Summary
In humans, several HLA-DRB loci (DRB1/3/4/5) encode diverse beta-chains which pair with alpha-chains to form DR molecules on the surface of APC. While DRB1 and DRB5 have been extensively studied, the role of DRB3/4 products of DR52/DR53 haplotypes has been largely neglected. To clarify the relative expression of DRB3, we quantified DRB3 mRNA levels in comparison with DRB1 mRNA from the same haplotype in both B cells and monocytes, observing quantitatively significant DRB3 synthesis. In CD19+ cells, DRB1*03/11/13 was 3.5-fold more abundant than DRB3, but in CD14+ this difference was only 2-fold. Monocytes also had lower overall levels of DR mRNA compared to B cells, which was confirmed by cell surface staining of DRB1 and DRB3. To evaluate the functional role of DRB3, tetramer-guided epitope mapping was used to detect T-cells against tetanus toxin and several influenza antigens presented by DRB3*0101/0202 or DRB1*03/11/13. None of the epitopes discovered were shared among any of the DR molecules. Quantitative assessment of DRB3-TT specific T-cells revealed that they are present at similar frequencies as those observed for DRB1. These results suggest that DRB3 plays a significant role in antigen-presentation with different epitopic preferences to DRB1. Therefore, DRB3, like DRB5, serves to extend and complement the peptide repertoire of DRB1 in antigen presentation.
Keywords: Antigen presentation/processing, MHC, T cells, Molecular immunology, Antigen presenting cells
1. Introduction
MHC class II molecules are expressed on the surface of antigen presenting cells such as B lymphocytes, macrophages and activated T cells. Three types of MHC class II molecules, HLA-DR, HLA-DQ, and HLA-DP, present peptides to CD4+ T cells. This recognition involves TCR binding to the MHC-peptide complex and triggers a cascade of downstream signaling events, culminating in T cell proliferation and the release of cytokines which shape the ensuing immune response [1, 2]. Structurally, MHC class II molecules are heterodimers formed by the association of homologous α and β chains. While the DR α chain is dimorphic, the β chain contains polymorphisms that influence the peptide binding properties of each variant. In addition to this diversity, certain DRB clusters contain a second “duplicate” DR β chain [3]. These duplications can be considered copy number polymorphisms (CNP), a re-discovered mechanism of gene variation [4]. In humans, next to DRB1, there are three functional secondary DRB chains: DRB3, DRB4 and DRB5. These secondary beta chains can also combine with the common DRα chain to form additional DR molecules on the cell surface [5]. Although the polymorphisms in all DRB loci are analogous, DRB1 is clearly the most polymorphic with over 500 alleles described. The other DRB loci are less polymorphic: DRB3 has 45 alleles, DRB4 has 14 alleles, and DRB5 has 18 alleles [6].
While DRB1 is present in all haplotypes, the secondary DRB loci are not found simultaneously in every individual. Rather, for any given HLA-DRB1 haplotype, only one secondary DRB gene such as DRB3, DRB4 or DRB5 may be present. This means that a maximum of 4 DRB products can be found in an individual. Furthermore, secondary DRB loci are in linkage disequilibrium with the DRB1, and therefore three main DRB haplotypes are defined: DR51, DR52, and DR53 [7]. These names originate from the serological classification of DR specificities created in the 1980s. The human HLA-DR52 haplotype comprises a single DRB1/DRB3 locus. However, several DRB1 alleles comprise this haplogroup: DRB1*03, DRB1*11, DRB1*12, DRB1*13, DRB1*14 (all of them named in this article HLA-DRB1-52). The role of DRB5*0101 in antigen presentation has been extensively studied due to the linkage disequilibrium with DRB1*15 and its association with multiple sclerosis (MS). Several works proved that the epitopes recognized by DRB1*1501 and DRB5*0101 are different [8–11], thus a complementary presentation exists. In favor of this complementarity, recent work in experimental autoimmune encephalomyelitis found that DRB5 attenuates MS severity, while encephalitogenic T cells are DRB1*1501 restricted [12].
Less work has been done in reference to the DRB3 locus. At present, based on the analysis of introns 1 to 4, it is believed that DRB3 and DRB1*03 appeared via gene duplication from a specific ancestral DRB1 gene, which was similar to DRB1*01/15 [3, 13]. On basis of exon 2 analysis, DRB3*0101 shows a higher similarity to DRB1*0301, so the generation of DRB3*0101 by gene conversion from DRB1*03 and an ancestral DRB3 appears feasible [14, 15]. Although 45 DRB3 alleles have been described, only B3*0101 and B3*0202 are frequent in all populations [16–19]. Interestingly, not all DRB1 alleles are associated with the same DRB3 alleles. Several population studies (mostly in ((AQ: North?)) American populations) have established the distribution and association of DRB3 and DRB1 alleles. These indicate that DRB1*1301 in caucasoid populations is primarily associated with DRB3*0101 (~63%) and DRB3*0202 (~37%) [16]; DRB1*0301 is associated mainly with DRB3*0101 (~83%) and less frequently with DRB3*0202 (~16%) or DRB3*0301 (~0.8%) [19]; DRB1*1101 is associated primarily with DRB3*0202 (~95%) [18]; DRB1*1401 has a 100% association with DRB3*0202, while DRB1*1402 is only associated with DRB3*0101 [17]; DRB1*1302 is directly associated with DRB3*0303. No study has specifically defined the distribution of DRB3 variants associated to DRB1*12, but this allele appears to be associated with DRB3*0101 and DRB3*0202. To simplify nomenclature, the abbreviation DRB1-52 will be used in this study in reference to all the DRB1 genes that are linked to DRB3.
It is well-known that the MHC-peptide-TCR interaction can lead either to T-cell activation or the induction of tolerance [20]. This function is modulated in part by the quantity of DR molecules presenting peptide to the T lymphocytes [21]. It is generally thought that DRB3 (like DRB4 and in contrast to DRB5) is expressed at significantly lower levels than DRB1 [5, 22]. However, the exact relationship between DRB3 and DRB1 expression levels remains unresolved. Traditional mRNA hybridization assays indicate a 4.5-fold difference in relative abundance [5, 22], but competitive PCR assays [23] and transcriptional activity analysis [24, 25] show similar levels for both. Also, only a limited set of DR52 haplotypes were evaluated in these studies. The functional ability of DRB3 to present peptides to T cells has been documented in several reports [26, 27]. In these, CD4+ T cells specific for several antigens (such as tetanus toxin) were found to respond in the context of DRB3. It has been suggested by some that DRB3*0101 and DRB1*03 could have similar binding preferences [28, 29] and that the DRB3*0101 epitopes could be cross-presented by DRB1*0301 [30]. Motivated by these apparent similarities, these authors suggest that the first and second molecules of the DR52 haplotype may have similar binding motifs and present redundant peptides. However, because the DRB3 loci shares no more homology with DRB1*0301 than the other DRB1-52 alleles [14], we hypothesized that the DRB3 loci may complement the repertoire of peptides that are presented by the DRB1 of the haplotype rather than cross-presenting similar peptides or duplicating its repertoire.
To more extensively characterize the expression of DRB3*0101 and DRB3*0202, we determined their relative expression by quantifying the amount of DRB3 mRNA in comparison to the amount of DRB1-52 mRNA in two different populations of APC, and by comparing cell surface staining by antibodies. Also, to better characterize the functional role of these proteins in antigen presentation, we developed DRB3 tetramers to identify T cell epitopes restricted by these DR molecules within tetanus toxin (TT) and several influenza antigens. We also determined the frequency of DRB3 restricted T cells specific for these antigens within the PBMC of pre-sensitized individuals. Our hypothesis for this work was that DRB3 plays a significant biological role in presenting antigens to T cells.
2. Results
2.1 Quantifying expression of DRB3 mRNA versus DRB1 (52) mRNA
To quantify DRB3 and DRB1-52 expression, we developed a real time PCR assay and tested its specificity, sensitivity and accuracy as indicated in the materials and methods section. We quantified the DRB3 and DRB1-52 mRNA in two different populations (CD19+ and CD14+) of APC using blood samples taken from seven healthy individuals representing four different haplotype patterns: two DRB3*0202-DRB1*11 individuals, two DRB3*0101-DRB1*03 individuals, two DRB3*0101-DRB1*13 and one homozygous DRB3*0101/0101-DRB1*03/13 (supplementary table 1).
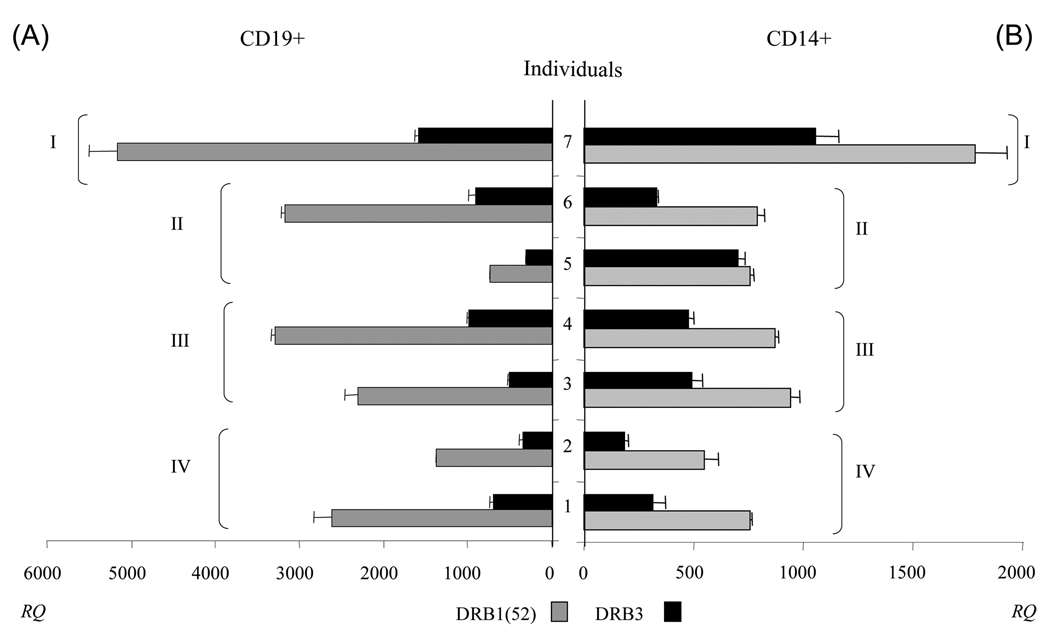
As expected, the quantity of mRNA measured in both cell populations indicated that DRB1-52 is expressed at significantly higher levels (p=0.0078) than DRB3 (see Fig 1). This trend was observed regardless of both DRB1-52 allele and DRB3 allele. Examining the individual APC populations, the mean of the DRB1/DRB3 ratio for CD19+ cells was 3.54 ± 0.65 indicating that DRB1 mRNA was clearly more abundant than DRB3 (figure 1A). When quantifying the mRNA abundance in CD14+ cells the mean of the DRB1/DRB3 ratio was 2.04 ± 0.62, indicating that DRB1-52 was also more abundant than DRB3 (figure 1B). These results differed from those observed using competitive PCR assays [23], which indicated similar levels of expression for both mRNA, but were somewhat consistent with mRNA hybridization assays, which indicated ~4.5-fold difference in expression [22]. It was evident for the subjects tested (see figure 1) that DRB1-52 mRNA levels were higher in CD19+ cells than in CD14+ cells: average mRNA levels were 2,294.26±936.03 and 809.66±130.57 ((AQ: Number of significant figures quoted varies, please make consistent (this recurs throughout the manuscript).)) respectively, while the average CD19/CD14 mRNA ratio (calculated for each individual and then averaged so that each would be weighed equally) was 2.85±1.07. DRB3 mRNA levels were also higher in CD19+ cells than in CD14+ cells: average mRNA levels were 647.55±270 and 448.43±199 respectively, while the average CD19/CD14 mRNA ratio was 1.69±0.81. However, for donorfive the overall DR mRNA expression was higher in monocytes than B cells (figure 1).
Figure 1. Level of expression of DRB1-52 and DRB3 on CD19+ cells and on CD14+ cells.
Level of DRB1-52 and DRB3 in CD19+ (A) and CD14+ (B) cells (see Supporting Information Table 1 for donor information). Data are expressed as ratio of DRB1 (grey) or DRB3 (black) to GAPDH × 105 copy numbers, and referred to as RQ. Data show mean + SD (n=3). I: donor with DRB1*0301/*1301 - DRB3*0101/*0101 typing; II: donors with DRB1*0301 and DRB3*0101 typing; III: donors with DRB1*1301 and DRB3*0101 typing; IV: donors with typing DRB1*1101 and DRB3*0202 typing. The paired comparison (two tailored Wilcoxon signed test) of DRB1 and DRB3 mRNA in both cell populations indicated that DRB1-52 is expressed at significantly higher levels (p=0.0078) than DRB3.
It might be expected that a “homozygous” DRB1-52/DRB3 individual would have increased levels of DRB1-52 and/or DRB3 mRNA. Indeed, donor seven had the highest mRNA levels for DRB1-52 and DRB3. However, the DRB1-52/DRB3 ratio was similar to the other donors. Notably, the mean DRB1/DRB3 ratio was significantly (p=0.0078) lower for the CD14+ population than mean ratio for the CD19+ population. These different DRB1/DRB3 ratios suggest that the contribution of DRB3 to the total pool of mRNA in resting state is quantitatively more important in monocytes than in B cells. It is also worth highlighting that for donor five, the DRB1-52/DRB3 ratio was close to 1. Together, these results suggest that while DRB3 mRNA is less abundant than DRB1-52 in all of the individuals and all of the cell types tested, the relative proportion can vary from being four times lower to nearly equal to the amount of DRB1 mRNA produced. Thus, it seems plausible that DRB3 molecule could play an important role in antigen presentation to T-cells.
2.2 Evaluation of DRB3 and DRB1 cell surface expression in B cells and monocytes
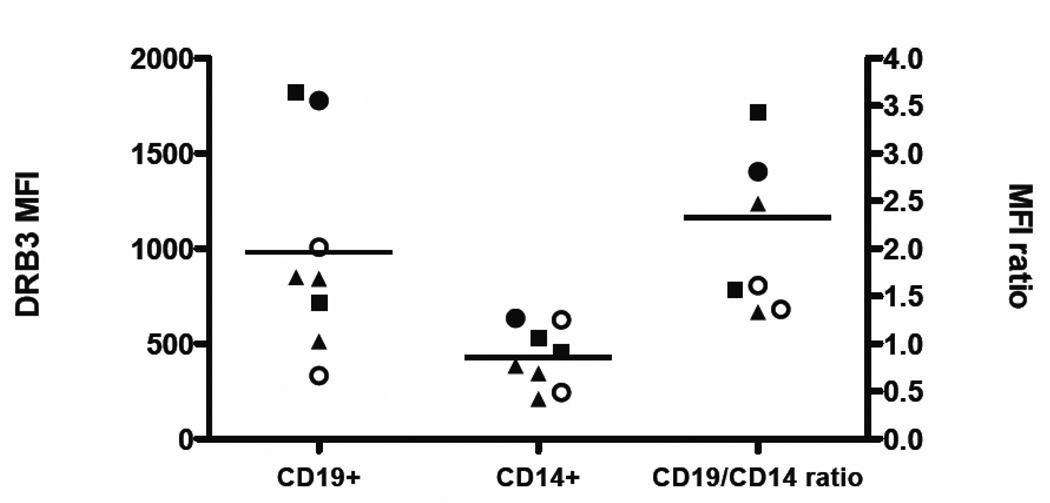
To corroborate our mRNA results at the protein level, we evaluated the expression of DRB3 on the surface of CD19+ B cells and CD14+ monocytes by flow cytometry. For these experiments, we used DRB3 (7.3.19.1) and DR11 (4i131) specific antibodies for cell surface staining. Because surface staining is only semi-quantitative, we compared the relative surface expression of DRB3 by dividing the mean fluorescence intensity (MFI) observed in CD19+ cells by the MFI observed in CD14+ cells. As expected, the level of MFI DRB3 was always higher on B cells than on monocytes (Figure 2) (P=0.0148). This was in agreement with observed mRNA levels: the mean ratio (CD19/CD14) of DRB3 surface expression was 2.33±1.02 (this level is not significantly different to the 1.6-fold difference seen in our mRNA results, p=0.21). For DR11, the mean ratio (CD19/CD14) was 1.59±0.18 (data not shown), and for the whole set of DR molecules was 1.9±0.36 (data not shown). These data suggest a modest correlation (r=0.647) between the observed mRNA levels and cell surface expression of protein ((AQ: The repeated reference to significance makes this sentence a little confusing. Pearson coefficients don’t come with an associated significance. By referencing significance of correlation are you commenting on a test of the null hypothesis that the slope of the best fit line is equal to zero? You need to quote a p-value for this test or give degrees of freedom as well as r-value if you want to indicate significance.)). The differences between the DRB3 mRNA and cell surface staining results could imply differences at the level of protein translation or cell surface recirculation.
Figure 2. Mean fluorescence intensity (MFI) observed for DR52 antibody staining.
The observed MFI for DRB3 antibody staining in seven donors is plotted for CD19+ cells (left), CD14+ cells (middle) with mean values indicated by horizontal lines. The CD19+/CD14+ ratio (right) is also plotted. Symbols in all the representations stand for donors typing, open circles for DRB1*13-DRB3*0101, filled triangles for DRB1*11-DRB3*0202, filled squares for DRB1*03-DRB3*0101 and filled circles for DRB1*03/13-DRB3*0101/0101. The level of DRB3 MFI was higher on B cells than on monocytes, p=0.0148 (two tailored Wilcoxon signed test).
2.3 Identification of HLA-DRB3 and HLA-DRB1-52 restricted T cell epitopes within influenza and tetanus toxin proteins
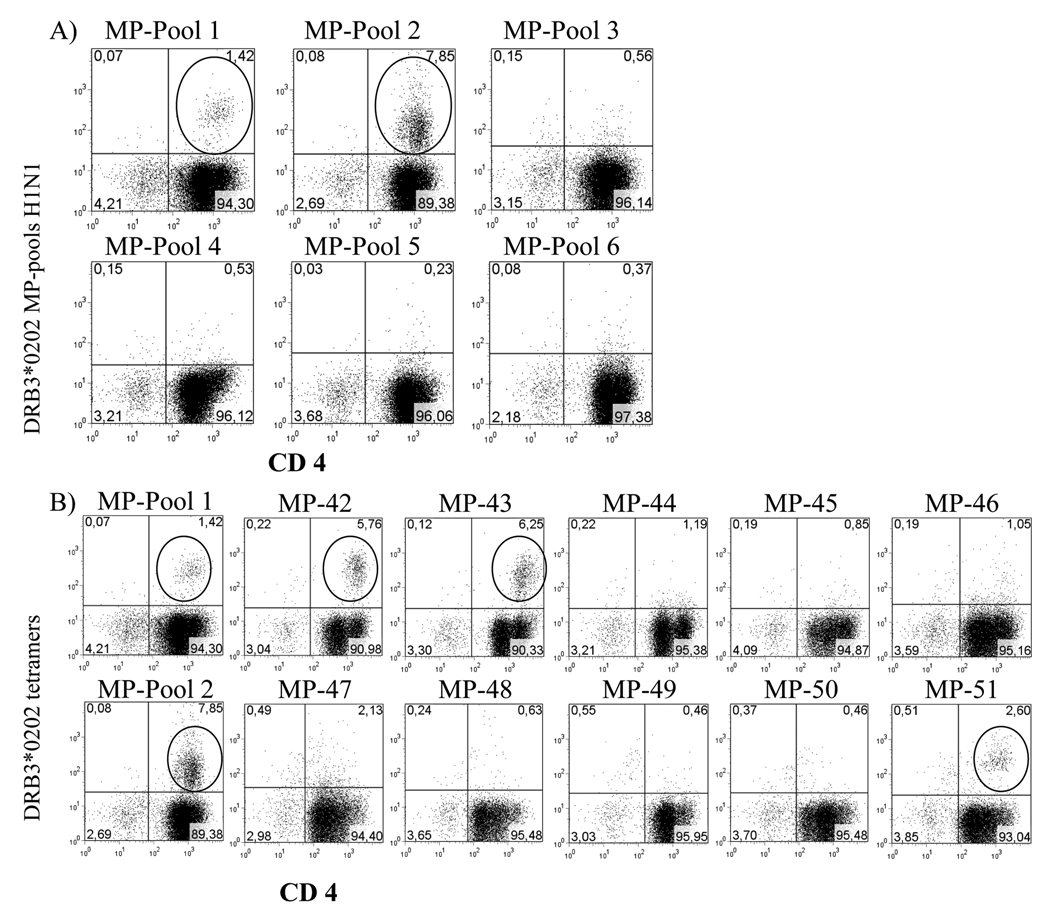
Tetramer Guided Epitope Mapping (TGEM) [31] approach was used to observe DRB3 restricted T cell responses and identify CD4+ T-cell epitopes within influenza virus matrix protein (MP) and Clostritium tetani toxin (TT) for individuals with HLA DRB3*0101 and DRB3*0202 haplotypes. For the mapping of MP epitopes, a total of 30 overlapping peptides (20 amino acids in length with an overlap of 12 residues) were synthesized. These peptides were divided into six pools with five consecutive peptides per pool. First, these peptide pools were used to stimulate the PBMC of a DRB3*0202 subject. Subsequent staining with the corresponding DRB3*0202 pooled peptide tetramers indicated a positive response for cells stimulated with pools 1 and 2 (figure 3A). These cells were analyzed again using tetramers loaded with the individual peptides from the positive pools, indicating positive responses to peptides MP1–20 (MSLLTEVETYVLSIIPSGPL) and MP9–28 (TYVLSIIPSGPLKAEIAQRL) from within pool 1 and MP73–92 (GLQRRRFVQNALNGNGDPNN) from within pool 2 (Figure 3B). Since MP1–20 and MP9–28 share residues 9–20, it is possible that these two peptides contain a single shared epitope. Identical experiments were performed for three additional DRB3*0202 individuals, showing similar positive results for MP1–20, MP9–28 and MP73–92. Similar experiments were carried out to examine MP responses in subjects with DRB3*0101, DRB1*0301, DRB1*1101 or DRB1*1301 typing. We did not find any epitopes for DRB3*0101, but we found one eptiope for DRB1*0301, three for DRB1*1101 and three for DRB1*1301. None of the epitopes were shared among any of the DR molecules; these results are summarized in Table 1. Subsequent experiments also identified the Influenza A panama H3N2 (HA3) derived peptide HA3271–291 (PIGKCNSECITPNGSIPNDK) as a DRB3*0202 restricted epitope.
Figure 3. Identification of DRB3*0202-restricted MP specific epitopes.
A) Representative staining profiles with DRB3*0202-MP tetramer pools. CD4+ T cells from a DRB3*0202 subject were stimulated with six pools of MP peptides, and stained with DRB3*0202 pooled peptide tetramers 14 days after stimulation. Circled events indicate populations that were tetramer positive. B) Fine mapping was performed for the tetramer positive pools (MP pool 1 and MP pool 2) using individual peptide tetramers. MP1–20 (MP42), MP9–28 (MP43), and MP73–92 (MP51) were identified as peptides that contain DRB3*0202 restricted epitopes, negative individual peptide tetramers from MP pools 1 and 2 are also represented. TGEM with MP pools was performed in two individuals.
Table 1.
Results of the TGEM analysis with DRB1 and DRB3 tetramers.
| Peptides | DRB1 positivities a | DRB3 positivities a | ||||||
|---|---|---|---|---|---|---|---|---|
| Antigen | Name | Position | Sequence | DRB1*0301 | DRB1*1101 | DRB1*1301 | DRB3*0101 | DRB3*0202 |
| MP | 42 | 1–20 | MSLLTEVETYVLSIIPSGPL | X | ||||
| MP | 43 | 9–28 | TYVLSIIPSGPLKAEIAQRL | X | ||||
| MP | 47 | 41–60 | VLMEWLKTRPILSPLTKGIL | X | ||||
| MP | 51 | 73–92 | GLQRRRFVQNALNGNGDPNN | X | ||||
| MP | 53 | 89–108 | DPNNMDKAVKLYRKLKREIT | X | ||||
| MP | 54 | 97–116 | VKLYRKLKREITFHGAKEIS | X | X | |||
| MP | 63 | 169–188 | TNPLIRHENRMVLASTTAKA | X | ||||
| MP | 67 | 201–220 | EAMEVASQARQMVQAMRTIG | X | X | |||
| TT | p5 | 490–509 | QDEIVSYNTKNKPLNFNYSL | X | ||||
| TT | p6 | 498–517 | TKNKPLNFNYSLDKIIVDYN | X | ||||
| TT | p7 | 506–525 | NYSLDKIIVDYNLQSKITLP | X | ||||
| TT | p16 | 578–597 | MTNSVDDALINSTKIYSYFP | X | ||||
| TT | p17 | 586–605 | LINSTKIYSYFPSVISKVNQ | X | ||||
| TT | P20 | 610–629 | ILFLQWVRDIIDDFTNESSQ | X | ||||
| TT | p27 | 666–685 | ETTGVVLLLEYIPEITLPVI | X | ||||
| TT | p28 | 674–691 | LEYIPEITLPVIAALSIAES | X | ||||
| TT | P36 | 738–757 | SYQMYRSLEYQVDAIKKIID | X | ||||
| TT | P37 | 746–765 | EYQVDAIKKIIDYEYKIYSG | X | ||||
| TT | p47 | 826–845 | NILMQYIKANSKFIGITELK | X | X | |||
| TT | p48 | 834–853 | ANSKFIGITELKKLESKINK | X | ||||
| TT | p62 | 946–965 | MFNNFTVSFWLRVPKVSASH | X | ||||
| TT | p63 | 954–973 | FWLRVPKVSASHLEQYGTNE | X | ||||
| TT | P64 | 962–981 | SASHLEQYGTNEYSIISSMK | X | ||||
| TT | p67 | 986–1005 | SIGSGWSVSLKGNNLIWTLK | X | ||||
| TT | p76 | 1058–1077 | ITGLGAIREDNNITLKLDRC | X | ||||
| TT | p83 | 1114–1133 | LRDFWGNPLRYDTEYYLIPV | X | ||||
| TT | p84 | 1122–1141 | LRYDTEYYLIPVASSSKDVQ | X | ||||
| TT | p85 | 1130–1148 | LIPVASSSKDVQLKNITDYM | X | ||||
| TT | p88 | 1154–1173 | APSYTNGKLNIYYRRLYNGL | X | ||||
| TT | p90 | 1170–1189 | YNGLKFIIKRYTPNNEIDSF | X | ||||
| TT | p95 | 1210–1229 | GYPKDGNAFNNLDRILRVGY | X | ||||
| TT | p99 | 1242–1261 | AVKLRDLKTYSVQLKLYDDK | X | ||||
| TT | p100 | 1250–1269 | TYSVQLKLYDDKNASLGLVG | X | ||||
| TT | P102 | 1266–1285 | GLVGTHNGQIGNDPNRDILI | X | ||||
X indicates peptides that elicited a positive tetramer staining result
For the mapping of tetanus heavy chain (TT) epitopes, a total of 106 peptides (20 amino acids long with an overlap of 12 residues) were synthesized. These peptides were divided into 21 pools with five consecutive peptides per pool (excepting pool 21, which had 6 peptides). Epitope mapping experiments were carried out to identify DRB3*0101, DRB3*0202 DRB1*1101, DRB1*1301 restricted TT responses (see supplementary figures 1, 2 and 3); these data, combined with the DRB1*0301 results from our previously published study [32] are summarized in Table 1. Again, none of the epitopes was shared among any of the DR molecules. Because these experiments were carried out using peptide pools, there is some possibility that some DRB3 restricted epitopes within these antigens were missed due to peptide competition. However, our results indicate that DRB3 presents MP and TT epitopes that are distinct from those presented by the corresponding DRB1-52 alleles. Thus, we were able to identify a total of four peptides that contain three DRB3*0101 restricted epitopes and a total of five peptides that contain four DRB3*0202 epitopes (summarized in figure 4).
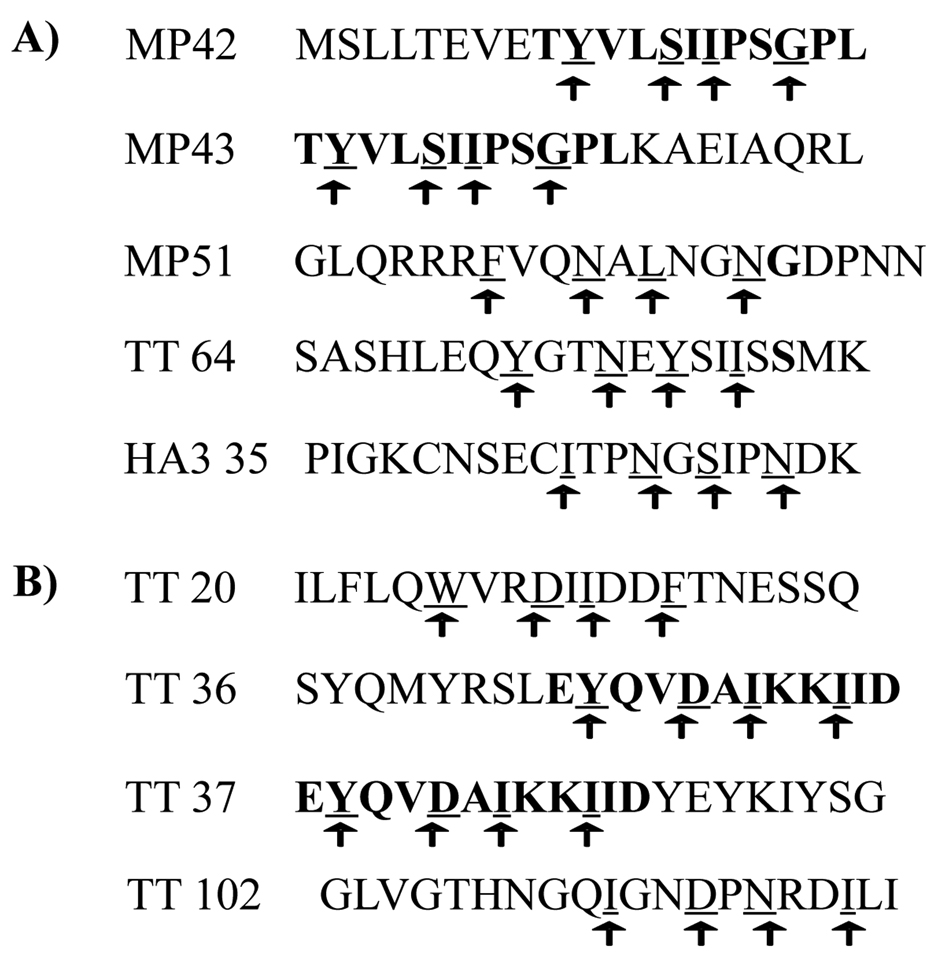
Figure 4. Peptides found to contain epitopes for DRB3*0101 and DRB3*0202.
The tentative anchoring residues are indicated by arrows. Bold residues are indicated based on overlapping peptides. A) MP, TT and HA3 peptides found to be presented and recognized in the context of DRB3*0202. b) TT peptides found to be presented and recognized in the context of DRB3*0101.
To more accurately define the DRB3 restricted epitopes within these antigenic peptides, truncated peptides (each 12 amino acids in length) were used to determine the minimal epitopes for three (two DRB3*0101 restricted and one DRB3*0202 restricted) tetanus toxoid peptides. Using these short peptides, tetramers were loaded and used to test the staining of peptide specific, tetramer positive CD4+ T cell lines. Based on these experiments (representative results shown in supplementary figure 4) precise epitopes for TT64 (LEQYGTNEYSII), TT36 (EYQVDAIKKIID) and TT102 (QIGNDPNRDILI) were defined. These minimal epitopes were consistent with previously reported binding preferences for DRB3*0101 [33, 34] and DRB3*0202 [35]. Therefore, we predicted minimal epitopes for the remaining peptides based on these postulated preferences: Y/F/W/I/L in P1, S/N/D in P4, I/L/Y/S/N in P6 and G/N/I/F/D in P9. These likely motifs are designated by arrows in figure 4. The fact that peptides MP1–20 and MP9–28 and peptides TT738–757 and TT746–765 are overlapping peptides aided our selection of their minimal epitope, since pairs of consecutive peptides often share a single motif. These shared residues are bolded in figure 4. Based on their respective protein sequences DRB3*0101 might be expected to prefer larger residues in pocket 9 than DRB3*0202 (B3*0101 has β37 Phe and β57 Val while B3*0202 has β37 Tyr and β57 Asp) and to accept acidic residues better in pocket 4 than DRB3*0202 (B3*0101 has β74 Arg while B3*0202 has β74 Gln). These differences may explain the lack of common epitopes between these alleles in spite of their general similarity. In total, we have identified four antigenic peptides (most likely representing three distinct epitopes) that are recognized in the context of DRB3*0101 and five antigenic peptides (most likely representing four distinct epitopes) that are recognized in the context of DRB3*0202.
For the corresponding DRB1-52 alleles, six epitopes were found for DRB1*0301, eleven epitopes (17 peptides) were found for DRB1*1101 and five epitopes were found for DRB1*1301. Thus the overall number of epitopes found in TT or MP for both DRB1-52 and DRB3 molecules is variable, with somewhat higher numbers of epitopes for DRB1.
2.4 Evidence of the processing and presentation of epitopes identified by TGEM
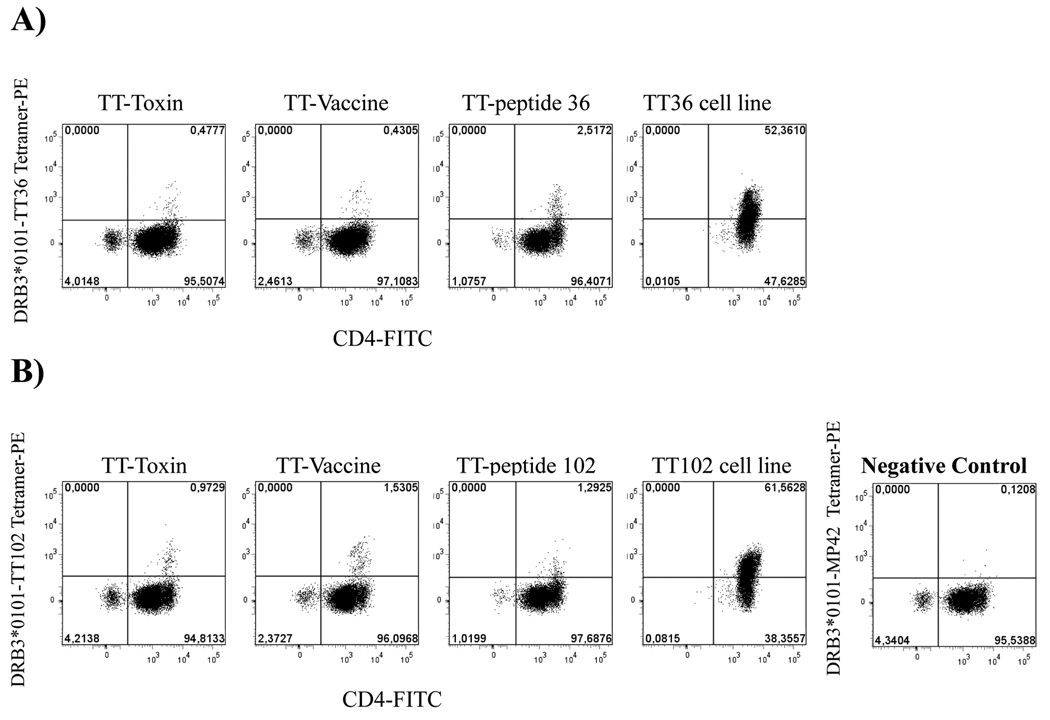
To assess the natural processing of tetanus epitopes restricted by DRB3*0101, T CD4+ cells were stimulated either with the tetanus-diphtheria vaccine (0.4 IU of the adsorbed tetanus toxoid) or the tetanus toxin (1µg/ml). Specific peptide stimulation (TT36, TT102) was used as a control. On day 14 of stimulation cells were stained with DRB3*0101-TT36 or DRB3*0101-TT102 tetramers. With all three stimuli (toxoid, toxin and peptide) tetramer positive DRB3*0101-TT36 and DRB3*0101-TT102 specific cells were detected (figure 5). To further assess the in vivo relevance of these epitopes, a proliferation assay was used to assess the dose-response of TT36 and TT102 specific cell lines with different concentrations of toxin or peptide. With both protein stimuli the level of proliferation was comparable (maximum proliferation reached at 4µg/ml), while the optimal concentration for the peptides was lower (see supplementary figure 5). Theis evidence of the natural processing and presentation of these epitopes support the notion that DRB3 can play a significant role in antigen presentation to CD4+ T cells.
Figure 5. Evidence of natural processing for DRB3*0101 restricted epitopes.
A) Tetramer staining of DRB3*0101-TT36 specific CD4+ T cells following stimulation with whole tetanus toxin (1µg/ml), tetanus-diphtheria vaccine (0.4 UI of the adsorbed tetanus toxoid) or 10 µg/ml of TT-36 peptide. DRB3*0101-TT36 specific cells were observed for the three different stimuli, implying natural processing and presentation of the epitope. Reference staining of a DRB3*0101-TT36 specific CD4+ T cell line is also shown. B) Tetramer staining of DRB3*0101-TT102 specific CD4+ T cells following stimulation with whole tetanus toxin, tetanus-diphtheria vaccine, or peptide in the same manner as panel A. Reference staining of a DRB3*0101-TT102 specific CD4+ T cell line is also shown. TGEM with TT pools was performed in two individuals.
2.5 Frequency of TT/MP-DRB3 epitope specific T cell precursors in the blood of vaccinated individuals
For a DRB3*0101 donor vaccinated 1 month prior to the experiment, the frequency of TT specific T-cells was evaluated by stimulating CFSE labeled cells with either TT738–757 or TT1266–1285 and subsequently co-staining with the corresponding DRB3*0101 tetramer. T cell frequencies were estimated based on the tetramer positive population and the number of in vitro cell divisions (based on CFSE dilution). Similarly, the frequency of MP specific T-cells was evaluated for a DRB3*0202 donor by stimulating with MP1–20 or MP73–92 and subsequently co-staining with the corresponding DRB3*0202 tetramer (supplementary figure 6). For the sake of comparison, the frequency of DRB1*1101 restricted MP specific T-cells was evaluated from the same donor by stimulating with MP97–116 and subsequently co-staining with the corresponding DRB1*1101 tetramer. As summarized in table 2, the observed frequencies ranged from 1 in 18,000 to 1 in 244,000 cells. These results are consistent with previously published frequencies observed for DRB1*0401 restricted responses to HA 307–319 peptide, which were approximately 1 in 25,000 [36]. To further demonstrate the relevance of these DRB3 restricted cells, direct ex vivo staining was carried out using DRB3*0101-TT102 tetramers and an irrelevant control tetramers using PBMC isolated from a previously vaccinated individual. The percentage of staining with DRB3*0101-TT102 was higher than that observed for the irrelevant tetramer (nearly triple the proportion) (see supplementary figure 7). These results indicate that the frequency of TT102 specific CD4+ T cells is 1 in 6,574 CD4+. The frequencies observed by these various methods imply that DRB3 can play a significant role in presenting antigens to T cells.
Table 2.
Frequencies of TT-MP specific CD4+ T-cellsa.
| Tetramer | Peptide | Frequency |
|---|---|---|
| DRB3*0101 | TT 738–757 | 1 in 20,000 |
| DRB3*0101 | TT 1266–1285 | 1 in 33,000 |
| DRB3*0202 | MP 1–20 | 1 in 19,000 |
| DRB3*0202 | MP 9–28 | 1 in 18,000 |
| DRB3*0202 | MP 73–92 | 1 in 100,000 |
Frequencies calculated based on CFSE dilution
3. Discussion
To more extensively characterize the relative expression of DRB3 andits functional role in antigen presentation, this work examined several aspects of the HLA-DRB3 biology, ranging from level of mRNA expression to identifying DRB3 restricted epitopes and the frequency of T-cells that respond to these epitopes. We found that the DRB3 message is present at lower levels than DRB1-52 in both CD14+ and CD19+ cells. There were distinct differences between the DRB1-52/DRB3 mRNA ratio and the total amount of DRB mRNA produced in these two cell types. B cells had higher levels of both DRB1-52 and DRB3 message than monocytes; however, the relative proportion of DRB3 message was lower (DRB1-52/DRB3 ratio of 3.5). Monocytes had lower levels of both DRB1-52 and DRB3 message; however, the relative proportion of DRB3 message was higher (DRB1-52/DRB3 ratio of 2). Comparing these observations to previous studies for different DR loci it can be stated that the DRB secondary loci behave differently: DRB5 mRNA was found to be more abundant than the DRB1*1501 [37], while the level of DRB4 mRNA may be as much as seven times lower than DRB1*07 [38, 39]. In DRB4 haplotypes, observed differences in expression levels have been attributed both to promoter polymorphisms and to post-transcriptional activity. Similarly, our observed differences in mRNA level (DRB3 vs DRB1-52) could be due to promoter polymorphism as has been suggested by an analysis of promoter activity [40]. The two HLA-DRB3 alleles studied here (DRB3*0101 and DRB3*0202) share the same sequence in their promoter regions (Hs6_111615; Hs6_111610). Therefore, it is not surprising that we did not observe obvious differences in their respective levels of transcription.
The differences seen at the mRNA level also appeared to be reflected at the level of protein expression on the cell surface. We detected roughly two-fold more total DR protein on CD19+ cells than in CD14+ cells, although our methodology (indirect immunofluorescence) did not allow a direct comparison of DRB1 and DRB3 protein expression on the cell surface. For DRB3, the calculated mean ratios (CD19+/CD14+) were 1.6 for mRNA versus 2.3 for protein. Although these mean ratios were statistically different, both measurements indicated that DRB3 was more abundant in CD19+ cells. While there could be important differences in post-transcriptional regulation or protein recirculation, there was some relationship (r=0.647) between the observed mRNA levels and cell surface staining of DRB3 protein. The observed level of DRB3 surface staining suggests that the level of DRB3 expression on the cell surface should be adequate to present peptides and to activate DRB3 restricted T cells.
The importance of DRB3 in antigen presentation was further highlighted by our epitope mapping studies. It is significant that a variety of DRB3*0101 and DRB3*0202 epitopes could be identified within MP and TT and for some of them, we proved that they are naturally processed. Our results indicate that the frequencies of DRB3 restricted T cells (as measured by CFSE or with direct staining) and the magnitude of their in vitro responses (as measured using tetramers) are similar to those previously observed for DRB1 restricted T cells. These similarities exist in spite of clear differences in relative expression. As summarized in Table 1, these DRB3 restricted responses are not merely directed against peptides that are cross presented by DRB1-52. Rather, there were no epitopes shared among any of the DRB1-52 or DRB3 alleles. While some previous reports suggested that the binding motifs of DRB1*0301 and DRB3*0101 may be nearly identical [29, 30, 33], our work indicates that the antigenic peptides presented by these two molecules are quite different. These observed differences are consistent with their amino acid sequences and existing knowledge of their peptide binding motifs [33, 35]. For example, DRB1*0301 and DRB3*0101 only share the same sequence in the P4 pocket [34], which confers a characteristic basic anchor preference, while having important differences (including an occluded P6) in the other binding pockets.
Thus, DRB3 and the associated DRB1-52 alleles appear to present complementary sets of antigenic peptides. The idea that DRB3 proteins are not a merely a duplication of their DRB1-52 counterparts is not completely new [30]. In fact Bontrop et al. [41] proposed that the less polymorphic DRB3 alleles have been retained in parallel with DRB1-52 because they present conserved bacterial heat shock protein epitopes so well, thus broadening the diversity of the responding T cells. Our current observation that DRB1-52 and DRB3 show no redundancy in their preferred tetanus and influenza epitopes further reinforces this concept.
It should be noted that the minor sequence changes among the three common DRB3 alleles account for clear differences in peptide selection and presentation [15]. It has been suggested that similarities between certain binding pockets can explain why some autoimmune diseases are associated with only two of the three DRB3 alleles:depending on the preferences of a binding pocket, a given peptide might bind to more than one allelic form of DRB3 [15]. The same reasoning can be applied to DRB1*0301 and DRB3*0101. While there alleles did not share any peptide in this study, their peptide binding pockets could have some overlap, allowing them to share a subset of peptides. Conversely, some peptides that have been assumed to have a DRB1*0301 restriction could actually be presented by DRB3*0101, since the haplotype which contains both alleles has been associated with several autoimmune diseases.
In reference to the cross presentation of epitopes by DRB1 and DRB3, previous reports [29, 42, 43] described the peptide TT102 (1272–1284) as either a DRB3*0101 or DRB1*0301 restricted epitope. However, binding of this peptide to DRB1*03 was two orders of magnitude weaker than binding exhibited by a typical natural ligand [42] ((AQ:Do you mean that the association constant is two orders of magnitude lower than that of the natural ligand?)). Our TGEM analysis indicated that TT102 acts as a high avidity epitope only when presented in the context of DRB3*0101. Thus the differences in the binding motifs of these molecules are relevant for the binding and presentation of peptides or possibly the positioning of TCR-contacting residues, in contrast to the conclusions of at least two reports [28, 29]. In general, claims about the class II MHC restriction of epitopes should be regarded carefully, because some experimental systems are not well suited to address this question. For example, methods that rely on “homozygous” EBV transformed cell lines (either as antigen presenting cells or as a source for DR protein) will be confounded by the inevitable presence of both primary secondary DR proteins. Studies based solely on peptide binding or elution should also be regarded carefully, because a peptide may be able to bind to a class II protein with appreciable affinity, and yet fail to be a legitimate T cell epitope.
Naturally, all the findings of this study must be weighed in light of the prevalence of DRB3 alleles in human populations. National Marrow Donor Program typing results [44] indicate that approximately 43% of caucasoid alleles are linked to some DRB3 allele, with nearly 35% having either DRB3*0101 or DRB3*0202. Based on these allele frequencies, it is expected that 2/3 of the caucasoid population has a DRB3 allele. Several diseases have been associated with DRB3 alleles, including neonatal alloimmune thrombocytopenia [30, 45] post-transfusion purpura [46] and inclusion body myositis [47]. In addition, the strong association between DRB3*0101 and DRB1*0301 (a member of the A1-B8-DR3 haplotype) could implicate this DRB3 allele in the presentation of peptides in certain DR3-linked autoimmune disorders. These associations may be partially masked by the high frequency of the allele and its strong DRB1 linkage.
In conclusion, our results demonstrate that, in spite of having significantly lower message and protein expression levels than HLA-DRB1-52, HLA-DRB3 is not merely a secondary, redundant locus. Rather, HLA-DRB3 presents peptides that can evoke measurable T cell responses to important viral antigens. These DRB3 restricted T cells exist at significant frequencies in the periphery and recognize antigenic patterns distinct from their DRB1-52 counterparts. Therefore, DRB3 serves to extend and complement the peptide repertoire of DRB1 in antigen presentation. Given its prevalence, further study of DRB3 in settings such as vaccine design or immune monitoring may be warranted.
4. Methods
4.1 Donor samples
Healthy blood donors were recruited with informed written consent. All donors used had been immunized (1 month to 2 years) with the Trivalent Type A and B Influenza Virus Fluzone Vaccine (Aventis, Bridgewater, NJ) or with a tetanus-diphtheria vaccine, and were HLA typed (see supplementary methods).
4.2 CD19+ and CD14+ cell separation, RNA extraction and Reverse transcription
From PBMC, monocytes and B cells were purified by positive selection using Human CD14 Selection Cocktail (Stem Cell Technologies) and CD19 MicroBeads (Miltenyi, Auburn, CA), respectively. RNA was extracted from dry pellets from these purified cells using Chomczynski-Sacchi method with minor modifications [48, 49]. Reverse transcription of RNA samples (1–2 µg) was completed as previously described [50].
4.4 Real time measurement of DRB1-52 and DRB3 mRNA
For the specific amplification of the DRB1 of the DR52 haplotype and the DRB3 genes, two pairs of primers were designed: the antisense primer was common to both amplifications and situated in the exon 3 (5’ AATGCTGCCTGGATAGAA 3’) of HLA-DRB genes to prevent amplification of contaminating genomic DNA, while the sense primers specific to DRB1 (in 52 haplotype) (5’ CACGTTTCTTGGAGTACTC 3’) or DRB3 (5’ CACGTTTCTYGGAGCT 3’) were placed in exon 2. Primer sequences for the housekeeping gene GAPDH (33s 5’ TCTTCTTTTGCGTCGCCAG 3’, 405as 5’ AGCCCCAGCCTTCTCCA 3’), were chosen to span exon junctions [51]. All primers were purchased from Sigma-Genosys (Haverhill, UK).
Specificity of PCR primers and Real Time measurement of mRNA
To assess primer specificity, the DRB1*1301 and DRB3*0101 exons 1 to 5 were cloned into a TOPO-TA vector (Invitrogen, California, US) using primers previously described [36]. The specificity of both amplification pairs of oligonucleotides was proven by the lack of amplification of 105 copies from DRB1*1301 linearized plasmid, for testing DRB3 primers, and the lack of amplification of 105 copies of DRB3*0101 for DRB1-52 primers. For the real-time measurement of mRNA expression for the DRB1-52 and DRB3 genes, standard curves were prepared using linearized plasmid DNA for HLA-DRB3*0101, HLA-DRB1*1301, and purified amplified product for GAPDH [51]. For further details of the protocol see supplementary methods.
4.5 Flow cytometry staining with anti-DRB3 and anti-DR11 antibodies
Specificity of antibody (Ab) clones 7.3.19.1 and 4i131 were defined as described elsewhere [37]. For the CD14+ or CD19+ populations we have only compared the mean of fluorescence in the same sample for the same Ab to avoid differences in affinity between both Ab (which could bias the fluorescence intensity (FI) levels measured by flow cytometry). We compared the ratio CD19+ MFI / CD14+ MFI in each sample for DRB3 with the ones obtained for DR11 and total DR (calculated using the CD3-FITC/DR-PE product of Becton-Dickinson (San Jose, CA)). PBMC where also labeled with anti-CD19-FITC (Becton-Dickinson, San Jose, CA) and anti CD14-APC (BD Biosciences, USA). Three-color analysis was performed on a FACScan using Flow-Jo software (Becton-Dickinson, San Jose, CA). The details of the staining protocol are described in supplementary methods.
4.6 Generation of HLA class II tetramers
For the generation of DRB3*0101, DRB3*0202, DRB1*0301, DRB1*1101 and DRB1*1301 as soluble class II molecules, the vector construction, expression in S2 Drosophila cells, purification by affinity chromatography, and assembly of class II tetramers were accomplished as previously described [36], modified only by the incorporation of an in vivo biotinylation step of DRB chain, as also described in a previous article [52]. Briefly, soluble class II molecules were purified from the supernatants of transfected insect cell cultures. Since the biotinylation was accomplished in vivo, purified class II molecules were immediately dialyzed into phosphate storage buffer (pH 6.0) and loaded with pooled peptide mixtures or individual peptides by combining 0.5 mg/ml class II molecule, 10 mg/ml of peptide (2 mg/ml of each individual peptide for pools) and 0.2% g-octyl-D-glucopyranoside. After incubating at 37°C for 48 h, tetramers were crosslinked using PE–streptavidin (Biosource, Camarillo, CA, USA).
4.7 Peptides
To study the epitopes of influenza, two panels of 20-amino-acid peptides with a 12-amino-acid overlap (previously described [53]) were used. The hemagglutinin (HA1) panel was 41 peptides, while the matrix protein (MP) panel was 30 peptides. To study the epitopes of the heavy chain of the Clostridium tetani toxin (TT) a previously described [32] panel of 106 peptides was used.
All individual peptides were dissolved in DMSO at 10 mg/ml. Peptides for each panel were grouped into pools of 5 peptides each for PBMC stimulation: 8 pools for the HA (with a final pool of 6 peptides), 6 pools for MP, and 21 pools for TT.
4.8 Mapping of CD4 T cell epitopes using class II tetramers
Peptide stimulation of CD4+ T cells or PBMC
For peptide stimulations, PBMC were isolated from the heparinized blood of immunized healthy donors by density centrifugation (Lymphoprep; Nycomed Pharma AS Diagnostics, Oslo, Norway). When T CD4+ cells were stimulated, these were isolated from PBMC using a negative selection method (CD4+ T Cell Isolation Kit II, Miltenyi Biotech, Auburn, CA) following manufacturer instructions and using the AutoMacs system (Miltenyi Biotech, Auburn, CA) to perform the separation. In both cases, peptide stimulation was performed adding with 10µg/ml of peptide. The same total amount of peptide was used for either individual or pooled peptides. On day 7, IL-2 was added to the culture at a final concentration of 20 U/ml. Tetramer staining was done on day 14 of the culture.
Tetramer staining
For tetramer staining, 0.1×106 cells in 50 µl were stained with 0.5 µg of the appropriate tetramers for 1 h at 37°C, 5% CO2. Cells were subsequently stained with anti-CD4-FITC at 4°C for 30 min, washed twice in PBS and analyzed using a FACSCalibur (BD biosciences, San Jose, CA). After the initial round of pooled tetramer staining (screening using tetramers loaded with peptide pools), cells from tetramer-positive wells were stained again using the five individual peptide loaded tetramers, each loaded with one individual peptide from within the corresponding peptide pool.
CFSE staining and specific stimulation of CD4+ cells
CD4+ T cells were obtained from healthy donors as previously described, and the CFSE staining protocol was followed as described elsewhere [36]. T CD4 cells isolated using beads labeled with CFSE, stimulated with autologous adherent cells and peptide, where stained on day 7 with PE-labeled tetramers and analyzed subsequently by flow cytometry. Cells derived from the same individual were stimulated with DRB3*0202 and DRB1*1101 antigens. . The number of cell divisions was calculated from the distinct CFSE fluorescence peaks produced by polyclonal stimulation with PHA and IL-2 as described elsewhere. Precursor frequency was estimated by dividing the number of tetramer-positive cells by 2x, where x is the average number of cell divisions, to determine the absolute number of precursors for the tetramer-positive cells, and then dividing this value by the total number of cells analyzed. In the DRB3*0101 case IL-2 was added to the culture on day 7 at a final concentration of 20 U/ml, and tetramer staining was done on day 9 of the culture.
4.9 Statistics
A non-parametric paired (Wilcoxon signed) test, was applied to paired non-normal distributed data, using the GraphPad Prism 5® software package (GraphPad Software, Inc. San Diego, US). A Mann Withney was used for non paired non-normal distributed data. A level of significance of 5% was used in all the statistical evaluations, and a one tailed pvalue assignation.
Supplementary Material
Acknowledgments
We thank Cristina Ambrós and Eduard Palou, Pilar Armengol and Roger Colobran, and Mariona Pascal for providing some SSO-PCR typed DNA samples, for their general support in the laboratory and for critical reading, respectively.
This study was supported by a grant from the “Fundación para la Investigación y la Prevención del Sida en España”, (project number 36487/05), by the Fondo de Investigaciones Sanitarias del Instituto de Salud Carlos III (PI07/0329), by the PROFIT (FIT 010000-2006-38) of the BST, and by the “Departament d’Educació i Universitats de la Generalitat de Catalunya” to Manel Juan and NIH contract HHSN 266200400028C to William W Kwok.
Abbreviations
- APC
Antigen presenting cell
- HLA-DRB1-52
HLA-DRB1-03/11/12/13/14
- MP
Matrix Protein
- TGEM
Tetramer Guided Epitope Mapping
- TT
Tetanus Toxin
Footnotes
Note
WW Kwok and M Juan share senior authorship.
Conflict of interest: The authors declare no financial or commercial conflict of interest.
References
- 1.Gonzalez PA, Carreno LJ, Coombs D, Mora JE, Palmieri E, Goldstein B, Nathenson SG, Kalergis AM. T cell receptor binding kinetics required for T cell activation depend on the density of cognate ligand on the antigen-presenting cell. Proc Natl Acad Sci U S A. 2005;102:4824–4829. doi: 10.1073/pnas.0500922102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Davis SJ, Ikemizu S, Evans EJ, Fugger L, Bakker TR, van der Merwe PA. The nature of molecular recognition by T cells. Nat Immunol. 2003;4:217–224. doi: 10.1038/ni0303-217. [DOI] [PubMed] [Google Scholar]
- 3.Bergstrom TF, Erlandsson R, Engkvist H, Josefsson A, Erlich HA, Gyllensten U. Phylogenetic history of hominoid DRB loci and alleles inferred from intron sequences. Immunol Rev. 1999;167:351–365. doi: 10.1111/j.1600-065x.1999.tb01404.x. [DOI] [PubMed] [Google Scholar]
- 4.Sebat J, Lakshmi B, Troge J, Alexander J, Young J, Lundin P, Maner S, Massa H, Walker M, Chi M, Navin N, Lucito R, Healy J, Hicks J, Ye K, Reiner A, Gilliam TC, Trask B, Patterson N, Zetterberg A, Wigler M. Large-scale copy number polymorphism in the human genome. Science. 2004;305:525–528. doi: 10.1126/science.1098918. [DOI] [PubMed] [Google Scholar]
- 5.Berdoz J, Gorski J, Termijtelen AM, Dayer JM, Irle C, Schendel D, Mach B. Constitutive and induced expression of the individual HLA-DR beta and alpha chain loci in different cell types. J Immunol. 1987;139:1336–1341. [PubMed] [Google Scholar]
- 6.Robinson J, Waller MJ, Parham P, Bodmer JG, Marsh SG. IMGT/HLA Database--a sequence database for the human major histocompatibility complex. Nucleic Acids Res. 2001;29:210–213. doi: 10.1093/nar/29.1.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gongora R, Figueroa F, Klein J. The HLA-DRB9 gene and the origin of HLA-DR haplotypes. Hum Immunol. 1996;51:23–31. doi: 10.1016/s0198-8859(96)00189-9. [DOI] [PubMed] [Google Scholar]
- 8.Jaraquemada D, Martin R, Rosen-Bronson S, Flerlage M, McFarland HF, Long EO. HLA-DR2a is the dominant restriction molecule for the cytotoxic T cell response to myelin basic protein in DR2Dw2 individuals. J Immunol. 1990;145:2880–2885. [PubMed] [Google Scholar]
- 9.Pette M, Fujita K, Wilkinson D, Altmann DM, Trowsdale J, Giegerich G, Hinkkanen A, Epplen JT, Kappos L, Wekerle H. Myelin autoreactivity in multiple sclerosis: recognition of myelin basic protein in the context of HLA-DR2 products by T lymphocytes of multiple-sclerosis patients and healthy donors. Proc Natl Acad Sci U S A. 1990;87:7968–7972. doi: 10.1073/pnas.87.20.7968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vergelli M, Kalbus M, Rojo SC, Hemmer B, Kalbacher H, Tranquill L, Beck H, McFarland HF, De Mars R, Long EO, Martin R. T cell response to myelin basic protein in the context of the multiple sclerosis-associated HLA-DR15 haplotype: peptide binding, immunodominance and effector functions of T cells. J Neuroimmunol. 1997;77:195–203. doi: 10.1016/s0165-5728(97)00075-1. [DOI] [PubMed] [Google Scholar]
- 11.Wilkinson D, de Vries RR, Madrigal JA, Lock CB, Morgenstern JP, Trowsdale J, Altmann DM. Analysis of HLA-DR glycoproteins by DNA-mediated gene transfer. Definition of DR2 beta gene products and antigen presentation to T cell clones from leprosy patients. J Exp Med. 1988;167:1442–1458. doi: 10.1084/jem.167.4.1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Khare M, Mangalam A, Rodriguez M, David CS. HLA DR and DQ interaction in myelin oligodendrocyte glycoprotein-induced experimental autoimmune encephalomyelitis in HLA class II transgenic mice. J Neuroimmunol. 2005;169:1–12. doi: 10.1016/j.jneuroim.2005.07.023. [DOI] [PubMed] [Google Scholar]
- 13.Doxiadis GG, de Groot N, Bontrop RE. Impact of endogenous intronic retroviruses on major histocompatibility complex class II diversity and stability. J Virol. 2008;82:6667–6677. doi: 10.1128/JVI.00097-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Doxiadis GG, de Groot N, de Groot NG, Doxiadis II, Bontrop RE. Reshuffling of ancient peptide binding motifs between HLA-DRB multigene family members: old wine served in new skins. Mol Immunol. 2008;45:2743–2751. doi: 10.1016/j.molimm.2008.02.017. [DOI] [PubMed] [Google Scholar]
- 15.Dai S, Crawford F, Marrack P, Kappler JW. The structure of HLA-DR52c: comparison to other HLA-DRB3 alleles. Proc Natl Acad Sci U S A. 2008;105:11893–11897. doi: 10.1073/pnas.0805810105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sintasath DM, Tang T, Slack R, Tilley EE, Ng J, Hartzman RJ, Hurley CK. Relative HLA-DRB1*13 allele frequencies and DRB3 associations of unrelated individuals from five US populations. Hum Immunol. 1999;60:1001–1010. doi: 10.1016/s0198-8859(99)00085-3. [DOI] [PubMed] [Google Scholar]
- 17.Gans CP, Tang TF, Slack R, Ng J, Hartzman RJ, Hurley CK. DRB1*14 diversity and DRB3 associations in four major population groups in the United States. Tissue Antigens. 2002;59:364–369. doi: 10.1034/j.1399-0039.2002.590502.x. [DOI] [PubMed] [Google Scholar]
- 18.Tang TF, Huang AY, Pappas A, Slack R, Ng J, Hartzman RJ, Hurley CK. Relative frequencies of DRB1*11 alleles and their DRB3 associations in five major population groups in a United States bone marrow registry. Hum Immunol. 2000;61:820–827. doi: 10.1016/s0198-8859(00)00145-2. [DOI] [PubMed] [Google Scholar]
- 19.Tang TF, Wang J, Slack R, Lin YS, Li L, Heine U, Ng J, Hartzman RJ, Katovich Hurley C. DRB1*03 diversity and DRB3 associations in five major population groups in the United States. Hum Immunol. 2002;63:221–228. doi: 10.1016/s0198-8859(01)00379-2. [DOI] [PubMed] [Google Scholar]
- 20.LeGuern C. Tolerogenic property of MHC class I and class II molecules: lessons from a gene therapy approach. Front Biosci. 2007;12:3133–3139. doi: 10.2741/2301. [DOI] [PubMed] [Google Scholar]
- 21.Bontrop R, Ottenhoff T, Van Miltenburg R, Elferink D, De Vries R, Giphart M. Quantitative and qualitative differences in HLA-DR molecules correlated with antigen-presentation capacity. Eur J Immunol. 1986;16:133–138. doi: 10.1002/eji.1830160205. [DOI] [PubMed] [Google Scholar]
- 22.Cotner T, Charbonneau H, Mellins E, Pious D. mRNA abundance, rather than differences in subunit assembly, determine differential expression of HLA-DR beta 1 and -DR beta 3 molecules. J Biol Chem. 1989;264:11107–11111. [PubMed] [Google Scholar]
- 23.Vincent R, Louis P, Gongora C, Papa I, Clot J, Eliaou JF. Quantitative analysis of the expression of the HLA-DRB genes at the transcriptional level by competitive polymerase chain reaction. J Immunol. 1996;156:603–610. [PubMed] [Google Scholar]
- 24.Louis P, Pinet V, Cavadore P, Kerlan-Candon S, Clot J, Eliaou JF. Differential expression of HLA-DRB genes according to the polymorphism of their regulatory region. C R Acad Sci III. 1994;317:161–166. [PubMed] [Google Scholar]
- 25.Louis P, Vincent R, Cavadore P, Clot J, Eliaou JF. Differential transcriptional activities of HLA-DR genes in the various haplotypes. J Immunol. 1994;153:5059–5067. [PubMed] [Google Scholar]
- 26.Tosi R, Tanigaki N, De Preval C, Gorski J, Mach B. Immunochemical analysis of a cell transfected with an HLA-DR gene reveals a new alloantigenic specificity within HLA-DRw52. Eur J Immunol. 1986;16:1603–1608. doi: 10.1002/eji.1830161221. [DOI] [PubMed] [Google Scholar]
- 27.Irle C, Jaques D, Tiercy JM, Fuggle SV, Gorski J, Termijtelen A, Jeannet M, Mach B. Functional polymorphism of each of the two HLA-DR beta chain loci demonstrated with antigen-specific DR3- and DRw52-restricted T cell clones. J Exp Med. 1988;167:853–872. doi: 10.1084/jem.167.3.853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Texier C, Pouvelle-Moratille S, Busson M, Charron D, Menez A, Maillere B. Complementarity and redundancy of the binding specificity of HLA-DRB1, -DRB3, -DRB4 and -DRB5 molecules. Eur J Immunol. 2001;31:1837–1846. doi: 10.1002/1521-4141(200106)31:6<1837::aid-immu1837>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 29.Wienhold W, Malcherek G, Jung C, Stevanovic S, Jung G, Schild H, Melms A. An example of immunodominance: engagement of synonymous TCR by invariant CDR3 beta. Int Immunol. 2000;12:747–756. doi: 10.1093/intimm/12.6.747. [DOI] [PubMed] [Google Scholar]
- 30.Nagvekar N, Corlett L, Jacobson LW, Matsuo H, Chalkley R, Driscoll PC, Deshpande S, Spack EG, Willcox N. Scanning a DRB3*0101 (DR52a)-restricted epitope cross-presented by DR3: overlapping natural and artificial determinants in the human acetylcholine receptor. J Immunol. 1999;162:4079–4087. [PubMed] [Google Scholar]
- 31.Reijonen H, Kwok WW. Use of HLA class II tetramers in tracking antigen-specific T cells and mapping T-cell epitopes. Methods. 2003;29:282–288. doi: 10.1016/s1046-2023(02)00350-x. [DOI] [PubMed] [Google Scholar]
- 32.James EA, Bui J, Berger D, Huston L, Roti M, Kwok WW. Tetramer-guided epitope mapping reveals broad, individualized repertoires of tetanus toxin-specific CD4+ T cells and suggests HLA-based differences in epitope recognition. Int Immunol. 2007;19:1291–1301. doi: 10.1093/intimm/dxm099. [DOI] [PubMed] [Google Scholar]
- 33.Wu S, Maslanka K, Gorski J. An integrin polymorphism that defines reactivity with alloantibodies generates an anchor for MHC class II peptide binding: a model for unidirectional alloimmune responses. J Immunol. 1997;158:3221–3226. [PubMed] [Google Scholar]
- 34.Parry CS, Gorski J, Stern LJ. Crystallographic structure of the human leukocyte antigen DRA, DRB3*0101: models of a directional alloimmune response and autoimmunity. J Mol Biol. 2007;371:435–446. doi: 10.1016/j.jmb.2007.05.025. [DOI] [PubMed] [Google Scholar]
- 35.Verreck FA, van de Poel A, Drijfhout JW, Amons R, Coligan JE, Konig F. Natural peptides isolated from Gly86/Val86-containing variants of HLA-DR1, -DR11, -DR13, and -DR52. Immunogenetics. 1996;43:392–397. doi: 10.1007/BF02199809. [DOI] [PubMed] [Google Scholar]
- 36.Novak EJ, Liu AW, Nepom GT, Kwok WW. MHC class II tetramers identify peptide-specific human CD4(+) T cells proliferating in response to influenza A antigen. J Clin Invest. 1999;104:R63–R67. doi: 10.1172/JCI8476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Prat E, Tomaru U, Sabater L, Park DM, Granger R, Kruse N, Ohayon JM, Bettinotti MP, Martin R. HLA-DRB5*0101 and -DRB1*1501 expression in the multiple sclerosis-associated HLA-DR15 haplotype. J Neuroimmunol. 2005;167:108–119. doi: 10.1016/j.jneuroim.2005.04.027. [DOI] [PubMed] [Google Scholar]
- 38.Leen MP, Gorski J. DRB4 promoter polymorphism in DR7 individuals: correlation with DRB4 pre-mRNA and mRNA levels. Immunogenetics. 1997;45:371–378. doi: 10.1007/s002510050218. [DOI] [PubMed] [Google Scholar]
- 39.Stunz LL, Karr RW, Anderson RA. HLA-DRB1 and -DRB4 genes are differentially regulated at the transcriptional level. J Immunol. 1989;143:3081–3086. [PubMed] [Google Scholar]
- 40.Emery P, Mach B, Reith W. The different level of expression of HLA-DRB1 and - DRB3 genes is controlled by conserved isotypic differences in promoter sequence. Hum Immunol. 1993;38:137–147. doi: 10.1016/0198-8859(93)90531-5. [DOI] [PubMed] [Google Scholar]
- 41.Bontrop RE, Elferink DG, Otting N, Jonker M, de Vries RR. Major histocompatibility complex class II-restricted antigen presentation across a species barrier: conservation of restriction determinants in evolution. J Exp Med. 1990;172:53–59. doi: 10.1084/jem.172.1.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Malcherek G, Gnau V, Stevanovic S, Rammensee HG, Jung G, Melms A. Analysis of allele-specific contact sites of natural HLA-DR17 ligands. J Immunol. 1994;153:1141–1149. [PubMed] [Google Scholar]
- 43.Demotz S, Lanzavecchia A, Eisel U, Niemann H, Widmann C, Corradin G. Delineation of several DR-restricted tetanus toxin T cell epitopes. J Immunol. 1989;142:394–402. [PubMed] [Google Scholar]
- 44.Schreuder GM, Hurley CK, Marsh SG, Lau M, Fernandez-Vina M, Noreen HJ, Setterholm M, Maiers M. The HLA Dictionary 2004: a summary of HLA-A, -B, -C, -DRB1/3/4/5 and -DQB1 alleles and their association with serologically defined HLA-A, -B, -C, -DR and -DQ antigens. Tissue Antigens. 2005;65:1–55. doi: 10.1111/j.1399-0039.2005.00350.x. [DOI] [PubMed] [Google Scholar]
- 45.Sukati H, Bessos H, Barker RN, Urbaniak SJ. Characterization of the alloreactive helper T-cell response to the platelet membrane glycoprotein IIIa (integrin-beta3) in human platelet antigen-1a alloimmunized human platelet antigen-1b1b women. Transfusion. 2005;45:1165–1177. doi: 10.1111/j.1537-2995.2005.00188.x. [DOI] [PubMed] [Google Scholar]
- 46.Gandemer V, Kaplan C, Quelvennec E, Poulain P, Laurent MC, Semana G, Renouard J, Le Gall E. Pregnancy-associated autoimmune neonatal thrombocytopenia: role of maternal HLA genotype. Br J Haematol. 1999;104:878–885. doi: 10.1046/j.1365-2141.1999.01270.x. [DOI] [PubMed] [Google Scholar]
- 47.Price P, Santoso L, Mastaglia F, Garlepp M, Kok CC, Allcock R, Laing N. Two major histocompatibility complex haplotypes influence susceptibility to sporadic inclusion body myositis: critical evaluation of an association with HLA-DR3. Tissue Antigens. 2004;64:575–580. doi: 10.1111/j.1399-0039.2004.00310.x. [DOI] [PubMed] [Google Scholar]
- 48.Chomczynski P, Sacchi N. The single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction: twenty-something years on. Nat Protoc. 2006;1:581–585. doi: 10.1038/nprot.2006.83. [DOI] [PubMed] [Google Scholar]
- 49.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 50.Armengol MP, Cardoso-Schmidt CB, Fernandez M, Ferrer X, Pujol-Borrell R, Juan M. Chemokines determine local lymphoneogenesis and a reduction of circulating CXCR4+ T and CCR7 B and T lymphocytes in thyroid autoimmune diseases. J Immunol. 2003;170:6320–6328. doi: 10.4049/jimmunol.170.12.6320. [DOI] [PubMed] [Google Scholar]
- 51.Sabater L, Ferrer-Francesch X, Sospedra M, Caro P, Juan M, Pujol-Borrell R. Insulin alleles and autoimmune regulator (AIRE) gene expression both influence insulin expression in the thymus. J Autoimmun. 2005;25:312–318. doi: 10.1016/j.jaut.2005.08.006. [DOI] [PubMed] [Google Scholar]
- 52.Yang J, Jaramillo A, Shi R, Kwok WW, Mohanakumar T. In vivo biotinylation of the major histocompatibility complex (MHC) class II/peptide complex by coexpression of BirA enzyme for the generation of MHC class II/tetramers. Hum Immunol. 2004;65:692–699. doi: 10.1016/j.humimm.2004.04.001. [DOI] [PubMed] [Google Scholar]
- 53.Yang J, James EA, Huston L, Danke NA, Liu AW, Kwok WW. Multiplex mapping of CD4 T cell epitopes using class II tetramers. Clin Immunol. 2006;120:21–32. doi: 10.1016/j.clim.2006.03.008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.