Abstract
Exosomes are 40–100-nm-diameter nanovesicles of endocytic origin that are released from diverse cell types. To better understand the biological role of exosomes and to avoid confounding data arising from proteinaceous contaminants, it is important to work with highly purified material. Here, we describe an immunoaffinity capture method using the colon epithelial cell-specific A33 antibody to purify colorectal cancer cell (LIM1215)-derived exosomes. LC-MS/MS revealed 394 unique exosomal proteins of which 112 proteins (28%) contained signal peptides and a significant enrichment of proteins containing coiled coil, RAS, and MIRO domains. A comparative protein profiling analysis of LIM1215-, murine mast cell-, and human urine-derived exosomes revealed a subset of proteins common to all exosomes such as endosomal sorting complex required for transport (ESCRT) proteins, tetraspanins, signaling, trafficking, and cytoskeletal proteins. A conspicuous finding of this comparative analysis was the presence of host cell-specific (LIM1215 exosome) proteins such as A33, cadherin-17, carcinoembryonic antigen, epithelial cell surface antigen (EpCAM), proliferating cell nuclear antigen, epidermal growth factor receptor, mucin 13, misshapen-like kinase 1, keratin 18, mitogen-activated protein kinase 4, claudins (1, 3, and 7), centrosomal protein 55 kDa, and ephrin-B1 and -B2. Furthermore, we report the presence of the enzyme phospholipid scramblase implicated in transbilayer lipid distribution membrane remodeling. The LIM1215-specific exosomal proteins identified in this study may provide insights into colon cancer biology and potential diagnostic biomarkers.
Exosomes represent a distinct class of membrane nanovesicles (40–100-nm diameter) of endocytic origin that are released from diverse cell types under both normal and pathological conditions (1). Although initial studies focused on exosomes released from various cell types in vitro, exosomes have also been reported in diverse body fluids such as urine (2), amniotic fluid (3, 4), malignant ascites (5–7), bronchoalveolar lavage fluid (8), synovial fluid (9), platelets (10), breast milk (11), and blood (12). Exosomes are formed through the inward budding of late endosomal membranes that give rise to intraluminal vesicles (ILVs)1 within intracellular multivesicular bodies (MVBs). MVBs have a well known intermediary function in the degradation of either proteins internalized from the cell surface (e.g. cell surface receptors) or intracellular proteins sorted from the trans-Golgi network. Proteins destined for degradation are sorted, typically in a ubiquitin-dependent manner, into the ILVs of the nascent MVBs, which then fuse with pre-existing lysosomes (13). An alternate fate for MVBs involves their fusion with the plasma membrane and ensuing release of ILVs into the extracellular environment as exosomes. The biogenesis of exosomes has been linked to the protein complex ESCRT machinery, which is required for both formation of MVBs and the recruitment of their endosome-derived cargo proteins (14).
Exosomes exhibit pleiotropic biological functions including immunomodulatory activity, mediation of cell-cell communication, and, possibly, the transport and propagation of infectious cargo such as prions and retroviruses (1, 15, 16). Despite these advances in our understanding of exosome function, the physiological significance of exosomes is still not fully understood. The observation that exosomes contains inactive RNA and microRNAs that can be transferred to another cell and be translated in the recipient suggest that exosomes may provide a novel vehicle for genetic exchange between cells (17). More recently, the finding of glioblastoma tumor cell-derived exosomes that contain mRNA mutant/variants and microRNAs characteristic of the glioma coupled with the finding of these microvesicles in serum of glioblastoma patients suggests that blood-based exosomes may provide important diagnostic information and aid in therapeutic decisions for cancer patients (18).
The molecular composition of exosomes purified from the cell culture medium from various cell types and diverse body fluids has been analyzed by proteomics as well as fluorescence-activated cell sorting, Western blot analysis, and immunohistochemistry (1, 19). In addition to displaying a protein composition that reflects their endosomal origin, these proteome profiling studies also indicate a unique protein fingerprint that reflects their cellular origin as well as possible physiological role and targeting properties. However, interpretation of exosomal proteome profiles in a biological context also highlights a cautionary note, especially if exosomes are not highly purified. For example, retroviruses such as HIV particles that bud from the cell surface using the same endocytic pathway machinery as exosomes to egress from hematopoietic cells can be a confounding factor in biochemical and physiological analyses of exosomes. Furthermore, exosomes and HIV-1 particles have similar biophysical properties such as size (40–100 and 100 nm, respectively) and buoyant density (1.13–1.21 g/liter (20) and 1.13–1.21 g/liter (21), respectively) as well as molecular composition and their ability to activate immune cells. Although earlier studies describe exosomes carrying virion cargo (22–24), recent exosome purification strategies deploying immunoaffinity capture (25) or a combination of immunoaffinity capture and density gradient centrifugation (26) demonstrate that exosomes from hematopoietic cells can be purified free of virions like HIV-1.
In-depth proteomics studies with large data sets that might contribute to the understanding of the biological function of exosomes are, to date, limited (2, 17). Moreover, strategies used to purify exosomes differ between laboratories (1) with little consensus concerning criteria of purity. Isolation strategies typically involve a combination of differential centrifugation, filtration, concentration, and flotation density gradient followed by characterization using electron microscopy, flow cytometry, and Western blotting (for a review, see Simpson et al. (1)). As a first step toward understanding the physiological role of exosomes in colon cancer biology, we describe here a robust strategy to isolate and characterize exosomes released from LIM1215 colorectal carcinoma cells (27) for the purpose of proteome analysis. This isolation strategy utilized the colon epithelial cell-specific A33 antibody (28–31) to immunoaffinity capture A33-containing exosomes using microbeads. Here, we report for the first time an in-depth proteomics analysis of A33-containing exosomes released from the LIM1215 colon carcinoma cell line. Using these data, we performed a comparative bioinformatics analysis with human urinary and mast cell-derived exosomes.
EXPERIMENTAL PROCEDURES
Cell Culture
Human colon carcinoma cell line LIM1215 (27) was cultured in RPMI 1640 medium (Invitrogen) containing 10% FCS, α-thioglycerol (10 μm), insulin (25 units/liter), and hydrocortisone (1 mg/liter) with 10% CO2 at 37 °C as described (32).
Isolation of Crude Exosomes from LIM1215-conditioned Medium
LIM1215 cells (∼1 × 109 cells) were grown to 80–90% confluence, washed (four times) with 10 ml of RPMI 1640 medium, and cultured for 24 h in 15 ml of serum-free RPMI 1640 medium supplemented with 0.8% insulin-transferrin-selenium solution (Invitrogen). Conditioned medium (CM; ∼600 ml) was harvested and centrifuged (480 × g for 5 min and then at 1,900 × g for 10 min) to remove cell debris. The CM was filtered through a VacuCap® 60 filter unit fitted with a 0.1-μm Supor® membrane (PALL Life Sciences) and then concentrated to ∼6 ml using an Amicon® Ultracel-5K (5,000) molecular weight cutoff centrifugal filter device (Millipore). This concentrated CM (CCM) was ultracentrifuged at 100,000 × g for 1 h, and the resulting pellet was washed twice with PBS to obtain the “crude exosomes.”
Rate Zonal Centrifugation through a Discontinuous Iodixanol (OptiPrepTM) Gradient
CCM from LIM1215 cells (500 μl) was overlaid on a discontinuous OptiPrep gradient (40, 20, 10, and 5% OptiPrep solution (Axis-Shield PoC, Oslo, Norway) in 0.25 m sucrose, 10 mm Tris, pH 7.5) and centrifuged at 100,000 × g for 16 h. Fractions (1 ml) were collected from the top of the gradient, diluted with 2 ml of 10 mm Tris buffer, and centrifuged at 100,000 × g for 3 h; the subsequent pellets were washed once with PBS and subjected to Western blot analysis. The density of each fraction was determined by absorbance at 244 nm using a duplicate parallel discontinuous OptiPrep gradient overlaid with 500 μl of 0.25 m sucrose, 10 mm Tris, pH 7.5 (33).
Electron Microscopy (EM)
EM imaging of crude exosomes was performed as described (34). Briefly, 2–4 μg of 1% (v/v) glutaraldehyde-fixed exosomes was spotted onto a Formvar-coated 200 mesh copper grid and dried at RT. The grids were washed twice with 0.1 m sodium cacodylate and twice with water for 5 min before staining with 5% (w/v) uranyl acetate for 10 min. For immunoelectron microscopy, 2% (w/v) paraformaldehyde-fixed exosomes were layered onto 200 mesh nickel grids. The grids were treated first for 30 min with 50 μl of blocking buffer (5% (w/v) BSA, 0.2% (w/v) cold water fish skin gelatin, 5% (v/v) normal goat serum (Aurion, Wageningen, Netherlands) in PBS) followed by 100 μl of incubation buffer (0.2% BSA-c (acetylated) in PBS) for 5 min (two times). Mouse anti-human A33 antibody (Ludwig Institute for Cancer Research) (100 μl of 5 μg/ml) in 2% BSA-c in PBS was incubated with the grid for a minimum of 4 h and then washed (five times) with washing buffer (0.5% BSA, 0.1% cold water fish skin gelatin, 15 mm sodium azide in PBS) at RT. The grid was then incubated with ultrasmall gold-conjugated goat anti-mouse antibody using the same procedure. Silver enhancement of NANOGOLDTM was performed according to the manufacturer's (Nanoprobes, Inc.) instructions. All EM reagents were obtained from ProSciTech (Queensland, Australia). Imaging was performed using a Siemens Elmiskop 102 or a Philips 10 electron microscope at 60 kV with 25,000× magnification.
Western Blot Analysis
Exosome samples (5–10 μg f protein) were solubilized in 2× lithium dodecyl sulfate sample loading buffer (Invitrogen) containing 50 mm DTT and electrophoretically separated on a precast Novex 4–12% Bis-Tris NuPAGE gel (Invitrogen) using MES running buffer according to the manufacturer's instructions. Proteins were electrotransferred onto nitrocellulose membranes (Osmonics), and the membranes were blocked in 5% (w/v) skim milk powder in Tris-buffered saline with 0.05% (v/v) Tween 20 (TBST) for 1 h at RT. Membranes were probed with mouse anti-CD9 (Santa Cruz Biotechnology; 1:1,000), mouse anti-TSG101 (BD Biosciences, 1:500), mouse anti-HSP70 (BD Biosciences; 1:1,000), mouse anti-Alix (Cell Signaling Technology; 1:1,000), and mouse anti-human A33 antibodies (1 μg/ml) for 1 h in TBST followed by incubation in horseradish peroxidase-conjugated goat anti-mouse IgG (Bio-Rad; 1:5,000) or IRDye 800CW goat anti-mouse IgG (LI-COR Biosciences; 1:15,000) for 1 h. All antibody incubations were carried out at RT with shaking, and blots were washed (three times) with TBST for 10 min after each incubation step. Antigen-antibody complexes were visualized by chemiluminescence (ECL Plus Western blotting detection reagents, GE Healthcare) on x-ray film or scanned using the Odyssey imager (LI-COR Biosciences).
Preparation of Immunoaffinity Capture Microbeads
Humanized A33 monoclonal antibody immunoaffinity capture microbeads (Protein G DynabeadsTM, Invitrogen) were prepared as follows. Briefly, 500 μl of Dynabeads (5 × 108 beads) in citrate-phosphate buffer, pH 5.0 were mixed with 100 μl of capture antibody (300 μg) and incubated for 40 min at RT with gentle rotation according to the manufacturer's instructions. The beads were placed on a magnet for 2 min, the supernatant was discarded, and the beads were washed (two times) with citrate-phosphate buffer, pH 5.0. The Dynabeads were washed twice with 1 ml of 0.2 m triethanolamine, pH 8.2 with the use of a magnet; then suspended in 1 ml of freshly prepared 20 mm dimethyl pimelimidate in 0.2 m triethanolamine, pH 8.2; and incubated for 30 min at RT with gentle agitation. The beads were placed on a magnet for 2 min, and the supernatant was discarded. The reaction was halted by resuspending the beads in 1 ml of 50 mm Tris, pH 7.5 for 15 min with gentle mixing. The cross-linked beads were harvested with the use of a magnet and washed (three times) with PBS containing 0.05% Tween 20.
Purification of LIM1215 Cell-derived Exosomes by Immunoaffinity Capture Using A33 Antibodies
CCM from LIM1215 cells (∼1.9 mg of protein in 3 ml) was preincubated with Dynabeads (5 × 108 beads) for 2 h at 4 °C with gentle rotation to reduce nonspecific binding. The beads were harvested using a magnet (2 min), and the recovered supernatant solution was then incubated with A33 antibody-coated Dynabeads (300 μg of antibody/5 × 108 beads) for 2 h at 4 °C with gentle rotation. The A33 antibody-coupled beads were washed for 5 min at RT in 1 ml of PBS (three times) with the use of a magnet (2 min), and A33-containing exosomes were eluted from the beads with 100 μl of lithium dodecyl sulfate sample loading buffer (Invitrogen) for SDS-PAGE analysis.
LC-MS/MS
Exosome samples (16 μg) were electrophoretically separated using SDS-PAGE, and proteins were visualized by staining with Imperial protein stain (Pierce). Gel lanes were cut into 20 × 2-mm bands using a GridCutter (The Gel Co., San Francisco, CA), and proteins were trypsinized as described previously (35). Extracted tryptic peptides from each gel band were concentrated to ∼10 μl by centrifugal lyophilization and analyzed by LC-MS/MS using an LTQ-Orbitrap mass spectrometer (Thermo Fisher Scientific) fitted with a nanoflow reversed-phase HPLC system (Model 1200, Agilent). Reversed-phase HPLC of peptide mixtures was performed on a nanoAcquity (C18) 150-mm × 0.15-mm-internal diameter reversed-phase UPLC column (Waters) developed using a linear 60-min gradient from 0 to 100% B (0.1% aqueous formic acid, 60% (v/v) aqueous ACN) with a flow rate of 0.8 μl/min at 45 °C. Survey MS scans were acquired with resolution set to a value of 30,000. Up to five of the most intense ions per cycle were fragmented and analyzed on the linear trap, and selected ions were then dynamically excluded from further analysis for 180 s.
Database Searching and Protein Identification
Parameters used to generate the peak lists, using extract-msn as part of Bioworks 3.3.1 (Thermo Fisher Scientific), were as follows: minimum mass, 700 Da; maximum mass, 5,000 Da; grouping tolerance, 0.01 Da; intermediate scans, 200; minimum group count, 1; minimum peaks, 10; and total ion current, 100. Peak lists for each LC-MS/MS run were merged into a single MASCOT generic format (MGF) file for MASCOT searches. Automatic charge state recognition was used because of the high resolution survey scan (30,000). LC-MS/MS spectra were searched against the Ludwig non-redundant database (36) using MASCOT (v2.2.01, Matrix Science, London, UK). The search parameters used were as follows: fixed modification, carboxymethylation of cysteine (+58 Da); variable modification, oxidation of methionine (+16 Da); three missed tryptic cleavages; 20-ppm peptide mass tolerance; and 0.8-Da fragment ion mass tolerance. Peptide identifications with ion scores greater than identity scores and proteins with at least two peptide identifications were considered to generate a preliminary protein list, resulting in a 0% false discovery rate (derived from corresponding decoy database search) for both purified and crude exosome preparations. Manual interrogation of the preliminary list was done to remove FCS contaminants.
Comparison of Crude and Pure Exosome Proteomes
In-house Perl scripts were written to map protein identifiers, obtained from crude and purified exosome preparations, to NCBI Entrez gene (37) identifiers. Entrez gene identifiers were then mapped to HUGO (Human Genome Organisation) gene nomenclature committee-approved gene symbols (38) to facilitate cross-comparison at the gene product level. Perl scripts were written to compare the gene symbols.
Comparison of Pure Exosome Proteome with Published Exosome Proteomes
Two studies that had detected more than 200 exosomal proteins were compared with the LIM1215-derived pure proteome described in this study. Pisitkun et al. (2) identified 295 proteins in human urinary exosomes that were mapped to Entrez gene identifiers. In a separate study, Valadi et al. (17) identified 272 proteins in both murine and human mast cell-derived exosomes. Those 272 protein identifiers were mapped to Entrez gene and later mapped to human orthologs through Homologene (39), resulting in 266 gene identifiers. The two gene lists were then compared with the LIM1215-derived pure exosome gene identifiers using Perl scripts.
Biological Process, Molecular Function, and Subcellular Localization
Gene ontology annotations for biological process, molecular function, and subcellular localization were downloaded from the Human Protein Reference Database (HPRD) (40). In-house Perl scripts were used to map the proteins with corresponding gene ontology terms.
Domain/Motif Enrichment
Protein sequence analysis of the identified exosomal proteins was performed to identify domains/motifs that are significantly enriched. In-house Perl scripts were used to submit the protein sequences individually to the web interface of SMART (41) with an interval of 10 s between the subsequent posts. The same was done for the entire human RefSeq (42) proteome (∼39,000 sequences). The χ2 test with Yates correction was used to detect the p value for the domain enrichment analysis. SignalP (43) was used in both the neural network and the hidden Markov model modes to predict the occurrence of signal peptides.
Tissue Signature Analysis of Human Colon Carcinoma (LIM1215 Cell)-, Urine-, and Mast Cell-derived Exosomes
Exosomal protein data with respect to the tissue from which they were derived were overlaid in a matrix. The score provided for the protein is 0 if not detected in exosome or 1 if detected. The same scoring was applied to all 759 unique protein identifiers (266 from mast cell exosomes, 295 from urine exosomes, and 394 from colorectal cancer (LIM1215-derived pure) exosomes). Human protein tissue expression profiles with respect to urine, mast cells, and colorectal cancer were downloaded from the Human Proteinpedia (44), HPRD, and Human Protein Atlas (45). In addition, a urinary proteome study (46) was also used to populate the urinary proteome catalogue. The obtained tissue expression profiles were added on to the exosomal protein matrix with the same scoring pattern of 0 (not detected in a specific tissue) or 1 (detected in a specific tissue). Each cell will have a value ranging from 0 to 2 depending on its identification in exosome and corresponding tissue. Hierarchical clustering was performed (scaled to row) to bring protein clusters that were exclusively found in a specific tissue and its corresponding exosome using R.
Cancer Hallmarks
Biomart was used to assign gene ontology annotations to the identified proteins in LIM1215-derived pure exosomes (to get extended gene ontology annotations). The gene ontology terms were grouped to seven different classes of cancer hallmarks such as sustained angiogenesis, insensitivity to antigrowth signals, an inflammatory microenvironment, tissue invasion and metastasis, limitless replicative potential, evading apoptosis, and self-sufficiency in growth signals. Up-regulated genes in colon cancer metastasis from a few studies were downloaded from Oncomine (47). Gene symbols commonly found between colon cancer metastasis and LIM1215-derived pure exosomes were included in the metastasis category.
RESULTS AND DISCUSSION
Production and Characterization of Exosomes Derived from LIM1215 Cells
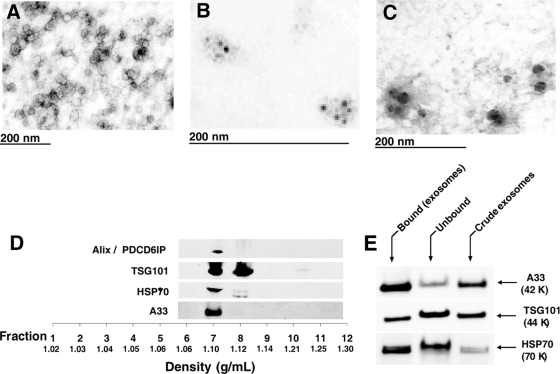
We have developed a strategy, using a combination of differential centrifugation and molecular weight cutoff membranes, for isolating crude exosomes (40–100-nm diameter) in high yield from the CM of LIM1215 cells grown for 24 h in serum-free RPMI 1640 medium supplemented with 0.8% insulin-transferrin-selenium. The recovery of crude exosomes from CCM by ultracentrifugation (100,000 × g for 1 h) was ∼50 μg from 5 × 108 cells. EM revealed that the crude exosomes were essentially homogeneous and 40–100 nm in diameter (Fig. 1A). Immunoelectron microscopy using a NANOGOLD probe showed that A33 and the lysosomal marker (1) CD63 antibodies localized to the exosome surface (Fig. 1, B and C, respectively). Crude exosomes in the CCM were further characterized by rate zonal centrifugation through a discontinuous iodixanol (OptiPrep) gradient (Fig. 1D). Fractions of increasing density were collected, and those containing exosomes were identified by Western blotting using A33 antibodies and the common exosome molecular markers HSP70, Alix/PDCD6IP, and TSG101. It can be seen in Fig. 1D that crude exosomes released from LIM1215 cells float at a density of 1.10–1.12 g/ml (Fractions 7 and 8), consistent with the density of exosomes from other cell types (15, 17, 18).
Fig. 1.
Morphological characterization and proteomics analysis of LIM1215 colorectal cancer-derived exosomes. Electron micrographs of crude exosomes negatively stained with uranyl acetate and examined at 60 kV (A), anti-A33 (B), and anti-CD63 immunogold/uranyl acetate staining (C) are shown. D, OptiPrep density gradient separation of crude exosomes using Alix/PDCD6IP, TSG101, HSP70, and A33 as exosomal markers to locate the fraction of exosome sedimentation. E, Western blot analysis of bound, unbound, and crude exosomes (5 μg/lane) for A33 antigen, TSG101, and HSP70.
To better characterize colon tumor cell-derived exosomes, we applied an additional purification step utilizing immunoaffinity capture with A33 antibody-coated Dynabeads. CCM containing crude exosomes, which had been pretreated with Dynabeads, was mixed with A33 antibody-coated beads (2 h at 4 °C) and recovered using a magnet. Western blot analysis using anti-A33 antibodies revealed that ∼80% of the A33-containing exosomes in the CCM can be immunoaffinity captured by this approach with an overall yield of ∼25 μg (i.e. ∼50% compared with that achieved using ultracentrifugation at 100,000 × g for 1 h); see Fig. 1E.
Proteome Profiling of Immunoaffinity Capture-purified LIM1215 Cell-derived Exosomes
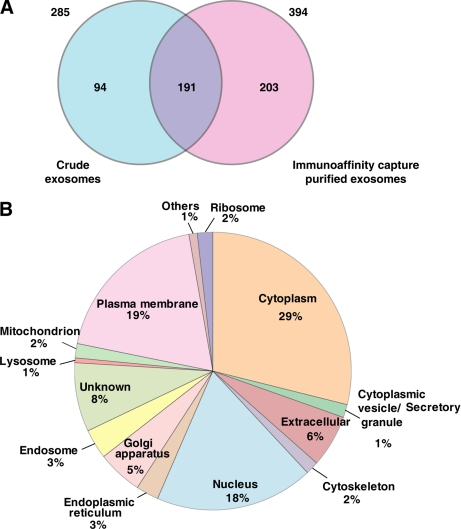
Next, we compared the proteome profiles of immune capture-purified exosomes (LIM1215-derived pure) with crude exosomes harvested directly from the CM by ultracentrifugation. Both exosome preparations were solubilized in SDS-PAGE sample buffer and analyzed using one-dimensional SDS-PAGE. Gel bands were excised and individually trypsinized, and extracted tryptic peptides were subjected to LC-MS/MS. Protein database searching of MS/MS data resulted in the identification of 285 and 394 unique proteins in the crude and pure exosome data sets, respectively (see supplemental Tables 1 and 2). As revealed in the Venn diagram (Fig. 2A), 191 proteins were common to the two data sets, whereas 94 and 203 proteins were uniquely identified in the crude and pure exosomes, respectively. Interestingly, the list of crude exosomal proteins depleted upon immunoaffinity purification is rich in protein subunits of high Mr complexes (e.g. 26 S proteasome complex (48) and the vault ribonucleoprotein complex (e.g. MVP and PRAP4) (49)) or proteins with a demonstrated ability to form high Mr oligomers (e.g. gel-forming mucins such as MUC5AC (50)) (see supplemental Table 3). Because of their ability to sediment at high centrifugal force, these proteins are, presumably, artifacts of the purification strategy. We also observed the reduction of several ribosomal and histone proteins identified in the pure exosomes upon immunoaffinity capture.
Fig. 2.
Overlap of crude and immunoaffinity capture-purified exosomal proteomes and subcellular localization distribution of LIM1215-derived pure exosomal proteins. A, Venn diagram depicting the overlap of exosomal proteomes. The numbers present outside the circle represent the total number of proteins identified in the particular data set. A two-way Venn diagram of the LIM1215 exosome proteome between crude exosomes and the immunoaffinity capture-based purified data set is depicted. 191 proteins are common between the two exosomal data sets. 394 proteins found in the immunoaffinity capture-based purified exosomes were used in further analysis. B, subcellular localization of the 394 proteins identified in the LIM1215-derived pure exosomes is shown. 29% of the proteins have a cytoplasmic localization, whereas 19% of the proteins have plasma membrane localization. 6% of the proteins are known to be extracellular, and 20% are known to be localized to the nucleus.
An inspection of the proteome data set for LIM1215-derived pure exosomes also confirmed the presence of many proteins common to exosomes studied to date. These include components of the ESCRT machinery such as Alix, a molecular marker of exosomes (20); TSG101, an endosomal marker; vacuolar protein sorting-associated proteins (VPS25, VPS28, and VPS37); charged multivesicular body proteins (CHMP2, CHMP4, and CHMP5); and tetraspanins (CD9, CD81, CD82, and tetraspanin-8) (51). Other exosome-associated proteins such as Rabs, Raps, annexins, guanine nucleotide-binding proteins, heat shock proteins, cytoskeletal proteins, metabolic enzymes, HLA class I antigen, integrins, lactadherin, claudins, and clathrin were also identified. Interestingly, several kinases (LYN, MINK1, and MAP4K4), proteases (ADAM10, DPEP1, and ST14), transporters (SLC1A4, SLC16A1, and CLIC1), and receptors (CD46, CD55, and NOTCH1) were also present. Cell type-specific proteins associated with the gastrointestinal tract were also present (A33 antigen, cadherin-17, carcinoembryonic antigen, and ephrin-B1 and -B2) (supplemental Tables 1 and 2). Additionally, this is the first study to report the presence of phospholipid scramblase 3 in exosomes. Phosphatidylserine externalization is thought to be due to the action of floppase, flippase, and scramblase activities (52); this is justified by the identification of phospholipid scramblase in our study. The data are available free for download from ExoCarta, a database for exosomal proteins and RNA (53). These data suggest possible multifunctional roles for exosomes. The biological process classification of the 394 proteins identified in LIM1215-derived pure exosomes is shown in supplemental Fig. 1.
As revealed in Fig. 2B, 19% of the total exosomal proteins are known to localize in the plasma membrane, 29% are known to localize in the cytoplasm, and 18% are known to localize in the nucleus. SMART database analysis of the LIM1215-derived pure exosome proteins revealed that ∼19% of the exosomal proteins have at least one transmembrane-spanning domain (TM). Of these, 57% (42 proteins) have one TM, 11% (eight proteins) have two TMs, 15% (11 proteins) have four TMs, 3% (two proteins) have five TMs, 1% (one protein) has seven TMs, and 13% (10 proteins) have >7 TMs (supplemental Table 4). Most four-TM-containing proteins can be classified as tetraspanins that are abundantly found in tumor-derived vesicles including exosomes (54). Interestingly, we found that the LIM125-derived pure exosome proteins are enriched with tetraspanin-containing proteins (p value 0.0001) when compared with the entire human proteome (RefSeq). Tetraspanins are a family of four-transmembrane proteins with relatively short N- and C-terminal tails, a small intracellular loop between TM regions 2 and 3, a small extracellular loop at TM1 and TM2, and a large extracellular loop (ECL2) at TM3 and TM4 (54). Highly conserved cysteines in ECL2 are characteristic of tetraspanins where palmitoylation modification is critical for interaction with other tetraspanins as well as linking cholesterol and gangliosides (55, 56). Tetraspanins assemble a network of complexes containing different tetraspanins and transmembrane and cytosolic proteins that are necessary for their functions in cell migration, metastasis, and angiogenesis (57, 58). Seven tetraspanins were detected in this study, including CD9, CD81, tetraspanin-1, -6, -8, -14, and -15. Importantly, tetraspanin-associated proteins such as integrins, ADAM10, SLC44A1, PTGFRN, IGSF8, CD44, EpCAM, and many others (56, 58–61) were identified in our analysis, creating an extensive tetraspanin network. Each individual tetraspanin protein performs differently via actions through their respective interactors; for example, CD9 and CD82 mediate metastasis inhibition by several mechanisms, whereas CD151 supports tumor progression by activating matrix metalloproteinases (54, 62). However, the actual role of these tetraspanins in exosomes awaits to be determined, although recent results indicate that tetraspanin-8-containing exosomes can induce angiogenesis via capillary sprouting (63).
Domain/Motif Enrichment Analysis of LIM1215-derived Pure Exosomal Proteins
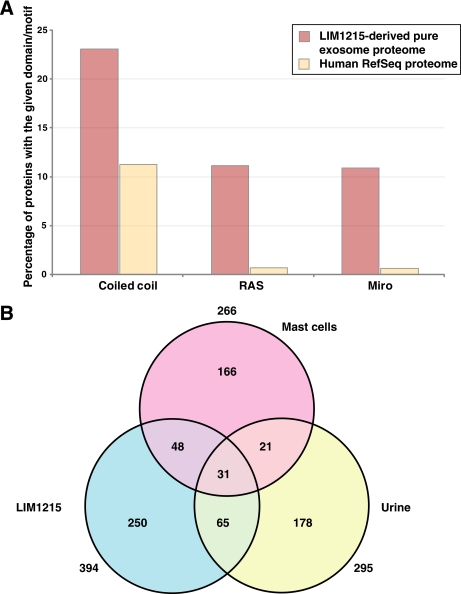
The identified 394 proteins were searched against the SMART database for the presence of domains/motifs that are significantly overrepresented in the LIM1215-derived pure exosome data set. This study revealed that coiled coil motifs were enriched (23% occurrence) in the LIM1215-derived pure exosomes compared with 11% of proteins in the entire human proteome (p value <0.0001) (Fig. 3A). Additionally, both RAS and MIRO domains were preferentially enriched in the LIM1215-derived pure exosomes (both were observed 11% of the exosome proteins compared with 0.7% in the entire human proteome (p value <0.0001)).
Fig. 3.
Protein domain distribution of LIM1215-derived pure exosomal proteins and overlap of urine-, mast cell-, and LIM1215-derived pure exosomal proteomes. A, distribution of protein domains that are preferentially enriched in the LIM1215-derived pure exosomes. Coiled coil domain is predicted to be present in ∼23% of the exosomal proteome, whereas it is present in only ∼11% of the entire human proteome (human RefSeq proteome). Likewise, RAS and MIRO domains are highly significant in the LIM1215-derived pure exosomal proteomes (11%) as compared with the entire human proteome (0.7%). B, three-way Venn diagram depicting the overlap between the exosomal proteomes derived from urine, mast cells, and LIM1215 cells. Here, two previous exosomal studies published in the scientific literature were used to find the overlap between the exosomal proteomes. 31 proteins were found to be identified in all three exosomal proteomes, whereas 96 and 79 proteins were found to be in common between LIM1215-urine and LIM1215-mast cell exosome data sets, respectively.
Coiled coil motifs are recognized to play a role in dimerization domains of several proteins, such as GCN4 (64), Fos, and Jun (65). Interestingly, coiled coil motifs have been shown to play a vital role in vesicular transport (66) and the localization of certain proteins to the early endosomes (67). As coiled coil motifs mediate protein-protein interactions and vesicular transport, it is interesting to speculate that coiled coil motifs are required for certain proteins to be engulfed in exosomes. Likewise, RAS domains (68) are found in Ras proteins (69, 70), which are small GTPases that perform GTP binding. These small GTPases are predominantly found in the exosomes and perform functions with respect to cell growth, proliferation, and differentiation. Despite interacting with a common set of proteins, distinct signal outputs are produced by different Ras isoforms such as H-Ras, N-Ras, and K-Ras (69). Preferential enrichment of RAS domains in exosomes suggests the dominance of GTPases in these vesicles. Exosomal proteins were also preferentially enriched with MIRO domain-containing proteins. MIRO domains are found in Rho GTPases (71) that are larger than the classical small GTPases. Rho GTPases perform roles in mitochondrial homeostasis, apoptosis, and trafficking (72).
Non-classical Secretory Mechanism
In total, ∼28% of the proteins were predicted to contain signal peptides using SignalP. Distribution of the subcellular localization of the 394 proteins revealed that 6% of the proteins are known to be extracellular (Fig. 2B). A majority of the LIM1215-derived pure exosome proteins (72%) did not contain a signal peptide. This finding suggests that exosomes might function as a non-classical secretory mechanism (2) wherein proteins are transported via the circulatory system to various tissues. It is interesting to speculate that non-classical secretory behavior of exosomes might perform a vital role in cell-cell communication either by stimulation via ligands expressed on their surface or through transfer of protein/RNA molecules. These exosomes might function as communicasomes wherein the molecule content of exosomes, apart from a common set of molecules, is influenced by the tissue/cell type and, indeed, the function performed.
Comparative Proteome Analysis of Urine-, Mast Cell-, and LIM125-derived Pure Exosomes
Exosomes are likely to harbor a common set of proteins that aid in various processes including biogenesis, cell growth, cell maintenance, cell communication, and signal transduction. With the availability of 394 proteins from colorectal cancer-derived exosomes, it would be interesting to compare them with the already reported exosomal proteomes in the literature. Pisitkun et al. (2) reported 295 proteins identified in human urine exosomes, whereas Valadi et al. (17) had reported 272 proteins from murine and human mast cells. We compared the two exosomal proteomes along with the LIM1215-derived pure exosomal proteome obtained from our study. We found 31 proteins to be common between all of three exosomal proteomes (Table I). As shown in Fig. 3B, 52 proteins are in common between the urine- and mast cell-derived exosomal proteomes, whereas 79 and 96 proteins were in common between LIM1215-mast cell and LIM1215-urine data sets, respectively. The 31 common proteins include Alix, transferrin, actins (α, β, and γ), RAB5B, RAB5C, EH-domain containing 4, heat shock proteins, annexins A6 and A11, and ADP-ribosylation factor 1 among others. Distribution of the biological process (supplemental Fig. 2A) for these 31 proteins revealed that 52% of them are involved in cell communication and signal transduction, whereas 19% of them are involved in cell growth and/or maintenance. 6% of the proteins have been shown to be involved in transport, whereas 3% have been shown to be involved in endosome transport. Distribution of the molecular function for the 31 proteins revealed that 23% of them are reported to have GTPase activity (supplemental Fig. 2B). 16% of the proteins are either involved in a structural constituent of the cytoskeleton or in cytoskeletal protein binding. These observations suggest that these 31 proteins form the crux or at least a part of it in the exosomes with respect to cell communication, signal transduction, cell growth, and maintenance.
Table I. List of 31 common exosomal proteins identified in this study and in exosomes derived from urine and mast cells.
| Gene symbol | Protein name | |
|---|---|---|
| 1 | PDCD6IP | Programmed cell death 6-interacting protein |
| 2 | CFL1 | Cofilin 1 (non-muscle) |
| 3 | CLIC1 | Chloride intracellular channel 1 |
| 4 | ENO1 | Enolase 1 (α) |
| 5 | GNAI3 | Guanine nucleotide-binding protein (G protein), α-inhibiting activity polypeptide 3 |
| 6 | GNB1 | Guanine nucleotide-binding protein (G protein), β polypeptide 1 |
| 7 | SFN | Stratifin |
| 8 | EHD4 | EH domain-containing 4 |
| 9 | ANXA6 | Annexin A6 |
| 10 | ANXA11 | Annexin A11 |
| 11 | HSPA8 | Heat shock 70-kDa protein 8 |
| 12 | HSP90AA1 | Heat shock protein 90 kDa α (cytosolic), class A member 1 |
| 13 | HSP90AB1 | Heat shock protein 90 kDa α (cytosolic), class B member 1 |
| 14 | ARF1 | ADP-ribosylation factor 1 |
| 15 | PFN1 | Profilin 1 |
| 16 | PGK1 | Phosphoglycerate kinase 1 |
| 17 | PKM2 | Pyruvate kinase, muscle |
| 18 | RAB5A | RAB5A, member RAS oncogene family |
| 19 | RAB5B | RAB5B, member RAS oncogene family |
| 20 | RAB5C | RAB5C, member RAS oncogene family |
| 21 | RAC1 | Ras-related C3 botulinum toxin substrate 1 (Rho family, small GTP-binding protein Rac1) |
| 22 | RAP1B | RAP1B, member of RAS oncogene family |
| 23 | ACTB | Actin, β |
| 24 | SDCBP | Syndecan-binding protein (syntenin) |
| 25 | ACTC1 | Actin, α, cardiac muscle 1 |
| 26 | TF | Transferrin |
| 27 | ACTG1 | Actin, γ 1 |
| 28 | YWHAB | Tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, β polypeptide |
| 29 | YWHAE | Tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, ϵ polypeptide |
| 30 | YWHAG | Tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, γ polypeptide |
| 31 | YWHAZ | Tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, ζ polypeptide |
Exosomal Protein Content Has Tissue-associated Signatures
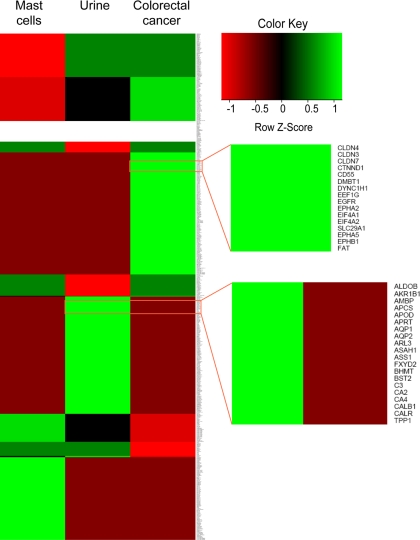
As proteins are engulfed in the MVBs in the cytoplasm, the ILVs incorporate proteins present in the parent cell/tissue. To examine this, we overlaid the exosomal protein data and protein tissue expression profile data in a matrix. Hierarchical clustering was performed using R, resulting in a heat map (Fig. 4) that displays the protein clusters with respect to the tissue. As shown in Fig. 4, bright green patterns represent those proteins that are identified in the exosomal data set and also in the specific tissue. Some of the proteins are expressed exclusively in colorectal cancer and its exosome but were not known to be expressed in urine or mast cells and were not detected in their respective exosomes. A subset of these molecules can be specific to the colorectal cancer cell types, whereas the rest are proteins that could also be found in many other tissues/cell types. Investigation of these protein molecules in the scientific literature revealed a subset of proteins that are indeed involved in colorectal cancer. These molecules include A33, cadherin-17, centrosomal protein 55 kDa (CEP55), claudin 1 (CLDN1), claudin 3 (CLDN3), claudin 7 (CLDN7), epidermal growth factor receptor, ephrin receptor A2 (EPHA2), keratin 18 (KRT18), mucin 13 (MUC13), proliferating cell nuclear antigen, POLD1, PPP2R1B, carcinoembryonic antigen, epithelial cell surface antigen (EpCAM), misshapen-like kinase 1, mitogen-activated protein kinase 4, ephrin-B1, ephrin-B2, and RUVBL1. Likewise, proteins exclusively observed in the urine-derived exosomes include specific proteins found in proximal tubules (aquaporin 1 (AQP1)), the thick ascending limb of Henle (type 2 Na-K-2Cl cotransporter (SLC12A1)), the collecting duct (aquaporin 2 (AQP2) and Rh family, C glycoprotein (RHCG)), and the transitional epithelium of the urinary bladder (uroplakin-1 and uroplakin-2). These findings suggest that exosomes resemble the tissue/cell type from which they are derived and confirm the heterogeneous roles of tissue/cell type-specific exosomes. Given the specific tissue signatures, exosomes might harbor diagnostic markers.
Fig. 4.
Tissue-specific signature of exosomal proteins. A heat map of exosomal proteomes along with the protein tissue expression is shown. Hierarchical clustering of the matrix plotted by the presence or absence of a specific protein in a particular exosomal data set and its corresponding protein tissue expression profiles shows proteins that are expressed in a particular tissue type as compared with the others. Deep red indicates that the protein is not known to be found in the specific tissue and not identified in the corresponding exosomal data set as well. Bright green indicates that the protein is identified in the exosomal data set and also in the specific tissue. Protein tissue profiles were downloaded from Human Proteinpedia, HPRD, Human Protein Atlas, and the urinary proteome data set. The top zoom panel displays a subset of proteins that are uniquely identified in the LIM1215 (colorectal cancer)-derived pure exosomes and are also known to be expressed in colorectal cancer tissues. The bottom zoom panel shows subset of proteins that are uniquely identified in urine-derived exosomes.
LIM1215-derived Pure Exosomes Have Attributes of Cancer
Cancer can be attributed by six categories (73) such as sustained angiogenesis, insensitivity to antigrowth signals, tissue invasion and metastasis, limitless replicative potential, evading apoptosis, and self-sufficiency in growth signals. Recently, the inflammatory microenvironment was also attributed to cancer (74) and included as the seventh hallmark of cancer (75). In this study, ∼50% (199 of 394 proteins) of the LIM1215-derived pure exosome proteins could be categorized in at least one of the seven cancer hallmark attributes (supplemental Fig. 3). 36% of these proteins belong to the category of self-sufficiency in growth signals, whereas 24% had limitless replicative potential. 17% of the proteins are up-regulated in colon cancer metastasis, whereas 6% evade apoptosis. Given the known cell-cell communication of exosomes, the presence of LIM1215-derived pure exosomes with cancer hallmark attributes suggests their importance in cancer biology.
Conclusions
We have shown that highly purified human colon tumor exosomes can be obtained by immunoaffinity capture using the colon epithelial cell-specific A33 antibody. Comparative proteomics analysis of LIM1215-derived pure exosomes with human urine- and murine mast cell-derived exosomes revealed the existence of proteins common between exosomes derived from different cell types. Interestingly, exosomes display tissue-associated protein signatures as well. Domain/motif enrichment analysis revealed the significant overrepresentation of coiled coil, RAS, and MIRO domains in the LIM1215-derived pure exosome data set. Our study indicates that more robust and purified exosome proteome profiles need to be obtained to clearly catalogue the common molecular signatures of exosomes in general and to better understand their underlying biological functions. The colon cancer-associated exosomal proteins identified in this study may provide diagnostic information and aid in the development of a blood-based test for colorectal cancer patients, especially when the exosomal fraction of the blood is targeted.
Supplementary Material
Acknowledgments
We thank Cong Dacchi Ho and Anna for assistance with the electron microscopy imaging at the Department of Pathology, University of Melbourne. We thank the Australian Proteomics Computational Facility for providing us with the computer resources for our research.
Footnotes
* This work was supported by National Health and Medical Research Council Program Grant 487922 (to R. J. S.) and a University of Melbourne Australian postgraduate award (to J. W. E. L.).
 The on-line version of this article (available at http://www.mcponline.org) contains supplemental Figs. 1–3 and Tables 1–4.
The on-line version of this article (available at http://www.mcponline.org) contains supplemental Figs. 1–3 and Tables 1–4.
1 The abbreviations used are:
- ILV
- intralumenal vesicle
- MVB
- multivesicular body
- ESCRT
- endosomal sorting complex required for transport
- CM
- conditioned medium
- CCM
- concentrated conditioned medium
- EM
- electron microscopy
- HPRD
- Human Protein Reference Database
- HIV-1
- human immunodeficiency virus, type 1
- RT
- room temperature
- Bis-Tris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- SMART
- simple modular architecture research tool
- TM
- transmembrane-spanning domain.
REFERENCES
- 1.Simpson R. J., Jensen S. S., Lim J. W. ( 2008) Proteomic profiling of exosomes: current perspectives. Proteomics 8, 4083– 4099 [DOI] [PubMed] [Google Scholar]
- 2.Pisitkun T., Shen R. F., Knepper M. A. ( 2004) Identification and proteomic profiling of exosomes in human urine. Proc. Natl. Acad. Sci. U.S.A 101, 13368– 13373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Taylor D. D., Akyol S., Gercel-Taylor C. ( 2006) Pregnancy-associated exosomes and their modulation of T cell signaling. J. Immunol 176, 1534– 1542 [DOI] [PubMed] [Google Scholar]
- 4.Sabapatha A., Gercel-Taylor C., Taylor D. D. ( 2006) Specific isolation of placenta-derived exosomes from the circulation of pregnant women and their immunoregulatory consequences. Am. J. Reprod. Immunol 56, 345– 355 [DOI] [PubMed] [Google Scholar]
- 5.Andre F., Schartz N. E., Movassagh M., Flament C., Pautier P., Morice P., Pomel C., Lhomme C., Escudier B., Le Chevalier T., Tursz T., Amigorena S., Raposo G., Angevin E., Zitvogel L. ( 2002) Malignant effusions and immunogenic tumour-derived exosomes. Lancet 360, 295– 305 [DOI] [PubMed] [Google Scholar]
- 6.Bard M. P., Hegmans J. P., Hemmes A., Luider T. M., Willemsen R., Severijnen L. A., van Meerbeeck J. P., Burgers S. A., Hoogsteden H. C., Lambrecht B. N. ( 2004) Proteomic analysis of exosomes isolated from human malignant pleural effusions. Am. J. Respir. Cell Mol. Biol 31, 114– 121 [DOI] [PubMed] [Google Scholar]
- 7.Mears R., Craven R. A., Hanrahan S., Totty N., Upton C., Young S. L., Patel P., Selby P. J., Banks R. E. ( 2004) Proteomic analysis of melanoma-derived exosomes by two-dimensional polyacrylamide gel electrophoresis and mass spectrometry. Proteomics 4, 4019– 4031 [DOI] [PubMed] [Google Scholar]
- 8.Admyre C., Grunewald J., Thyberg J., Gripenbäck S., Tornling G., Eklund A., Scheynius A., Gabrielsson S. ( 2003) Exosomes with major histocompatibility complex class II and co-stimulatory molecules are present in human BAL fluid. Eur. Respir. J 22, 578– 583 [DOI] [PubMed] [Google Scholar]
- 9.Skriner K., Adolph K., Jungblut P. R., Burmester G. R. ( 2006) Association of citrullinated proteins with synovial exosomes. Arthritis Rheum 54, 3809– 3814 [DOI] [PubMed] [Google Scholar]
- 10.Heijnen H. F., Schiel A. E., Fijnheer R., Geuze H. J., Sixma J. J. ( 1999) Activated platelets release two types of membrane vesicles: microvesicles by surface shedding and exosomes derived from exocytosis of multivesicular bodies and alpha-granules. Blood 94, 3791– 3799 [PubMed] [Google Scholar]
- 11.Admyre C., Johansson S. M., Qazi K. R., Filén J. J., Lahesmaa R., Norman M., Neve E. P., Scheynius A., Gabrielsson S. ( 2007) Exosomes with immune modulatory features are present in human breast milk. J. Immunol 179, 1969– 1978 [DOI] [PubMed] [Google Scholar]
- 12.Caby M. P., Lankar D., Vincendeau-Scherrer C., Raposo G., Bonnerot C. ( 2005) Exosomal-like vesicles are present in human blood plasma. Int. Immunol 17, 879– 887 [DOI] [PubMed] [Google Scholar]
- 13.Raposo G., Fevrier B., Stoorvogel W., Marks M. S. ( 2002) Lysosome-related organelles: a view from immunity and pigmentation. Cell Struct. Funct 27, 443– 456 [DOI] [PubMed] [Google Scholar]
- 14.Babst M. ( 2005) A protein's final ESCRT. Traffic 6, 2– 9 [DOI] [PubMed] [Google Scholar]
- 15.Fevrier B., Vilette D., Archer F., Loew D., Faigle W., Vidal M., Laude H., Raposo G. ( 2004) Cells release prions in association with exosomes. Proc. Natl. Acad. Sci. U.S.A 101, 9683– 9688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wiley R. D., Gummuluru S. ( 2006) Immature dendritic cell-derived exosomes can mediate HIV-1 trans infection. Proc. Natl. Acad. Sci. U.S.A 103, 738– 743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Valadi H., Ekström K., Bossios A., Sjöstrand M., Lee J. J., Lötvall J. O. ( 2007) Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol 9, 654– 659 [DOI] [PubMed] [Google Scholar]
- 18.Skog J., Würdinger T., van Rijn S., Meijer D. H., Gainche L., Sena-Esteves M., Curry W. T., Jr., Carter B. S., Krichevsky A. M., Breakefield X. O. ( 2008) Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat. Cell Biol 10, 1470– 1476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van Niel G., Porto-Carreiro I., Simoes S., Raposo G. ( 2006) Exosomes: a common pathway for a specialized function. J. Biochem 140, 13– 21 [DOI] [PubMed] [Google Scholar]
- 20.Théry C., Boussac M., Véron P., Ricciardi-Castagnoli P., Raposo G., Garin J., Amigorena S. ( 2001) Proteomic analysis of dendritic cell-derived exosomes: a secreted subcellular compartment distinct from apoptotic vesicles. J. Immunol 166, 7309– 7318 [DOI] [PubMed] [Google Scholar]
- 21.Wang J. J., Horton R., Varthakavi V., Spearman P., Ratner L. ( 1999) Formation and release of virus-like particles by HIV-1 matrix protein. AIDS 13, 281– 283 [DOI] [PubMed] [Google Scholar]
- 22.Février B., Vilette D., Laude H., Raposo G. ( 2005) Exosomes: a bubble ride for prions? Traffic 6, 10– 17 [DOI] [PubMed] [Google Scholar]
- 23.Février B., Laude H., Raposo G., Vilette D. ( 2005) Exosomes: carriers of prions? Med. Sci 21, 132– 133 [DOI] [PubMed] [Google Scholar]
- 24.Robertson C., Booth S. A., Beniac D. R., Coulthart M. B., Booth T. F., McNicol A. ( 2006) Cellular prion protein is released on exosomes from activated platelets. Blood 107, 3907– 3911 [DOI] [PubMed] [Google Scholar]
- 25.Coren L. V., Shatzer T., Ott D. E. ( 2008) CD45 immunoaffinity depletion of vesicles from Jurkat T cells demonstrates that exosomes contain CD45: no evidence for a distinct exosome/HIV-1 budding pathway. Retrovirology 5, 64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cantin R., Diou J., Bélanger D., Tremblay A. M., Gilbert C. ( 2008) Discrimination between exosomes and HIV-1: purification of both vesicles from cell-free supernatants. J. Immunol. Methods 338, 21– 30 [DOI] [PubMed] [Google Scholar]
- 27.Whitehead R. H., Macrae F. A., St John D. J., Ma J. ( 1985) A colon cancer cell line (LIM1215) derived from a patient with inherited nonpolyposis colorectal cancer. J. Natl. Cancer Inst 74, 759– 765 [PubMed] [Google Scholar]
- 28.Heath J. K., White S. J., Johnstone C. N., Catimel B., Simpson R. J., Moritz R. L., Tu G. F., Ji H., Whitehead R. H., Groenen L. C., Scott A. M., Ritter G., Cohen L., Welt S., Old L. J., Nice E. C., Burgess A. W. ( 1997) The human A33 antigen is a transmembrane glycoprotein and a novel member of the immunoglobulin superfamily. Proc. Natl. Acad. Sci. U.S.A 94, 469– 474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ji H., Moritz R. L., Reid G. E., Ritter G., Catimel B., Nice E., Heath J. K., White S. J., Welt S., Old L. J., Burgess A. W., Simpson R. J. ( 1997) Electrophoretic analysis of the novel antigen for the gastrointestinal-specific monoclonal antibody, A33. Electrophoresis 18, 614– 621 [DOI] [PubMed] [Google Scholar]
- 30.Ritter G., Cohen L. S., Nice E. C., Catimel B., Burgess A. W., Moritz R. L., Ji H., Heath J. K., White S. J., Welt S., Old L. J., Simpson R. J. ( 1997) Characterization of posttranslational modifications of human A33 antigen, a novel palmitoylated surface glycoprotein of human gastrointestinal epithelium. Biochem. Biophys. Res. Commun 236, 682– 686 [DOI] [PubMed] [Google Scholar]
- 31.Garin-Chesa P., Sakamoto J., Welt S., Real F. X., Rettig W. J., Old L. J. ( 1996) Organ-specific expression of the colon cancer antigen A33, a cell surface target for antibody based therapy. Int. J. Oncol 9, 465– 471 [DOI] [PubMed] [Google Scholar]
- 32.Simpson R. J., Connolly L. M., Eddes J. S., Pereira J. J., Moritz R. L., Reid G. E. ( 2000) Proteomic analysis of the human colon carcinoma cell line (LIM 1215): development of a membrane protein database. Electrophoresis 21, 1707– 1732 [DOI] [PubMed] [Google Scholar]
- 33.Schröder M., Schäfer R., Friedl P. ( 1997) Spectrophotometric determination of iodixanol in subcellular fractions of mammalian cells. Anal. Biochem 244, 174– 176 [DOI] [PubMed] [Google Scholar]
- 34.Mathias R. A., Lim J. W., Ji H., Simpson R. J. ( 2009) in Proteomics Analysis of Membrane Proteins: Methods and Protocols ( Peirce M. J., Wait R. eds) pp. 227– 242Humana Press, Totowa, NJ [Google Scholar]
- 35.Moritz R. L., Eddes J. S., Reid G. E., Simpson R. J. ( 1996) S-Pyridylethylation of intact polyacrylamide gels and in situ digestion of electrophoretically separated proteins: a rapid mass spectrometric method for identifying cysteine-containing peptides. Electrophoresis 17, 907– 917 [DOI] [PubMed] [Google Scholar]
- 36.Eddes J. S., Kapp E. A., Frecklington D. F., Connolly L. M., Layton M. J., Moritz R. L., Simpson R. J. ( 2002) CHOMPER: a bioinformatic tool for rapid validation of tandem mass spectrometry search results associated with high-throughput proteomic strategies. Proteomics 2, 1097– 1103 [DOI] [PubMed] [Google Scholar]
- 37.Maglott D., Ostell J., Pruitt K. D., Tatusova T. ( 2007) Entrez Gene: gene-centered information at NCBI. Nucleic Acids Res 35, D26– D31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Eyre T. A., Ducluzeau F., Sneddon T. P., Povey S., Bruford E. A., Lush M. J. ( 2006) The HUGO Gene Nomenclature Database, 2006 updates. Nucleic Acids Res 34, D319– D321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wheeler D. L., Barrett T., Benson D. A., Bryant S. H., Canese K., Chetvernin V., Church D. M., Dicuccio M., Edgar R., Federhen S., Feolo M., Geer L. Y., Helmberg W., Kapustin Y., Khovayko O., Landsman D., Lipman D. J., Madden T. L., Maglott D. R., Miller V., Ostell J., Pruitt K. D., Schuler G. D., Shumway M., Sequeira E., Sherry S. T., Sirotkin K., Souvorov A., Starchenko G., Tatusov R. L., Tatusova T. A., Wagner L., Yaschenko E. ( 2008) Database resources of the National Center for Biotechnology Information. Nucleic Acids Res 36, D13– D21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mishra G. R., Suresh M., Kumaran K., Kannabiran N., Suresh S., Bala P., Shivakumar K., Anuradha N., Reddy R., Raghavan T. M., Menon S., Hanumanthu G., Gupta M., Upendran S., Gupta S., Mahesh M., Jacob B., Mathew P., Chatterjee P., Arun K. S., Sharma S., Chandrika K. N., Deshpande N., Palvankar K., Raghavnath R., Krishnakanth R., Karathia H., Rekha B., Nayak R., Vishnupriya G., Kumar H. G., Nagini M., Kumar G. S., Jose R., Deepthi P., Mohan S. S., Gandhi T. K., Harsha H. C., Deshpande K. S., Sarker M., Prasad T. S., Pandey A. ( 2006) Human protein reference database—2006 update. Nucleic Acids Res 34, D411– D414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schultz J., Milpetz F., Bork P., Ponting C. P. ( 1998) SMART, a simple modular architecture research tool: identification of signaling domains. Proc. Natl. Acad. Sci. U.S.A 95, 5857– 5864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pruitt K. D., Tatusova T., Maglott D. R. ( 2007) NCBI reference sequences (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res 35, D61– D65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bendtsen J. D., Nielsen H., von Heijne G., Brunak S. ( 2004) Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol 340, 783– 795 [DOI] [PubMed] [Google Scholar]
- 44.Mathivanan S., Ahmed M., Ahn N. G., Alexandre H., Amanchy R., Andrews P. C., Bader J. S., Balgley B. M., Bantscheff M., Bennett K. L., Björling E., Blagoev B., Bose R., Brahmachari S. K., Burlingame A. S., Bustelo X. R., Cagney G., Cantin G. T., Cardasis H. L., Celis J. E., Chaerkady R., Chu F., Cole P. A., Costello C. E., Cotter R. J., Crockett D., DeLany J. P., De Marzo A. M., DeSouza L. V., Deutsch E. W., Dransfield E., Drewes G., Droit A., Dunn M. J., Elenitoba-Johnson K., Ewing R. M., Van Eyk J., Faca V., Falkner J., Fang X., Fenselau C., Figeys D., Gagné P., Gelfi C., Gevaert K., Gimble J. M., Gnad F., Goel R., Gromov P., Hanash S. M., Hancock W. S., Harsha H. C., Hart G., Hays F., He F., Hebbar P., Helsens K., Hermeking H., Hide W., Hjernø K., Hochstrasser D. F., Hofmann O., Horn D. M., Hruban R. H., Ibarrola N., James P., Jensen O. N., Jensen P. H., Jung P., Kandasamy K., Kheterpal I., Kikuno R. F., Korf U., Körner R., Kuster B., Kwon M. S., Lee H. J., Lee Y. J., Lefevre M., Lehvaslaiho M., Lescuyer P., Levander F., Lim M. S., Löbke C., Loo J. A., Mann M., Martens L., Martinez-Heredia J., McComb M., McRedmond J., Mehrle A., Menon R., Miller C. A., Mischak H., Mohan S. S., Mohmood R., Molina H., Moran M. F., Morgan J. D., Moritz R., Morzel M., Muddiman D. C., Nalli A., Navarro J. D., Neubert T. A., Ohara O., Oliva R., Omenn G. S., Oyama M., Paik Y. K., Pennington K., Pepperkok R., Periaswamy B., Petricoin E. F., Poirier G. G., Prasad T. S., Purvine S. O., Rahiman B. A., Ramachandran P., Ramachandra Y. L., Rice R. H., Rick J., Ronnholm R. H., Salonen J., Sanchez J. C., Sayd T., Seshi B., Shankari K., Sheng S. J., Shetty V., Shivakumar K., Simpson R. J., Sirdeshmukh R., Siu K. W., Smith J. C., Smith R. D., States D. J., Sugano S., Sullivan M., Superti-Furga G., Takatalo M., Thongboonkerd V., Trinidad J. C., Uhlen M., Vandekerckhove J., Vasilescu J., Veenstra T. D., Vidal-Taboada J. M., Vihinen M., Wait R., Wang X., Wiemann S., Wu B., Xu T., Yates J. R., Zhong J., Zhou M., Zhu Y., Zurbig P., Pandey A. ( 2008) Human Proteinpedia enables sharing of human protein data. Nat. Biotechnol 26, 164– 167 [DOI] [PubMed] [Google Scholar]
- 45.Berglund L., Björling E., Oksvold P., Fagerberg L., Asplund A., Szigyarto C. A., Persson A., Ottosson J., Wernérus H., Nilsson P., Lundberg E., Sivertsson A., Navani S., Wester K., Kampf C., Hober S., Pontén F., Uhlén M. ( 2008) A genecentric human protein atlas for expression profiles based on antibodies. Mol. Cell. Proteomics 7, 2019– 2027 [DOI] [PubMed] [Google Scholar]
- 46.Adachi J., Kumar C., Zhang Y., Olsen J. V., Mann M. ( 2006) The human urinary proteome contains more than 1500 proteins, including a large proportion of membrane proteins. Genome Biol 7, R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rhodes D. R., Kalyana-Sundaram S., Mahavisno V., Varambally R., Yu J., Briggs B. B., Barrette T. R., Anstet M. J., Kincead-Beal C., Kulkarni P., Varambally S., Ghosh D., Chinnaiyan A. M. ( 2007) Oncomine 3.0: genes, pathways, and networks in a collection of 18,000 cancer gene expression profiles. Neoplasia 9, 166– 180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang X., Chen C. F., Baker P. R., Chen P. L., Kaiser P., Huang L. ( 2007) Mass spectrometric characterization of the affinity-purified human 26S proteasome complex. Biochemistry 46, 3553– 3565 [DOI] [PubMed] [Google Scholar]
- 49.van Zon A., Mossink M. H., Scheper R. J., Sonneveld P., Wiemer E. A. ( 2003) The vault complex. Cell. Mol. Life Sci 60, 1828– 1837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hollingsworth M. A., Swanson B. J. ( 2004) Mucins in cancer: protection and control of the cell surface. Nat. Rev. Cancer 4, 45– 60 [DOI] [PubMed] [Google Scholar]
- 51.Escola J. M., Kleijmeer M. J., Stoorvogel W., Griffith J. M., Yoshie O., Geuze H. J. ( 1998) Selective enrichment of tetraspan proteins on the internal vesicles of multivesicular endosomes and on exosomes secreted by human B-lymphocytes. J. Biol. Chem 273, 20121– 20127 [DOI] [PubMed] [Google Scholar]
- 52.Hugel B., Martínez M. C., Kunzelmann C., Freyssinet J. M. ( 2005) Membrane Microparticles: two sides of the coin. Physiology 20, 22– 27 [DOI] [PubMed] [Google Scholar]
- 53.Mathivanan S., Simpson R. J. ( 2009) ExoCarta: a compendium of exosomal proteins and RNA. Proteomics 9, 4997– 5000 [DOI] [PubMed] [Google Scholar]
- 54.Zöller M. ( 2009) Tetraspanins: push and pull in suppressing and promoting metastasis. Nat. Rev. Cancer 9, 40– 55 [DOI] [PubMed] [Google Scholar]
- 55.Todeschini A. R., Dos Santos J. N., Handa K., Hakomori S. I. ( 2008) Ganglioside GM2/GM3 complex affixed on silica nanospheres strongly inhibits cell motility through CD82/cMet-mediated pathway. Proc. Natl. Acad. Sci. U.S.A 105, 1925– 1930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Berditchevski F., Odintsova E., Sawada S., Gilbert E. ( 2002) Expression of the palmitoylation-deficient CD151 weakens the association of alpha 3 beta 1 integrin with the tetraspanin-enriched microdomains and affects integrin-dependent signaling. J. Biol. Chem 277, 36991– 37000 [DOI] [PubMed] [Google Scholar]
- 57.Hemler M. E. ( 2005) Tetraspanin functions and associated microdomains. Nat. Rev. Mol. Cell Biol 6, 801– 811 [DOI] [PubMed] [Google Scholar]
- 58.André M., Le Caer J. P., Greco C., Planchon S., El Nemer W., Boucheix C., Rubinstein E., Chamot-Rooke J., Le Naour F. ( 2006) Proteomic analysis of the tetraspanin web using LC-ESI-MS/MS and MALDI-FTICR-MS. Proteomics 6, 1437– 1449 [DOI] [PubMed] [Google Scholar]
- 59.Le Naour F., André M., Greco C., Billard M., Sordat B., Emile J. F., Lanza F., Boucheix C., Rubinstein E. ( 2006) Profiling of the tetraspanin web of human colon cancer cells. Mol. Cell. Proteomics 5, 845– 857 [DOI] [PubMed] [Google Scholar]
- 60.Stipp C. S., Kolesnikova T. V., Hemler M. E. ( 2001) EWI-2 is a major CD9 and CD81 partner and member of a novel Ig protein subfamily. J. Biol. Chem 276, 40545– 40554 [DOI] [PubMed] [Google Scholar]
- 61.Murayama Y., Shinomura Y., Oritani K., Miyagawa J., Yoshida H., Nishida M., Katsube F., Shiraga M., Miyazaki T., Nakamoto T., Tsutsui S., Tamura S., Higashiyama S., Shimomura I., Hayashi N. ( 2008) The tetraspanin CD9 modulates epidermal growth factor receptor signaling in cancer cells. J. Cell. Physiol 216, 135– 143 [DOI] [PubMed] [Google Scholar]
- 62.Shiomi T., Okada Y. ( 2003) MT1-MMP and MMP-7 in invasion and metastasis of human cancers. Cancer Metastasis Rev 22, 145– 152 [DOI] [PubMed] [Google Scholar]
- 63.Gesierich S., Berezovskiy I., Ryschich E., Zöller M. ( 2006) Systemic induction of the angiogenesis switch by the tetraspanin D6.1A/CO-029. Cancer Res 66, 7083– 7094 [DOI] [PubMed] [Google Scholar]
- 64.O'Shea E. K., Klemm J. D., Kim P. S., Alber T. ( 1991) X-ray structure of the GCN4 leucine zipper, a two-stranded, parallel coiled coil. Science 254, 539– 544 [DOI] [PubMed] [Google Scholar]
- 65.O'Shea E. K., Rutkowski R., Kim P. S. ( 1992) Mechanism of specificity in the Fos-Jun oncoprotein heterodimer. Cell 68, 699– 708 [DOI] [PubMed] [Google Scholar]
- 66.Nair J., Müller H., Peterson M., Novick P. ( 1990) Sec2 protein contains a coiled-coil domain essential for vesicular transport and a dispensable carboxy terminal domain. J. Cell Biol 110, 1897– 1909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Raiborg C., Bremnes B., Mehlum A., Gillooly D. J., D'Arrigo A., Stang E., Stenmark H. ( 2001) FYVE and coiled-coil domains determine the specific localisation of Hrs to early endosomes. J. Cell Sci 114, 2255– 2263 [DOI] [PubMed] [Google Scholar]
- 68.Rubio I. ( 2005) Use of the Ras binding domain of c-Raf for biochemical and live-cell analysis of Ras activation. Biochem. Soc. Trans 33, 662– 663 [DOI] [PubMed] [Google Scholar]
- 69.Hancock J. F. ( 2003) Ras proteins: different signals from different locations. Nat. Rev. Mol. Cell Biol 4, 373– 384 [DOI] [PubMed] [Google Scholar]
- 70.Stenmark H., Olkkonen V. M. ( 2001) The Rab GTPase family. Genome Biol 2, REVIEWS3007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Fransson S., Ruusala A., Aspenström P. ( 2006) The atypical Rho GTPases Miro-1 and Miro-2 have essential roles in mitochondrial trafficking. Biochem. Biophys. Res. Commun 344, 500– 510 [DOI] [PubMed] [Google Scholar]
- 72.Fransson A., Ruusala A., Aspenström P. ( 2003) Atypical Rho GTPases have roles in mitochondrial homeostasis and apoptosis. J. Biol. Chem 278, 6495– 6502 [DOI] [PubMed] [Google Scholar]
- 73.Hanahan D., Weinberg R. A. ( 2000) The hallmarks of cancer. Cell 100, 57– 70 [DOI] [PubMed] [Google Scholar]
- 74.Kim S., Takahashi H., Lin W. W., Descargues P., Grivennikov S., Kim Y., Luo J. L., Karin M. ( 2009) Carcinoma-produced factors activate myeloid cells through TLR2 to stimulate metastasis. Nature 457, 102– 106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mantovani A. ( 2009) Cancer: inflaming metastasis. Nature 457, 36– 37 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.