Abstract
Mutations in the functionally uncharacterized protein SH3TC2 are associated with the severe hereditary peripheral neuropathy, Charcot–Marie–Tooth disease type 4C (CMT4C). Similarly, to other proteins mutated in CMT, a role for SH3TC2 in endocytic membrane traffic has been previously proposed. However, recent descriptions of the intracellular localization of SH3TC2 are conflicting. Furthermore, no clear functional pathogenic mechanisms have so far been proposed to explain why both nonsense and missense mutations in SH3TC2 lead to similar clinical phenotypes. Here, we describe our intracellular localization studies, supported by biochemical and functional data, using wild-type and mutant SH3TC2. We show that wild-type SH3TC2 targets to the intracellular recycling endosome by associating with the small GTPase, Rab11, which is known to regulate the recycling of internalized membrane and receptors back to the plasma membrane. Furthermore, we demonstrate that SH3TC2 interacts preferentially with the GTP-bound form of Rab11, identifying SH3TC2 as a novel Rab11 effector. Of clinical pathological relevance, all SH3TC2 constructs harbouring disease-causing mutations are shown to be unable to associate with Rab11 with consequent loss of recycling endosome localization. Moreover, we show that wild-type SH3TC2, but not mutant SH3TC2, influences transferrin receptor dynamics, consistent with a functional role on the endocytic recycling pathway. Our data therefore implicate mistargeting of SH3TC2 away from the recycling endosome as the fundamental molecular defect that leads to CMT4C.
INTRODUCTION
Charcot–Marie–Tooth disease (CMT) encompasses a large heterogeneous group of inherited progressive sensorimotor peripheral neuropathies. CMT has a prevalence of 1 in 2500 (1), making it the most common hereditary peripheral nerve disorder. Clinical, electrophysiological and pathological observations have led to CMT being categorized into ‘axonal’ and ‘demyelinating’ forms, reflecting the presumed site of pathology in the axon or Schwann cell, respectively. Recent studies have identified a large number of genes associated with CMT (2). Moreover, a significant number of these genes are thought to encode proteins known to play important roles in endocytic membrane traffic (3,4).
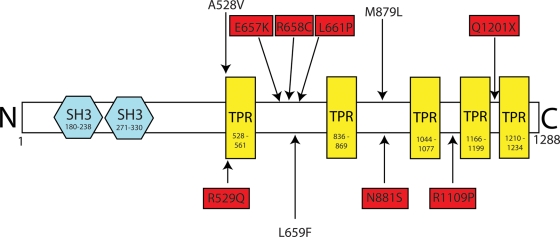
CMT4C is an autosomal recessive demyelinating form of CMT characterized by a severe early-onset sensorimotor neuropathy with scoliosis an apparent prominent clinical feature (5,6). Pathologically, CMT4C can be distinguished from other forms of CMT by the identification of specific Schwann cell membranous protrusions seen in electron micrographs of nerve biopsy specimens taken from affected patients (7). CMT4C was shown to be associated with the gene SH3TC2 (previously known as KIAA1985) found on chromosome 5 (8). SH3TC2 encodes a predicted 144 kDa protein with 2 N-terminal SH3 domains and 5 C-terminal TPR motifs, suggesting a role in forming protein–protein interactions (Fig. 1). Interestingly, CMT4C is associated with both nonsense and missense mutations found throughout the gene. Although recent reports have proposed conflicting intracellular localizations for SH3TC2 at the plasma membrane and in the endocytic pathway (9,10), no SH3TC2-interacting proteins have so far been described and the SH3TC2 protein has, until now, remained functionally uncharacterized at the molecular level. Most significantly, a consistent hypothesis to explain why both missense and nonsense mutations in SH3TC2 lead to clinical disease has hitherto not been proposed.
Figure 1.
Schematic diagram showing predicted domain organization of SH3TC2. The sites of CMT4C-associated pathogenic mutations used in this study are shown in red boxes. The locations of conservative mutations introduced adjacent to CMT4C-associated mutation sites are also shown (without red boxes). Amino acid residue numbers are shown within the predicted domains. Domains were predicted using SMART (simple modular architecture research tool - http://smart.embl-heidelberg.de).
Here we describe the recycling endosome as the precise intracellular compartment to which wild-type SH3TC2 targets. Furthermore, we show that a GTP-dependent association with the small GTPase, Rab11, mediates SH3TC2 localization. Conversely, we show that all CMT4C-associated mutations in SH3TC2 prevent Rab11 binding and lead to intracellular mistargeting away from the recycling endosome. We also describe the functional effects of pathogenic SH3TC2 mutations on transferrin receptor dynamics. These data allow us to put forward a consistent hypothesis to explain the dysfunctional cellular mechanisms that may lead to the development of CMT4C in affected patients.
RESULTS
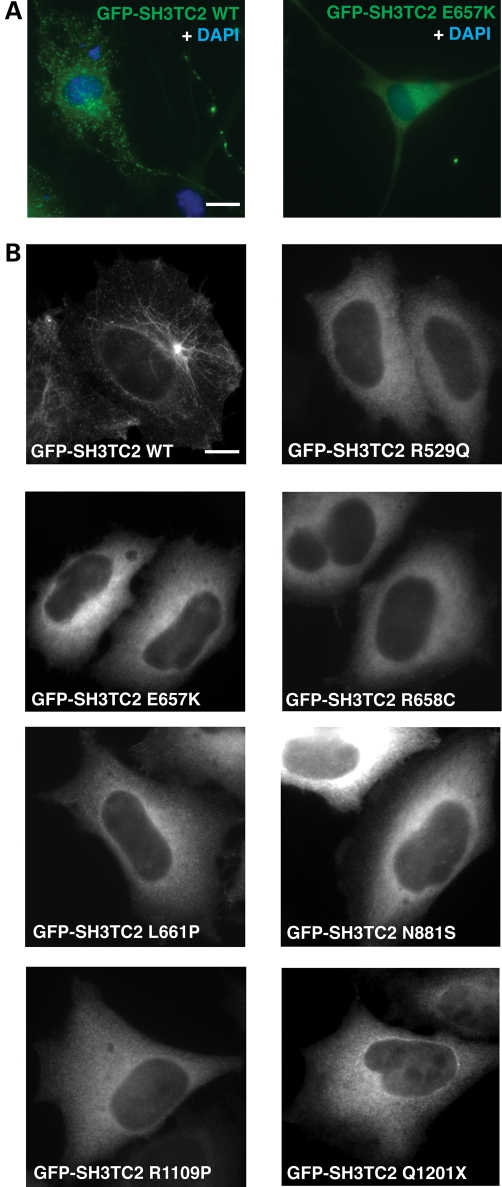
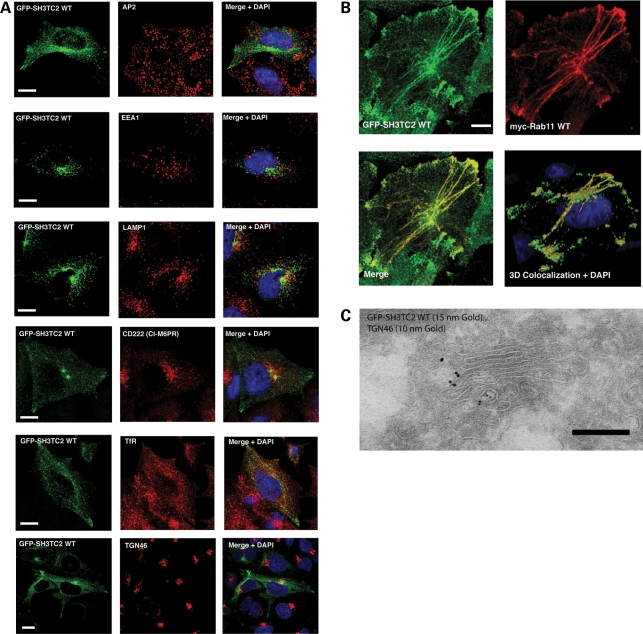
To see whether SH3TC2 localized to a specific intracellular compartment, we transiently expressed wild-type SH3TC2 tagged with GFP (GFP-SH3TC2 WT) in purified primary rat Schwann cells. GFP-SH3TC2 WT localized to intracellular tubulovesicular structures concentrated near the nucleus but distributed throughout the cell (Fig. 2A). On the contrary, GFP-SH3TC2 harbouring CMT4C-associated mutations failed to localize and remained cytosolic (Fig. 2A). Given these interesting findings, we turned to HeLa cells, in which the secretory and endocytic pathways are much better described and characterized. GFP-SH3TC2 WT transiently expressed in HeLa cells displayed a distinct tubulovesicular localization emanating from a perinuclear compartment and extending to the plasma membrane (Fig. 2B). Similarly to Schwann cells, we confirmed that all GFP-SH3TC2 constructs containing known CMT4C-associated pathogenic mutations failed to localize to any specific intracellular structure when also transiently expressed in HeLa cells, and instead displayed a non-specific cytosolic distribution (Fig. 2B). Using a battery of known endocytic marker proteins, we found that GFP-SH3TC2 WT showed partial colocalization with transferrin receptors (Fig. 3A) and a high degree of colocalization with the small GTPase Rab11 (Fig. 3B), indicating that SH3TC2 targets to the recycling endosome. In order to corroborate our fluorescent microscopic findings, we used immuno-electron microscopy of HeLa cells stably expressing GFP-SH3TC2 WT, which displayed an identical tubulovesicular intracellular localization by fluorescent microscopy to that seen in cells transiently expressing this construct. GFP-SH3TC2 WT (15 nm gold particle) was observed in tubulovesicular structures, some of which were close to the Golgi stack (Fig. 3C). However, these structures were always distinct from the trans Golgi network seen labelled with antibodies to TGN46 (10 nm gold particle), consistent with its localization at the recycling endosome.
Figure 2.
Intracellular localization of SH3TC2. (A) Rat Schwann cells transiently expressing GFP-SH3TC2 WT or GFP-SH3TC2 E657K visualized by widefield fluorescent microscopy. Scale bar denotes 10 µm. (B) HeLa cells transiently expressing GFP-SH3TC2 WT or GFP-SH3TC2 harbouring CMT4C-associated pathogenic mutations visualized by widefield fluorescent microscopy. Scale bar denotes 10 µm.
Figure 3.
(A) Colocalization immunofluorescence studies in HeLa cells by confocal microscopy. Endogenous marker proteins known to target distinct locations on the endocytic pathway were visualized to determine the intracellular localization of SH3TC2. The left-side panels show HeLa cells transiently expressing GFP-SH3TC2 WT (Green). The middle panels show the location of endogenous marker proteins in the same cells labelled with specific antibodies (Red). Merged images are shown on the right with nuclei stained with DAPI. The figure shows that GFP-SH3TC2 partially colocalized with transferrin receptors (TfR), but did not colocalize with AP-2 (clathrin coated vesicles), EEA1 (early endosome), LAMP1 (lyzosome), cation-independent mannose 6-phosphate receptor (CD222/M6PR, late endosome and Golgi) or TGN46 (trans Golgi network). Scale bars denote 10 µm. (B) Confocal microscopic images of a HeLa cell stably expressing GFP-SH3TC2 WT and transiently expressing myc-Rab11 WT, visualized by immunofluorescence. All images are confocal maximum-intensity z projections. Scale bar denotes 10 µm. The bottom right panel shows a three-dimensional render of the confocal z stacks to show areas of colocalization between myc-Rab11 and GFP-SH3TC2 WT using simulated fluorescence (Volocity 5.2, PerkinElmer). (C) Immuno-electron microscopy of HeLa cells stably expressing GFP-SH3TC2 WT. Scale bar denotes 200 nm.
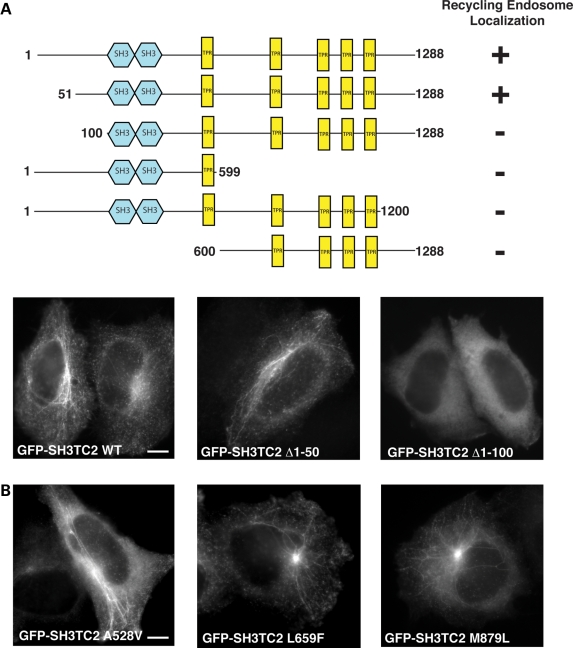
To identify the region of GFP-SH3TC2 WT that targets to the endocytic recycling compartment, we transiently expressed a number of truncated GFP-tagged constructs in HeLa cells. All the truncated constructs examined, apart from GFP-SH3TC2 missing the first 50 amino acid residues (GFP-SH3TC2 Δ1-50), failed to localize to any intracellular structures (Fig. 4A). In contrast, GFP-SH3TC2 constructs containing conservative amino acid substitutions adjacent to sites mutated in CMT4C localized to the recycling endosome (Fig. 4B); an important finding in view of the mistargeting seen with GFP-SH3TC2 constructs harbouring pathogenic point mutations.
Figure 4.
(A) Schematic showing the effects of truncations on the intracellular localization of SH3TC2. Lower panels show the intracellular localization of N-terminal SH3TC2 deletion mutants visualized by widefield fluorescent microscopy. GFP-SH3TC2 WT is shown for comparison. Scale bar denotes 10 µm. (B) Intracellular localization of GFP-SH3TC2 constructs containing conservative mutations adjacent to sites of known CMT4C-associated pathogenic amino acid substitutions. Scale bar denotes 10 µm.
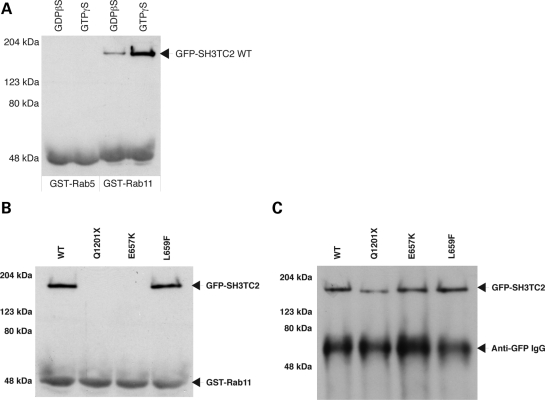
Rab proteins are a family of small GTPases known to regulate specific steps in the secretory and endocytic pathway (11). In particular, Rab11 regulates the recycling of internalized proteins and membrane back to the cell surface. As the colocalization of SH3TC2 with Rab11 was so striking, we investigated whether these proteins were able to form complexes. Figure 5A shows that GST-Rab11 was able to associate with GFP-SH3TC2 WT. Conversely, Rab5, a small GTPase known to function at the early endosome, was unable to associate with GFP-SH3TC2 WT. Rab proteins are activated when bound to GTP, and proteins that bind to the GTP-form of Rabs are known as effectors. A number of Rab11 effectors have been described previously (12). Therefore, we next investigated whether SH3TC2 also behaved as a Rab11 effector. Figure 5A shows that the amount of SH3TC2 isolated from cell cytosol was significantly increased when GST-Rab11 was pre-incubated with the non-hydrolyzable GTP analogue, GTPγS. Furthermore, GFP-SH3TC2 WT displayed a high degree of colocalization with myc-Rab11 Q70L in HeLa cells (Supplementary Material, Fig. S1). The Q70L mutation renders Rab11 constitutively active, mimicking the GTP-bound form of the small GTPase. In contrast, co-expression of the dominant-negative form of Rab11, myc-Rab11 S25N, leads to a non-specific cytosolic distribution of both myc-Rab11 S25N and GFP-SH3TC2 WT. These data strongly support the hypothesis that SH3TC2 is a novel Rab11 effector.
Figure 5.
(A) Western blot of pull-down experiments using cell lysates from HeLa cells stably expressing GFP-SH3TC2 WT. GFP-SH3TC2 WT was detected using affinity-purified rabbit anti-GFP antibody raised against recombinant GST-GFP. Note that the antiserum also detects GST with lower affinity and confirms equal loading of recombinant protein in each experiment. (B) Western blot of GST-Rab11 pull-down experiments using cell lysates from HeLa cells transiently expressing GFP-SH3TC2 constructs. GFP protein was detected using affinity-purified rabbit anti-GFP as in (A). (C) Immunoprecipitation of GFP-SH3TC2 constructs used in (B) to confirm full-length transient expression in HeLa cells.
Having confirmed that SH3TC2 associates with Rab11 in a GTP-dependent manner, we investigated whether CMT4C-associated pathogenic mutations could disrupt this interaction. Indeed, Figure 5B (and Supplementary Material, Fig. S2A) shows that Rab11 was able to associate with SH3TC2 WT but could not associate with SH3TC2 constructs harbouring CMT4C-associated pathogenic mutations. Immunoblotting clearly demonstrated that full-length proteins were expressed (Fig. 5C). Constructs containing the conservative mutation (L659F), adjacent to E657K, and SH3TC2 Δ1-50 remained able to associate with Rab11, consistent with their ability to localize to the recycling endosome.
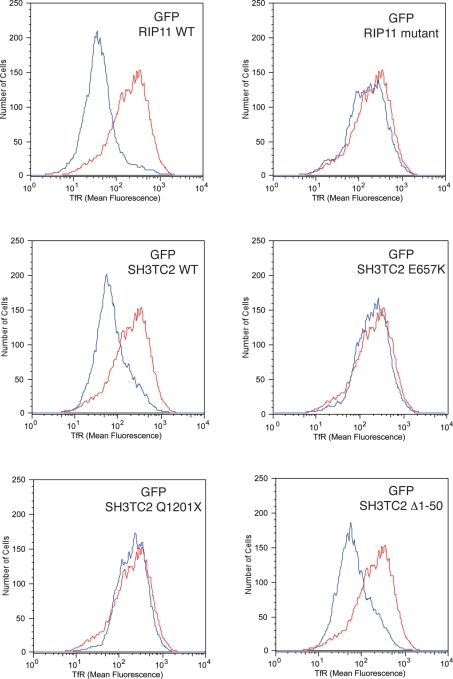
Finally, we investigated whether SH3TC2 regulated protein recycling. To test this, we used fluorescence activated cell sorting (FACS) analysis to measure surface TfR in cells transiently expressing wild-type and mutated constructs of GFP-SH3TC2. Overexpression of Rab11 binding domains of known Rab11-interacting proteins inhibit transferrin recycling by sequestering the small GTPase, leading to reduced concentration of TfR on the cell surface (13–15). In cells transfected with GFP-SH3TC2 WT, we also found decreased cell surface concentration of TfR (Fig. 6). In contrast, overexpression of GFP-SH3TC2 constructs containing CMT4C-associated pathogenic mutations had no effect on cell surface TfR concentration (Fig. 6 and Supplementary Material, Fig. S3). These data imply that disease-causing mutations in SH3TC2 render the resulting protein non-functional. Figure 6 also shows that GFP-SH3TC2 Δ1-50 behaved identically to wild-type SH3TC2. This finding, along with the ability of GFP-SH3TC2 Δ1-50 to localize to tubules and to associate with Rab11, is significant as protein prediction programs have suggested that the glycine residue at position 2 of the full-length protein may be myristoylated (9,10). Our data, therefore, strongly suggest that myristoylation is not required for SH3TC2's function at the recycling endosome. Behaving similarly to other known Rab11 effectors, we also found that depletion of endogenous full-length SH3TC2 by RNA inhibition increased surface TfR concentration, consistent with its proposed role in regulating endocytic protein recycling (Supplementary Material, Fig. S4).
Figure 6.
Cell surface TfR measured by FACS analysis. Cells transfected with GFP alone as control are represented by the red line, while the blue line represents cells transiently expressing GFP-RIP11 or GFP-SH3TC2 constructs as labelled. The histograms show that GFP-SH3TC2 WT and GFP-SH3TC2 Δ1-50 decreased the amount of surface TfR to a similar extent as cells expressing GFP-RIP11 WT. In contrast, GFP-SH3TC2 harbouring the CMT4C-associated pathogenic mutations E657K and Q1201X did not affect surface TfR concentration, analogous to overexpression of a mutated GFP-RIP11 construct (GFP-RIP11 I629E) that cannot bind to Rab11.
DISCUSSION
Defects in membrane traffic are thought to play key roles in many human diseases (16,17). By scrutinizing the genes known to be associated with demyelinating CMT, it becomes apparent that a significant number of these genes encode proteins known to play roles in the endocytic pathway (3,4). The characterization of these genes will not only improve our understanding of the pathogenesis of CMT, but will also lead to the identification of potential therapeutic targets aiming to stabilize or even reverse the peripheral nerve degeneration seen in CMT patients.
Before SH3TC2 was identified as the gene responsible for CMT4C, it was recognized that CMT4C patients displayed a specific Schwann cell pathology when nerve biopsy specimens were visualized by electron microscopy (7). It is tempting to speculate that the membranous extensions seen may reflect the consequence of a specific underlying defect in Schwann cell intracellular membrane transport. Many mutations in SH3TC2 have now been described in humans leading to CMT4C (www.molgen.ua.ac.be/CMTMutations). Interestingly, these mutations lead to premature truncations at the N- and C-termini and also amino acid substitutions found throughout the predicted SH3TC2 protein. In addition, we now know that the absence of Sh3tc2 in mice also leads to Schwann cell pathology and a neuropathy similar to that seen in human patients (10).
A recent report has suggested a generalized localization for SH3TC2 at the plasma membrane and endocytic pathway (9), including colocalization with cellular markers such as EEA1 (early endosomes), M6PR (late endosomes and trans Golgi network) and clathrin coated vesicles. Surprisingly, this report by Lupo et al. (9) also states that a GFP-tagged SH3TC2 construct, which appears to be the same as we describe in our manuscript, localizes to the endoplasmic reticulum, although unfortunately no data to support this claim are included. Moreover, no definitive biochemical supporting evidence is offered to support their immunofluorescence studies and no functional data are presented. However, in agreement with the data that we present here, Arnaud et al. (10) have very recently suggested that Sh3tc2 targets to the endocytic recycling compartment.
Our work goes further than previous reports in characterizing the wild-type SH3TC2 protein by showing definitive targeting of SH3TC2 to the recycling endosome, by identifying SH3TC2 as a novel Rab11 effector and by confirming functional activity on the endocytic recycling pathway. In our hands, we found that targeting of SH3TC2 to the recycling endosome occurred whether the GFP tag was located at either the N- or C-terminus (data not shown). Furthermore, we did not observe SH3TC2 at the early endosome, the late endosome, trans Golgi network, nor associated with clathrin-coated vesicles. We also never observed SH3TC2 localized at the endoplasmic reticulum. We acknowledge that SH3TC2 may undergo myristoylation on glycine 2, which may partly explain the discrepancy in localization seen between the different reports, and this modification is also likely to account for the partial plasma membrane localization seen when SH3TC2 is tagged at the C-terminus (data not shown; 9,10). However, we provide evidence by means of localization studies, Rab11-association and TfR dynamics, that removal of the first 50 amino acids containing the predicted myristoylation site had no apparent effect on SH3TC2 function at the recycling endosome.
Most significantly, especially from a clinical perspective, we have shown that all SH3TC2 constructs harbouring missense and nonsense CMT4C-associated pathogenic mutations fail to localize to the recycling endosome, fail to associate with Rab11 and have no effects on TfR dynamics when overexpressed. Lupo et al. did attempt to characterize the effects of a small number of truncations and pathogenic point mutations on SH3TC2 localization, but their results were inconclusive and therefore no unifying hypothesis was proposed to explain how all mutations in SH3TC2 lead to CMT4C.
The universal effects of pathogenic CMT4C-associated mutations that we describe offer novel insights into the pathology of CMT4C. The Sh3tc2 knock-out mouse model of CMT4C described recently confirms that the absence of Sh3tc2, as found in a proportion of CMT4C patients, leads to Schwann cell dysfunction and defects in myelination (10). However, Arnaud et al.'s (10) work does not explain the reasons why all pathogenic amino acid substitutions found throughout the same protein lead to the same clinical phenotype. With mutations spread throughout the SH3TC2 protein (Fig. 1), and no clearly defined intracellular targeting domain observed (Fig. 4), it is likely that the SH3TC2 protein is a highly complex molecule with a possible propensity to becoming unfolded as a result of mutations at key sites leading to the loss of specific interacting and/or stabilizing regions. However, we saw no degradation of the SH3TC2 protein harbouring CMT4C-associated mutations when expressed in cells. Therefore, an alternative explanation would be that the mutated residues and truncated regions are all found on the surface of the protein, forming a Rab11-interacting surface. The resulting mutated protein would then be unable to associate with Rab11, would not target to the recycling endosome and thus would also be unable to influence endocytic protein recycling. Although SH3TC2 does not appear to contain a known Rab11 binding motif, such as the highly conserved Rab11/25 binding domain (RBD) found in Class I and Class II Rab11-family interacting proteins (FIPs) (18), the presence of such a domain is not an absolute prerequisite to function as a Rab11 effector since it is also not seen in the known Rab11-interacting proteins, Sec15, Evi5 and D-AKAP2 (19–21).
In summary, we present data showing that wild-type SH3TC2 targets to the recycling endosome, binds to Rab11 in a GTP-dependent manner and is functionally active as shown by its effects on TfR dynamics. In complete contrast, SH3TC2 mutants containing CMT4C-associated pathogenic mutations did not associate with Rab11 and therefore failed to localize intracellularly and did not affect TfR dynamics when overexpressed. While the transferrin receptor is the most widely studied reporter molecule for assessing endocytic recycling within cultured cells, it is probable that SH3TC2 is involved in regulating the recycling of other cargos. We now know that there are a large number of receptors that require endocytic recycling by clathrin-dependent and also clathrin-independent mechanisms (12). As CMT4C leads to progressive demyelination of peripheral nerves, we propose that SH3TC2 may regulate the recycling of specific, hitherto unidentified, membrane receptors in Schwann cells. Since the endocytic recycling of these proteins is likely to be critical for the maintenance of myelination, the next step will be to identify these receptors to improve our understanding of Schwann cell membrane traffic both in health and disease.
We conclude that mistargeting of the novel Rab11 effector, SH3TC2, away from the endocytic recycling compartment is likely to be ultimately responsible for the development of CMT4C.
MATERIALS AND METHODS
Cell culture and reagents
Primary rat Schwann cells were prepared from postnatal day 5 (P5) rats as previously described (22).
HeLaM cells were used throughout this work. HeLa cells were grown at 37°C in RPMI (Sigma-Aldrich) containing 10% FCS and 2 mm l-glutamine in a 5% CO2 humidified atmosphere. Cells stably expressing GFP-SH3TC2 WT were generated by transfecting HeLa cells with pIRESneo2 GFP-SH3TC2 WT and applying continuous selection with 500 µg/ml G418 (Gibco). Transfected cells were sorted according to GFP expression levels using FACS.
For transfection of rat Schwann cells, we used FuGene6 (Roche). FuGene6 and Lipofectamine2000 (Invitrogen) were used to transfect HeLa cells.
Antibodies used for immunofluorescence include Rabbit anti-GFP (Invitrogen); Mouse anti-GFP (Invitrogen); Mouse anti-myc tag (Millipore); Mouse anti-AP2 (AP6, Affinity Bioreagents); Mouse anti-EEA1 (BD Biosciences); Mouse anti-LAMP1 (H4A3, Developmental Studies, Hybridoma Bank, University of Iowa); Mouse anti-human CD222 (Cation-independent M6PR) (Serotec); Mouse anti-TfR (Zymed); Sheep anti-human TGN46 (Serotec); Alexa Fluor 488- and Alexa Fluor 568-conjugated Goat anti-rabbit, anti-mouse and anti-sheep secondary antibodies (Invitrogen). Affinity-purified polyclonal anti-GFP antibodies were produced by inoculating rabbits with purified recombinant GST-GFP or His-tagged GFP (for western blotting and immunoprecipitation, respectively). HRP-conjugated goat anti-rabbit and goat anti-mouse antibodies were used as secondary antibodies for western blotting (Sigma-Aldrich).
Plasmids
Full-length SH3TC2 cDNA was obtained from the IMAGE consortium (clone ID 40081845) and full-length Rab11 was obtained from the cDNA resource centre, University of Missouri-Rolla. SH3TC2 and Rab11 constructs were amplified by PCR with oligonucleotides containing restriction enzyme sites, and the purified DNA was ligated into the appropriate plasmid. Plasmids used in this study included pEGFP-C1 and pEGFP-C2 (Clontech), pCMV-myc (Clontech), pIRESneo2 (Clontech) and pGEX-4T1 (GE Healthcare). pGEX-KG Rab11a WT was provided by Dr Rytis Prekeris (University of Colorado). GFP constructs of RIP11 WT and RIP11 I629E have been reported previously (14). Site-directed mutagenesis was performed using a QuikChange II XL kit (Stratagene). All DNA constructs were sequenced and validated by Geneservice (Cambridge, UK).
Immunofluorescence
For immunofluorescence, cells were fixed with 4% (w/v) paraformaldehyde at 37°C for 20 min. The cells were then permeabilized with 0.2% Triton X-100 for 5 min before quenching in 10 mm Glycine for 15 min. Fixed cells were blocked in 1% BSA in PBS before indirect immunofluorescence using primary antibodies (indicated in the Figure legends) followed by antibodies coupled to Alexa Fluor 488 or Alexa Fluor 568 (Molecular Probes). Widefield fluorescent images were obtained using a Zeiss Axioplan Fluorescent Microscope and confocal images were obtained using a Zeiss LSM510 META confocal microscope (Carl Zeiss). Data obtained from the confocal microscope were analysed using Volocity 5.2 (PerkinElmer).
Electron microscopy
Protein A conjugated to 10 and 15 nm colloidal gold was purchased from the Department of Cell Biology, University of Utrecht. Rabbit anti-TGN46 was a gift from Dr Vas Ponnambalam (University of Leeds, UK). Rabbit anti-GFP (ab6556) was purchased from Abcam (Cambridge, UK). HeLa cells stably expressing GFP-tagged SH3TC2 WT were washed with PBS, fixed with 4% paraformaldehyde/0.1% glutaraldehyde in 0.1 M sodium cacodylate buffer (pH 7.2) and pelleted in an Eppendorf tube (16 000g for 5 min). The fixative was aspirated and the cell pellet was re-suspended in warm 10% gelatin in PBS. The cells were then pelleted (16 000g for 5 min) and the gelatin-enrobed cells were set on ice, trimmed into 1 mm3 blocks and infused with 1.7 M sucrose/15% poly vinyl pyrolidone for 24 h at 4°C. The blocks were subsequently mounted on cryostubs and snap-frozen in liquid nitrogen. Frozen ultrathin sections were cut using a diamond knife in an ultramicrotome with a cryochamber attachment (Leica, Milton Keynes, UK) at −120°C, collected from the knife-edge with 50:50 2% methyl cellulose: 2.3 M sucrose and mounted on formvar-carbon coated EM grids.
Sequential immunolabelling of TGN46 and GFP was performed using the protein A-gold technique at room temperature. The sections were contrasted by embedding in 1.8% methyl cellulose/0.3% uranyl acetate and air-dried prior to observation in a Philips CM100 transmission electron microscope at an operating voltage of 80 kV (23,24).
GST pull-down assays
Purified GST-Rab11a or GST-Rab5 were coupled to Glutathione Sepharose 4B beads (GE Healthcare) and washed twice with 20 mm Tris (pH 7.5), 150 mm NaCl, 0.5 mm EDTA. The GST fusion proteins attached to beads were then incubated with 20 mm Tris (pH 7.5), 150 mm NaCl, 0.5 mm EDTA, 1 mg/ml BSA containing either 0.2 mm GDPβS or GTPγS (Sigma-Aldrich) for 15 min at room temperature. MgCl2 was then added to a final concentration of 30 mm before mixing vigorously and washing twice with lysis buffer [20 mm HEPES (pH 7.5), 120 mm NaCl, 10 mm Na4P2O7, 20 mm NaF, 1% Ipegal (v/v) and Complete protease inhibitor cocktail (Roche)]. Cell lysates were prepared by scraping HeLa cells into a small volume of lysis buffer and transferring into a 1.5 ml microcentrifuge tube. The lysate was then sheared by gently passing the solution back and forth through a 21 G needle followed by a 25 G needle, before centrifugation at 16 000g for 30 min at 4°C. GDPβS or GTPγS was then added to the lysis supernatant before incubation with the purified GST fusion proteins for 2 h at 4°C with gentle agitation. Following incubation for 2 h, the beads were extensively washed with lysis buffer followed by PBS before boiling in SDS Sample Buffer [6 M Urea, 1% SDS (w/v), 1 M 2-mercaptoethanol and 150 mm Tris (pH 6.7)]. Samples were separated by 7.5% SDS–PAGE. Proteins were then transferred to a nitrocellulose membrane for western blotting. HRP-conjugated secondary antibodies were used following incubation with primary antibodies and bands were detected using ECL (GE Healthcare).
Immunoprecipitation
Cell lysates were prepared as above. The lysates were incubated with protein A sepharose CL 4B (GE Healthcare) for 1 h at 4°C. The unbound supernatant was transferred to a new microcentrifuge tube and incubated with affinity-purified rabbit anti-GFP antibodies for 2 h at 4°C. Fresh protein A Sepharose CL 4B was then added and allowed to mix for 1 h at 4°C. The beads were washed extensively with lysis buffer followed by PBS before boiling in SDS sample buffer. Proteins were separated by 7.5% SDS–PAGE and transferred to nitrocellulose in preparation for western blotting.
FACS-based surface TfR assays
FACS-based surface TfR assays were carried out as previously described (14). Briefly, HeLa cells were transfected with either GFP alone, GFP-SH3TC2 WT or GFP-SH3TC2 harbouring CMT4C-associated pathogenic mutations. GFP-RIP11-F1 and GFP-RIP11-F1(I629E) were used as positive and negative controls, respectively (14). The transfected cells were trypsinized, washed with ice-cold opti-MEM (Gibco) and incubated with APC-conjugated mouse anti-CD71 (TfR) (BD Biosciences) for 20 min on ice. The cells were washed again twice with ice-cold opti-MEM before analysis using a BD FACS Calibur flow cytometer (BD Biosciences). 7-aminoactinomycin D (final concentration 0.3 mg/ml, Invitrogen) was used to exclude non-viable cells from further analysis. FACS data were analysed using FloJo software (Tree Star, Inc). A total of 10 000 transfected cells were analysed in each experimental run.
SUPPLEMENTARY MATERIAL
FUNDING
This work was supported by a Next Generation Fellowship to R.C.R. as part of the Wellcome Trust Strategic Award to the Cambridge Institute for Medical Research (No. 079895) and an MRC Programme grant to J.P.L. A.A.P. and J.K.-J. are funded by the MRC. F.B. is funded by the Wellcome Trust.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Professor Rhona Mirsky and Professor Kristjan Jessen (University College London) for their help with preparing purified Schwann cells and Dr Rytis Prekeris (University of Colorado) for kindly supplying GST-Rab11 constructs. We acknowledge Matthew Gratian and Mark Bowen (Cambridge Institute for Medical Research) for technical assistance with microscopy and image analysis.
Conflict of Interest statement. The authors declare no conflicts of interest.
REFERENCES
- 1.Martyn C.N., Hughes R.A. Epidemiology of peripheral neuropathy. J. Neurol. Neurosurg. Psychiatry. 1997;62:310–318. doi: 10.1136/jnnp.62.4.310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Reilly M.M. Sorting out the inherited neuropathies. Pract. Neurol. 2007;7:93–105. [PubMed] [Google Scholar]
- 3.Niemann A., Berger P., Suter U. Pathomechanisms of mutant proteins in Charcot–Marie–Tooth disease. Neuromolecular Med. 2006;8:217–242. doi: 10.1385/nmm:8:1-2:217. [DOI] [PubMed] [Google Scholar]
- 4.Scherer S.S., Wrabetz L. Molecular mechanisms of inherited demyelinating neuropathies. Glia. 2008;56:1578–1589. doi: 10.1002/glia.20751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Azzedine H., Ravise N., Verny C., Gabreels-Festen A., Lammens M., Grid D., Vallat J.M., Durosier G., Senderek J., Nouioua S., et al. Spine deformities in Charcot–Marie–Tooth 4C caused by SH3TC2 gene mutations. Neurology. 2006;67:602–606. doi: 10.1212/01.wnl.0000230225.19797.93. [DOI] [PubMed] [Google Scholar]
- 6.Houlden H., Laura M., Ginsberg L., Jungbluth H., Robb S.A., Blake J., Robinson S., King R.H., Reilly M.M. The phenotype of Charcot–Marie–Tooth disease type 4C due to SH3TC2 mutations and possible predisposition to an inflammatory neuropathy. Neuromuscul. Disord. 2009;19:264–269. doi: 10.1016/j.nmd.2009.01.006. [DOI] [PubMed] [Google Scholar]
- 7.Gabreels-Festen A., van Beersum S., Eshuis L., LeGuern E., Gabreels F., van Engelen B., Mariman E. Study on the gene and phenotypic characterisation of autosomal recessive demyelinating motor and sensory neuropathy (Charcot–Marie–Tooth disease) with a gene locus on chromosome 5q23-q33. J. Neurol. Neurosurg. Psychiatry. 1999;66:569–574. doi: 10.1136/jnnp.66.5.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Senderek J., Bergmann C., Stendel C., Kirfel J., Verpoorten N., De Jonghe P., Timmerman V., Chrast R., Verheijen M.H., Lemke G., et al. Mutations in a gene encoding a novel SH3/TPR domain protein cause autosomal recessive Charcot–Marie–Tooth type 4C neuropathy. Am. J. Hum. Genet. 2003;73:1106–1119. doi: 10.1086/379525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lupo V., Galindo M.I., Martinez-Rubio D., Sevilla T., Vilchez J.J., Palau F., Espinos C. Missense mutations in the Sh3tc2 protein causing Charcot–Marie–Tooth Disease Type 4c affect its localization in the plasma membrane and endocytic pathway. Hum. Mol. Genet. 2009;18:4603–4614. doi: 10.1093/hmg/ddp427. [DOI] [PubMed] [Google Scholar]
- 10.Arnaud E., Zenker J., de Preux Charles A.S., Stendel C., Roos A., Medard J.J., Tricaud N., Weis J., Suter U., Senderek J., et al. SH3TC2/KIAA1985 protein is required for proper myelination and the integrity of the node of Ranvier in the peripheral nervous system. Proc. Natl Acad. Sci. USA. 2009;106:17528–17533. doi: 10.1073/pnas.0905523106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stenmark H. Rab GTPases as coordinators of vesicle traffic. Nat. Rev. Mol. Cell. Biol. 2009;10:513–525. doi: 10.1038/nrm2728. [DOI] [PubMed] [Google Scholar]
- 12.Grant B.D., Donaldson J.G. Pathways and mechanisms of endocytic recycling. Nat. Rev. Mol. Cell. Biol. 2009;10:597–608. doi: 10.1038/nrm2755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tarbutton E., Peden A.A., Junutula J.R., Prekeris R. Class I FIPs, Rab11-binding proteins that regulate endocytic sorting and recycling. Methods Enzymol. 2005;403:512–525. doi: 10.1016/S0076-6879(05)03045-4. [DOI] [PubMed] [Google Scholar]
- 14.Junutula J.R., Schonteich E., Wilson G.M., Peden A.A., Scheller R.H., Prekeris R. Molecular characterization of Rab11 interactions with members of the family of Rab11-interacting proteins. J. Biol. Chem. 2004;279:33430–33437. doi: 10.1074/jbc.M404633200. [DOI] [PubMed] [Google Scholar]
- 15.Peden A.A., Schonteich E., Chun J., Junutula J.R., Scheller R.H., Prekeris R. The RCP-Rab11 complex regulates endocytic protein sorting. Mol. Biol. Cell. 2004;15:3530–3541. doi: 10.1091/mbc.E03-12-0918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Howell G.J., Holloway Z.G., Cobbold C., Monaco A.P., Ponnambalam S. Cell biology of membrane trafficking in human disease. Int. Rev. Cytol. 2006;252:1–69. doi: 10.1016/S0074-7696(06)52005-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Olkkonen V.M., Ikonen E. Genetic defects of intracellular-membrane transport. N. Engl. J. Med. 2000;343:1095–1104. doi: 10.1056/NEJM200010123431507. [DOI] [PubMed] [Google Scholar]
- 18.Prekeris R., Davies J.M., Scheller R.H. Identification of a novel Rab11/25 binding domain present in Eferin and Rip proteins. J. Biol. Chem. 2001;276:38966–38970. doi: 10.1074/jbc.M106133200. [DOI] [PubMed] [Google Scholar]
- 19.Wu S., Mehta S.Q., Pichaud F., Bellen H.J., Quiocho F.A. Sec15 interacts with Rab11 via a novel domain and affects Rab11 localization in vivo. Nat. Struct. Mol. Biol. 2005;12:879–885. doi: 10.1038/nsmb987. [DOI] [PubMed] [Google Scholar]
- 20.Westlake C.J., Junutula J.R., Simon G.C., Pilli M., Prekeris R., Scheller R.H., Jackson P.K., Eldridge A.G. Identification of Rab11 as a small GTPase binding protein for the Evi5 oncogene. Proc. Natl Acad. Sci. USA. 2007;104:1236–1241. doi: 10.1073/pnas.0610500104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eggers C.T., Schafer J.C., Goldenring J.R., Taylor S.S. D-AKAP2 interacts with Rab4 and Rab11 through its RGS domains and regulates transferrin receptor recycling. J. Biol. Chem. 2009;284:32869–32880. doi: 10.1074/jbc.M109.022582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Morgan L., Jessen K.R., Mirsky R. The effects of cAMP on differentiation of cultured Schwann cells: progression from an early phenotype (04+) to a myelin phenotype (P0+, GFAP-, N-CAM-, NGF-receptor-) depends on growth inhibition. J. Cell. Biol. 1991;112:457–467. doi: 10.1083/jcb.112.3.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liou W., Geuze H.J., Slot J.W. Improving structural integrity of cryosections for immunogold labeling. Histochem. Cell Biol. 1996;106:41–58. doi: 10.1007/BF02473201. [DOI] [PubMed] [Google Scholar]
- 24.Slot J.W., Geuze H.J., Gigengack S., Lienhard G.E., James D.E. Immuno-localization of the insulin regulatable glucose transporter in brown adipose tissue of the rat. J. Cell. Biol. 1991;113:123–135. doi: 10.1083/jcb.113.1.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.