Abstract
The nuclear poly(A)-binding protein 1 (PABPN1) is a ubiquitously expressed protein that plays a critical role in polyadenylation. Short expansions of the polyalanine tract in the N-terminus of PABPN1 lead to oculopharyngeal muscular dystrophy (OPMD), which is an adult onset disease characterized by eyelid drooping, difficulty in swallowing and weakness in the proximal limb muscles. Although significant data from in vitro biochemical assays define the function of PABPN1 in control of poly(A) tail length, little is known about the role of PABPN1 in mammalian cells. To assess the function of PABPN1 in mammalian cells and specifically in cells affected in OPMD, we examined the effects of PABPN1 depletion using siRNA in primary mouse myoblasts from extraocular, pharyngeal and limb muscles. PABPN1 knockdown significantly decreased cell proliferation and myoblast differentiation during myogenesis in vitro. At the molecular level, PABPN1 depletion in myoblasts led to a shortening of mRNA poly(A) tails, demonstrating the cellular function of PABPN1 in polyadenylation control in a mammalian cell. In addition, PABPN1 depletion caused nuclear accumulation of poly(A) RNA, revealing that PABPN1 is required for proper poly(A) RNA export from the nucleus. Together, these experiments demonstrate that PABPN1 plays an essential role in myoblast proliferation and differentiation, suggesting that it is required for muscle regeneration and maintenance in vivo.
INTRODUCTION
The nuclear poly(A)-binding protein 1 (PABPN1) is a ubiquitous nuclear protein that binds with high affinity to polyadenosine RNA and plays an important role in mRNA polyadenylation (1). PABPN1 contains an RNA recognition motif (RRM) in its central region that mediates RNA binding (2) and a C-terminal arginine-rich domain, which mediates oligomerization (2–4) and contains a nuclear localization sequence (5). The function of PABPN1 in polyadenylation has been extensively characterized using in vitro biochemical approaches (1). The protein is part of the polyadenylation complex through direct interaction with cleavage and polyadenylation specificity factor (CPSF) and poly(A) polymerase (1). This interaction stimulates a rapid, processive polyadenylation reaction, and the control of poly(A) tail length is due to the termination of this processive elongation. Consistent with 250 nt being the average poly(A) length observed in mammals, extension of mRNA poly(A) tails beyond 250 adenine residues is largely distributive and therefore slow (1). PABPN1 is predominantly nuclear at steady state and is mostly concentrated in nuclear speckles (6). However, it shuttles between the nucleus and cytoplasm, with export to the cytoplasm via a facilitated transport pathway that is independent of RNA synthesis (5).
Oculopharyngeal muscular dystrophy (OPMD) is characterized by a trinucleotide repeat expansion in the coding sequence of the PABPN1 gene (7). The N terminus of PABPN1 contains a stretch of 10 alanines encoded by a (GCG)6 repeat, which is expanded to 12–17 alanines in the mutant protein. OPMD is a late onset, autosomal dominant disease characterized primarily by progressive eyelid drooping (ptosis) and difficulties in swallowing (8). Additional weakness is noted in proximal limb, facial and other extraocular muscles (EOM) (9,10). Disease progression is variable and complications include choking, regurgitation, aspiration and pneumonia. The pathological hallmark of the disease is the presence of nuclear aggregates of PABPN1 in muscle (11,12). However, why expansion of the N-terminal polyalanine tract in PABPN1 causes OPMD is unknown.
Although the function of PABPN1 in polyadenylation has been precisely defined using in vitro biochemical approaches, limited information exists about the role of PABPN1 in mammalian cells. The in vivo function of PABPN1 has been analyzed in Drosophila and in fission yeast (Pab2), where PABPN1 proved to be essential for viability and required for proper polyadenylation (13,14). Furthermore, PABPN1 is recruited to nuclear speckles in HeLa cells, and this recruitment is dependent on poly(A) RNA binding (6). A study of the influenza virus NS1 protein provides indirect evidence that PABPN1 is important for polyadenylation in HEK 293 cells (15). This study revealed that NS1 forms a ternary complex with PABPN1 and CPSF, which inhibits PABPN1-stimulated poly(A) addition in an in vitro biochemical assay (15). Furthermore, influenza virus-infected HEK 293 cells showed accumulation of pre-RNAs with 12 adenosine tails, indicative of a defect in PABPN1-dependent elongation (15). However, the effect of directly depleting PABPN1 from mammalian cells has not been examined, much less for skeletal muscle, which is the tissue specifically affected in OPMD.
Skeletal muscle may be particularly dependent on PABPN1 function, as it is one of the few tissues that display extensive regenerative capacity. Regeneration is due to the ability of myogenic stem cells called satellite cells to undergo myogenesis (16). Satellite cells are quiescent muscle precursor cells that lie underneath the basal lamina that surrounds each myofiber. In response to injury, satellite cells begin to proliferate and give rise to progeny called myoblasts. During muscle regeneration, myoblasts differentiate and fuse to form new myofibers, restoring normal tissue structure (17). Alterations of PABPN1 function could lead to defects in myoblast proliferation, differentiation and/or fusion, resulting in impaired regeneration.
To begin to understand the function of PABPN1 in muscle, we examined the effects of PABPN1 depletion in primary mouse myoblasts. Our results indicate that PABPN1 is important for normal myoblast proliferation as well as differentiation. At the molecular level, depletion of PABPN1 in myoblasts leads to a shortening of mRNA poly(A) tails and nuclear accumulation of poly(A) RNA. Although the involvement of PABPN1 in polyadenylation is expected, considering the large amount of data from in vitro biochemical assays (1), we demonstrate that loss of PABPN1 causes defects in mRNA polyadenylation in mammalian cells. Furthermore, we provide experimental evidence that PABPN1 is required for efficient poly(A) RNA export from the nucleus. These experiments reveal that PABPN1 plays an important role during myoblast proliferation and differentiation, and consequently probably also in muscle regeneration and maintenance.
RESULTS
PABPN1 expression is not altered during in vitro myogenesis
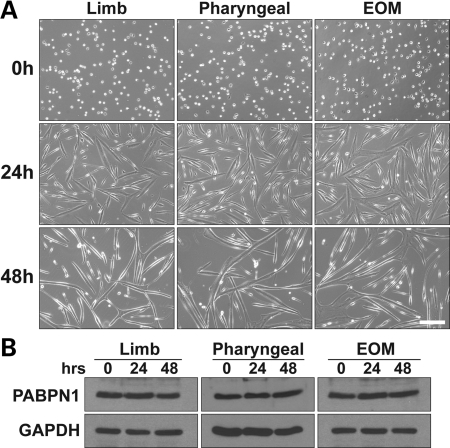
During skeletal muscle myogenesis, myoblasts undergo a phase of rapid proliferation, followed by cell-cycle exit, differentiation and finally cell fusion (16). These events lead to the formation of multinucleated myofibers in vivo and myotubes in vitro. To begin to assess the function of PABPN1 in skeletal muscle and to determine whether a specific stage of myogenesis is particularly dependent on PABPN1, we analyzed the steady-state levels of PABPN1 during in vitro myogenesis. Because EOM and pharyngeal muscles, and to a lesser extent proximal limb muscles, are the most affected muscles in OPMD (8–10), primary mouse myoblasts from EOM, pharyngeal and limb muscles were isolated and expanded in cell culture. Myoblasts were allowed to differentiate for 0, 24 and 48 h and PABPN1 levels were determined by immunoblotting using a polyclonal antibody. As shown in Figure 1A, no obvious differences were observed in differentiation and fusion of myoblasts from different muscle origins. In addition, PABPN1 levels were very similar among the three types of muscle cells and the levels of PABPN1 did not change during myogenesis (Fig. 1B), suggesting that PABPN1 is critical throughout myogenesis rather than functioning only at a specific stage.
Figure 1.
PABPN1 expression is not altered during in vitro myogenesis. Pure cultures of primary mouse myoblasts from limb, pharyngeal and extraocular (EOM) muscles were differentiated for 0, 24 and 48 h. (A) Representative phase-contrast images of muscle cells after 0, 24 and 48 h of differentiation are shown (bar, 100 µm). (B) Protein extracts were analyzed by immunoblotting for PABPN1 expression. GAPDH was used as a loading control. Similar results were obtained from at least three independent assays.
PABPN1 is required for normal myoblast proliferation and differentiation
To determine the role of PABPN1 in skeletal muscle cells, we assessed myoblast proliferation and differentiation in cells in which PABPN1 expression was knocked down. To determine what effect loss of PABPN1 has on cell proliferation, we transfected primary myoblasts isolated from limb, pharyngeal and EOM muscles or 3T3 fibroblasts with one of two siRNA oligonucleotides for PABPN1 or a scrambled control siRNA, labeled cells with bromodeoxyuridine (BrdU) and subsequently immunostained for BrdU. Significant knockdown of PABPN1 was achieved with each PABPN1 siRNA oligonucleotide (Fig. 2A). Both siRNAs target sequences outside the conserved RRM region on the PABPN1 transcript, which minimizes cross-targeting of other RNA-binding proteins. Immunoblotting for PABPC, the cytoplasmic poly(A)-binding protein with a related RRM domain (1), revealed no changes in the levels of this protein after PABPN1 knockdown (data not shown). Representative images of limb myoblasts immunostained for BrdU after siRNA transfection are presented in Figure 2B. A 40–50% decrease occurred in the percentage of BrdU+ cells in all myoblast types, indicating that PABPN1 is required for normal myoblast proliferation (Fig. 2C). Similar results were obtained with PABPN1 knockdown in fibroblasts, suggesting that the function of PABPN1 in regulating proliferation may be conserved in different cell types.
Figure 2.
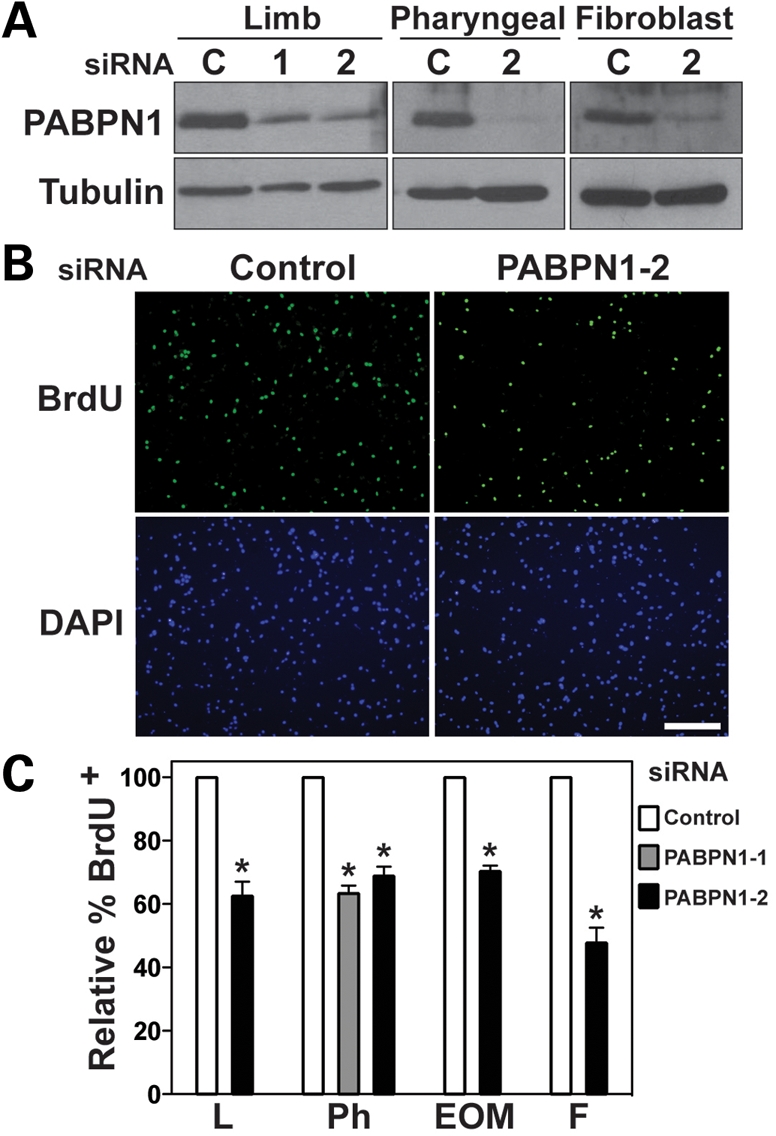
PABPN1 is required for normal myoblast proliferation. Pure cultures of primary mouse myoblasts from limb, pharyngeal and extraocular (EOM) muscles were transfected with one of two siRNA oligonucleotides for PABPN1 [PABPN1-1 (1) or PABPN1-2 (2)] or a scrambled control (C). Cells were labeled with BrdU and immunostained with anti-BrdU antibody. (A) Protein extracts were analyzed by immunoblotting for PABPN1 to confirm knockdown. Alpha-tubulin was used as a loading control. (B) Representative BrdU immunostaining after siRNA transfection (bar, 100 µm). DAPI staining was used for nuclear visualization. (C) The relative percentage of BrdU+ nuclei was determined in limb (L), pharyngeal (Ph) and extraocular (EOM) myoblasts and fibroblasts (F) transfected with siRNA. PABPN1 knockdown results in ∼40% decrease in the percentage of BrdU+ cells. Data are mean ± SE, *P < 0.05, n = 3.
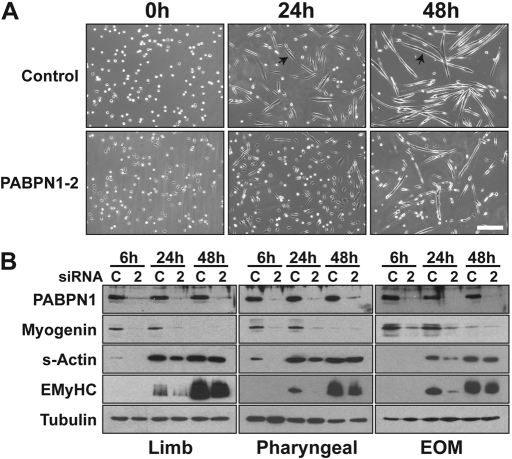
Transfection with PABPN1 siRNA was also performed in order to analyze the impact of loss of PABPN1 function on terminal differentiation of myoblasts. After siRNA oligonucleotide transfection, myoblasts were plated at equal density and immediately differentiated for 6, 24 and 48 h. Defects in myotube formation, as evidenced by fewer myotubes as well as a decrease in the number of large myotubes, were noted at 24 and 48 h in cells transfected with PABPN1 siRNA (Fig. 3A). In contrast, after transfection of control siRNA, cells fused normally over the 48 h time period (Fig. 3A). Immunoblotting for PABPN1 confirmed depletion at 6, 24 and 48 h of differentiation (Fig. 3B). Myoblast differentiation was also assessed by immunoblotting for known myogenic differentiation markers. Biochemical differentiation was markedly diminished after PABPN1 knockdown with decreases in myogenin, an early differentiation marker (18), noted at 6 and 24 h in all three cell types (Fig. 3B). Decreases in later differentiation markers, such as sarcomeric actin (s-actin) and embryonic myosin heavy chain (eMyHC), were also observed at 24 h with smaller differences noted at 48 h. The defect in myoblast differentiation did not seem to differ among limb, pharyngeal and EOM myoblasts. These results suggest that PABPN1 is also required for normal myoblast differentiation in this in vitro myogenesis model.
Figure 3.
PABPN1 is required for proper myoblast differentiation. Pure cultures of primary mouse myoblasts from limb, pharyngeal and extraocular (EOM) muscles were transfected with PABPN1-2 (2) or control scrambled (C) siRNA. (A) Representative phase-contrast images of limb muscle cells after 0, 24 and 48 h of differentiation (bar, 100 µm). PABPN1 siRNA cells exhibit defects in myotube formation at 24 and 48 h. Arrows indicate myotubes in control siRNA cells. (B) Protein extracts were analyzed by immunoblotting after 6, 24 and 48 h of differentiation for the differentiation markers myogenin, sarcomeric actin and embryonic myosin heavy chain (eMyHC). Knockdown of PABPN1 was also determined and alpha-tubulin was used as a loading control. Biochemical differentiation is defective in all three types of muscle cells following PABPN1 knockdown. Similar effects were observed in three independent assays.
PABPN1 is required for proper polyadenylation in muscle cells
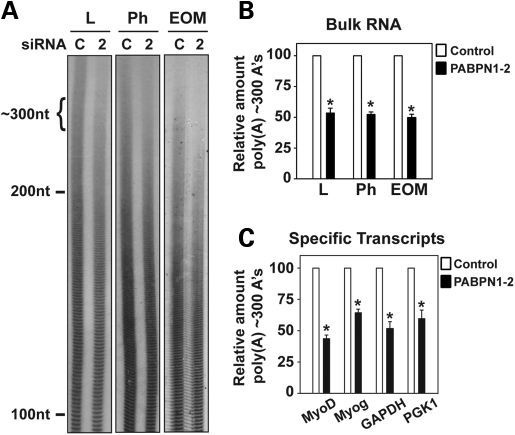
The main function of PABPN1 demonstrated biochemically in vitro is stimulating mRNA polyadenylation and controlling poly(A) tail length (1,19). Studies performed in Drosophila and fission yeast also indicate the involvement of PABPN1 in polyadenylation in vivo (13,14). To address whether the depletion of PABPN1 affects the length of mRNA poly(A) tails on bulk RNA, myoblasts were transfected with control or PABPN1 siRNA and total RNA was isolated and labeled with [32P] cytidine 3′,5′-bis(phosphate) using T4 RNA ligase and digested with RNase A/T1. As seen in Figure 4A, limb, pharyngeal and EOM myoblasts treated with PABPN1-2 siRNA displayed a significant decrease in the amount of poly(A) tails with tract length of 200–300 nt. Densitometric quantification of poly(A) tracts of ∼300 nt showed a 50% decrease in the amount of this polyadenylated RNA population after PABPN1 knockdown in all three types of myoblasts (Fig. 4B). Similar effects were obtained using PABPN1-1 siRNA (data not shown). Slot-blot analysis showed that PABPN1 depletion did not cause a significant reduction in total poly(A) RNA content relative to rRNA (Supplementary Material). These results demonstrate, in a mammalian system, a biological function of PABPN1 in poly(A) tail length control.
Figure 4.
PABPN1 is required for proper poly(A) tail length in muscle cells. Pure cultures of primary mouse myoblasts from limb (L), pharyngeal (Ph) and extraocular (EOM) muscles were transfected with PABPN1-2 (2) or control scrambled (C) siRNA. Cells were collected and total RNA was isolated. (A) Representative distribution of bulk poly(A) tails from different types of myoblasts following PABPN1 knockdown. Total RNA was labeled with [32P] cytidine 3′,5′-bis(phosphate) using T4 RNA ligase and digested with RNase A/T1. Samples were resolved by electrophoresis in denaturing polyacrylamide gels and exposed to radiographic film. Cells treated with PABPN1 siRNA show a significant decrease in poly(A) tract length of ∼300 nt. (B) Densitometric quantification of poly(A) tracts of ∼300 nt as normalized to poly(A) tracts of ∼100 nt for control siRNA versus PABPN1-2 siRNA. A 50% decrease in the relative number of ∼300 nt poly(A) tails is observed following PABPN1 knockdown. Data are mean ± SD, *P < 0.05, n = 5. (C) Linker ligation-mediated poly(A) tail (LLM-PAT) assays were used to determine poly(A) tail length of specific transcripts following myoblast transfection with control or PABPN1-2 siRNA. mRNA transcripts of myogenin, MyoD, GAPDH and PGK1 showed a significant decrease in poly(A) tract length of ∼300 nt. Data are mean ± SD, *P < 0.05, n = 3.
To determine if PABPN1 depletion affects mRNA polyadenylation generally or could be restricted to genes specific for myoblast differentiation, we used a linker ligation-mediated poly(A) tail (LLM-PAT) assay to determine changes in poly(A) tail length of specific transcripts following myoblast transfection with control or PABPN1-2 siRNA. This method is more sensitive than bulk mRNA poly(A) length analysis, because significant changes in specific transcripts could be diluted and masked when analyzing overall changes in poly(A) tail length. We analyzed the distribution of the poly(A) tail length of the myogenic differentiation factor 1 (MyoD), myogenin, glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and phosphoglycerate kinase 1 (PGK1) transcripts. MyoD and myogenin are muscle-specific transcription factors that are essential for the development of functional skeletal muscle (20), whereas GAPDH and PGK1 are housekeeping genes. All four transcripts analyzed demonstrated a significant shortening in their poly(A) tails after PABPN1 depletion, at levels comparable to those observed for bulk poly(A) (Fig. 4C). Similar effects on the poly(A) tail lengths of specific transcripts were obtained after transfection with PABPN1-1 siRNA (data not shown). These results suggest that the effect of PABPN1 depletion on mRNA poly(A) tail size is unlikely to be restricted to specific transcripts, but rather PABPN1 plays a more general role in polyadenylation.
PABPN1 is required for proper poly(A) RNA export from the nucleus
Although the involvement of PABPN1 in mRNA export has not been directly analyzed, multiple lines of evidence suggest a possible role for PABPN1 in this process (5,15). Considerable data reveal that mRNA 3′-end processing and nuclear export are tightly coupled in cells (21). Given that both the poly(A) tail and the active process of polyadenylation are required for mRNA export (22), the proper recruitment and positioning of factors onto transcripts by the polyadenylation process is most likely necessary for mRNA export. As PABPN1 plays a essential role in polyadenylation and binds directly to poly(A), associates with mRNA during docking at the nuclear pore (23), and shuttles from the nucleus to the cytoplasm (5), a possible function of PABPN1 in bulk mRNA nuclear export should be investigated. To determine whether PABPN1 is involved in mRNA nuclear export in myoblasts, we analyzed poly(A) RNA localization by fluorescent in situ hybridization (FISH) using an oligo(dT) probe following PABPN1 knockdown. Figure 5A displays a representative image of mRNA distribution in myoblasts transfected with control or PABPN1-2 siRNA. Although no alterations in poly(A) RNA localization were observed in cells transfected with control siRNA, significant nuclear accumulation of poly(A) RNA was observed after depletion of PABPN1. Analysis of poly(A) RNA localization demonstrated that poly(A) RNA nuclear export is impaired in >50% of cells after transfection with either of two different PABPN1 siRNAs (Fig. 5B), revealing that PABPN1 is required for proper export of bulk poly(A) RNA from the nucleus to the cytoplam.
Figure 5.
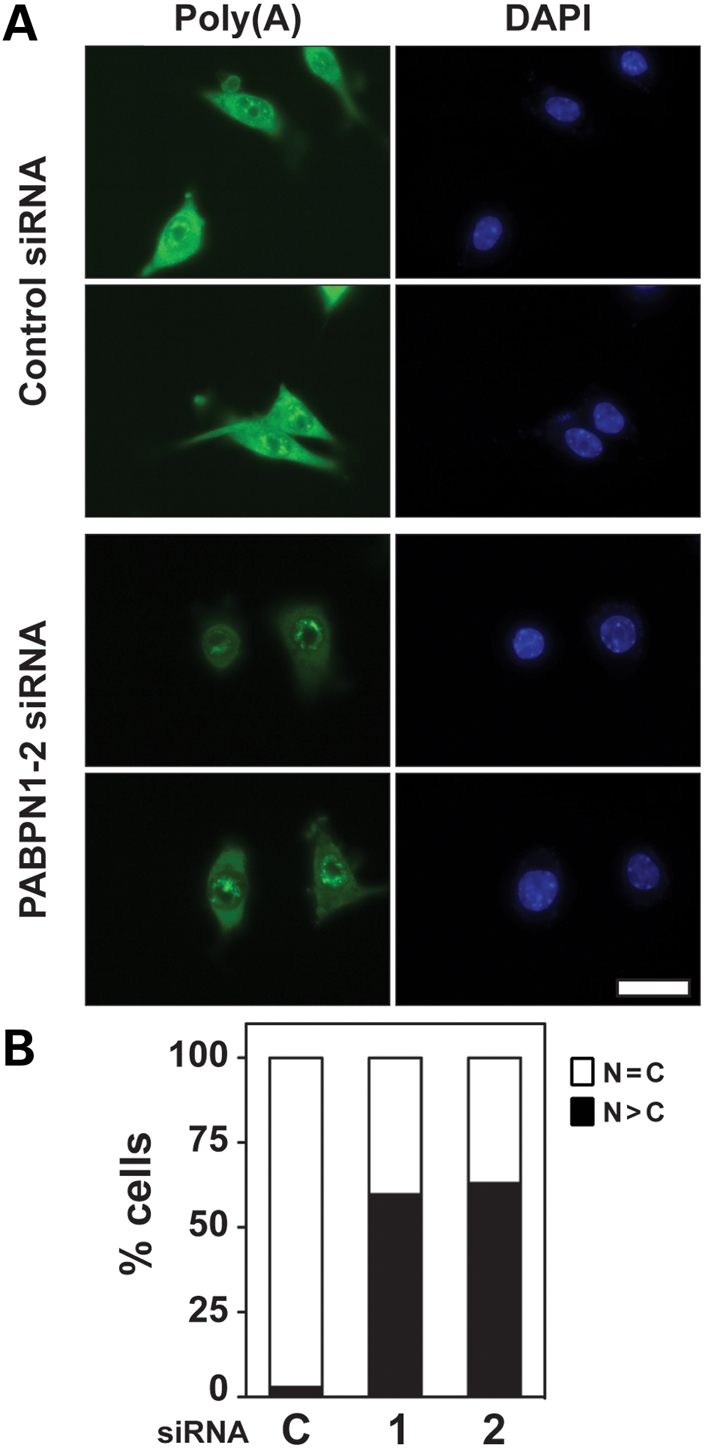
PABPN1 is required for proper poly(A) RNA export from the nucleus. Pure cultures of primary mouse myoblasts from limb were transfected with PABPN1-1 or 2 or control scrambled (C) siRNA and poly(A) localization was determined by fluorescent in situ hybridization (FISH) using an oligo(dT) probe. (A) Representative image of poly(A) RNA localization after siRNA transfection (bar, 20 µm). DAPI staining was used for nuclear visualization. (B) Relative distribution of cells showing normal diffuse nuclear (N) and cytoplasmic (C) localization (n = C) or nuclear accumulation (n > C) of poly(A) RNA after siRNA transfection. Following transfection with either PABPN1-1 or -2 siRNA, more than 50% of myoblasts displayed nuclear poly(A) RNA accumulation. Data are the percentage of nuclear poly(A) RNA distribution for at least 70 cells. Similar results were obtained in three independent experiments.
DISCUSSION
Our experiments define the key functions of PABPN1 in mRNA biogenesis and myogenesis. Depletion of PABPN1 with siRNA led to defective myogenesis, by impairing proliferation and differentiation of myoblasts isolated from different muscle groups affected in OPMD. The molecular mechanism for such impairment may be due to improper mRNA biogenesis, as after PABPN1 knockdown, myoblasts had shorter mRNA poly(A) tails and showed accumulation of poly(A) RNA within the nucleus. Since myogenesis is essential for muscle regeneration and maintenance, these data suggest a critical role for PABPN1 activity in normal muscle physiology.
A significant amount of data precisely dissects the function of PABPN1 in polyadenylation, however, most of the work describing PABPN1 function derives from in vitro biochemical observations (1) or from in vivo studies in Drosophila or in fission yeast (Pab2) (13,14). However, PABPN1 function had not been directly analyzed in mammalian cells or, importantly, in the cell type affected in OPMD. We analyzed the functions of PABPN1 in primary mouse myoblasts. We demonstrate that the proliferative capacity of mouse myoblasts from different muscle origins is significantly diminished following PABPN1 depletion. These results suggest an essential cellular function of PABPN1 in cell proliferation, because comparable results were obtained for fibroblasts. In addition, depletion of PABPN1 also delayed myoblast differentiation, revealing another important requirement of this protein in myogenesis.
At the molecular level, we observed that the depletion of PABPN1 in myoblasts leads to shortening in mRNA poly(A) tails. Polyadenylation is a highly regulated step in mRNA biogenesis and the poly(A) tail is crucial for gene expression (24,25). In the cytoplasm, mRNAs are degraded from the 3′-end and addition of the poly(A) tail is an important mechanism for mRNA stability control in mammalian cells (24,25). Furthermore, polyadenylation enhances the translation of mRNA (24,25), therefore, changes in mRNA poly(A) tail length could alter the expression levels of different genes. The shortening in the length of poly(A) tails observed after PABPN1 knockdown in myoblasts may lead to downstream effects on the expression of genes necessary for cellular functions, possibly explaining the decrease in myoblast proliferation and differentiation observed following PABPN1 knockdown.
We also show that poly(A) RNA accumulates in the nucleus of myoblasts depleted of PABPN1, demonstrating that PABPN1 is required for proper export of poly(A) RNA from the nucleus. There is significant evidence that mRNA 3′-end processing and nuclear export are coordinated and mechanistically coupled rather than merely sequential (21). Defects in mRNA 3′-end processing can inhibit nuclear export of transcripts and mutations in mRNA export factors can also lead to aberrations in 3′-end formation. Mutations in mRNA export factors in the yeast Saccharomyces cerevisiae result in mRNA hyperadenylation (26,27). Conversely, transcripts resulting from improper processing are retained in the nucleus near the transcription site or at the nuclear pore (28). Also in yeast, the poly(A)-binding proteins Pab1 and Nab2 appear to have activity in both mRNA polyadenylation regulation and export (1). Similarly, the protein dZC3H3 is required for proper mRNA polyadenylation and nuclear export in Drosophila (29). Depletion of PABPN1 could affect mRNA nuclear export by two non-mutually exclusive mechanisms. Given that poly(A) tails are essential for mRNA export (22), shorter poly(A) tails resulting from decreased mRNA polyadenylation stimulation could indirectly affect mRNA nuclear export. Alternatively, the PABPN1–poly(A) complex may play a direct role in mRNA export, thus transcripts with poly(A) tails not fully coated with PABPN1 would fail to be exported. PABPN1, together with poly(A), forms compact spherical particles of uniform size, where each particle can accommodate 200–300 nt of poly(A) (30). On very long poly(A) tracts, several of these particles of uniform size are arranged in a beads-on-a-string pattern, but shorter poly(A) tracts support the formation of smaller, probably incomplete particles (30). In this scenario, both shorter poly(A) tails and decreased amounts of PABPN1 would impair the formation of such particles, which may be the structure required for mRNA export. Further analysis will be necessary to learn more about PABPN1 involvement in mRNA nuclear export.
PABPN1 function is likely necessary for muscle regeneration and maintenance, because myoblast proliferation and differentiation are crucial for the regenerative ability of skeletal muscle. Mutations causing alanine expansion in PABPN1 lead to OPMD, but why these changes cause OPMD is unknown. The predominant hypothesis suggests that a toxic gain-of-function caused by the alanine expansion is causative of OPMD through the formation of nuclear aggregates of PABPN1 (31). Evidence in favor of a pathological role for the aggregates comes from positive results obtained with anti-aggregation therapy in both cell culture and transgenic mouse models (32–34). Furthermore, mutations in PABPN1 that render it incapable of aggregating do not lead to muscle pathology in a Drosophila model of OPMD (35). Conversely, other findings suggest that the alanine expansion may decrease availability of PABPN1 at the protein level and cause a relative loss-of-function in OPMD (36). Evidence that brings the contribution of nuclear aggregates into question include the observation that even wild-type PABPN1 can form reversible aggregates in neurons in response to changes in cell physiology or development without overt pathology (37). In addition, PABPN1 aggregates are only detected in up to 5% of myofiber nuclei in muscle sections from OPMD patients (38). Furthermore, overexpression of wild-type PABPN1 reduces toxicity caused by the expression of mutant PABPN1 in both cell and mouse models of OPMD (39). Thus, toxicity of PABPN1 nuclear aggregates is unlikely to be the sole basis of disease etiology. As we demonstrated, a decrease in PABPN1 levels results in defective myogenesis in vitro, which strengthens the hypothesis that a relative loss-of-function of PABPN1 caused by the alanine expansion may contribute to the development of OPMD by impairing muscle maintenance and regeneration. Consistent with our results, pharyngeal myoblasts isolated from OPMD patients display premature proliferative arrest in vitro (40). Although poly(A) tail length is not altered in cultured myoblasts isolated from OPMD deltoid muscles (4), the range of poly(A) tails analyzed by these researchers (10–180 nt) did not cover the tract length where we observed shortening of poly(A) tails (∼300 nt) after PABPN1 knockdown. In addition, as disease progression is variable among OPMD patients, whether these myoblasts came from muscles that were clinically affected in these particular patients is unknown. Finally, whether cultured myoblasts from OPMD muscle display PABPN1 nuclear aggregates has not been studied, which makes establishing a correlation between culture studies and muscle tissue difficult. Thus, further experiments are needed to determine whether polyadenylation is altered in muscles from OPMD patients and verify if the alanine expansion leads to loss of PABPN1 function in muscle tissue.
In conclusion, our results demonstrate that PABPN1 is essential for both mRNA biogenesis and myogenic activity of primary mouse myoblasts. Myoblast proliferation and differentiation are crucial for the regenerative ability of skeletal muscle, thus PABPN1 function is likely necessary for muscle regeneration and maintenance.
MATERIALS AND METHODS
Primary muscle cell culture
Primary myoblasts were derived from the hindlimb, pharyngeal or EOM of 6- to 24-week-old Balb/C mice and cultured to >99% purity as previously described (41). Cells were maintained in growth media (GM: Ham's F10, 20% FBS, 5 ng/ml bFGF, 100 U/ml penicillin G, 100 mg/ml streptomycin) in a humidified 5% CO2 incubator at 37°C on collagen-coated dishes. To induce differentiation, cells were plated on dishes coated with Entactin–Collagen IV–Laminin (ECL; Upstate Biotechnology) in GM and shortly thereafter switched to differentiation media [DM: DMEM, 1% Insulin–Transferrin–Selenium-A supplement (Invitrogen), 100 U/mL penicillin G and 100 µg/mL streptomycin] for the indicated times. For all experiments, at least three independent isolates were analyzed.
Immunoblotting
Cells were lysed with RIPA-2 (50 mm Tris–HCl pH 8.0, 150 mm NaCl, 1% NP-40, 0.5% deoxycholic acid, 0.1% SDS) containing protease inhibitors (Mini Complete, Roche) and equal amounts of total protein (5–20 µg) were resolved by SDS–PAGE, transferred to nitrocellulose and the desired protein was detected by immunoblotting with appropriate antibodies. Recombinant His-tagged PABPN1 was purified as previously described (2) and used for immunization of rabbits to obtain a polyclonal anti-PABPN1 antibody (1:5000). Other antibodies used were anti-GAPDH (1:1000, Santa Cruz), anti-alpha-tubulin (1:5000, Sigma), anti-myogenin (1:100, F5D Developmental Studies Hybridoma Bank), anti-s-actin (1:2000, Sigma) and anti-eMyHC (1:100, F1652).
Transfection of primary myoblasts
Stealth RNAi (Invitrogen) was used to knockdown PABPN1 expression in primary myoblasts. Myoblasts were plated in GM at a density of 1 × 105 cells per well on collagen-coated six-well plates and after 6 h, duplexed siRNAs at the final concentration of 80 nm were used to transfect cells using Lipofectamine 2000 (Invitrogen) in GM according to the manufacturer's instructions. After overnight incubation, medium containing transfection complexes was replaced by fresh GM. PABPN1 knockdown was assessed by immunoblotting using anti-PABPN1 40 h after transfection. Cells were transfected with either scrambled control or one of two PABPN1 siRNAs (Invitrogen) (PABPN1-1, ACGAGGUAGAGAAGCAGAUGAAUAU; PABPN1-2, UCCCGAUCUCGAUUCUACAGUGGUU).
BrdU assay
Scrambled control or PABPN1 siRNAs were used to transfect cells as described previously. Myoblasts were incubated in GM with 25 µm BrdU (Sigma) starting 39 h after transfection. Following 1 h incubation with BrdU, cells were fixed and analyzed by immunostaining as described (42) except that cells were incubated in 1 N HCl for 30 min prior to blocking. Fluorescence images were acquired using a microscope (Axiovert 200 M; Carl Zeiss MicroImaging, Inc.) with a 0.3 NA 10× Plan-Neofluar objective (Carl Zeiss MicroImaging, Inc.) and camera (QImaging) with OpenLab 5.5.0 (Improvision) and the percentage of BrdU+ nuclei in six random fields (>1000 total nuclei) was determined for each condition. At least three independent experiments were performed.
Poly(A) tail length analysis
Bulk poly(A) tails were analyzed as described previously (43). In brief, total RNA (10 µg) was 3′-end labeled with [32P]-pCp (cytidine 3′,5′ bisphosphate) and T4 RNA ligase followed by digestion with RNase A and T1. Products were separated on TBE-Urea (90 mm Tris-borate, 2 mm EDTA, 8 m urea) 7% polyacrylamide gels and imaged by autoradiography. Poly(A) tail length of specific transcripts was analyzed by LLM-PAT as described previously (44). Total RNA was ligated to a 5′ pppRNA linker (Linker 3, Integrated DNA Technologies) and the RNA was then reverse transcribed using a reverse transcription primer specific to the RNA linker (44). The resulting cDNA was then amplified by PCR using the reverse transcription primer and a primer specific to the transcript to be analyzed (MyoD, TCTCCCAGGCATGCTGTG; myogenin, GTAATTCTTTTGCTAACTTATTTGG; GAPDH, TAACAGGAGGGGCCTAGG; PGK1, TCTGGTTAGCTTCGTCACTC). PCR products were separated on a 5% non-denaturing polyacrylamide gel and visualized with SYBR Green I nucleic acid gel stain (Invitrogen).
FISH
The protocol used for poly(A) RNA localization was previously described (29). Briefly, myoblast cells were allowed to adhere to ECL-coated coverslips in DM for 6 h. Cells were fixed with 4% paraformaldehyde in PBS for 10 min, permeabilized with 100% cold methanol for 10 min and 70% ethanol at 4°C overnight. Cells were hybridized for 1–2 h at 37°C with DIG-Oligo-dT(50) probe (Integrated DNA Technologies) at 0.2 mm in hybridization buffer (25% formamide, 10% dextran sulfate, 0.005% BSA, 1 mg/mL yeast tRNA in 2× SSC). Following washes with 2× SSC, cells were incubated with FITC-conjugated anti-DIG antibody (Molecular Probes) at RT for 1 h and with Hoechst 33325 dye (Sigma) for 5 min. Images were obtained using an Olympus IX81 microscope with a 0.3 NA 100× Zeiss Plan-Neofluar objective. Images were captured using a Hamamatsu digital camera with Slidebook software (version 5.0). At least three independent experiments were performed.
SUPPLEMENTARY MATERIAL
FUNDING
This work was supported by the Muscular Dystrophy Association [68022 to G.K.P.] and the National Institutes of Health [NS069234 to G.K.P. and A.H.C.].
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr Julia Chekanova, for assistance with bulk poly(A) tail length assay.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Kuhn U., Wahle E. Structure and function of poly(A) binding proteins. Biochim. Biophys. Acta. 2004;1678:67–84. doi: 10.1016/j.bbaexp.2004.03.008. [DOI] [PubMed] [Google Scholar]
- 2.Kuhn U., Nemeth A., Meyer S., Wahle E. The RNA binding domains of the nuclear poly(A)-binding protein. J. Biol. Chem. 2003;278:16916–16925. doi: 10.1074/jbc.M209886200. [DOI] [PubMed] [Google Scholar]
- 3.Fan X., Dion P., Laganiere J., Brais B., Rouleau G.A. Oligomerization of polyalanine expanded PABPN1 facilitates nuclear protein aggregation that is associated with cell death. Hum. Mol. Genet. 2001;10:2341–2351. doi: 10.1093/hmg/10.21.2341. [DOI] [PubMed] [Google Scholar]
- 4.Calado A., Tome F.M., Brais B., Rouleau G.A., Kuhn U., Wahle E., Carmo-Fonseca M. Nuclear inclusions in oculopharyngeal muscular dystrophy consist of poly(A) binding protein 2 aggregates which sequester poly(A) RNA. Hum. Mol. Genet. 2000;9:2321–2328. doi: 10.1093/oxfordjournals.hmg.a018924. [DOI] [PubMed] [Google Scholar]
- 5.Calado A., Kutay U., Kuhn U., Wahle E., Carmo-Fonseca M. Deciphering the cellular pathway for transport of poly(A)-binding protein II. RNA. 2000;6:245–256. doi: 10.1017/s1355838200991908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Calado A., Carmo-Fonseca M. Localization of poly(A)-binding protein 2 (PABP2) in nuclear speckles is independent of import into the nucleus and requires binding to poly(A) RNA. J. Cell Sci. 2000;113:2309–2318. doi: 10.1242/jcs.113.12.2309. [DOI] [PubMed] [Google Scholar]
- 7.Brais B., Bouchard J.P., Xie Y.G., Rochefort D.L., Chretien N., Tome F.M., Lafreniere R.G., Rommens J.M., Uyama E., Nohira O., et al. Short GCG expansions in the PABP2 gene cause oculopharyngeal muscular dystrophy. Nat. Genet. 1998;18:164–167. doi: 10.1038/ng0298-164. [DOI] [PubMed] [Google Scholar]
- 8.Victor M., Hayes R., Adams R.D. Oculopharyngeal muscular dystrophy. A familial disease of late life characterized by dysphagia and progressive ptosis of the evelids. N. Engl. J. Med. 1962;267:1267–1272. doi: 10.1056/NEJM196212202672501. [DOI] [PubMed] [Google Scholar]
- 9.Ruegg S., Lehky Hagen M., Hohl U., Kappos L., Fuhr P., Plasilov M., Muller H., Heinimann K. Oculopharyngeal muscular dystrophy - an under-diagnosed disorder? Swiss Med. Wkly. 2005;135:574–586. doi: 10.4414/smw.2005.11221. [DOI] [PubMed] [Google Scholar]
- 10.Van Der Sluijs B.M., Hoefsloot L.H., Padberg G.W., Van Der Maarel S.M., Van Engelen B.G. Oculopharyngeal muscular dystrophy with limb girdle weakness as major complaint. J. Neurol. 2003;250:1307–1312. doi: 10.1007/s00415-003-0201-6. [DOI] [PubMed] [Google Scholar]
- 11.Tome F.M., Fardeau M. Nuclear inclusions in oculopharyngeal dystrophy. Acta Neuropathol. 1980;49:85–87. doi: 10.1007/BF00692226. [DOI] [PubMed] [Google Scholar]
- 12.Tome F.M., Chateau D., Helbling-Leclerc A., Fardeau M. Morphological changes in muscle fibers in oculopharyngeal muscular dystrophy. Neuromuscul. Disord. 1997;7(Suppl. 1):S63–S69. doi: 10.1016/s0960-8966(97)00085-0. [DOI] [PubMed] [Google Scholar]
- 13.Benoit B., Mitou G., Chartier A., Temme C., Zaessinger S., Wahle E., Busseau I., Simonelig M. An essential cytoplasmic function for the nuclear poly(A) binding protein, PABP2, in poly(A) tail length control and early development in Drosophila. Dev. Cell. 2005;9:511–522. doi: 10.1016/j.devcel.2005.09.002. [DOI] [PubMed] [Google Scholar]
- 14.Perreault A., Lemieux C., Bachand F. Regulation of the nuclear poly(A)-binding protein by arginine methylation in fission yeast. J. Biol. Chem. 2007;282:7552–7562. doi: 10.1074/jbc.M610512200. [DOI] [PubMed] [Google Scholar]
- 15.Chen Z., Li Y., Krug R.M. Influenza A virus NS1 protein targets poly(A)-binding protein II of the cellular 3′-end processing machinery. EMBO J. 1999;18:2273–2283. doi: 10.1093/emboj/18.8.2273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Charge S.B., Rudnicki M.A. Cellular and molecular regulation of muscle regeneration. Physiol. Rev. 2004;84:209–238. doi: 10.1152/physrev.00019.2003. [DOI] [PubMed] [Google Scholar]
- 17.Horsley V., Pavlath G.K. Forming a multinucleated cell: molecules that regulate myoblast fusion. Cells Tissues Organs. 2004;176:67–78. doi: 10.1159/000075028. [DOI] [PubMed] [Google Scholar]
- 18.Costelli P., Almendro V., Figueras M.T., Reffo P., Penna F., Aragno M., Mastrocola R., Boccuzzi G., Busquets S., Bonelli G., et al. Modulations of the calcineurin/NF-AT pathway in skeletal muscle atrophy. Biochim. Biophys. Acta. 2007;1770:1028–1036. doi: 10.1016/j.bbagen.2007.03.003. [DOI] [PubMed] [Google Scholar]
- 19.Kuhn U., Gundel M., Knoth A., Kerwitz Y., Rudel S., Wahle E. Poly(A) tail length is controlled by the nuclear poly(A)-binding protein regulating the interaction between poly(A) polymerase and the cleavage and polyadenylation specificity factor. J. Biol. Chem. 2009;284:22803–22814. doi: 10.1074/jbc.M109.018226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Berkes C.A., Tapscott S.J. MyoD and the transcriptional control of myogenesis. Semin. Cell Dev. Biol. 2005;16:585–595. doi: 10.1016/j.semcdb.2005.07.006. [DOI] [PubMed] [Google Scholar]
- 21.Vinciguerra P., Stutz F. mRNA export: an assembly line from genes to nuclear pores. Curr. Opin. Cell Biol. 2004;16:285–292. doi: 10.1016/j.ceb.2004.03.013. [DOI] [PubMed] [Google Scholar]
- 22.Huang Y., Carmichael G.G. Role of polyadenylation in nucleocytoplasmic transport of mRNA. Mol. Cell Biol. 1996;16:1534–1542. doi: 10.1128/mcb.16.4.1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bear D.G., Fomproix N., Soop T., Bjorkroth B., Masich S., Daneholt B. Nuclear poly(A)-binding protein PABPN1 is associated with RNA polymerase II during transcription and accompanies the released transcript to the nuclear pore. Exp. Cell Res. 2003;286:332–344. doi: 10.1016/s0014-4827(03)00123-x. [DOI] [PubMed] [Google Scholar]
- 24.Lewis J.D., Gunderson S.I., Mattaj I.W. The influence of 5′ and 3′ end structures on pre-mRNA metabolism. J. Cell Sci. Suppl. 1995;19:13–19. doi: 10.1242/jcs.1995.supplement_19.2. [DOI] [PubMed] [Google Scholar]
- 25.Wickens M., Anderson P., Jackson R.J. Life and death in the cytoplasm: messages from the 3′ end. Curr. Opin. Genet. Dev. 1997;7:220–232. doi: 10.1016/s0959-437x(97)80132-3. [DOI] [PubMed] [Google Scholar]
- 26.Hilleren P., Parker R. Defects in the mRNA export factors Rat7p, Gle1p, Mex67p, and Rat8p cause hyperadenylation during 3′-end formation of nascent transcripts. RNA. 2001;7:753–764. doi: 10.1017/s1355838201010147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jensen T.H., Patricio K., McCarthy T., Rosbash M. A block to mRNA nuclear export in S. cerevisiae leads to hyperadenylation of transcripts that accumulate at the site of transcription. Mol. Cell. 2001;7:887–898. doi: 10.1016/s1097-2765(01)00232-5. [DOI] [PubMed] [Google Scholar]
- 28.Fasken M.B., Corbett A.H. Mechanisms of nuclear mRNA quality control. RNA Biol. 2009;6:237–241. doi: 10.4161/rna.6.3.8330. [DOI] [PubMed] [Google Scholar]
- 29.Farny N.G., Hurt J.A., Silver P.A. Definition of global and transcript-specific mRNA export pathways in metazoans. Genes Dev. 2008;22:66–78. doi: 10.1101/gad.1616008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Keller R.W., Kuhn U., Aragon M., Bornikova L., Wahle E., Bear D.G. The nuclear poly(A) binding protein, PABP2, forms an oligomeric particle covering the length of the poly(A) tail. J. Mol. Biol. 2000;297:569–583. doi: 10.1006/jmbi.2000.3572. [DOI] [PubMed] [Google Scholar]
- 31.Abu-Baker A., Rouleau G.A. Oculopharyngeal muscular dystrophy: recent advances in the understanding of the molecular pathogenic mechanisms and treatment strategies. Biochim. Biophys. Acta. 2007;1772:173–185. doi: 10.1016/j.bbadis.2006.10.003. [DOI] [PubMed] [Google Scholar]
- 32.Abu-Baker A., Messaed C., Laganiere J., Gaspar C., Brais B., Rouleau G.A. Involvement of the ubiquitin–proteasome pathway and molecular chaperones in oculopharyngeal muscular dystrophy. Hum. Mol. Genet. 2003;12:2609–2623. doi: 10.1093/hmg/ddg293. [DOI] [PubMed] [Google Scholar]
- 33.Davies J.E., Wang L., Garcia-Oroz L., Cook L.J., Vacher C., O'Donovan D.G., Rubinsztein D.C. Doxycycline attenuates and delays toxicity of the oculopharyngeal muscular dystrophy mutation in transgenic mice. Nat. Med. 2005;11:672–677. doi: 10.1038/nm1242. [DOI] [PubMed] [Google Scholar]
- 34.Davies J.E., Berger Z., Rubinsztein D.C. Oculopharyngeal muscular dystrophy: potential therapies for an aggregate-associated disorder. Int. J. Biochem. Cell Biol. 2006;38:1457–1462. doi: 10.1016/j.biocel.2006.01.016. [DOI] [PubMed] [Google Scholar]
- 35.Chartier A., Benoit B., Simonelig M. A Drosophila model of oculopharyngeal muscular dystrophy reveals intrinsic toxicity of PABPN1. EMBO J. 2006;25:2253–2262. doi: 10.1038/sj.emboj.7601117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Klein A.F., Ebihara M., Alexander C., Dicaire M.J., Sasseville A.M., Langelier Y., Rouleau G.A., Brais B. PABPN1 polyalanine tract deletion and long expansions modify its aggregation pattern and expression. Exp. Cell Res. 2008;314:1652–1666. doi: 10.1016/j.yexcr.2008.02.005. [DOI] [PubMed] [Google Scholar]
- 37.Berciano M.T., Villagra N.T., Ojeda J.L., Navascues J., Gomes A., Lafarga M., Carmo-Fonseca M. Oculopharyngeal muscular dystrophy-like nuclear inclusions are present in normal magnocellular neurosecretory neurons of the hypothalamus. Hum. Mol. Genet. 2004;13:829–838. doi: 10.1093/hmg/ddh101. [DOI] [PubMed] [Google Scholar]
- 38.Brais B., Rouleau G.A., Bouchard J.P., Fardeau M., Tome F.M. Oculopharyngeal muscular dystrophy. Semin. Neurol. 1999;19:59–66. doi: 10.1055/s-2008-1040826. [DOI] [PubMed] [Google Scholar]
- 39.Davies J.E., Sarkar S., Rubinsztein D.C. Wild-type PABPN1 is anti-apoptotic and reduces toxicity of the oculopharyngeal muscular dystrophy mutation. Hum. Mol. Genet. 2008;17:1097–1108. doi: 10.1093/hmg/ddm382. [DOI] [PubMed] [Google Scholar]
- 40.Perie S., Mamchaoui K., Mouly V., Blot S., Bouazza B., Thornell L.E., St Guily J.L., Butler-Browne G. Premature proliferative arrest of cricopharyngeal myoblasts in oculo-pharyngeal muscular dystrophy: Therapeutic perspectives of autologous myoblast transplantation. Neuromuscul. Disord. 2006;16:770–781. doi: 10.1016/j.nmd.2006.07.022. [DOI] [PubMed] [Google Scholar]
- 41.Rando T.A., Blau H.M. Primary mouse myoblast purification, characterization, and transplantation for cell-mediated gene therapy. J. Cell Biol. 1994;125:1275–1287. doi: 10.1083/jcb.125.6.1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mitchell P.O., Pavlath G.K. Skeletal muscle atrophy leads to loss and dysfunction of muscle precursor cells. Am. J. Physiol. Cell Physiol. 2004;287:C1753–C1762. doi: 10.1152/ajpcell.00292.2004. [DOI] [PubMed] [Google Scholar]
- 43.Chekanova J.A., Shaw R.J., Belostotsky D.A. Analysis of an essential requirement for the poly(A) binding protein function using cross-species complementation. Curr. Biol. 2001;11:1207–1214. doi: 10.1016/s0960-9822(01)00371-2. [DOI] [PubMed] [Google Scholar]
- 44.Garneau N.L., Sokoloski K.J., Opyrchal M., Neff C.P., Wilusz C.J., Wilusz J. The 3′ untranslated region of sindbis virus represses deadenylation of viral transcripts in mosquito and Mammalian cells. J. Virol. 2008;82:880–892. doi: 10.1128/JVI.01205-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.