Abstract
The global prevalence of obesity has increased significantly in recent decades, mainly due to excess calorie intake and increasingly sedentary lifestyle. Here, we test the association between obesity measured by body mass index (BMI) and one of the best-known genetic variants showing strong selective pressure: the functional variant in the cis-regulatory element of the lactase gene. We tested this variant since it is presumed to provide nutritional advantage in specific physical and cultural environments. We genetically defined lactase persistence (LP) in 31 720 individuals from eight European population-based studies and one family study by genotyping or imputing the European LP variant (rs4988235). We performed a meta-analysis by pooling the β-coefficient estimates of the relationship between rs4988235 and BMI from the nine studies and found that the carriers of the allele responsible for LP among Europeans showed higher BMI (P = 7.9 × 10−5). Since this locus has been shown to be prone to population stratification, we paid special attention to reveal any population substructure which might be responsible for the association signal. The best evidence of exclusion of stratification came from the Dutch family sample which is robust for stratification. In this study, we highlight issues in model selection in the genome-wide association studies and problems in imputation of these special genomic regions.
INTRODUCTION
The lactase gene [LCT (MIM 603202)] is expressed in the intestinal epithelial cells and its protein product contributes to carbohydrate metabolism via digestion of the milk sugar lactose. The activity of the lactase enzyme in intestinal cells normally declines during childhood. Individuals with lactase non-persistence show varying degrees of abdominal pain, diarrhoea, bloating and gas formation after consumption of lactose-containing foods, in particular milk and other non-fermented dairy products. Molecular studies of lactose intolerance led to the identification of specific variations in a cis-regulatory element of the LCT gene resulting in lactase persistence (LP) which is an autosomal dominant condition (MIM 223100). The LP single-nucleotide polymorphism (SNP) most common among individuals of European descent (C/T-13910, rs4988235) was first identified by Enattah et al. (1) in Finnish lactose intolerance families. The T allele correlated perfectly with the LP phenotype of intestinal cells and functional studies have shown that the T allele is associated with the disrupted down-regulation of the LCT gene during childhood (2,3). The enhancer function of this mutation has also been shown in vitro (4,5). Subsequently, this region has yielded a spectrum of variants, some showing population specificity (6–8). These variants have been under strong positive selection regionally and seem to reflect independent events of cattle domestication, as exemplified by the two different allelic backgrounds of the Arabic and European LP alleles (6,9). The European LP allele has been associated with higher milk consumption in Finnish, Estonian and Austrian populations, one of which being the Finnish YF sample used in this study (10–15).
We hypothesized that since lactase non-persistent individuals are on a more restricted diet when compared with the LP individuals, this may have an impact on body mass index (BMI) and/or height. Lactose-containing dairy products are often high in energy content potentially increasing daily calorie intake, and for the lactase non-persistent, they may cause diarrhoea which can prevent the absorption of nutrients. Further, milk is rich in calcium, required for skeletal growth.
BMI is calculated as weight in kilograms divided by the square of height in meters. BMI has been studied extensively due to the health effects of obesity and its relative ease of measurement. Excessive weight increases the risk of type II diabetes, coronary artery disease and hypertension, and is associated with many forms of cancer along with adverse social and psychological consequences (16,17). The need for more effective clinical strategies has become evident with the increasing prevalence of obesity in recent years (18). This has put pressure on genetic studies to find variants responsible for differences in response to positive energy balance. Recent genome-wide association studies (GWASs) have been able to show association between BMI and genetic variants in a total of 13 genomic regions (19–22). In this study, we show the association between genetically defined LP and BMI.
RESULTS
The effect of LP was evaluated in five Finnish representative population cohorts, all non-ascertained for BMI or height: The North Finland Birth Cohort 1966 (NFBC 1966), The Health 2000 Health Examination Survey (Health2000), The Cardiovascular Risk of Young Finns Study (YF), the Alpha-Tocopherol, Beta-Carotene Cancer Prevention (ATBC) study and the FINRISK which is a cross-sectional population survey targeting coronary risk factors collected every 5 years. The cohort-specific analyses were performed on corrected phenotype comparing the C-13910 allele homozygotes (lactase non-persistent, CC genotypes) against T-13910 allele carriers (LP, CT and TT genotypes) using linear regression. There was evidence for association with BMI and the LP in the NFBC 1966 (n = 5498, C allele frequency = 0.39, β = −0.11, P = 0.002) and in ATBC cohorts (n = 2126, C allele frequency = 0.40, β = −0.12, P = 0.04) and the same direction of association in Health2000 (n = 5320, C allele frequency = 0.42, β = −0.06, P = 0.10), YF (n = 2165, C allele frequency = 0.42, β = −0.04, P = 0.51) and FINRISK (n = 2265, C allele frequency = 0.45, β = −0.09, P = 0.09) cohorts. Table 1 summarizes the cohort-specific analyses.
Table 1.
Cohort-specific results
| n (males/females) | Results |
||||
|---|---|---|---|---|---|
| LP prevalence (%) | β | SE | P-value | ||
| NFBC 1966 | 5498 (2636/2862) | 85 | −0.11 | 0.04 | 0.002 |
| ATBC | 2126 (2126/0) | 84 | −0.12 | 0.06 | 0.04 |
| FINRISK | 2265 (1555/710) | 80 | −0.09 | 0.05 | 0.09 |
| Health2000 | 5320 (2437/2883) | 82 | −0.06 | 0.03 | 0.10 |
| YF | 2165 (985/1180) | 83 | −0.04 | 0.06 | 0.51 |
| BWHHS | 3109 (0/3109) | 94 | −0.06 | 0.07 | 0.40 |
| ERF | 2104 (909/1195) | 90 | −0.08 | 0.06 | 0.16 |
| Rotterdam | 5689 (3320/2369) | 91 | −0.02 | 0.05 | 0.69 |
| KORA S3 | 1578 (783/795) | 87 | −0.001 | 0.07 | 0.99 |
| KORA S4 | 1755 (859/896) | 87 | 0.08 | 0.06 | 0.24 |
n, number of individuals; SE, standard error; LP, lactase persistence determined by the frequency of T-13910 allele carriers, β (SE) are from the association analysis between standardized BMI and LP (rs4988235 CC against CT/TT genotypes). The nationwide Finnish cohort analyses of Health2000, YF and FINRISK were corrected for region of residence (Southwest, East and Oulu region), age and sex effects in Health2000 cohort and FINRISK cohort, with sex in NFBC 1966 cohort, with age in ATBC cohort, with age and birth place coordinates in BWHHS, with sex and age in ERF study (family-based sample), with age, sex and first 10 principal components in Rotterdam and KORA studies. Outliers deviating more than three standard deviations were removed from the analyses.
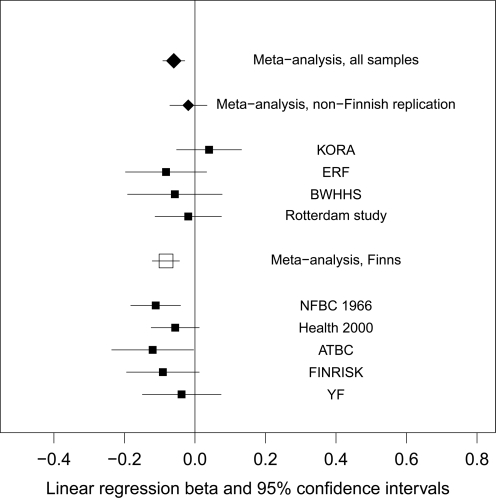
The CC genotype was associated with decreased BMI compared with CT/TT genotypes in the meta-analysis of 17 374 Finns (β = −0.08, P = 1.5 × 10−5). The individuals carrying the LP allele had 0.3 kg/m2 higher BMI than the non-persistent individuals which corresponds to ∼1 kg for an average person. The single study results from linear regression and the meta-analyses are summarized as a forest plot (Fig. 1).
Figure 1.
The forest plot of the linear regression analyses and meta-analyses.
We also performed a sex-specific meta-analysis and found that males (n = 9739, β = −0.09, P = 1.6 × 10−4) had somewhat higher effect estimate in the model than females (n = 7635, β = −0.06, P = 0.02), but there was no robust statistical evidence of a gender difference (P = 0.6).
Genome-wide SNP data were available for two of the cohorts, NFBC 1966 (n = 4911) and Health2000 (n = 2145, metabolic syndrome case–control sample). The first 14 eigenvectors from the principal components analysis of the GWA SNP panel explained relatively the largest proportion of the variance of the principal components in the NFBC 1966; therefore, we tested for the correlation of BMI between these eigenvectors in both genome-wide SNP data sets. We used linear regression to evaluate evidence of correlation between the eigenvectors and BMI. No correlation was found in either NFBC 1966 or the Health2000 (P > 0.05). We tested for the correlation of LP and eigenvectors using logistic regression since the response variable was dichotomous. The 5th (P = 0.02) and 10th (P = 0.01) principal components showed most evidence of correlation with LP in NFBC 1966, and the 2nd (P = 0.02), 7th (P = 0.008) and 12th (P = 0.008) eigenvectors with LP in Health2000. Including these eigenvectors as covariates in the linear regression did not change association between BMI and LP in the NFBC 1966 data or in the Health2000 subsample. In NFBC 1966, the linear regression β changed marginally (β = −0.12, P = 0.003), and in the Health2000 subsample, the effect size remained the same (β = −0.13, P = 0.03). Since the Health2000 subsample was ascertained for metabolic syndrome, we tested for the effect in both cases and controls separately, and found the linear regression β to be equal in both groups.
Frayling et al. (19) have shown that the FTO rs9939609 A allele is positively associated with BMI. We had both LCT and FTO genotypes available in NFBC 1966 cohort for 4955 individuals. The difference between FTO AA and TT homozygotes was 0.5 kg/m2. We compared the proportion of variance in BMI explained by FTO and the proportion explained by LP. The additive model (n = 4955, P = 6.7 × 10−4) of the FTO gene association was found to explain 0.2% of the variance in BMI in the NFBC 1966 cohort. Similarly, the dominant effect of LP was found to account for 0.2% of the variance in BMI in this cohort.
We tested for the association with height and LP marker but we did not find evidence of a meaningful association in the meta-analysis in Finns (n = 17 374, β = −0.004, P = 0.41). We also performed a weight analysis which—as expected, given the association with BMI but lack of association with height—demonstrated robust association (n = 17 374, β = −0.08, P = 1.4 × 10−5). Both height and weight analyses were performed in the same way as the BMI analyses.
Since the initial GWASs have not identified this association in the LCT region, we tested the effect of using an additive model in the analyses instead of the correct dominant model and combined the results with meta-analysis. The additive model decreased the association (P = 0.001) compared with the dominant model (P = 1.5 × 10−5). We simulated the size of the power decrease resulting from using the incorrect additive model instead of the correct dominant model through bootstrapping. The results of this power comparison are summarized in Table 2.
Table 2.
The results of power calculation for using the additive model instead of correct dominant model where sample size (n = 17 374), effect size (β = −0.082) and MAF (0.39) the same as the LCT variant had in combined Finnish sample of this study
| Significance level | Power (%) |
|
|---|---|---|
| Dominant model | Additive model | |
| 0.05 | 98 | 83 |
| 0.01 | 91 | 63 |
| 0.001 | 73 | 35 |
| 1 × 10−4 | 51 | 16 |
| 1 × 10−5 | 30 | 7 |
| 1 × 10−6 | 16 | 2 |
| 1 × 10−7 | 8 | 1 |
| 5 × 10−8 | 6 | 1 |
We performed a dominance deviation test from additivity in each cohort separately and pooled the β-estimates with meta-analysis. The dominance term suggested that we could reject the additive model (P = 0.001, β = 0.047).
The linear regression analysis of the British Women's Heart and Health Study (BWHHS) showed that the difference between the LP and lactase non-persistent was 0.30 kg/m2 (n = 3109, β = −0.06, P = 0.4, C allele frequency = 0.25) which is in line with our finding in the Finnish population but underpowered due to smaller sample size. The Dutch analysis was performed with the Erasmus Rucphen Family (ERF) study with extended pedigrees and Rotterdam study. The effect size estimate from the family data was again in line with the Finnish and British data (β = −0.082, n = 1952, P = 0.16, C allele frequency = 0.32) but the population-based Rotterdam study sample did not provide evidence for association (β = −0.02, n = 5745, P = 0.69, C allele frequency = 0.29) and showed smaller effect size. The German analysis was performed with KORA S3 and KORA S4 study samples. The effect size was not in the same direction in the combined KORA samples (β = 0.04, n = 3333, P = 0.20, C allele frequency = 0.36). We combined the effect sizes of non-Finnish samples in meta-analysis (β = −0.019, n = 14 346, P = 0.24) which did not prove to be significant.
Since we had the rs4988235 genotyped for individuals in Health2000 subsample and NFBC 1966 and additionally we had genome-wide SNP data, we were able to impute the marker and compare the imputed genotypes with genotyped ones. The imputation quality score was 0.91 and r2 was 0.81. The phenotype-defining C/C genotype had 23% discordance between the imputed versus genotyped calls. Of the 1060 C/C calls in the genotyped data, 245 were not defined C/C in the imputed data. In 15 cases, the imputation gave C/C genotype where the genotyped individuals had C/T genotype. In total, 11% of all calls were discordant between imputed and genotyped data.
We combined the effect sizes of all nine cohorts in meta-analysis. The association with BMI remained robust (β = −0.06, P = 7.9 × 10−5) in the combined analysis of 31 720 European individuals. There was some evidence of heterogeneity in the meta-analysis of all samples (I2 = 10), although the Q statistics was not significant (P = 0.35). Results of the individual study linear regression analyses and the meta-analyses are summarized as a forest plot (Fig. 1) and in Table 1.
DISCUSSION
In this study, we present novel evidence of association between genetically defined LP and BMI (P = 7.9 × 10−5) in 31 720 individuals from four European populations. The LCT C/T-13910 marker frequency shows strong population differences. Campbell et al. (23), sampling from an European American population, tested for an association between height and the C/T-13910 marker and found robust association between the marker and height if they did not correct for the ancestral origin of the samples. After the correction for ancestral origin, the association was considerably attenuated, thus indicating that the association was most likely an incidental reflection of population stratification of this variant. Similar generation of a link between the LCT C/T-13910 marker and self-rated health has been shown to be due to population stratification in a British study (24). We tested for the association with height and the LP marker and we did not find this marker to be associated with height. The NFBC 1966 cohort and a subset of the Health2000 cohort had genome-wide SNP data available with which we could assess the effect of population substructure. The first two principal components describe the geographical origin of the individuals in NFBC 1966 cohort, as has been shown by Jakkula et al. (25). We showed that none of the first 14 eigenvectors had a material effect on the association analysis in both the regional NFBC 1966 sample and a country-wide Health2000 subsample, which strongly suggests that the association was not due to population stratification. The best evidence which would exclude the effect of stratification came from the ERF study from the Netherlands. Although the QTDT (26) analyses were not significant, the effect size was in line with the population-based studies which is a good indicator that the population-based results were not due to stratification. All five Finnish study cohorts showed the same trend of increased BMI (heterogeneity I2 = 0), although the association was not conventionally significant in all cohorts individually. However, after combining the cohorts in the meta-analysis, the association was robust (Fig. 1). We showed that the increase in BMI was mediated through the increase in body weight (P = 1.4 × 10−5) but not through stature (P = 0.41). The non-Finnish population samples did not replicate the association significantly (P = 0.24). The marker was genotyped in the British study which showed similar effect size than the Finnish sample. The cohorts which show smaller effect size have all been imputed. In our Finnish sample, we found that the imputation of this region proves to be difficult and the erroneous genotypes concentrate on the phenotype-defining genotype. This effect is shown in Finnish population and it is not possible to assess whether it is the same case in our European replication cohorts. The ERF study is more robust for this kind of error since it benefits from the family structure in the imputation. Should the same phenomenon exist in the imputed European replication samples, the effect size would be a drastic underestimate and would explain why the European replication cohorts show smaller effect size.
We had FTO genotypes available for the NFBC 1966 cohort and were able to compare the effect size of these two variants. The LP variant had a similar effects size and explained a similar proportion of variance to FTO in this cohort.
A GWAS by Meyre et al. (27) has previously shown association in this region. They showed an association (P = 9.2 × 10−4) with a marker which is in LD (HapMap, r2 = 0.58, D′ = 0.80, rs2011946) with the LP variant and BMI in 135 obese adults and 794 lean adults of French origin. It is not possible to determine whether this association was due to stratification or not. They could not show association in an independent population-based cohort possibly because the cohort was underpowered (n = 4417). The effect in the study by Meyre et al. was in the same direction as in our study in all of the three tested adult cohorts. Any further comparison is not possible since they did not have the same marker genotyped.
There are several possible explanations as to why the LP association with BMI may have remained undetected in rest of the GWA analyses. First, the functional variant is not included in any currently used SNP arrays and the power has been reduced due to insufficient linkage disequilibrium (LD). The recently published GWASs were genotyped either by Affymetrix 5.0 or Illumina array which has less coverage in this area than the HumanHap 650 array (19–22). We found that the strongest LD (r2) with the LCT C/T-13910 and any marker within 500 000 bp in the HapMap CEPH (Utah residents with ancestry from northern and western Europe) sample included in the Illumina HumanHap 650 or Affymetrix 5.0 array was 0.78 (rs309160) (28). Second, the first four GWASs had used an additive model, which given the dominant inheritance model of LP and based on what we show here is inappropriate, and results in a considerable decrease in power as shown in Table 2. The strength of evidence of association was also reduced in our data set when the incorrect model was used (additive P = 0.001 versus dominant P = 1.5 × 10−5). Third, the imputation accuracy might be compromised in this region as was shown by our Finnish sample. Together, the incomplete tagging or inaccurate imputation combined with the use of an inappropriate genetic model may explain why this association has remained undetected. Furthermore, the association of LP with milk consumption varies between populations, being stronger in settings of intermediate allele frequency than when one allele is rare (24). One cannot reject cultural influences on milk consumption overriding the generally rather minor discomfort consequent on milk ingestion by non-persistent individuals and lead to the expectation that the association of LP genotype with BMI will be context-specific (29). The effect size of this variant may be large in Finland where dairy product consumption is very common and there is a correlation with liquid dairy product consumption and the LP variant (10,11). Further, the effect size may have been overestimated in the initial Finnish discovery sample.
The mechanism by which LP affects body composition may be related to the more restricted diet the lactase non-persistent individuals may choose or perhaps the negative symptoms such as diarrhoea after using lactose-containing food products play some role. This finding may also reflect the positive selection the LP allele has been subject to in recent history. Our study highlights the fact that the recent GWASs have not exhaustively revealed the common variants affecting BMI. Although explanation at a functional level is a matter for future studies and finding is not significant in the GWAS established genome-wide level of significance, we propose that the European variant behind LP, and perhaps other variants affecting the regulatory region of LCT, contributes to human obesity.
MATERIALS AND METHODS
Study samples
Informed consent was obtained from all participants and study was conducted according to the principles expressed in the Declaration of Helsinki and the studies were reviewed and accepted by local ethics committees. Table 3 shows the cohort characteristics. More detailed description of the study cohorts can be found at the National Biobank of Finland website (see Web Resources) and from the University of Turku website (YF cohort, see Web Resources). In brief, the NFBC 1966 includes individuals born in the Northernmost provinces of Finland with expected date of birth in 1966 who underwent a health examination at age 31. The Health2000 was conducted in 2000 to explore the health of the Finnish population. Our Health2000 cohort consists of individuals aged 30 and over. The ATBC study was initially collected to test whether α-tocopherol and β-carotene supplements would reduce the incidence of lung and other cancers. The participants were male smokers aged 50–69 years from South-western Finland. FINRISK is a cross-sectional population survey targeting coronary risk factors collected every 5 years. The current study contains individuals recruited during years 1992 and 1997 and were selected for cardiovascular study. The YF is a population-based prospective cohort study conducted at five university departments of medical schools in Finland (i.e. Turku, Helsinki, Kuopio, Tampere and Oulu), with the aim of studying the levels of cardiovascular risk factors in children and adolescents in different parts of the country. The latest follow-up was conducted in 2007. The present analysis included 2165 subjects with data on height, weight and the gene variant. The study and data collection protocols have been described in detail by Raitakari et al. (30). The Health2000, YF and FINRISK participants were collected across Finland, whereas NFBC 1966 was sampled within the Northern region and the ATBC within the South-western region of Finland. The same analyses were performed in the BWHHS. Full details of the selection of participants and measurements have been reported earlier (31,32). Women aged 60–79 years were randomly selected from primary care lists in 23 British towns. Women were recognized as non-white in the BWHHS and excluded from further analyses.
Table 3.
Sex-specific cohort characteristics
| Males |
Females |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| n | BMI |
Age |
n | BMI |
Age |
|||||
| Mean | SD | Mean | SD | Mean | SD | Mean | SD | |||
| NFBC 1966 | 2636 | 25.2 | 3.6 | 31 | 0 | 2862 | 23.3 | 3.2 | 31 | 0 |
| ATBC | 2126 | 26.9 | 4.2 | 64 | 5 | NA | NA | NA | NA | NA |
| FINRISK | 1555 | 28.0 | 4.1 | 58 | 10 | 710 | 28.2 | 5.2 | 58 | 9 |
| Health2000 | 2437 | 27.0 | 3.9 | 52 | 14 | 2883 | 26.7 | 4.9 | 55 | 16 |
| YF | 985 | 26.7 | 4.2 | 38 | 5 | 1180 | 25.4 | 5.1 | 38 | 5 |
| BWHHS | NA | NA | NA | NA | NA | 3109 | 27.3 | 4.5 | 69 | 5 |
| ERF | 909 | 27.3 | 4.3 | 50 | 15 | 1195 | 26.4 | 4.9 | 49 | 15 |
| Rotterdam | 3320 | 26.5 | 3.7 | 70 | 10 | 2369 | 25.6 | 2.9 | 68 | 8 |
| KORA S3 | 783 | 27.6 | 3.4 | 53 | 10 | 795 | 26.7 | 4.2 | 52 | 10 |
| KORA S4 | 859 | 27.8 | 3.5 | 54 | 9 | 896 | 27.2 | 4.6 | 54 | 9 |
NA, not available; SD, standard deviation; BMI, body mass index; n, number of individuals.
The ERF study is an extended-pedigree study (n = 2700) focused on the identification of quantitative traits loci related to neuropsychiatric, cardiovascular, endocrinological, ophthalmological and musculoskeletal disorders. The ERF sample has been described in more detail by Aulchenko et al. (33).
The Rotterdam study (34) is an ongoing prospective, population-based cohort study among 7983 persons aged 55 years and older, living in Ommoord, a district of Rotterdam, The Netherlands. The study was designed to investigate the incidence and determinants of chronic disabling diseases. The study is composed of an outbred ethnically homogeneous population of Dutch Caucasian origin. Rationale and design have been described previously. Informed consent was obtained from each participant, and the Medical Ethics Committee of the Erasmus Medical Centre Rotterdam approved the study. At baseline (1990–1993), all participants were interviewed and underwent extensive physical examination. These examinations were performed according to the same protocol at the second follow-up (1997–1999).
The KORA [Cooperative Health Research in the Augsburg Region; Wichmann et al. (35)] studies are population-based samples from the general population living in the region of Augsburg, Southern Germany. The KORA S3 survey (4856 subjects, response 75%) has been examined in the years 1994–1995. The KORA S4 survey was conducted in 1999–2001. The KORA S3 and S4 samples do not overlap. The study has been approved by the local ethical committee. Genome-wide data are available from Affymetrix 500K chips in KORA S3 and Affymetrix 6.0 chips in KORA S4.
Methods
In Finland, the C allele homozygosity of the LCT C/T-13910 polymorphism (rs4988235) serves as a diagnostic marker for lactase non-persistence, whereas T homozygotes and heterozygotes can be classified as LP. We tested this SNP in a set of representative population cohorts and measured its potential association with BMI and height. The SNP LCT C/T-13910 was genotyped using iPlex assay on the MassARRAY System (Sequenom, San Diego, CA, USA) using standard protocols in a total of 17 374 Finnish individuals. The YF cohort LCT C/T-13910 genotyping was performed by using the 5′-nuclease assay and fluorogenic, allele-specific, TaqMan probes (Applied Biosystems, Foster City, CA, USA) and primers, with the ABI Prism 7000 sequence detection system (Applied Biosystems). The genotype frequencies followed Hardy–Weinberg equilibrium (HWE) in all five cohorts (YF, P = 0.814; ATBC, P = 0.84; FR, P = 0.77; NFBC, P = 0.93; Health2000, P = 0.54) and call rate was >95% in all samples. BWHHS genotypes for LCT C/T-13910 (rs4988235) were generated using the KASPar chemistry which is a competitive allele-specific PCR SNP genotyping system using FRET quencher cassette oligos using standard protocols. The rs4988235 marker was imputed to the ERF, Rotterdam and KORA studies. The genetic data for ERF, the Rotterdam study and KORA S4 was imputed using MACH with HAPMAP rel 22 CEU build 36 as a reference (ERF imputation, r2 = 0.94, Rotterdam imputation study, r2 = 0.92, KORA S4 observed versus expected variance = 0.89), whereas KORA S3 was imputed with MACH based on HAPMAP rel 21 build 35 (observed versus expected variance = 0.91). The genotype frequencies followed HWE in all four non-Finnish cohorts (BWHHS, P = 0.88; ERF, P = 0.23; Rotterdam study, P = 0.09; KORA S3, P = 0.75; KORA S4, P = 0.27).
The association analyses were performed in parallel by three collaborating groups, hence NFBC 1966 and Health2000 were analysed with PLINK 1.04, FINRISK, KORA and ATBC cohorts were analysed with R, and YF and Rotterdam study cohort with SPSS, using linear regression. The ERF study was analysed using QTDT.
Lahti-Koski et al. (36) have shown that there are no regional differences in BMI within Finland in males, but a slight difference in females between southern and western Finland. Therefore, we corrected the nationwide Health2000, YF and FINRISK analyses for the area of residence (Southwest, East and Northern Finland). The NFBC 1966 cohort was collected from Northern Finland and ATBC from southwest Finland, and therefore, no regional correction was applied. The phenotypes were corrected for age and sex effects in Health2000, YF and FINRISK cohorts, with sex in NFBC 1966 cohort (all 31 years old) and age in ATBC cohort (all men). BMI outliers deviating more than three standard deviations from the mean within a cohort were removed prior to analysis, resulting in approximately normal distribution (kurtosis and skewness both between −1 and 1). The Rotterdam study and KORA were corrected for sex, age and first 10 principal components from the genome-wide data and outliers deviating more than four standard deviations from principal components or three standard deviations from standardized corrected phenotype means were removed. The ERF cohort was corrected for sex and age. The family-based analyses were performed with QTDT (26). The association method was orthogonal association which is very robust to stratification. Dominance effect was included in the analysis model.
The meta-analysis was performed by pooling the β-estimates and standard errors from the individual studies using the inverse variance method.
We had genome-wide SNP data (Illumina HumanHap 370) and phenotype data available for 4911 individuals from the NFBC 1966 cohort. Similarly, we had genome-wide SNP data (llumina HumanHap 610) available for a subset of the Health2000 cohort (2145 individuals) ascertained for matched case–control study for metabolic syndrome. We tested for hidden population substructure by performing a principal components analysis using EIGENSOFT (37). The imputation of the marker in these cohorts was performed with MACH using phased HAPMAP rel 22 CEU build 36 as a reference.
The power calculations were performed via bootstrapping. We performed 10 000 simulations where we simulated a sample of 17 374 individuals where the minor allele frequency (MAF = 0.39) and effect size (β = −0.082) were the same as LCT variant found in this study. Each sample was analysed with both additive and dominant models. The simulations were performed with R.
The dominance deviation test was performed by fitting jointly the additive term and a heterozygote indicator.
The gender interaction analysis was performed by using the meta-analysis summary statistics. The method is described further in a book by Hardy (38).
WEB RESOURCES
National Biobank of Finland: http://www.nationalbiobanks.fi; Description of YF cohort: http://med.utu.fi/cardio/youngfinnsstudy/; The International HapMap Project: http://www.hapmap.org.
FUNDING
Northern Finland Birth Cohort 1966 (NFBC 1966) is supported by the European Commission; contract number QLG1-CT-2000-01643, Biocenter Oulu, University of Oulu, Finland, and the Academy of Finland, and by NHLBI, Stampeed (Genetics of cardiovascular risk factors in large founder population birth cohorts) NIH/NHLBI 1-R01-HL087679-01. V.A. was supported by the Finnish Culture Foundation. V.S. is supported by Sigrid Juselius Foundation and the Finnish Foundation for Cardiovascular Research. L.P. and K.S. are supported by the Academy of Finland Centre of Excellence in Complex Disease Genetics, the Biocentrum Helsinki Foundation, Helsinki, Finland, and the Finnish Foundation for Cardiovascular Research. The ATBC study was supported by US Public Health Service contracts N01-CN-45165, N01-RC-45035 and N01-RC-37004 from the National Cancer Institute, Department of Health and Human Services. The Young Finns cohort was supported by the Emil Aaltonen Foundation (T.L.), the Academy of Finland (grants 53392 and 34316), the Social Insurance Institution of Finland, the Turku University Foundation, the Juho Vainio Foundation, the Finnish Foundation of Cardiovascular Research, the Finnish Cultural Foundation, and the Tampere and Turku University Central Hospital Medical Fund. We would like to acknowledge the International HapMap Consortium for the linkage disequilibrium data. The KORA research platform (KORA: Kooperative Gesundheitsforschung in der Region Augsburg) was initiated and financed by the Helmholtz Zentrum München—National Research Center for Environmental Health, which is funded by the German Federal Ministry of Education, Science, Research and Technology and by the State of Bavaria. Part of this work was financed by the German National Genome Research Network (NGFN-2 and NGFNPlus: 01GS0823) and within the Munich Center of Health Sciences (MC Health) as part of LMUinnovativ. Funding to pay the Open Access Charge was provided by Wellcome Trust Sanger Institute.
ACKNOWLEDGEMENTS
We warmly thank all the Finnish volunteers who participated in the studies. Thanks are due to Siv Knaappila and Minna Suvela for excellent laboratory work.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Enattah N.S., Sahi T., Savilahti E., Terwilliger J.D., Peltonen L., Jarvela I. Identification of a variant associated with adult-type hypolactasia. Nat. Genet. 2002;30:233–237. doi: 10.1038/ng826. [DOI] [PubMed] [Google Scholar]
- 2.Rasinpera H., Kuokkanen M., Kolho K.L., Lindahl H., Enattah N.S., Savilahti E., Orpana A., Jarvela I. Transcriptional downregulation of the lactase (LCT) gene during childhood. Gut. 2005;54:1660–1661. doi: 10.1136/gut.2005.077404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Enattah N.S., Kuokkanen M., Forsblom C., Natah S., Oksanen A., Jarvela I., Peltonen L., Savilahti E. Correlation of intestinal disaccharidase activities with the C/T-13910 variant and age. World J. Gastroenterol. 2007;13:3508–3512. doi: 10.3748/wjg.v13.i25.3508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Troelsen J.T., Olsen J., Moller J., Sjostrom H. An upstream polymorphism associated with lactase persistence has increased enhancer activity. Gastroenterology. 2003;125:1686–1694. doi: 10.1053/j.gastro.2003.09.031. [DOI] [PubMed] [Google Scholar]
- 5.Olds L.C., Sibley E. Lactase persistence DNA variant enhances lactase promoter activity in vitro: functional role as a cis-regulatory element. Hum. Mol. Genet. 2003;12:2333–2340. doi: 10.1093/hmg/ddg244. [DOI] [PubMed] [Google Scholar]
- 6.Enattah N.S., Jensen T.G., Nielsen M., Lewinski R., Kuokkanen M., Rasinpera H., El-Shanti H., Seo J.K., Alifrangis M., Khalil I.F., et al. Independent introduction of two lactase-persistence alleles into human populations reflects different history of adaptation to milk culture. Am. J. Hum. Genet. 2008;82:57–72. doi: 10.1016/j.ajhg.2007.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ingram C.J., Elamin M.F., Mulcare C.A., Weale M.E., Tarekegn A., Raga T.O., Bekele E., Elamin F.M., Thomas M.G., Bradman N., et al. A novel polymorphism associated with lactose tolerance in Africa: multiple causes for lactase persistence? Hum. Genet. 2007;120:779–788. doi: 10.1007/s00439-006-0291-1. [DOI] [PubMed] [Google Scholar]
- 8.Tishkoff S.A., Reed F.A., Ranciaro A., Voight B.F., Babbitt C.C., Silverman J.S., Powell K., Mortensen H.M., Hirbo J.B., Osman M., et al. Convergent adaptation of human lactase persistence in Africa and Europe. Nat. Genet. 2007;39:31–40. doi: 10.1038/ng1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bersaglieri T., Sabeti P.C., Patterson N., Vanderploeg T., Schaffner S.F., Drake J.A., Rhodes M., Reich D.E., Hirschhorn J.N. Genetic signatures of strong recent positive selection at the lactase gene. Am. J. Hum. Genet. 2004;74:1111–1120. doi: 10.1086/421051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lehtimaki T., Hemminki J., Rontu R., Mikkila V., Rasanen L., Laaksonen M., Hutri-Kahonen N., Kahonen M., Viikari J., Raitakari O. The effects of adult-type hypolactasia on body height growth and dietary calcium intake from childhood into young adulthood: a 21-year follow-up study–the Cardiovascular Risk in Young Finns Study. Pediatrics. 2006;118:1553–1559. doi: 10.1542/peds.2006-0542. [DOI] [PubMed] [Google Scholar]
- 11.Enattah N., Valimaki V.V., Valimaki M.J., Loyttyniemi E., Sahi T., Jarvela I. Molecularly defined lactose malabsorption, peak bone mass and bone turnover rate in young Finnish men. Calcif. Tissue Int. 2004;75:488–493. doi: 10.1007/s00223-004-0029-9. [DOI] [PubMed] [Google Scholar]
- 12.Obermayer-Pietsch B.M., Bonelli C.M., Walter D.E., Kuhn R.J., Fahrleitner-Pammer A., Berghold A., Goessler W., Stepan V., Dobnig H., Leb G., et al. Genetic predisposition for adult lactose intolerance and relation to diet, bone density, and bone fractures. J. Bone Miner. Res. 2004;19:42–47. doi: 10.1359/JBMR.0301207. [DOI] [PubMed] [Google Scholar]
- 13.Gugatschka M., Dobnig H., Fahrleitner-Pammer A., Pietschmann P., Kudlacek S., Strele A., Obermayer-Pietsch B. Molecularly-defined lactose malabsorption, milk consumption and anthropometric differences in adult males. Q. J. Math. 2005;98:857–863. doi: 10.1093/qjmed/hci140. [DOI] [PubMed] [Google Scholar]
- 14.Lember M., Torniainen S., Kull M., Kallikorm R., Saadla P., Rajasalu T., Komu H., Jarvela I. Lactase non-persistence and milk consumption in Estonia. World J. Gastroenterol. 2006;12:7329–7331. doi: 10.3748/wjg.v12.i45.7329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Obermayer-Pietsch B.M., Gugatschka M., Reitter S., Plank W., Strele A., Walter D., Bonelli C., Goessler W., Dobnig H., Hogenauer C., et al. Adult-type hypolactasia and calcium availability: decreased calcium intake or impaired calcium absorption? Osteoporos. Int. 2007;18:445–451. doi: 10.1007/s00198-006-0251-6. [DOI] [PubMed] [Google Scholar]
- 16.Must A., Spadano J., Coakley E.H., Field A.E., Colditz G., Dietz W.H. The disease burden associated with overweight and obesity. J. Am. Med. Assoc. 1999;282:1523–1529. doi: 10.1001/jama.282.16.1523. [DOI] [PubMed] [Google Scholar]
- 17.Calle E.E., Rodriguez C., Walker-Thurmond K., Thun M.J. Overweight, obesity, and mortality from cancer in a prospectively studied cohort of U.S. adults. N. Engl. J. Med. 2003;348:1625–1638. doi: 10.1056/NEJMoa021423. [DOI] [PubMed] [Google Scholar]
- 18.Ogden C.L., Carroll M.D., Curtin L.R., McDowell M.A., Tabak C.J., Flegal K.M. Prevalence of overweight and obesity in the United States, 1999–2004. J. Am. Med. Assoc. 2006;295:1549–1555. doi: 10.1001/jama.295.13.1549. [DOI] [PubMed] [Google Scholar]
- 19.Frayling T.M., Timpson N.J., Weedon M.N., Zeggini E., Freathy R.M., Lindgren C.M., Perry J.R., Elliott K.S., Lango H., Rayner N.W., et al. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science. 2007;316:889–894. doi: 10.1126/science.1141634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Loos R.J., Lindgren C.M., Li S., Wheeler E., Zhao J.H., Prokopenko I., Inouye M., Freathy R.M., Attwood A.P., Beckmann J.S., et al. Common variants near MC4R are associated with fat mass, weight and risk of obesity. Nat. Genet. 2008;40:768–775. doi: 10.1038/ng.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thorleifsson G., Walters G.B., Gudbjartsson D.F., Steinthorsdottir V., Sulem P., Helgadottir A., Styrkarsdottir U., Gretarsdottir S., Thorlacius S., Jonsdottir I., et al. Genome-wide association yields new sequence variants at seven loci that associate with measures of obesity. Nat. Genet. 2009;41:18–24. doi: 10.1038/ng.274. [DOI] [PubMed] [Google Scholar]
- 22.Willer C.J., Speliotes E.K., Loos R.J., Li S., Lindgren C.M., Heid I.M., Berndt S.I., Elliott A.L., Jackson A.U., Lamina C., et al. Six new loci associated with body mass index highlight a neuronal influence on body weight regulation. Nat. Genet. 2009;41:25–34. doi: 10.1038/ng.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Campbell C.D., Ogburn E.L., Lunetta K.L., Lyon H.N., Freedman M.L., Groop L.C., Altshuler D., Ardlie K.G., Hirschhorn J.N. Demonstrating stratification in a European American population. Nat. Genet. 2005;37:868–872. doi: 10.1038/ng1607. [DOI] [PubMed] [Google Scholar]
- 24.Smith G.D., Lawlor D.A., Timpson N.J., Baban J., Kiessling M., Day I.N., Ebrahim S. Lactase persistence-related genetic variant: population substructure and health outcomes. Eur. J. Hum. Genet. 2008;17:357–367. doi: 10.1038/ejhg.2008.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jakkula E., Rehnstrom K., Varilo T., Pietilainen O.P., Paunio T., Pedersen N.L., deFaire U., Jarvelin M.R., Saharinen J., Freimer N., et al. The genome-wide patterns of variation expose significant substructure in a founder population. Am. J. Hum. Genet. 2008;83:787–794. doi: 10.1016/j.ajhg.2008.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Abecasis G.R., Cardon L.R., Cookson W.O. A general test of association for quantitative traits in nuclear families. Am. J. Hum. Genet. 2000;66:279–292. doi: 10.1086/302698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meyre D., Delplanque J., Chevre J.C., Lecoeur C., Lobbens S., Gallina S., Durand E., Vatin V., Degraeve F., Proenca C., et al. Genome-wide association study for early-onset and morbid adult obesity identifies three new risk loci in European populations. Nat. Genet. 2009;41:157–159. doi: 10.1038/ng.301. [DOI] [PubMed] [Google Scholar]
- 28.The International HapMap Consortium. The International HapMap Project. Nature. 2003;426:789–796. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- 29.Savaiano D.A., Boushey C.J., McCabe G.P. Lactose intolerance symptoms assessed by meta-analysis: a grain of truth that leads to exaggeration. J. Nutr. 2006;136:1107–1113. doi: 10.1093/jn/136.4.1107. [DOI] [PubMed] [Google Scholar]
- 30.Raitakari O.T., Juonala M., Ronnemaa T., Keltikangas-Jarvinen L., Rasanen L., Pietikainen M., Hutri-Kahonen N., Taittonen L., Jokinen E., Marniemi J., et al. Cohort profile: the cardiovascular risk in Young Finns Study. Int. J. Epidemiol. 2008;37:1220–1226. doi: 10.1093/ije/dym225. [DOI] [PubMed] [Google Scholar]
- 31.Lawlor D.A., Bedford C., Taylor M., Ebrahim S. Geographical variation in cardiovascular disease, risk factors, and their control in older women: British Women's Heart and Health Study. J. Epidemiol. Community Health. 2003;57:134–140. doi: 10.1136/jech.57.2.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lawlor D.A., Ebrahim S., Davey Smith G. Socioeconomic position in childhood and adulthood and insulin resistance: cross-sectional survey using data from British women's heart and health study. Br. Med. J. 2002;325:805–810. doi: 10.1136/bmj.325.7368.805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Aulchenko Y.S., Heutink P., Mackay I., Bertoli-Avella A.M., Pullen J., Vaessen N., Rademaker T.A., Sandkuijl L.A., Cardon L., Oostra B., et al. Linkage disequilibrium in young genetically isolated Dutch population. Eur. J. Hum. Genet. 2004;12:527–534. doi: 10.1038/sj.ejhg.5201188. [DOI] [PubMed] [Google Scholar]
- 34.Hofman A., Breteler M.M., van Duijn C.M., Janssen H.L., Krestin G.P., Kuipers E.J., Stricker B.H., Tiemeier H., Uitterlinden A.G., Vingerling J.R., et al. The Rotterdam Study: 2010 objectives and design update. Eur. J. Epidemiol. 2009;24:553–572. doi: 10.1007/s10654-009-9386-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wichmann H.E., Gieger C., Illig T. KORA-gen—resource for population genetics, controls and a broad spectrum of disease phenotypes. Gesundheitswesen. 2005;67(Suppl. 1):S26–S30. doi: 10.1055/s-2005-858226. [DOI] [PubMed] [Google Scholar]
- 36.Lahti-Koski M., Taskinen O., Simila M., Mannisto S., Laatikainen T., Knekt P., Valsta L.M. Mapping geographical variation in obesity in Finland. Eur. J. Public Health. 2008;18:637–643. doi: 10.1093/eurpub/ckn089. [DOI] [PubMed] [Google Scholar]
- 37.Price A.L., Patterson N.J., Plenge R.M., Weinblatt M.E., Shadick N.A., Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 38.Hardy M.A. Regression with Dummy Variables (Sage University Paper Series on Quantitative Applications in the Social Sciences, 07-093) Newbury Park, CA: SAGE Publications; 1993. [Google Scholar]