doi: 10.1182/blood-2009-05-224915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Crawley JT, Lam JK, Rance JB, Mollica LR, O'Donnell JS, Lane DA. Proteolytic inactivation of ADAMTS13 by thrombin and plasmin. Blood. 2005;105(3):1085–1093. doi: 10.1182/blood-2004-03-1101. [DOI] [PubMed] [Google Scholar]
- 34.Tjernberg P, Vos HL, Spaargaren-van Riel CC, et al. Differential effects of the loss of intrachain- versus interchain-disulfide bonds in the cystine-knot domain of von Willebrand factor on the clinical phenotype of von Willebrand disease. Thromb Haemost. 2006;96(6):717–724. doi: 10.1160/th06-08-0460. [DOI] [PubMed] [Google Scholar]
- 35.Chion CK, Doggen CJ, Crawley JT, Lane DA, Rosendaal FR. ADAMTS13 and von Willebrand factor and the risk of myocardial infarction in men. Blood. 2007;109(5):1998–2000. doi: 10.1182/blood-2006-07-038166. [DOI] [PubMed] [Google Scholar]
- 36.Zanardelli S, Crawley JT, Chion CK, Lam JK, Preston RJ, Lane DA. ADAMTS13 substrate recognition of von Willebrand factor A2 domain. J Biol Chem. 2006;281(3):1555–1563. doi: 10.1074/jbc.M508316200. [DOI] [PubMed] [Google Scholar]
- 37.van den Biggelaar M, Bierings R, Storm G, Voorberg J, Mertens K. Requirements for cellular co-trafficking of factor VIII and von Willebrand factor to Weibel-Palade bodies. J Thromb Haemost. 2007;5(11):2235–2242. doi: 10.1111/j.1538-7836.2007.02737.x. [DOI] [PubMed] [Google Scholar]
- 38.Dong JF, Moake JL, Bernardo A, et al. ADAMTS-13 metalloprotease interacts with the endothelial cell-derived ultra-large von Willebrand factor. J Biol Chem. 2003;278(32):29633–29639. doi: 10.1074/jbc.M301385200. [DOI] [PubMed] [Google Scholar]
- 39.Zhang P, Pan W, Rux AH, Sachais BS, Zheng XL. The cooperative activity between the carboxyl-terminal TSP1 repeats and the CUB domains of ADAMTS13 is crucial for recognition of von Willebrand factor under flow. Blood. 2007;110(6):1887–1894. doi: 10.1182/blood-2007-04-083329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schneppenheim R, Budde U, Oyen F, et al. von Willebrand factor cleaving protease and ADAMTS13 mutations in childhood TTP. Blood. 2003;101(5):1845–1850. doi: 10.1182/blood-2002-08-2399. [DOI] [PubMed] [Google Scholar]
- 41.McKinnon TA, Chion AC, Millington AJ, Lane DA, Laffan MA. N-linked glycosylation of VWF modulates its interaction with ADAMTS13. Blood. 2008;111(6):3042–3049. doi: 10.1182/blood-2007-06-095042. [DOI] [PubMed] [Google Scholar]
- 42.van den Biggelaar M, Bouwens EA, Kootstra NA, Hebbel RP, Voorberg J, Mertens K. Storage and regulated secretion of factor VIII in blood outgrowth endothelial cells. Haematologica. 2009;94(5):670–678. doi: 10.3324/haematol.13427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tao Z, Wang Y, Choi H, et al. Cleavage of ultralarge multimers of von Willebrand factor by C-terminal-truncated mutants of ADAMTS-13 under flow. Blood. 2005;106(1):141–143. doi: 10.1182/blood-2004-11-4188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Akiyama M, Takeda S, Kokame K, Takagi J, Miyata T. Crystal structures of the noncatalytic domains of ADAMTS13 reveal multiple discontinuous exosites for von Willebrand factor. Proc Natl Acad Sci U S A. 2009;106(46):19274–19279. doi: 10.1073/pnas.0909755106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang X, Halvorsen K, Zhang CZ, Wong WP, Springer TA. Mechanoenzymatic cleavage of the ultralarge vascular protein von Willebrand factor. Science. 2009;324(5932):1330–1334. doi: 10.1126/science.1170905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang Q, Zhou YF, Zhang CZ, Zhang X, Lu C, Springer TA. Structural specializations of A2, a force-sensing domain in the ultralarge vascular protein von Willebrand factor. Proc Natl Acad Sci U S A. 2009;106(23):9226–9231. doi: 10.1073/pnas.0903679106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Furlan M, Robles R, Lamie B. Partial purification and characterization of a protease from human plasma cleaving von Willebrand factor to fragments produced by in vivo proteolysis. Blood. 1996;87(10):4223–4234. [PubMed] [Google Scholar]
- 48.Tsai HM. Physiologic cleavage of von Willebrand factor by a plasma protease is dependent on its conformation and requires calcium ion. Blood. 1996;87(10):4235–4244. [PubMed] [Google Scholar]
- 49.Kokame K, Nobe Y, Kokubo Y, Okayama A, Miyata T. FRETS-VWF73, a first fluorogenic substrate for ADAMTS13 assay. Br J Haematol. 2005;129(1):93–100. doi: 10.1111/j.1365-2141.2005.05420.x. [DOI] [PubMed] [Google Scholar]
- 50.Groot E, Fijnheer R, Sebastian SA, de Groot PG, Lenting PJ. The active conformation of von Willebrand factor in patients with thrombotic thrombocytopenic purpura in remission. J Thromb Haemost. 2009;7(6):962–969. doi: 10.1111/j.1538-7836.2009.03433.x. [DOI] [PubMed] [Google Scholar]
- 51.Banno F, Chauhan AK, Kokame K, et al. The distal carboxyl-terminal domains of ADAMTS13 are required for regulation of in vivo thrombus formation. Blood. 2009;113(21):5323–5329. doi: 10.1182/blood-2008-07-169359. [DOI] [PMC free article] [PubMed] [Google Scholar]