Abstract
Recent discovery of the copy number variation (CNV) in normal individuals has widened our understanding of genomic variation. However, most of the reported CNVs have been identified in Caucasians, which may not be directly applicable to people of different ethnicities. To profile CNV in East-Asian population, we screened CNVs in 3578 healthy, unrelated Korean individuals, using the Affymetrix Genome-Wide Human SNP array 5.0. We identified 144 207 CNVs using a pooled data set of 100 randomly chosen Korean females as a reference. The average number of CNVs per genome was 40.3, which is higher than that of CNVs previously reported using lower resolution platforms. The median size of CNVs was 18.9 kb (range 0.2–5406 kb). Copy number losses were 4.7 times more frequent than copy number gains. CNV regions (CNVRs) were defined by merging overlapping CNVs identified in two or more samples. In total, 4003 CNVRs were defined encompassing 241.9 Mb accounting for ∼8% of the human genome. A total of 2077 CNVRs (51.9%) were potentially novel. Known CNVRs were larger and more frequent than novel CNVRs. Sixteen percent of the CNVRs were observed in ≥1% of study subjects and 24% overlapped with the OMIM genes. A total of 476 (11.9%) CNVRs were associated with segmental duplications. CNVS/CNVRs identified in this study will be valuable resources for studying human genome diversity and its association with disease.
INTRODUCTION
Comprehensive mapping of human genome variation is expected to facilitate the understanding of individual phenotypic differences including disease susceptibility and drug responsiveness (1,2). In addition to single-nucleotide polymorphisms (SNPs), the recent discovery of the DNA structural variation in normal individuals has widened our understanding of genomic variation (3,4). Since the two pioneering studies reported the existence of large-scale DNA structural variation, more than 38 000 DNA copy number variations (CNVs) have been identified in various populations (5–9) and included in public databases such as the Database of Genomic Variants (DGV; http://projects.tcag.ca/variation/) (10). Since CNV is reported to comprise 5–12% of the human genome and to have the potential to affect gene expression levels, it becomes acknowledged as a major contributor to genetic diversity. Also, associations between CNV and various diseases have been reported, which makes CNV look more promising (9,11–13). However, studying CNV–disease associations has been impeded by incomplete knowledge on reference human CNV, mainly due to the lack of standardization in terms of CNV detection and statistical analysis. Also, most of the reported CNVs have been identified in Caucasians, which may not be directly applicable to people of different ethnicities including Koreans.
To profile CNV patterns in East-Asian population, we screened CNVs in 3578 Korean individuals, using the Affymetrix Genome-Wide Human SNP array 5.0 and explored their characteristics. We uncovered 2077 novel CNVRs in addition to 1926 known ones, which can be valuable clues to human genetic diversity. The ontology data for genes enriched in our CNV loci and the sequence properties in and around them will also provide insights to functional and evolutional implications of CNVs.
RESULTS
General characteristics of CNVs and CNVRs
We used genotyping data of 3678 healthy, unrelated Korean people (1808 men, 1870 women; mean age of 52.1 years) provided by the Korea National Institute of Health (KNIH). KNIH recruited and genotyped 10 038 healthy Koreans aged 40–69 to study genotype–phenotype/disease associations. When we examined the quality of the arrays, we found the inter-chip discrepancy in values of signal intensities. We adopted the average standard deviation (SD) of signal intensities and the median of the absolute values of all pairwise differences (MAPD) as a per-chip variability estimate. We chose 3678 arrays, of which both average SD and MAPD values rank the 50th percentile or below. CNVs were defined using two types of references: the genomic DNA of a single reference individual, a European-American male (NA10851) from the HapMap study; a pooled data set of 100 randomly chosen Korean females as described in Materials and Methods. When using the pooled reference, CNVs were called from 3578 individuals, excluding 100 samples used for the reference. The number of CNVs defined using NA10851 as a reference was 131 052 and that of CNVs defined using the pooled reference was 144 207. The list of CNVs and related information are available on our (website http://ircgp.com/cnv_RND2.html). We performed further analyses using the CNV calls from 3578 Koreans defined against the pooled reference, because >99% of the CNVs (131 015/131 052) identified using NA10851 were included in the CNVs identified using the pooled reference, and also because it is possible to assign gain or loss status to CNVs/CNVRs identified using the pooled reference. General characteristics of the CNVs in our data set are summarized in Table 1 and Fig. 1A. The average number of CNVs per individual genome was 40.3 (SD 69.7, range 6–993; Supplementary Material, Fig. S1), and the median size was 18.9 kb (range 0.2–5406 kb). Copy number losses (CN-losses) were 4.7 times more frequent than copy number gains (CN-gains) (25 347 gains versus 118 860 losses). The median size of CN-losses (20.0 kb) is slightly larger than that of CN-gains (13.4 kb).
Table 1.
General characteristics of CNVs and CNVRs in this study
| CNV | CNVR | |
|---|---|---|
| Total count | 144 207 | 4003 |
| CN-gain count | 25 347 | 112 |
| CN-loss count | 118 860 | 3553 |
| Complex count | — | 338 |
| Average number per genome | 40.3 | 37.5 |
| Median size (range) (kb) | 18.9 (0.2–5406) | 30.3 (0.4–5521) |
| Median size of CN-gains | 13.4 (0.2–2263) | 17.7 (1.2–345) |
| Median size of CN-losses | 20.0 (0.2–5406) | 29.9 (0.4–5521) |
| Genome coverage | — | 241.9 Mb (∼8%) |
CN-gain/loss, copy number gain/loss CNVs.
Figure 1.
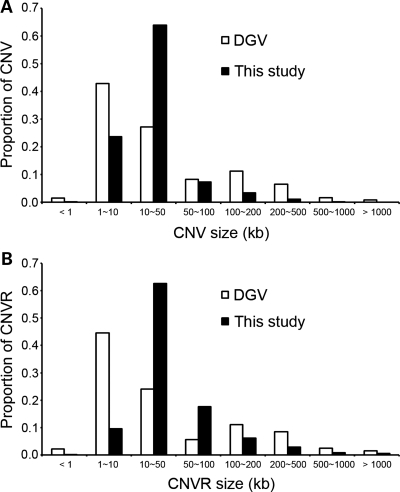
Size distribution of CNVs (A) and CNVRs (B) from this study with corresponding CNV/CNVRs from the DGV (August 2009 version). X-axis, the sizes in kilobases. Y-axis, the proportions of CNV/CNVRs within each size bin.
We defined CNV regions (CNVRs) using 144 207 CNVs by merging overlapping CNVs identified in two or more samples as described previously (9). A total of 3076 CNVs called in a single individual were excluded from defining CNVRs. In total, 4003 CNVRs were defined that encompass 241.9 Mb across the whole chromosomes except for Y, accounting for ∼8% of the human genome. CNVRs showed characteristics similar to CNVs; CNVR-losses were more frequent than CNVR-gains; the size of CNVR-losses was larger than that of CNVR-gains. More information on CNVRs is also presented in Table 1 and in Supplementary Material, Table S1. CNVR sizes ranged from 0.4 to 5521 kb, with a median size of 30.3 kb. Although the proportion of the CNVR <10 kb in our data set was lower than that of the DGV CNVRs (August 2009 version), the proportion of CNVRs <50 kb was higher in our data set (Fig. 1B). In other words, the size distribution of CNV/CNVRs in our data set appeared to be less right-skewed. Sixteen percent of the CNVRs (656/4003) were observed in ≥1% of 3578 study subjects, which satisfy a criteria for polymorphism (Supplementary Material, Table S1). Among the CNVRs with an allele frequency ≥1%, 130 CNVRs (3.2% of total CNVRs) were observed in ≥5% of study subjects. A majority of CNVRs (91.5%, 3665/4003) showed the same direction of change, i.e. gain or loss, whereas 8.5% (338/4003) contain both gains and losses in the same CNVR (hereinafter referred to as complex CNVRs). Figure 2 shows examples of gain only, loss only and complex CNVRs. To examine the consistency of the boundaries of CNVs belonging to each CNVR, we measured the coefficient of variation (CV), which is the ratio of SD to the mean of linear position of each CNV. Most of the CNVRs in this study had common boundaries; 3967 out of 4003 CNVRs showed CV <1%. Among all 4003 CNVRs, 972 (24.3%) CNVRs overlapped with the OMIM genes, 127 (3.2%) with the OMIM morbid map (data source: UCSC database, hg18, omimGene and omimMorbidMap) (Supplementary Material, Table S1).
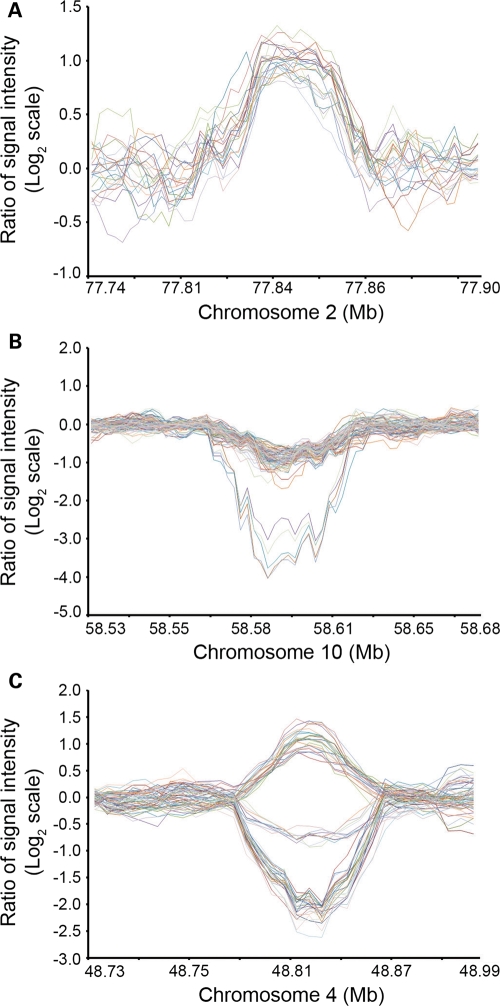
Figure 2.
Examples of signal intensity ratio plots of CNVRs. (A) A CNVR on 2p12, gain only. (B) A CNVR on 10g21.1, loss only. (C) A CNVR on 4p11, gain/loss complex. X-axis, genomic coordinates (Mb). Y-axis, signal intensity ratios (test/reference) in log2 scale.
Known and novel CNVRs
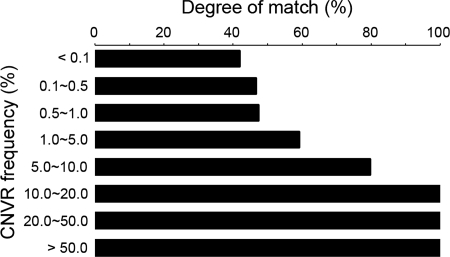
All 4003 CNVRs identified by the current study were compared with the 8410 CNVRs in the DGV (hereinafter referred to as DGV CNVR). If a CNVR overlapped any DGV CNVR, we considered it a known CNVR. Otherwise, it was classified as a novel CNVR. In total, 1926 CNVRs (48.1%) were known ones, and remaining 2077 CNVRs (51.9%) were potentially novel. In total, 2881 out of 3578 subjects contained at least one novel CNVRs. The median size of known CNVRs (36.7 kb) was significantly larger than that of novel CNVRs (27.1 kb) (P < 0.0001). The mean allele frequency of known CNVRs (1.5%) was significantly higher than that of novel CNVRs (0.4%) (P < 0.0001). Accordingly, the frequency of CNVRs showed a positive correlation with the degree of match between our CNVRs and DGV ones (Fig. 3). All the CNVRs with an allele frequency ≥10% overlapped the DGV CNVRs, whereas 55% of the CNVRs with an allele frequency <1% did not. Interestingly, among 74 CNVRs with an allele frequency of 5–10%, 15 CNVRs (20.3%) did not overlap the DGV CNVRs. Table 2 shows 15 potentially Korean-specific novel CNVRs with an allele frequency of 5–10%.
Figure 3.
Degree of match between CNVRs from this study and DGV CNVRs with respect to the allele frequency. X-axis, the degree of match (%). Y-axis, the allele frequency of CNVRs.
Table 2.
Potential Korean-specific CNVRs with the allele frequency ≥5%
| Chr | Cytoband | Start (bp) | End (bp) | Size (kb) | Frequency (%) | Status | Genes |
|---|---|---|---|---|---|---|---|
| 1 | 1q23.3 | 162009464 | 162082267 | 72.804 | 6.74 | L | — |
| 1 | 1q31.2 | 189272340 | 189325007 | 52.668 | 5.76 | L | — |
| 2 | 2q22.1 | 140652118 | 140854714 | 202.597 | 6.99 | L | LRP1B |
| 3 | 3p24.2 | 24363248 | 24448859 | 85.612 | 5.09 | L | THRB |
| 3 | 3p13 | 72398646 | 72444156 | 45.511 | 5.28 | L | — |
| 4 | 4q13.1 | 61078573 | 61119445 | 40.873 | 6.99 | C | — |
| 5 | 5q11.2 | 50838014 | 50898889 | 60.876 | 6.09 | L | — |
| 5 | 5q14.3 | 89233437 | 89297026 | 63.59 | 5.48 | L | — |
| 7 | 7p21.3 | 11648289 | 11768235 | 119.947 | 9.03 | L | THSD7A |
| 7 | 7q21.3 | 92910385 | 93006702 | 96.318 | 5.25 | L | CALCR, MIR653,MIR489 |
| 8 | 8q21.12 | 78570181 | 78643662 | 73.482 | 6.65 | L | — |
| 11 | 11p15.1 | 21345470 | 21395190 | 49.721 | 5.25 | L | NELL1 |
| 12 | 12q21.1 | 70034230 | 70095777 | 61.548 | 7.04 | L | — |
| 13 | 13q21.1 | 55173808 | 55234854 | 61.047 | 7.57 | L | — |
| 17 | 17q21.33 | 47334364 | 47468516 | 134.153 | 5.81 | L | CA10 |
L, CNVRs containing CN-losses only; C, complex CNVRs containing both gains and losses in the same loci.
Intrachromosomal distribution and sequence properties of CNVR
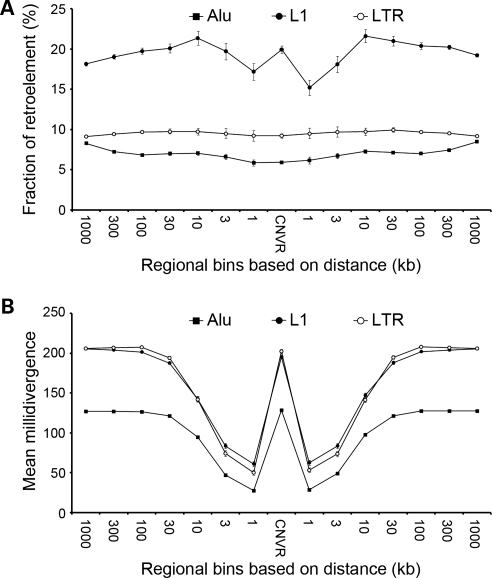
We also interrogated the intrachromosomal distribution of CNVRs across the whole genome. CNVRs identified in this study distributed evenly across each chromosome arm from centromere to telomere, but novel CNVRs were relatively uncommon in pericentromeric and subtelomeric regions (Supplementary Material, Fig. S2). The average GC content of sequences in 4003 CNVRs was 37.7%. In terms of SNP, the average number of SNPs per 100 kb-sized segment in the CNVRs was 561, which was lower than the average number of SNPs across the whole genome (628 per 100 kb) (data source: UCSC database, hg18, snp130). Among the 4003 CNVRs, 476 (11.9%) were associated with segmental duplications, and 82% (391/476) of them were known CNVRs. Highly repetitive sequences such as retrotransposons were also investigated for their correlation with CNVRs. The frequencies of three major retrotransposon families, LINE-1 (L1), long terminal repeat (LTR) and Alu, were assessed according to the separating distance from the CNVRs (Fig. 4A). In the case of Alu and LTR elements, the regional fractions around CNVRs were constant (7.0 and 9.5%, respectively), which are similar to the whole-genome averages (8.3 and 10.6%, respectively). In contrast, the regional fraction of L1 elements showed fluctuations around CNVRs. L1 elements were relatively enriched in the flanking regions 10 kb away from CNVRs, but sparse in the immediate vicinity of CNVRs. The average divergence rates for these retrotransposons were also measured by the separating distance from the CNVRs (Fig. 4B). Alu-repetitive sequences, which are relatively young elements, showed constantly lower divergence rates than L1 and LTR elements. However, for all three elements, divergence rates showed the lowest levels around 1 kb of flanking regions from the CNVRs and increased in up to 30 kb of flanking regions from the CNVRs.
Figure 4.
Association of retrotransposons with CNVRs. (A) The mean regional fractions of the three retrotransposons around CNVRs by distance from the CNVRs. X-axis, the separating distance from the CNVRs. Y-axis, the fractions (%) of the retroelements. 95% confidence intervals are shown with error bars. (B) Divergence rates of the three retrotransposons around the CNVRs. X-axis, the separating distance from the CNVRs. Y-axis, the mean milliDivergence of the retroelements. 250 milliDivergence corresponds to 25.0% of divergence rates. The orientation of repeat sequences was determined with respect to the corresponding CNVRs; concordant elements are those whose tails are more closely located to CNVRs; divergent elements are those with opposite orientation. Calculation of milliDivergence was performed following the description at the RepeatMaskers (http://www.repeatmasker.org/).
Genes enriched in CNVRs
A total of 1598 (7.9%) out of 20 119 RefSeq genes were found to be included in 4003 CNVRs in our data set. Among the 1598 genes, 971 (60.7%) were located in the known CNVRs and 627 (39.3%) in the novel CNVRs. To explore the functional implications of structural variations, we performed the gene set enrichment analysis for the CNVRs. We analyzed genes located in known and novel CNVRs separately. The annotated functions are summarized in Table 3. Enriched functions in known CNVRs include diverse metabolisms, immune reaction, nervous system development and cell-adhesion-related molecular functions. The most significantly enriched function in novel CNVRs was neuroactive ligand–receptor interaction functions including glutamate signaling pathway.
Table 3.
Functional enrichment analysis of genes associated with CNVRs
| Annotated functions | Genes | Gene number | Significance (P-value) | |
|---|---|---|---|---|
| Known CNVR | Pentose and glucuronate interconversions | 14 | 25 | 9.20E − 13 |
| Starch and sucrose metabolism | 23 | 83 | 4.72E − 12 | |
| Metabolism of xenobiotics by cytochrome P450 | 21 | 70 | 7.42E − 12 | |
| Porphyrin and chlorophyll metabolism | 15 | 41 | 3.14E − 10 | |
| Antigen processing and presentation | 18 | 75 | 1.22E − 08 | |
| Androgen and estrogen metabolism | 14 | 54 | 1.80E − 07 | |
| Nervous system development | 37 | 382 | 4.73E − 05 | |
| Cell adhesion molecules | 18 | 132 | 6.54E − 05 | |
| Xenobiotic metabolic process | 5 | 11 | 9.38E − 05 | |
| Response to xenobiotic stimulus | 5 | 12 | 0.000154 | |
| Heparan sulfate biosynthesis | 6 | 19 | 0.000196 | |
| Glutathione transferase activity | 5 | 15 | 0.000519 | |
| Starch and sucrose metabolism | 7 | 31 | 0.000567 | |
| Extracellular structure organization and biogenesis | 7 | 32 | 0.000695 | |
| Type I diabetes mellitus | 8 | 43 | 0.00092 | |
| Synaptogenesis | 5 | 18 | 0.001311 | |
| N-Acetylglucosamine metabolic process | 4 | 12 | 0.001955 | |
| Classic pathway | 4 | 13 | 0.002716 | |
| Novel CNVR | Glutamate signaling pathway | 6 | 17 | 8.25E − 06 |
| Neuroactive ligand receptor interaction | 21 | 239 | 2.06E − 05 | |
| 3,5-Cyclic nucleotide phosphodiesterase activity | 5 | 13 | 3.02E − 05 | |
| Cyclic nucleotide phosphodiesterase activity | 5 | 14 | 4.58E − 05 | |
| Receptor activity | 36 | 571 | 5.45E − 05 | |
| Ionotropic glutamate receptor activity | 4 | 10 | 0.000169 | |
| Axon guidance | 13 | 127 | 0.00017 | |
| Cell adhesion molecules | 13 | 132 | 0.00025 | |
| Glutamate receptor activity | 5 | 20 | 0.000304 | |
| Long-term depression | 9 | 74 | 0.000476 | |
| Neurotransmitter secretion | 4 | 13 | 0.000534 | |
| Regulated secretory pathway | 4 | 15 | 0.000969 | |
| Transmembrane receptor activity | 25 | 410 | 0.001168 |
DISCUSSION
We identified CNVs in 3578 unrelated Korean individuals, using high-resolution Affymetrix SNP 5.0 arrays and explored the biological implications of them. The average number of CNVs per genome in this study (40.3 per genome) is higher than that of CNVs called in the HapMap populations using lower resolution platforms such as Affymetrix 500K EA platform (24 per genome) or human tiling BAC array (15.1 per genome) (8,9). The median size of CNVs in our data set (18.9 kb) is much smaller than the sizes reported by Redon et al. (81 kb by 500K and 228 kb by whole genome tile path BAC array) (9). In terms of the frequency and size of CNVs, our data look generally consistent with recent reports using higher resolution platforms including McCarroll et al.'s (6,7,14). Among CNVRs in our data set, 51.9% were potentially novel. Given that the median size of novel CNVRs was smaller by ∼10 kb than that of known ones, they might have been missed by older, lower resolution platforms. Size difference is more prominent if we compare the size of novel and known CNVRs with an allele frequency >5% (63.5 versus 78.6 kb). To estimate a false discovery rate, we validated 16 randomly selected loci using genomic quantitative PCR (qPCR) and found 14 (87.5%) loci showed corresponding copy number changes (Supplementary Material, Table S2). Our estimated cumulative CNVR length of 241.9 Mb (8% of the genome) is relatively high compared with recent reports (8,14,15), but lower than what we expected when the sample size was taken into consideration. This might be due to our stringent criteria for CNV detection and the limited CNV coverage of the platform we used (14). The other possibility is that the study population we used is relatively homogeneous than the HapMap samples.
Although half the CNVRs identified in this study did not overlap any DGV CNVRs, all the CNVRs with an allele frequency ≥10% overlapped them. There was a positive correlation between the allele frequency and the degree of match between our CNVRs and DGV CNVRs, which reflects that common CNVRs in Koreans are also common in other populations of different ethnicities. Interestingly, 15 out of 74 relatively common CNVRs with an allele frequency of 5–10% did not overlap any known DGV CNVRs. Taking their relatively high allele frequencies into consideration, it is possible that some of them are Korean-specific novel CNVRs which need to be considered for CNV-disease association studies in East-Asian populations (16).
We assessed the GC content of the CNVRs to test the hypothesis that CNVs are located in gene-poor regions (17,18). The average GC content of the CNVRs in our data set (37.7%) was slightly lower than the genome-wide average GC contents (41%) reported in human (19), which suggests that CNVs are likely to coincide with gene-poor regions. In terms of SNPs, common CNVRs in our study are located in relatively SNP-poor regions compared with the genome average number of SNPs (561 versus 628 per 100 kb), which agrees with McCarroll et al.'s observation that common CNVs corresponded to physically bald spots in SNP arrays (14). CNVRs we identified in this study overlapped 7.9% of total RefSeq genes and 972 CNVRs overlapped OMIN genes. Our CNVRs also overlapped 36 disease-associated CNV loci recently reported (20,21), which suggests that our results will be useful resources for studying the association between CNVs and disease/complex traits in East-Asian population. Through the gene set enrichment analysis, we found that gene functions significantly enriched in the known CNVRs include starch metabolism, metabolism of xenobiotics by cytochrome P450, antigen presentation, nervous system development and cell-adhesion-related molecular functions, which is consistent with the previous observations (6,22). However, the gene functions enriched in novel CNVRs include new ones such as glutamate signaling pathway, which was reported to be involved in bipolar disorder and schizophrenia (23). Since the enrichment of genes does not necessarily indicate the functional consequences, further studies are required to interpret the results.
To understand the physical properties of CNV, we examined the association of CNVRs with segmental duplications and repetitive sequences. In our study, 11.9% of CNVRs overlapped segmental duplications, which is smaller than that reported in previous studies. However, a majority of CNVRs associated with segmental duplications (82%) are known ones, which agrees with previous studies. It can support the hypothesis that non-allelic homologous recombination plays a role for CNV formation in human, especially for common ones (9,13,14,24,25). Among the repetitive sequence elements, L1 elements showed a relative enrichment (regional fraction >20%) in the flanking regions of CNVRs, which supports Hastings et al.'s suggestion that recurrent CNVs are likely to arise between repeated sequences (13). All three major retrotransposons (Alu, L1 and LTR) showed characteristic patterns of divergence rates in the flanking region of CNVRs, being lowest around 1 kb of flanking regions from the CNVRs and increased in up to 30 kb of flanking regions from the CNVRs. As divergence rates are generally proportional to the insertional age of the corresponding elements (19), low divergence rates around CNVRs imply that relatively young retrotransposons are dominant near the CNVRs. This active turnover of genetic elements in the vicinity of CNVs could be associated with the formation of structural variants (26,27).
There are several limitations in our study. First, due to the innate limitations of SNP 5.0 arrays, we may have missed many common CNVs. McCarroll et al. reported that common CNVs tend to be missed by earlier SNP arrays, including 5.0, because common CNVs tend to cause SNP data to fail the quality checks, which makes corresponding SNP probes excluded for the commercial array (14). Second, although it is known that SNPs in Koreans were similar to those in Japanese and Chinese (28), we did not examine it in terms of CNVs. To generalize our results and complete the CNV map of East-Asian populations, further studies using samples of various ethnicities will be required.
Nevertheless, CNV/CNVRs identified in this study will be valuable resources for studying human genome diversity including formation mechanisms of structural variation and its associations with various diseases.
MATERIALS AND METHODS
Study subjects
We used genotyping data of 3678 healthy, unrelated Korean people (1808 men, 1870 women; mean age of 52.1 years) provided by KNIH. KNIH recruited and genotyped 10 038 healthy Koreans aged 40–69 to study genotype–phenotype/disease associations under the Korean Genome Epidemiology Study (KoGES). We examined the quality of 10 038 arrays and found the inter-chip discrepancy in values of signal intensities. We adopted the average SD of signal intensities and the MAPD as a per-chip variability estimate, which Affymetrix introduced as quality control measure for SNP Array 6.0, but had not for 5.0 (MAPD and Quality Control in GTC 2.0, http://www.affymetrix.com/products_services/software/specific/genotyping_console_software.affx#1_4). We chose 3678 arrays, of which average SD and MAPD values both rank the 50th percentile or below simultaneously. The 50th percentile MAPD value from our 10 038 arrays was 0.41, which is close to the cutoff range Affymetrix recommended being 0.3–0.4.
This study was approved by the Institutional Review Board of the Catholic University of Korea School of Medicine (CUMC07U047). Details of genotyping procedures were described elsewhere (28). In brief, 500 ng of genomic DNA was extracted from peripheral blood leukocytes and assayed on the Affymetrix Genome-Wide Human SNP array 5.0 (Affymetrix, Santa Clara, CA, USA).
CNV discovery
For intensity pre-processing, i.e. background correction, normalization and probe set summarization, we adopted the robust multiarray average algorithm with slight modifications (29). In brief, background-corrected data were normalized using quantile normalization and summarized by median polish. Finally, CNV was called on the basis of test/reference signal intensity ratio in log2 scale thorough the modified SW-ARRAY algorithm (30). For CNV detection, we set up three criteria for CNV calling: the number of minimum consecutive probes; a threshold cutoff; a consecutive probe (island) score cutoff. To obtain reliable CNV calls, we considered only CNVs that involved at least six consecutive probe sets. To optimize the other two parameters, we used the Affymetrix SNP array 5.0 data generated from three replicates of two HapMap cell line DNAs, NA10851 and NA15510 (Coriell, Camden, NJ, USA). ‘>50% reproducibility’, a percentage of CNVs called >50% of time in all nine pairwise comparisons, was calculated as an index of reproducibility following the methods of Komura et al. (31). We calculated it for 30 different threshold cutoff levels ranging from 2 to 5 median absolute deviations (MAD) and for 5 different island score cutoff levels ranging from 1 to 5 MAD (Supplementary Material, Fig. S3). Although the highest >50% reproducibility appeared at 3.6 MAD threshold–5 MAD island score cutoff combination, this combination produced just a small number of CNVs (only 18 CNVs identified from the comparison between NA10851 and NA15510). To decide more practical but reliable threshold and island score cutoff values, a self-versus-self analysis using the two HapMap samples was also performed under the same threshold and island score combinations described earlier. At 2.8 MAD threshold–1 MAD island score and above, there was no CNV called from the self-versus-self analysis (data not shown). Since CNVs called from self-versus-self tests are likely to be false positives, we set 2.8 and 1 MAD as the minimum cutoff for CNV calling to minimize the false-positive calls. We further optimized our criteria through CNV validation. We validated all the CNVs called at least once from the triplicate experiments between NA10851 and NA15510 using genomic qPCR. At 2.8 MAD threshold–1 MAD island score cutoffs, a total of 88 CNVs were defined from nine pairwise comparisons. It was possible to validate 67 CNV loci, and 55 CNVs among them (82.1%) showed consistent copy number changes with the array results. When we increased the cutoffs to 3 MAD threshold–5 MAD island score levels, a false-positive rate decreased; 55 CNVRs were defined, and 44 out of the 48 testable CNVs (91.7%) showed consistent results (Supplementary Material, Table S3). After validation results being considered, we chose 3 MAD for the threshold cutoff, 5 MAD for the island score cutoff and six for the minimum number of consecutive probes for CNV calling in our study. Copy number status of individual CNVs, i.e. gains or losses, was determined on the basis of the median signal intensity ratios of the probes located in the CNVRs as described previously (31). We used two types of references to define CNVs in our samples: the genomic DNA of a single reference individual, a European-American male (NA10851) from the HapMap study; a pooled data set of 100 randomly chosen Korean females. For the pooled reference, we randomly selected 100 female genotype data and used the median intensity values of each data point as a pooled reference value. Since those 100 samples were excluded from CNV calling, we defined CNVs from a total of 3578 Koreans (1808 men and 1770 women). When using the NA10851 reference, we considered only the CNVs reliable which were called more than twice out of three comparisons using the triplicate genotyping data of NA10851. We defined CNVRs by merging overlapping CNVs identified in two or more samples as described previously (9). CNVs called in a single individual were excluded from defining CNVRs.
Validation of CNVs by genomic qPCR
To estimate the false discovery rate of our CNV-calling algorithm, CNVs were validated by genomic real-time qPCR. Genomic qPCR was performed using the Mx3000P qPCR system (Stratagene, La Jolla, CA, USA). To validate the CNVs identified in the current study, we randomly selected 16 CNV loci and performed genomic qPCR using DNAs from study subjects who showed corresponding CNVs on that loci. Information of all the primers for the validation is available in Supplementary Material, Table S4. Twenty microliters of reaction mixtures contained 20 ng of genomic DNA, SYBR® Premix Ex Taq TM II (TaKaRa Bio, Shiga, Japan), 1×ROX (TaKaRa Bio, Shiga, Japan) and 10 pmol of primers. Thermal cycling conditions consisted of one cycle of 30 s at 95°C, followed by 45 cycles of 5 s at 95°C, 10 s at 55–65°C and 30 s at 72°C. All PCR experiments were repeated twice, and amplification efficiencies for both target and reference genes were evaluated using a standard curve over 1:5 serial dilutions. Relative copy number was calculated by the ΔΔCt method using the Ct values (32).
Chromosomal distribution and sequence context of CNVR
Genomic coordinates of segmental duplications and repetitive sequences were downloaded from the UCSC genome browser, which is of the same version for array probe-mapping (Human Build 36.1). We also downloaded genomic coordinates and related information on Alu, L1 and LTR elements from the RepeatMaskers (http://www.repeatmasker.org/). Flanking regions of the CNVRs were segmented into the regional bins according to the separating distance from the midpoint of CNVRs up to 1 Mb. Regional fractions of repetitive sequences were measured and averaged by regional bin. The orientation of repeat sequences were determined with respect to the corresponding CNVRs; concordant elements are those whose tails are more closely located to CNVRs; divergent elements those with opposite orientation.
Gene set enrichment analysis
We downloaded the list of 20 119 RefSeq genes from the UCSC Genome Browser. Gene set enrichment analysis was performed using a total of 1598 RefSeq genes included in CNVRs in our data set. For that, we downloaded three functional gene sets from the Molecular Signature Database (MSigDB; http://www.broad.mit.edu/gsea/msigdb). Three sets include canonical gene sets (cp_c2), GO biological process (GO_bp) and GO molecular functions (GO_mf). A total of 1831 gene sets were used after removing sets with less than 5 genes or more than 1000 genes. Significance of gene enrichment in CNVRs was assessed using hypergeometric distribution. The functional enrichment analysis was performed using the GEAR software (http://www.systemsbiology.co.kr/GEAR/) (33).
Consistency of CNV boundaries
The consistency of CNV boundaries in each CNVR was measured by CV, which is the ratio of the SD to the mean of liner position of each CNV which belongs to the same CNVR.
SUPPLEMENTARY MATERIAL
FUNDING
This work was supported by the Korea Research Foundation Grant funded by the Korean Government (KRF-2008-220-E00025). The Consortium for Large Scale Genome Wide Association Study was supported by genotyping data (genome wide association analysis of community based cohort study, 2007) from the Korean Genome Analysis Project (4845-301) that was funded by a grant from the Korea National Institute of Health (Korea Center for Disease Control, Ministry for Health, Welfare and Family Affairs), Republic of Korea. Funding to pay the Open Access publication charges for this article was provided by Korea Research Foundation Grant (KRF-2008-220-E00025).
Conflict of Interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Feuk L., Carson A.R., Scherer S.W. Structural variation in the human genome. Nat. Rev. Genet. 2006;7:85–97. doi: 10.1038/nrg1767. [DOI] [PubMed] [Google Scholar]
- 2.Estivill X., Armengol L. Copy number variants and common disorders: filling the gaps and exploring complexity in genome-wide association studies. PLoS Genet. 2007;3:1787–1799. doi: 10.1371/journal.pgen.0030190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Iafrate A.J., Feuk L., Rivera M.N., Listewnik M.L., Donahoe P.K., Qi Y., Scherer S.W., Lee C. Detection of large-scale variation in the human genome. Nat. Genet. 2004;36:949–951. doi: 10.1038/ng1416. [DOI] [PubMed] [Google Scholar]
- 4.Sebat J., Lakshmi B., Troge J., Alexander J., Young J., Lundin P., Månér S., Massa H., Walker M., Chi M., et al. Large-scale copy number polymorphism in the human genome. Science. 2004;305:525–528. doi: 10.1126/science.1098918. [DOI] [PubMed] [Google Scholar]
- 5.Simon-Sanchez J., Scholz S., Fung H.C., Matarin M., Hernandez D., Gibbs J.R., Britton A., de Vrieze F.W., Peckham E., Gwinn-Hardy K., et al. Genome-wide SNP assay reveals structural genomic variation, extended homozygosity and cell-line induced alterations in normal individuals. Hum. Mol. Genet. 2007;16:1–14. doi: 10.1093/hmg/ddl436. [DOI] [PubMed] [Google Scholar]
- 6.de Smith A.J., Tsalenko A., Sampas N., Scheffer A., Yamada N.A., Tsang P., Ben-Dor A., Yakhini Z., Ellis R.J., Bruhn L., et al. Array CGH analysis of copy number variation identifies 1284 new genes variant in healthy white males: implications for association studies of complex diseases. Hum. Mol. Genet. 2007;16:2783–2794. doi: 10.1093/hmg/ddm208. [DOI] [PubMed] [Google Scholar]
- 7.Perry G.H., Ben-Dor A., Tsalenko A., Sampas N., Rodriguez-Revenga L., Tran C.W., Scheffer A., Steinfeld I., Tsang P., Yamada N.A., et al. The fine-scale and complex architecture of human copy-number variation. Am. J. Hum. Genet. 2008;82:685–695. doi: 10.1016/j.ajhg.2007.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Ståhl T.D., Sandgren J., Piotrowski A., Nord H., Andersson R., Menzel U., Bogdan A., Thuresson A.C., Poplawski A., von Tell D., et al. Profiling of copy number variations (CNVs) in healthy individuals from three ethnic groups using a human genome 32 K BAC-clone-based array. Hum. Mutat. 2008;29:398–408. doi: 10.1002/humu.20659. [DOI] [PubMed] [Google Scholar]
- 9.Redon R., Ishikawa S., Fitch K.R., Feuk L., Perry G.H., Andrews T.D., Fiegler H., Shapero M.H., Carson A.R., Chen W., et al. Global variation in copy number in the human genome. Nature. 2006;444:444–454. doi: 10.1038/nature05329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Feuk L., Marshall C.R., Wintle R.F., Scherer S.W. Structural variants: changing the landscape of chromosomes and design of disease studies. Hum. Mol. Genet. 2006;15:R57–R66. doi: 10.1093/hmg/ddl057. [DOI] [PubMed] [Google Scholar]
- 11.Beckmann J.S., Estivill X., Antonarakis S.E. Copy number variants and genetic traits: closer to the resolution of phenotypic to genotypic variability. Nat. Rev. Genet. 2007;8:639–646. doi: 10.1038/nrg2149. [DOI] [PubMed] [Google Scholar]
- 12.Stranger B.E., Forrest M.S., Dunning M., Ingle C.E., Beazley C., Thorne N., Redon R., Bird C.P., de Grassi A., Lee C., et al. Relative impact of nucleotide and copy number variation on gene expression phenotypes. Science. 2007;315:848–853. doi: 10.1126/science.1136678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hastings P.J., Lupski J.R., Rosenberg S.M., Ira G. Mechanisms of change in gene copy number. Nat. Rev. Genet. 2009;10:551–564. doi: 10.1038/nrg2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McCarroll S.A., Kuruvilla F.G., Korn J.M., Cawley S., Nemesh J., Wysoker A., Shapero M.H., de Bakker P.I., Maller J.B., Kirby A., et al. Integrated detection and population-genetic analysis of SNPs and copy number variation. Nat. Genet. 2008;40:1166–1174. doi: 10.1038/ng.238. [DOI] [PubMed] [Google Scholar]
- 15.Lin C.H., Lin Y.C., Wu J.Y., Pan W.H., Chen Y.T., Fann C.S. A genome-wide survey of copy number variations in Han Chinese residing in Taiwan. Genomics. 2009;94:241–246. doi: 10.1016/j.ygeno.2009.06.004. [DOI] [PubMed] [Google Scholar]
- 16.McCarroll S.A., Altshuler D.M. Copy-number variation and association studies of human disease. Nat. Genet. 2007;39:S37–S42. doi: 10.1038/ng2080. [DOI] [PubMed] [Google Scholar]
- 17.Conrad D.F., Andrews T.D., Carter N.P., Hurles M.E., Pritchard J.K. A high-resolution survey of deletion polymorphism in the human genome. Nat. Genet. 2006;38:75–81. doi: 10.1038/ng1697. [DOI] [PubMed] [Google Scholar]
- 18.Freeman J.L., Perry G.H., Feuk L., Redon R., McCarroll S.A., Altshuler D.M., Aburatani H., Jones K.W., Tyler-Smith C., Hurles M.E., et al. Copy number variation: new insights in genome diversity. Genome Res. 2006;16:949–961. doi: 10.1101/gr.3677206. [DOI] [PubMed] [Google Scholar]
- 19.Lander E.S., Linton L.M., Birren B., Nusbaum C., Zody M.C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., et al. International Human Genome Sequencing Consortium. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 20.Zhang F., Gu W., Hurles M.E., Lupski J.R. Copy number variation in human health, disease, and evolution. Annu. Rev. Genomics Hum. Genet. 2009;10:451–481. doi: 10.1146/annurev.genom.9.081307.164217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Conrad D.F., Pinto D., Redon R., Feuk L., Gokcumen O., Zhang Y., Aerts J., Andrews T.D., Barnes C., Campbell P., et al. Origins and functional impact of copy number variation in the human genome. Nature. 2009 doi: 10.1038/nature08516. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cooper G.M., Nickerson D.A., Eichler E.E. Mutational and selective effects on copy-number variants in the human genome. Nat. Genet. 2007;39:S22–S29. doi: 10.1038/ng2054. [DOI] [PubMed] [Google Scholar]
- 23.Wilson G.M., Flibotte S., Chopra V., Melnyk B.L., Honer W.G., Holt R.A. DNA copy-number analysis in bipolar disorder and schizophrenia reveals aberrations in genes involved in glutamate signaling. Hum. Mol. Genet. 2006;15:743–749. doi: 10.1093/hmg/ddi489. [DOI] [PubMed] [Google Scholar]
- 24.Kidd J.M., Cooper G.M., Donahue W.F., Hayden H.S., Sampas N., Graves T., Hansen N., Teague B., Alkan C., Antonacci F., et al. Mapping and sequencing of structural variation from eight human genomes. Nature. 2008;453:56–64. doi: 10.1038/nature06862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Carvalho C.M., Lupski J.R. Copy number variation at the breakpoint region of isochromosome 17q. Genome Res. 2008;18:1724–1732. doi: 10.1101/gr.080697.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Boissinot S., Entezam A., Furano A.V. Selection against deleterious LINE-1-containing loci in the human lineage. Mol. Biol. Evol. 2001;18:926–935. doi: 10.1093/oxfordjournals.molbev.a003893. [DOI] [PubMed] [Google Scholar]
- 27.Sheen F.M., Sherry S.T., Risch G.M., Robichaux M., Nasidze I., Stoneking M., Batzer M.A., Swergold G.D. Reading between the LINEs: human genomic variation induced by LINE-1 retrotransposition. Genome Res. 2000;10:1496–1508. doi: 10.1101/gr.149400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cho Y.S., Go M.J., Kim Y.J., Heo J.Y., Oh J.H., Ban H.J., Yoon D., Lee M.H., Kim D.J., Park M., et al. A large-scale genome-wide association study of Asian populations uncovers genetic factors influencing eight quantitative traits. Nat. Genet. 2009;41:527–534. doi: 10.1038/ng.357. [DOI] [PubMed] [Google Scholar]
- 29.Irizarry R.A., Hobbs B., Collin F., Beazer-Barclay Y.D., Antonellis K.J., Scherf U., Speed T.P. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 30.Price T.S., Regan R., Mott R., Hedman A., Honey B., Daniels R.J., Smith L., Greenfield A., Tiganescu A., Buckle V., et al. SW-ARRAY: a dynamic programming solution for the identification of copy-number changes in genomic DNA using array comparative genome hybridization data. Nucleic Acids Res. 2005;33:3455–3464. doi: 10.1093/nar/gki643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Komura D., Shen F., Ishikawa S., Fitch K.R., Chen W., Zhang J., Liu G., Ihara S., Nakamura H., Hurles M.E., et al. Genome-wide detection of human copy number variations using high-density DNA oligonucleotide arrays. Genome Res. 2006;16:1575–1584. doi: 10.1101/gr.5629106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 33.Kim T.M., Jung Y.C., Rhyu M.G., Jung M.H., Chung Y.J. GEAR: genomic enrichment analysis of regional DNA copy number changes. Bioinformatics. 2008;24:420–421. doi: 10.1093/bioinformatics/btm582. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.